Abstract
Background
N6-methyladenosine (m6A) is the most abundant mRNA modification. Whether m6A regulators can determine tumor aggressiveness and risk of immune evasion in pancreatic ductal adenocarcinoma (PDAC) remains unknown.
Methods
An integrated model named “m6Ascore” is constructed based on RNA-seq data of m6A regulators in PDAC. Association of m6Ascore and overall survival is validated across several different datasets. Overlaps of m6Ascore and established molecular classifications of PDAC is examined. Immune infiltration, enriched pathways, somatic copy number alterations (SCNAs), mutation profiles and response to immune checkpoint inhibitors are compared between m6Ascore-high and m6Ascore-low tumors.
Findings
m6Ascore is associated with dismal overall survival and increased tumor recurrence in PDAC as well as several other solid tumors including colorectal cancer and breast cancer. Basal-like (Squamous) PDAC has higher m6Ascore than that in the classical PDAC. Mechanism study showed m6Ascore-high tumors are characterized with reduced immune infiltration and T cells exhaustion. Meanwhile, m6Ascore is associated with genes regulating cachexia and chemoresistance in PDAC. Furthermore, distinct SCNAs patterns and mutation profiles of KRAS and TP53 are present in m6Ascore-high tumors, indicating immune evasion. m6Ascore-low tumors have higher response rates to immune checkpoint inhibitors (ICIs).
Interpretation
These findings indicate m6Ascore can predict aggressiveness and immune evasion in pancreatic cancer. This model has implications for pancreatic cancer prognosis and treatment response to ICIs.
Funding
This work was supported in part by National Institutes of Health (NIH) grants to M. Li (R01 CA186338, R01 CA203108, R01 CA247234 and the William and Ella Owens Medical Research Foundation) and NIH/National Cancer Institute Q39 award P30CA225520 to Stephenson Cancer Center.
Keywords: Pancreatic cancer, m6A regulators; Immune evasion; Immunotherapy; RNA modification
Graphical abstract
Research in context.
Evidence before this study
Pancreatic cancer is the third leading cause of cancer related death in the United States. The 5-year overall survival rate is only 9%. Epitranscriptome is an emerging field focusing on RNA modification. m6A is the most abundant mRNA modification. m6A regulators play critical roles in cancer progression and immune evasion.
Added value of this study
This study identified a signature (m6Ascore) consisting of nine m6A regulators. High m6Ascore is associated with worse prognosis in PDAC. Highly aggressive PDAC subtypes (Basal subtype, Squamous subtype, QM-PDA) have higher m6Ascore than less aggressive counterparts. m6Ascore-high tumors had distinct mutation profiles and reduced immune infiltration. m6Ascore can predict response and treatment outcome of immune checkpoint blockade.
Implications of all the available evidence
This study indicates mRNA modification as a potential therapeutic target to increase the efficacy of immunotherapy. m6Ascore could serve as a potential tool to identify patients that are more likely to respond to immunotherapy.
Alt-text: Unlabelled box
Introduction
Immunotherapy has emerged as a promising tumor treatment strategy. Immune checkpoint inhibitors (ICIs) showed impressive anti-tumor efficacy in a subset of solid tumors. However, currently we are still in lack of reliable biomarkers to predict the response to ICIs [1]. RNA modifications grant tumor cells the abilities to rapidly and reversibly alter the transcriptional profiles in order to survive in the fast changing and stressful microenvironment [2]. Among all the mRNA modification, N6-methyladenosine (m6A) is the most common modification, regulated by “writers”, “erasers” and “readers” [2], [3], [4]. m6A is catalyzed by methyltransferase complex (known as m6A writers), interpreted by reader proteins (known as m6A readers), and removed by demethylases (known as m6A erasers) [4]. m6A modification on mRNA suppressed the ability of classical dendritic cells (DCs) on antigen presenting and T cell priming by enhancing translating lysosomal cathepsin [5]. Deficiency of m6A modification could increase the infiltration of CD8+ T cells and reduce the recruitment of MDSCs [5].
Pan-cancer analysis has advanced our understanding on the landscapes of m6A regulators across different tumors [6]. A pan-cancer analysis identified a gene signature consisting of six m6A regulators and showed that high-risk patients was associated with mesenchymal subtype and tumor metastasis [7]. A signature was developed to predict the survival of gastric cancer, based on the m6A modification pattern, which included the expression of differentially expressed genes (DEGs) instead of the m6A regulators [8]. Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer related death in the United States, with a median 5-year survival rate of 9% [9]. However, the role of m6A in PDAC immune evasion remains uncharacterized. Currently, several molecular classification systems have been proposed in PDAC, including the Moffitt subtypes [10], Collisson subtypes [11], and Bailey subtypes [12]. However, these classifications are not intended for the selection of patients for immunotherapy. Identifying optimal biomarkers is key to maximizing the treatment efficacy of ICIs [13].
In this study, we constructed a model (termed as “m6Ascore”) which consists of nine m6A regulators and proposed it as a potential molecular classification for PDAC, which can identify distinct immune infiltration and mutation pattern. We also demonstrated m6Ascore as a practical tool to access the risk of immune evasion and predict response to ICIs immunotherapy.
Methods
Datasets
We collected data from three independent databases, including the TCGA (The Cancer Genome Atlas) database (https://portal.gdc.cancer.gov/), the International Cancer Genome Consortium (ICGC) database (https://dcc.icgc.org/) and Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) for the following tumors including Pancreatic ductal adenocarcinoma (PDAC), Breast Invasive Carcinoma (BRCA) and Colorectal adenocarcinoma (COAD and READ). Data were normalized by the “RSEM” pipeline and log2 transformed [14]. Ethical approval was waived by institutional ethics committee because data are obtained from public databases and all the patients are de-identified. Basic information of these datasets is listed in Table 1.
Table 1.
Basic information of the datasets included in this study.
| Dataset | No. of cases | Gender | Age | T Stage | N Stage | M Stage | Stage | Platform |
|---|---|---|---|---|---|---|---|---|
| TCGA- PDAC | 150 | M: 81 F: 69 | <60: 47, ≥60: 103 | 1-2: 21 3-4: 128 NA: 1 |
0: 39 1: 110 NA: 1 |
0: 68 1: 4 NA:78 |
I/II:142 III/IV:8 |
Illumina |
| Hugo cohort | 26 | NA | NA | NA | NA | NA | NA | Illumina HiSeq2000 |
| Van Allen cohort | 42 | M: 14 F: 28 | <60: 19, ≥60: 23 | NA | NA | 0: 0 1: 42 |
IV: 42 | Illumina |
| ICGC-PACA-AU | 89 | M: 46 F: 43 | <60:21, ≥60: 68 | NA | NA | NA | NA | Illumina HiSeq 2500 |
| GSE21501 | 102 | NA | NA | NA | NA | NA | NA | Agilent- Microarray |
| GSE17891 | 25 | NA | <60: 7, ≥60: 18 | 1-2: 6 3-4: 14 NA: 5 |
0: 6 1: 14 NA: 5 |
0: 20 NA: 5 | I/II: 20 NA: 5 | Affymetrix U133 Plus 2.0 Array |
| GSE28735 | 42 | NA | NA | NA | NA | NA | NA | Affymetrix 1.0 ST Array |
| TCGA-BRCA | 1090 | M: 12 F:1078 | <60: 579, ≥60: 511 | 1–2: 910 3–4: 177 NA: 3 | 0: 514 1–3: 556 NA:20 |
0: 907 1:22 NA:161 | I/II:798 III/IV:268 NA: 24 | Illumina |
| TCGA-COADREAD | 373 | M: 206 F: 167 | <60: 129, ≥60: 244 | 1–2: 67 3–4: 305 NA: 1 |
0: 205 1-2: 165 NA: 3 |
0: 252 1: 51 NA:70 |
I/II: 191 III/IV:164 NA: 18 | Illumina |
M: Male; F: Female; NA: Not Available.
