Abstract
“Functional wasteland,” “Nonrecombining desert,” and “Gene-poor chromosome” are only some examples of the different definitions given to the Y chromosome in the last decade. In comparison to the other chromosomes, the Y is poor in genes, being more than 50% of its sequence composed of repeated elements. Moreover, the Y genes are in continuous decay probably due to the lack of recombination of this chromosome. But the human Y chromosome, at the same time, plays a central role in human biology. The presence or absence of this chromosome determines gonadal sex. Thus, mammalian embryos with a Y chromosome develop testes, while those without it develop ovaries (Polani [1]). What is responsible for the male phenotype is the testis-determining SRY gene (Sinclair [2]) which remains the most distinguishing characteristic of this chromosome. In addition to SRY, the presence of other genes with important functions has been reported, including a region associated to Turner estigmata, a gene related to the development of gonadoblastoma and, most important, genes related to germ cell development and maintenance and then, related with male fertility (Lahn and Page [3]). This paper reviews the structure and the biological functions of this peculiar chromosome.
STRUCTURE OF THE Y CHROMOSOME
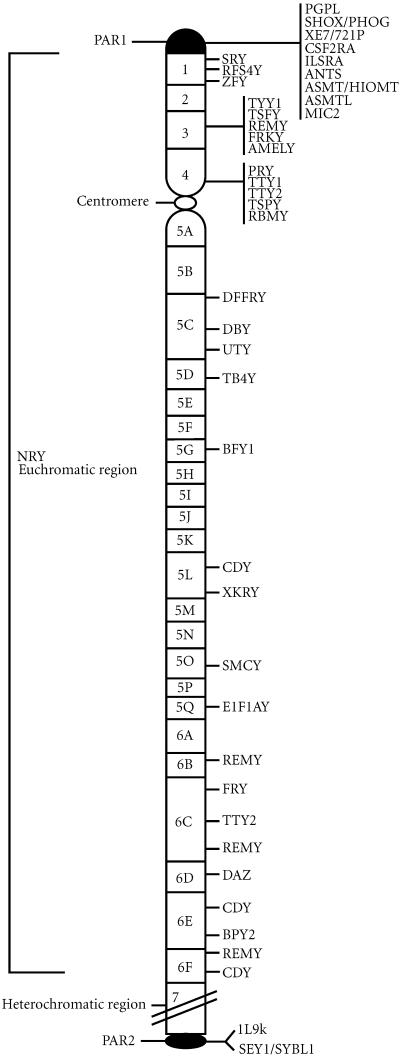
The Y is one of the smallest chromosomes in the human genome (∼ 60 Mb) and represent around 2%–3% of a haploid genome. Cytogenetic observations based on chromosome-banding studies allowed different Y regions to be identified: the pseudoautosomal portion (divided into two regions: PAR1 and PAR2) and the euchromatic and heterochromatic regions (Figure 1).
Figure 1.
Schematic representation of the Y chromosome. Genes in the two pseudoautosomal regions (PAR1 and PAR2) as well as those in the nonrecombining Y region (NRY) are illustrated.
The Pseudoautosomal regions (PAR): PAR1 is located at the terminal region of the short arm (Yp), and the PAR2 at the tip of the long arm (Yq). PAR1 and PAR2 cover approximately 2600 and 320 kb of DNA, respectively. The pseudoautosomal regions, and in particular PAR1, are where the Y chromosome pairs and exchanges genetic material with the pseudoautosomal region of the X chromosome during male meiosis. Consequently, genes located within the PAR are inherited in the same manner as autosomal genes. The euchromatic region is distal to the PAR1 and consists of the short arm paracentromeric region, the centromere and the long arm paracentromeric region. Finally, the heterochromatic region comprises distal Yq corresponding to Yq12. This region is assumed to be genetically inert and polymorphic in length in different male populations, since it is composed mainly of two highly repetitive sequences families, DYZ1 and DYZ2, containing about 5000 and 2000 copies of each respectively.
Whereas PAR1 and PAR2 represent the 5% of the entire chromosome, the majority of the length of the Y (95%) is made by the so-called “Non-Recombining Y” (NRY). This includes the euchromatic and heterochromatic regions of the chromosome. Whereas the heterochromatic region is considered genetically inert, the euchromatic region has numerous highly repeated sequences but also contains some genes responsible for important biological functions that we will review here.
