Abstract
Scope:
Serum metabolomic markers of the Dietary Approaches to Stop Hypertension (DASH) diet were previously reported. We investigated if urine metabolomic markers were similar in an independent clinical trial.
Methods and Results:
In the DASH-Sodium trial, participants were randomly assigned to the DASH diet or control diet, and received three sodium interventions (high, intermediate, low) within each randomized diet group in random order for 30 days each. Urine samples were collected at the end of the intervention period and analyzed for 938 metabolites. We conducted two comparisons of metabolomic profiles: 1) DASH-high sodium (n=199) vs. control-high sodium (n=193), and 2) DASH-low sodium (n=196) vs. control-high sodium. We compared significant metabolites identified using multivariable linear regression and the top 10 influential metabolites identified using partial least-squares discriminant analysis to the results from a previous analysis of the DASH trial. Nine out of 10 predictive metabolites of the DASH-high sodium and DASH-low sodium diets were identical. Most candidate biomarkers from the DASH trial replicated. N-methylproline, chiro-inositol, stachydrine, and theobromine replicated as influential metabolites of DASH diets.
Conclusions:
Candidate biomarkers of the DASH diet identified in serum replicated in urine. Replicated influential metabolites are likely to be objective biomarkers of the DASH diet.
Keywords: dietary patterns, metabolomics, biomarkers, feeding study
Graphical Abstract
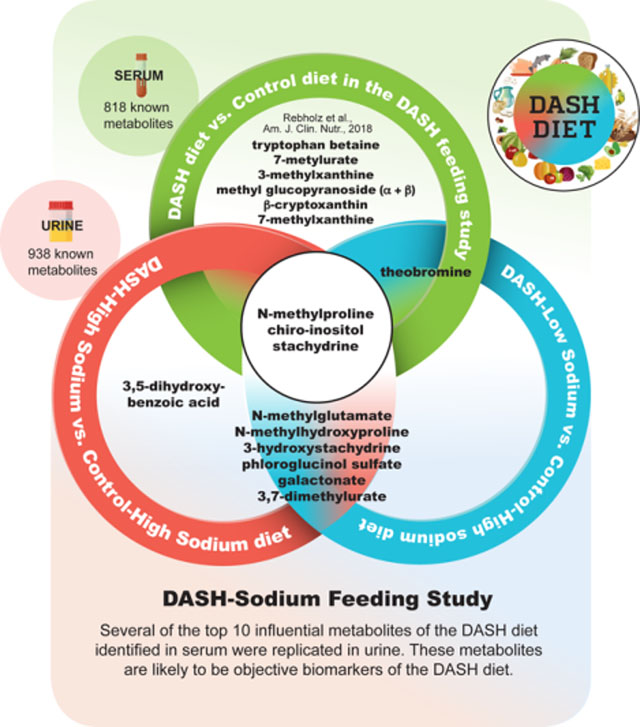
The Dietary Approaches to Stop Hypertension (DASH) diet is recommended as a healthy diet. A recent study identified candidate biomarkers of the DASH diet in serum. The present study revealed that several serum metabolites identified as the top 10 predictive metabolites of the DASH diet replicated as predictive urine metabolites of the DASH diet in an independent population.
