Abstract
Persulfides (R-SSH) have been hypothesized as potent redox modulators and signaling compounds. Reported here is the synthesis, characterization, and in vivo evaluation of a persulfide donor that releases N-acetyl cysteine persulfide (NAC-SSH) in response to the prokaryote-specific enzyme nitroreductase. The donor, termed NDP-NAC, decomposed in response to E. coli nitroreductase, resulting in release of NAC-SSH. NDP-NAC elicited gastroprotective effects in mice that were not observed in animals treated with control compounds incapable of persulfide release or in animals treated with Na2S. NDP-NAC induced these effects by the upregulation of beneficial small and medium chain fatty acids and through increasing growth of Turicibacter sanguinis, a beneficial gut bacterium. It also decreased the populations of Synergistales bacteria, opportunistic pathogens implicated in gastrointestinal infections. This work represents the first example of targeted delivery of persulfides and reveals the possibility of maintaining gut health or treating microbiome-related diseases with reactive sulfur species such as persulfides.
Keywords: Gasotransmitter, microbiome, stimuli-responsive, inflammation, oxidative stress
Graphical Abstract

Introduction
Hydrogen sulfide (H2S) has been under investigation as a biological signaling gas (gasotransmitter) since its physiological signaling capacity was discovered in 1996.[1] To aid in understanding its (patho)physiological roles, many different types of donors have been synthesized with a variety of triggers, including water,[2] nucleophiles,[3] light,[4] and enzymes.[5] Some may hold therapeutic value in the form of prodrugs and drug conjugates.[6] More recently, additional work has been conducted in the synthesis of compounds that release a variety of related reactive sulfur species (RSS) including carbonyl sulfide (COS),[7] sulfur dioxide (SO2),[8] and persulfides (R-SSH),[9] with the goal of understanding the specific roles of each of these RSS in the greater reactive species interactome.[10] Of these RSS, persulfides have received considerable interest lately because of their role as likely H2S signaling products, coupled with advances in redox biology showing that native persulfides carry out specific, anti-oxidative physiological functions.[11] Several persulfide-releasing compounds (termed persulfide donors) release their payload under a variety of conditions, including changes in pH,[9d] and addition of fluoride,[12] hydrogen peroxide,[9b] or esterases.[9a, 9c] These donors provide insight into persulfide interactions from a systemic perspective, but it is difficult to determine the role(s) persulfides may serve in specific areas of the body due to a lack of persulfide donors with targeting capabilities.
One particular area where persulfides may play important physiological and/or pathophysiological roles is in the gut.[13] The gut microbiome is a promising drug target because many links have been discovered connecting microbiome health and composition to a variety of human diseases. The effects of exogenously delivered H2S in the microbiome are controversial; some studies have shown beneficial effects of H2S in the regeneration of colon epithelial tissues by decreasing local inflammation,[14] while others have found harmful effects, such as inhibiting colonocyte respiration and increasing inflammation.[15] The discrepancies in the observed outcomes of exogenously delivered H2S may be a result of inconsistent delivery methods and concentrations, combined with the use of H2S donors capable of reaching the bloodstream and exerting off-target effects. As for other RSS, little is known; for example, there exist no studies evaluating the effect of persulfides in the microbiome due to a lack of persulfide donors specifically activated in the gut.
We sought to address this lack of targeted persulfide donors and gap in knowledge in how persulfide delivery affects the gut microbiome by synthesizing a persulfide donor that responds to the prokaryote enzyme nitroreductase (NR). NRs are located specifically in bacteria, sequestered mostly in the mouth, skin, and intestines in mammals. Pharmacologically, NRs have been utilized to trigger the release of a variety of compounds ranging from prodrug systems[16] to chemical probes used in imaging.[17] For example, Dubikovskaya developed an NR-responsive caged luciferin probe for use in gene-directed enzyme prodrug therapy. The probe fluoresced exclusively in the presence of NR, yielding a novel method to view NR transformed cancer cells.[17a] Chakrapani et al. reported the synthesis and characterization of an NR-responsive nitric oxide (NO)-releasing prodrug.[16a] The donor was stable in PBS buffer, but released NO selectively in response to NR. Chakrapani also synthesized an NR-responsive H2S prodrug by combining p-nitrobenzyl thiol with a ketone to form a thioketal.[18] The donor released H2S in the presence of bacteria and mediated cytoplasmic redox potential upon exposure to H2O2; however, it also significantly increased antibiotic resistance. Inspired by these studies and intrigued by the possibility of generating persulfides specifically in the gut, we set out to synthesize and evaluate an NR-triggered persulfide donor.
Results and Discussion
Prodrug design and synthesis
Using a similar design strategy to a previously reported H2O2-reponsive persulfide donor,[9b] we synthesized a new persulfide donor termed NDP-NAC for Nitroreductase Disulfide Prodrug- N-acetylcysteine (Scheme 1). NRs utilize a 2-electron reduction mechanism employing nicotinamide adenine dinucleotide (NADH) as a cofactor. The para positioned nitro (R-NO2) group is first reduced to a nitroso (R-NO) intermediate, then reduced to a hydroxylamine (R-NHOH), and finally to an amine (R-NH2) group.[19] In the case of NDP-NAC, we envisioned that this free amine (or hydroxylamine) could undergo a 1,6-elimination mechanism, resulting in the concomitant release of N-acetylcysteine persulfide (NAC-SSH) and p-aminobenzyl alcohol (pABA).
