Abstract
Proteins in the tripartite motif-containing protein (TRIM) family participate in carcinogenesis. However, little attention was paid to the role of TRIM6 in development of breast cancer. Our results showed that the expression level of TRIM6 was found to be markedly enhanced in breast cancer cells and tissues. Functional assays demonstrated that overexpression of TRIM6 promoted breast cancer progression through increasing the expression of YAP1 (Yes-associated Protein 1), while knockdown of TRIM6 suppressed in vitro breast cancer progression and in vivo tumor growth through decreasing of YAP1. Co-Immunoprecipitation showed that TRIM6 interacted with STUB1 (stress induced phosphoprotein 1 homology and U-box containing protein 1). TRIM6 promoted ubiquitination-mediated degradation of STUB1 thereby promoting YAP1 signaling. Overexpression of STUB1 attenuated TRIM6-induced promotion of breast cancer growth. In conclusion, TRIM6 contributed to breast cancer progression through ubiquitination-dependent proteasomal degradation of STUB1 and provocation of YAP1 pathway, providing potential therapeutic target for breast cancer.
Key words: TRIM6, STUB1, ubiquitination, breast cancer, growth, migration
Introduction
Breast cancer is one of the most common tumors and the second leading cause of cancer-related deaths in women.1 Although the development of antitumor drugs and diagnostic methods have greatly improved the overall survival rates of patients with breast cancer, the metastatic breast cancer hampered the effectiveness of antitumor drugs.2 Considering that plenty of factors, including chemokines, cytokines or growth factors, have been shown to be responsible for the metastasis of breast cancer,3 the molecular mechanisms involved in tumor growth, migration and invasion have not been fully elucidated. Strategies to retard metastasis of breast cancer are crucial for the treatment of patients with metastatic breast cancer. Proteins in tripartite motif-containing protein (TRIM) family are also known as RING-B-box-coiled-coil proteins, and function as E3 ubiquitin ligase to participate in biological process, including cell development, apoptosis and autophagy.4 Increasing evidence showed that TRIM was implicated in various pathological conditions through ubiquitination of downstream targets. 5 Dysregulation of TRIM can lead to a variety of diseases, including developmental disorders, immunological disease, and cancer.6 TRIM6, belonging to TRIM protein family, contains conventional tripartite motif and acts as an E3 ubiquitin ligase during virus replication.7 TRIM6 demonstrated antiviral response through activation of IKKε,8 and interacted with Myc to reduce its transcriptional activity during differentiation of stem cells.9 Recently, TRIM6 was reported to be upregulated in colorectal cancer cells, and promoted ubiquitination of anti-proliferative gene, TIS21, to facilitate the tumor progression.10 However, the role and mechanism of TRIM6 in breast cancer remain unclear.
In the present study, we firstly investigated the expression level of TRIM6 in breast cancer cells and tissues, and found that the expression of TRIM6 was significantly enhanced in breast cancer. Further functional assays were performed to evaluate the role of TRIM6 in breast cancer progression, and the results suggested the pro-proliferative role of TRIM6 in breast cancer growth. Moreover, interference of TRIM6 demonstrated suppressive effects on in vivo breast cancer growth. TRIM6 functioned as an E3 ligase to promote the ubiquitination-mediated degradation of STUB1 (stress induced phosphoprotein 1 homology and U-box containing protein 1). Results in this study might provide novel potential therapeutic target for breast cancer.
Materials and Methods
Tumor tissues and immunohistochemistry
Eighty-five paired breast tumor tissues and adjacent non-tumor tissues were collected from patients who underwent surgery at Institute of Fudan-Minhang Academic Health System, Minhang Hospital, Fudan University from March 2014 to September 2018. Informed consents were obtained from all the patients included in this study, and patients with chemotherapy or radiation therapy were excluded. The procedure was approved by Fudan-Minhang Academic Health System, Minhang Hospital, Fudan University, and in accordance with the 1964 Helsinki Declaration and its later amendments for ethical research involving human subjects. For immunohistochemical analysis of TRIM6 expression in breast tumor tissues and normal tissues, formalin-fixed and paraffinembedded tissues were cut into 5 μm sections. Following deparaffinization in xylene and rehydration in series of ethanol solutions, the sections were incubated with 0.01 M sodium citrate (pH 6.0) for antigen retrieval, and 3% hydrogen peroxide for blocking of endogenous peroxidase activity. Sections were preincubated with 5% normal goat serum before incubation with antibodies against TRIM6 (1:100 diluted in phosphate buffered saline; Abcam, Cambridge, MA, USA) at 4°C overnight. Following incubation with biotinylated secondary antibody (1:500 diluted in phosphate buffered saline; Abcam) at 37 °C for 20 min and streptavidin- biotin peroxidase complex solution, the sections were performed with Pierce™ DAB Substrate Kit (Thermo Fisher Scientific, Carlsbad, CA, USA) and counterstaining with haematoxylin solution. The representative images were observed under optical microscope (Carl Zeiss, Jena, Germany).
