Abstract
Reactive oxygen species play a central role in the regulation of plant responses to environmental stress. Under prolonged iron (Fe) deficiency, increased levels of hydrogen peroxide (H2O2) initiate signaling events, resulting in the attenuation of Fe acquisition through the inhibition of FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT). As this H2O2 increase occurs in a FIT-dependent manner, our aim was to understand the processes involved in maintaining H2O2 levels under prolonged Fe deficiency and the role of FIT. We identified the CAT2 gene, encoding one of the three Arabidopsis catalase isoforms, as regulated by FIT. CAT2 loss-of-function plants displayed severe susceptibility to Fe deficiency and greatly increased H2O2 levels in roots. Analysis of the Fe homeostasis transcription cascade revealed that H2O2 influences the gene expression of downstream regulators FIT, BHLH genes of group Ib, and POPEYE (PYE); however, H2O2 did not affect their upstream regulators, such as BHLH104 and ILR3. Our data shows that FIT and CAT2 participate in a regulatory loop between H2O2 and prolonged Fe deficiency.
Keywords: bHLH, catalase, FIT, H2O2, iron uptake, ROS
We demonstrate that CAT2 modulates root H2O2 levels and forms a regulatory loop with the central Fe acquisition transcription factor FIT to influence Arabidopsis response to prolonged Fe deficiency.
Introduction
Plants are constantly confronted with a varying array of stress conditions to which they need to respond and adapt in order to survive. Aerobic metabolic processes, such as photosynthesis and respiration, as well as chemical reactions in the cell, lead to the production of reactive oxygen species (ROS), the most prominent of them being superoxide (O2˙ −), hydrogen peroxide (H2O2), hydroxyl radical (OH·), and singlet oxygen (1O2). ROS can be produced in different cellular compartments and have different reactivity and stability. They can increase in amount under stress conditions and damage proteins, nucleic acids, and lipids, eventually triggering cell death. On the other hand, controlled and spatially defined ROS production can trigger signaling events and initiate or contribute to specific plant stress responses (Noctor and Foyer, 2016; Mittler, 2017; Czarnocka and Karpinski, 2018; Smirnoff and Arnaud, 2019). To maintain ROS homeostasis, plants employ ROS-scavenging enzymes, such as catalases (CATs), superoxide dismutases, ascorbate peroxidases, glutathione peroxidases, and peroxiredoxins (Mittler et al., 2004; Inupakutika et al., 2016; Khedia et al., 2019). Arabidopsis thaliana (Arabidopsis) has three catalase genes, CAT1, CAT2, and CAT3, whose products dismutate H2O2 to H2O and O2, with varying contributions of the three proteins in the different plant organs (Mhamdi et al., 2010). CAT1 is mainly expressed in pollen and seeds, CAT2 in photosynthetic tissues, roots, and seeds, and CAT3 in leaves and vascular tissues (Frugoli et al., 1996; McClung, 1997; Mhamdi et al., 2010). The differential contribution and partial functional redundancy of the three catalases in different organs has been recently studied (Su et al., 2018; Yang et al., 2019). CAT loss-of-function mutants have been successfully employed as a model system to study the role of ROS in abiotic and biotic stress responses (Mhamdi et al., 2010; Sewelam et al., 2014; Schmidt et al., 2020).
An abiotic stress with a profound impact on plant health and survival is the lack of sufficient iron (Fe). Fe deficiency-induced anemia affects billions of people worldwide, necessitating the study and deeper understanding of the mechanisms that plants utilize to acquire Fe from the soil, in order to combat Fe malnutrition through the development of Fe-fortified crops (Naranjo-Arcos and Bauer, 2016). Most flowering plants, except members of the Poaceae family, acquire Fe by employing a reduction-based mechanism, involving the function of a root surface ferric reductase and a ZIP-family bivalent metal transporter. In Arabidopsis, the Fe reductase activity is carried by the FERRIC REDUCTASE-OXIDASE2 (FRO2) protein and transport is achieved through the action of the IRON-REGULATED TRANSPORTER1 (IRT1), these being encoded by genes up-regulated under Fe deficiency (Ivanov et al., 2012; Kobayashi and Nishizawa, 2012; Brumbarova et al., 2015). Transcriptionally, Fe acquisition is controlled by a series of regulatory events representing a hierarchical cascade of mainly basic helix–loop–helix (bHLH) family transcription factors (Gao et al., 2019). Gene co-expression analysis defines different Fe-related regulatory modules (Ivanov et al., 2012; Brumbarova and Ivanov, 2019; Schwarz and Bauer, 2020). One module consists of genes involved in the acquisition of Fe, such as FRO2 and IRT1, regulated by FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT). Another module comprises genes with functions in Fe homeostasis, regulated by and co-expressed with POPEYE (PYE) (Long et al., 2010). A third module contains mainly Fe storage-related genes, such as FERRITIN genes, also controlled by IAA-LEUCINE RESISTANT3 (ILR3) and bHLH121/UPSTREAM REGULATOR OF IRT1 (URI) (Samira et al., 2018; Brumbarova and Ivanov, 2019; Kim et al., 2019; Tissot et al., 2019; Gao et al., 2020a, b; Lei et al., 2020). At the same time, ILR3, together with URI, is involved in the regulation of PYE and the bHLH subgroup Ib genes BHLH038, BHLH039, BHLH100, and BHLH101 (Kim et al., 2019; Gao et al., 2020a; Lei et al., 2020). The subgroup Ib bHLHs interact with FIT to regulate its expression, as well as the expression of FRO2 and IRT1 (Wang et al., 2007; Yuan et al., 2008; Wang et al., 2013; Trofimov et al., 2019). Among Ib bHLHs, BHLH039 is a robust Fe deficiency marker gene up-regulated by Fe deficiency (Wang et al., 2007; Ivanov et al., 2012). Its expression is not regulated by FIT; however, when FIT is lacking, BHLH039 undergoes additional up-regulation, following a strong Fe deficiency signal in the plant up-regulating the bHLH cascade (Wang et al., 2007; Schwarz and Bauer, 2020). The result of this complex interplay of regulators is the dynamic transcriptional response ensuring the balanced acquisition, redistribution, and safe storage of Fe.
With the help of hormonal signaling, the Fe acquisition domain is dynamically regulated not only in time but also in space. Thus, the expression domain of IRT1 is adjusted around the early differentiation zone of the root according to the Fe requirements of the plant through crosstalk between the phytohormones ethylene and auxin (Blum et al., 2014). ROS, and specifically H2O2, were previously shown to influence the activity of FIT through an interaction with the oxidative stress-responsive transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) (Le et al., 2016). The interaction between the two occurs under prolonged Fe deficiency, leading to FIT inactivation and attenuation of the Fe deficiency response. The specific increase in H2O2 observed under these conditions was FIT dependent and a central prerequisite for the observed major switch in the response strategy of the plant (Brumbarova et al., 2016b; Le et al., 2016, 2019; Brumbarova and Ivanov, 2018).
A question that remains open is which processes are involved in the maintenance of the increased H2O2 levels under prolonged Fe deficiency and in which way FIT participates in this regulation. Here, we show through histochemical co-staining of H2O2 and O2˙ − in the root that lack of functional FIT leads to spatially deregulated ROS distribution under prolonged Fe deficiency. FIT is involved in the transcriptional regulation of CAT genes, specifically CAT2. CAT2 loss-of-function mutants display severely hampered Fe deficiency responses. CAT2 is a major contributor to the H2O2-scavenging capacity of roots under prolonged Fe deficiency. Lack of CAT2 leads to a marked increase of H2O2 in root hair cells of the early differentiation zone. Gene expression analysis showed that CAT2 activity affects the regulation of PYE and FIT, together with genes from their target modules. At the same time, their upstream regulators ILR3 and bHLH104 are not affected transcriptionally by the lack of CAT2, suggesting post-translational effects of CAT2 and H2O2 on ILR3 and bHLH104 activity. Our data suggest the existence of a FIT- and CAT2-mediated feedback loop controlling the upstream regulators of Fe uptake and homeostasis under prolonged Fe deficiency.
Materials and methods
Plant material and growth conditions
Arabidopsis wild-type Col-0, the FIT loss-of-function fit-3 mutant (GABI_108C10, Jakoby et al., 2004), and the CAT2 loss-of-function cat2-1 mutant SALK_076998, N576998, named cat2-2 in Queval et al. (2007) and later renamed cat2-1 as in Bueso et al. (2007), were used in this study. T-DNA insertion lines were in the Col-0 genetic background. Plants were grown upright for 10 d on half-strength Hoagland medium agar plates (Jakoby et al., 2004) either supplemented with 50 µM FeNaEDTA (Fe-sufficient condition) or without Fe (Fe-deficient condition).
Root length measurement
Images of plants grown upright on half-strength Hoagland medium agar plates were acquired and root lengths were measured using the JMicroVision software, version 1.2.7 (http://www.jmicrovision.com). Following spatial calibration, length measurements were made of freehand drawn lines covering the root on the image. The obtained lengths were averaged, and the SD was calculated.