Scheme 1.
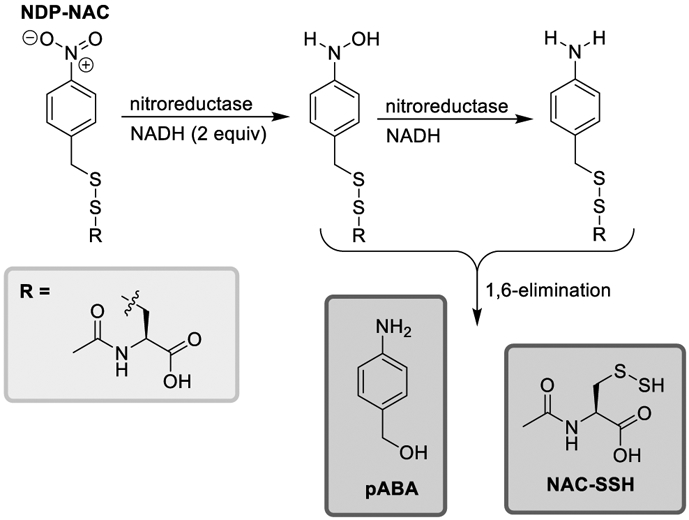
Proposed mechanism of persulfide release from NDP-NAC. 1,6-elimination may proceed via the hydroxylamine or amine intermediates.
NDP-NAC was synthesized in 4 steps (Scheme 2). First, p-nitrotoluene was brominated using N-bromosuccinimide (NBS), yielding p-nitrobenzyl bromide (1). Next, thiourea was added, and subsequent aminolysis using hexylamine yielded p-nitrobenzyl thiol (2). The resulting thiol was added to the activated disulfide of NAC (NACpyDS) in a disulfide exchange reaction to afford the prodrug in a 35% overall yield with only one column purification step required throughout the synthesis.
Scheme 2.
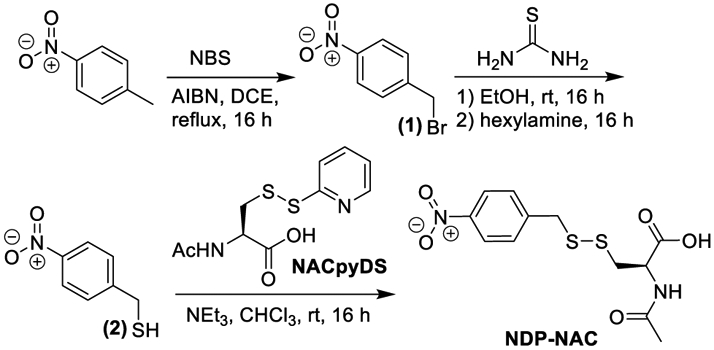
Synthesis of NDP-NAC.
Persulfide release kinetics
Due to their inherent instability, persulfides are difficult to detect directly by high resolution mass spectrometry (HRMS), so electrophilic trapping reagents such as N-ethylmaleimide (NEM), iodoacetamide (IAM),[20] or dinitrofluorobenzene (DNFB)[9a] are commonly used. The reaction of a persulfide with these trapping agents forms a disulfide that typically can withstand MS ionization conditions, whereas the persulfide itself mostly disproportionates into H2S and a thiol or forms trisulfides when directly analyzed.[9e] We attempted to trap the persulfide using NEM, IAM, and DNFB, and we found that DNFB worked best in this system.
In the experimental setup, NDP-NAC, DNFB, and NADH were dissolved in pH 8.0 PBS and added to a vial with a screwcap lid, and an aliquot was removed for a zero timepoint (t = 0). Exact procedures for dilution and analysis are described in the supporting information. NR was then added to the reaction mixture, and aliquots were removed every 30 min. After a brief workup step to remove excess NADH, each aliquot was analyzed by analytical HPLC (Figures 1, S15). The peak corresponding to NDP-NAC (elution time 4.0 min) decreased in intensity over time as a new peak (6.1 min) evolved, which we determined corresponded to the trapped persulfide adduct (DNB-NAC, Figure S10) based on comparison to an authentic sample. After 2 h, NDP-NAC had been consumed, and DNB-NAC remained. The conversion of NDP-NAC into DNB-NAC indicated that NDP-NAC efficiently released its persulfide payload in the presence of NR.
Figure 1.