Construction of m6Ascore model
We obtained the RNA-seq data of 20 m6A regulators, including 7 “Writers” (METTL3, METTL14, WTAP, VIRMA, RBM15, RBM15B and ZC3H13), 11 “Readers” (YTHDC1, YTHDC2, IGF2BP1, IGF2BP2, IGF2BP3, YTHDF1, YTHDF2, YTHDF3, HNRNPA2B1, HNRNPC and RBMX), and 2 “Erasers” (FTO, ALKBH5) [6]. Then we constructed the m6Ascore model using the least absolute shrinkage and selection operator machine learning algorithm [15]. Cross-validation was used to tune the parameter lambda. Finally, 9 regulators were included in the m6Ascore model, including 6 “Readers” (IGF2BP2, IGF2BP3, HNRNPC, YTHDF1, YTHDF2 and YTHDC1) and 3 “Writers” (METTL3, WTAP and VIRMA). Cut-off value was determined by maximally selected rank statistics in maxstat package.
Gene set enrichment analysis and construction of enrichment map
Hallmark gene sets were obtained from MSigDB database (https://www.gsea-msigdb.org/gsea/index.jsp) and analyzed in GSEA software (v4.0.1) [16]. EdgeR algorithm was applied to determine the DEGs [17]. The network of the top 50 DEGs was constructed with GeneMANIA [18]. For the enrichment map, the c2: curated gene sets with all canonical pathways were also downloaded from MSigDB database and analyzed in Cytoscape [19]. Number of permutations was 1000. Each gene set had 50 to 200 genes. The cutoff values of FDR q-value and similarity was 0.01 and 0.375 respectively. AutoAnnotate was used to annotate each cluster [19].
Somatic copy number alterations (SCNAs) and mutation analysis
SNP6 array data was obtained from Firehose Broad Institute GDAC (http://firebrowse.org/). Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm was utilized to identify significantly aberrant regions [20]. Custom settings were applied according to GISTIC2.0 module of GenePattern. Thresholds of amplifications and deletions were 0.10. Confidence level and focal length cutoff were 0.90 and 0.50, respectively. Regions with q-values < 0.25 were defined as significantly aberrant regions with recurrent copy number variation [20]. GRCh37 (hg19) was applied as human genome reference. Mutation frequency were analyzed with the maftools package [21]. The most significantly different mutations were listed in forest plot (P<0.01).
Immune signature and cell types abundance
The 66 immune markers was manually curated to represent the broad landscape of immune profile [22]. Specific immune cells markers were manually curated for Natural killer cells (NK cells) and DCs [23,24]. Immune cell types abundance was analyzed in CIBERSORTx and xCell by imputing the RNA-seq data from bulk tumor tissues [25,26].
Univariate and multivariate analysis
Several clinicopathological parameters were included for the univariate analysis, including age, gender, tumor location, depth (T stage), lymph node status (N stage), metastasis status (M stage), pathological grade and m6Ascore. Variables that are significantly associated with overall survival were further included in multivariate analysis.
Receiver operating characteristic (ROC) analysis
ROC analysis was performed using package “timeROC”, based on the 6-months survival or response to anti-PD-L1/anti-CTLA-4 immunotherapy, and the RNA-seq data. Area Under the Curve (AUC) was calculated to summarize the diagnostic accuracy. The 95% Confidence Interval was calculated in SPSS 20.0 (IBM).
Survival analysis
Cox proportional hazards model was applied for survival analysis. When patients were stratified into 3 groups, cut-off values were determined at the top 75% and the low 25%. When patients were stratified into 2 groups, cut-off value was determined by maximally selected rank statistics. Log-rank test was applied.
Statistics analysis
Statistics analysis was performed in R (https://www.bioconductor.org/), Prism (https://www.graphpad.com/), and SPSS (https://www.ibm.com/analytics/spss-statistics-software) using two-tailed unpaired student's t-test or log-rank test, unless otherwise specified. Wilcoxon test was applied when the data was not normally distributed [27]. Chi square test was applied to compare the response rates of immunotherapy between m6Ascore-high and m6Ascore-low group. When the total number of cases is less than 40, and/or more than 2 theoretical frequencies are less than 5, we used the Fisher's exact test. Otherwise, we used the Pearson's Chi square test. P < 0.05 was considered as significant difference.
Results
Construction of m6Ascore model
m6Ascore was constructed based on the transcriptome data of twenty m6A regulators, including “Writers”, “Readers” and “Erasers” in TCGA database. This algorithm finally enrolled nine m6A regulators, including six m6A “readers” (IGF2BP2, IGF2BP3, HNRNPC, YTHDF1, YTHDF2 and YTHDC1) and three m6A “writers” (VIRMA, METTL3 and WTAP).
m6Ascore is correlated with overall survival of multiple malignancies
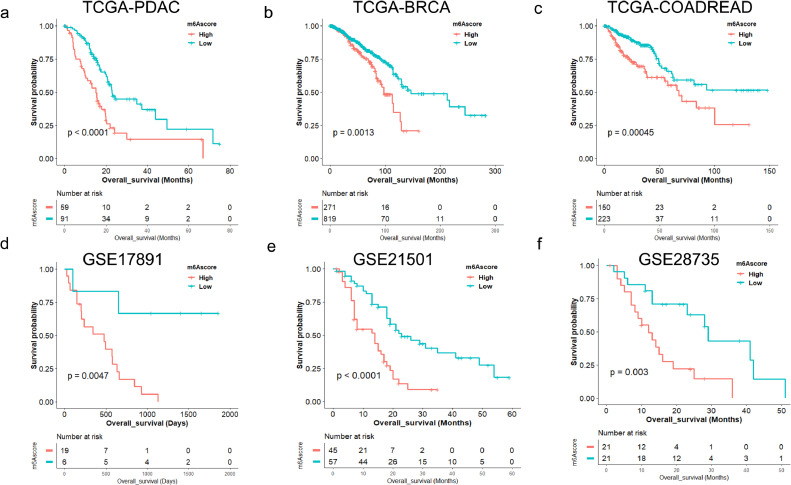
m6Ascore is correlated with overall survival of PDAC in TCGA cohort (P<0.0001). Then we validated the prognostic value of this m6Ascore model in Breast Invasive Carcinoma (BRCA, P = 0.0013) and Colorectal Adenocarcinoma (COAD and READ, P = 0.00038) (Fig. 1a-c). Furthermore, we demonstrated that m6Ascore is correlated with PDAC survival in three independent PDAC cohorts including GSE21501 cohort (P < 0.0001), GSE28735 cohort (P = 0.0030), and GSE17891 cohort (P = 0.0047) (Fig. 1d-f).
Fig. 1.
m6Ascore is associated with overall survival across multiple independent datasets. (a) Kaplan-Meier survival analysis based on m6Ascore in TCGA-PDAC cohort (P < 0.0001, log-rank test); (b) Kaplan-Meier survival analysis based on m6Ascore in TCGA-BRCA cohort (P = 0.0013, log-rank test); BRCA, breast cancer; (c) Kaplan-Meier survival analysis based on m6Ascore in TCGA-COADREAD cohort (P = 0.00038, log-rank test); COADREAD, colorectal cancer. (d) Kaplan-Meier survival analysis based on m6Ascore in GSE17891 cohort (P = 0.0047, log-rank test); (e) Kaplan-Meier survival analysis based on m6Ascore in GSE21501 cohort (P < 0.0001, log-rank test); (f) Kaplan-Meier survival analysis based on m6Ascore in GSE28735 cohort (P = 0.0030, log-rank test).