PHYSICAL AND MOLECULAR MAPPING
The physical mapping of the Y chromosome has mainly depended on naturally occurring deletions on this chromosome. The creation of a deletion map, and the resultant ordering of DNA loci along the chromosome, is very useful not only in locating genes but also in studying the structural diversity of the Y within and among human populations and primates. This allows information on the evolution of human species through paternal lineages to be obtained.
The first attempts at mapping the Y were based on cytogenetically detectable deletions on this chromosome and suffer, then, from the limited accuracy and resolution of chromosome banding patterns. However, these preliminary studies led, for the first time, to the hypothesis that a gene or genes located on Yq were related to spermatogenic failure (Tiepolo and Zuffardi [4]). Similar studies defined also a region associated with sex determination (Jacobs and Ross [5], Buhler [6]).
Vergnaud et al. [7] performed the first molecular map of the Y in 1986. By using different Y-specific probes on patients with microscopically detectable Y anomalies, they subdivided the Y chromosome into 7 intervals, corresponding with naturally occurring deletions of this chromosome. Later in 1992, Vollrath et al. [8] constructed a more precise deletion map of the Y chromosome based on the detection of about 200 sequence-tagged sites (STS's). The presence or absence of these STS's on a large set of patients with a wide range of Y anomalies subdivided the euchromatic into 43 ordered intervals, all defined by naturally occurring chromosomal breakpoints. These 43 deletion intervals further refined the seven-interval map of Vergnaud et al. [7]. This collection of ordered STS's along the Y chromosome have been extensively used in order to define shortest deleted regions associated with particular phenotypes and then, in identifying Y chromosomal genes and exploring the origin of Y chromosome disorders. Moreover, the same group in Boston led by David Page prepared a library of yeast artificial clones (YAC) from a human XYYYY male. The clones were screened with the Y-specific STS's in order to identify those containing the corresponding sequences. Finally, an essentially complete physical map of the Y chromosome was generated with 196 overlapping DNA clones, which covered 98 percent of the euchromatic region (Foote et al. [9]). These Y physical maps have certainly accelerated the search for new genes and made it much easier to explore the biology of this chromosome.
GENES ON THE Y CHROMOSOME
Compared to the other human chromosomes, the Y chromosome has a limited number of genes. The Y gene poverty may have been the result of the known the tendency of Y chromosome's genes to degenerate during evolution, being nowadays the relic of an ancient common ancestry with the X chromosome (Graves [10]). Both mammalian X and Y chromosomes evolved from ancestral autosomes. The most ancestral gene functions were retained on the nascent X chromosome but deteriorated on NRY portion of the emerging Y (Bull [11]) giving females with two copies but males with only one copy of many genes. The gene dosage problem has been solved through inactivation of one X chromosome in females.
In spite of the limited make-up of genes, different transcription units or families of closely related transcription units have been identified in the NRY region during the past decade (see [12, 13, 14, 2, 15, 16, 17, 18]). Recently, Lahn and Page [3] identified 12 novel genes or gene families and assessed their expression in diverse human tissues. The different genes identified so far throughout both the NRY region and the two pseudoautosomal regions are summarised in Table I, together with some information on their location and possible pathological implications. According to the same authors, all NRY genes can be divided into two different categories. The first comprises those genes which are ubiquitously expressed, have X homologues, appear in a single copy on the NRY and exhibit housekeeping cell functions. The second category include genes expressed specifically in the testes, exist in multiple copies (with the exception of SRY) on the NRY and encode proteins which more specialised functions. It is worth mentioning the finding of X-homologous NRY genes, which suggest an alternative solution for the gene dosage compensation. It has been proposed that these genes should escape X-inactivation and encoded proteins functionally interchangeable (Lahn and Page [3]).
Table 1.
Genes of the human Y chromosome PAR1, PAR2, and NRY.