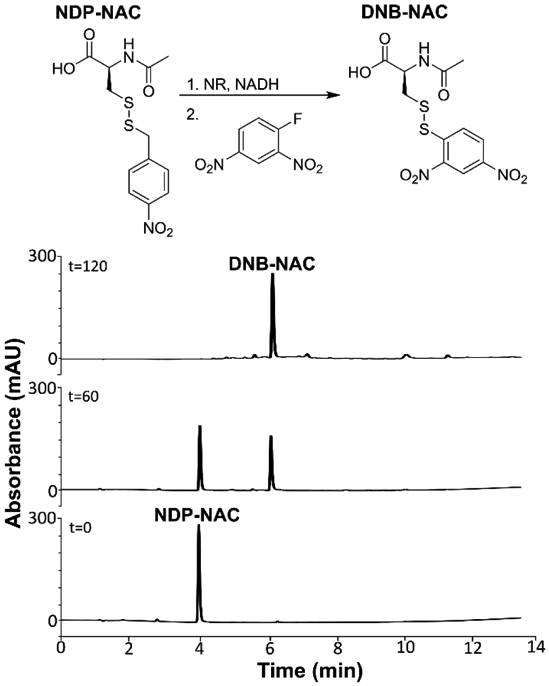
Analytical HPLC chromatogram series depicting persulfide release from NDP-NAC over time using DNFB as a trapping reagent. The peak eluting at 4.0 min corresponds to NDP-NAC while the peak at 6.1 min corresponds to the persulfide adduct DNB-NAC. Reactions were performed at 22 °C in pH 8.0 phosphate buffer (the pH level stabilized at 7.8) at 100 μM NDP-NAC. Monitoring wavelength was 282 nm.
Next, we set out to determine the persulfide release half-life from NDP-NAC using 1H NMR spectroscopy (Figure 2). NDP-NAC and NADH were dissolved in a mixture of DMSO-d6 and deuterated phosphate buffer (dPB) at pD = 8 (1:4 v/v). This pD value was chosen to mimic the mildly basic conditions of the intestines, where most gut bacteria are located. A zero timepoint was taken, NR was then added to the NMR tube, and subsequent scans were taken over the course of several hours. As shown in the zero timepoint spectrum, a clear doublet (a) corresponding to NDP-NAC was observed at 7.42 ppm. After the addition of NR, a new doublet (a’), which we attribute to pABA, was observed at 7.15 ppm. Other signals in the aromatic region were due to NADH and NAD+, the oxidation product of NADH, including the three new signals above 8.6 ppm. By 10.7 h, complete disappearance of NDP-NAC was observed. A decomposition half-life (t1/2) of 1.5 h ± 0.3 h (n=3) was calculated from these experiments (Figure S16). We and others have observed similar discrepancies between the half-lives observed in 1H NMR and HPLC experiments previously,[21] and we attributed them to slower 1,6-elimination rates in organic solvents relative to aqueous solvent mixtures.[22] Although replication of the complex gut microbiome environment is not possible in these NMR and HPLC experiments, taken together they show that NDP-NAC generates NAC-SSH in response to nitroreductase over the course of hours under the mildly basic conditions present in the intestines.
Figure 2.
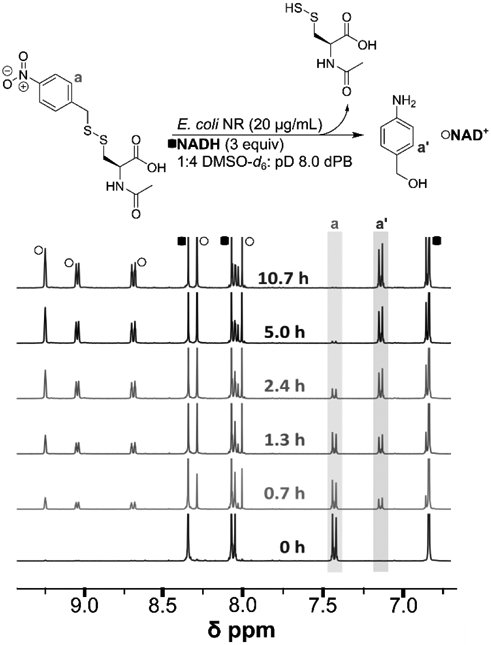
1H NMR analysis of NDP-NAC decomposition kinetics. The half-life was calculated to be 1.5 h ± 0.3 h (n = 3). Reactions were performed at 22 °C in pD 8.0 phosphate buffer (the pD level stabilized at 8.1) at 3.6 mM NDP-NAC.
Furthermore, we next evaluated the reactivity of NDP-NAC with Cys to assess the potential for biological reductants to hamper the prodrug’s ability to release persulfides. NDP-NAC and Cys (2 eq) were dissolved in 1:4 DMSO-d6:pD 8.0 dPB or 1:4 DMSO-d6:pD 2.5 dPB. In both cases, no substantial changes to the 1H NMR spectrum of NDP-NAC were observed after 24 h at rt (Figures S17 and S18), suggesting that NDP-NAC is stable in the presence of thiols under these conditions.