Cell lines
Human normal breast cell line (Hs 578Bst) and breast cancer cell lines (BT-20, MDA-MB-231, MDA-MB-436, MDA-MB-157) were acquired from Genechem (Shanghai, China). Following identification via short tandem repeat analysis, cells were cultured in RPMI 1640 medium (PAA Laboratories GmbH, Cölbe, Germany) containing 10% fetal bovine serum (Gibco, Mississauga, ON, Canada) in a 37°C incubator.
Plasmid construction and cell transfection
Full length of TRIM6 and STUB1 were constructed into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). shRNAs targeting TRIM6 (sh-TRIM6 #1 and #2) were synthesized by RiboBio (Guangzhou, China). Stable MDA-MB-231 cells transfected with sh-TRIM6 #1 or negative control (shNC) were obtained from Genechem. Briefly, lentiviral-mediated silence of TRIM6 by shTRIM6 or the negative control (shNC) were generated by Genechem, and then transfected into HEK-293T cells to harvest the lentiviruses. The lentiviruses were transfected into MDAMB- 231 cells with 8 mg/mL polybrene through ViraPower™ Packaging Mix. Cells with stable silence of TRIM6 was obtained following treatment with 5 μg/mL puromycin for seven days. MDA-MB-231 or MDA-MB-157 cells were transfected with pcDNA vectors or shRNAs via Lipofectamine 2000 (Invitrogen).
Cell viability and proliferation
MDA-MB-231 or MDA-MB-157 with indicated transfections were seeded into 96-well plates for 24 h. Cells were then incubated with 20 μL CCK8 solution (Dojindo, Tokyo, Japan) at an every 24 h intervals (0, 24, 48, 72 h). 2 h later, the absorbance at 450 nm of each well was measured by Microplate Autoreader (Thermo Fisher). For cell proliferation assay, MDA-MB-231 or MDA-MB- 157 cells with indicated transfections were seeded into 6-well plates and cultured in medium for two weeks. Paraformaldehydefixed and crystal violet-stained colonies were observed under microscope (Carl Zeiss).
Cell migration and invasion
MDA-MB-231 or MDA-MB-157 cells with indicated transfections were seeded into 6-well plates and scratched by pipette tips. 24 h later, the wounds were photographed and calculated by microscope (Carl Zeiss). MDA-MB-231 or MDA-MB-157 cells with indicated transfections were suspended in fetal bovine serum free medium and seeded into the upper chamber of Matrigel-coated (BD Biosciences, Bedford, MA, USA) well (Merck KGaA, Darmstadt, Germany). Medium containing 20% fetal bovine serum was placed into the lower chamber; 24 h later, the invasive cells in the lower chamber were fixed with paraformaldehyde and stained with crystal violet before observation at microscope (Carl Zeiss).
Co-immunoprecipitation (Co-IP) and ubiquitination assays
Flag-labeled TRIM6 or HA-labeled STUB1 were constructed into pcDNA3.1 vector (Invitrogen), and then transfected into HEK-293T cells. Cells were lysed in Pierce™ IP Lysis Buffer (Thermo Fisher Scientific), and incubated with specific antibodies against Flag or HA (Abcam). Antibody against normal IgG was used as negative control. Following incubation with protein G Sepharose beads (GE Healthcare, Eindhoven, The Netherlands), the mixture was performed with Western blot analysis via anti- TRIM and anti-STUB1 antibodies. For ubiquitination assay, MDA-MB-231 cells with or without transfection with pcDNATRIM6 were incubated with 10 μM MG132 for 4 h. Cells were lysed in the Lysis Buffer (Thermo Fisher Scientific), and incubated with specific antibody against STUB1 (Abcam). Following incubation with protein G Sepharose beads (GE Healthcare), the mixture was performed with Western blot analysis via anti-ubiquitin antibody.
Cell treatment
MDA-MB-231 cells with transfection with pcDNA-TRIM6 or empty vector were treated with 5 μM cycloheximide (CHX) for 1, 2, 4 or 8 h before Western blot analysis via anti-STUB1 antibody.