Fe reductase activity measurement
The activity of the root surface ferric reductase was measured as described in Le et al. (2016). Briefly, roots were rinsed in 100 mM Ca(NO3)2 solution and incubated in a solution containing 100 μM FeNaEDTA and 300 μM ferrozine for 1 h at room temperature in the dark. Enzymatic activity was calculated using the ferrozine molar extinction coefficient of 28.6 mM−1 cm−1. The experiment was performed in six replicates. Each replicate comprised a pool of five seedlings. The obtained data were normalized to the sum of the root length for each replicate. Fe reductase activity of the six replicates per condition was averaged and the SD was calculated.
Photosynthetic pigment measurement
For plant pigment extraction, pre-weighed rosettes of 10-day-old seedlings were shock-frozen in liquid N2 prior to tissue rupture using 1.4 mm ceramic beads and a Precellys 24 tissue homogenizer (Bertin Instruments) three times for 20 s, at 6500 rpm. Samples were frozen in between homogenization steps in liquid N2. Pigment was extracted adding 500 µl of pure acetone immediately after grinding, followed by a centrifugation step (3 min, 13 000 rpm). The extraction was repeated once. The supernatant was collected and absorption was measured at 646, 663, and 750 nm in a 96-well plate reader (Infinite 200 PRO, Tecan). Concentrations (c) of Chl a, Chl b, and carotenoids (in μg ml–1) were calculated as described in Lichtenthaler and Wellburn (1983):
Data were normalized to the sample fresh weight to obtain the pigment content of the rosette. Six independent replicates per condition were measured, averaged, and the SD was calculated.
Catalase activity measurement
Catalase activity was measured following the protocol described in Noctor et al. (2016) with some modifications, as follows. Total root and shoot extracts were prepared by grinding the frozen tissue using 1.4 mm ceramic beads and a Precellys 24 tissue homogenizer (Bertin Instruments) three times for 20 s, at 6500 rpm. The powder was homogenized in 0.1 M phosphate buffer, 1 mM EDTA (pH 7.5). The buffer volume was adjusted according to the fresh weight of the samples, so that 10 μl of buffer were added to each 1 mg of sample. The suspension was centrifuged for 10 min at 4 °C, at 13 000 rpm. H2O2 at 40 mM was added to 180 μl of each extract. The samples were immediately placed in a 96-well plate reader (Infinite 200 PRO, Tecan) and the disappearance of H2O2 absorption at 240 nm was recorded over time. The data from the first 1 min of the reaction were used to calculate catalase activity, applying the H2O2 molar extinction coefficient of 40 mM−1 cm−1. The catalase activity for three independent replicates per condition was measured, averaged, and the SD was calculated.
H2O2 content measurement
H2O2 in root samples was measured using the Amplex Red H2O2–peroxidase assay kit (Molecular Probes) as described in detail in Brumbarova et al. (2016a). The H2O2 content values for three independent replicates per condition were measured, averaged, and the SD was calculated.
Seed Fe content measurement
The Fe content of seeds was measured using a colorimetric assay, adapted for plant seeds from the protocol of Tamarit et al. (2006). Seeds were dried overnight at 100 °C, ground in an agate mortar (Merck), and weighed. A 10–15 mg aliquot of material was resuspended in 500 µl of 3% HNO3 and incubated for 16 h at 100 °C, followed by centrifugation for 5 min at 12 000 rpm. Then 400 µl of 3% HNO3 containing either FeNaEDTA standard or seed material were mixed with 160 µl of sodium ascorbate (38 mg ml–1), 320 µl of Fe chelator BPDS (1.7 mg ml–1), and 126 µl ammonium acetate solution (diluted 1:3 from saturated solution). The specific absorbance of the Fe–chelator complex was recorded at 535 nm after 5 min, together with a reference measurement at 680 nm for non-specific absorbance, using a 96-well plate reader (Infinite 200 PRO, Tecan). The obtained Fe concentration was normalized to the dry weight of the sample. The Fe content values for three independent replicates per genotype were obtained, averaged, and the SD was calculated.
Gene expression analysis by quantitative reverse transcription–PCR (RT–qPCR)
Gene expression analysis was performed as described by Gratz et al. (2019). In brief, total RNA was prepared from roots of Arabidopsis plants using the Spectrum Plant Total RNA kit (Sigma-Aldrich). cDNA was prepared using oligo(dT) primer and a RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific). For quantitative PCR (qPCR), reactions were prepared with a DyNAmo Color-Flash SYBR Green qPCR Kit (Thermo Fisher Scientific) and recorded in a C100 Touch PCR Cycler containing the CFX96 Real-Time System (Bio-Rad). The CFX Manager software (Bio-Rad) was used for data analysis. Primer pairs used in this study are listed in Supplementary Table S1. Mass standards for each gene were used to calculate absolute gene expression, which was normalized to the expression of the EF1Ba reference gene. The gene expression was assayed on three independent replicates, averaged and SD was calculated.
Histochemical staining of ROS
To compare H2O2 accumulation in seedlings grown under Fe-sufficient or -deficient conditions, 3,3′-diaminobenzidine (DAB; Sigma-Aldrich) staining was performed. After a short vacuum infiltration (5 min, –400 mbar), seedlings were incubated for 1 h in the dark under gentle agitation in DAB solution (1 mg ml–1 DAB in 50 mM MES buffer, pH 6.5). Samples were rinsed once with ddH2O. EtOH (80%) was added and seedlings were heated to 80 °C for 20 min. Staining was documented on an Axio Imager.M2 microscope (Zeiss).
The distribution of H2O2 and O2˙ − in roots was visualized by co-staining using the fluorescent dyes 3′-O-acetyl-6′-O-pentafluoroben-zenesulfonyl-2′,7′-difluorofluorescein (BES-H2O2-Ac, Wako Pure Chemical Corporation) and dihydroethidium (DHE; Sigma-Aldrich). Seedlings grown under Fe-sufficient (50 mM FeNaEDTA, +Fe) or -deficient (0 mM FeNaEDTA, –Fe) conditions for 10 d were incubated for 30 min in the dark at room temperature in liquid half-strength Hoagland medium containing 50 µM BES-H2O2-Ac and 10 µM DHE.
Fluorescence microscopy and image analysis
Images of BES-H2O2-Ac- and DHE-stained roots were taken immediately after the 30 min incubation. For imaging, an Axio Imager.M2 microscope (Zeiss) was used. BES-H2O2-Ac dye was visualized using Filter set 38 [HE eGFP shift free (E) (EX BP 470/40, BS FT 495, EM BP 525/50)] and DHE using Filter set 43 [HE Cy 3 shift free (E) (EX BP 550/25, BS FT 570, EM BP 605/70)]. All roots were pictured at ×20 magnification in tiles mode, and single images were stitched to produce a composite image of the root. The same imaging settings were employed for all genotypes and replicates.
Images obtained from fluorescent ROS staining were imported into ImageJ (http://rsbweb.nih.gov/ij/) for further analysis. To mark the region of interest (ROI) for signal intensity measurement, a segmented line, synchronized for DHE and BES-H2O2-Ac channels, was superimposed onto the root. Absolute signal intensity was measured and values were imported into Microsoft Excel. Signal intensity of both channels was normalized to the maximum signal intensity measured for BES-H2O2-Ac to ensure correct representation of the DHE to BES-H2O2-Ac signal ratio. This maximum value was set to 100 and all other values are plotted relative to it. Relative signal intensity plots represent the signal either along the root or in virtual cross-sections across three zones. Each plot is an average of three biological replicates with their respective SD.
Statistical analysis
Data were analyzed using one-way ANOVA followed by Fisher’s least statistical difference post-hoc test in the SPSS Statistics software (IBM). Statistically significant differences were estimated based on P-values (P<0.05).