Effects of NDP-NAC on mammalian and bacterial cells in culture
We then aimed to determine the effects of exogenous delivery of persulfides in the microbiome. However, we first needed to establish whether NDP-NAC was toxic to human cells. NDP-NAC cell viability was conducted on H9C2 cardiomyocytes as a representative normal mammalian cell line (Figure 3A). NDP-NAC was nontoxic up to 400 μM, inducing proliferation to a small degree at lower levels compared with untreated controls. Of note, cell viability greater than 100% in these experiments is likely due to artifacts of the CCK-8 assay.[23]
Figure 3.
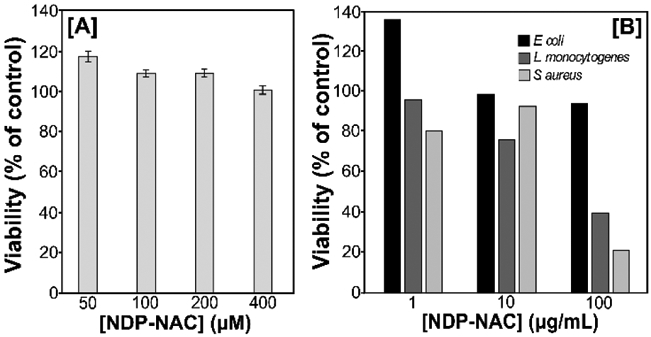
Summary of eukaryote and bacteria viability after treatment with NDP-NAC. A) H9C2 viability after treatment with NDP-NAC at various concentrations. Viability was quantified using cell counting kit 8 (CCK-8), comparing treatment groups to an untreated control. Results are expressed as the mean ± SEM (n = 3, 3–5 wells per experiment). B) Bacteria viability in response to NDP-NAC at various concentrations after 24 h culture in agar plates. Viability is reported with respect to control plates with no NDP-NAC.
Next, the toxicity of NDP-NAC to prokaryotes that are commonly located in the gut was evaluated (Figure 3B). An aqueous solution of NDP-NAC at either 1, 10, or 100 μg/mL was added to molten agar and stirred until homogeneous. Agar plates were prepared, and then E. coli, L. monocytogenes, and S. aureus were then used to inoculate the plates. After 24 h in culture, NDP-NAC exhibited no toxicity towards E. coli, a common inhabitant of the human gut. However, NDP-NAC did elicit a dose-dependent decrease in viability in L. monocytogenes and S. aureus, two opportunistic pathogens.
In vivo effects of NDP-NAC on the mouse microbiome
With these results in hand, we set out to investigate effects on general bacteria populations elicited by NDP-NAC and various controls in the mouse microbiome. Two compounds that are similar in structure to NDP-NAC were synthesized as control compounds for use in these studies (Figure 4A). Benz-NAC, which is analogous to NDP-NAC but lacks the nitro group, was prepared as a control compound that could not release a persulfide but was otherwise structurally similar. NDP-TE, which possesses a thioether linkage instead of a disulfide linkage, was synthesized as a second control compound that could undergo decomposition in response to nitroreductases but would release a thiol (NAC) instead of a persulfide. Despite potentially low bioavailability due to the oral gavage method used for delivery, Na2S was utilized as a model H2S donor control in these experiments due to its widespread use as a simple, instantaneous H2S donor that lacks byproducts. After mice (n = 5 with five treatment groups: NDP-NAC, Benz-NAC, NDP-TE, Na2S, and PBS vehicle only) were given single daily gavages of NDP-NAC (100 μL, 100 μg/mL), control compounds (100 μL, 100 μg/mL) or PBS vehicle (100 μL) over 5 d, feces were harvested and plated on agar. Morphology assessments were conducted, and individual, morphologically unique colonies were picked and cultured to logarithmic growth phase in this biased screening approach. Each specimen was identified to at least the genera level using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry.
Figure 4.
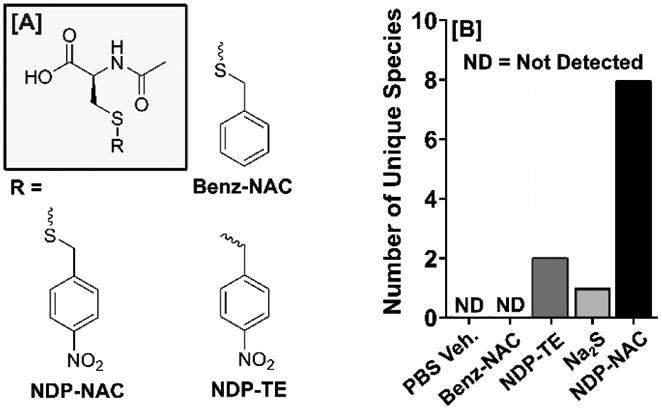
Summary of structural analogs of NDP-NAC used in MALDI-TOF. A) Chemical structures NDP-NAC and controls Benz-NAC (lacking nitro group) and NPD-TE (a thioether in place of a disulfide) used in MALDI-TOF analysis. B) Broad identification of the bacteria present in the mouse gastrointestinal microbiome upon treatment with NDP-NAC or various controls using biased sampling and MALDI-TOF.