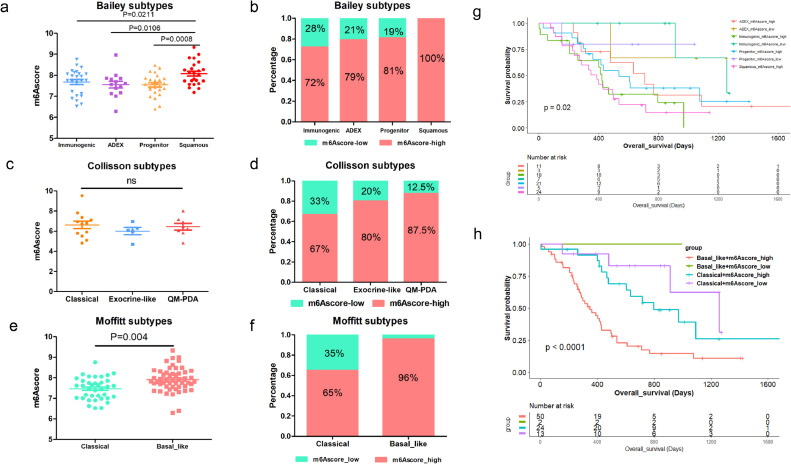
Correlation between m6Ascore model and other molecular classifications of PDAC
m6Ascore is correlated with Bailey et al defined molecular subtypes (Immunogenic, ADEX, Pancreatic Progenitor, Squamous). The Squamous subtype, which had the worst prognosis in Bailey's cohort also showed the highest m6Ascore compared to Immunogenic subtype (P = 0.0211), ADEX subtype (P = 0.0106) and Progenitor subtype (P = 0.0008) (Fig. 2a). Meanwhile, those defined as Squamous subtype were all annotated as m6Ascore-high (Fig. 2b, P = 0.060) The m6Ascore was comparable among the Collisson et al defined subtypes: Classical, Exocrine-like, QM-PDA (P = 0.550). But the QM-PDA subtype, which had the worst prognosis in Collisson's cohort, had the highest proportion of m6Ascore-high patients (Fig. 2c-d,). We further compared the m6Ascore in Moffitt et al defined PDAC subtypes: Classical and Basal-like (P < 0.001). Basal-like tumors had higher m6Ascore and 96% of these tumors were also annotated as m6Ascore-high (Fig. 2e-f). Combination of Bailey subtypes and m6Ascore in Kaplan-Meier survival analysis showed that Squamous subtype combined with m6Ascore-high patients had the most dismal prognosis (Fig. 2g). Combination of m6Ascore and Moffitt subtypes can stratify PDAC patients into four groups with distinct prognosis. m6Ascore-high and Basal-like tumors had the worst prognosis (Fig. 2h).
Fig. 2.
Correlation between m6Ascore model and other molecular classifications of PDAC. (a) Comparison of m6Ascore in molecular subtypes (Immunogenic, ADEX, Pancreatic Progenitor, Squamous) defined by Bailey et al; (b) The proportion of m6Ascore-high and m6Ascore-low in Bailey et al defined molecular subtypes; (c) Comparison of m6Ascore in molecular subtypes (Classical, Exocrine-like, QM-PDA) defined by Collisson et al; (d) Proportion of m6Ascore-high and m6Ascore-low in Collisson et al defined molecular subtypes; (e) Comparison of m6Ascore in molecular subtypes (Classical, Basal-like) defined by Moffitt et al; (f) Proportion of m6Ascore-high and m6Ascore-low in Moffitt et al defined molecular subtypes; (g) Kaplan-Meier survival analysis based on the combination of m6Ascore and Bailey et al defined molecular subtypes; (h) Kaplan-Meier survival analysis based on the combination of m6Ascore and Moffitt et al defined molecular subtypes.
m6Ascore is associated with metastasis and tumor recurrence
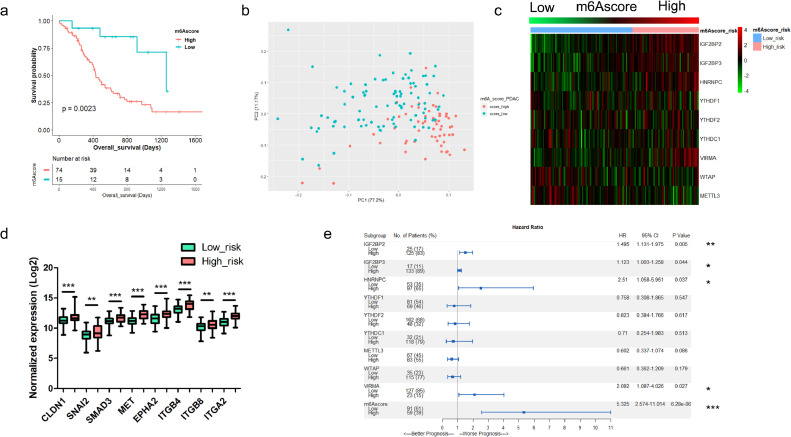
The m6Ascore-high tumors had worse prognosis than the m6Ascore-low tumors in ICGC-PDAC cohort (P = 0.0023) (Fig. 3a). Principal component analysis (PCA) showed distinct expression pattern between m6Ascore-high and m6Ascore-low tumors (Fig. 3b). Heatmap showed the expression pattern of the nine m6A regulators in PDAC tissue (Fig. 3c). The expression of each m6A regulator varied between the two groups (Fig. S1). Even if we set the cutoff value of m6Ascore as 25% and 75% percentile of all samples, m6Ascore-high was still associated with inferior survival (P < 0.0001) (Fig. S2a-b). The expression of each regulator was also correlated to m6Ascore (Fig. S2c-l). PCA analysis also showed distinct patterns of these groups (Fig. S2m). Meanwhile, we found that m6Ascore-high tumors had increased expression of several key regulators of tumor metastasis (Fig. 3d). Then we examined tumor recurrence and found that recurrent PDAC tumors had higher m6Ascore than those without recurrence (Fig. S3a). 74.6% (44/59) of m6Ascore-high tumors would develop recurrence, compared to 52.7% (48/91) in m6Ascore-low tumors (P = 0.007) (Fig. S3b). Metastasis upregulated genes are enriched in the m6Ascore-high tumors, which also had inferior prognosis independent of recurrence status (P = 0.0139 for the recurrent patients; P = 0.0301 for the recurrence-free patients) (Fig. S3c-f). Combination of recurrence status and m6Ascore can predict the survival of PDAC patients more precisely (P < 0.0001) (Fig. S3g).
Fig. 3.
Establishment and evaluation of m6Ascore model in PDAC patients. (a) Kaplan-Meier survival analysis based on m6Ascore in ICGC-PDAC cohort (P = 0.0023, log-rank test); (b) Principal component analysis (PCA) of TCGA-PDAC patients based on the expression of the m6Ascore model; (c) Heatmap showed the expression profiles of nine m6A regulators that enrolled in the m6Ascore model in TCGA-PDAC patients. m6Ascore risk was listed as column annotations; Gradient m6Ascore was listed from left to right; (d) Expression of several key regulators of tumor metastasis were upregulated in m6Ascore-high patients than in m6Ascore-low patients; (e) COX regression and forest plot were applied to examine the hazard ratio (HR) and 95% confidence interval of each m6A regulators and the m6Ascore model.
m6Ascore had better performance on predicting survival than each m6A regulator alone
We examined the prognostic value of each m6A regulator in the m6Ascore model (Fig. S4). Then, we drew the receiver operating characteristic (ROC) curve and compared the area under curve (AUC) of m6Ascore to each regulator in this model. AUC of 6-month survival in m6Ascore model was 0.738, larger than each m6A regulator alone (Fig. S5). The 95% confidence intervals were listed in Table 2.
Table 2.
