Abstract
Cytosine-phosphate-guanine (CpG) oligonucleotides are commonly-used vaccine adjuvants to promote the activation of antigen-presenting cells (APCs). To mount an effective immune response, CpG needs to be internalized and bind to its endosomal Toll-like receptor 9 (TLR-9) inside the APCs. Using flow cytometry and fluorescence microscopy, this article presents the cellular uptake data of the amino-dextran nanoparticle (aDNP) and aDNP loaded with CpG immobilized on its surface by either electrostatic adsorption or covalent conjugation. The uptake of fluorescently-labelled aDNPs by murine splenic dendritic cells and macrophages was determined by flow cytometry and uptake by murine bone-marrow-derived dendritic cells was evaluated by fluorescence microscopy. The data presented in this paper correlates with the in vitro immune-stimulatory activity observed for the two different CpG loading methods in the research article “Nanoparticle system based on amino-dextran as a drug delivery vehicle: immune-stimulatory CpG-oligonucleotide loading and delivery” (Nguyen et al., 2020) [1]. The data provide experimental evidence for a better understanding how the nanoparticle surface loading method of CpG influences the uptake of these nanoparticles by antigen-presenting cells as a step guide in the design of more effective vaccine formulations.
Keywords: Dextran nanoparticle, CpG oligonucleotides, Electrostatic adsorption, Covalent conjugation, Flow cytometry, Fluorescence microscopy, Cellular uptake, Vaccine formulation
Specifications Table
| Subject | Pharmaceutical Science, Immunology |
| Specific subject area | Vaccine delivery system, Uptake of nanoparticles by immune cells |
| Type of data | Graph, Image |
| How data was acquired | Flow cytometry, Gallios flow cytometer by Beckman Coulter Fluorescence microscopy, IX53 inverted fluorescence microscope by Olympus |
| Data format | Raw and analyzed |
| Parameters for data collection | Fluorescently-labelled CpG-loaded amino-dextran nanoparticles (25 µg/mL) were incubated with splenocytes or bone-marrow-derived dendritic cells (BMDCs) generated from C57BL/6 mice at 4 or 37 °C. After 4 h or 24 h incubation, the sample was then analyzed by either flow cytometry or fluorescence microscopy. |
| Description of data collection | For flow cytometry, the live cells were determined by a live-dead dye, and the dendritic cell or macrophage population was identified using fluorescently-labelled antibodies, anti-CD11c-FITC and anti-F4/80-PE, respectively. The median fluorescence intensity (MFI) of the fluorescent aDNP associated with cells was then measured by flow cytometry. For fluorescence microscopy, BMDCs was fixed by 4% paraformaldehyde and BMDC nucleus was stained with Hoestch 34322. The fixed cells were observed by fluorescence microscopy. |
| Data source location | University of Otago, Dunedin, New Zealand |
| Data accessibility | Data is within this article. Raw data is within the supplementary material |
| Related research article | H.V. Nguyen, K. Campbell, G.F. Painter, S.L. Young, G.F. Walker, Nanoparticle system based on amino-dextran as a drug delivery vehicle: immune-stimulatory CpG-oligonucleotide loading and delivery, Pharmaceutics 12 (2020) 1150; https://doi.org/10.3390/pharmaceutics12121150. |
Value of the Data
-
•
The data provide a greater understanding of why the method of CpG immobilization to the surface of a nanoparticle influences its immune response.
-
•
The data will be useful for researchers who are designing and developing nanoparticle systems for the delivery of nucleic acids.
-
•
The data highlight the value of investigating the cellular uptake of nanoparticles by the target cells when considering the method of immobilization of nucleic acids to a nanoparticle surface.