1. INTRODUCTION
The Dietary Approaches to Stop Hypertension (DASH) diet is recommended as a healthy dietary pattern in numerous guidelines, including the 2015 Dietary Guidelines for Americans.[1] The DASH diet emphasizes high intake of fruits, vegetables, and low-fat dairy; includes moderate intake of protein sources such as poultry, fish, legumes, nuts, and seeds; and limits intake of red meats, saturated fats, and sweets.[2] In the original DASH feeding study and the subsequent DASH-Sodium feeding study, individuals who were randomly assigned to the DASH diet had significantly lower blood pressure at the end of the study than those assigned to the control diet. In the DASH-Sodium trial, reducing sodium intake also lowered blood pressure, with greatest blood pressure reductions occurring on the DASH diet with low sodium intake. Subsequently, many observational studies have reported that greater adherence to the DASH diet is associated with lower risk of major chronic diseases and mortality.[3–6]
In observational studies, assessment of adherence to the DASH diet relies on self-reported data. However, self-reported data are prone to systematic biases, such as recall bias, social desirability bias, coding bias, or inaccuracies in nutrient estimation from food composition databases.[7,8] Objective biomarkers of the DASH diet which are not influenced by these systematic errors and that can replace or supplement self-reported data are highly desirable. Metabolomics, an approach which allows for the simultaneous measurement of a large number of molecules and metabolites in biospecimens, can be used to identify biomarkers of foods, nutrients, and dietary patterns.[9–14]
Recently, untargeted metabolomics was used to identify candidate biomarkers of the DASH diet using serum samples from the original DASH feeding study.[7] Identified were 97 potential biomarkers that differed significantly between the DASH and control diets, and 10 metabolites [N-methylproline, stachydrine, tryptophan betaine, theobromine, 7-methylurate, chiro-inositol, 3-methylxanthine, methyl glucopyranoside (α + β), β-cryptoxanthin, and 7-methylxanthine] that were highly influential in discriminating between the DASH and control diets. However, a limitation of this prior metabolomic study was the absence of replication in an independent cohort. Replication is essential if these metabolites are to be adopted for use as biomarkers of dietary intake for future studies. Many established biomarkers of dietary intake are currently measured in urine (e.g., urea nitrogen, sodium, potassium).[15] Thus, it is useful to determine if metabolites reported to be biomarkers of the DASH diet in serum are replicated in urine.
Leveraging the rigorous design of the DASH-Sodium trial, our aim was to determine if candidate biomarkers identified in serum for the DASH diet from the original DASH feeding study[7] replicate in urine in the DASH-Sodium trial, and assess the potential of identifying additional biomarkers of the DASH diet.
2. MATERIALS AND METHODS
2.1. Study design and study population
The DASH-Sodium trial was a multicenter, randomized feeding study conducted in 1997–1999 which aimed to evaluate the effects of dietary sodium levels and dietary patterns, alone and combined, on blood pressure.[16] Details on the trial design have been previously published.[17] Briefly, adults (≥ 22 years of age) who were prehypertensive or had stage 1 hypertension [systolic blood pressure (SBP) 120–160 mm Hg and diastolic blood pressure (DBP) 80–95 mm Hg] were eligible for the study. After a 2-week run-in period, 412 participants were randomly assigned to either a DASH diet or a control diet (typical US diet) intervention for 12 weeks (parallel arm design). Within each randomized diet, participants consumed 3 different levels of sodium [high, intermediate, low] in random order for 30 days each (crossover design). The trial was approved by the Institutional Review Boards at each study center, and participants provided written informed consent.
For the present metabolomics study, we used 24-hour urine specimens obtained from the National Health, Lung, and Blood Institute Biologic Specimen and Data Repository Information Coordinator Center (BioLINCC).[18] Among the 412 participants of the DASH-Sodium trial, 17 participants were excluded because biospecimens were unavailable or no informed consent was obtained for further use of biospecimens (Figure 1). Thus, metabolomics data were available in 202 participants from the DASH diet intervention and 193 participants from the control diet intervention. For the present metabolomics study, we conducted two comparisons of the urine metabolome: 1) DASH-high sodium diet (n=199) vs. control-high sodium diet (n=193), and 2) DASH-low sodium diet (n=196) vs. control-high sodium diet (n=193). The first comparison, which tested the DASH diet alone, was used to replicate the comparison in the original DASH trial in which the DASH and control groups had similar sodium intake levels.[2] The second comparison was an assessment of the combined effects of the DASH diet and low sodium compared to a typical American diet (control diet and high sodium). We did not compare metabolites associated with the sodium effect (e.g., DASH-high sodium diet vs. DASH-low sodium diet or DASH-intermediate sodium diet vs. DASH-low sodium diet) because this was reported in an earlier study using data from the DASH-Sodium trial[19] and given the stated the purpose of the current study to focus on urine biomarkers of the overall DASH diet relative to our previous research on blood biomarkers of the overall DASH diet.[7]
Figure 1.