Area Under the ROC Curve of 6-months survival
| Variables | Area | 95% CI |
|
|---|---|---|---|
| Lower | Upper | ||
| m6Ascore | 0.738 | 0.585 | 0.892 |
| IGF2BP2 | 0.624 | 0.465 | 0.783 |
| IGF2BP3 | 0.658 | 0.505 | 0.812 |
| YTHDF1 | 0.495 | 0.326 | 0.666 |
| YTHDF2 | 0.578 | 0.400 | 0.756 |
| YTHDC1 | 0.544 | 0.386 | 0.702 |
| HNRNPC | 0.628 | 0.441 | 0.815 |
| VIRMA | 0.719 | 0.558 | 0.800 |
| METTL3 | 0.526 | 0.372 | 0.680 |
| WTAP | 0.612 | 0.455 | 0.769 |
aUnder the nonparametric assumption.
bNull hypothesis true area = 0.5
To further determine the hazard ratio (HR) of these potential prognostic factors, we performed Cox regression analysis, and found the m6Ascore model had the highest HR (5.325, 95%CI 2.574-11.014, P = 6.28E−6) (Fig. 3e). These results indicated that m6Ascore had better performance than each m6A regulator alone on predicting survival in PDAC. Univariate (P < 0.001) and multivariate analysis (P < 0.001) showed that m6Ascore is an independent risk factor for PDAC (Table 3).
Table 3.
Univariate and Multivariate analysis in TCGA-PDAC cohort.
| Characteristics | Univariate analysis |
Multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95%CI |
P | HR | 95%CI |
P | |||
| Lower | Upper | Lower | Upper | |||||
| Age (≤60 as reference) | ||||||||
| >60 | 1.245 | 0.772 | 2.010 | 0.369 | ||||
| Gender (Male as reference) | ||||||||
| Female | 1.275 | 0.823 | 1.974 | 0.276 | ||||
| Location (Head as reference) | ||||||||
| Body | 0.657 | 0.264 | 1.634 | 0.366 | ||||
| Tail | 1.186 | 0.543 | 2.593 | 0.669 | ||||
| Others | 0.157 | 0.021 | 1.157 | 0.069 | ||||
| Depth (T1 as reference) | ||||||||
| T2 | 3.759 | 0.461 | 30.681 | 0.216 | ||||
| T3 | 5.074 | 0.703 | 36.627 | 0.107 | ||||
| T4 | 2.608 | 0.162 | 41.914 | 0.499 | ||||
| Lymph node status (N0 as reference) | ||||||||
| N1 | 1.893 | 1.094 | 3.274 | 0.022 | 1.861 | 1.076 | 3.218 | 0.026 |
| Metastasis (M0 as reference) | ||||||||
| M1 | 0.959 | 0.230 | 3.998 | 0.954 | ||||
| Mx | 1.007 | 0.647 | 1.569 | 0.974 | ||||
| Pathological grade (Grade 1 as reference) | ||||||||
| Grade 2 | 1.333 | 0.646 | 2.751 | 0.436 | ||||
| Grade 3 | 1.623 | 0.756 | 3.484 | 0.214 | ||||
| Grade 4 | 1.659 | 0.208 | 13.224 | 0.633 | ||||
| m6Ascore (Low as reference) | ||||||||
| High | 2.344 | 1.509 | 3.641 | <0.001 | 2.315 | 1.493 | 3.590 | <0.001 |
m6Ascore is associated with genes regulating cachexia and chemoresistance in PDAC
To further explore why m6Ascore-high tumors had shorter survival, we examined the expression of zinc transporters and genes regulating cachexia and chemoresistance in PDAC. Previously, we have identified a zinc transporter, Solute Carrier Family 39 Member 4 (SLC39A4, ZIP4), which plays a critical role in PDAC progression and metastasis [28,29]. ZIP4 can induce cachexia by increasing CAMP Responsive Element Binding Protein 1 (CREB1) regulated RAB27B, resulting in the release of extracellular vesicle containing heat shock protein 70 (HSP70) and heat shock protein 90 (HSP90) [30]. Here we found that ZIP4 expression was higher in m6Ascore-high tumors (Fig. S6a). We also found that m6Ascore-high tumors had higher expression of RAB27B, Mitogen-Activated Protein Kinase 14 (MAPK14), HSP70 (HSPA4) and HSP90 (HSP90AA1), indicating m6Ascore-high tumors had high potential to develop muscle wasting and cachexia. Meanwhile, we also found that the expression of several metastasis associated genes also increased in m6Ascore-high tumors, including Integrin Subunit Alpha 3 (ITGA3), Integrin Subunit Beta 1 (ITGB1), CREB1, and Yes-associated protein 1 (YAP1) (Fig. S6b-i) [29,31]. These results suggested the m6Ascore-high tumors have more progressive phenotype.
Gene set enrichment analysis (GSEA) identified activated and inactivated pathways
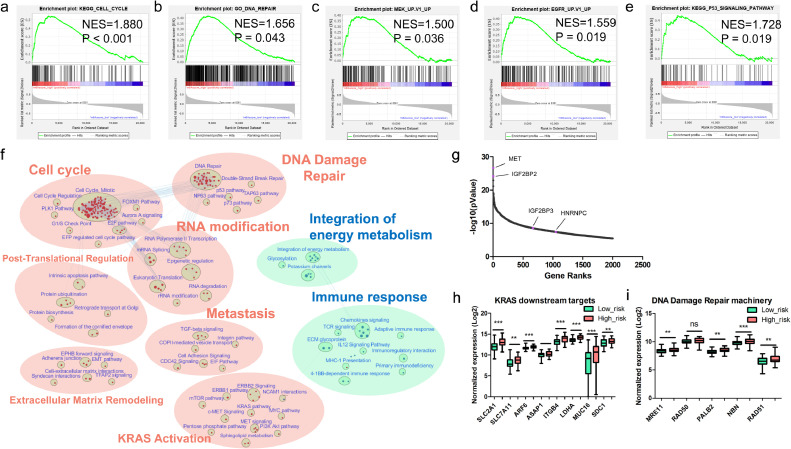
GSEA analysis showed that m6Ascore-high tumors had increased expression of several gene sets including cell cycle, KRAS, p53, DNA damage repair, etc. (Fig. 4a-e). The network of the top 50 DEGs between m6Ascore-high and m6Ascore-low tumors showed MET pathway was amplified in m6Ascore-high tumors (Fig. S7). To further elucidate the differentially enhanced or suppressed cellular processes between the m6Ascore-high and m6Ascore-low PDAC tumors, we constructed an enrichment map. It showed that RNA modification, DNA damage repair and Cell cycle are the three most significantly enhanced processes in m6Ascore-high tumors (Fig. 4f). Meanwhile, the activity of several other processes involving tumor progression also increased, including extracellular matrix remodeling, metastasis, KRAS Proto-Oncogene (KRAS) activation and post-translational regulation (Fig. 4f). MET is the most significantly different gene between the two groups. Intriguingly, there are three m6A regulators that are among the top 2000 DEGs, including IGF2BP2, IGF2BP3 and HNRNPC, which are all correlated with overall survival in PDAC (Fig. 4g and Fig. S4). m6Ascore-high tumors showed increased activity of downstream targets of KRAS mutation (Fig. 4h). Key regulators of DNA damage repair (DDR) are enriched in m6Ascore-high tumors compared to that in the m6Ascore-low tumors (Fig. 4i). More intriguingly, the activity of immune response pathways was relatively lower in the m6Ascore-high group, indicating the presence of immune evasion.
Fig. 4.
