Abstract
Cryptosporidium is a protozoan parasite that is transmitted to both humans and animals through zoonotic or anthroponotic means. When a host is infected with this parasite, it causes a gastrointestinal disease known as cryptosporidiosis. To understand the transmission dynamics of Cryptosporidium, the small subunit (SSU or 18S) rRNA and gp60 genes are commonly studied through PCR analysis and conventional Sanger sequencing. However, analyzing sequence chromatograms manually is both time consuming and prone to human error, especially in the presence of poorly resolved, heterozygous peaks and the absence of a validated database. For this study, we developed a Cryptosporidium genotyping tool, called CryptoGenotyper, which has the capability to read raw Sanger sequencing data for the two common Cryptosporidium gene targets (SSU rRNA and gp60) and classify the sequence data into standard nomenclature. The CryptoGenotyper has the capacity to perform quality control and properly classify sequences using a high quality, manually curated reference database, saving users' time and removing bias during data analysis. The incorporated heterozygous base calling algorithms for the SSU rRNA gene target resolves double peaks, therefore recovering data previously classified as inconclusive. The CryptoGenotyper successfully genotyped 99.3% (428/431) and 95.1% (154/162) of SSU rRNA chromatograms containing single and mixed sequences, respectively, and correctly subtyped 95.6% (947/991) of gp60 chromatograms without manual intervention. This new, user-friendly tool can provide both fast and reproducible analyses of Sanger sequencing data for the two most common Cryptosporidium gene targets.
Keywords: Genotyping tool, Sanger sequencing, SSU rRNA gene, gp60 gene, Validated database, Mixed infections
Graphical abstract
Highlights
-
•
Developed CryptoGenotyper to analyze Cryptosporidium sequencing chromatograms.
-
•
Uses a high quality, manually curated database to report results accurately.
-
•
Provides timely results in standard notation for ease of reporting.
-
•
Correctly identified 99.3% SSU rRNA and 95.1% mixed SSU rRNA.
-
•
Subtyped 95.6% of gp60 samples in concordance with manual analysis.
1. Introduction
The protozoan parasite Cryptosporidium is an emerging pathogen that causes gastrointestinal disease worldwide. Existing as an infectious, thick-walled oocyst in the environment, this enteric pathogen can infect a wide range of hosts, including wildlife, domestic livestock and humans (Morris et al., 2019). This apicomplexan is transmitted via the fecal-oral route either directly from infected people or animals, or through contaminated water or food (Vanathy et al., 2017). With the number of recognized Cryptosporidium species increasing and their transmission dynamics still unclear, further molecular research is required as interactions between humans and animals are steadily increasing (Zahedi et al., 2015).
Two genetic loci are commonly used to characterize Cryptosporidium positive samples: the small subunit ribosomal RNA (SSU rRNA) gene (also known as the 18S rRNA gene), and the 60-kDa glycoprotein (gp60) gene. The SSU rRNA gene contains variable regions interspersed amongst conserved regions, allowing for effective species and genotype identification (Morris et al., 2019). This region is AT-rich, ranging from 57 to 61% (Xiao et al., 1999). Each oocyst contains twenty copies of the gene, evenly split amongst its four haploid genomes (Morris et al., 2019). The highly polymorphic gp60 gene, which encodes a protein responsible for attachment and invasion in the small intestine, can further determine genotypic families and subgenotypes (Alves et al., 2003). The 3′ conserved region defines the allelic family, whereas the 5′ hyper-variable microsatellite region defines the subtype (Chalmers et al., 2009).
Traditional methods to molecularly characterize these two regions include combining conventional PCR with Sanger sequencing. These methods are still commonly used (Firoozi et al., 2019; Chalmers et al., 2009), despite the increasing availability of next-generation sequencing methods, due to their associated low costs, readily available equipment and standardized analysis tools. However, there are many challenges faced when analyzing the Sanger sequencing results from the SSU rRNA and gp60 loci.
Analyzing Sanger sequencing chromatograms can be time consuming and is subject to human error. For instance, the presence of mixed infections or paralogs within a sequenced sample (Stensvold et al., 2015) result in heterozygous peaks throughout the sequenced region, causing difficulty in analyzing sequences. Once sequences are obtained, they are often misclassified due to inconsistent naming within public databases. Therefore, without a validated database alongside the difficulties in sequence assembly, data for transmission studies may be misinterpreted or left inconclusive.
To address these issues, we developed an automated tool specifically to analyze Sanger sequencing data of two Cryptosporidium gene targets. This tool, named CryptoGenotyper, accepts raw Sanger sequencing data for the SSU rRNA or gp60 loci and accurately classifies the data in a fast and reproducible way using a validated manually curated database. The CryptoGenotyper assesses data quality and resolves abnormal peaks appropriately, capturing all information within the raw Sanger sequencing data. This tool is available on the web-based platform Galaxy (Afgan et al., 2018) from the public ToolShed repository (https://toolshed.g2.bx.psu.edu/, tool id: cryptogenotyper, owner: nml) and from the public Galaxy server at https://usegalaxy.eu/, or as a standalone Bioconda package (https://anaconda.org/bioconda/cryptogenotyper). The CryptoGenotyper source code and documentation are available at https://github.com/phac-nml/CryptoGenotyper. We are currently in discussions with members of CryptoDB (Puiu et al., 2004) for potentially hosting the CryptoGenotyper on the CryptoDB.org Galaxy instance.
2. Materials and Methods
2.1. CryptoGenotyper Development
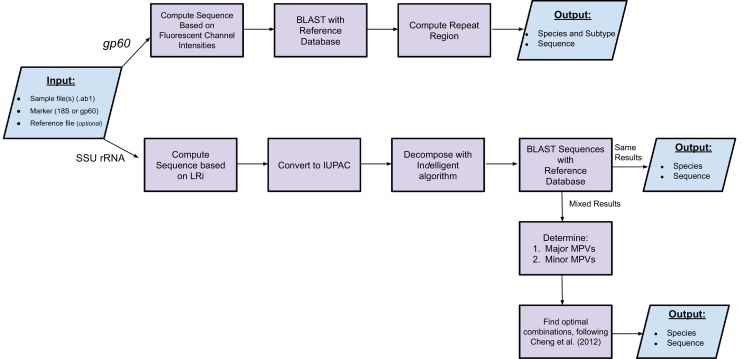
We developed the CryptoGenotyper, a Python (v3.6) program, to perform a fast, accurate and reproducible analysis on raw Sanger sequencing data. Due to common heterozygous peaks in the SSU rRNA gene sequence, we included a heterogeneity detection algorithm into the portion of the tool that targets the SSU rRNA region. This algorithm was inspired by the Mixed Sequence Reader developed by Chang et al. (2012), which utilizes heterozygous base-calling to distinguish mixed sequences within human papilloma virus (HPV) samples by using comparisons against reference sequences to identify indels (insertions-deletions), single nucleotide polymorphisms and sequence repeats. The program's workflow is shown in Fig. 1.
Fig. 1.
CryptoGenotyper schematic workflow.
The program begins with the user inputting the gene target, sample chromatograms and database (optional). (a) If chromatograms correspond to the gp60 gene target, the sequence is retrieved by analyzing the fluorescent channel intensities. A homology search is performed against the reference database and the repeat region is calculated. (b) If chromatograms correspond to the SSU rRNA gene, the sequence is computed based on the log ratio of intensity and converted to IUPAC based nucleotide code where double peaks appear. Afterwards, the sequence is decomposed with Indelligent (Dmitriev and Rakitov, 2008) and a homology search is performed using BLAST against the reference database. If mixed sequences are determined, they are classified by following the protocol outlined in Chang et al. (2012) for determining the most possible variances (MPVs) and optimal combinations. For both markers, the sequence and species and/or subtype information is outputted.