| Gene symbol | Location | Gene name | Associate pathology/function | X-homologs | Reference |
| CSFR2Rα | PAR1 | GM-CSF receptor α subunit | unknown | + | Disteche et al. 1992 [19] |
| SHOX | " | Short stature homebox-containing | short stature, Lerf-Weill syndrome | + | Rao et al. 1997 [20] |
| IL3RA | " | Interleukin-3 receptor α subunit | unknown | + | Ellison et al. 1996 [21] |
| ANT3 | " | Adenine nucleotide translocase | unknown | + | Shiebel et al. 1993 [22] |
| ASMTL | " | Acetylserotonine methyltransferase-like | unknown | + | Ried et al. 1998 [23] |
| ASMT | " | Acetylserotonine methyltransferase | unknown | + | Rappold 1993 [24] |
| XE7 | " | X-escapee | unknown | + | Ellison et al. 1996 [21] |
| PGPL | " | Pseudoautosomal GTP-binding protein-like | unknown | + | Gianfrancesco et al. 1998 [25] |
| MIC2 | " | unknown | + | Goodfellow et al. 1988 [26] | |
| SRY★ | Yp: 1A1A | Sex Reversal Y | Sex reversal | − | Sinclair et al. 1990 [2] |
| RPS4Y | Yp: 1A1B | Ribosomal protein S4, Y | Turner syndrome? | + | Fisher et al. 1990 [14] |
| ZFY | Yp: 1A2 | Zinc-finger Y | Turner syndrome? | + | Page et al. 1987 [13] |
| PRKY | Yp: 3C-4A | protein kinase, Y | unknown | + | Schiebel et al. 1997 [27] |
| TTY1★ | Yp: 4A | testis transcript, Y1 | unknown | − | Reijo et al. 1995 [17] |
| TSPY★ | Yp: 3C + 5 | testis-specific protein, Y | gonadoblastoma? | − | Arnemann et al. 1987 [12] |
| AMELY | Yp: 4A | Amelogenin, Y | unknown | + | Nakahori et al. 1991 [28] |
| PRY★ | Y: 4A, 6E | putative tyrosine phosphatase protein-related Y | infertility? | + | Reijo et al. 1995 [17] |
| TTY2★ | Y: 4A, 6C | testis transcript, Y2 | unknown | − | Reijo et al. 1995 [17] |
| USP9Y (or DFFRY) | Yq: 5C | ubiquitin-specific protease (or Drosophila fat-facets related, Y) | azoospermia? | + | Lahn and Page 1997 [3] |
| DBY | Yq: 5C | DEAD box, Y | infertility? | + | Lahn and Page 1997 [3] |
| UTY | Yq: 5C | Ubitiquitous TRY motif, Y | infertility | + | Lahn and Page 1997 [3] |
| TB4Y | Yq: 5D | Thymosin ?4, Y isoform | infertility | + | Lahn and Page 1997 [3] |
| BPY1★ | Yq: 5G | basic protein, Y1 | Turner? | + | Reijo et al. 1995 [17] |
| CDY | Yq: 5L, 6F | chromodomain, Y | infertility? | − | Lahn and Page 1997 [3] |
| XKRY★ | Yq: 5L | XK-related, Y | infertility? | − | Reijo et al. 1995 [17] |
| RBM★ | Yp + q | RNA-binding motif, Y | infertility? | − | Ma et al. 1993 [15] |
| SMCY | Yq: 5P | Selected Mouse cDNA, Y | unknown | + | Agulnik et al. 1994 [16] |
| EIF1AY | Yq: 5Q | Translation initiation factor 1A, Y | infertility? | + | Lahn and Page 1997 [3] |
| DAZ★ | Yq: 6F | Deleted in azoospermia | infertility? | − | Reijo et al. 1995 [17] |
| VCY2 | Yq: 6A | variably charged protein, Y2 | infertility | − | Reijo et al. 1995 [17] |
| IL9R | PAR2 | Interleukin 9 receptor | unknown | + | Vermeesh et al. 1997 [29] |
| SYBL1 | " | Synaptobrevin-like 1 | unknown | + | Kermouni et al. 1995 [30] |
| HSPRY3 | " | Human-sprouty 3 | unknown | + | Ciccododicola et al. 2000 [31] |
| CXYorf1 | " | CXYorf1 | unknown | + | Ciccododicola et al. 2000 [31] |
★Testis-specific genes or families.
Note: All genes expressed specifically in the testis are present in multiple copies dispersed throughout the euchromatic portion of the Y chromosome. Exceptional is SRY, which is expressed specifically in the testis but present in single copy.
BIOLOGICAL FUNCTIONS OF THE HUMAN Y CHROMOSOME
Several phenotypes have been associated with the nonrecombining portion of the Y chromosome. For obvious reasons, most of these are male-specific and make the Y a specialised chromosome during human evolution. The most characterising features of this chromosome remain its implication in human sex determination and in male germ cell development and maintenance.