Results
Root H2O2 content increases under prolonged Fe limitation
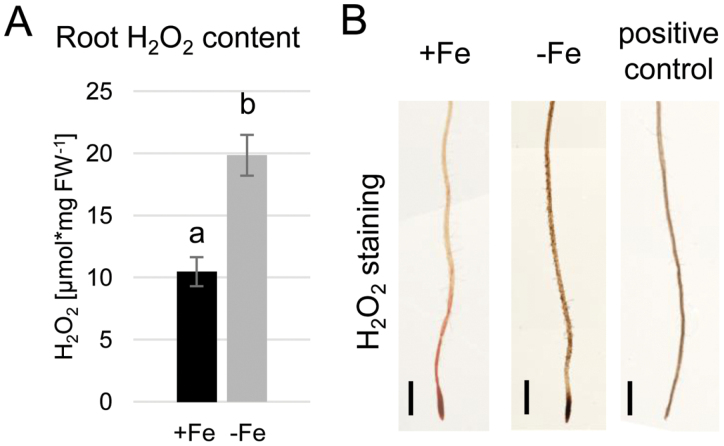
Increased H2O2 content in the root was previously shown to influence responses to prolonged Fe deficiency (Le et al., 2016). We aimed to understand whether this increase affected the whole root equally or whether the observation could be linked to certain root zones, such as those responsible for the acquisition of Fe. First, we performed a quantitative measurement of endogenous H2O2 in plants grown for 10 d under either Fe-sufficient or Fe-deficient conditions. As quantified in Fig. 1A, the overall H2O2 content of Fe-deficient plants was significantly increased compared with the control Fe condition. We next performed DAB staining for visualizing H2O2 in the root (Fig. 1B). A general increase in staining could be observed in Fe-deficient roots, consistent with the quantitative data. To confirm the specificity of the staining, cotyledons of a control plant were manually wounded with forceps. The elicited stress response caused an increase in H2O2 levels in the whole seedling, resulting in strong DAB staining (Fig. 1B). The data suggested a general increase in H2O2 content throughout the root under prolonged Fe deficiency. However, due to the low dynamic range of the staining method, it did not provide sufficient detail on potential fine differences between different root zones.
Fig. 1.
Root H2O2 content is increased under prolonged Fe deficiency. (A) Quantitative H2O2 measurement in wild-type roots grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply (n=3). Bars represent mean values ±SD. Different letters indicate statistically significant differences (P<0.05). (B) Histochemical H2O2 staining of wild-type roots grown for 10 d under +Fe or –Fe. Positive control: seedling damaged by wounding to induce H2O2 accumulation. Size bars=0.5 cm.
ROS distribution depends on Fe availability and the plant’ capacity to take up Fe
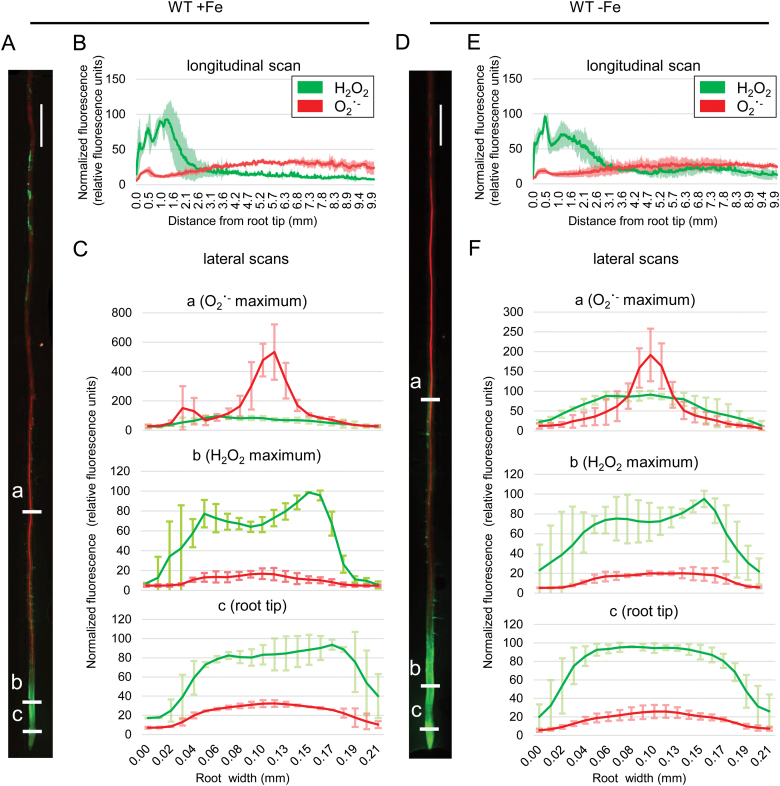
Previously, it was shown that the ratio between H2O2 and O2˙ − along the root is important for defining the differentiation state of the root cells (Tsukagoshi et al., 2010; Reyt et al., 2015). Therefore, in order to obtain a detailed map of ROS distribution within the root under prolonged Fe deficiency, we performed a dual histochemical staining for H2O2 and superoxide (O2˙ −; Fig. 2), using BES-H2O2-Ac and DHE, respectively. This allows the distinction of the tissue-level localization of the two ROS species simultaneously and in vivo. In wild-type Fe-sufficient roots, we observed strong H2O2 staining in the root tip and a gradual decrease of signal towards the root base. In the differentiation zone, the signal was observed in both the central cylinder and the epidermis (Fig. 2A–C, green channel). Signal quantification confirmed this, showing that the fluorescent intensity peaked in the transition zone between the meristem and the elongation zone (Fig. 2A, B, green channel). This observation is consistent with previous reports (Tsukagoshi et al., 2010; Reyt et al., 2015). The O2˙ − pattern, on the other hand, differed greatly along the root. We observed a small maximum at the tip of the meristem above the stem cell niche. However, the majority of the signal was present in the differentiation zone, especially in the central cylinder (Fig. 2A–C, red channel). Prolonged Fe deficiency resulted in the expansion of the H2O2 domain within the meristem together with a marked increase in signal intensity throughout the root (Fig. 2D–F, green channel), consistent with our DAB staining and the quantitative H2O2 measurement.
Fig. 2.
Histochemical H2O2 and O2˙ − staining in wild-type roots grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply. (A) Staining example of a root grown under +Fe. (B) Longitudinal signal intensity scan of roots grown under +Fe, ±SD. (C) Lateral signal intensity scans at three different positions, indicated as ‘a’, ‘b’, and ‘c’ in (A), of roots grown under +Fe. (D) Staining example of a root grown under –Fe. (E) Longitudinal signal intensity scan of roots grown under –Fe. (F) Lateral signal intensity scans at three different positions, indicated as ‘a’, ‘b’, and ‘c’ in (D), of roots grown under –Fe. All graphs represent averaged data from five roots, ±SD. Size bars in (A) and (D): 0.5 cm.
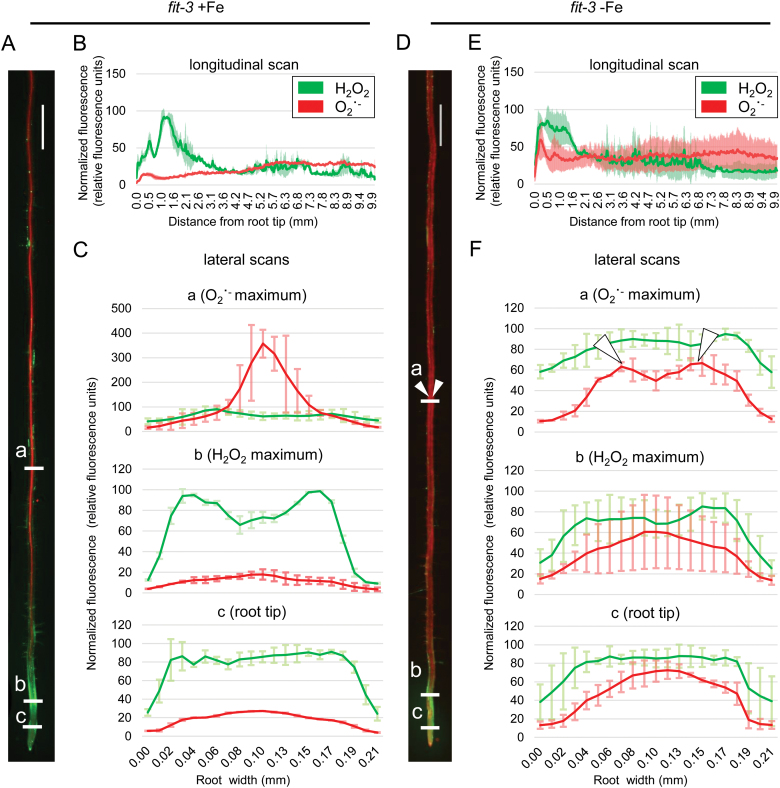
The FIT loss-of-function mutant fit-3 (Jakoby et al., 2004) was previously reported to lack prolonged Fe deficiency-induced H2O2 accumulation, suggesting that ROS homeostasis might be under the control of FIT (Le et al., 2016). Roots of fit-3 grown under control Fe conditions showed distribution patterns for both H2O2 and O2˙ − comparable with those of the wild type (Fig. 3A–C). As expected, under Fe deficiency, the H2O2 pattern remained unchanged compared with the sufficient Fe condition (Fig. 3D–F). A prominent effect occurred under Fe deficiency, where the lateral distribution of the O2˙ − signal shifted from the central cylinder and was instead very prominent in the fit-3 root periphery (Fig. 3D, F). Our findings suggest that in the absence of FIT, the H2O2 to O2˙ − ratio cannot adjust to the prolonged Fe deficiency conditions, implying a major role for the Fe deficiency regulator FIT in ROS signaling as part of the Fe deficiency response cascade.
Fig. 3.