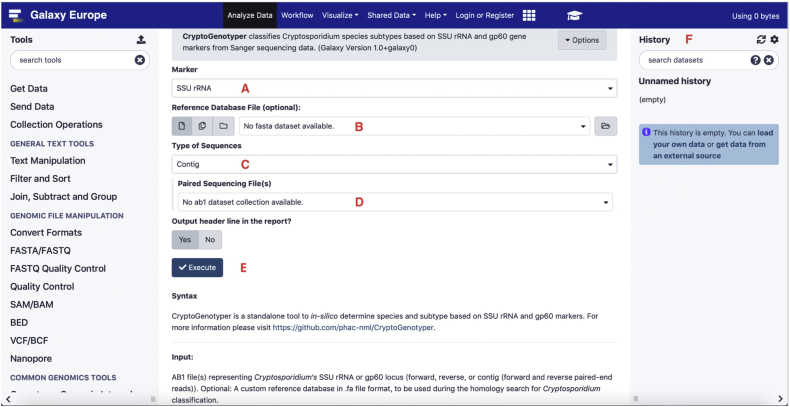
The CryptoGenotyper is available on two interfaces: web-based (Galaxy) and command-line (GitHub and Bioconda). The tool's Galaxy interface can either be accessed at the Galaxy Europe server (https://usegalaxy.eu/) or can be installed through a user's own Galaxy instance admin interface. Workflows (available from https://github.com/phac-nml/CryptoGenotyper/tree/main/CryptoGenotyper/GalaxyWorkflows) can then be imported to easily analyze multiple samples at once. Once setup, the user uploads their sequencing data to Galaxy using the Get Data feature. To analyze more than one sample, the user must create a dataset list (forward or reverse mode) or a list of dataset pairs (contig mode). From the CryptoGenotyper graphical user interface, the user can select the corresponding marker (SSU rRNA or gp60), reference database (custom or default), and samples (single, collection, paired collection) before executing the tool (Fig. 2A–E). The corresponding results then appear in the Galaxy History panel (Fig. 2F). The command-line version has the same functionality, except the user must define the directory that contains all samples to be analyzed or specify a path to a sample file. If the user wishes to perform a contig mode analysis on a directory with multiple files, the user will also need to define both forward and reverse primer names that are included in the chromatogram filenames so the tool can identify the forward and reverse sequence for each sample. Alternatively, if a directory contains sample files from the same sequencing primer pair (e.g. SSUF and SSUR) or forward or reverse analysis mode is set, the primer names can be omitted.
Fig. 2.
Graphical user interface of Galaxy tool implementation.
The user must upload Sanger sequence reads (.ab1) using the Get Data feature under Galaxy Tools. The sequence names will appear in the history on the right. To build a contig, forward and reverse reads must be inputted as a dataset pair. For multiple samples to be processed at once, the reads must be inputted as a list. Then the following must be selected: (A) the gene marker (SSU rRNA or gp60), (B) reference database (default or custom) and (C) type of sequences (forward only, reverse only, or forward and reverse). Afterwards the appropriate sequencing files (D) must be selected. When all inputted information is entered, the execute button (E) will launch the analysis. Final typing results appear in the History (F) as two entries corresponding to extracted FASTA sequence(s) and tab-delimited text report file for easy reporting. Workflows have been created to concatenate results from multiple samples available at https://github.com/phac-nml/CryptoGenotyper. The Galaxy tool implementation can be accessed at https://usegalaxy.eu/.
2.1.1. CryptoGenotyper Inputs
2.1.1.1. Raw Sanger Sequencing Data
The CryptoGenotyper accepts raw Sanger sequencing chromatogram data file(s) in .ab1 file format as input. This data can correspond to three different sequencing read formats: forward, reverse, or contig. Both forward and reverse mode accept either forward (5′–3′) or reverse (3′–5′) reads, whereas the contig mode requires two files containing both forward and reverse reads. The gene target, either SSU rRNA or gp60, must then be selected.
2.1.1.2. Reference Database
The CryptoGenotyper tool contains two validated reference databases: an SSU rRNA database and a gp60 database. We manually curated these databases to ensure they contain representative Cryptosporidium reference sequences for species (Table S1) or subtypes (Table S2) that have been described to date on NCBI. The current version of the SSU rRNA database (v1.0) contains unique reference genotypes that were selected based on the criteria set out by Ruecker et al. (2012). To elaborate, each representative sequence contains the polymorphic region within the 613–810 base pair region of C. parvum (AF164102.1), measures at least 400 base pairs in length, contains no more than two ambiguities, has a definitive source, and does not contain any cloned PCR products. The gp60 database contains a representative sequence of all accepted subtypes to date, collected through personal communication with Prof. Lihua Xiao in addition to those described by Xiao and Feng (2017). These subtypes were chosen to be included in the database as they are complete sequences (the trinucleotide repeat region, repetitive sequence (if applicable), and conserved region are present), contain no ambiguities, and were previously used as a reference sequence in other studies.
If a user wishes to use their own database, a corresponding FASTA (.fa) file must be provided as input. The user can either download the existing CryptoGenotyper database available from https://github.com/phac-nml/CryptoGenotyper and append any new sequences or create their own database entirely. The tool will build a BLAST database from the sequences in the imported FASTA file and perform any homologous searches against that database. If no database file is inputted, the tool will default to the most current version of the validated database.
2.1.2. CryptoGenotyper Algorithm
Once all parameters are inputted into the program, the relevant sequencing information is extracted from each .ab1 file. Both the fluorescent channel intensities and Phred quality score for each base location are processed and recorded using the Biopython library functions (Cock et al., 2009). To remove the low-quality bases that inherently exist on the ends of Sanger sequences, both 5′ and 3′ regions are trimmed by scanning the read with a 5-base sliding window and cut when the average Phred quality drops below 20 (99% base call accuracy).
2.1.2.1. SSU rRNA Algorithm
To reflect the heterozygous peaks in the sequence from the chromatogram, the log ratio of intensity (LRi) is calculated for each base location by taking the log base 2 of the quotient of the major fluorescence intensity to the minor fluorescence intensity as described by Chang et al. (2012). According to Chang et al. (2012), an LRi value close to zero reflects two heterozygous peaks with similar intensities. We use an LRi cutoff value of 2.0 in the CryptoGenotyper to reflect the minimum cutoff to determine a ‘mixed’ peak based on fluorescent ratios (Chang et al., 2012). Those peaks with LRi values less than or equal to 2.0 are then converted in IUPAC nucleotide code to represent the multiple nucleotides present at that location.
Once the entire sequence is parsed, it is decomposed using the Indelligent algorithm (Dmitriev and Rakitov, 2008). This algorithm converts the IUPAC code to two corresponding bases, outputting a major and minor sequence (Dmitriev and Rakitov, 2008). For those bases that were ambiguous to the algorithm, major and minor bases are determined based on the intensities of the fluorescent signals.
The major and minor sequences are then queried using BLAST against the SSU rRNA reference database using the following parameters: word_size = 11, match = 1, mismatch = −2, gap_open = 5, gap_extend = 2. This determines the most possible variances (MPVs), which are then categorized as mixed sequences or indels and the optimal combinations are calculated as described by Chang et al. (2012).
2.1.2.2. gp60 Algorithm
To determine the gp60 sequence, the algorithm records the channel with the highest fluorescence at each base location. Afterwards, the gp60 sequence is divided into two regions: the microsatellite region and the varying region.
The program searches the microsatellite region linearly, using a sliding window of three nucleotides, to determine which repeating trinucleotide sequence is encountered based on an array of previously determined trinucleotide repeat sequences. Once the repeat sequence is determined, the program searches linearly and counts the number of times each repeat appears. Finally, the repeat region is reported in standard notation as described in detail in Xiao and Feng (2017) and illustrated in Chalmers et al. (2009).
The varying region of the sequence undergoes a homology search with the following BLAST parameters: match = 1, mismatch = −2, gap_open = 5, gap_extend = 2. The subtype family is determined based on the result with the highest bit score and percent identity.
