Abstract
At least six small alternative-frame open reading frames (ORFs) overlapping well-characterized SARS-CoV-2 genes have been hypothesized to encode accessory proteins. Researchers have used different names for the same ORF or the same name for different ORFs, resulting in erroneous homological and functional inferences. We propose standard names for these ORFs and their shorter isoforms, developed in consultation with the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. We recommend calling the 39 codon Spike-overlapping ORF ORF2b; the 41, 57, and 22 codon ORF3a-overlapping ORFs ORF3c, ORF3d, and ORF3b; the 33 codon ORF3d isoform ORF3d-2; and the 97 and 73 codon Nucleocapsid-overlapping ORFs ORF9b and ORF9c. Finally, we document conflicting usage of the name ORF3b in 32 studies, and consequent erroneous inferences, stressing the importance of reserving identical names for homologs. We recommend that authors referring to these ORFs provide lengths and coordinates to minimize ambiguity caused by prior usage of alternative names.
Keywords: Accessory protein, Alternative reading frame, Nomenclature, Open reading frame, ORF3b, ORF3d, ORF9a, ORF9b, Overlapping ORF, SARS-CoV-2, ORF3c, ORF2b
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the recently identified strain (F.Wu et al., 2020a; Zhou et al., 2020; Zhu et al., 2020) of the species Severe acute respiratory syndrome-related coronavirus in the family Coronaviridae (subgenus Sarbecovirus, genus Betacoronavirus, subfamily Orthocoronavirinae) (Gorbalenya et al., 2020) that is the causative agent of coronavirus disease 2019 (COVID-19). Characterization of the SARS-CoV-2 proteome is vital for understanding its molecular biology and for development of countermeasures against the COVID-19 pandemic. Of particular interest are proteins that are unique to SARS-CoV-2, differ substantially from their SARS-CoV homologs, or have not been well characterized in other viruses of this species.
Coronaviruses have positive-sense single-stranded RNA genomes that encode proteins expressed from genomic and subgenomic RNAs using complex regulation at the transcriptional, translational, and post-translational levels (Fung et al., 2016; Fung and Liu, 2018; Sola et al., 2015). Some of the protein-coding open reading frames (ORFs) are conserved across coronaviruses, with homologs in all strains, and were named according to a uniform coronavirus-wide nomenclature (de Groot et al., 2012). At the 5′ end are two large ORFs, ORF1a and ORF1b. ORF1a encodes polyprotein pp1a, and the combination of ORF1a and ORF1b encodes polyprotein pp1ab via a programmed frameshift. Polyproteins pp1a and pp1ab are proteolytically processed to yield 11 and 15 non-structural proteins (“nsp’s”), respectively (16 unique, nsp1-nsp16). These include the 3C-like cysteine proteinase (nsp5), RNA-dependent RNA polymerase (nsp12), helicase (nsp13), and exonuclease (nsp14) (Snijder et al., 2003). The name ORF1ab is sometimes used to refer to the two ORFs combined via the frameshift. However, we refer to ORF1a and ORF1b as separate ORFs following common practice in the nidovirus field motivated by their large sizes and small overlap, despite the fact that ORF1b begins at a frameshift site rather than a start codon, unlike the other ORFs we discuss here. The other ORFs conserved across coronaviruses encode, from 5′ to 3′, S (Spike protein), E (Envelope), M (Membrane), and N (Nucleocapsid). Other “accessory” ORFs, located in the region downstream of ORF1b, may be species-specific or present only in some strains of a species.
SARS-CoV-2 has a full complement of ORFs previously identified in other viruses of the species Severe acute respiratory syndrome-related coronavirus, which includes the prototype SARS-CoV, the causative agent of the 2002–2003 SARS outbreak. In addition to the ORFs common to all coronaviruses these include, from 5′ to 3′, the accessory genes ORF3a, ORF6, ORF7a, ORF7b, and ORF8 (split into ORF8a and ORF8b in some SARS-CoV isolates) (Cui et al., 2019; Liu et al., 2014; Wu et al., 2020a). Because of the unprecedented interest in SARS-CoV-2, its proteome has been extensively investigated by various experimental and computational techniques. One additional independent ORF, ORF10, and several additional ORFs overlapping S, ORF3a, and N in alternative positive-sense reading frames have been hypothesized to encode functional proteins. Some of these alternative-frame ORFs are unique to SARS-CoV-2, some are completely or partially homologous to ones already described for SARS-CoV, and one is present in SARS-CoV but was not identified until the SARS-CoV-2 genome was investigated. Alternative-frame ORFs could be translated from the same subgenomic RNA as the main ORF via leaky scanning or internal ribosome entry, from a subgenomic RNA specific to the alternative-frame ORF, or via a translational frameshift (Di et al., 2017; Firth and Brierley, 2012; Irigoyen et al., 2016; Kim et al., 2020b; Liu and Inglis, 1992; O’Connor and Brian, 2000; Thiel and Siddell, 1994). Due in part to the biological and evolutionary complexity of these ORFs and the incremental nature of scientific discovery, different research groups have assigned different names to the alternative-frame ORFs, which has complicated clear scientific communication.