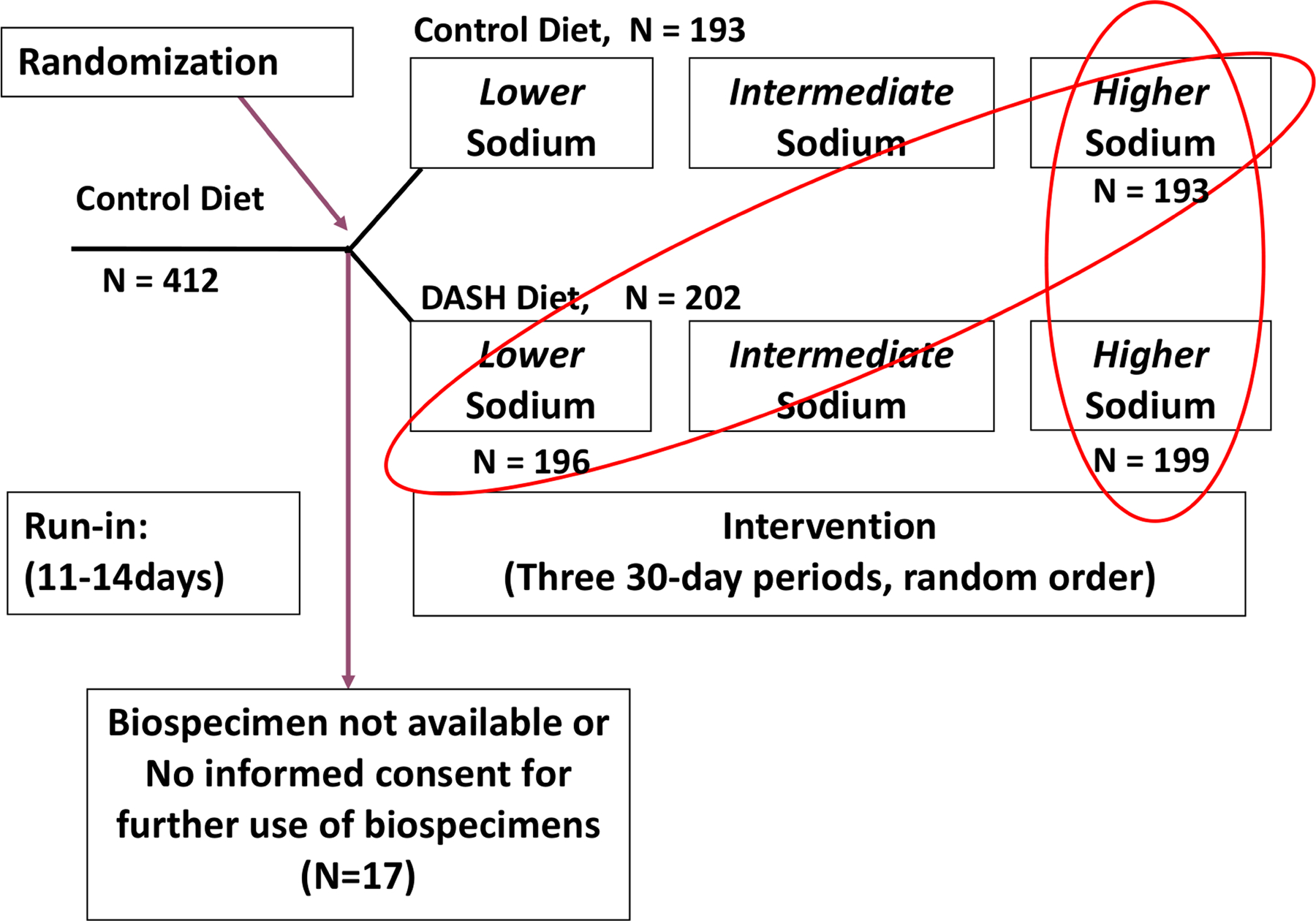
Flow diagram of participant selection
DASH, Dietary Approaches to Stop Hypertension
2.2. Dietary exposures
The DASH diet is high in fruits, vegetables, and low-fat dairy products, and moderate in different protein foods such as poultry, fish, legumes, and nuts. Intakes of red meat, sugar-sweetened beverages, and desserts were limited.[17] Compared to the control diet, the DASH diet had higher carbohydrate and protein as a percentage of energy; and were higher in fiber and micronutrients (potassium, calcium, magnesium, phosphorus, folate, iron, zinc, vitamins), but lower in total fat, specifically saturated fat and cholesterol (Table 1). The control diet was designed to mirror the average macronutrient and fiber intake of American diets, and the 25th percentile of potassium, magnesium, and calcium intake of US consumption.