Xenograft tumor assay
Experimental procedures involved in animal were followed the criteria of National Institutes of Health Laboratory Animal Care and Use Guidelines. The procedure was approved by Fudan- Minhang Academic Health System, Minhang Hospital, Fudan University Twelve BALB/c nude mice (female, 5-week-old, 20-22 g weight; SLAC Laboratory Animal Co. Ltd, Shanghai, China) were separated into two groups. Mice in each group were subcutaneously injected with stable MDA-MB-231 cells transfected with sh- TRIM6 #1 or shNC suspended in 100 μL phosphate-buffered saline. Tumor volume was monitored every 3 days. Twenty-seven days later, all mice were sacrificed and tumor tissues were dissected, weighed and photographed. The tumor tissues were then subjected with immunohistochemistry analysis of TRIM6, STUB1, YAP1 and Ki-67 according to the aforementioned method.
qRT-PCR
RNAs were extracted from tumor and normal tissues by TRIzol reagent (Invitrogen), and then converted into cDNAs through PrimeScript™ RT-PCR kit (Takara, Tokyo, Japan). Relative expression of TRIM6 was performed by SYBR Green PCR Master Mix (Takara) with procedure: 95°C for 3 min, 40 cycles with 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. β-actin was used as endogenous control. The primer sequences are shown in Table 1.
Western blot
Cells or tissues were lysed in the Lysis Buffer (Thermo Fisher Scientific), and the protein concentration was determined by BCA Protein Assay Kit (Beyotime, Jiangsu, China). Proteins were separated with SDS-PAGE and transferred to nitrocellulose membranes (Beyotime). Membranes were blocked with 5% skim milk in PBS and then incubated with specific antibodies against TRIM6 (1:2500, Abcam), PCNA (1:2500, Abcam), p21 (1:2500, Abcam), E-cadherin (1:3000, Abcam), MMP2 (1:3000, Abcam), STUB1 (1:3000, Abcam), ubiquitin (1:3000, Abcam), YAP1 (1:3500, Abcam) and β-actin (1:3500, Abcam). Following incubation with horseradish peroxidase-conjugated secondary antibodies (1:5000, Beyotime), the membranes were performed with Pierce™ DAB Substrate Kit (Thermo Fisher Scientific) and the images were analyzed by ImageJ 1.50i (National Institutes of Health, Bethesda, MD, USA).
Table 1.
Primer used in this study.
| ID | Sequence (5’- 3’) |
|---|---|
| β-actin F | GGGCATCCTGACCCTCAAG |
| β-actin R | TCCATGTCGTCCCAGTTGGT |
| TRIM6 F | CTTTCCCACTACTCTTTGTC |
| TRIM6 R | TAAGCCTCAGGGTACTTATC |
| STUB1 F | GAGCTGGACGGGCAGTCTGTG |
| STUB1 R | CAAAGTTGAGTCGCTGCTCCTTGG |
Statistical analysis
Data, presented as the mean ± SD, were analyzed via GraphPad Prism version 6.0 (GraphPad, San Diego, CA, USA). Statistical difference was determined through one-way analysis of variance and student’s t-test, and p<0.05 was considered statistically significant.
Results
Aberrant expression of TRIM6 in breast cancer
To evaluate possible role of TRIM6 in breast cancer, the expression level was determined by qRT-PCR, Western blot and immunohistochemistry analysis. Results showed that both of mRNA (Figure 1A) and protein (Figure 1 B,C) expression of TRIM6 were markedly increased in breast cancer tissues compared to adjacent non-tumor tissue. Similarly, a significant up-regulation of TRIM6 was verified in breast cancer cell lines (BT-20, MDAMB- 231, MDA-MB-436, MDA-MB-157) compared to human normal breast cell line (Hs 578Bst) through Western blot analysis (Figure 1D), suggesting the possible regulatory role of TRIM in breast cancer.