Here we propose a standard set of names for these overlapping SARS-CoV-2 ORFs for use by the scientific community in order to facilitate unambiguous communication and minimize confusion while the coding potential and biological function of these ORFs continues to be investigated.
2. Results and discussion
2.1. SARS-CoV-2 overlapping ORFs and their ambiguous names
The term “open reading frame” or “ORF” has been used with slightly different meanings by different authors. Here we use the term to mean any contiguous stretch of RNA codons beginning with a start codon, ending with a stop codon, and with no intermediate in-frame stop codons. Given appropriate evidence, the 5′ end of the ORF might be moved to a site with a known stop codon readthrough or frameshift signal, as in the case of ORF1b, in order to accommodate the complexity of genome expression in viruses. (Note that, although we require an ORF to end with a stop codon, we do not include the stop codon when we report the lengths and coordinates of the ORF.) We do not require that an ORF exceeds some minimum length or that undisputed evidence is available for its translation into a protein. In what follows, we will only be discussing ORFs with AUG start codons, but our definition would include ORFs with other start codons (typically near-cognate to AUG, such as CUG). By this definition, the conceptual translation of the nucleotide sequence using a codon table determines whether a genome region is an ORF, whereas experimental or computational evidence is needed to determine if an ORF is indeed translated and encodes a functional protein during virus infection. This evidence may come from, but is not limited to, ribosome profiling, protein or peptide detection, and observation of evolutionary signals. Although a large number of ORFs satisfy our definition, we will only be discussing ORFs for which some evidence has suggested translation. Their consideration would benefit from having agreed nomenclature, even if for some of them this evidence may not pass the test of time.
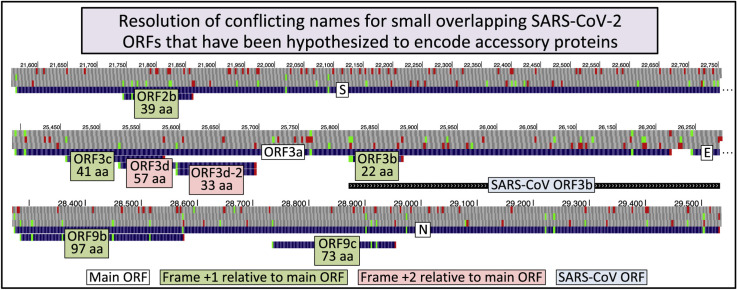
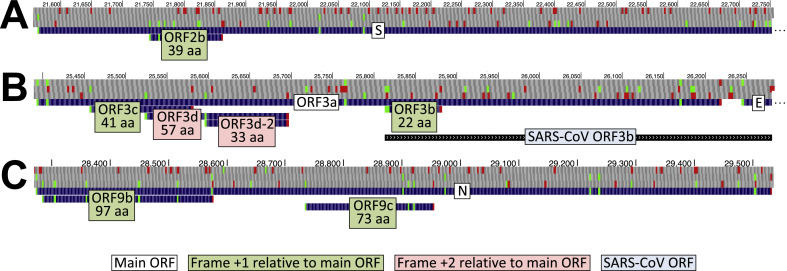
At least six ORFs overlapping S, ORF3a, and N in alternative reading frames have been hypothesized to encode functional proteins. These ORFs are detailed in Fig. 1 and Table 1 , and issues relating to their naming are discussed in the following paragraphs.
Fig. 1.
Browser image of recommended names for overlapping ORFs. UCSC Genome Browser images showing our recommended names and the number of amino acids (below name) for overlapping ORFs (light green or pink background for ORFs whose codons are shifted 1 or 2 nucleotides, respectively, in the 3′ direction from those of the main ORF, white background). AUG (green) and stop codons (red) are shown in each of three positive-sense genomic reading frames. (A.). 5′ end of Spike ORF (S) containing ORF2b (39 codons). B. ORF3a containing ORFs 3c (41 codons), 3d (57 codons), 3d-2 (33 codons), and 3b (22 codons). The region homologous to SARS-CoV ORF3b, which overlaps the 3′ half of ORF3a and the 5′ end of the envelope protein ORF (E) is also shown (light blue background). Note that ORFs 3a, 3c, and 3d are in different reading frames (+0, +1, and +2, respectively), so the 59 nucleotide region common to all three could be a rare example of RNA translated in three different reading frames. C. Nucleocapsid ORF (N) containing ORFs 9b (97 codons) and 9c (73 codons). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Recommended standard names. Recommended standard names for each of six ORFs overlapping S, ORF3a, or N, in 5′–3′ order, and the shorter isoform of ORF3d, with number of codons, coordinates, and a list of other names that have been used in previous publications or preprints. Codon counts and coordinates do not include the stop codon. Coordinates are with respect to the Wuhan-Hu-1 reference genome (NCBI: NC_045512.2). Frame +1 or +2 indicates that codons are shifted one or two nucleotides, respectively, in the 3′ direction from codons in the main (larger) ORF, which occupies frame +0.