Gene set enrichment analysis based on the m6Ascore signature. (a-e) Enrichment plot on cell cycle, DNA damage repair, MEK pathway, EGFR pathway and TP53 pathway; (f) Enrichment map showed the upregulated pathways (marked in red) and downregulated pathways (marked in blue) in m6Ascore-high TCGA-PDAC tumors. The FDR cutoff value is 0•01; (g) Gene ranks of the top 2000 DEGs between m6Ascore-high and m6Ascore-low tumors; (h) Compare the expression of several downstream targets of KRAS mutation between m6Ascore-high and m6Ascore-low tumors. (i) Examined the expression of key regulators of DDR machinery in m6Ascore-high and m6Ascore-low tumors.
m6Ascore-high tumors showed “immune-cold” characteristics
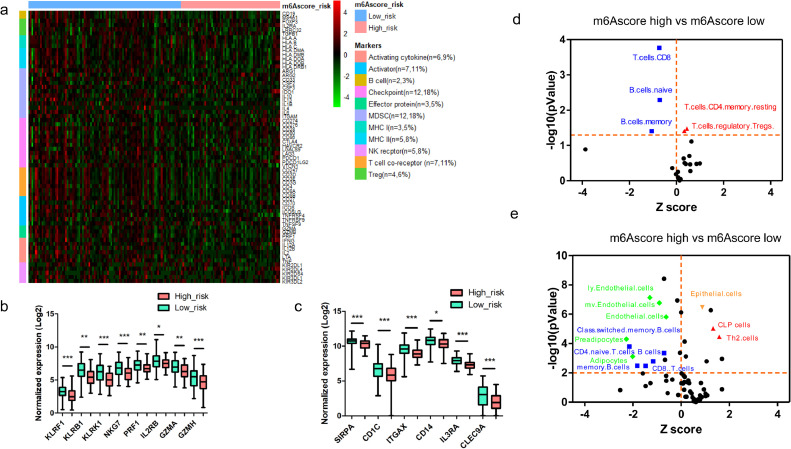
To further investigate tumor immune evasion, we evaluated 66 immune markers [22] including markers of T cells, B cells, NK cells, Myeloid-Derived Suppressor Cells (MDSC), Antigen-presenting cells (APCs), and immune checkpoints in PDAC (Fig. 5a). We found that cytotoxic T cells markers (CD8A and CD8B), helper T cells marker (CD4) and B cells marker (CD19) were lower in the m6Ascore-high group, indicating these tumors might have less infiltration of CD8+, CD4+ T cells and less B cells. We also evaluated the expression of several immune checkpoints, including PD-L1 (CD274), PD1 (PDCD1) and CTLA4, and found that PD1 and CTLA4 were lower and PD-L1 was higher in the m6Ascore-high group, suggesting that the m6Ascore-high tumors might have T cells exhaustion, which may contribute to immune evasion (Fig. 5a). Meanwhile, we found that m6Ascore-high tumors had lower expression of NK cells markers (Fig. 5b) [23]. This result indicated that m6Ascore-high tumors might have lower activity and infiltration of NK cells. Furthermore, we also found that m6Ascore-high tumors had lower expression of DCs markers [24], including Signal Regulatory Protein Alpha (SIRPA), CD1c Molecule (CD1C), Integrin Subunit Alpha X (ITGAX), cluster of differentiation 14 (CD14), Interleukin 3 Receptor Subunit Alpha (IL3RA) and C-Type Lectin Domain Containing 9A (CLEC9A), indicating m6Ascore-high tumors might have impaired DCs functions (Fig. 5c).
Fig. 5.
m6Ascore-high tumors showed “immune-cold” characteristics. (a) Hierarchical clustering showed distinct expression patterns of manually curated 66 immune markers in m6Ascore-high and m6Ascore-low tumors; (b) Comparison of curated markers of NK cells between m6Ascore-high and m6Ascore-low tumors; (c) Comparison of curated markers of DCs between m6Ascore-high and m6Ascore-low tumors; (d) The CIBERSORTx estimated fractions of immune cell population in PDAC tumor tissue. The blue dots represent lower estimated fractions, while red dots represent higher estimated fractions in m6Ascore-high tumors; (e) The xCell estimated fractions of immune cell population in PDAC tumor tissue. The blue and green dots represent lower estimated fractions, while red dots represent higher estimated fractions in m6Ascore-high tumors.
To further evaluate the infiltration of immune cells, we applied the CIBERSORTx algorithm which could examine the abundance of 22 immune cells and myeloid cells in tumor microenvironment [26]. Interestingly, we found that m6Ascore-high tumors had lower proportions of CD8+ T cells, Naïve B cells, and Memory B cells, but had higher Regulatory T cells (Tregs) and Memory resting CD4+ T cells compared to those in m6Ascore-low tumors (Fig. 5d). We also examined proportion of 64 stromal and immune cell types according to a previous established gene signature [25], and found that m6Ascore-high tumors had lower proportions of CD8+ T cells, CD4+ T cells, B cells, and memory B cells, and higher proportions of Th2 cells and CLP cells, indicating decreased immune surveillance in these tumors (Fig. 5e). Furthermore, we found that m6Ascore-high tumors had lower Immune-score, Stromal-score and ESTIMATE score (Fig. S8). Taken together, these results indicated that reduced immune surveillance may contribute to immune evasion in m6Ascore-high tumors.
Mutation and somatic copy number alterations (SCNAs)
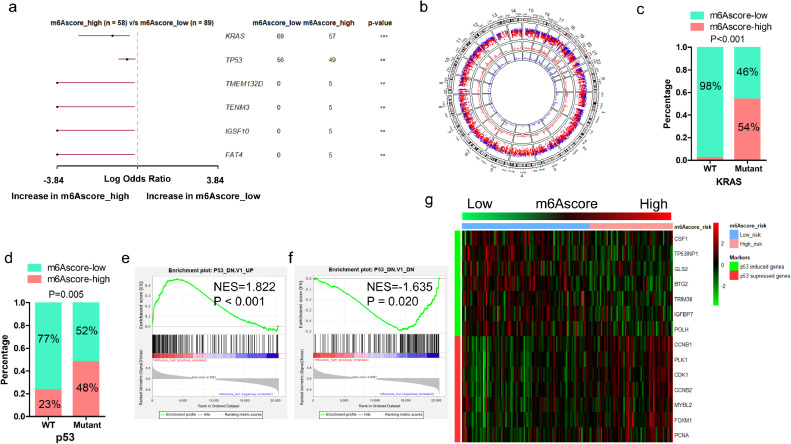
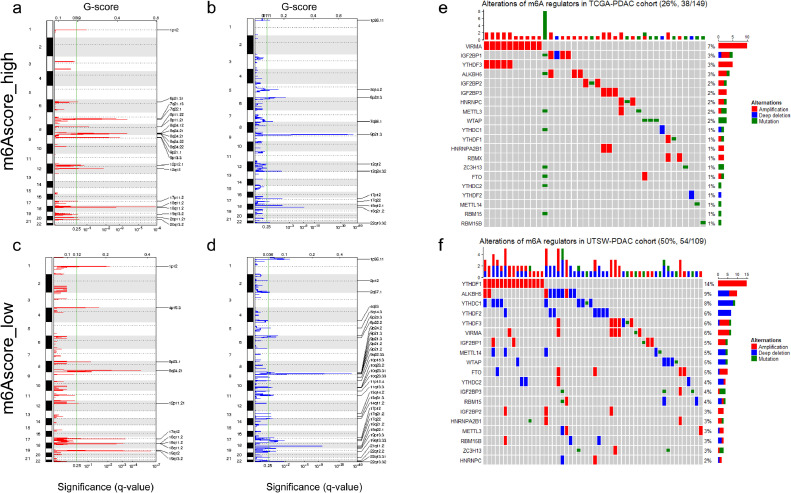
We compared the mutation profiles of m6Ascore-high and m6Ascore-low tumors, and identified that KRAS and TP53 are among the most differentially mutant genes (Fig. 6a-b). m6Ascore-high tumors are associated with higher prevalence of TP53 and KRAS mutation. Those with KRAS mutation are more likely to be m6Ascore-high (54%, 58/107) compared to tumors with wild type KRAS (2%, 1/40). Meanwhile, those with TP53 mutation are more likely to be m6Ascore-high (48%, 48/100) compared to tumors with wild type TP53 (23%, 11/47). (Fig. 6c-d). m6Ascore is positively correlated with the activation of mutant TP53 induced genes (Fig. 6e-f). p53 induced genes had lower expression in m6Ascore-high tumors, while the p53 suppressed genes had higher expression (Fig. 6g). This suggests that the differences of mutation patterns, especially KRAS and TP53 mutation, may contribute to the distinct prognosis between m6Ascore-high and m6Ascore-low tumors. List of the mutant genes occurred in more than 3 mutant samples in each group was shown in a map (Fig. S9). We also compared SCNAs between the m6Ascore-high and m6Ascore-low tumors. We found that m6Ascore-high tumors had more regions of amplification and less regions of deletion compared with m6Ascore-low tumors (Fig. 7a-b). For m6Ascore-high tumors, the peak of amplification fell in the cytoband of 18q11.2, involving Laminin Subunit Alpha 3 (LAMA3), RIO Kinase 3 (RIOK3) and Intracellular Cholesterol Transporter 1 (NPC1), while the peak of deletion fell in the cytoband of 9p21.3, involving Cyclin Dependent Kinase Inhibitor 2A (CDKN2A) and Cyclin Dependent Kinase Inhibitor 2B (CDKN2B). m6Ascore-high tumors were characterized with several regions of recurrent amplifications including 7q21, 7q22, 8p11, 8q24, 9q11 and 9q23, while the deletion regions were relatively less. The m6Ascore-low tumors had less regions of recurrent amplifications on 8p11, 8q24, but had more regions of recurrent deletions, including 9q22, 10p15, etc. The peak of amplification for m6Ascore-low tumors fell in the cytoband of 19q13.2, enclosing NUMB Like Endocytic Adaptor Protein (NUMBL), etc., while the peak of deletion fell in 9p21.3, identifying Methylthioadenosine Phosphorylase (MTAP) and CDKN2A (Fig. 7c-d). Raw copy numbers of m6Ascore-high and m6Ascore-low tumors were showed in heatmaps (Fig. S10). Genetic variation landscape of m6A regulators showed that 26% (38/149) PDAC patients in TCGA cohort and 50% (54/109) PDAC patients in the UTSW cohort had gene alterations of m6A regulators (Fig. 7e-f).