1. Data Description
These data add value to our research article “Nanoparticle system based on amino-dextran as a drug delivery vehicle: immune-stimulatory CpG-oligonucleotide loading and delivery” which have previously demonstrated that CpG conjugated to the surface of aDNP had a higher level of immune-stimulation for BMDCs in vitro compared to CpG adsorbed onto aDNP [1]. To establish the concentration of the fluorescently-labelled aDNP for the uptake experiments, splenocytes were incubated with various concentrations of Dylight-labelled unloaded aDNP and then analyzed by flow cytometry. Supplementary Fig. S1 shows the flow cytometry histograms of splenocytes which were incubated with either 5, 10, 25, or 50 µg/mL at 37 °C for 24 h or left untreated.
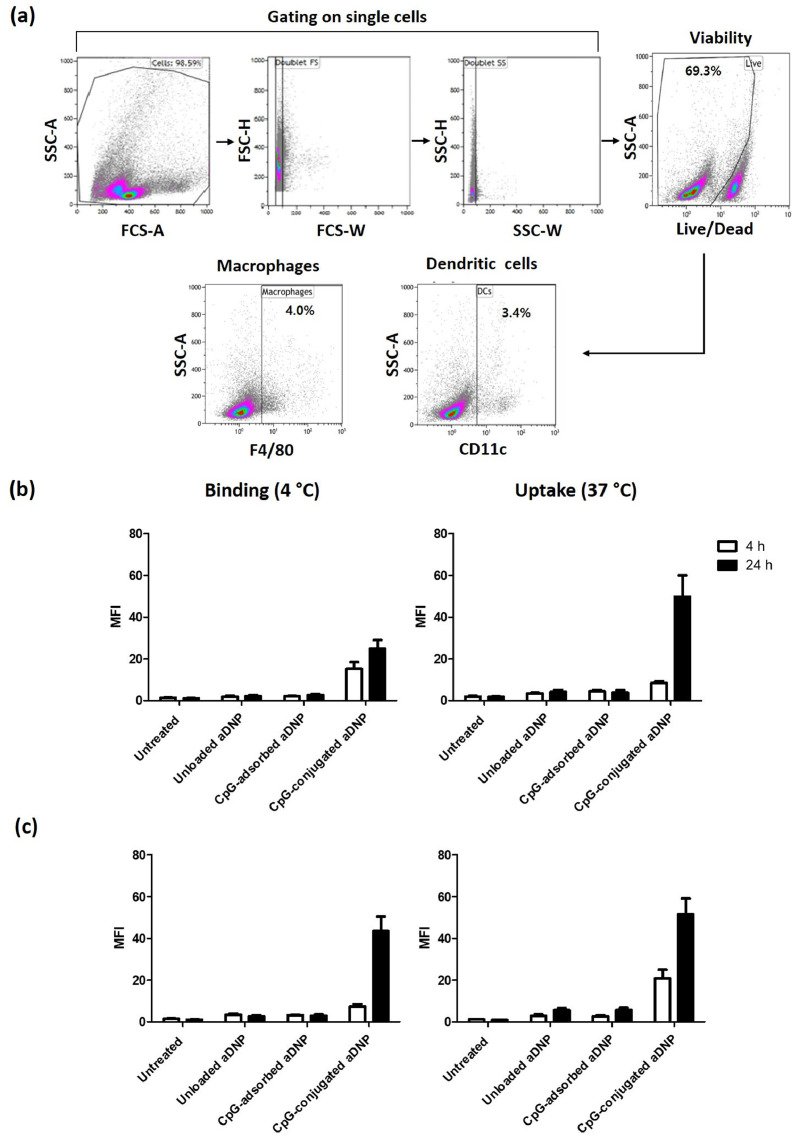
Using the aDNP dose of 25 µg/mL, Fig. 1 shows the flow cytometry data for the association of aDNP, CpG-adsorbed aDNP, or CpG-conjugated aDNP to murine splenic dendritic cells and macrophages at either 4 or 37 °C at 4 h and 24 h time points. To identify fluorescently-labelled aDNPs association with dendritic cells or macrophages, splenocytes were gated on single cells, viability, and CD11c+ dendritic cells or F4/80+ macrophages (Fig. 1a). The level of binding (4 °C) or uptake (37 °C) of the fluorescently-labelled aDNPs at 4 and 24 h time points by dendritic cells is presented in Fig. 1b, the raw data are summarized in the supplementary Table S1 and Table S2. The data for the cellular association by macrophages are shown in Fig. 1c, with the raw data being presented in the supplementary Table S3 and Table S4.