The types of bacteria discovered in this approach were quite different in animals treated with NDP-NAC versus all others (Figure 4B). As expected, animals that received PBS only were rich in Bacillus species with no unique or non-Bacillus species. Bacillus species are quite prevalent in a healthy gut microbiome. Mice treated with Benz-NAC also elicited no microbiome change, whereas NDP-TE and Na2S elicited minor changes (two and one unique or non-Bacillus species, respectively). However, in animals treated with NDP-NAC, we recovered 8 unique or non-Bacillus species, including Enterococcus faecalis, Bacillus cereus, and Staphylococcus lentus, which are all common probiotic bacteria found in commercial supplements. While we recognize this prodrug may react in unintended ways in the complex gut environment, these findings suggested that NDP-NAC was capable of regulating the microbiome, warranting a deeper search into possible effects on metabolism within the gut.
To expand upon the findings associated with the microbiome population changes, we also sought to determine the effects of the altered microbiome on functional and metabolic processes of biological and physiological relevance by evaluating short and medium chain fatty acids (SCFAs and MCFAs) in the fecal specimens using gas chromatography.[24] SCFA/MCFA analysis revealed mostly minor differences in metabolites evaluated using NDP-NAC, Benz-NAC, NDP-TE, and vehicle-treated groups. However, we did observe a nine-fold increase in heptanoic acid production between NDP-NAC and PBS-treated groups (Figure S19). In the gastrointestinal tract, heptanoic acid plays key roles in lipid transport, peroxidation, and metabolism; decreased levels have been found in patients with Crohn’s disease, ulcerative colitis, and pouchitis.[25] Together, these data suggest that NDP-NAC may exhibit gastroprotective and anti-inflammatory effects.[26] Mouse blood plasma samples were also analyzed using ultra high performance liquid chromatography with tandem mass spectrometry (UPLC-MS/MS). No trace of NDP-NAC or pABA was detected in any of the six mouse plasma samples (LOD = 0.1 ng/mL, see Supporting Information). These results suggest that the NDP-NAC does not pass through the intestinal epithelial cell barrier and its effects appear to be contained within the gastrointestinal tract, likely indicating that the prodrug is metabolized directly by the bacteria or at least in close proximity. Likewise, no adverse effects were observed in animals treated with NDP-NAC. In summary, NDP-NAC induced changes in the mouse microbiome, affecting the types of bacterial species present and the production of metabolites, that were not observed in either of the control molecules or Na2S.
Building upon these findings, we next repeated our animal studies (same administration method, dose, and numbers of animals, but only using NDP-NAC and PBS-vehicle treatment groups) and utilized a more robust, non-biased screening approach to better define microbiome population differences between the NDP-NAC-treated and untreated animals. Feces were harvested from mice at the end of the 5-day study, with collection directly from the colon at necropsy; tissue scraping was conducted to also recover adherent populations. Fecal specimens were flash frozen upon sterile collection, followed by nucleic acid extraction for shotgun metagenomics analysis.
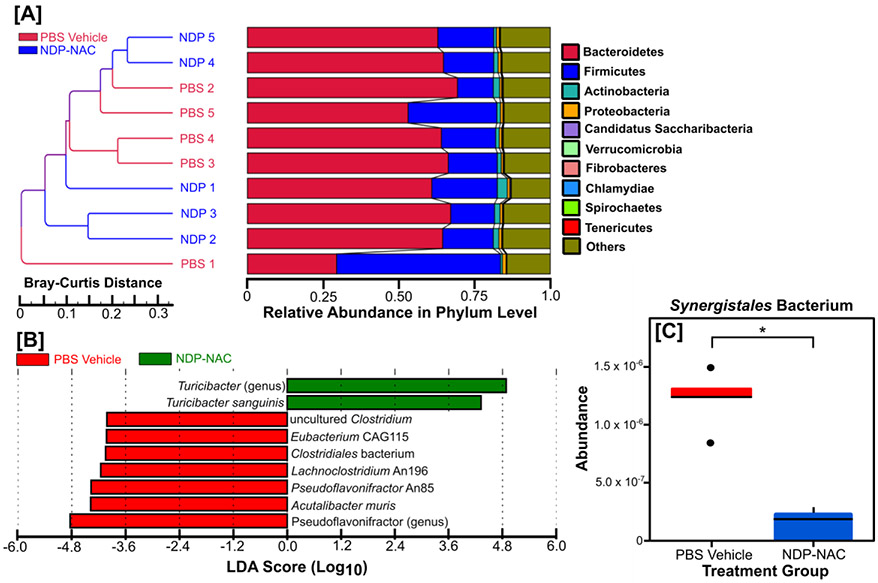
Consistent with the biased screening assessments noted above, treatment with NDP-NAC resulted in changes in microbiome populations, with significant differences noted between the PBS vehicle- and NDP-NAC-treated animals. Sample clustering with taxonomic abundance revealed that the majority of the PBS vehicle-treated animals clustered together (Figure 5A). However, treatment with NDP-NAC generated two distinct groups that were significantly different in composition at the Phylum level compared to the PBS vehicle (Figure 5B). Further assessments by LEfSe (linear discriminant analysis effect size) analysis revealed that the differences between the PBS vehicle- and NDP-NAC-treated animals were due to decreased members of the genus Pseudoflavonifractor and at the species level decreased clostridium, eubacterium, clostridiales, lachnoclostridium, pseudoflavonifractor, and acutalibacter species in the microbiome isolated from the vehicle-treated animals. Conversely, the microbiome from the NDP-NAC-treated animals had a significant increase in members of the Turicibacter genus, specifically Turicibacter sanguinis species. Intriguingly, T. sanguinis has recently been identified as a highly relevant gut bacterium that naturally expresses a neurotransmitter sodium symporter-related protein homologous to mammalian SERT, which is a host 5-HT (serotonin) transporter.