SRY gene and sex determination
The first indices that the Y chromosome was involved in male sex determination came from the observation that XY or XYY (Klinefelter syndrome) individuals develop testes whereas XX or XO (Turner's syndrome) individuals develop ovaries (Jacobs and Strong [32]). Later, studies showing that mice XX presenting a male phenotype carried a small portion of the Y chromosome supported the proposition that a master gene involved in male sex determination was carried by the Y chromosome (Goodfellow and Darling [33]). In 1990, the gene responsible for testicular determination, named SRY (Sex-determining Region on the Y chromosome), was finally identified (Sinclair et al. [2]). SRY was cloned by isolation of small fragments of translocated Y on XX sex-reversed patients. This gene is located on the short arm of the Y chromosome close to the pseudoautosomal boundary. It comprises a single exon encoding a protein of 204 amino acids which presents conserved DNA-binding domain (the HMG-box: High Mobility Group), suggesting this protein regulates gene expression. This gene has been shown to be essential for initiating testis development and the differentiation of the indifferent, bipotential, gonad into the testicular pathway. Moreover, SRY has been proposed to be the master gene regulating the cascade of testis determination. Although many genes and loci have been proposed to interact with SRY protein, such as WT-1 (Wilm's tumour gene), SF-1 (Steroidogenic Factor 1) and SOX-9, the question of how these genes are regulated, if so, by SRY is still unanswered.
Anti-Turner syndrome effect
Turner syndrome is characterised by a female 45 X karyotype or monosomy X. The principal manifestations of this syndrome are growth failure, infertility, anatomic abnormalities, and selective cognitive deficits. This human genetic disorder is ascribed to haplo-insufficiency of genes of the X chromosome that are common to both X and Y. These genes must escape X-inactivation because otherwise no difference will be observed between 45, X and 46, XX females. Secondly, in 46, XY these genes must have a male counterpart on the Y responsible to simulate the effects of their X homologues. Although there is no formal identification of genes involved in Turner syndrome, there appear to be different loci on the X and Y chromosome associated with Turner characteristic features, such as SHOX/PHOG (Rao et al. [20], Ellison et al. [21]), ZFX/ZFY (Page et al. [34]), GCY and TCY (Barbaux et al. [35]).
Genes controlling spermatogenesis
Tiepolo and Zuffardi [4] reported the occurrence of grossly cytogenetically detectable de novo deletions in six azoospermic individuals, describing for the first time the role of the Y chromosome in spermatogenesis. These observations led the authors to postulate the existence of a locus, called AZoospermia Factor (AZF), on Yq11 required for a complete spermatogenesis since the seminal fluid of these patients did not contain mature spermatozoa. The location of AZF in Yq11 was further confirmed by numerous studies at cytogenetic and molecular level (see [36, 37, 38]). Once the molecular map by Vergnaud et al. [7] became available, AZF was localised to the deletion interval 6, a region in band q11.23. The publication of about 200 Y-specific STS, allowed by Vollrath et al. [8], allowed a much simpler Y chromosome screening for microdeletions to be performed. Thus, the original AZF region was further subdivided into three different nonoverlapping subregions in Yq11 associated with male infertility, named AZFa, AZFb, and AZFc (Vogt et al. [39]). Each one of these regions contains several genes proposed as candidate genes involved in male infertility.
The AZFa region is located in proximal Yq within the deletion interval 5 and its molecular extension has been roughly estimated between 1 and 3Mb. Several genes have been identified in this region; Dead Box Y (DBY), Ubiquitous TPR motif Y (UTY), Tymosin B4Y isoform (TB4Y), and the homologue of the Drosophila Developmental gene Fats Facets (DFFRY). The first three genes have no apparent specialised functions and they seem to be involved in cellular “housekeeping.” By contrast, the DFFRY gene has been proposed to play a role in gametogenesis. It encodes a protein involved in desubiquitination (the process by which proteins are tagged for degradation) and mutations in the Drosophila homologue of the gene causes a sterile phenotype (Fischer-Vize et al. [40], Brown et al. [41]).
The AZFb region is located between deletion interval 5 and proximal deletion interval 6, and its molecular extension has been estimated to be similar to that of the AZFa region (1–3 Mb). Five genes have been so far described within this interval; RNA-binding motif (RBM), Chromodomain Y (CDY), XK Related Y (XKRY), eukaryotic translation initiation factor 1A (eIF-1A), and Selected Mouse cDNA on the Y (SMCY). The RBM gene encode germ cell specific nuclear proteins containing RNA-binding motif and it is present in multiple copies along the Y. However, not all of these copies are functional and most may be pseudogenes. It has been strongly proposed as a candidate infertility gene since its expression is testis-specific, it is recurrently deleted in azoospermic men and is seems to be specifically expressed in spermatogonia and primary spermatocytes (Ma et al. [15], Elliot et al. [42]). Other two genes are expressed specifically in adult testis and are recurrently deleted in infertile males, the CDY and the XKRY.