Fig. 1.
Cellular association of the unloaded amino-dextran nanoparticle (aDNP) and CpG-loaded aDNPs by splenocytes. Splenocytes were incubated with fluorescently-labelled unloaded aDNP, CpG-adsorbed-aDNP, CpG-conjugated-aDNP (25 µg/mL) or PBS as control at 4 or 37 °C for 4 h or 24 h. a) Gating strategy for flow cytometry analysis of uptake studies. The cellular association of aDNPs by (b) dendritic cells and (c) macrophages is presented as median fluorescence intensity (MFI) determined by flow cytometry for each cell population. Data are the mean of MFI ± the standard error of the mean (SEM) obtained from three independent replicates.
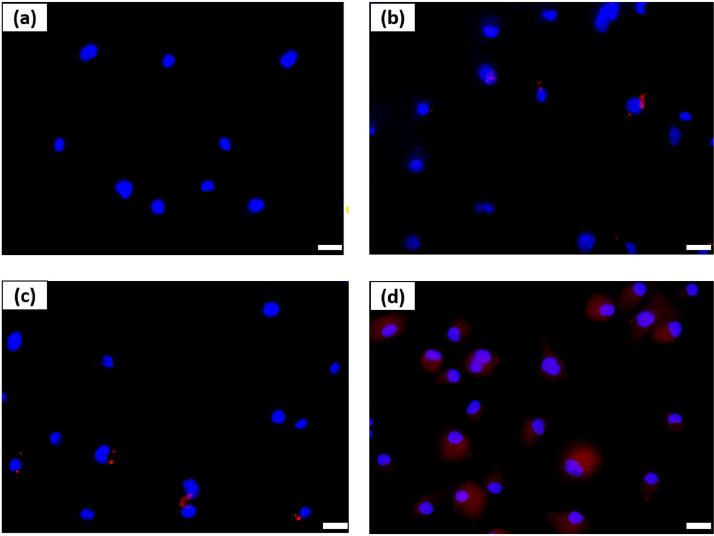
In addition to evaluating the cellular uptake of fluorescently-labelled aDNPs by flow cytometry, the fluorescence microscopy was used to determine the cellular uptake and the localization of nanoparticles. The BMDCs were used for the fluorescence microscopy studies instead of splenocytes, as BMDCs provide a higher percentage of the vaccine-target dendritic cells (greater than 75%) compared to splenocytes (typically 4%). Fig. 2 shows the fluorescence microscopic pseudo-colour images of BMDCs which have been incubated with fluorescently-labelled aDNPs at 37 °C for 24 h. The blue colour is the stained nuclei of BMDCs and the red colour is the fluorescently-labelled aDNPs.
Fig. 2.
Fluorescence microscopic images of bone-marrow-derived dendritic cells incubated with 25 µg/mL fluorescently-labelled amino-dextran nanoparticles (aDNPs) for 24 h at 37 °C; (a) no treatment, (b) unloaded amino-dextran nanoparticle (aDNP), (c) CpG-adsorbed aDNP, and (d) CpG-conjugated aDNP. The nucleus was stained with Hoechst 33342 (blue), and fluorescently-labelled aDNP (red). Optical maginification, 40 ×. Scale bar, 10 µm.