[27] T. sanguinis blooms in the GI tract can alter a broad range of host genes impacting multiple biochemical pathways, including increases in those important for lipid and steroid metabolism and decreasing systemic triglyceride levels and inguinal adipocyte size.[27] In addition to T. sanguinis being increased in the NDP-NAC-treated animals, additional metastats analysis was conducted, which relies on a non-parametric t-test for detecting differentially abundant features in metagenomic studies. Results revealed a significant decrease in the relative abundance of Synergistales bacterium in NDP-NAC-treated animals (Figure 5C). Synergistales bacteria are a recently identified family of anaerobic, gram-negative, opportunistic pathogens that are implicated in the formation and progression of cysts, abscesses, periodontal disease, and gastrointestinal infections.[28]
Figure 5. Sample clustering with taxonomic abundance.
A) Sample clustering analysis based on Bray-Curtis distance highlighting the similarity of samples from each animal. The distance was calculated according to relative taxonomic abundance, and the data shown were generated by combining the clustering results and relative abundances of different samples at the phylum level. B) LEfSe analysis identifying genomic features (taxa) characterizing the significant differences between the microbiome populations among treatment groups. The histogram of the LDA scores shows the species whose LDA scores are larger than the set threshold (4 set by default). C) Metastats analysis demonstrating a significant decrease in the relative abundance of Synergistales bacterium in mice treated with NDP-NAC. *p<0.05
Finally, to gain additional insight into the microbial metabolic processes impacted by NDP-NAC, we also utilized shotgun metagenomics and to assemble protein sequence data to predict microbial protein function. This analysis identified changes in only three functions following NDP-NAC treatment: genetic information processing/translation, lipid metabolism, and endocrine and metabolic diseases (Figure 6). The increased translation and lipid metabolism differences are highly consistent with the findings associated with the increased Turicibacter sanguinis levels (Figure 5B) as this bacterium significantly impacts lipid metabolism, triglyceride levels, and host genetic functions.[27] These findings are also consistent with the observed changes in heptanoic acid (Figure S19), which is critical for lipid transport and metabolism. We also found that in contrast to previous reports on H2S delivery to the gut,[18] genetic markers for antibiotic resistance did not increase in the NDP-NAC treatment group (Figures S23, S24, and S25).
Figure 6. Metastats analysis of functional differences.
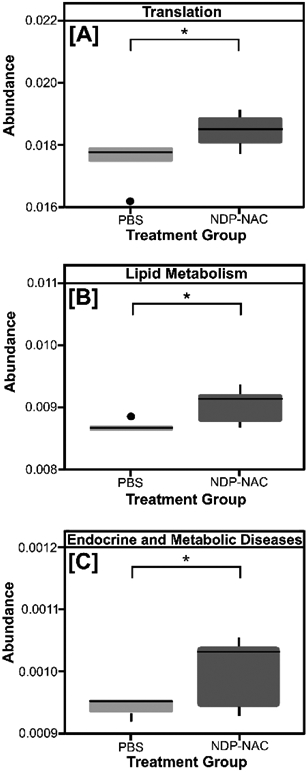
Three functions, whose abundances were significantly different between the PBS vehicle- and NDP-NAC-treatment groups, were identified from shotgun metagenomic analyses: (A) genetic information processing/translation, (B) lipid metabolism, (C) endocrine and metabolic diseases. *p<0.05
Conclusions
In summary, NDP-NAC, a nitroreductase-responsive persulfide prodrug, which to our knowledge represents the first persulfide donor with targeting capabilities, released persulfides in the presence of microbial nitroreductase with a half-life on the order of hours. NDP-NAC significantly impacted the composition and function of the host microbiome in mice treated with the prodrug compared with mice treated with non-persulfide releasing control compounds and Na2S, an H2S donor. Changes elicited by NDP-NAC created favorable niches for the expansion of a limited repertoire of bacteria species, including Turicibacter sanguinis, that appear to have robust host effects impacting