Fig. 6.
Comparison of mutation landscape between m6Ascore-high and m6Ascore-low tumors. (a) Forest plot shows the most significantly mutant genes that vary between m6Ascore-high and m6Ascore-low tumors (**P < 0.01; ***P < 0.001); (b) A circular plot shows the mutation profile of m6Ascore-high and m6Ascore-low tumors. From the most inner to the most outer circles listed the genomic density of mutation genes in m6Ascore-high tumors, in m6Ascore-low tumors, and genomic rainfall of mutation genes in all samples; (c) Proportion of m6Ascore-high and m6Ascore-low group in patients harbored wildtype or mutant KRAS; (d) Proportion of m6Ascore-high and m6Ascore-low group in patients harbored wildtype or mutant TP53; (e) GSEA analysis of genes up-regulated in NCI-60 panel of cell lines with mutated TP53; (f) GSEA analysis of genes down-regulated in NCI-60 panel of cell lines with mutated TP53; (g) Correlation of m6Ascore and the expression of p53-induced genes or p53-suppressed genes in PDAC patients.
Fig. 7.
Amplificated and deleted regions and somatic copy number alterations of m6A regulators. (a) Amplification regions were delineated by GISTIC_2.0 algorithm in m6Ascore-high PDAC tumors; (b) Regions of deletion were delineated by GISTIC_2.0 algorithm in m6Ascore-high PDAC tumors; (c) Regions of amplification were delineated by GISTIC_2.0 algorithm in m6Ascore-low PDAC tumors; (d) Regions of deletion were delineated by GISTIC_2.0 algorithm in m6Ascore-low PDAC tumors. q-value (bottom) and G-score (Top) were listed as X-axis. The green line indicated the cutoff value of q-value (0.25). Chromosome numbers were labeled on the left and regions with recurrent copy number variation were labeled on the right of each plot. Red indicated amplification and blue indicated deletion; (e) OncoPrint of all the m6A regulators in TCGA-PDAC cohort; (f) OncoPrint of all the m6A regulators in UTSW-PDAC cohort.
m6Ascore serves as a promising indicator to predict response to ICIs immunotherapy
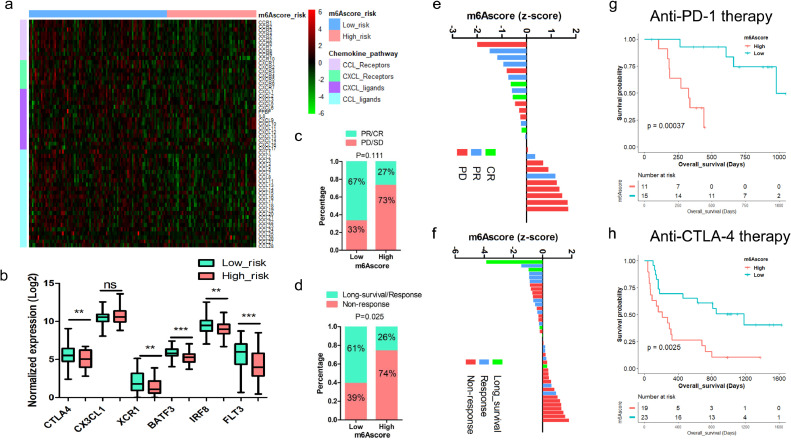
The results above indicate m6Ascore is correlated to tumor immune evasion. We investigated the expression of ligands and receptors in Chemokine Ligand (CCL) and chemokine (C-X-C motif) ligand (CXCL) chemokine family, which has been reported to regulate CD8+ T cells infiltration in PDAC [32], and found that most ligands and receptors in CCL and CXCL family decreased in m6Ascore-high tumors, including CCL4 and CCL5 (Fig. 8a). These results suggest that suppressed chemokine signature in m6Ascore-high tumors may lead to immune evasion in PDAC. We also found downregulation of CTLA4 (P = 0.0036), X-C Motif Chemokine Receptor 1 (XCR1) (P = 0.0034), Basic leucine zipper transcription factor ATF-like 3 (BATF3) (P < 0.0001), Interferon Regulatory Factor 8 (IRF8) (P = 0.0037), FMS-like tyrosine kinase 3 (FLT3) (P = 0.0002) and upregulation of C-X3-C Motif Chemokine Ligand 1 (CX3CL1) (P = 0.2203) in m6Ascore-high tumors, which are all proposed indicators for immune evasion (Fig. 8b). The result above prompts us to evaluate the efficacy of m6Ascore in predicting the response to ICIs immunotherapy.
Fig. 8.
Association of m6Ascore and response to ICIs. (a) Expression of ligands and receptors of CCL and CXCL chemokine families were examined in the m6Ascore-high and m6Ascore-low PDAC tumors; (b) Curated biomarkers for immunotherapy, including CTLA4, CX3CL1, XCR1, BATF3, IRF8 and FLT3; (c) The response rates of patients receiving anti-PD-1 treatment in GSE78220 cohort were compared between the m6Ascore-high and m6Ascore-low tumors (PD, progressive disease; SD, stable disease; PR, partial response; CR, complete response); (d) The response rates of patients receiving anti-CTLA4 treatment in Van Allen's cohort were compared between the m6Ascore-high and m6Ascore-low tumors; (e) Waterfall plot showed the correlation between m6Ascore and the response to anti-PD-1 therapy; (f) Waterfall plot showed the correlation between m6Ascore and treatment response to anti-CTLA4 therapy; (g) Kaplan-Meier survival analysis based on m6Ascore in patients treated with anti-PD-1 therapy (P = 0.00037, log-rank test); (h) Kaplan-Meier survival analysis based on m6Ascore in patients treated with anti-CTLA4 therapy (P = 0.0025, log-rank test).