TRIM6 contributed to breast cancer cell growth
The biological effects of TRIM6 on breast cancer was evaluated by gain- and loss- of functional assays. Western blot analysis confirmed the transfection efficiency of pcDNA-TRIM6 and sh- TRIM6 in MDA-MB-231 and MDA-MB-157 cells (Figure 2A). Meanwhile, sh-TRIM6#1 showed lower expression than sh- TRIM6#2 (Figure 2A), which was used in the subsequently loss-of functional assays. Forced TRIM6 expression promoted cell viability (Figure 2B) and proliferation (Figure 2C) of MDA-MB-231 and MDA-MB-157 cells, while the cell viability (Figure 2B) and proliferation (Figure 2C) of MDA-MB-231 and MDA-MB-157 cells was repressed by transfection with sh-TRIM6#1. Transfection with pcDNA-TRIM6 demonstrated higher expression of PCNA (proliferating cell nuclear antigen) and lower expression of p21 than that of vector (Figure 2D), while sh-TRIM6#1 transfection indicated opposite effect (Figure 2D), suggesting that TRIM6 contributed to breast cancer cell proliferation and cell cycle. Moreover, cell migration (Figure 3A) and invasion (Figure 3B) of MDA-MB-231 and MDA-MB-157 cells were also promoted by over-expression of TRIM6, while inhibited by silencing of TRIM. Protein expression of E-cadherin was suppressed, while MMP2 was increased, by over-expression of TRIM6 (Figure 3C). Transfection with sh-TRIM6#1 enhanced E-cadherin and reduced MMP2 (Figure 3C), suggesting that TRIM6 contributed to breast cancer cell progression.
TRIM6 contributed to ubiquitination-mediated degradation of STUB1
To elucidate molecular mechanism of TRIM6-mediated breast cancer progression, UbiBrowser (http://ubibrowser.ncpsb.org/ubibrowser/) was used to predict the target of TRIM6 and the analysis showed that STUB1 might be the binding target of TRIM6 (Figure 4A). Although TRIM6 had no significant effect on mRNA expression of STUB1 in MDA-MB- 231 and MDA-MB-157 cells (Supplementary Figure S1), TRIM6 negatively regulated protein expression of STUB1 in MDA-MB- 231 and MDA-MB-157 cells (Figure 4B). Co-IP validated the direct binding between TRIM6 and STUB1 (Figure 4C). Forced TRIM6 expression promoted the ubiquitination of STUB1 in MDA-MB-231 cells under MG132 treatment (Figure 4D). Treatment with Cycloheximide (CHX) showed a time-dependent down-regulation of STUB1 in MDA-MB-231 cells (Figure 4E), and transfection with pcDNA-TRIM6 aggravated the degradation of STUB1 (Figure 4E). These results revealed that TRIM6 binds to STUB1 and contributes to ubiquitination-mediated degradation of STUB1 in breast cancer cell.
TRIM6 promoted YAP1 pathway to destabilize STUB1
To investigate the downstream signaling pathway involved in TRIM6/STUB1-mediated breast cancer progression, Western blot was performed and showed that YAP1 pathway was validated to be involved in TRIM6-mediated STUB1 degradation. TRIM6 positively modulated YAP1 expression in MDA-MB-231 and MDAMB- 157 cells (Figure 5A). Over-expression of STUB1 attenuated TRIM6-induced increase in YAP1 in MDA-MB-231 and MDAMB- 157 cells (Figure 5B), suggesting that TRIM6 promoted YAP1 pathway to destabilize STUB1 in breast cancer cells.
STUB1 participated in TRIM6-mediated breast cancer progression
Functional assays were performed to investigate role of TRIM6/STUB1 axis in breast cancer progression. Data from CCK8 assay showed that over-expression of STUB1 attenuated TRIM6-induced increase in MDA-MB-231 cell viability (Figure 6A). Moreover, TRIM6-induced promotion of cell migration (Figure 6B) and invasion (Figure 6C) in MDA-MB-231 cells was also reversed by STUB1 over-expression, demonstrating that TRIM6 contributed to breast cancer progression through regulation of STUB1.
Figure 1.
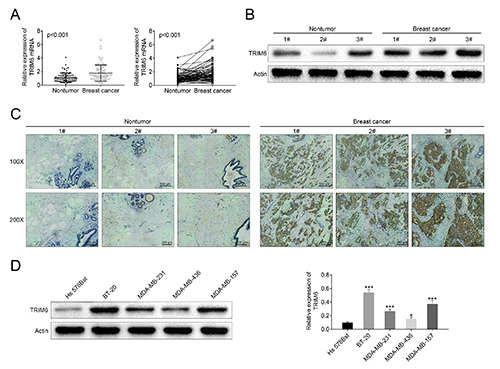
Aberrant expression of TRIM6 in breast cancer. A) mRNA expression of TRIM6 was increased in breast cancer tissues compared to adjacent non-tumor tissue. B) Protein expression of TRIM6 was increased in breast cancer tissues compared to adjacent nontumor tissue. C) Immunohistochemistry analysis showed upregulation of TRIM6 in breast cancer tissues compared to normal tissues; scale bars: 200 μm. D) TRIM6 was upregulated in breast cancer cell lines (BT-20, MDA-MB-231, MDA-MB-436, MDA-MB-157) compared to human normal breast cell line (Hs 578Bst); *p<0.05, ***p<0.001.