2. Experimental Design, Materials and Methods
2.1. Materials
Amino-dextran (70 kDa) functionalized with 50 amines per dextran molecule was obtained from Fina Biosolutions (Rockville, MD). Phosphorothioate-modified class-B CpG-1668 oligodeoxynucleotide (5′-TCCATGACGTTCCTGATGCT-3′) modified with an amino group at 3’-end was purchased from Integrated DNA Technologies (Coralville, IA). Complete Iscove's Modified Dulbecco's Medium (cIMDM) comprising of IMDM and GlutaMAX, heat-inactivated fetal bovine serum (FBS), Dylight-633 NHS ester, Hoechst 33342 (10 mg/mL) solution in water, and LIVE/DEAD™ Fixable Yellow Dead Cell Stain Kit were obtained from Thermo Fisher Scientific (Pittsburg, PA). Purified anti-mouse CD16/32 antibody (Fc block), fluorescently-tagged monoclonal antibodies CD11c (clone N418) and F4/80 (clone BM8), granulocyte-macrophage colony-stimulating factor (mGM-CSF) were purchased from BioLegend (San Diego, CA). Glutaraldehyde solution 25% (w/v) in water was obtained from Sigma-Aldrich (St Louis, MO). Succinimidyl-6-hydrazino-nicotinamide (HyNic) and succinimidyl 4-formyl benzoate (4FB) were obtained from TriLink Biotechnologies (San Diego, CA).
2.2. Preparation of fluorescently-labelled amino-dextran nanoparticles
2.2.1. Preparation of amino-dextran nanoparticles
Amino-dextran solution (10 mg/mL) was prepared in deionized water (1 mL) and filtered through a 0.22-µm syringe filter (Millipore Corp., Billerica, MA). The filtered amino-dextran solution (1 mL) was added to acetone (5 mL) with a flow speed of 1 mL/min using a syringe pump (200 syringe pump; Chemyx Stafford, TX) while being magnetically stirred at 500 rpm. Glutaraldehyde (1.8% (w/v) in acetone, 100 µL) was added dropwise to the dextran suspension and was continuously stirred at 500 rpm for 6 h. The suspension was then dialyzed against 2 L of deionized water at room temperature for 24 h with three exchanges of media.
2.2.2. Fluorescent labelling of amino-dextran nanoparticles
aDNP (800 µg dry mass equivalent) was buffer-exchanged into 100 µL of 0.1 M NaHCO3 buffer (pH 8.3) by centrifugal filtration (Vivaspin 500, MWCO 10 kDa, Satorius, Göttingen, Germany) at 12,000 × g and 4 °C. The nanosuspension (100 µL) was reacted with a 10-fold molar excess of Dylight-633 NHS-ester in dry dimethylformamide (10 mg/mL, 12.2 µL) at room temperature. After 3 h incubation, the unreacted dye was removed by centrifugal filtration (Vivaspin 500, MWCO 10 kDa, Satorius) with five repeated cycles at 12,000 × g and 4 °C using the PBS buffer (pH 7.4), the retentate was then resuspended into 1 mL of PBS.
2.3. CpG loading to fluorescently-labelled amino-dextran nanoparticles
2.3.1. CpG adsorption to fluorescently-labelled amino-dextran nanoparticles
Fluorescent aDNP suspended in PBS (300 µg dry weight equivalent, 375 µL) was mixed with the CpG solution in PBS (5 µg/µL, 6 µL). The mixture was gently shaken (100 rpm) at room temperature for 4 h using an orbital shaker (Ratek Instruments, Melbourne, Australia). The unbound CpG was then removed by repeated centrifugal filtration (Vivaspin 500, MWCO 10 kDa, Sartorius) with five cycles at 12,000 × g and 4 °C using PBS buffer (pH 7.4).
2.3.2. CpG conjugation to fluorescently-labelled amino-dextran nanoparticles
The amino-modified CpG (4.9 µg/µL, 134 µL) prepared in the modification buffer (0.1 M NaPO4, 0.15 M NaCl, pH 8.0) was mixed with a 20-fold molar excess of 4FB linker (40 µg/µL in DMSO, 12.5 µL) at room temperature. After 4 h incubation, the unreacted linker was washed with the reaction buffer (0.1 M NaPO4, 0.15 M NaCl, pH 6.0) by repeated centrifugal filtration (Vivaspin 500, MWCO 3 kDa, Satorius) with five cycles at 12,000 × g and 4 °C. The 4FB-modified CpG was then resuspended into 100 µL of the reaction buffer.