2.1.3. CryptoGenotyper Outputs
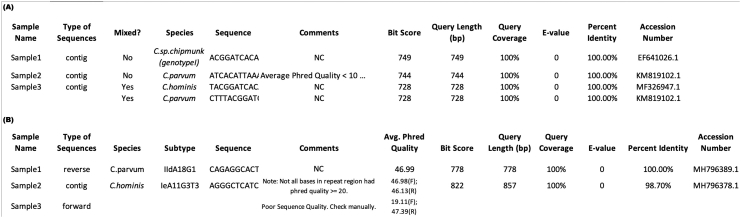
Once the species and/or subtype is determined, the program outputs the results in FASTA and tab-delimited file formats. The FASTA file (Fig. 3) contains a header indicating the tool's parameters (gene target, mode, version of the reference database, primer names) that were used to produce the results. Following the FASTA format guidelines, the sample name and the species (SSU rRNA) or species and subtype (gp60) is written to the output FASTA file followed by the sequence. This output file is written in a way that it could be directly inputted into BLAST for further investigation.
Fig. 3.
The CryptoGenotyper FASTA output file.
One of the results file the CryptoGenotyper generates is a FASTA (.fa) file. For both gene target analyses, a header is outputted at the beginning of the file indicating the run parameters (reference file, program mode, forward and reverse primer names). (A) For the SSU rRNA gene target analysis, the sample name and species identified along with its corresponding sequence are outputted. (B) For the gp60 gene target analysis, the output consists of the same name, species, and subtype, followed by the sequence. This file is designed to allow the user to input it directly into BLAST for further analysis, if desired.
The second output file contains the results in a tab-delimited manner (Fig. 4). For each sample analyzed, the following information is recorded and outputted: sample name, sequence type, species, subtype (if applicable), sequence, comments (Table 1), average Phred quality (gp60 only), bit score, query length (bp), query coverage (%), e-value, percent identity and accession number.
Fig. 4.
The CryptoGenotyper tab-delimited (.txt) output file.
The CryptoGenotyper also generates a text file (.txt) that is tab-delimited with each analysis. (A) For the SSU rRNA gene target analysis, the sample name, analysis mode (forward, reverse, contig), whether the chromatogram had mixed sequences detected, species, sequence, comments, and the BLAST statistics (bit score, query length, query coverage, e-value, percent identity, and accession number of the nearest BLAST hit) is recorded. (B) For the gp60 gene target analysis, the sample name, analysis mode (forward, reverse, contig), species, subtype, sequence, comments, average Phred quality of the chromatograms and the BLAST statistics (similar to the SSU rRNA described) are outputted.
Table 1.
CryptoGenotyper warning messages and their explanations.
| Gene target | Warning message | Explanation |
|---|---|---|
| SSU and gp60 | Could not analyze chromatogram. Please check manually | Program cannot interpret the chromatogram file |
| Poor Sequence Quality. Check manually | The sequence has low Phred score and cannot decipher the data | |
| No blast hits. | The sequence does not represent a Cryptosporidium gene target in the database | |
| NC | No comment | |
| SSU specific | Average Phred Quality <10, could be other potential mixed seqs. Check manually | A Cryptosporidium species was identified, however due to low Phred quality, further mixed infections could be present. |
| gp60 specific | Not all bases in repeat region had Phred quality ≥ 20 | The repeat region is not of the highest quality. Often a result of mixed peaks or sequencing artifacts. |
| Could not classify repeat region. Check manually. | The program could not classify the repeat region with certainty. Most likely a cause of sequencing artifacts or the sequence was trimmed into the repeat region. | |
| BLAST percent identity is less than 99.1%. Check manually in case of new gp60 family | The program may have identified a new subfamily that is not described in the database. Check the sequence manually to confirm. |
If the CryptoGenotyper is unable to decipher the raw data from the Sanger sequence chromatograms, whether due to bad quality or the presence of unusual artifacts, the tool will output a warning message for the sample in question to be manually interpreted (Table 1).
2.2. Data Generation for CryptoGenotyper Validation
2.2.1. SSU rRNA
Sequence data from Canada, the United Kingdom and Sweden were used for the development and validation of the CryptoGenotyper tool. To test the SSU rRNA marker on the CryptoGenotyper program, Sanger chromatograms of 456 samples were analyzed on the contig mode setting from various Canadian studies at the National Microbiology Laboratory (NML, Public Health Agency of Canada, Guelph, Ontario, Canada). This includes 175 animal specimens (138 cattle, 22 dogs, 7 cats, 1 pig, 6 snakes and 1 chicken) collected from 2009 to 2011 and 2015 (unpublished), 119 clinical samples collected between 2006 and 2017 (Guy et al., 2021) and 162 water samples collected between 2008 and 2015 (unpublished). These samples were broken down to 275 samples (68 animal, 64 clinical, 143 water) containing single species and 181 samples (107 animal, 55 clinical, 19 water) containing either mixed species or variants (heterogeneous copies) of one species. In total, the single-species SSU rRNA test samples represent 18 distinct species and 9 wildlife genotypes of Cryptosporidium, and the mixed SSU rRNA test samples represent 7 different combinations of 2 species and 2 unique double variants of the same species (Table S3).
Additionally, chromatograms from 187 samples genotyped at the National Veterinary Institute (SVA, Uppsala, Sweden) between 2013 and 2020 were inputted into the tool using both forward and reverse sequences for each sample. These samples were collected from several studies (Pettersson et al., 2020; Åberg et al., 2020; Åberg et al., 2019; Björkman et al., 2018; Björkman et al., 2015; Alsmark et al., 2018; unpublished), outbreak investigations and sporadic cases sent for routine diagnosis. The samples were comprised of 123 cattle, 34 pigs, 20 squirrels, 4 sheep, 2 horses, 2 chital deer and 1 roe deer. In total, the 167 single-species SSU rRNA test samples represent 11 species and genotypes of Cryptosporidium (Table S3). The remaining 20 samples represent known mixed infections, including two different mixed species and one mixed variant (Table S3).
All samples were tested regardless of whether or not species could be determined manually from both forward and reverse sequences in order to test how well the program would deal with poor as well as good quality sequences. To generate the results table, we checked the results manually if the CryptoGenotyper was indicating a result with a bit score less than 400 (indicating suboptimal percent identity and percent coverage values) or had a check manually or no blast hits comment in the Comments header. The remaining automated results were compared against results obtained manually for validation.
2.2.2. gp60
To test the gp60 marker, the sequence data from 147 samples, representing 100 clinical specimens collected between 2006 and 2017 along with 47 cattle isolates collected between 2009 and 2011 from the NML were tested. The clinical data was comprised of six different species with 32 distinct gp60 subtypes (15 C. hominis, 12 C. parvum, 2 C. ubiquitum, 1 C. cuniculus, 1 C. meleagridis and 1 C. felis) and the cattle data represented 5 subtypes of C. parvum. Of these samples, 36 were tested on the reverse setting only as the forward primer binding site is very close to the microsatellite region, resulting in the initial messy sequence running into the repeat region.
Additionally, chromatograms from 693 samples tested at the Cryptosporidium Reference Unit (CRU, Swansea, UK) between 2018 and 2020, and comprising 52 different gp60 subtypes (10 C. hominis, 40 C. parvum and 2 C. cuniculus) were tested using the reverse only setting in the program. A further 249 samples tested at the CRU between 2012 and 2015, comprising of 38 different gp60 subtypes (6 C. hominis, 31 C. parvum and 1 C. cuniculus) were tested using the contig mode setting of the CryptoGenotyper.
All samples were tested regardless of whether a species or gp60 subtype could be generated manually or not in order to test how well the program would deal with poor as well as good quality sequences. From the results table generated by the CryptoGenotyper, we separated those results with bit scores lower than 200 (for reverse mode) or less than 400 (for contig mode), flagged by the program to check manually, or having some bases in the repeat region with a Phred quality less than 20, to examine manually. The remaining samples had their automated genotype compared with original manual analysis.