Knockdown of TRIM6 repressed in vivo breast cancer growth
To elucidate in vivo regulatory role of TRIM6 in breast cancer, stable MDA-MB-231 cells transfected with sh-TRIM6 #1 or shNC were subcutaneously injected into nude mice. The breast tumor growth was inhibited by sh-TRIM6 with reduced tumor volume and weight (Figure 7A). Moreover, immunohistochemical analysis showed that TRIM6, YAP1 and Ki-67 were reduced in tumor tissues isolated from mice injected with sh-TRIM6 (Figure 7B), while STUB1 was enhanced by sh-TRIM6 (Figure 7B). These results further verified the positively regulatory role of TRIM6 in breast cancer growth.
Figure 2.
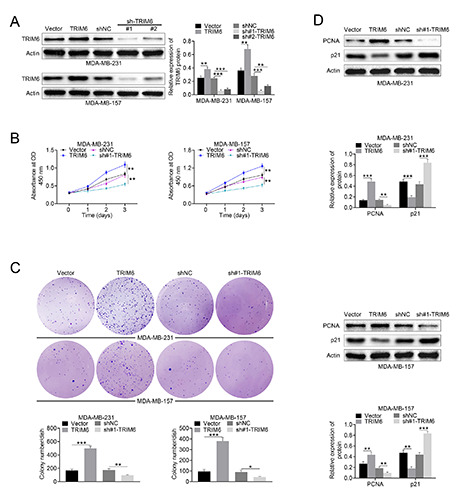
TRIM6 contributed to breast cancer cell growth. A) Transfection efficiency of pcDNA-TRIM6 and sh-TRIM6 in MDA-MB- 231 and MDA-MB-157 cells was detected by Western blot analysis. B) Forced TRIM6 expression promoted cell viability of MDA-MB- 231 and MDA-MB-157 cells, while knockdown of TRIM decreased the cell viability. C) Forced TRIM6 expression promoted cell proliferation of MDA-MB-231 and MDA-MB-157 cells, while knockdown of TRIM decreased the cell proliferation. D) Forced TRIM6 expression promoted protein expression of PCNA and inhibited p21 in MDA-MB-231 and MDA-MB-157 cells, while knockdown of TRIM decreased PCNA and increased p21; *p<0.05, **p<0.01, ***p<0.001.
Figure 3.
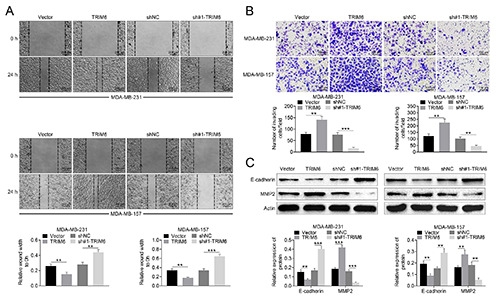
TRIM6 contributed to breast cancer cell migration and invasion. A) Forced TRIM6 expression promoted cell migration of MDA-MB-231 and MDA-MB-157 cells, while knockdown of TRIM decreased the cell migration; scale bars: 200 μm. B) Forced TRIM6 expression promoted cell invasion of MDA-MB-231 and MDA-MB-157 cells, while knockdown of TRIM decreased the cell invasion; scale bars: 100 μm. C) Forced TRIM6 expression promoted protein expression of MMP2 and inhibited E-cadherin in MDA-MB-231 and MDA-MB-157 cells, while knockdown of TRIM decreased MMP2 and increased E-cadherin; **p<0.01, ***p<0.001.
Figure 4.
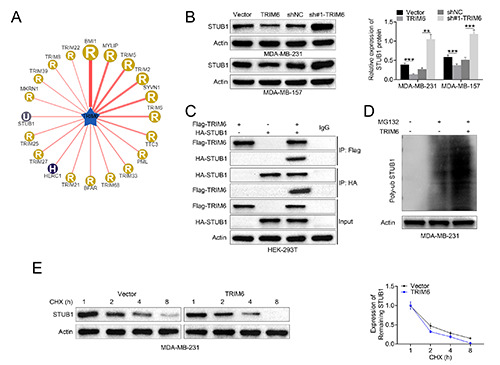
TRIM6 contributed to ubiquitination-mediated degradation of STUB1. A) Potential binding target of TRIM6 was predicted through UbiBrowser (http://ubibrowser.ncpsb.org/ubibrowser/) analysis. B) TRIM6 negatively regulated protein expression of STUB1 in MDA-MB-231 and MDA-MB-157. C) TRIM6 directly binds to STUB1 via Co-IP analysis. D) Forced TRIM6 expression promoted the ubiquitination of STUB1 in MDA-MB-231 under MG132 treatment. E) Transfection with pcDNA-TRIM6 aggravated CHXinduced degradation of STUB1; **p<0.01, ***p<0.001.