Fluorescent aDNP (300 µg aDNP dry weight equivalent, 375 µL) was buffer-exchanged into 60 µL of the modification buffer by centrifugal filtration (Vivaspin 500, MWCO 10 kDa, Satorius). The fluorescent aDNP suspension (100 µL) was reacted with the HyNic linker (20 µg/µL in DMSO, 2.5 µL) with a molar ratio of 1:40 at room temperature. After 3 h incubation, the unreacted linker was washed with the reaction buffer by repeated centrifugal filtration (Vivaspin 500, MWCO 10 kDa, Satorius). The HyNic-modified fluorescent aDNP was then resuspended into 60 µL of the reaction buffer.
The HyNic-modified fluorescent aDNP (60 µL) was mixed with a five-fold molar excess of 4FB-modified CpG (21.5 µL) at room temperature. After 3.5 h incubation, the fluorescent CpG-conjugated aDNP was purified and resuspended into 50 µL of PBS buffer (pH 7.4) by five cycles of centrifugal filtration (Vivaspin 500, MWCO 10 kDa, Satorius) at 12,000 × g and 4 °C.
aDNP concentration of the fluorescent CpG-loaded aDNP was determined by measuring the fluorescence intensity at the excitation wavelength (Ex) of 584 nm and emission wavelength (Em) of 620 nm using a POLARStar Omega microplate reader (BMG Labtech GmbH, Ortenberg, Germany). The concentration was then calculated using a standard curve of Dylight-labelled aDNP (0.1-2 mg/mL).
2.4. Cellular association of CpG-loaded amino-dextran nanoparticles
2.4.1. Preparation of murine splenocytes
Female C57BL/6 mice (2-3 month old) were purchased from the Biomedical Research Facility, University of Otago, Dunedin, New Zealand. The spleen was dissected from a mouse, a single cell suspension of splenocytes was then generated by gently pressing the spleen through a 70-µm cell strainer (Falcon, Becton Dickinson Labware, NJ) with a 3-mL syringe plunger. The strainer was then washed with PBS supplemented with 5% FBS to flush the remaining cells on the strainer. The single cell suspension was centrifuged (300 × g, 4 °C, 10 min), the cell pellet was resuspended in 10 mL of warm ACK (Ammonium-Chloride-Potassium) red blood cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) and incubated at room temperature for 3 min. Lysis buffer was then neutralized with 20 mL of PBS containing 5% FBS and passed through a 70-µm cell strainer (Falcon). The cell suspension was centrifuged (300 × g, 4 °C, 10 min) and the cell pellet was resuspended at 1 × 106 cells/mL in cIMDM supplemented with 5% FBS. The cell suspension (200 µL) was added to each well of a 96-well plate (Greiner Bio-One GmbH, Frickenhausen, Germany) and incubated at either 4 or 37 °C for 30 min.
2.4.2. Cellular association by splenocytes using flow cytometry
For the dose titration experiment, the pre-incubated cells at 37 °C were treated with either 5, 10, 25 or 50 µg/mL of fluorescently-labelled unloaded aDNP at 37 °C and 5% CO2 or left untreated. Following 24 h incubation, the cells were washed by PBS and stained with a LIVE/DEAD™ fixable dye. Fluorescence was then measured by a flow cytometer (Gallios, Beckman Coulter, Miami Lakes, FL) and the flow cytometric data for a total of 50,000 events per sample were analyzed using Kaluza software version 1.3 (Beckman Coulter).