2.2.3. DNA Extraction
We extracted the clinical and animal fecal samples, collected from the NML, directly from stool using the QIAamp Fast DNA Mini Stool kit (Qiagen, Hilden, Germany) with modifications as previously described (Guy et al., 2021). River and lake water samples (10L) collected from 2009 to 2011 were concentrated using continual flow centrifugation using the CFC Express (Scientific Methods, Granger, IN), plus/minus immunomagnetic bead separation (unpublished). We extracted the DNA using the MoBio Power Water kit (MoBio, Carlsberg, CA). River and lake water samples (20L) from 2011 to 2015 were concentrated using Filta-max (IDEXX, Westbrook, ME) filtration, immunomagnetic bead separation and enumeration on glass slides. DNA for genotyping was extracted directly from glass slides as previously described (Ruecker et al., 2005).
Samples tested from SVA were purified using a saturated salt/glucose flotation method and DNA was extracted as described in their respective studies (Pettersson et al., 2020; Åberg et al., 2020; Åberg et al., 2019; Björkman et al., 2018; Björkman et al., 2015; Alsmark et al., 2018), with samples from unpublished data processed by the same method as described in Pettersson et al. (2020).
Samples tested from the CRU that were received before 2018 were extracted by saturated salt float, boiling and QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and those between 2018 and 2020 by QIAamp DNA Mini Stool kit (Qiagen, Hilden, Germany), as previously described (Pre-2018 - Chalmers et al., 2009; 2018–2020 - Chalmers et al., 2019).
2.2.4. PCRs and Sanger Sequencing
Samples for SSU rRNA and gp60 gene targets at the NML were amplified and purified as previously described by Guy et al. (2021) whereas the SSU rRNA samples from the SVA were processed as described by Pettersson et al. (2020). For both laboratories, the SSU rRNA gene was amplified following the assay conditions as previously described (Xiao et al., 1999, Xiao et al., 2000).
In terms of the gp60 gene target, we used at the NML species-specific gp60 primers, depending on the species identified using SSU rRNA nPCR: C. parvum, C. hominis and C. cuniculus (Xiao et al., 2009), C. felis (Rojas-Lopez et al., 2020), C. meleagridis (Stensvold et al., 2014), and C. ubiquitum (Li et al., 2014). Samples included for gp60 testing from the CRU were amplified, purified and sequenced as previously described (Chalmers et al., 2009). This entailed using either a cocktail of single-round PCR primers that generates a short amplicon of ~300–400 bp (Chalmers et al., 2009) for use in the reverse only setting of the CryptoGenotyper, or a nested gp60 assay (Alves et al., 2003) that generates a longer (~800–900 bp) fragment when testing the contig mode setting.
3. Results
3.1. SSU rRNA Gene Target
The CryptoGenotyper's SSU rRNA function can either detect single species (classified as single peaks throughout the chromatogram) or mixed species or mixed variants of the same species (classified as heterozygous peaks throughout the chromatogram). For accurate testing, these two capabilities were tested individually using both Canadian and Swedish samples.
3.1.1. Single Species
3.1.1.1. Canadian Human, Animal and Water Isolates
In total, we used 275 samples with a single Cryptosporidium species detected from various Canadian studies (animal, clinical and environmental) to test the SSU rRNA locus on the CryptoGenotyper program using the contig mode setting. In total, the CryptoGenotyper produced a contig for 260 of the samples, with 259 of those samples having an accurate species identified with no comments (Table 2). The remaining sample was flagged for manual analysis; however, the final result agreed with the manual result. Of the 15 samples where the tool was unable to produce a contig, 11 were not typable via manual analysis due to poor sequence quality, and four corresponded to uncultured eukaryotes, which was in agreement with the CryptoGenotyper results of ‘No blast hits’. Therefore, this tool accurately identified the species in the corresponding chromatograms for all 275 (100%) chromatograms that represented a single species.
Table 2.
Validation of SSU using Canadian and Swedish isolates.
| Sample origin | Number of samples | Number (%) contigs produced | Number (%) called correctly | Number (%) called incorrectly | Number (%) flagged requiring manual analysis | Number (%) flagged in agreement with manual analysis | Number (%) flagged that are further resolved with manual analysis | Number (%) samples that a contig was unable to be produced | Number (%) non-contig samples in agreement with manual analysis in one or both directions | Number (%) non-contig samples not in agreement with manual analysis in one or both directions |
|---|---|---|---|---|---|---|---|---|---|---|
| Canada: | ||||||||||
| Human | 64 | 64 (100%) | 64 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Animala | 67b | 67 (100%) | 66 (98.5%) | 0 (0%) | 1 (1.5%) | 1 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Water | 133c | 129 (97.0%) | 129 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 (3.0%) | 4 (100%) | 0 (0%) |
| Sweden: | ||||||||||
| Animald | 167 | 157 (94.0%) | 154 (98.1%) | 2 (1.3%)e | 1 (0.6%) | 1 (100%) | 0 (0%) | 10 (6.0%) | 9 (90%) | 1 (10%)f |
| Total: | 431 | 417 (96.8%) | 413 (99.0%) | 2 (1.0%) | 2 (0.5%) | 2 (0.5%) | 0 (0%) | 14 (3.2%) | 13 (92.9%) | 1 (7.1%) |
Animal sample sources from 43 cattle, 20 dogs, 1 cat, 1 chicken, 2 snakes.
1 sample was not typable either by CryptoGenotyper or manually so have been removed from this total.
10 samples were not typable either by CryptoGenotyper or manually so have been removed from this total.
Animal sample sources from 117 cattle, 34 pigs, 10 squirrels, 3 sheep, 2 horses, 1 roe deer.
2 samples were called as C. parvum (Type A) and C. parvum (Type B) although this could not be confirmed in manual analysis.
One sample called as mixed infection in the reverse sequence although only one species could be identified in manual analysis.
3.1.1.2. Swedish Animal Isolates
A total of 167 Swedish samples representing singular species were inputted into the tool using the contig mode setting. The CryptoGenotyper correctly identified all but 3 samples (Table 2). For two of the sequences, two variants of C. parvum (Type A and Type B) were reported. Manual analysis could not determine if this was correct or not since the potential mix was too low and peaks were indistinguishable from the background in the chromatograms. In addition, one sample was identified as a mixed infection in a reverse sequence of a pig sample. This sample was partially incorrect as it reported a C. parvum/C. sp. ground squirrel (genotype II) mix in the reverse sequence but instead only C. parvum could be verified manually. Hence, this tool accurately identified the species for 164/167 (98.2%) single-species chromatograms, with 3/167 (1.8%) being partially incorrect as an additional species/variant was called alongside the correct single-species.
3.1.1.3. Overall SSU rRNA Single Species Performance Results
Combining both the Canadian and Swedish single infection isolates, there were a total of 431 samples (234 animal, 64 clinical, 133 water) that were used to test the SSU rRNA single-species functionality. The CryptoGenotyper reported 96.8% (417/431) of the Cryptosporidium genotypes as a contig (Table 2). Of the 417 contigs produced, 413 (99.0%) were correctly identified. Of the remaining four contig samples, two were flagged for manual analysis (of which both agreed with manual analysis) and two were partially incorrect as the CryptoGenotyper reported a mix of two C. parvum variants (where only one variant could be verified manually). Furthermore, the CryptoGenotyper was unable to produce a contig for 3.2% of the samples (14/431), but 92.9% (13/14) of those samples agreed with manual analysis and 7.1% (1/14) were partially misidentified. Altogether, the CryptoGenotyper identified the correct Cryptosporidium species in 99.3% (428/431) of the sample chromatograms that represented a single species.