Histochemical H2O2 and O2˙ − staining in fit-3 roots grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply. (A) Staining example of a root grown under +Fe. (B) Longitudinal signal intensity scan of roots grown under +Fe supply, ±SD. (C) Lateral signal intensity scans at three different positions, indicated as ‘a’, ‘b’, and ‘c’ in (A), of roots grown under +Fe. (D) Staining example of a root grown under –Fe. White arrowheads highlight the predominant O2˙ − signal in the peripheral root zones. (E) Longitudinal signal intensity scan of roots grown under –Fe. (F) Lateral signal intensity scans at three different positions, indicated as ‘a’, ‘b’, and ‘c’ in (D), of roots grown under –Fe. White arrowheads in the top graph highlight the predominant O2˙ − signal in the peripheral root zones and correspond to the arrowheads in (D). All graphs represent averaged data from five roots, ±SD. Size bars in (A) and (D): 0.5 cm.
FIT is required for the transcriptional regulation of H2O2 dismutases in roots
One possible explanation for the observed H2O2 localization in the root under Fe deficiency is differential activity of H2O2-metabolizing enzymes. FIT may affect the formation of ROS at the transcriptional level, for example through controlling gene expression of catalases, enzymes that eliminate H2O2.
We analyzed the expression of three catalase gene isoforms, CAT1, CAT2, and CAT3, due to their very prominent activity and the dependence of their products on the Fe state of the plant, being Fe cofactor-dependent enzymes (Nicholls et al., 2000). For identifying potential Fe and FIT transcriptional targets among the three catalase genes, we reasoned that the candidate regulatory gene should be up-regulated under Fe deficiency and behave oppositely in the absence of functional FIT.
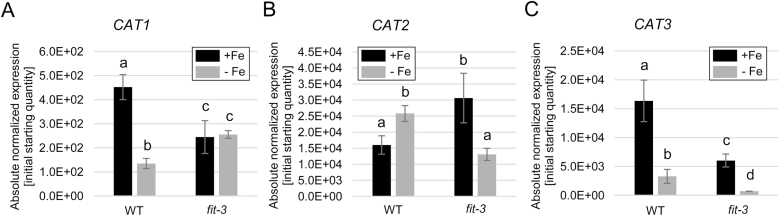
Gene expression profiling revealed that CAT1 underwent down-regulation under prolonged Fe deficiency in the wild type, whereas, in the fit-3 mutant, a reduced and non-Fe-responsive expression was observed (Fig. 4A). CAT2 showed a significant up-regulation under prolonged Fe deficiency in the wild type. Interestingly, in a FIT loss-of-function situation, its regulation was exactly opposite to that observed in the wild type (Fig. 4B), suggesting a strong dependence of CAT2 expression on the function of FIT. Furthermore, CAT2 abundance under low Fe supply is most probably a compensatory mechanism to counteract excess H2O2 levels, hence preventing toxic H2O2 effects. CAT3, similarly to CAT1, was down-regulated by prolonged low Fe availability in the wild type. (Fig. 4C) This tendency remained in the fit-3 mutant; however, in the absence of FIT, the overall expression of the gene was strongly reduced (Fig. 4C).
Fig. 4.
Expression of the genes encoding three Arabidopsis catalase isoforms, (A) CAT1, (B) CAT2, and (C) CAT3, in roots of 10-day-old wild-type and fit-3 mutant plants grown under sufficient (+Fe) or deficient (–Fe) Fe supply (n=3). Bars represent mean values ±SD. Different letters indicate statistically significant differences (P<0.05).
The expression analysis showed that all three catalase-encoding genes were dynamically regulated in response to prolonged Fe deprivation and their expression depended to a different extent on the presence of the central Fe acquisition transcriptional regulator FIT. The above-described CAT2 expression changes under prolonged Fe deficiency suggested that the CAT2 protein might play a role in modulating H2O2 levels in these growth conditions.
Perturbance of intracellular H2O2 homeostasis compromises root Fe acquisition
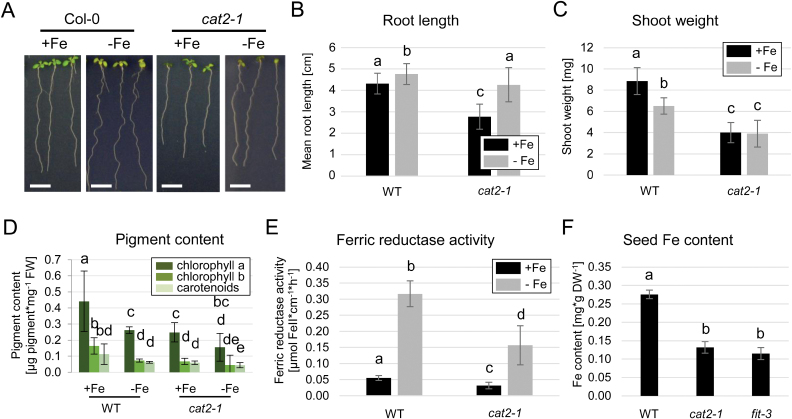
Our gene expression analysis revealed CAT2 as a potential FIT-dependent regulator of H2O2 levels under prolonged Fe deficiency. To understand how CAT2 participates in the regulation of Fe deficiency responses, we investigated the Fe response in the previously characterized CAT2 loss-of-function mutant cat2-1 (Bueso et al., 2007; Queval et al., 2007). In wild-type plants, Fe deficiency resulted in the well-documented elongation of the primary root (Gruber et al., 2013; Ivanov et al., 2014; Gratz et al., 2019), which was also the case for the cat2-1 mutant. However, the length of the primary cat2-1 root was reduced by 13% compared with that of the wild type. Under sufficient Fe supply, cat2-1 root length was reduced even more, by 35% compared with the wild type (Fig. 5A, B). This could be due to the combined deleterious effects of reduced H2O2-scavenging capacity of the mutant and the available Fe.
Fig. 5.
Fe-related phenotypes of cat2-1 mutant plants. (A) Representative images of wild-type and cat2-1 plants grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply. Size bars=1 cm. (B) Root length quantification (n=27–32). (C) Shoot weight quantification (n=6). (D) Photosynthetic pigment quantification (n=6). (E) Ferric reductase activity (n=6). (F) Seed Fe content of soil-grown wild-type, cat2-1, and fit-3 plants (n=3). (B–E) Bars represent mean values ±SD. Different letters indicate statistically significant differences (P<0.05).
Although cat2-1 roots were able to elongate under prolonged Fe deficiency, the penalty on the shoot weight of the mutant was strong under both Fe supply conditions (Fig. 5C), probably due to the photorespiratory phenotype of cat2-1, and consistent with the documented major role of CAT2 in controlling ROS homeostasis in leaves (Mhamdi et al., 2010).
The degradation of photosynthetic pigments is a well-documented effect under Fe deficiency (Ivanov et al., 2012), and we could observe it in our test system (Fig. 5D). In cat2-1 plants, the overall levels of chlorophyll and carotenoids were significantly lower than in the wild type and were further reduced under prolonged Fe limitation (Fig. 5D). Root surface ferric reductase activity was tested as an indicator for the plant capacity to take up Fe. Activity levels were reduced in cat2-1 even though the ferric reductase responded to Fe limitation (Fig. 5E). We then grew plants on soil and measured the Fe content in their seeds, as a direct consequence of the plant Fe status (Fig. 5F). As confirmation to our observations so far, cat2-1 seeds contained low levels of Fe, comparable with the strongly Fe-deficient fit-3 mutant (Fig. 5F). Overall, we characterized cat2-1 loss-of-function plants as strongly Fe deficient and unable to induce Fe uptake to counteract Fe deficiency.