| Recommended ORF name | Length (codons) | Coordinates | Frame relative to main (+0) ORF | Other names used (not recommended) | SARS-CoV homolog |
|---|---|---|---|---|---|
| ORF2b | 39 | 21744-21860 | +1 (S) | S.iORF1 | None |
| ORF3c | 41 | 25457-25579 | +1 (ORF3a) | ORF3h, 3a.iORF1, ORF3b | Unnamed |
| ORF3d ORF3d-2 |
57 33 |
25524-25694 25596-25694 |
+2 (ORF3a) +2 (ORF3a) |
ORF3b, ORF3c 3a.iORF2 |
None None |
| ORF3b | 22 | 25814-25879 | +1 (ORF3a) | 5′ end of ORF3b | |
| ORF9b | 97 | 28284-28574 | +1 (N) | ORF9a, N.iORF1 | ORF9b |
| ORF9c | 73 | 28734-28952 | +1 (N) | ORF9b, ORF14 | ORF9c, ORF14 |
UniProt (The UniProt Consortium, 2019) annotates two ORFs overlapping N in a different reading frame, namely a 97 codon ORF with coordinates 28284-28574, which they call ORF9b, and a 73 codon ORF with coordinates 28734-28952, which they call ORF14. (As a result of our recommendation, the 73 codon ORF is called ORF9c beginning with UniProt release 2021_01.) The name ORF14, which is out of sequence from the other SARS-CoV-2 ORF names, dates back to the 2003 paper that introduced the SARS-CoV genome (Marra et al., 2003), which numbered all ORFs sequentially, including overlapping ORFs. Later papers renumbered so that overlapping ORFs were distinguished using different letters following a shared number, but the name ORF14 continued to be used by some authors whereas others used the name ORF9c. Various authors have referred to the 97 and 73 codon SARS-CoV-2 ORFs overlapping N, respectively, as ORF9a and ORF9b (Cagliani et al., 2020; Davidson et al., 2020; Wu et al., 2020a), ORF9b and ORF9c (Gordon et al., 2020; Michel et al., 2020; Nelson et al., 2020b), or ORF13 and ORF14 (Lu et al., 2020), resulting in ambiguity about whether ORF9b refers to the 97 or 73 codon ORF.
Biological and evolutionary complexity have engendered even greater confusion about the names of ORFs overlapping ORF3a. SARS-CoV contains an alternative-frame 154 codon ORF, ORF3b, that partially overlaps both ORF3a and E (Chan et al., 2005), but the homologous 155 codon region in SARS-CoV-2 contains several in-frame stop codons. The longest alternative-frame ORF overlapping SARS-CoV-2 ORF3a is the 57 codon ORF with coordinates 25524-25694 that overlaps a 5′-proximal portion of ORF3a that has no homology to SARS-CoV ORF3b (Fig. 1), though this ORF is truncated to 13 codons in a substantial fraction of isolates (Lu et al., 2020). Because there is no SARS-CoV-2 ORF of comparable length in the region homologous to SARS-CoV ORF3b, Chan et al. (2020) referred to this 57 codon ORF as ORF3b (the paper does not explicitly state the length or coordinates, and ORF3b is not included in the corresponding NCBI record, accession MN975262, but the ORF can be inferred from the amino acid sequence specified in their Fig. 4). However, Konno et al. used the name ORF3b to refer to the 22 codon ORF with coordinates 25814-25879 at the 5′ end of the region homologous to SARS-CoV ORF3b, which they reported to be an interferon antagonist when expressed from a plasmid (Konno et al., 2020b), a property that had previously been reported for the much longer SARS-CoV ORF3b (Kopecky-Bromberg et al., 2007). Adding to the potential for confusion, the non-homologous 57 codon ORF overlapping ORF3a has also been reported to function as an interferon antagonist in a paper that also referred to it as ORF3b (Lu et al., 2020). The 57 codon ORF was predicted to be translated and functional based on a statistical test (Schlub et al., 2018) for unexpectedly long overlapping ORFs, a ribosome profiling analysis, and a d N/d S analysis comparing SARS-CoV-2 to pangolin-CoV GX/P5L, and was named ORF3c in an early preprint (Nelson et al., 2020a), but its name was changed to ORF3d in the final published version (Nelson et al., 2020b) to reflect the consensus reported here. It was also predicted to be a bona fide gene using an independent sequence composition analysis method, but left unnamed (Pavesi, 2020). Complicating matters further, a ribosome profiling study reported evidence for translation of a 33 codon isoform of the 57 codon ORF that starts at a downstream in-frame AUG (coordinates 25596-25694), calling it ORF3a.iORF2, but did not obtain evidence that the full 57 codon isoform is translated (Finkel et al., 2020). As this example illustrates, some proteins have more than one potential start site and it can be difficult to determine which are functionally important. Other studies that discuss the 22 or 57 codon ORFs are listed in Table 2 and Supplementary Table 1.