3.1.2. Mixed Species or Variants of the Same Species
3.1.2.1. Canadian Human, Animal and Water Isolates
Altogether, 180 Canadian samples with heterozygous peaks in the chromatograms, representing mixed species or multiple variants of the same species, were inputted into the CryptoGenotyper to test the mixed SSU rRNA function. For 142 of the mixed samples, the tool accurately identified the mixed species or variants on at least the forward or reverse strand (Table 3). For one water sample, the CryptoGenotyper detected an incorrect mixed infection of C. wrairi/C. sp. W1; this was not detected during manual analysis and could not be confirmed through re-analysis. Furthermore, the tool detected the dominant species in 37 of the samples in addition to outputting a comment that another mixed sequence may be present. Only 4 of these samples could be further resolved with manual analysis as the other 33 samples were suspected to be mixed but could not be resolved manually. Finally, the CryptoGenotyper was unable to produce a contig for one sample and flagged it for manual analysis. This sample corresponded to a chromatogram that was uninterpretable manually due to poor sequence quality. Removing the 33 sample chromatograms with heterozygous peaks that could not be resolved manually and the poor quality sequence, the CryptoGenotyper identified the two correct sequences present, whether from two distinct species or two variants from the same species, for 142/147 (96.6%) mixed specimens that were determined manually.
Table 3.
Validation of mixed SSU rRNA using Canadian and Swedish isolates.
| Sample origin | Number samples with mixed sequences | Number (%) samples with mixed species/genotypes correctly identified on at least one strand | Number (%) samples with mixed species/genotypes incorrectly identified on at least one strand | Number (%) samples identified as a single species/genotype | Number (%) samples with one species/genotype correctly identified | Number (%) samples with one species/genotype incorrectly identified | Number (%) samples with one species/genotype correctly identified but second species/genotype further resolved with manual analysis |
|---|---|---|---|---|---|---|---|
| Canada: | |||||||
| Human | 55 | 38 (69.1%) | 0 (0%) | 17 (30.9%) | 13 (76.5%) | 0 (0%) | 4 (23.5%) |
| Animala | 107 | 94 (87.9%) | 0 (0%) | 13 (12.1%) | 13 (100%) | 0 (0%) | 0 (0%) |
| Water | 18b | 10 (55.6%) | 1 (5.6%) | 7 (38.9%) | 7 (100%) | 0 (0%) | 0 (0%) |
| Sweden: | |||||||
| Animalc | 20 | 12 (60.0%) | 1 (5.0%) | 7 (35.0%) | 5 (71.4%) | 0 (0%) | 2 (28.6%) |
| Total: | 200 | 154 (77.0%) | 2 (1.0%) | 44 (22.0%) | 38 (86.4%) | 0 (0%) | 6 (13.6%) |
Animal sample sources from 95 cattle, 2 dogs, 6 cats, 1 pig, 3 snakes.
1 sample was not typable either by CryptoGenotyper or manually so have been removed from this total.
Animal sample sources from 7 cattle, 10 squirrels, 2 chital deer, 1 sheep.
3.1.2.2. Swedish Isolates
In addition, we evaluated the results of the CryptoGenotyper's mixed SSU rRNA function using 20 Swedish samples that were manually identified to contain mixed species or multiple variants of the same species. For 12 of the 20 samples, the tool accurately identified the mixed species or variants on at least the forward or reverse strand (Table 3). One of the samples was detected as a mixed infection, but only one of the species was correctly called as the CryptoGenotyper reported a C. ferret genotype/C. hominis mixed infection; however manual analysis revealed a C. ferret genotype/C. chipmunk genotype I infection. In the remaining seven samples, the tool detected one correct species and one of those samples was flagged as a potential mix for further manual analysis. For five of the samples where one species was correctly determined, the manual analysis was aided by complementary gp60 analysis confirming a low grade mix, although the heterozygous peaks were not clear. Hence, since the manual analysis of the SSU rRNA marker alone could not identify the mixed infection, we conclude the CryptoGenotyper correctly identified 12/15 (80.0%) of the Swedish samples with mixed species compared to the manual analysis.
3.1.3. Overall SSU rRNA Mixed Species or Variants of Same Species Performance Results
Combining the Canadian and Swedish results, 200 samples (127 animal, 55 clinical, 18 water) were of chromatograms representing mixed species or mixed variants of the same species. The CryptoGenotyper reported 77.0% (154/200) of the mixed sequences correctly at one or both strands (Table 3). However, the tool gave an incorrect mixed result for 1.0% (2/200) of the samples. For 22% (44/200) of the samples, the CryptoGenotyper was unable to identify the mixed species but correctly identified the dominant species present in the chromatogram. Of the 44 samples, 13.6% (6/44) were able to be further resolved manually. Of the remaining 38 samples where a single species was determined but could not be further resolved through manual analysis, 33 were suspected to be mixed when analyzed manually, but the two individual sequences could not be identified. In addition, 5 of those samples were determined to have mixed species through gp60 analysis but heterozygous peaks were not visible by manual inspection. Excluding these 38 samples where two species could not be identified in the chromatograms manually, the CryptoGenotyper determined the correct mixed species or variants in 95.1% (154/162) of mixed chromatograms that were decipherable through manual analysis, with 1.2% (2/162) reported in error.
3.2. Validation of the gp60 Gene Target
3.2.1. Canadian Clinical and Animal Isolates
In total, 147 samples (36 reverse only and 111 contig) were used to validate the gp60 portion of the CryptoGenotyper. Of the 36 samples tested using the reverse-only setting, 16.7% (6/36) were flagged for manual analysis (poor sequence quality in the repeat region identified) but were all in agreement with manual analysis (Table 4). The remaining 83.3% (30/36) of samples were correctly identified. For the 111 samples that were tested in the contig mode setting, the CryptoGenotyper produced contigs for 86.5% (96/111) of them, with 67.7% (65/96) correctly identified. The other 32.3% (31/96) of contigs produced were flagged for manual analysis, with 93.5% (29/31) in agreement with the results determined manually and 6.5% (2/31) further resolved. The tool was unable to produce a contig for 13.5% (15/111) of samples. Of these samples, 13.3% (2/15) could not be typed manually due to messy chromatograms, 73.3% (11/15) agreed with manual analysis but were flagged due to low quality repeat regions (Phred quality <20), and 13.3% (2/15) were further resolved with manual analysis. Overall, the CryptoGenotyper accurately subtyped 141 (97.2%) of the 145 Canadian samples that were typable.
Table 4.
Validation of gp60 gene target using Canadian and UK isolates.
| Sample origin | Number of samples | Number (%) contigs produced | Number (%) called correctly and able to produce contig | Number (%) called incorrectly | Number (%) flagged requiring manual analysis | Number (%) flagged in agreement with manual analysis | Number (%) flagged that are further resolved with manual analysis | Number (%) samples that a contig was unable to be produceda | Number (%) non-contig samples in agreement with manual analysis in one or both directionsa | Number (%) non-contig samples further resolved with manual analysisa |
|---|---|---|---|---|---|---|---|---|---|---|
| Canada: | ||||||||||
| Reverse | 36 | N/A | 30 (83.3%) | 0 (0%) | 6 (16.7%) | 6 (100%) | 0 (0%) | N/A | N/A | N/A |
| Contig | 109b | 96 (88.1%) | 65 (67.7%) | 0 (0%) | 31 (32.3%) | 29 (93.5%) | 2 (6.5%) | 13 (11.9%) | 11 (84.6%) | 2 (15.4%) |
| UK: | ||||||||||
| Reverse | 610c | N/A | 550 (90.2%) | 0 (0%) | 60 (9.8%) | 31 (51.7%) | 29 (48.3%) | N/A | N/A | N/A |
| Contig | 236d | 213 (90.3%) | 207 (97.2%) | 0 (0%) | 6 (2.8%) | 0 (0%) | 6 (100%) | 23 (9.7%) | 18 (78.3%) | 5 (21.7%) |
| Total: | ||||||||||
| Reverse | 646 | N/A | 580 (89.8%) | 0 (0%) | 66 (10.2%) | 37 (56.1%) | 29 (43.9%) | N/A | N/A | N/A |
| Contig | 345 | 309 (89.6%) | 272 (88.0%) | 0 (0%) | 37 (12.0%) | 29 (78.4%) | 8 (21.6%) | 36 (10.4%) | 29 (80.6%) | 7 (19.4%) |
Contig mode only.