The AZFc region is located in the proximity of the heterochromatin region distal to Yq11 and its molecular extension is about 500 kb (Reijo et al. [17]). This region contains the DAZ (Deleted in AZoospermia) gene cluster, two copies of the PTP-BL Related Y (PRY), Basic Protein Y2 (BPY2), as well as copies of CDY and RBM. DAZ encodes a testis-specific RNA binding protein (Reijo et al. [17]) and contains seven tandem repeats of 24 aa unit. DAZ is present in at least six to nine copies, all being located within AZFc. It is homologous to an autosomal gene on chromosome 3, with a single DAZ repeat, named DAZL1 (DAZ like-autosomal 1) which is also specifically expressed in the testis. It has been hypothesized that DAZ originated from a translocation and subsequent amplification of this ancestral autosomal gene. (Reijo et al. [43], Saxena et al. [44]). Cooke et al. [45] described the homologue of the human Y-linked DAZ gene, named Dazla (DAZ like autosomal), in the mouse where is located on chromosome 17. Dazla presents an RNA-binding domain with 89% of homology with DAZ and is expressed specifically in the testis and ovaries (see [45], [43]). Knockout mice for this gene have been shown to be infertile in both the two sexes (Ruggiu et al. [46]). These observations suggested Dazla as an important gene in mouse gametogenesis. Although DAZ has been proposed as the cause of the AZFc phenotype, other genes must be involved since deletions within AZFc region without including DAZ have been recently reported (see [47]–[49]). The other genes identified within this region, PRY, BPY2, and TTY2, all also present a testis-specific expression and are present in multiple copies on the Y.
Many of the AZF genes have been proposed as candidate genes involved in human male fertility on the basis of their expression profiles (testis-specific or highly expressed in testis) and sterile phenotypes from targeted disruption of their homologues in mice. However, no direct relation between a Y chromosome gene and male infertility has been demonstrated. In a recent paper, Page and coworkers (Sun et al. [50]) relate spematogenic failure to a single mutation in a Y-linked gene in AZFa: the USP9Y or, also called, DFFRY. They found a de novo 4bp deletion in a splice-donor site of this gene present in a patient with nonobstructive azoospermia but absent in his fertile brother. This mutation causes protein truncation leading to spermatogenic arrest. These findings lead the authors to conclude that the USP9Y gene has a role in human spermatogenesis.
Oncogenic role of the Y chromosome
The implication of the Y chromosome in cancer remains still speculative. Y chromosome loss and rearrangements have been associated with different types of cancer, such as bladder cancer (Sauter et al. [51]), male sex cord stroma tumours (de Graaff et al. [52]), lung cancer (Center et al. [53]) and esophageal carcinoma (Hunter et al. [54]). Although loss and rearrangements of this chromosome are relatively frequent in different types of cancer, there is no direct evidence for a role of Y in tumour progression since no proto-oncogenes, tumour suppresser genes or mismatch repair genes have been localised to the Y chromosome.
However, it is well presumable that both oncogenes and tumour supressor genes must lay on this chromosome, having a pathogenic significance mainly in male-specific organs such as testis. One cancer predisposition locus has been assigned to this chromosome, the gonadoblastoma locus on the Y chromosome (GBY). The gonadoblastoma is a rare form of cancer that consists of aggregates of germ cells and sex cord elements. It develops in more than 30% of dysgenetic gonads from sex-reversed females (Swyers syndrome) who harbour some Y-chromosomal material. This observation led to postulate the existence of a predisposing locus on the Y (GBY) that enhance dysgenetic gonads to develop gonadoblastoma. This locus could act as an oncogene in dysgenetic gonads, having a normal function in the testis, and it would have a pathogenic effect when is expressed out of its natural environment (normal testis). This locus would expand over a region of 1–2 Mb on the short arm of the Y chromosome, in the region 4A-4B (Figure 1) Several genes have been proposed as candidates for GBY according to their location, function, and expression profile. Among them, the most likely candidate seems to be TSPY. This gene, present in several copies, is located in the critical region where GBY has been mapped and is expressed in gonadoblastoma, in spermatogonias at early stages of testicular tumorigenesis, in carcinoma in situ of the testis, in seminoma and prostate cancers. These observations strongly suggest that this Y-linked gene may predispose germ cells to other oncogenic events in the multistep process of tumorigenesis.