Table 2.
ORF3b studies. Thirty two studies that use the name “ORF3b”, but do not distinguish the 22 codon and 57 codon ORFs as separate entities. Information about what each study was investigating and how we determined the ORF referred to is provided in Supplementary Table 1.
| ORF called “ORF3b” in study | Recommended name | Study type | Studies (first author and citation) |
|---|---|---|---|
| 22 codon ORF |
ORF3b |
Genome report | (Wu et al., 2020a) |
| Empirical | (Konno et al., 2020b; Lokugamage et al., 2020; Xia et al., 2020; Zhang et al., 2020) | ||
| Review |
Sa Ribero et al. (2020) |
||
| 57 codon ORF |
ORF3d |
Genome Report | Chan et al. (2020) |
| Empirical | (Banerjee et al., 2020; Gordon et al., 2020; Hachim et al., 2020; Hayn et al., 2020; Lam et al., 2020; Laurent et al., 2020; Samavarchi-Tehrani et al., 2020.; St-Germain et al., 2020) | ||
| Laboratory resource - sequence clone collection | (Kim et al., 2020a) | ||
| Computational | (Michel et al., 2020; Pasquier and Robichon, 2020; Sadegh et al., 2020) | ||
| Review |
(Celik et al., 2020; Garofalo et al., 2020; Helmy et al., 2020; Taefehshokr et al., 2020; Wu et al., 2020b; Yang et al., 2020; Yi et al., 2020; Yoshimoto, 2020; Zinzula, 2020) |
||
| Unclear | Unclear | Empirical | (Lei et al., 2020; Nabeel-Shah et al., 2020; Yuen et al., 2020) |
| Computational | Sun (2020) |
A third ORF overlapping ORF3a, the 41 codon ORF with coordinates 25457-25579, was proposed to be translated based on synonymous constraint across 6 closely-related strains of the species and called ORF3h (Cagliani et al., 2020). This ORF was independently identified using ribosome profiling (Finkel et al., 2020), by synonymous constraint in a larger group of strains (Firth, 2020), and using evolutionary signatures of protein-coding regions (Jungreis et al., 2021), and referred to as ORF3a.iORF1, ORF3c, and ORF3c in these three respective studies, with additional evidence of purifying selection reported by comparing different SARS-CoV-2 isolates (Nelson et al., 2020b). Adding to the confusion, this ORF has also been referred to as 3b protein (Pavesi, 2020). Interestingly, the 41 codon and 57 codon ORFs have a 59-nucleotide overlap (including the stop codon of the former), so if both encode functional proteins then this region of ORF3a would be translated in the main reading frame and both alternative reading frames, three frames in total (ORFs 3a, 3c and 3d, Fig. 1B). Translation of three genes overlapping the same sites in different reading frames is rare but known to occur in at least one other virus, namely Env, Tat, and Rev in HIV-2 (Bakouche et al., 2013).
Lastly, the 39 codon ORF with coordinates 21744-21860 overlapping the Spike protein was found to show evidence of translation in a ribosome profiling experiment (Finkel et al., 2020). The sequence of this ORF displays evidence of purifying selection between human hosts, using a π N/π S method intended for use with overlapping genes (Nelson et al., 2020b).