Therefore, we examined whether m6Ascore can predict response to ICIs in two independent cohorts of patients with advanced tumor who had received either anti-PD-1 therapy or anti-CTLA4 therapy [33,34]. For those receiving anti-PD-1 therapy, partial/complete response (PR/CR) rate is higher in m6Ascore-low group (67%, 10/15) than that in m6Ascore-high group (27%, 3/11). m6Ascore-low group shows extended survival (P = 0.00037). (Fig. 8c, e, g). For those receiving anti-CTLA4 treatment, response rate is higher in m6Ascore-low group (61%, 14/23) than that in m6Ascore-high group (26%, 5/19). m6Ascore-low group also shows extended survival (P = 0.0025) (Fig. 8d, f, h). We further compared the m6Ascore with another potential indicator, named immuno-predictive score (IMPRES) in predicting response to ICIs. [35,36] We found that IMPRES-high patients have higher response rates to anti-PD1 (85%, 11/13, VS 15%, 2/13) and anti-CTLA4 therapy (55%, 17/31, VS 15%, 2/13) than that of IMPRES-low patients (P = 0.001 and P = 0.006, respectively). 67% achieved CR/PR in the m6Ascore-low group compared to 85% in the IMPRES-high group receiving anti-PD-1 treatment. On the other hand, 61% are responsive in the m6Ascore-low group compared to 55% in the IMPRES-high group receiving anti-CTLA4 treatment. These results indicate that m6Ascore might be better at selecting those who might respond to anti-CTLA4 treatment, while IMPRES might be better at selecting patients who might respond to anti-PD-1 treatment (Fig. 8c-d and Fig. S11).
Discussion
m6A plays critical roles in tumorigenesis and tumor progression [37], [38], [39]. Studies showed m6A can regulate the maturation and the ability of neoantigens presentation in immune cells, and regulate response to immunotherapy [5,40]. However, how m6A mediates immune infiltration and tumor survival remains uncharacterized. Here we reported that m6Ascore model can stratify PDAC patients into two subtypes, with distinct patterns of immune infiltration, mutation landscapes and survival. m6A regulators play different roles on tumor immune surveillance. For example, METTL3 plays critical roles in the activation of several immune cells, including T cells and DCs. [40,41] However, another m6A regulator, YTHDF1, contributes to tumor immune evasion by regulating the activity of DCs. Deletion of YTHDF1 can increase the response to immune checkpoint blockade [5]. Considering that integration of these m6A regulators may perform better on predicting the survival and response to immunotherapy, we constructed a m6Ascore model which finally enrolled nine regulators. It doesn't necessarily mean these nine regulators are more important than other m6A regulators. It indicates that the combination of these nine m6A regulators can adequately predict PDAC prognosis and immune evasion risk. Other m6A regulators may also play critical roles during tumor progression. For example, as a m6A “Eraser”, fat mass- and obesity-associated protein (FTO) can promote tumor progression by activating MYC/CEBPA pathway [42].
We found that markers of NK cells were significantly lower in the m6Ascore-high tumors, which is consistent to another study suggesting the infiltration of NK cells was associated with better survival in melanoma patients [23]. Restoration of both the quantity and quality of Dendritic cells (DCs) could increase the immune surveillance in PDAC [43]. m6Ascore was inversely correlated with the markers of DCs, indicating dampened activity and infiltration of these immune cells, which is consistent to previous studies that showed m6A can suppress the maturation and function of antigen presenting in DCs [5,40]. Furthermore, the expression of phagocytosis checkpoint, SIRPα, was lower in the m6Ascore-high group, which may contribute to immune evasion. A chemokine signature which consists of four chemokines (CCL4, CCL5, CXCL9 and CXCL10) was reported to be associated with CD8+ T cells infiltration in PDAC [32]. The current study also found that m6Ascore-low tumors are characterized with increased profiles of CCL and CXCL chemokine ligands, similar to the “T cell-inflamed” phenotype proposed by Romero and colleagues, which may partially explain the distinct immune infiltration profiles between m6Ascore-low and m6Ascore-high tumors. Our findings are also consistent to another study showing that deletion of CXCL9/CXCL10 will suppress the efficacy of immune checkpoint inhibitors [44]. Previous study has found that the upregulation of CTLA4 and absence of CX3CL1 were associated with the response to ICIs [45]. Meanwhile, cumulative score of Dendritic cells’ signature including the expression of XCR1, BATF3, IRF8, and FLT3 is associated with anti-PD-L1 treatment in non–small cell lung cancer (NSCLC) and renal cell carcinoma (RCC) [46]. All these markers are correlated to m6Ascore, indicating potential connections between m6A and immune evasion.
DNA damage repair (DDR) is a double-edged sword. DDR can suppress tumorigenesis by maintaining genome integrity. On the other hand, during treatment induced tumor evolution, DDR sufficient cancer cells can outgrow the DDR deficient tumor clones, thus leading to the development of treatment resistance. MRN complex (MRE11, RAD50 and NBN) is the initial core of DNA double-strand breaks (DSB) repair machinery, while PALB2, RAD51 among several other key regulators contribute to the later stages of DDR [47]. Our study identifies the enrichment of several key regulators of DDR in m6Ascore-high tumors, indicating a potential link between m6A, DDR, and immune evasion in PDAC.
Furthermore, previous data showed that SCNAs were correlated with tumor immune evasion and resistance to immunotherapy [48]. We found that recurrent amplifications were more frequent in m6Ascore-high tumors, while the recurrent deletions were more frequent in m6Ascore-low tumors. GISTIC analysis identified recurrent gain of LAMA3, RIOK3 and NPC1, which are critical in driving tumor progression in m6Ascore-high tumors. We also found recurrent gain of NUMBL, a tumor suppressor in m6Ascore-low tumors, which may contribute to suppression of tumor growth. In addition to SCNAs, m6Ascore is also associated with more events of KRAS and TP53 mutation. Dual inhibition of KRAS and cell cycle pathways can overcome immune evasion [49,50]. Mutation induced tumor heterogeneity and clonal evolution leads to dynamic immune landscape in tumor tissue, which may mediate efficacy of immunotherapy [51]. Besides, hypermethylation is associated with nearly 20% of silenced neoantigens, indicating that immune surveillance at the early stages of cancers may result in the selection of evolving subclones which had silencing neoantigens due to promoter hypermethylation [52].
To evaluate the role of this m6Ascore model in predicting response to immunotherapy, we analyzed two independent cohorts of patients received ICIs treatment [33,34]. m6Ascore-low patients had higher PR and CR rates. Long term survivors of pancreatic cancer showed increased T cell infiltration in tumor microenvironment, indicating enhanced tumor immune surveillance [53]. Chemokines and tumor derived extracellular vesicles (EVs) play critical roles in tumor progression and cachexia [30,54,55]. Genes that are involved in epithelial-mesenchymal transition (EMT) and release of EVs, including CREB, ZEB1 and RAB27B were associated with inferior prognosis in PDAC [56]. Intriguingly, chemokine ligands and receptors were much lower in m6Ascore-high tumors, which is consistent with the non-T-cell inflamed phenotype. Reduced response to immunotherapy may also be attributed to the decrease of chemokines in PDAC. Further studies are warranted to explore how m6A regulates response to ICIs, thus charting paths towards the development of effective strategies to improve the efficacy of immunotherapy.
This study also has limitations. How these m6A regulators cooperate with each other and drive tumor immune evasion remains unexplored. Meanwhile, the mechanisms of m6A modification regulated tumor metastasis and DDR remains unknown. Further studies are warranted to test the efficacy of m6Ascore on predicting response to ICIs in larger clinical trials, and to explore the interactions between these m6A regulators in orchestrating immune evasion.