2 samples were not typable either by CryptoGenotyper or manually so have been removed from this total.
88 samples were not typable either by CryptoGenotyper or manually so have been removed from this total.
13 samples were not typable either by CryptoGenotyper or manually so have been removed from this total.
3.2.2. UK Isolates
Of the 698 UK isolates that were tested using the reverse-only setting of the gp60 function on the CryptoGenotyper, 21.2% (148/698) were flagged for manual analysis (either a bit score below 200 or poor quality sequence identified) and 78.8% (550/698) produced a result (Table 4). All of these 550 samples generated the correct result in the CryptoGenotyper when compared with the original result at the time of submission. Of the samples flagged for manual analysis, 59.5% (88/148) could not be typed manually due to failed sequencing or uninterpretable chromatograms. The remaining 40.5% (60/148) of samples were manually typable, with 51.7% (31/60) of these also being called correctly by the program. Therefore, of the 610 samples with good quality chromatograms that could result in a subtype, 95.2% (581/610) were correctly subtyped by the program.
Of the 249 isolates that were tested using the contig mode setting of the gp60 function on the CryptoGenotyper, 85.5% (213/249) were able to produce a contig. Of the contigs produced, 97.2% (207/213) had the correct result outputted without further analysis and 2.8% (6/213) were flagged for manual checking, and were resolved that way (Table 4). The remaining 14.5% (36/249) isolates could not produce a contig, where 36.1% (13/36) were not typable by either the CryptoGenotyper or manual analysis and of the remaining 63.9% (23/36) of isolates that were manually typable, 78.3% (18/23) were also reported correctly by the program in one or both directions. Therefore, 95.3% (225/236) of the typable UK samples that were evaluated using the contig mode setting were correctly subtyped by the program, and the remaining 4.7% (11/236) were flagged and identified by manual confirmation.
3.2.3. Overall gp60 Performance
In total, 1094 samples were inputted into the CryptoGenotyper (734 in reverse mode and 360 in contig mode). Of these samples, 22.1% (242/1094) were flagged for manual analysis: 42.6% (103/242) of these were not typable either manually or automatically due to poor chromatogram quality, 39.2% (95/242) had results in agreement with manual analysis despite being flagged due to peak(s) in the repeat region having a Phred score of <20, and 18.2% (44/242) were further resolved with manual analysis. Removing the samples with poor quality chromatograms that were not typable either manually or by using the CryptoGenotyper, the CryptoGenotyper reported the correct result for 95.6% (947/991) of samples with 100% accuracy and the remaining 4.4% (44/991) of samples were further resolved through manual analysis.
4. Discussion
Obtaining usable results for all genotyping and sub-genotyping experiments of Cryptosporidium is critical for fully understanding how this pathogen is transmitted. The CryptoGenotyper program automatically and consistently alleviates many of the difficulties encountered when analyzing Cryptosporidium sequences: obtaining reproducible and accurate results in a timely manner, detecting and distinguishing between mixed variants of the same species and mixed infections, and reporting results in standard nomenclature.
The main benefit to utilizing the CryptoGenotyper during Sanger sequence analysis is obtaining accurate and complete results. This tool uses a high quality, manually curated reference database for molecular characterization, removing the potential for misclassifying or misnaming of the species due to errors in the public databases, a common human error when analyzing Cryptosporidium sequences. Combined with the ability to analyze large datasets more efficiently, the results are reported in a timelier manner and with less systematic biases compared to manual analysis. To ensure accuracy and consistent results are obtained with the CryptoGenotyper, the tool was evaluated and fine-tuned using a comprehensive and diverse dataset that included Sanger sequencing chromatograms from laboratories in three different countries.
From the final evaluation of the CryptoGenotyper's performance on the SSU rRNA gene target, the tool correctly identified 99.3% of the chromatograms inputted that contained single peaks. For the remaining 0.7% of samples, the CryptoGenotyper outputted partially incorrect results as two species or two variants of the same species were identified but only one of species or variants could be verified manually. These partially incorrect results could be a result of the tool detecting peaks just above the background noise that is not discernable through manual analysis. Of the 99.3% samples that were correctly identified, the CryptoGenotyper was unable to produce a contig for 3.2% of those samples. The main reason why the tool was unable to produce a contig is due to an uninterpretable chromatogram for one of the strands being inputted. When the tool encounters this situation, both forward and reverse strands are analyzed separately, giving the results for a single direction despite contig mode being selected. For all cases, the result in the single direction corresponded to the manual findings obtained.
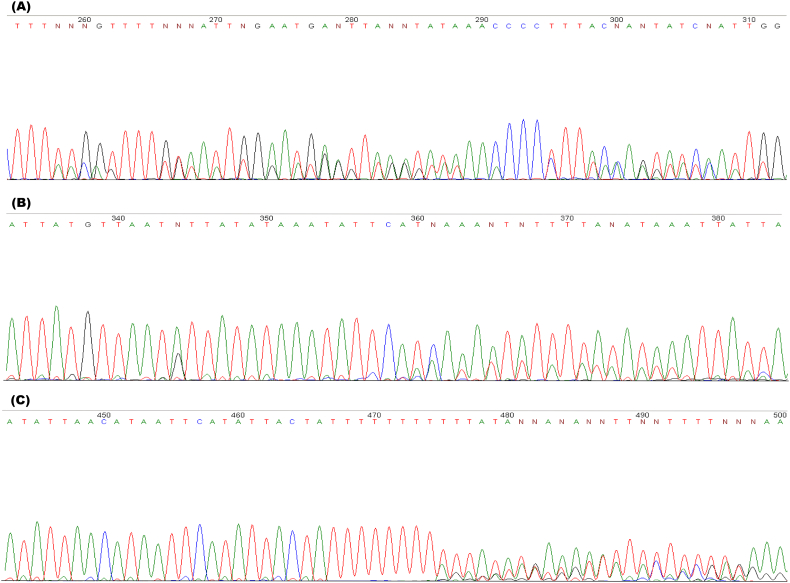
Further to the SSU rRNA gene target analysis, one of the most common issues in analyzing this target for Cryptosporidium is the presence of heterozygous peaks within the chromatogram. This is usually the result of multiple sequences overlapping due to either a mixed infection of two or more species or the presence of variant copies of the gene in the genome of the infecting species (Fig. 5) (Xiao et al., 1999). It has been reported that mixed Cryptosporidium samples are not uncommon (Fig. 5A) (Cama et al., 2006) and variant copies of the SSU rRNA gene in a single isolate have been reported, such as TGA polymorphisms between Type A C. parvum and Type B C. parvum (Fig. 5B) (Carraway et al., 1995; Gharieb et al., 2019), a thymine insertion between Type A C. andersoni and Type B C. andersoni (Ikarashi et al., 2013), and varying number of thymines (6, 8, 10, 11) in the poly-T repeat region in C. hominis (Fig. 5C) (Xiao et al., 1999; Sulaiman et al., 2001). Fortunately, the CryptoGenotyper is programmed to differentiate the sequence populations and identify up to two mixed sequences within a chromatogram, if present.
Fig. 5.
Heterozygous peaks due to Mixed SSU rRNA populations.