Ambiguity in the usage of the name ORF3b has been particularly confusing. Two of the earliest papers about the SARS-CoV-2 genome used the term ORF3b to mean different genomic regions: Wu et al. show ORF3b as the region homologous to SARS-CoV ORF3b in their Fig. 1 and their Supplementary Table 6 without noting the in-frame stop codons (F. Wu et al., 2020a), whereas Chan et al. use the name ORF3b to refer to the 57 codon ORF, which has no homology to SARS-CoV ORF3b (Chan et al., 2020). At the time of writing, at least 30 subsequent published papers or preprints have used the term ORF3b to refer to one or both of these two ORFs, many of which provide little if any information from which the reader might deduce which ORF is being referred to (Table 2, Supplementary Table 1). For example, a recent report about the antibody response to SARS-CoV-2 ORF3b (Hachim et al., 2020) does not state which ORF3b is referred to, only that proteins were chosen based on previous studies, and citing the two aforementioned papers (Chan et al., 2020; Wu et al., 2020a), which have different definitions of ORF3b. We were able to infer from the PCR primers in their Supplementary Table 5 (Hachim et al., 2020) (confirmed by personal communication) that it refers to the 57 amino acid protein. A subsequent preprint about the 22 amino acid protein cited the Hachim et al. study (Hachim et al., 2020) of the 57 amino acid protein as evidence of expression (Konno et al., 2020a), though this was corrected in the published version (Konno et al., 2020b). On the other hand, a report about interferon evasion (Xia et al., 2020) refers to one of the investigated ORFs as ORF3b, without providing coordinates, but we were able to infer from the PCR primers in their Supplementary Table S1 that the amplified region was the 155 codon SARS-CoV-2 region homoloous to SARS-CoV ORF3b, so presumably in this case it was the 22 codon ORF that was expressed, since that is at the 5′ end of this region. Furthermore, at least five review papers discuss the 57 codon and 22 codon ORFs overlapping ORF3a as if they were the same entity (Supplementary Table 1). Particularly confusing is one review that mentions ORF3b without providing coordinates and depicts it graphically as the ORF at the 3′ end of the SARS-CoV-2 region homologous to SARS-CoV ORF3b (overlapping the 3′ end of ORF3a and the 5′ end of E), but we can infer from the cited reference that they are referring to the 22 codon ORF at the 5′ end of the homologous region (Sa Ribero et al., 2020). We know of at least one instance of similar confusion for ORF9b: Davidson et al. (2020) report that they “could not detect peptides from ORF9b as described by Bojkova et al., 2020”, but “peptides corresponding to the ORF9a protein were identified”; however, Davidson's ORF9a and Bojkova's ORF9b are different names for the same 97 amino acid protein, and Davidson's ORF9b is the 73 amino acid protein, for which Bojkova et al. also did not find peptides (Bojkova et al., 2020), so the two studies detected the same N-overlapping protein after all. Examples of confusion caused by inconsistent naming continue to accumulate rapidly.
2.2. Consensus nomenclature for SARS-CoV-2 overlapping ORFs and isoforms
After discussions with many of the researchers that have published evidence related to overlapping ORFs in SARS-CoV-2, and in consultation with members of the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, we propose consensus nomenclature for the six aforementioned overlapping ORFs, and the shorter isoform of ORF3d (Table 1, Fig. 1).
Our naming decisions were motivated by the following considerations. First, we strongly recommend using the same name as the SARS-CoV homolog where one exists. This rule is in agreement with the prevailing practice for ORF and protein naming by the Coronaviridae Study Group. This facilitates the transfer of knowledge in the coronavirus field and cross-communication between research on SARS-CoV-2, SARS-CoV, and other viruses of the species Severe acute respiratory syndrome-related coronavirus. The importance of this rule can be appreciated by imagining a scenario in which researchers use the name “hemoglobin” to refer to hemoglobin (homologous) in one eukaryotic species, but to insulin (not homologous) in another — accurate transfer of knowledge would be error-prone indeed. Practically, homology recognition is based on analysis of sequence and structure similarity which is not always straightforward for small ORFs unless assisted by other considerations like genome collinearity (synteny). However, we recommend that researchers naming ORFs make every possible effort to determine whether there is homology to ORFs in related strains or species, and avoid names that could lead to mistaken assumptions of homology or lack thereof. At least eight studies using the name ORF3b to refer to the 57 codon SARS-CoV-2 ORF overlapping ORF3a (following Chan et al. (2020)) have mistakenly assumed homology and a possible functional relationship to SARS-CoV ORF3b due to this departure from the homology rule (Supplementary Table 1). The application of the same-name-for-homologs rule to ORF9b is unambiguous, because SARS-CoV and SARS-CoV-2 encode full-length homologs. On the other hand, there are several small ORFs within the region of the SARS-CoV-2 genome homologous to SARS-CoV ORF3b, but to our knowledge only the ORF at the 5′ end of this region, beginning at the AUG codon homologous to the start codon of SARS-CoV ORF3b, has been proposed to be protein-coding (Konno et al., 2020b). Thus, we assign the name ORF3b to the 22 codon ORF, in line with prior studies of ORFs of various lengths in bat viruses of this species homologous to the 5′ end of SARS-CoV ORF3b (Zhou et al., 2012).
Second, we maintain the convention of naming overlapping ORFs as ORF{Number1}{letter}, where “Number1” is the number of the main ORF (note that the numeric names of S and N are ORF2 and ORF9a, respectively (Inberg and Linial, 2004)) and “letter” is a lower case letter. We reserve “a” for the main ORF and default to sequential (alphabetical) letters to name additional overlapping ORFs in 5′–3′ order, but retain the flexibility to accommodate the same-name-for-homologs rule or historical usage. In the case of ORFs overlapping ORF3a, “a” is taken by the main ORF and “b” by the SARS-CoV homolog. Thus, we have named the remaining two overlapping ORFs 3c and 3d in 5′–3′ order even though they occur 5′ of ORF3b. In the case of ORF9c, we choose to use “c” because it is 3′ of ORF9b, and because the homologous ORF in SARS-CoV has sometimes been called ORF9c (though it has also been called ORF14).