Discussion
TRIMs have been shown to promote or suppress tumorigenicity of breast cancer cells, thus enhancing or reducing colonization, survival, and invasive capacities.11 TRIM44 enhanced NF-κB signaling to promote breast cancer progression,12 and TRIM59 stabilized programmed cell death protein 10 to promote the motility of breast cancer cell.13 Considering that TRIM6 contributed to embryonic stem cell pluripotency,9 and promoted cell proliferation of colorectal cancer,10 the regulatory role of TRIM6 in breast cancer progression was then investigated in this study.
Upregulation of TRIM6 was verified in breast cancer cells and tissues through qRT-PCR, Western blot and immunohistochemistry analysis in this study. Functional assays revealed that TRIM6 contributed to breast cancer cell proliferation, migration and invasion. Knockdown of TRIM6 repressed in vivo breast cancer growth. Interestingly, TRIM6 suppressed E-cadherin in breast cancer cells to promote epithelial-to-mesenchymal transition and invasiveness. Since epithelial-to-mesenchymal transition contributes to stem cell-like properties,11 the effects of TRIM6 on other epithelial-tomesenchymal transition biomarkers and breast cancer stem cell phenotype should be investigated in the further research.
TRIM6 functioned as an E3-ubiquitin ligase to destabilize TIS21 during colorectal cancer progression.10-13 STUB1 was verified as binding target of TRIM6 in this study through Co-IP analysis, and TRIM6 contributed to ubiquitination-mediated degradation of STUB1 in breast cancer cells. STUB1 was also verified as E3 ligase to participate in protein homeostasis involved in neurodegenerative diseases.14 STUB1 promoted ubiquitination-mediated degradation of FOXP3 during gastric cancer progression.15 STUB1 suppressed stemness and tumorigenicity of oral carcinoma cell,16 and downregulation of STUB1 promoted epithelial-mesenchymal transition of ovarian cancer.17 Recent study has shown that STUB1 was aberrantly expressed in breast cancer,18 and STUB1 promoted ubiquitin/proteasome-dependent degradation of ERBB2-mediated oncogenes in breast cancer.19 Our results showed that over-expression of STUB1 attenuated TRIM6-induced increase in breast cancer cell proliferation, migration and invasion. Moreover, reduction of STUB1 was found to release pro-oncogenic pathway, including matrix-degrading enzyme, to promote breast cancer progression.19 Our result also confirmed that forced TRIM6 expression promoted MMP2 expression in breast cancer cells, thus unleashing the pro-oncogenic pathway.
Figure 5.
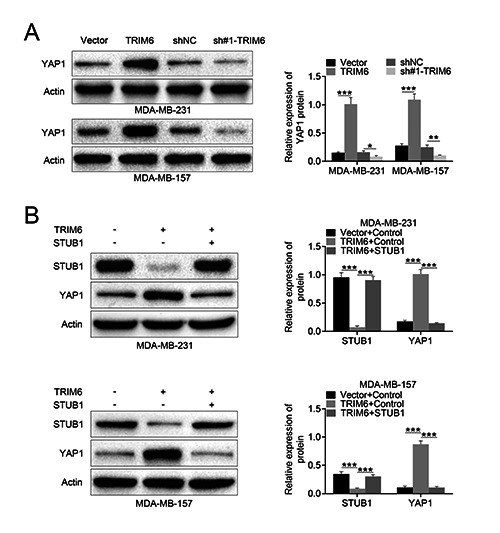
TRIM6 promoted YAP1 pathway to destabilize STUB1. A) TRIM6 positively modulated YAP1 expression in MDA-MB-231 and MDA-MB-157. B) Over-expression of STUB1 attenuated TRIM6-induced increase of YAP1 in MDA-MB-231 and MDA-MB-157; *p<0.05, **p<0.01, ***p<0.001.