CAT2 participates in modulating root ROS accumulation under prolonged Fe deficiency
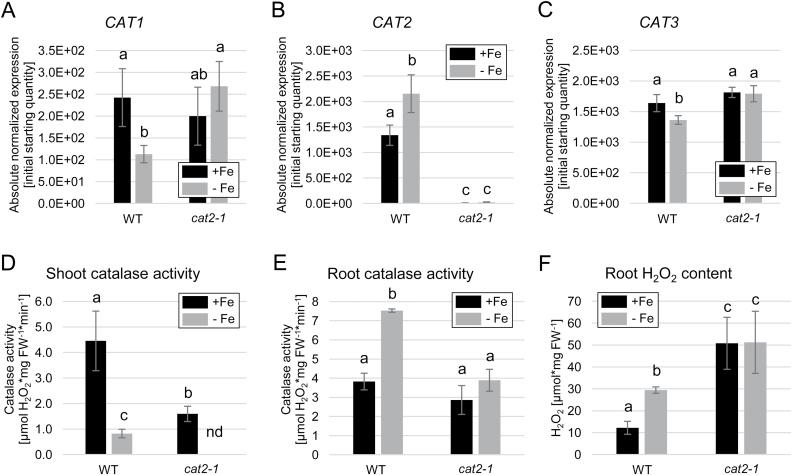
The impaired Fe mobilization under prolonged Fe deficiency in the cat2-1 mutant suggests that enhanced H2O2 was responsible for this effect. In order to analyze the significance of CAT2 for ROS accumulation under prolonged Fe deficiency, we first evaluated the expression of all CAT genes in cat2-1 plants (Fig. 6A–C). As expected, CAT1 expression was reduced under Fe deficiency in the wild type, while the levels in cat2-1 remained comparable with the wild-type sufficient Fe condition. As expected, CAT2 transcript was not detected in the cat2-1 mutant, and CAT3 showed a similar expression tendency to CAT1 (Fig. 6A–C). This indicated that the expression of CAT1 and CAT3 was higher than usual in the absence of CAT2 under prolonged Fe deficiency and pointed towards potential compensatory regulation of CAT1 and CAT3 for the missing CAT2, most probably in response to enhanced ROS levels. To test the effect of this potential compensation, we performed a total catalase activity measurement in shoots and roots of wild-type and cat2-1 plants. In shoots, lack of CAT2 led to a dramatic reduction in overall shoot catalase activity under both sufficient Fe and prolonged Fe deficiency (Fig. 6D), consistent with available data (Queval et al., 2007). In roots, the effect was also significant, but less prominent (Fig. 6E). Fe-deficient wild-type root extracts showed enhanced catalase activity compared with those from sufficient Fe-grown roots. Compared with the wild type, in cat2-1, the catalase activity under sufficient Fe was similar; however, it was significantly reduced under prolonged Fe deficiency (Fig. 6E). At the same time, the fit-3 mutant showed strongly reduced catalase activity (Supplementary Fig. S1), suggesting that the presence of a functional FIT positively influences the catalase function. We then tested the net outcome of these effects by measuring the total H2O2 content of the root. As already shown, prolonged Fe deficiency led to a marked increase in H2O2 levels in the wild type (Fig. 6F). In cat2-1, the overall H2O2 accumulation was increased compared with the wild type under both conditions, indicating a prominent role for CAT2 in maintaining the ROS balance in roots (Fig. 6F). The data suggest that despite potential compensation from CAT1 and CAT3, CAT2 is the isoform utilized by roots to control the levels of intracellular H2O2 under prolonged Fe limitation. One must note that additional ROS-scavenging enzymes might play a role in this process, explaining, for example, the pronounced H2O2 increase in cat2-1 under sufficient Fe supply.
Fig. 6.
Role of CAT2 in root catalase activity and H2O2 accumulation under prolonged Fe deficiency. (A–C) Gene expression of (A) CAT1, (B) CAT2, and (C) CAT3 in wild-type and cat2-1 plants grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply (n=3). (D) Shoot catalase activity in the wild type and cat2-1 (n=3). (E) Root catalase activity in the wild type and cat2-1 (n=3). (F) Root H2O2 content in the wild type and cat2-1 (n=3). Bars represent mean values ±SD. Different letters indicate statistically significant differences (P<0.05).
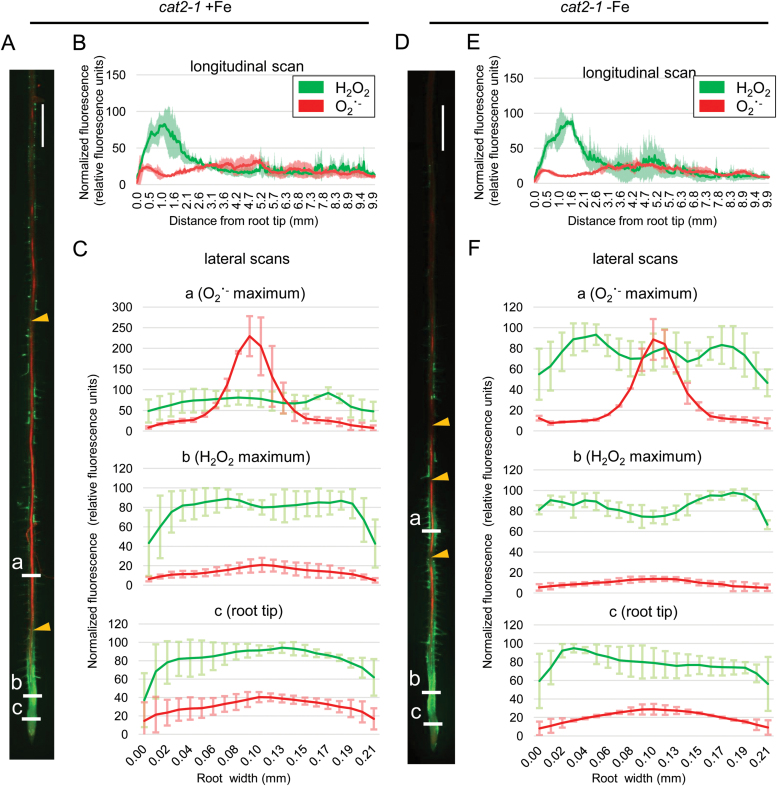
To estimate the effect of CAT2 at the organ level, we stained cat2-1 roots for H2O2 and O2˙ − (Fig. 7). Compared with the wild type (Fig. 2), cat2-1 showed a similar staining pattern, with a H2O2 peak in the transition zone and a small O2˙ − peak in the meristem; however, we observed major changes in the differentiation zone (Fig. 7A–C, green channel). Here, strong H2O2 signal was observed in the epidermis, particularly prominent in some but not all root hair cells. O2˙ − signal was confined mainly in the central cylinder; however, along the length of the root, we observed zones without staining (Fig. 7A, B, red channel). These zones lacking O2˙ − were more prominent and more frequent under prolonged Fe deficiency, where H2O2 staining was also strong, especially in root hair cells close to the root tip (Fig. 7D–F). The overall picture in the absence of CAT2 showed a disturbed ROS homeostasis, with over-pronounced H2O2 accumulation in the differentiation zone across the root, masking O2˙ − levels, and hence resulting in a disturbed balance between the ROS species under prolonged Fe deficiency. These enhanced H2O2 levels could provoke stress response signals due to ROS toxicity, overpowering the signaling role of H2O2 in activating the Fe deficiency response network.
Fig. 7.
Histochemical H2O2 and O2˙ − staining in cat2-1 roots grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply. (A) Staining example of a root grown under +Fe. (B) Longitudinal signal intensity scan of roots grown under +Fe, ±SD. (C) Lateral signal intensity scans at three different positions, indicated as ‘a’, ‘b’, and ‘c’ in (A), of roots grown under +Fe. (D) Staining example of a root grown under –Fe. (E) Longitudinal signal intensity scan of roots grown under –Fe. (F) Lateral signal intensity scans at three different positions, indicated as ‘a’, ‘b’, and ‘c’ in (D), of roots grown under –Fe. All graphs represent averaged data from five roots ±SD. Yellow arrowheads in (A) and (D) indicate regions along the central cylinder lacking O2˙ − signal. Size bars in (A) and (D): 0.5 cm.
Coordination of transcriptional Fe homeostasis response is compromised in the absence of a functional CAT2 gene
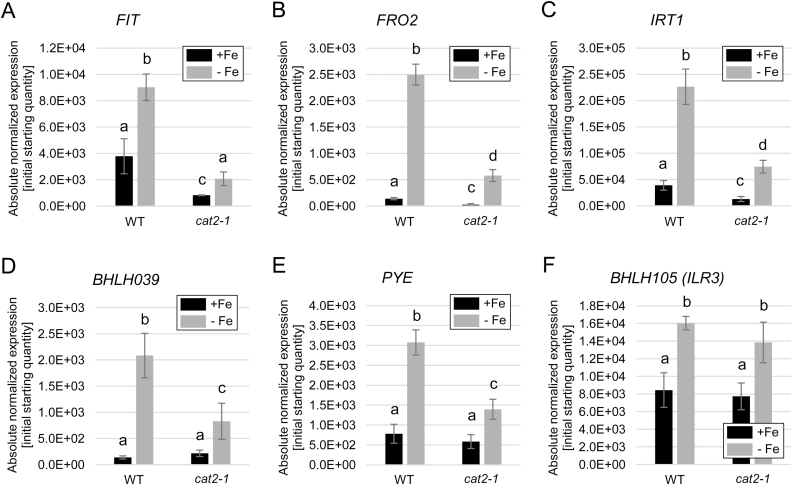
Based on the observed phenotypes of deregulated Fe mobilization in cat2-1 loss-of-function plants, we next aimed to uncover the transcriptional regulation that underlies this susceptibility to prolonged Fe deficiency. We first investigated the expression of the Fe uptake-related genes that are under the control of FIT, namely FIT itself, FRO2, and IRT1 (Fig. 8A–C). In the wild type, all three genes performed as previously reported, with expression up-regulated under Fe deficiency. However, in the cat2-1 mutant, the expression was markedly decreased in both Fe conditions relative to the wild type (Fig. 8A–C). This suggests that together with the increased H2O2 levels in the absence of CAT2, Fe uptake genes undergo repression or are hardly expressed, consistent with the reported effects of external H2O2 application and the action of ZAT12 as a FIT inhibitor (Le et al., 2016). The ROS-responsive ZAT12 gene showed strong up-regulation in cat2-1, as expected because of enhanced H2O2 levels in this mutant (Supplementary Fig. S2). We then tested the expression of the BHLH039 gene encoding a FIT partner and positive Fe deficiency response regulator (Wang et al., 2007; Yuan et al., 2008; Wang et al., 2013). BHLH039 transcriptional up-regulation does not require FIT protein but, instead, it is directly coupled to an Fe deficiency signaling cascade upstream of FIT, mediated by bHLH transcription factors of the subgroups IVb and IVc, such as URI and ILR3 (Samira et al., 2018; Kim et al., 2019; Tissot et al., 2019; Lei et al., 2020; Gao et al., 2020a). In the absence of FIT, the BHLH039 gene is strongly up-regulated; however, in the absence of ZAT12, BHLH039 expression is reduced. These expression patterns can be explained by the different Fe levels, caused by different FIT activity, in wild-type and zat12 mutant plants (Le et al., 2016). Interestingly, BHLH039 expression in cat2-1 was strongly reduced (Fig. 8D), similarly to FIT. This suggested that the link coordinating the expression of the two genes is dependent on the function of CAT2. The genes encoding the closest bHLH039 homologs, BHLH038, BHLH100, and BHLH101, were similarly down-regulated in cat2-1 in comparison with the wild type (SupplementaryFig. S3A–C). Thus, the upstream bHLH signaling cascade involving the bHLH subgroup IVb and IVc transcription factors is also dependent on ROS regulation.