Finally, we extend our convention by naming smaller isoforms of overlapping ORFs using alternative start codons according to the template ORF{Number1}{letter}{-}{Number2}. Specifically, we introduce the name ORF3d-2 for the 33 codon isoform of ORF3d. Whether either, both, or neither of these two isoforms encode a functional protein has yet to be determined, so we have chosen to name the shorter isoform in case it is the only functional isoform, but use a name that relates it to ORF3d in case both are functional. There are several other sub-ORFs that have been proposed to be translated (Finkel et al., 2020) but we have not assigned names to them because, as far as we know, ORF3d-2 is the only one for which anyone has proposed that the shorter form is translated but the longer one is not. If, in the future, names are needed for other smaller isoforms of overlapping ORFs using alternative start codons, we suggest using a naming strategy that is analogous to ORF3d-2.
While researchers have presented experimental or computational evidence of translation or function for each of the six overlapping ORFs discussed here, a final consensus in the community has not yet been achieved. We would like to emphasize that in choosing names we are not intending to imply anything about the strength of evidence for translation or function of any of these ORFs, or parts thereof. With the humbling recognition that our knowledge of coronavirus biology in general and the SARS-CoV-2 genome in particular is far from complete, we have tried to suggest naming rules with sufficient flexibility to handle future discoveries.
3. Conclusions
We have proposed standard names for six SARS-CoV-2 ORFs and one shorter isoform that have been hypothesized to encode accessory proteins. The ORF names we have recommended here have been endorsed by several members of the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, namely Stanley Perlman, Bart L Haagmans, and Benjamin W Neuman, and two coauthors John Ziebuhr and Alexander E. Gorbalenya; by the other coauthors of this paper, who represent many of the groups that initially proposed or reported additional evidence for the protein-coding status of some of these ORFs; and by the virus curator of SwissProt/UniProt, Philippe Lemercier. We hope that future publications will adopt the recommended names, including the published versions of any current preprints that refer to these ORFs, in order to facilitate unambiguous communication and minimize confusion. We also recommend that authors referring to any of these ORFs explicitly provide the length or genome coordinates with respect to the reference SARS-CoV-2 genome Wuhan-Hu-1 (NCBI: NC_045512.2), and report the name used in any cited paper if it is different. These practices should help to resolve ambiguities caused by names that have already appeared in the literature.
Author contributions
Irwin Jungreis: Conceptualization, Writing - Original Draft, Writing - Review & Editing, Project administration. Chase W. Nelson: Conceptualization, Writing - Review & Editing. Zachary Ardern: Data Curation, Writing - Review & Editing. Yaara Finkel: Writing - Review & Editing. Nevan J. Krogan: Writing - Review & Editing. Kei Sato: Writing - Review & Editing. John Ziebuhr: Writing - Review & Editing. Noam Stern-Ginossar: Writing - Review & Editing. Angelo Pavesi: Writing - Review & Editing. Andrew E. Firth: Conceptualization, Writing - Review & Editing. Alexander E. Gorbalenya: Conceptualization, Supervision, Writing - Review & Editing. Manolis Kellis: Supervision, Writing - Review & Editing.
Acknowledgments
We thank Stanley Perlman (Department of Microbiology and Immunology, University of Iowa, Iowa City), Bart L. Haagmans (Department of Viroscience, Erasmus Medical Center, Rotterdam) and Benjamin W. Neuman (Biology Department, Texas A&M University, Texarkana) for critical reading of the manuscript. This work was supported by the National Human Genome Research Institute of the National Institutes of Health under Award Number U41HG007234 (IJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was provided by grant number NIH grant R01 HG004037 (IJ), an Academia Sinica Postdoctoral Research Fellowship (CWN), Leiden University Fund (AEG), Wellcome Trust grant 106207 (AEF), the German Federal Ministry of Education and Research (BMBF-COVINET) (JZ). This work has benefited from the equipment and framework of the COMP-HUB Initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022) (AP).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.02.013.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Bakouche N., Vandenbroucke A.-T., Goubau P., Ruelle J. Study of the HIV-2 Env cytoplasmic tail variability and its impact on Tat, Rev and Nef. PloS One. 2013;8:e79129. doi: 10.1371/journal.pone.0079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A.K., Blanco, M.R., Bruce, E.A., Honson, D.D., Chen, L.M., Chow, A., Bhat, P., Ollikainen, N., Quinodoz, S.A., Loney, C., Thai, J., Miller, Z.D., Lin, A.E., Schmidt, M.M., Stewart, D.G., Goldfarb, D., De Lorenzo, G., Rihn, S.J., Voorhees, R.M., Botten, J.W., Majumdar, D., Guttman, M., 2020. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell.. 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani, R., Forni, D., Clerici, M., Sironi, M., 2020. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infection, Genetics and Evolution. 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed]