Overlapping peaks are present throughout the SSU rRNA region, indicating mixed populations. (A) C. parvum and C. canis mixed infection. (B) Reverse Sanger sequence chromatogram representing the variant copies of the Type A and Type B (TGA polymorphism) SSU rRNA gene in a C. parvum isolate. (C) Forward Sanger sequence chromatogram representing the variant copies of the SSU rRNA gene in a C. hominis isolate with varying numbers of thymines.
When the CryptoGenotyper was given chromatograms representing mixed species or mixed variants of the same species, it correctly determined the two sequences in the chromatograms for 95.1% of samples. Of this testing dataset, only 1.2% had the second species called in error with the dominant species called correctly. Furthermore, 33 additional samples with chromatograms containing heterozygous peaks in which the second species or variant could not be deciphered manually were inputted into the CryptoGenotyper. Similarly to the manual analysis results, the tool was unable to determine the second species or variant causing the heterozygous peaks; however, it still outputted the correct dominant species along with a comment indicating manual analysis is required as a second species or variant may be present. Manually reading mixed chromatograms is not straightforward and requires experience, thus the CryptoGenotyper was valuable in identifying mixed samples that may have been misinterpreted. The tool's ability to decipher heterozygous peaks was especially beneficial in samples from impacted waters where a diversity of different species and wildlife genotypes are found.
The performance of the CryptoGenotyper on the gp60 gene target was similar to the SSU rRNA gene target. In this case however, the tool was only designed to report the dominant subtype as it is less common to obtain mixed sequences with this gene target. Overall, the CryptoGenotyper reported the correct result for 95.6% of samples with 100% accuracy. The remaining 4.4% of samples were further resolved through manual analysis. For the majority of these cases, the CryptoGenotyper failed to report the full subtype due to either a sequencing artifact present or peak(s) with Phred quality less than 20 within the repeat region. Often, the beginning of the repeat region on the forward strand has poor quality due to the forward primer binding site being very close to this microsatellite region. If the CryptoGenotyper encounters these scenarios, it will fail to provide a complete subtype and will identify the sample for manual analysis to ensure accurate results are produced.
It is important to note the limitations of the program. First, the CryptoGenotyper relies heavily on an accurate and well-curated database. It is important to regularly update the database to ensure the most accurate results are being reported. The CryptoGenotyper's databases are planned to be updated as new information becomes available, though users are able to upload their own updated databases if desired. Furthermore, Sanger sequencing data may not be reflecting all the populations within the sample, but rather the species for which the primers have best affinity to or the one or few most popular alleles (Morris et al., 2019). There is strong evidence that sub-populations of Cryptosporidium can be present within individual hosts due to genetic re-assortment during the sexual phase of the life cycle (Morris et al., 2019). It was demonstrated that Next Generation Sequencing (NGS) is more effective at identifying and resolving multiple populations within a sample (Zahedi et al., 2017). However, these methods are not commonly used due to costs and the inability to extract sufficient high quality DNA. Therefore, this program provides an immediate solution to resolve most Sanger chromatograms that show mixed populations within the data. However, it is important to note that due to the algorithm implemented, only two sequences can be resolved within a single chromatogram. The primary limitation of the program is poor quality chromatograms, as the program is unable to decipher these and will instead inform the user to perform a manual check. To ensure the CryptoGenotyper could handle poor quality data, a subset of samples that could not be genotyped manually due to failed sequencing or messy chromatogram were included in both the SSU rRNA and gp60 gene target datasets. For all cases, the tools failed to genotype the samples and outputted a comment to check the sample manually. It was important to test poor quality data as one of the powers of the program is to process samples without prior screening of the chromatograms. Furthermore, it is critical for the user to check the comments section of the final results file outputted by the tool as the CryptoGenotyper was designed to output a message if it encountered something unfamiliar, or suspected there are mixed sequences it cannot resolve, or a bad quality chromatogram was inputted to ensure accurate results. Most importantly, if the CryptoGenotyper encounters a novel genotype that is not described in its database, the tool will output a message indicating no significant hits were found and that manual analysis is required. However, if a novel gp60 family were to be inputted, the CryptoGenotyper will misclassify the sequence to that of its most closely related family due to high sequence similarities between gp60 families. Though if the percent identity of this match is below 99.1%, the tool will flag the sample and output a message for the user to check manually as this may be indicative that a new gp60 family has been identified. This further demonstrates the importance for the user to analyze the results file thoroughly.
5. Conclusion
We have developed a freely available and user-friendly tool, called the CryptoGenotyper, to help Cryptosporidium research groups analyze their Sanger sequencing results and triage complex samples for further manual processing. The CryptoGenotyper resolves heterozygous peaks and uses reputable reference alignments to characterize the genotype and subgenotype based on the SSU rRNA or gp60 locus, respectively, in the presence of mixed infections or artifacts from the sequencing itself.
Evaluating the CryptoGenotyper in three independent laboratories demonstrated that the tool correctly classified 99.3% (428/431) of SSU rRNA chromatograms with a single species present and 95.1% (154/162) with mixed species or variants identified. For the single species analysis, 0.7% (3/431) of samples were partially incorrectly called as mixed variants or species when only one species could be determined through manual analysis for these samples. For the mixed species or variants of the same species analysis, 3.7% (6/162) of the samples could be further resolved with manual analysis and the tool misidentified 1.2% (2/162) of the mixed species in the entire testing dataset. For the gp60 gene target, the tool accurately subtyped 95.6% (947/991) of Cryptosporidium positive samples, with 44/991 (4.4%) of samples being further resolved with manual analysis, and none misclassified.
With the ability to accurately identify genotypes (SSU rRNA) and subtypes (gp60) in Sanger sequencing chromatograms, the CryptoGenotyper will provide further insights into the transmission dynamics of Cryptosporidium as valuable data is recovered in a fast and reproducible way. Our tool will be useful for high throughput processing for labs with minimal bioinformatics expertise thanks to web-based accessibility options through the publicly accessible Galaxy server. Future developments will include the addition of analyzing other gene targets that could further aid in rapid identification and subtyping future Cryptosporidium infections.
Data statement
The CryptoGenotyper is available on the web-based platform Galaxy (public ToolShed repository (https://toolshed.g2.bx.psu.edu/, tool id: cryptogenotyper, owner: nml) or from the public Galaxy server at https://usegalaxy.eu/root?tool_id=toolshed.g2.bx.psu.edu/repos/nml/cryptogenotyper/CryptoGenotyper/. In addition, the CryptoGenotyper can be installed as a standalone Bioconda package (https://anaconda.org/bioconda/cryptogenotyper). The tool's reference databases, source code and documentation are available at https://github.com/phac-nml/CryptoGenotyper.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Prof. Rachel Chalmers at the Cryptosporidium Reference Unit, Swansea, for allowing the use of the UK isolate data in the evaluation. We would also like to thank Laura Martin at the National Microbiology Laboratory in Guelph for helping to develop the SSU rRNA reference database and Prof. Lihao Xiao for sharing his gp60 reference database. We thank Prof. Jessica Kissinger and Dr. Susanne Warrenfeltz of the University of Georgia for evaluating the CryptoGenotyper for potentially hosting the tool on the CryptoDB.org Galaxy instance. Finally, we would like to thank Stephanie Murphy for testing the initial versions of the CryptoGenotyper during early development. This project was funded by the Public Health Agency of Canada.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fawpar.2021.e00115.