CONCLUDING REMARKS
The Y is unique under many aspects. It is always in the haploid state, is full of repeated sequences but it is responsible for important biological roles such as sex determination and male fertility. Moreover, the Y chromosome is a powerful tool to study human populations and evolutionary pathways. The nonrecombining portion of the Y retains a record of the mutational events that have occurred along male lineages throughout evolution. This is because it is holoandrically transmitted, from father to son, without recombination at meiosis. Thus, the study of the different mutations this molecule has accumulated along its evolution may be highly informative in deducing the histories of human populations (see [55, 56, 57, 58, 59, 60, 61]). In conclusion, it is time to change the opinion about this singular chromosome. Although the Y chromosome has been studied for more than 30 years, it is considered by many to have little relevance, except in very limited circumstances. The Y has not only demonstrated to be extremely informative in disentangling the history of human populations but it also has essential biological roles that make this chromosome an important component of the human genome.
References
- Polani PE. Experiments on chiasmata and nondisjunction in mice. Hum Genet Suppl. 1981;2:145–146. doi: 10.1007/978-3-642-68006-9_10. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page D. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Ross A. Structural abnormalities of the Y chromosome in man. Nature. 1966;210:352–354. doi: 10.1038/210352a0. [DOI] [PubMed] [Google Scholar]
- Buhler EM. A synopsis of the human Y chromosome. Hum Genet. 1980;55:145–175. doi: 10.1007/BF00291764. [DOI] [PubMed] [Google Scholar]
- Vergnaud G, Page DC, Simmler MC, et al. A deletion map of the human Y chromosome based on DNA hybridization. Am J Hum Genet. 1986;38:109–124. [PMC free article] [PubMed] [Google Scholar]
- Vollrath D, Foote S, Hilton A, et al. The human Y chromosome: a 43-interval map based on naturally occuring deletions. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- Foote S, Vollrath D, Hilton A, et al. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Evolution of sex determining mechanism. Menlo Park: Benjamin Cummings.
- Arnemann AJ, Epplen J, Cooke H, et al. A human Y chromosomal DNA sequence expressed in testicular tissue. Nucl Acids Res. 1987;15:8713–8724. doi: 10.1093/nar/15.21.8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DC, Mosher R, Simpson, et al. The sex-determining region of the human Y chromosome encodes a finger protein. Cell. 1987;51:1091–1104. doi: 10.1016/0092-8674(87)90595-2. [DOI] [PubMed] [Google Scholar]
- Fisher EMC, Beer-Romero P, Brown LG, et al. Homologous ribosomal protein genes on the human X and Y chromosome: escape from X inactivation and possible implication for Turner Syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- Ma K, Inglis JD, Sharkey A, et al. A Y chromosome gene family with RNA-binding protein homology: candidates for the azoospermia factor AZF controlling spermatogenesis. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- Agulnik AI, Mitchell MJ, Mattei MG, et al. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet. 1994;3:879–884. doi: 10.1093/hmg/3.6.879. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–395. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Affara N, Bishop C, Brown W, et al. Report of the second international workshop on Y chromosome mapping 1995. Cytogenet Cell Genet. 1996;73:33–76. doi: 10.1159/000134310. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Brannan CI, Larsen A, et al. The human pseudoautosomal GM-CSF receptor a subunit gene is autosomal in mouse. Nat Genet. 1992;1:333–336. doi: 10.1038/ng0892-333. [DOI] [PubMed] [Google Scholar]
- Rao E, Weiss B, Fukami M, et al. Pseudoautosomal deletions encompassing a novel gene cause growth failure in idiopatic short stature and Turner syndrome. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- Ellison J, Li X, Francke U, et al. Rapid evolution of pseudoautosomal genes and their mouse homologs. Mammal Genome. 1996;7:25–30. doi: 10.1007/s003359900007. [DOI] [PubMed] [Google Scholar]