- Celik O., Saglam A., Baysal B., Derwig I.E., Celik N., Ak M., Aslan S.N., Ulas M., Ersahin A., Tayyar A.T., Duran B., Aydin S. Factors preventing materno-fetal transmission of SARS-CoV-2. Placenta. 2020;97:1–5. doi: 10.1016/j.placenta.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.S., Wu C., Chow S.C.S., Cheung T., To K.-F., Leung W.-K., Chan P.K.S., Lee K.-C., Ng H.-K., Au D.M.Y., Lo A.W.I. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019 doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12(68) doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R.J., Baker, S.C., Baric, R., Enjuanes, L., Gorbalenya, A.E., Holmes, K.V., Perlman, S., Poon, L., Rottier, P.J.M., Talbot, P.J., Others, 2012. Family Coronaviridae, p 806–828. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- Di H., Madden J.C., Jr., Morantz E.K., Tang H.-Y., Graham R.L., Baric R.S., Brinton M.A. Expanded subgenomic mRNA transcriptome and coding capacity of a nidovirus. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8895–E8904. doi: 10.1073/pnas.1706696114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Firth, A.E., 2020. A putative new SARS-CoV protein, 3c, encoded in an ORF overlapping ORF3a. Journal of General Virology. 10.1099/jgv.0.001469. [DOI] [PMC free article] [PubMed]
- Firth A.E., Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. Regulation of stress responses and translational control by coronavirus. Viruses. 2016;8 doi: 10.3390/v8070184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo, M., Staniszewska, M., Salmaso, S., Caliceti, P., Pancer, K.W., Wieczorek, M., Kuryk, L., 2020. Prospects of Replication-Deficient Adenovirus Based Vaccine Development against SARS-CoV-2. Vaccines (Basel) 8. 10.3390/vaccines8020293. [DOI] [PMC free article] [PubMed]
- Gorbalenya, A.E., Baker, S.C., Baric, R.S., de Groot, R.J., Drosten, C., Gulyaeva, A.A., Haagmans, B.L., Lauber, C., Leontovich, A.M., Neuman, B.W., Penzar, D., Perlman, S., Poon, L.L.M., Samborskiy, D.V., Sidorov, I.A., Sola, I., Ziebuhr, J., 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., Peiris J.S.M., Valkenburg S.A. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Hayn M., Hirschenberger M., Koepke L., Straub J.H., Nchioua R., Christensen M.H., Klute S., Bozzo C.P., Aftab W., Zech F., Conzelmann C., Müller J.A., Badarinarayan S.S., Stürzel C.M., Forne I., Stenger S., Conzelmann K.-K., Münch J., Sauter D., Schmidt F.I., Imhof A., Kirchhoff F., Sparrer K.M.J. Imperfect innate immune antagonism renders SARS-CoV-2 vulnerable towards IFN-γ and -λ. Cold Spring Harb. Lab. 2020 doi: 10.1101/2020.10.15.340612. [DOI] [Google Scholar]
- Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of Taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. Res. 2020;9 doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inberg A., Linial M. Evolutional insights on uncharacterized SARS coronavirus genes. FEBS Lett. 2004;577:159–164. doi: 10.1016/j.febslet.2004.09.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N., Firth A.E., Jones J.D., Chung B.Y.-W., Siddell S.G., Brierley I. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016;12:e1005473. doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.-K., Knapp, J.J., Kuang, D., Cassonnet, P., Samavarchi-Tehrani, P., Abdouni, H., Rayhan, A., Sheykhkarimli, D., Coyaud, E., van der Werf, S., Others, 2020a. A Flexible Genome-Scale Resource of SARS-CoV-2 Coding Sequence Clones. [DOI] [PMC free article] [PubMed]
- Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 Gene Content and COVID-19 Mutation Impact by Comparing 44 Sarbecovirus Genomes. Nat. Commun. 2021 doi: 10.1038/s41467-021-22905-7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, Y., Kimura, I., Uriu, K., Fukushi, M., Irie, T., 2020a. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Further Increased by a Naturally Occurring Elongation Variant. bioRxiv. [DOI] [PMC free article] [PubMed]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., USFQ-COVID19 Consortium, Nakagawa, S., Sato, K SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;108185 doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and Nucleocapsid proteins function as interferon antagonists. J. Virol. 2007 doi: 10.1128/jvi.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J.-Y., Yuen C.-K., Ip J.D., Wong W.-M., To K.K.-W., Yuen K.-Y., Kok K.-H. Loss of orf3b in the circulating SARS-CoV-2 strains. Emerg. Microb. Infect. 2020:1–678. doi: 10.1080/22221751.2020.1852892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent, E.M.N., Sofianatos, Y., Komarova, A., Gimeno, J.P., 2020. Global BioID-Based SARS-CoV-2 Proteins Proximal Interactome Unveils Novel Ties between Viral Polypeptides and Host Factors Involved in Multiple COVID19 …. bioRxiv.