STUB1 directly interacted with stemness factor, OCT4, to destabilize OCT4 and promote breast cancer stem cell phenotype and tumorigenesis.20 The downstream target involved in TRIM6/STUB1 axis-mediated breast cancer progression was also validated in this study. Our results showed that YAP1 signaling was promoted by TRIM6 in breast cancer. Previous study has shown that YAP1 was a component of Hippo pathway and participated in tumor growth.21 YAP1 demonstrated oncogenic activities in breast cancer,22 and over-expression of YAP1 was found to be associated with poor prognosis of patients with breast cancer.23 YAP1 functioned as an oncogene to promote breast cancer survival, 24 and repression of YAP1 contributed to suppression of breast cancer progression.25,26 YAP1 was reported to be the target of STUB1 in gastric cancer cells,27 and our results showed that forced STUB1 attenuated TRIM6-induced increase in YAP1. In addition, knockdown of TRIM6 repressed in vivo breast cancer growth through increasing of STUB1 and decreasing of YAP1. Therefore, results in this study suggested that TRIM6 contributed to breast cancer progression through ubiquitin-dependent degradation of STUB1, and promoting of oncogene, YAP1. TRIM6 was reported to interact with IKKε,6 and STUB1 retarded NF-κB-targeted oncogenes in colorectal cancer.28 STUB1 also promoted the degradation of TRAF2 and inactivation of NF-κB to suppress breast cancer cell invasion.29 Whether NF-κB is also implicated in TRIM6/STUB1-mediated breast cancer progression needs to be further investigated.
Figure 6.

STUB1 participated in TRIM6-mediated breast cancer progression. A) Over-expression of STUB1 attenuated TRIM6-induced increase of MDA-MB-231 cell viability. B) Over-expression of STUB1 attenuated TRIM6-induced increase of MDA-MB-231 cell migration.; scale bars: 200 μm. C) Over-expression of STUB1 attenuated TRIM6-induced increase of MDA-MB-231 cell invasion; scale bars: 100 μm; **p<0.01, ***p<0.001.
Figure 7.
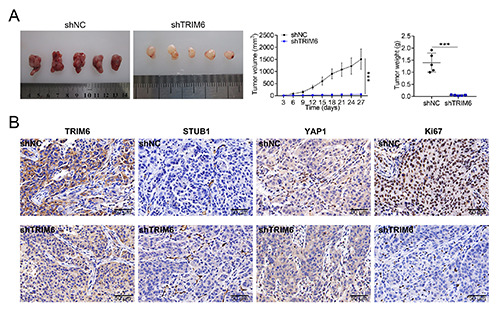
Knockdown of TRIM6 repressed in vivo breast cancer growth. A) Injection with stable MDA-MB-231 cells transfected with sh-TRIM6 #1 suppressed in vivo breast cancer growth with reduced tumor volume and weight compared to that of shNC. B) Immunohistochemical analysis showed that injection with stable MDA-MB-231 cells transfected with sh-TRIM6 #1 increased TRIM6, YAP1 and Ki-67 while decreased STUB1 compared to that of shNC; ***p<0.001.
In conclusion, enhanced TRIM6 expression in breast cancer cells and tissues promoted the degradation of STUB1 and promoted YAP1 signaling to facilitates breast cancer growth and migration, providing potential therapeutic target for breast cancer.
Funding Statement
Funding: This work was supported by the National Natural Science Foundation of China (NSFC). (Grant No. 81703881).
References
- 1.Tong D, Zhang J, Wang X, Li Q, Liu L, Yang J, et al. MeCP2 facilitates breast cancer growth via promoting ubiquitinationmediated P53 degradation by inhibiting RPL5/RPL11 transcription. Oncogenesis 2020;9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dizon DS, Krilov L, Cohen E, Gangadhar T, Ganz PA, Hensing TA, et al. Clinical cancer advances 2016: Annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol 2016;34:987-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronin PA, Wang JH, Redmond HP. Hypoxia increases the metastatic ability of breast cancer cells via upregulation of CXCR4. BMC Cancer 2010;10:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esposito D, Koliopoulos MG, Rittinger K. Structural determinants of TRIM protein function. Biochem Soc Trans 2017;45:183-91. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem 2017;161:135-44. [DOI] [PubMed] [Google Scholar]
- 6.Hatakeyama S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci 2017;42:297-311. [DOI] [PubMed] [Google Scholar]
- 7.Bharaj P, Atkins C, Luthra P, Giraldo MI, Dawes BE, Miorin L, et al. The host E3-ubiquitin ligase TRIM6 ubiquitinates the Ebola virus VP35 protein and promotes virus replication. J Virol 2017;91:e00833-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajsbaum R, Versteeg Gijs A, Schmid S, Maestre Ana M, Belicha-Villanueva A, Martínez-Romero C, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity 2014;40:880-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato T, Okumura F, Ariga T, Hatakeyama S. TRIM6 interacts with Myc and maintains the pluripotency of mouse embryonic stem cells. J Cell Sci.2012;125:1544. [DOI] [PubMed] [Google Scholar]