Fig. 8.
Regulation of Fe acquisition and homeostasis genes in response to increased H2O2 caused by the absence of CAT2. (A–F) Gene expression of (A) FIT, (B) FRO2, (C) IRT1, (D) BHLH039, (E) PYE, and (F) BHLH105 (ILR3) in wild-type and cat2-1 plants grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply (n=3). Bars represent mean values, ±SD. Different letters indicate statistically significant differences (P<0.05).
This was confirmed when we tested additional regulatory genes, encoding the Fe homeostasis regulator PYE (Long et al., 2010), from subgroup IVb, and the upstream regulators ILR3 and BHLH104 (Zhang et al., 2015), from subgroup IVc. PYE gene expression is co-regulated with that of BHLH039, and, similarly to BHLH039, the expression of PYE was reduced in the cat2-1 mutant (Fig. 8E), confirming that the upstream regulation is affected by cellular H2O2 levels. In contrast, ILR3 (Fig. 8F) and BHLH104 (Supplementary Fig. S3D) expression remained unaffected in cat2-1 compared with the wild type, suggesting that a H2O2-dependent post-translational event inhibits the activity of ILR3 and its homologs in response to prolonged Fe deficiency.
Discussion
ROS signaling is one of the key determinants underlying plant environmental responses (He et al., 2018). Among ROS, H2O2 is directly involved in the regulation of responses to prolonged Fe deficiency through promoting the expression of the transcription factor ZAT12 and regulating its stability (Brumbarova et al., 2016b; Le et al., 2016). In this study, we demonstrate that the accumulation of H2O2 in Arabidopsis roots under prolonged Fe deficiency is dependent on the activity of CAT2 (Figs 6F, 7). With the help of a CAT2 loss-of-function mutant, we were able to demonstrate that H2O2-dependent regulation affects not only the Fe uptake-related FIT target genes (Fig. 8A–C) but also the genes encoding the transcription factors from subgroup Ib (bHLH038, bHLH039, bHLH100, and bHLH101) and PYE (Fig. 8D, E; Supplementary Fig. S3A–C). The genes encoding the upstream regulators ILR3 and bHLH104 are themselves not affected by the increased H2O2 levels in the cat2-1 mutant (Fig. 8F; Supplementary Fig. S3D). However, ILR3 and bHLH104 protein activity is affected, a fact that points in the direction of an H2O2-dependent post-translational regulation of these proteins (Fig. 9).
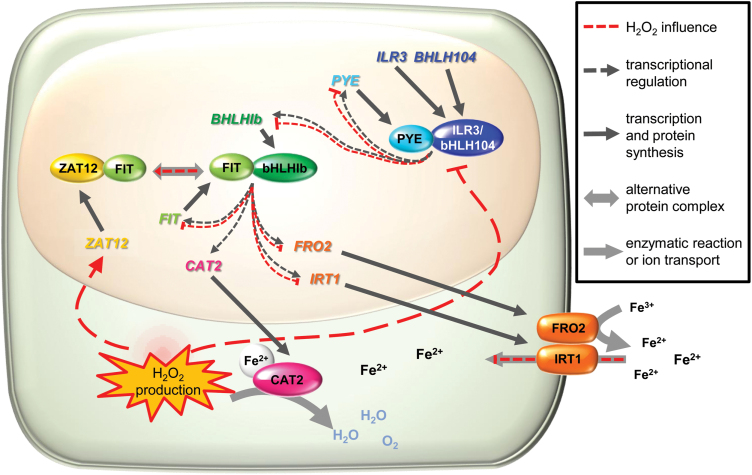
Fig. 9.
Proposed model for the role of FIT, CAT2, and H2O2 in the regulation of Fe acquisition and homeostasis under prolonged Fe deficiency. Subgroup IVc proteins ILR3 and bHLH104, together with other transcription factors (not depicted), form a complex with PYE and regulate the expression of downstream Fe homeostasis genes (dark dotted arrows), including PYE itself and subgroup Ib BHLHgenes (BHLH038, BHLH039, BHLH100, and BHLH101). Subgroup Ib bHLHs interact with FIT, a complex which positively regulates FIT, Fe acquisition genes, such as FRO2 and IRT1, and CAT2. CAT2 protein is a heme-containing H2O2 dismutating enzyme, which requires Fe, imported with the help of FRO2 ferric reductase and the transporter IRT1. Under prolonged Fe deficiency, H2O2 production is increased and acts in two directions (red dotted lines). First, high H2O2 levels negatively influence the positive regulation by PYE–ILR3/bHLH104. Thus, transcription of PYE and subgroup Ib BHLH genes is down-regulated. This depletion of subgroup Ib bHLH proteins is the first factor that negatively influences the formation of the positive Fe acquisition transcriptional complex FIT–bHLH Ib and results in attenuation of FRO2 and IRT1 expression, and therefore Fe import. The second effect of H2O2 accumulation is the activation of the ROS-responsive transcription factor ZAT12. ZAT12 can interact with FIT, forming an inactive FIT complex. This is the second factor that prevents the formation of the FIT–bHLH Ib positive regulatory complex. The role of CAT2 would be to limit the H2O2 levels by dismutating the excess H2O2 to balance the efficiency at which the transcriptional regulatory cascade functions.
Our data suggest that the role of CAT2 in this complex regulatory interplay most probably is to control the upper limit of available H2O2 under prolonged Fe deficiency. Under this condition, an as yet unidentified H2O2 source is activated and CAT2 activity is enhanced to maintain a balance of higher than usual cellular H2O2 levels. The data suggest that even though in roots CAT2 shares activity with the two other isoforms, CAT1 and CAT3, it is still the predominant catalase isoform determining H2O2 degradation under prolonged Fe deficiency. This is supported by the fact that cat2-1 plants exhibit decreased catalase activity and increased H2O2 cellular levels compared with the wild type under these conditions (Fig. 6E, F). In terms of gene expression, previous studies reported no compensatory effect of CAT1 and CAT3 expression in the absence of CAT2 in leaves (Queval et al., 2007). Under Fe deficiency, however, the expression of CAT1 and CAT3 was increased in roots of cat2-1 compared with the wild type, probably as an attempt at compensation for the missing CAT2 (Fig. 6A–C).
The obtained data support a model in which CAT2 is deeply integrated in the cellular regulatory events under prolonged Fe limitation (Fig. 9). The CAT2 gene closely responds to the Fe status of the root and the presence of FIT, suggesting that it is either a direct FIT target or a strongly FIT-dependent gene. Indirectly, through H2O2 availability and potentially additional players, including FIT, the activity of CAT2 modulates the functionality of the Fe response transcriptional cascade. As the CAT2 gene is dependent on the function of this cascade, this also functions as a feedback loop for the regulation of CAT2. An additional level of interplay between CAT2 and Fe response comes from the end result of the transcriptional cascade, namely the production of the IRT1 transporter, that provides Fe necessary for the enzymatic activity of CAT2.