In conclusion, m6Ascore is a potential molecular classification for PDAC, which can identify distinct immune infiltration and mutation pattern. It provides a practical tool to access the risk of immune evasion and predict response to ICIs immunotherapy.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
Contributors
Z.Z. was involved in conceptualization, data curation, project administration, resources, formal analysis, software, visualization and writing – original draft; J.Z. was involved in data curation, formal analysis, investigation and validation; C.X. was involved in methodology, project administration and writing – review & editing; J.Y. was involved in resources, supervision and writing – review & editing. Y.Z. was involved in resources, supervision and visualization; M.L. was involved in project administration, resources, visualization and writing – original draft; X.S. was involved in project administration, software and resources; X.L. was involved in resources, visualization and project administration; H.Z. was involved in project administration, software and resources; W.C. was involved in project administration, resources and software; L.M. was involved in project administration, resources and writing – review & editing; K.F. was involved in resources and investigation; W.L. was involved in resources, software and supervision; C.W. was involved in resources and supervision; Y.H. was involved in supervision and writing – review & editing; C.Z was involved in conceptualization, resources, supervision and writing – review & editing; M.L. was involved in conceptualization, funding acquisition, investigation, resources, supervision and writing – review & editing. Z.Z. and J.Z. have accessed verified the underlying data.
Acknowledgements
We would like to thank Prof. Qianghu Wang for providing valuable suggestions for this study. This work was supported in part by National Institutes of Health (NIH) grants to M. Li (R01 CA186338, R01 CA203108, R01 CA247234 and the William and Ella Owens Medical Research Foundation) and NIH/National Cancer Institute Q39 award P30CA225520 to Stephenson Cancer Center.
Data Sharing Statement
All data are available on public repositories, the accession numbers of which are listed in Table 1.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103271.
Contributor Information
Changhua Zhang, Email: zhchangh@mail.sysu.edu.cn.
Min Li, Email: Min-Li@ouhsc.edu.
Appendix. Supplementary materials
References
- 1.Liu L, Bai X, Wang J, Tang XR, Wu DH, Du SS. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin Cancer Res. 2019;25(24):7413–7423. doi: 10.1158/1078-0432.CCR-19-0558. [DOI] [PubMed] [Google Scholar]
- 2.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21(5):552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 3.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18(1):137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kandimalla R, Gao F, Li Y, Huang H, Ke J, Deng X. RNAMethyPro: a biologically conserved signature of N6-methyladenosine regulators for predicting survival at pan-cancer level. NPJ Precis Oncol. 2019;3:13. doi: 10.1038/s41698-019-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 10.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 13.Zou Y, Hu X, Zheng S, Yang A, Li X, Tang H. Discordance of immunotherapy response predictive biomarkers between primary lesions and paired metastases in tumours: a multidimensional analysis. EBioMedicine. 2020;63 doi: 10.1016/j.ebiom.2020.103137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network Electronic address WBE, cancer genome Atlas research n. comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol Res. 2019;7(7):1162–1174. doi: 10.1158/2326-6066.CIR-18-0500. [DOI] [PubMed] [Google Scholar]
- 24.Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology. 2013;140(1):22–30. doi: 10.1111/imm.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552(7683):126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M, Yang J, Zhang Y, Zhou Z, Cui X, Zhang L. ZIP4 Promotes Pancreatic Cancer Progression by Repressing ZO-1 and Claudin-1 through a ZEB1-Dependent Transcriptional Mechanism. Clin Cancer Res. 2018;24(13):3186–3196. doi: 10.1158/1078-0432.CCR-18-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Zhang Y, Yang J, Cui X, Zhou Z, Zhan H. ZIP4 increases expression of transcription factor ZEB1 to promote integrin alpha3beta1 signaling and inhibit expression of the gemcitabine transporter ENT1 in pancreatic cancer cells. Gastroenterology. 2020;158(3):679–692. doi: 10.1053/j.gastro.2019.10.038. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Zhang Z, Zhang Y, Ni X, Zhang G, Cui X. ZIP4 promotes muscle wasting and cachexia in mice with orthotopic pancreatic tumors by stimulating RAB27B-regulated release of extracellular vesicles from cancer cells. Gastroenterology. 2019;156(3):722–734. doi: 10.1053/j.gastro.2018.10.026. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Yang J, Cui X, Chen Y, Zhu VF, Hagan JP. A novel epigenetic CREB-miR-373 axis mediates ZIP4-induced pancreatic cancer growth. EMBO Mol Med. 2013;5(9):1322–1334. doi: 10.1002/emmm.201302507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero JM, Grunwald B, Jang GH, Bavi PP, Jhaveri A, Masoomian M. A four-chemokine signature is associated with a T-cell-inflamed phenotype in primary and metastatic pancreatic cancer. Clin Cancer Res. 2020;26(8):1997–2010. doi: 10.1158/1078-0432.CCR-19-2803. [DOI] [PubMed] [Google Scholar]
- 33.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter JA, Gilbo P, Atwal GS. IMPRES does not reproducibly predict response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2019;25(12):1833–1835. doi: 10.1038/s41591-019-0671-4. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11(1):2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020;39(23):4507–4518. doi: 10.1038/s41388-020-1303-7. [DOI] [PubMed] [Google Scholar]
- 39.Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X. Dynamic m(6)A mRNA methylation reveals the role of METTL3-m(6)A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38(24):4755–4772. doi: 10.1038/s41388-019-0755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. 2019;10(1):1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172(1-2):90–105. doi: 10.1016/j.cell.2017.11.031. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegde S, Krisnawan VE, Herzog BH, Zuo C, Breden MA, Knolhoff BL. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell. 2020;37(3):289–307. doi: 10.1016/j.ccell.2020.02.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26(2):487–504. doi: 10.1158/1078-0432.CCR-19-1868. [DOI] [PubMed] [Google Scholar]
- 45.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayoux M, Roller A, Pulko V, Sammicheli S, Chen S, Sum E. Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy. Sci Transl Med. 2020;12(534) doi: 10.1126/scitranslmed.aav7431. [DOI] [PubMed] [Google Scholar]
- 47.Perkhofer L, Gout J, Roger E, Kude de Almeida F, Baptista Simoes C, Wiesmuller L. DNA damage repair as a target in pancreatic cancer: state-of-the-art and future perspectives. Gut. 2020 doi: 10.1136/gutjnl-2019-319984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322) doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knudsen ES, Kumarasamy V, Chung S, Ruiz A, Vail P, Tzetzo S. Targeting dual signalling pathways in concert with immune checkpoints for the treatment of pancreatic cancer. Gut. 2020 doi: 10.1136/gutjnl-2020-321000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J, Chen P, Liu K, Liu J, Zhou B, Wu R. CDK1/2/5 inhibition overcomes IFNG-mediated adaptive immune resistance in pancreatic cancer. Gut. 2020 doi: 10.1136/gutjnl-2019-320441. [DOI] [PubMed] [Google Scholar]
- 51.Xu LX, He MH, Dai ZH, Yu J, Wang JG, Li XC. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol. 2019;30(6):990–997. doi: 10.1093/annonc/mdz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567(7749):479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z, Xia G, Xiang Z, Liu M, Wei Z, Yan J. A C-X-C chemokine receptor type 2-dominated cross-talk between tumor cells and macrophages drives gastric cancer metastasis. Clin Cancer Res. 2019;25(11):3317–3328. doi: 10.1158/1078-0432.CCR-18-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin C, He H, Liu H, Li R, Chen Y, Qi Y. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut. 2019;68(10):1764–1773. doi: 10.1136/gutjnl-2018-316324. [DOI] [PubMed] [Google Scholar]
- 56.Liu M, Zhang Y, Yang J, Zhan H, Zhou Z, Jiang Y. Zinc dependent regulation of ZEB1 and YAP1 co-activation promotes emt plasticity and metastasis in pancreatic cancer. Gastroenterology. 2021 doi: 10.1053/j.gastro.2020.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.