- Lei Q., Li Y., Hou H., Wang F., Ouyang Z., Zhang Y., Lai D., Banga Ndzouboukou J., Xu Z., Zhang B., Chen H., Xue J., Lin X., Zheng Y., Yao Z., Wang X., Yu C., Jiang H., Zhang H., Qi H., Guo S., Huang S., Sun Z., Tao S., Fan X. Antibody dynamics to SARS-CoV-2 in asymptomatic COVID-19 infections. Allergy. 2020;26(1193) doi: 10.1111/all.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014 doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 1992;66:6143–6154. doi: 10.1128/JVI.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020;94 doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Michel C.J., Mayer C., Poch O., Thompson J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17(131) doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeel-Shah, S., Lee, H., Ahmed, N., Marcon, E., 2020. SARS-CoV-2 Nucleocapsid Protein Attenuates Stress Granule Formation and Alters Gene Expression via Direct Interaction with Host mRNAs. bioRxiv.
- Nelson, C.W., Ardern, Z., Goldberg, T.L., Meng, C., Kuo, C.H., 2020a. A Previously Uncharacterized Gene in SARS-CoV-2 Illuminates the Functional Dynamics and Evolutionary Origins of the COVID-19 Pandemic. bioRxiv.
- Nelson C.W., Ardern Z., Goldberg T.L., Meng C., Kuo C.-H., Ludwig C., Kolokotronis S.-O., Wei X. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. Elife. 2020;9 doi: 10.7554/eLife.59633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J.B., Brian D.A. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology. 2000;269:172–182. doi: 10.1006/viro.2000.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C., Robichon A. Computational search of hybrid human/SARS-CoV-2 dsRNA reveals unique viral sequences that diverge from other coronavirus strains. Cold Spring Harb. Lab. 2020 doi: 10.1101/2020.04.08.031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology. 2020;546:51–66. doi: 10.1016/j.virol.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh S., Matschinske J., Blumenthal D.B., Galindez G., Kacprowski T., List M., Nasirigerdeh R., Oubounyt M., Pichlmair A., Rose T.D., Salgado-Albarrán M., Späth J., Stukalov A., Wenke N.K., Yuan K., Pauling J.K., Baumbach J. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat. Commun. 2020 doi: 10.1038/s41467-020-17189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Abdouni H., Knight J.D.R., Astori A., Samson R., Lin Z.-Y., Kim D.-K., Knapp J.J., St-Germain J., Go C.D., Larsen B., Wong C.J., Cassonnet P., Demeret C., Jacob Y., Roth F.P., Raught B., Gingras A.-C. [DOI]
- Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlub T.E., Buchmann J.P., Holmes E.C. A simple method to detect candidate overlapping genes in viruses using single genome sequences. Mol. Biol. Evol. 2018;35:2572–2581. doi: 10.1093/molbev/msy155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003:865–869. doi: 10.1016/s0022-2836(3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Germain, J.R., Astori, A., Samavarchi-Tehrani, P., Abdouni, H., Macwan, V., Kim, D.-K., Knapp, J.J., Roth, F.P., Gingras, A.-C., Raught, B., 2020. A SARS-CoV-2 BioID-Based Virus-Host Membrane Protein Interactome and Virus Peptide Compendium: New Proteomics Resources for COVID-19 Research. Cold Spring Harbor Laboratory. 10.1101/2020.08.28.269175. [DOI]
- Sun, W., 2020. The Discovery of Gene Mutations Making SARS-CoV-2 Well Adapted for Humans: Host-Genome Similarity Analysis of 2594 Genomes from China, the USA and Europe. Cold Spring Harbor Laboratory. 10.1101/2020.09.03.280727. [DOI]
- Taefehshokr N., Taefehshokr S., Hemmat N., Heit B. Covid-19: perspectives on innate immune evasion. Front. Immunol. 2020;11(580641) doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 1994;75(Pt 11):3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Yuan X., Wang B., Gu R., Li W., Xiang X., Tang L., Sun H. Severe acute respiratory syndrome coronavirus 2: from gene structure to pathogenic mechanisms and potential therapy. Front. Microbiol. 2020;11(1576) doi: 10.3389/fmicb.2020.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33(108234) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiao Z., Ye K., He X., Sun B., Qin Z., Yu J., Yao J., Wu Q., Bao Z., Zhao W. SARS-CoV-2: characteristics and current advances in research. Virol. J. 2020;17(117) doi: 10.1186/s12985-020-01369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Lagniton P.N.P., Ye S., Li E. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020 doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., Cai J.-P., Jin D.-Y., To K.K.-W., Chan J.F.-W., Yuen K.-Y., Kok K.-H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Cruz-cosme R., Zhuang M.-W., Liu D., Liu Y., Teng S., Wang P.-H., Tang Q. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Cold Spring Harb. Lab. 2020 doi: 10.1101/2020.08.02.233023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Li H., Wang H., Wang L.-F., Shi Z. Bat severe acute respiratory syndrome-like coronavirus ORF3b homologues display different interferon antagonist activities. J. Gen. Virol. 2012;93:275–281. doi: 10.1099/vir.0.033589-0. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L. Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.