Table 1.
Nutritional composition of the DASH diets and control diet at the 2100-kcal intake amount
| Control diet | DASH-high sodium | DASH-low sodium | |
|---|---|---|---|
| Energy intake, kcal | 2094 | 2077 | 2090 |
| Carbohydrate, % of total energy | 49 | 58 | 58 |
| Protein, % of total energy | 14 | 18 | 18 |
| Fat, % of total energy | 29 | 21 | 21 |
| Saturated fat, % of total energy | 12 | 5 | 5 |
| PUFAs, % of total energy | 6 | 6 | 6 |
| MUFAs, % of total energy | 10 | 9 | 9 |
| Sodium, mg | 3605 | 3727 | 1226 |
| Potassium, mg | 1741 | 4629 | 4538 |
| Calcium, mg | 455 | 1241 | 1260 |
| Magnesium, mg | 1734 | 478 | 498 |
| Phosphorus, mg | 873 | 1466 | 1662 |
| Fiber, g/1000 kcal | 5 | 14 | 14 |
| Cholesterol, mg/1000 kcal | 128 | 64 | 61 |
| Folate, μg | 154 | 367 | 391 |
| Iron, mg | 16 | 22 | 20 |
| Zinc, mg | 8 | 11 | 12 |
| Vitamin A, IU | 5008 | 14447 | 15754 |
| Thiamin, mg | 1 | 1 | 2 |
| Riboflavin, mg | 1 | 2 | 2 |
| Niacin, mg | 22 | 21 | 24 |
| Pantothenic acid, mg | 3 | 5 | 5 |
| Vitamin B-6, mg | 1 | 3 | 3 |
| Vitamin B-12, μg | 3 | 4 | 4 |
| Vitamin C, mg | 140 | 258 | 300 |
| Vitamin E, mg | 8 | 13 | 14 |
Bolded values indicate nutrients altered to be different between the DASH diet and control diet.
DASH, Dietary Approaches to Stop Hypertension; IU, international units; MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids.
High sodium level was set at 3450 mg, intermediate sodium level was set at 2300 mg, and low sodium level was set at 1150 mg for a diet consisting of 2100 kcal/d. The highest sodium level reflected typical sodium consumption in the US; intermediate sodium level reflected the upper limit of the national guidelines; and low sodium level reflected the amount which was hypothesized to have additional blood pressure lowering benefit.[16] Participants’ total energy intake was adjusted to maintain constant body weight during the study period, and daily sodium amount was proportionate to body size and physical activity. All meals were cooked in research kitchens using standardized menus. Participants received all cooked meals, including snacks. Participants ate lunch or dinner at study sites during weekdays, and received foods to eat for other meals and weekends.
2.3. Metabolomic profiling
Untargeted metabolomic profiling was conducted by Metabolon, Inc. (Durham, North Carolina). Samples were extracted using an automated liquid handling robot (Hamilton Labstar, Hamilton Robotics), and processed by adding recovery standards and precipitating proteins using methanol. The extract was divided into five parts. For analyses, Metabolon used two different reverse phase ultra-high performance liquid chromatography mass spectrometry with a positive ion mode electrospray ionization (ESI; “LC/MS Pos Early”, “LC-MS/MS Pos Late”), another reverse phase ultra-high performance liquid chromatography mass spectrometry with a negative ion mode ESI (“LC/MS Neg”), and a hydrophilic interaction ultra-performance liquid chromatography with negative ion mode ESI (“LC/MS Polar”).[20,21] For liquid chromatography mass spectrometry, a Waters ACQUITY liquid chromatographer and a ThermoFisher Scientific Q-Exactive high resolution mass spectrometer was used with Thermo Scientific Orbitrap mass analyzer. All metabolites had either a “tier 1” level or “tier 2” level of certainty. Except for metabolites with a footnote indicating a lower level of certainty (tier 2), all metabolites were considered to have “tier 1” level of certainty when at least two orthogonal measurements (e.g., retention time, accurate mass, and fragmentation patterns) were matched to an authentic reference using the same methodology.[22,23] Metabolon categorized metabolites to the following chemical classes: amino acids, carbohydrates, cofactors and vitamins, energy, lipids, nucleotides, partially characterized molecules, peptides, and xenobiotics. Participants’ biospecimens were de-identified and laboratory technicians were not aware of randomized diet assignments or sodium interventions. Samples were analyzed in a single batch and were in a random order. For quality control, 20 blind duplicate pairs were included. Blind duplicate samples showed that the metabolomics data are highly reproducible. Spearman’s correlation was ≥0.8 for 91.9% of metabolites, and 81.5% of metabolites had coefficients of variation <20%.