metabolism and overall gastrointestinal health. Conversely, NDP-NAC also demonstrated clear anti-microbial effects on a diverse range of opportunistic and pathogenic bacteria in the gut. This includes limiting Synergistales bacteria, which are associated with a variety of human health issues. Thus, NDP-NAC enhances several health-promoting bacteria while limiting the growth of various pathogenic bacteria. This work adds to the body of knowledge in the rapidly growing field of gut microbiome health, showing that controlled delivery of RSS to the gut may be useful for modulating the gastrointestinal microbiome during disease states, shifting the balance from dysbiosis to homeostasis.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R01GM123508), the National Science Foundation (DMR-1454754), the Virginia Tech Center for Drug Discovery, and the Dreyfus foundation. The authors thank Dr. Mehdi Ashraf-Khorassani for assistance with LCMS, Dr. Kristin Eden for her animal model expertise, and Elizabeth Smiley for fatty acid analysis. The authors also acknowledge Novogene Co., Ltd. for their genome sequencing and bioinformatics work.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- [1].Abe K, Kimura H, J. Neurosci 1996, 16, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].(a) Li L, Whiteman M, Guan YY, Neo KL, Cheng Y, Lee SW, Zhao Y, Baskar R, Tan C-H, Moore PK, Circulation 2008, 117, 2351; [DOI] [PubMed] [Google Scholar]; (b) Nicolau LAD, Silva RO, Damasceno SRB, Carvalho NS, Costa NRD, Aragão KS, Barbosa ALR, Soares PMG, Souza MHLP, Medeiros JVR, Braz. J. Med. Biol. Res 2013, 46, 708–714; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhao W, Zhang J, Lu Y, Wang R, EMBO J. 2001, 20, 6008–6016; [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kang J, Li Z, Organ CL, Park C-M, Yang C.-t., Pacheco A, Wang D, Lefer DJ, Xian M, J. Am. Chem. Soc 2016, 138, 6336–6339. [DOI] [PubMed] [Google Scholar]
- [3].(a) Zhao Y, Wang H, Xian M, J. Am. Chem. Soc 2011, 133, 15–17; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Foster JC, Powell CR, Radzinski SC, Matson JB, Org. Lett 2014, 16, 1558–1561. [DOI] [PubMed] [Google Scholar]
- [4].(a) Fukushima N, Ieda N, Sasakura K, Nagano T, Hanaoka K, Suzuki T, Miyata N, Nakagawa H, Chem. Commun 2014, 50, 587–589; [DOI] [PubMed] [Google Scholar]; (b) Devarie-Baez NO, Bagdon PE, Peng B, Zhao Y, Park C-M, Xian M, Org. Lett 2013, 15, 2786–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].(a) Zheng Y, Yu B, Ji K, Pan Z, Chittavong V, Wang B, Angew. Chem. Int. Ed 2016, 55, 4514–4518; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chauhan P, Bora P, Ravikumar G, Jos S, Chakrapani H, Org. Lett 2017, 19, 62–65; [DOI] [PubMed] [Google Scholar]; (c) Li Z, Organ CL, Zheng Y, Wang B, Lefer DJ, Circulation 2016, 134, 17903. [Google Scholar]
- [6].Powell CR, Dillon KM, Matson JB, Biochem. Pharmacol 2018, 149, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].(a) Powell CR, Foster JC, Okyere B, Theus MH, Matson JB, J. Am. Chem. Soc 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Steiger AK, Pardue S, Kevil CG, Pluth MD, J. Am. Chem. Soc 2016, 138, 7256–7259; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Powell CR, Foster JC, Swilley SN, Kaur K, Scannelli SJ, Troya D, Matson JB, Polym. Chem 2019, 10, 2991–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].(a) Wang W, Ji X, Du Z, Wang B, Chem. Commun 2017, 53, 1370–1373; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Day JJ, Yang Z, Chen W, Pacheco A, Xian M, ACS Chem. Biol 2016, 11, 1647–1651; [DOI] [PubMed] [Google Scholar]; (c) Malwal SR, Sriram D, Yogeeswari P, Konkimalla VB, Chakrapani H, J. Med. Chem 2012, 55, 553–557. [DOI] [PubMed] [Google Scholar]
- [9].(a) Zheng Y, Yu B, Li Z, Yuan Z, Chelsea L, Rishi K, Wang S, Lefer David J, Wang B, Angew. Chem 2017, 129, 11911–11915; [Google Scholar]; (b) Powell CR, Dillon KM, Wang Y, Carrazzone RJ, Matson JB, Angew. Chem. Int. Ed 2018, 57, 6324–6328; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yuan Z, Zheng Y, Yu B, Wang S, Yang X, Wang B, Organic Letters 2018, 20, 6364–6367; [DOI] [PubMed] [Google Scholar]; (d) Artaud I, Galardon E, Chembiochem 2014, 15, 2361–2364; [DOI] [PubMed] [Google Scholar]; (e) Khodade VS, Toscano JP, J. Am. Chem. Soc 2018, 140, 17333–17337. [DOI] [PubMed] [Google Scholar]