For the main uptake experiment, the pre-incubated cells were treated with 25 µg/mL of fluorescently-labelled aDNP or PBS at 4 °C or 37 °C and 5% CO2. Following 4 and 24 h incubation, the cells were washed by PBS, stained with a LIVE/DEAD™ fixable dye, incubated with Fc block and stained with antibodies (anti-CD11c-FITC and anti-F4/80-PE). The stained cells were analyzed by flow cytometry using the same methods as for the dose titration experiment.
2.4.3. Generation of murine bone-marrow-derived dendritic cells
BMDCs were generated from femurs and tibiae of a mouse, as previously described by Inaba et al. [2]. The bone marrow was isolated from the bone using PBS buffer supplemented with 5% FBS. The cell suspension was passed through a 70-µm cell strainer (Falcon) and centrifuged (250 × g, 4 °C) for 7 min. The supernatant was discarded, the cell pellet was resuspended in 3 mL of warm ACK lysis buffer and incubated at room temperature for 2 min. After lysing red blood cells, the ACK lysis buffer was neutralized with 20 mL PBS containing 5% FBS and passed through a 70-µm cell strainer. The cell suspension was centrifuged (250 × g and 4 °C) for 7 min, and the cell pellet was resuspended at 0.5 × 106 cells/mL in cIMDM containing 5% FBS and 20 ng/mL mGM-CSF. The cell suspension (4 mL) was then seeded onto each well of a six-well plate (Greiner Bio-One GmbH) and cultured at 37 °C and 5% CO2 for 7 days to allow differentiation into dendritic cells.
2.4.4. Cellular association by bone-marrow-derived dendritic cells using fluorescence microscopy
After 7 days of culture, BMDCs were harvested, resuspended at 5 × 105 cells/mL in cIMDM containing 5% FBS and 20 ng/mL mGM-CSF. The cells were grown on a 13-mm glass coverslip (Marienfeld GmbH, Lauda‐Koenigshofen, Germany) coated with poly-D-Lysine in a 24-well plate (Greiner Bio-One GmbH) at 37 °C and 5% CO2. Following 24 h incubation, the cell suspension (1 mL) was treated with either fluorescently-labelled aDNP (25 µg/mL) or left untreated at 37 °C and 5% CO2. After 24 h, cells were washed thrice with PBS (1 mL), fixed with 4% (w/v) paraformaldehyde in PBS (0.5 mL) at 37 °C for 30 min and then washed thrice with PBS (1 mL). The fixed cells were stained with Hoechst 33342 (4 µg/mL, 0.5 mL) at 37 °C for 30 min and then washed thrice with PBS (1 mL). The coverslip was taken out from the 24-well plate, washed twice with deionized water, then mounted on a glass microscope slide with 10 µL mounting media (Fluoromount-G™, Thermo Fisher Scientific). The fluorescence image was observed using a fluorescence microscope (Olympus IX53, Olympus Corporation, Tokyo, Japan) with an optical magnification of 40 × and a filter set (DAPI: 360/40 nm, TXRED: 525/36 nm). The image was cropped and processed using the ImageJ software (National Institutes of Health, Bethesda, MD).
Ethics Statement
Animal studies were conducted in accordance with the ethics approved by the University of Otago Animal Ethics Committee (ethics code: DET17/17). Cervical dislocation technique was used to euthanize mice before collecting their spleens and bones.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Acknowledgements
This research was financially supported by the New Zealand's Ministry of Business, Innovation and Employment (MBIE) research grant (grant number: RTVU1603). The first author (H.V.N.) thanks the University of Otago-School of Pharmacy for providing the Doctoral Scholarship.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.106883.
Appendix. Supplementary materials
References
- 1.Nguyen H.V., Campbell K., Painter G.F., Young S.L., Walker G.F. Nanoparticle system based on amino-dextran as a drug delivery vehicle: immune-stimulatory CpG-oligonucleotide loading and delivery. Pharmaceutics. 2020;12:1150. doi: 10.3390/pharmaceutics12121150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inaba K., Inaba M., Romani N., Aya H., Deguchi M., Ikehara S., Muramatsu S., Steinman R.M. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.