- 10.Zheng S, Zhou C, Wang Y, Li H, Sun Y, Shen Z. TRIM6 promotes colorectal cancer cells proliferation and response to thiostrepton by TIS21/FoxM1. J Exp Clin Cancer Res 2020;39:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaworska AM, Wlodarczyk NA, Mackiewicz A, Czerwinska P. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 2020;38:165-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawabata H, Azuma K, Ikeda K, Sugitani I, Kinowaki K, Fujii T, et al. TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-κB signaling. Int J Mol Sci 2017;18:1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan P, Ye Y, He L, Xie J, Jing J, Ma G, et al. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol 2018;16:e3000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayer SN, Deconinck T, Bender B, Smets K, Züchner S, Reich S, et al. STUB1/CHIP mutations cause Gordon Holmes syndrome as part of a widespread multisystemic neurodegeneration: evidence from four novel mutations. Orphanet J Rare Dis 2017;12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Li Q, Xu J, Sun G, Xu Z. Cimetidine promotes STUB1-mediated degradation of tumoral FOXP3 by activating PI3K-Akt pathway in gastric cancer. Ann Transl Med 2020;8:1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C-M, Yu C-C, Lin T, Liao Y-W, Hsieh P-L, Yu C-H, et al. E3 ligase STUB1 attenuates stemness and tumorigenicity of oral carcinoma cells via transglutaminase 2 regulation. J Formos Med Assoc 2020;119:1532-8. [DOI] [PubMed] [Google Scholar]
- 17.Park S-M, Park S-H, Ryu K-J, Kim I-K, Han H, Kim H-J, et al. Downregulation of CHIP promotes ovarian cancer metastasis by inducing Snail-mediated epithelial-mesenchymal transition. Mol Oncol 2019;13:1280-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res 2006;4:695. [DOI] [PubMed] [Google Scholar]
- 19.Luan H, Mohapatra B, Bielecki TA, Mushtaq I, Mirza S, Jennings TA, et al. Loss of the nuclear pool of ubiquitin ligase CHIP/STUB1 in Breast cancer unleashes the MZF1-cathepsin pro-oncogenic program. Cancer Res 2018;78:2524-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho Y, Kang H, Kim S-J, Lee S, Jee S, Ahn S, et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ 2018;25:1781-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Xu R, Li X, Ren W, Ou C, Wang Q, et al. Prognostic value of Yes-associated protein 1 (YAP1) in various cancers: A meta-analysis. PloS One 2015;10:e0135119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maugeri-Saccà M, Barba M, Pizzuti L, Vici P, Di Lauro L, Dattilo R, et al. The hippo transducers TAZ and YAP in breast cancer: oncogenic activities and clinical implications. Expert Rev Mol Med 2015;17:e14. [DOI] [PubMed] [Google Scholar]
- 23.Guo L, Chen Y, Luo J, Zheng J, Shao G. YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio 2019;9:437-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J-G, Chen X-W, Zhang L-P, Wang J, Diehn M. Yap1 promotes the survival and self-renewal of breast tumor initiating cells via inhibiting Smad3 signaling. Oncotarget 2016;7:9692-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu S, Zhang M, Huang L, Ma Z, Gong X, Liu W, et al. ERK1 indicates good prognosis and inhibits breast cancer progression by suppressing YAP1 signaling. Aging (Albany NY) 2019;11:12295-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimei M, Alrouh S, Saber-Ayad M, Hafezi SA, Vinod A, Rawat S, et al. Inhibition of Yes-associated protein-1 (YAP1) enhances the response of invasive breast cancer cells to the standard therapy. Breast Cancer (Dove Med Press) 2020;12:189-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang D-E, Dai Y, Lin L-W, Xu Y, Liu D-Z, Hong X-P, et al. STUB1 suppresseses tumorigenesis and chemoresistance through antagonizing YAP1 signaling. Cancer Sci 2019;110:3145-56. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wang Y, Ren F, Wang Y, Feng Y, Wang D, Jia B, et al. CHIP/Stub1 functions as a tumor suppressor and represses NF- κB-mediated signaling in colorectal cancer. Carcinogenesis 2014;35:983-91. [DOI] [PubMed] [Google Scholar]
- 29.Jang KW, Lee KH, Kim SH, Jin T, Choi EY, Jeon HJ, et al. Ubiquitin ligase CHIP induces TRAF2 proteasomal degradation and NF-κB inactivation to regulate breast cancer cell invasion. J Cell Biochem 2011;112:3612-20. [DOI] [PubMed] [Google Scholar]