An interesting question concerns the localization of the CAT2-derived effects, on both the subcellular and whole-organ level. CAT2 was known as an exclusively peroxisomal protein; however, recent studies clearly demonstrate the presence, function, and significance of catalases outside the peroxisome. Examples are available for additional CAT roles in the cytoplasm and the nucleus (He et al., 2018). It has also been proposed that CATs can relocate in the cell in response to redox signals (Foyer et al., 2020). At this point, it is not possible to speculate if a specific compartment might harbor catalase activity orchestrating the responses to prolonged Fe limitation. At the same time, however, our data clearly suggest the domains within the root where the described regulatory events might take place. In wild-type plants under prolonged Fe deficiency, the H2O2 level in the differentiation zone of the root was clearly increased, especially in regions where O2˙ −, visible in the central cylinder, reached a plateau (Fig. 2D–F). The presence of H2O2 in this region, which we refer to as the early differentiation zone, is very pronounced in root hair cells, coinciding very well with the reported expression of the principal Fe transporter IRT1 (Barberon et al., 2011; Blum et al., 2014). These observations suggest that the root hair cells of the early differentiation zone are an important location where the described regulatory crosstalk occurs. This is supported by the fact that under prolonged Fe deficiency, cat2-1 plants show strong H2O2 signal in root hair cells of this region (Fig. 7D–F), suggesting that in root hair cells the role of CAT2 is to metabolize excess H2O2 to maintain ROS balance and coordinate Fe responses.
Excess Fe supply is another condition that requires adjustment of the Fe uptake capacity of the plant to avoid Fe toxicity. It was previously demonstrated that the balance between H2O2 and O2˙ − in the root meristematic and differentiation zones partially mediated the response of the root system architecture to Fe excess (Reyt et al., 2015).
At present, the details of the mechanistic role of H2O2 in the process remain unclear. FIT and its target genes are dependent on the presence of bHLH039. BHLH39 and PYE are co-expressed, and their promoters are up-regulated in response to Fe deficiency by the bHLH subgroup IVc transcription factors (Gao et al., 2019). Subgroup IVc bHLHs ILR3 and bHLH104 are transcriptionally up-regulated by Fe deficiency (Wang et al., 2017; Samira et al., 2018), similarly to FIT and subgroup Ib bHLHs. Very interestingly, however, their gene expression was not affected by ROS in the cat2-1 mutant (Fig. 8F; Supplementary Fig. S3D), while their protein activity, judged by the expression of their direct target genes BHLH039 and PYE (Fig. 8D, E), was inhibited by ROS in cat2-1. Low subgroup Ib transcription factor activity leads to lower FIT activity and Fe acquisition downstream of FIT in cat2-1 (Figs 5E, F, 8A–C). Our data conform with previous observations from BHLH039 overexpression in plants (Naranjo-Arcos et al., 2017). It was shown that increased Fe levels due to BHLH039 overexpression resulted in a down-regulation of all BHLH subgroup Ib genes and PYE, indicating that the Fe deficiency response cascade acting upstream of these genes was not active. At the same time, BHLH039-overexpressing plants had high levels of ROS and, accordingly, elevated ZAT12 expression along with symptoms of oxidative stress. Here, we show that the presence of ROS, even in the absence of Fe, is responsible for switching off the Fe deficiency response. ZAT12 may be one of the regulators mediating the inhibition through FIT interaction.
Therefore, in our model, we propose that H2O2 post-translationally inhibits transcriptional regulators, such as ILR3 and bHLH104, and therefore causes the observed downstream transcriptional changes. Redox-dependent conformational changes have been shown to affect protein activity and nuclear localization, for example of NONEXPRESSOR OF PATHOGENESIS-RELATED GENE 1 (NPR1), involved in pathogen response (Mou et al., 2003; Tada et al., 2008). Alternatively, or additionally, redox-sensing mechanisms may involve differential binding to a partner protein, or altered protein stability and DNA binding activity (He et al., 2018). Further studies are needed to determine whether the transcription factor proteins ILR3, bHLH104, or other subgroup IVc proteins or URI are directly controlled by ROS. Their activity is regulated by Fe-dependent post-translational control mechanisms. These include, for example, E3 ligases, determining bHLH transcription factor protein abundance, and their activity might also be ROS controlled (Hindt et al., 2017).
Previous studies have proposed that ROS regulation displays considerable interplay with other compounds, such as reactive nitrogen species (Wang and Chu, 2020), among which nitric oxide has been shown to affect plant responses to Fe limitation (Graziano and Lamattina, 2007; Meiser et al., 2011). Another interesting aspect of ROS is their interplay with calcium (Ca2+) signaling (Kollist et al., 2019). The signal relay between ROS and Ca2+ in the central cylinder may result in the generation of O2˙ − through the action of RESPIRATORY BURST OXIDASE HOMOLOGS (RBOHs). As this radical is short-lived and is quickly converted to H2O2 (Jimenez-Quesada et al., 2016), it is of great interest as a potential H2O2 donor for the above-described signaling events. The subcellular origin of H2O2 production is important, as it may influence the transcriptional outcome (Sewelam et al., 2014). Excess H2O2 might inhibit the Ca2+–ROS cascade and such an inhibition might be the reason for the ‘gaps’ we observed in the O2˙ − staining in the central cylinder of cat2-1 plants. Due to this, we cannot exclude that disturbed Ca2+ signaling might contribute to the Fe response phenotypes of CAT2 loss-of-function plants, as Ca2+ is documented to play a role in both transcriptional (Gratz et al., 2019) and post-translational regulation of Fe import (Dubeaux et al., 2018; Khan et al., 2019), targeting FIT and IRT1 activity, respectively.
In summary, here we described a role for CAT2 in modulating the root responses to prolonged Fe deficiency. This is achieved through the regulation of H2O2 levels in root epidermis cells, where the Fe deficiency-induced increase of generated H2O2 is counteracted by CAT2 to a level that prevents toxic H2O2 effects and yet allows the maintenance of sufficient H2O2 amounts to coordinate the transcriptional Fe response. Through mutual regulation, CAT2 and FIT represent components of a regulatory loop for regulation of plant responses to Fe deficiency, where H2O2 functions as an adjustable signaling intermediate.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Catalase activity in fit-3 mutant plants.
Fig. S2. Regulation of the ZAT12 gene in wild-type and cat2-1 plants grown for 10 d under sufficient (+Fe) or deficient (–Fe) Fe supply.
Fig. S3. Regulation of Fe acquisition and homeostasis genes in response to increased H2O2 caused by the absence of CAT2.
Table S1. List of primers used for RT-qPCR.
Acknowledgements
We thank Elke Wieneke for help with RT-qPCR and plant growth, and Ginte Matthäi for help with establishing the Fe content measurement. This work received funding from Germany’s Excellence Strategy, EXC 2048/1, Project ID: 390686111.
Author contributions
RI, TB, and PB conceived the project; CvdM, RI, ME, and TB performed the experiments; CvdM, RI, and TB analyzed data; all authors discussed the results; CvdM, RI, and TB wrote the original draft and prepared the figures; all authors contributed to finalizing the manuscript; PB acquired funding.
Data availability
Sequence data from this article can be found in the TAIR and GenBank data libraries under accession numbers: BHLH038 (AT3G56970), BHLH039 (AT3G56980), BHLH047 (PYE, AT3G47640) BHLH100 (AT2G41240), BHLH101 (AT5G04150), BHLH104 (AT4G14410), BHLH105 (ILR3, AT5G54680), CAT1 (AT1G20630), CAT2 (AT4G35090), CAT3 (AT1G20620), FIT (AT2G28160), FRO2 (AT1G01580), IRT1 (AT4G19690), and ZAT12 (AT5G59820).
The data supporting the findings of this study are available from the corresponding author, Tzvetina Brumbarova, upon request.