- Schiebel K, Winkelmann M, Mertz A, et al. Abnormal XY interchange between a novel isolated protein kinase gene, PRKY, and its homologue, PRKX, accounts for one third of all (Y+)XX males and (Y−)XY females. Hum Mol Genet. 1997;6:1985–1989. doi: 10.1093/hmg/6.11.1985. [DOI] [PubMed] [Google Scholar]
- Ried K, Rao E, Schiebel K, et al. Gene duplications as a recurrent theme in the evolution of the human pseudoautosomal region 1: isolation of the gene ASMTL. Hum Mol Genet. 1998;7:1771–1778. doi: 10.1093/hmg/7.11.1771. [DOI] [PubMed] [Google Scholar]
- Rappold G. The pseudoautosomal regions of the human sex chromosomes. Hum Genet. 1993;92:315–324. doi: 10.1007/BF01247327. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco F, Esposito T, Montanini L, et al. A novel pseudoautosomal gene encoding a putative GTP-binding protein resides in the vicinity of the Xp/Yp telomere. Hum Mol Genet. 1998;7:407–414. doi: 10.1093/hmg/7.3.407. [DOI] [PubMed] [Google Scholar]
- Goodfellow PN, Pym B, Pritchard C, et al. MIC2: a human pseudoautosomal gene. Phil Trans R Soc Lond B Biol Sci. 1988;322:145–154. doi: 10.1098/rstb.1988.0122. [DOI] [PubMed] [Google Scholar]
- Schiebel K, Weiss B, Wohrle D, et al. A pseudoautosomal gene, ADP/ATP translocase, escapes X-inactivation whereas a homologue on Xq is subject to X-inactivation. Nat Genet. 1993;3:82–87. doi: 10.1038/ng0193-82. [DOI] [PubMed] [Google Scholar]
- Nakahaori Y, Takenaka O, Nakagome Y, et al. A human X-Y homologous region encodes “amelogenin”. Genomics. 1991;9:264–269. doi: 10.1016/0888-7543(91)90251-9. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Petit P, Kermouni A, et al. The IL-9 receptor gene, located in the Xq/Xp pseudoautosomal region, has an autosomal origin, escapes X inactivation and is expressed from the Y. Hum Mol Genet. 1997;1:1–8. doi: 10.1093/hmg/6.1.1. [DOI] [PubMed] [Google Scholar]
- Kermouni A, Vanroose E, Arden KC, et al. The IL9 receptor gene (IL9R) genomic structure, chromosomal localization in the pseudoautosomal region of the long arm of the sex chromosomes, and identification of IL9R pseudogenes at 9qter, 10pter, 16qter and 18pter. Genomics. 1995;29:371–382. doi: 10.1006/geno.1995.9992. [DOI] [PubMed] [Google Scholar]
- Ciccodicola A, D'Esposito M, Esposito T, et al. Differentially regulated and evolved genes in the fully sequenced Xp/Yq pseudoautonomal region. Hum Mol Genet. 2000;9:395–401. doi: 10.1093/hmg/9.3.395. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Strongs JA. A case of human intersexuality having a possible XXY sex determining machanism. Nature. 1959;183:302–303. doi: 10.1038/183302a0. [DOI] [PubMed] [Google Scholar]
- Goodfellow PN, Darling SM. Genetics of sex determination in man and mouse. Development. 1988;102:251–258. doi: 10.1242/dev.102.2.251. [DOI] [PubMed] [Google Scholar]
- Page DC. Is ZFY the sex-determining gene on the human Y chromosome? Philos Trans R Soc Lond B Biol Sci. 1988;322:155–157. doi: 10.1098/rstb.1988.0123. [DOI] [PubMed] [Google Scholar]
- Barbaux S, Vilain E, Raoul O, et al. Proxiaml deletions on the long arm of the Y chromosome suggest a critical region associated with a specific subset of characteristic Turner stigmata. Hum Mol Genet. 1995;4:1565–1568. doi: 10.1093/hmg/4.9.1565. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Affara NA, Magenis RE. Ordering of Y-specific sequences by deletion mapping and analysis of X-Y interchange males and females. Development Suppl. 1987;101:41–50. doi: 10.1242/dev.101.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Andersson M, Page DC, Pettay D, et al. Y; autosome translocations and mosaicism in the aetiology of 45, X maleness: assigmenbt of fertility factor to distal Yq11. Hum Genet. 1988;79:2–7. doi: 10.1007/BF00291700. [DOI] [PubMed] [Google Scholar]