Appendix A. Supplementary data
Supplementary material
References
- Åberg M., Emanuelson U., Troell K., Björkman C. Infection dynamics of Cryptosporidium bovis and Cryptosporidium ryanae in a Swedish dairy herd. Vet. Parasitol. X. 2019;1:100010. doi: 10.1016/j.vpoa.2019.100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åberg M., Emanuelson U., Troell K., Camilla Björkman C. A single-cohort study of Cryptosporidium bovis and Cryptosporidium ryanae in dairy cattle from birth to calving. Vet. Parasit. Reg. Stud. Rep. 2020;20:100400. doi: 10.1016/j.vprsr.2020.100400. [DOI] [PubMed] [Google Scholar]
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Grüning B., Guerler A., Hillman-Jackson J., Hiltemann S., Jalili V., Rasche H., Soranzo N., Goecks J., Taylor J., Nekrutenko A., Blankenberg D. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsmark C., Nolskog P., Lindqvist Angervall A., Toepfer M., Winiecka-Krusnell J., Bouwmeester J., Bjelkmar P., Troell K., Lahti E., Beser J. Two outbreaks of cryptosporidiosis associated with cattle spring pasture events. Vet. Parasit. Reg. Stud. Rep. 2018;14:71–74. doi: 10.1016/j.vprsr.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Alves M., Xiao L., Sulaiman I., Lal A.A., Matos O., Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 2003;41(6):2744–2747. doi: 10.1128/jcm.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman C., Lindström L., Oweson C., Ahola H., Troell K., Axén C. Cryptosporidium infection in suckler herd beef calves. Parasitology. 2015;142(8):1108–1114. doi: 10.1017/S0031182015000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman C., von Brömssen C., Troell K., Svensson C. Disinfection with hydrated lime may help manage cryptosporidiosis in calves. Vet. Parasitol. 2018;264:58–63. doi: 10.1016/j.vetpar.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama V., Gilman R.H., Vivar A., Ticona E., Ortega Y., Bern C., Xiao L. Mixed Cryptosporidium infections and HIV. Emerg. Infect. Dis. 2006;12(6):1025–1028. doi: 10.3201/eid1206.060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway M., Tzipori S., Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl. Environ. Microbiol. 1995;62(2):712–716. doi: 10.1128/AEM.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R.M., Elwin K., Thomas A.L., Guy E.C., Mason B. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 2009;14(2) doi: 10.2807/ese.14.02.19086-en. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M., Robinson G., Elwin K., Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit. Vectors. 2009;12(95) doi: 10.1186/s13071-019-3354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.T., Tsai C.N., Tang C.Y., Chen C.H., Lian J.H., Hu C.Y., Tsai C.L., Chao A., Lai C.H., Wang T.H., Lee Y.S. Mixed sequence reader: a program for analyzing DNA sequences with heterozygous base calling. Sci. World J. 2012;2012:365104. doi: 10.1100/2012/365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J.L. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev D.A., Rakitov R.A. Decoding of superimposed traces produced by direct sequencing of heterozygous Indels. PLoS Comput. Biol. 2008;4(7) doi: 10.1371/journal.pcbi.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoozi Z., Sazmand A., Zahedi A., Astani A., Fattahi-Bafghi A., Kiani-Salmi N., Ebrahimi B., Dehghani-Tafti A., Ryan U., Akrami-Mohajeri F. Prevalence and genotyping identification of Cryptosporidium in adult ruminants in Central Iran. Parasit. Vectors. 2019;12:510. doi: 10.1186/s13071-019-3759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharieb R.M.A., Bowman D.D., Liotta J.L., Xiao L. Isolation, genotyping and subtyping of single Cryptosporidium oocysts from calves with special reference to zoonotic significance. Vet. Parasitol. 2019;271:80–86. doi: 10.1016/j.vetpar.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Guy R.A., Yanta C.A., Muchaal P.K., Rankin M.A., Thivierge K., Lau R., Boggild A.K. Molecular characterization of Cryptosporidium isolates from humans in Ontario, Canada. Parasit. Vectors. 2021;14:69. doi: 10.1186/s13071-020-04546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikarashi M., Fukuda Y., Honma H., Kasai K., Kaneta Y., Nakai Y. First description of heterogeneity in 18S rRNA genes in the haploid genome of Cryptosporidium andersoni Kawatabi type. Vet. Parasitol. 2013;196(1–2):220–224. doi: 10.1016/j.vetpar.2012.12.053. [DOI] [PubMed] [Google Scholar]
- Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., Santin M., Fayer R., Kvac M., Ryan U., Sak B., Stanko M., Guo Y., Wang L., Zhang L., Cai J., Roellig D., Feng Y. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014;20(2):217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A., Robinson G., Swain M.T., Chalmers R.M. Direct sequencing of Cryptosporidium in stool samples for public health. Front. Public Health. 2019;7:360. doi: 10.3389/fpubh.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson E.I., Ahola H., Frössling J., Wallgren P., Troell K. Detection and molecular characterisation of Cryptosporidium spp. in Swedish pigs. Acta Vet. Scand. 2020;62:40. doi: 10.1186/s13028-020-00537-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puiu D., Enomoto S., Buck G.A., Abrahamsen M.S., Kissinger J.C. CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res. 2004;32(Database issue):D329–D331. doi: 10.1093/nar/gkh050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Lopez L., Elwin K., Chalmers R.M., Enemark H.L., Beser J., Troell K. Development of a gp60-subtyping method for Cryptosporidium felis. Parasit. Vectors. 2020;13:39. doi: 10.1186/s13071-020-3906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruecker N.J., Bounsombath N., Wallis P., Ong C.S.L., Isaac-Renton J.L., Neumann N.N. Molecular forensic profiling of Cryptosporidium species and genotypes in raw water. Appl. Environ. Microbiol. 2005;71(12):8991–8994. doi: 10.1128/AEM.71.12.8991-8994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruecker N.J., Matsune J.C., Wilkes G., Lapen D.R., Topp E., Edge T.A., Sensen C.W., Xiao L., Neumann N.F. Molecular and phylogenetic approaches for assessing sources of Cryptosporidium contamination in water. Water Res. 2012;46(16):5135–5150. doi: 10.1016/j.watres.2012.06.045. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Beser J., Axén C., Lebbad M. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J. Clin. Microbiol. 2014;52(7):2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensvold C.R., Elwin K., Winiecka-Krusnell J., Chalmers R.M., Xiao L., Lebbad M. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J. Clin. Microbiol. 2015;53(6):1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I.M., Lal A.A., Xiao L. A population genetic study of Cryptosporidium parvum human genotype parasites. J. Eukaryot. Microbiol. 2001;48:24s–27s. doi: 10.1111/j.1550-7408.2001.tb00441.x. [DOI] [PubMed] [Google Scholar]
- Vanathy K., Parija S.C., Mandal J., Hamide A., Krishnamurthy S. Cryptosporidiosis: a mini review. Trop. Parasitol. 2017;7(2):72–80. doi: 10.4103/tp.TP_25_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Alderisio K., Limor J., Royer M., Lal A.A. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Applied and environmental microbiology. 2000;66(12):5492–5498. doi: 10.1128/aem.66.12.5492-5498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food and Waterborne Parasitology. 2017;8–9:14–32. doi: 10.1016/j.fawpar.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L.H., Limor J.R., Li L.X., Morgan U., Thompson R.C.A., Lal A.A. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 1999;46:44S–45S. [PubMed] [Google Scholar]
- Xiao L., Hlavsa M.C., Yoder J., Ewers C., Dearen T., Yang W., Nett R., Harris S., Brend S.M., Harris M., Onischuk L., Valderrama A.L., Cosgrove S., Xavier K., Hall N., Romero S., Young S., Johnston S.P., Arrowood M., Roy S., Beach M.J. Subtype analysis of Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa in 2007: widespread occurrence of one Cryptosporidium hominis subtype and case history of an infection with the Cryptosporidium horse genotype. J. Clin. Microbiol. 2009;47(9):3017–3020. doi: 10.1128/JCM.00226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi A., Paparini A., Jian F., Robertson I., Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int. J. Parasitol. Parasites Wildl. 2015;5(1):88–109. doi: 10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi A., Gofton A.W., Jian F., Paparini A., Oskam C., Ball A., Roberston I., Ryan U. Next generation sequencing uncovers within-host differences in the genetic diversity of Cryptosporidium gp60 subtypes. Int. J. Parasitol. 2017;47(10−11):601–607. doi: 10.1016/j.ijpara.2017.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material