- [10].Cortese-Krott MM, Koning A, Kuhnle GGC, Nagy P, Bianco CL, Pasch A, Wink DA, Fukuto JM, Jackson AA, van Goor H, Olson KR, Feelisch M, Antioxid. Redox. Signal 2017, 27, 684–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].(a) Kasamatsu S, Nishimura A, Morita M, Matsunaga T, Abdul Hamid H, Akaike T, Molecules 2016, 21; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yadav PK, Martinov M, Vitvitsky V, Seravalli J, Wedmann R, Filipovic MR, Banerjee R, J. Am. Chem. Soc 2016, 138, 289–299; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cuevasanta E, Möller MN, Alvarez B, Arch. Biochem. Biophys 2017, 617, 9–25. [DOI] [PubMed] [Google Scholar]
- [12].Kang J, Xu S, Radford Miles N, Zhang W, Kelly Shane S, Day Jacob J, Xian M, Angew. Chem. Int. Ed 2018, 57, 5893–5897. [DOI] [PubMed] [Google Scholar]
- [13].(a) Barton LL, Ritz NL, Fauque GD, Lin HC, Dig. Dis. Sci 2017, 62, 2241–2257; [DOI] [PubMed] [Google Scholar]; (b) Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR, Front. Physiol 2012, 3, 448–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wallace JL, Motta J-P, Buret AG, Am. J. Physiol. Gastrointest. Liver Physiol 2018, 314, G143–G149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Beaumont M, Andriamihaja M, Lan A, Khodorova N, Audebert M, Blouin J-M, Grauso M, Lancha L, Benetti P-H, Benamouzig R, Tomé D, Bouillaud F, Davila A-M, Blachier F, Free Radical Biol. Med 2016, 93, 155–164. [DOI] [PubMed] [Google Scholar]
- [16].(a) Sharma K, Sengupta K, Chakrapani H, Bioorg. Med. Chem. Lett 2013, 23, 5964–5967; [DOI] [PubMed] [Google Scholar]; (b) Searle PF, Chen M-J, Hu L, Race PR, Lovering AL, Grove JI, Guise C, Jaberipour M, James ND, Mautner V, Young LS, Kerr DJ, Mountain A, White SA, Hyde EI, Clin. Exp. Pharmacol. Physiol 2004, 31, 811–816. [DOI] [PubMed] [Google Scholar]
- [17].(a) Vorobyeva AG, Stanton M, Godinat A, Lund KB, Karateev GG, Francis KP, Allen E, Gelovani JG, McCormack E, Tangney M, Dubikovskaya EA, PLoS One 2015, 10, e0131037; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhu D, Xue L, Li G, Jiang H, Sens. Actuators B Chem 2016, 222, 419–424. [Google Scholar]
- [18].Shukla P, Khodade VS, SharathChandra M, Chauhan P, Mishra S, Siddaramappa S, Bulagonda EP, Singh A, Chakrapani H, Chem. Sci 2017, 8, 4967–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Miller A-F, Park TJ, Ferguson LK, Pitsawong W, Bommarius SA, Molecules 2018, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chatterji T, Keerthi K, Gates KS, Bioorg. Med. Chem. Lett 2005, 15, 3921–3924. [DOI] [PubMed] [Google Scholar]
- [21].Dillon KM, Powell CR, Matson JB, Synlett 2019, 30, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schmid KM, Jensen L, Phillips ST, J. Org. Chem 2012, 77, 4363–4374. [DOI] [PubMed] [Google Scholar]
- [23].Lin J, Akiyama M, Bica I, Long FT, Henderson CF, Goddu RN, Suarez V, Baker B, Ida T, Shinkai Y, Nagy P, Akaike T, Fukuto JM, Kumagai Y, Chem. Res. Toxicol 2019, 32, 447–455. [DOI] [PubMed] [Google Scholar]
- [24].Ringel-Scaia VM, Qin Y, Thomas CA, Huie KE, McDaniel DK, Eden K, Wade PA, Allen IC, J. Innate Immun. 2019, 11, 416–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S, Rutgeerts P, Verbeke K, Gut 2015, 64, 447. [DOI] [PubMed] [Google Scholar]
- [26].(a) Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR, Nature 2009, 461, 1282–1286; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wlodarska M, Kostic AD, Xavier RJ, Cell Host Microbe 2015, 17, 577–591; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kim YI, Nutr. Rev 1998, 56, 17–24. [DOI] [PubMed] [Google Scholar]
- [27].Fung TC, Vuong HE, Luna CDG, Pronovost GN, Aleksandrova AA, Riley NG, Vavilina A, McGinn J, Rendon T, Forrest LR, Hsiao EY, Nat. 2019, 4, 2064–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].(a) Jumas-Bilak E, Carlier J-P, Jean-Pierre H, Citron D, Bernard K, Damay A, Gay B, Teyssier C, Campos J, Marchandin H, Int. J. Syst. Evol. Microbiol 2007, 57, 2743–2748; [DOI] [PubMed] [Google Scholar]; (b) Horz H-P, Citron DM, Warren YA, Goldstein EJC, Conrads G, J. Clin. Microbiol 2006, 44, 2914–2920; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Vartoukian SR, Palmer RM, Wade WG, Anaerobe 2007, 13, 99–106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.