- Bardoni B, Zuffardi O, Guioli S, et al. A deletion map of the human Yq11 region: implications for the evolution of the Y chromosome and tentative mapping of a locus involved in spermatogenesis. Genomics. 1991;11:443–451. doi: 10.1016/0888-7543(91)90153-6. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsh S, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize JA, Rubin GM, Lehmann R. The fat tacets gene is required for Drosophila eye and embryo development. Development. 1992;116:985–1000. doi: 10.1242/dev.116.4.985. [DOI] [PubMed] [Google Scholar]
- Brown GM, Furlong RA, Sargent CA, et al. Characterisation of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum Mol Genet. 1998;7:97–107. doi: 10.1093/hmg/7.1.97. [DOI] [PubMed] [Google Scholar]
- Eliott DJ, Millar MR, Oghene K, et al. Expression of RBM in the nuclei of human germ cells is dependent on a critical region of the Y chromosome long arm. Proc Natl Acad Sci USA. 1997;94:3848–3853. doi: 10.1073/pnas.94.8.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo R, Alagappan RK, Patrizio P, et al. Severe oligospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- Saxena R, Brown LG, Hawkins T, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Cooke HJ, Lee M, Kerr S, et al. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet. 1996;5:513–516. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Stuppia L, Mastroprimiano G, Calabrese G, et al. Microdeletions in interval 6 of the Y chromosome detected by STS-PCR in 6 of 33 patients with idiopathic oligo- and azoospermia. Cytogenet Cell Genet. 1996;72:155–158. doi: 10.1159/000134174. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Huang V, Yen P, et al. Substantial prevalence of microdeletions of the Y-chromosome in infertile men with idiopathic azoospermia and oligospermia detected using a sequence tagged site based mapping strategy. J Clin Endocrinol Metabol. 1996;81:1347–1352. doi: 10.1210/jcem.81.4.8636331. [DOI] [PubMed] [Google Scholar]
- Foresta C, Ferlin A, Garolla A, et al. Y-chromosome deletions in idiopathic severe testiculopathies. J Clin Endocrinol Metabol. 1997;82:1075–1080. doi: 10.1210/jcem.82.4.3798. [DOI] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Birren B, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Sauter G, Moch H, Wagner U, et al. Y chromosome loss detected by FISH in bladder cancer. Cancer Genet Cytogenet. 1995;82:163–169. doi: 10.1016/0165-4608(95)00030-s. [DOI] [PubMed] [Google Scholar]
- de Graaff WE, van Echten J, van der Veen AY, et al. Loss of the Y-chromosome in the primary metastasis of a male sex cord stromal tumor: pathogenetic implications. Cancer Genet Cytogenet. 1999;112:21–25. doi: 10.1016/s0165-4608(98)00245-3. [DOI] [PubMed] [Google Scholar]
- Center R, Lukeis R, Vrazas V, et al. Y chromosome loss and rearrangement in non-small lung cancer. Int J Cancer. 1993;55:390–393. doi: 10.1002/ijc.2910550309. [DOI] [PubMed] [Google Scholar]
- Hunter S, Gramlich T, Abbott K, et al. Y chromosome loss in esophageal carcinoma: an in situ hybridization study. Genes Chromosomes Cancer. 1993;8:172–177. doi: 10.1002/gcc.2870080306. [DOI] [PubMed] [Google Scholar]
- Jobling M, Tyler-Smith C. Fathers and sons: the Y chromosome and human evolution. Trends Genet. 1995;11:449–455. doi: 10.1016/s0168-9525(00)89144-1. [DOI] [PubMed] [Google Scholar]
- Mitchell RJ, Hammer MF. Human evolution and the Y chromosome. Curr Opin Genet Dev. 1996;6:737–742. doi: 10.1016/s0959-437x(96)80029-3. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A, Naya K, Goldstein DB, et al. Geographic clustering of human Y-chromosome haplotypes. Ann Hum Genet. 1996;60:401–408. doi: 10.1111/j.1469-1809.1996.tb00438.x. [DOI] [PubMed] [Google Scholar]
- Santachiara-Benerecetti AS, Semino O. Y-chromosome polymorphisms and history of populations. Cell Pharmacology. 1996;3:199–204. [Google Scholar]
- Hammer MF, Spurdle AB, Karafet T, et al. The geographic distribution of human Y chromosome variation. Genetics. 1997;145:787–805. doi: 10.1093/genetics/145.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, et al. Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol. 1998;15:427–441. doi: 10.1093/oxfordjournals.molbev.a025939. [DOI] [PubMed] [Google Scholar]
- Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]