References
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G. 2011. Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proceedings of the National Academy of Sciences, USA 108, E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A, Brumbarova T, Bauer P, Ivanov R. 2014. Hormone influence on the spatial regulation of IRT1 expression in iron-deficient Arabidopsis thaliana roots. Plant Signaling & Behavior 9, e28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R. 2015. Molecular mechanisms governing Arabidopsis iron uptake. Trends in Plant Science 20, 124–133. [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Ivanov R. 2018. SNX1-mediated protein recycling: piecing together the tissue-specific regulation of Arabidopsis iron acquisition. Plant Signaling & Behavior 13, e1411451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Ivanov R. 2019. The nutrient response transcriptional regulome of Arabidopsis. iScience 19, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Le CT, Bauer P. 2016a. Hydrogen peroxide measurement in Arabidopsis root tissue using Amplex Red. Bio-Protocol 6, doi: 10.21769/BioProtoc.1999. [Google Scholar]
- Brumbarova T, Le CT, Ivanov R, Bauer P. 2016b. Regulation of ZAT12 protein stability: the role of hydrogen peroxide. Plant Signaling & Behavior 11, e1137408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Alejandro S, Carbonell P, Perez-Amador MA, Fayos J, Bellés JM, Rodriguez PL, Serrano R. 2007. The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. The Plant Journal 52, 1052–1065. [DOI] [PubMed] [Google Scholar]
- Czarnocka W, Karpiński S. 2018. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radical Biology & Medicine 122, 4–20. [DOI] [PubMed] [Google Scholar]
- Dubeaux G, Neveu J, Zelazny E, Vert G. 2018. Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Molecular Cell 69, 953–964.e5. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Baker A, Wright M, Sparkes IA, Mhamdi A, Schippers JHM, Van Breusegem F. 2020. On the move: redox-dependent protein relocation in plants. Journal of Experimental Botany 71, 620–631. [DOI] [PubMed] [Google Scholar]
- Frugoli JA, Zhong HH, Nuccio ML, McCourt P, McPeek MA, Thomas TL, McClung CR. 1996. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiology 112, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Robe K, Bettembourg M, et al. 2020. a. The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. The Plant Cell 32, 508–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Robe K, Dubos C. 2020b. Further insights into the role of bHLH121 in the regulation of iron homeostasis in Arabidopsis thaliana. Plant Signaling & Behavior 15, 1795582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Robe K, Gaymard F, Izquierdo E, Dubos C. 2019. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Frontiers in Plant Science 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz R, Manishankar P, Ivanov R, et al. 2019. CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling. Developmental Cell 48, 726–740 e710. [DOI] [PubMed] [Google Scholar]
- Graziano M, Lamattina L. 2007. Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. The Plant Journal 52, 949–960. [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N. 2013. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiology 163, 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Van Breusegem F, Mhamdi A. 2018. Redox-dependent control of nuclear transcription in plants. Journal of Experimental Botany 69, 3359–3372. [DOI] [PubMed] [Google Scholar]
- Hindt MN, Akmakjian GZ, Pivarski KL, Punshon T, Baxter I, Salt DE, Guerinot ML. 2017. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inupakutika MA, Sengupta S, Devireddy AR, Azad RK, Mittler R. 2016. The evolution of reactive oxygen species metabolism. Journal of Experimental Botany 67, 5933–5943. [DOI] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P. 2012. Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Molecular Plant 5, 27–42. [DOI] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Blum A, Jantke AM, Fink-Straube C, Bauer P. 2014. SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. The Plant Cell 26, 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. 2004. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Letters 577, 528–534. [DOI] [PubMed] [Google Scholar]
- Jiménez-Quesada MJ, Traverso JÁ, Alché Jde D. 2016. NADPH oxidase-dependent superoxide production in plant reproductive tissues. Frontiers in Plant Science 7, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Gratz R, Denezhkin P, et al. 2019. Calcium-promoted interaction between the C2-domain protein EHB1 and metal transporter IRT1 inhibits Arabidopsis iron acquisition. Plant Physiology 180, 1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khedia J, Agarwal P, Agarwal PK. 2019. Deciphering hydrogen peroxide-induced signalling towards stress tolerance in plants. 3 Biotech 9, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, LaCroix IS, Gerber SA, Guerinot ML. 2019. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proceedings of the National Academy of Sciences, USA 116, 24933–24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2012. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology 63, 131–152. [DOI] [PubMed] [Google Scholar]
- Kollist H, Zandalinas SI, Sengupta S, Nuhkat M, Kangasjarvi J, Mittler R. 2019. Rapid responses to abiotic stress: priming the landscape for the signal transduction network. Trends in Plant Science 24, 25–37. [DOI] [PubMed] [Google Scholar]
- Le CT, Brumbarova T, Bauer P. 2019. The interplay of ROS and iron signaling in plants. In: Panda S, Yamamoto Y, eds. Redox homeostasis in plants. Signaling and communication in plants. Cham: Springer, 43–66. [Google Scholar]
- Le CT, Brumbarova T, Ivanov R, Stoof C, Weber E, Mohrbacher J, Fink-Straube C, Bauer P. 2016. Zinc finger of Arabidopsis thaliana12 (ZAT12) interacts with Fer-like iron deficiency-induced transcription factor (FIT) linking iron deficiency and oxidative stress responses. Plant Physiology 170, 540–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei R, Li Y, Cai Y, Li C, Pu M, Lu C, Yang Y, Liang G. 2020. bhlh121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis. Molecular Plant 13, 634–649. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. 1983. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochemical Society Transactions 11, 591–592. [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. 2010. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. The Plant Cell 22, 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. 1997. Regulation of catalases in Arabidopsis. Free Radical Biology & Medicine 23, 489–496. [DOI] [PubMed] [Google Scholar]
- Meiser J, Lingam S, Bauer P. 2011. Posttranslational regulation of the iron deficiency basic helix–loop–helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiology 157, 2154–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G. 2010. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. Journal of Experimental Botany 61, 4197–4220. [DOI] [PubMed] [Google Scholar]
- Mittler R. 2017. ROS are good. Trends in Plant Science 22, 11–19. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. 2003. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Naranjo-Arcos MA, Bauer P. 2016. Iron nutrition, oxidative stress, and pathogen defense. In: Erkekoglu P, Kocer-Gumusel B, eds. Nutritional deficiency. InTech, 63–98. [Google Scholar]
- Naranjo-Arcos MA, Maurer F, Meiser J, Pateyron S, Fink-Straube C, Bauer P. 2017. Dissection of iron signaling and iron accumulation by overexpression of subgroup Ib bHLH039 protein. Scientific Reports 7, 10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls P, Fita I, Loewen PC. 2000. Enzymology and structure of catalases. In: Advances in Inorganic Chemistry, Vol. 51. Academic Press, 51–106. [Google Scholar]
- Noctor G, Foyer CH. 2016. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiology 171, 1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Foyer CH. 2016. Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant, Cell & Environment 39, 1140–1160. [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G. 2007. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. The Plant Journal 52, 640–657. [DOI] [PubMed] [Google Scholar]
- Reyt G, Boudouf S, Boucherez J, Gaymard F, Briat JF. 2015. Iron- and ferritin-dependent reactive oxygen species distribution: impact on Arabidopsis root system architecture. Molecular Plant 8, 439–453. [DOI] [PubMed] [Google Scholar]
- Samira R, Li B, Kliebenstein D, Li C, Davis E, Gillikin JW, Long TA. 2018. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Molecular Biology 97, 297–309. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Mächtel R, Ammon A, Engelsdorf T, Schmitz J, Maurino VG, Voll LM. 2020. Reactive oxygen species dosage in Arabidopsis chloroplasts can improve resistance towards Colletotrichum higginsianum by the induction of WRKY33. New Phytologist 226, 189–204. [DOI] [PubMed] [Google Scholar]
- Schwarz B, Bauer P. 2020. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. Journal of Experimental Botany 71, 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N, Jaspert N, Van Der Kelen K, Tognetti VB, Schmitz J, Frerigmann H, Stahl E, Zeier J, Van Breusegem F, Maurino VG. 2014. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Molecular Plant 7, 1191–1210. [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Arnaud D. 2019. Hydrogen peroxide metabolism and functions in plants. New Phytologist 221, 1197–1214. [DOI] [PubMed] [Google Scholar]
- Su T, Wang P, Li H, et al. 2018. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. Journal of Integrative Plant Biology 60, 591–607. [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. 2008. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit J, Irazusta V, Moreno-Cermeño A, Ros J. 2006. Colorimetric assay for the quantitation of iron in yeast. Analytical Biochemistry 351, 149–151. [DOI] [PubMed] [Google Scholar]
- Tissot N, Robe K, Gao F, et al. 2019. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytologist 223, 1433–1446. [DOI] [PubMed] [Google Scholar]
- Trofimov K, Ivanov R, Eutebach M, Acaroglu B, Mohr I, Bauer P, Brumbarova T. 2019. Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT. Plant Direct 3, e00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. [DOI] [PubMed] [Google Scholar]
- Wang C, Yao X, Yu D, Liang G. 2017. Fe-deficiency-induced expression of bHLH104 enhances Fe-deficiency tolerance of Arabidopsis thaliana. Planta 246, 421–431. [DOI] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. 2007. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908. [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ. 2013. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Molecular Plant 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chu C. 2020. S-Nitrosylation control of ROS and RNS homeostasis in plants: the switching function of catalase. Molecular Plant 13, 946–948. [DOI] [PubMed] [Google Scholar]
- Yang Z, Mhamdi A, Noctor G. 2019. Analysis of catalase mutants underscores the essential role of CATALASE2 for plant growth and day length-dependent oxidative signalling. Plant, Cell & Environment 42, 688–700. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. 2008. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research 18, 385–397. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB. 2015. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. The Plant Cell 27, 787–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in the TAIR and GenBank data libraries under accession numbers: BHLH038 (AT3G56970), BHLH039 (AT3G56980), BHLH047 (PYE, AT3G47640) BHLH100 (AT2G41240), BHLH101 (AT5G04150), BHLH104 (AT4G14410), BHLH105 (ILR3, AT5G54680), CAT1 (AT1G20630), CAT2 (AT4G35090), CAT3 (AT1G20620), FIT (AT2G28160), FRO2 (AT1G01580), IRT1 (AT4G19690), and ZAT12 (AT5G59820).
The data supporting the findings of this study are available from the corresponding author, Tzvetina Brumbarova, upon request.