Of 1,425 metabolites identified in this study, we excluded 12 metabolites with >80% missing values. Then, for the remaining metabolites, we imputed missing values at the minimum detectable level for each metabolite. After rescaling each metabolite to a median of 1 by dividing by the batch-specific median and log-transformation (loge), we additionally excluded 44 metabolites with variance <0.01 on log scale. Outliers were capped at ± 5 standard deviations. After excluding 431 unknown metabolites, we analyzed 938 known metabolites.
2.4. Statistical Analysis
We compared baseline characteristics using means [standard deviation (SD)] for continuous variables and proportions for categorical variables according to randomly assigned dietary pattern (DASH diet vs. control diet).
We used three methods to examine if candidate biomarkers of the DASH diet replicated in our study.[7] First, we used multivariable linear regression models to evaluate the associations between diet interventions [DASH-high sodium vs. control-high sodium; DASH-low sodium vs. control-high sodium] and individual metabolites, adjusting for age (5 year increments), sex (men/women), race (African American/non-African American), total energy intake (continuous), body mass index (BMI) (continuous), income (<$29,999, $30,000-$59,999, ≥$60,000), and 24-hour urine creatinine (continuous). We adjusted for these baseline covariates to be consistent with the original DASH metabolomics study and to improve precision. Urine creatinine level was adjusted as a covariate to account for inter-individual differences in urine dilution levels.[24] We used Bonferroni-adjusted P values to account for the large number of statistical tests conducted (0.05/938 metabolites = 5.33 × 10−5). Then, we assessed if any of these significant metabolites overlapped with the significant metabolites identified in the DASH feeding study.[7]
Second, we aimed to assess if any of the 10 serum metabolites which were highly influential in discriminating between the DASH and control diets replicated as top 10 urine biomarkers of the DASH diet in our study, and to identify additional novel biomarkers of the DASH diets. We selected the top 10 metabolites because we identified the top 10 serum metabolites in the original DASH trial. Further, in the present study, more than 100 metabolites were associated with the two DASH diets compared to the control-high sodium diet even after Bonferroni adjustment. We narrowed down the significant metabolites to a reasonable number so that they may be more feasible to assess as biomarkers of the DASH diet in the future. We conducted partial least-squares discriminant analysis (PLS-DA) to detect the top 10 metabolites that represent the DASH-high sodium diet and DASH-low sodium diet compared to the control-high sodium diet. Variable importance in projection (VIP) scores from PLS-DA, which estimate the importance of each metabolite in predicting the DASH diets relative to the control-high sodium diet, were used to determine the top 10 influential metabolites. We validated all PLS-DA models using permutation testing, and the results from permutation testing showed that models were at low risk of overfitting (P <0.05). To assess the ability of the top 10 metabolites to predict DASH dietary patterns, we calculated C-statistics by building logistic regression models with incremental addition of each of the top 10 metabolites and diet interventions as the outcome. We calculated C-statistics in a random sample of two-thirds of the population (testing sample), and then validated in the other one-third of the sample (validation sample). As a secondary analysis, we examined the top 15 metabolites of the two DASH diets in our study to assess if more serum biomarkers of the DASH diet replicated as influential urine metabolites in the present study.
Third, we calculated C-statistics again using the top 10 serum candidate biomarkers identified in the original DASH trial.[7] We did not include methyl glucopyranoside (α + β) and β-cryptoxanthin for this analysis, because these two metabolites were not available in the DASH-Sodium metabolomics dataset.
All analyses were conducted in R version 3.6.2 (R Foundation for Statistical Computing, Vienna, Austria) and Stata version 15.0 (StataCorp, College Station, Texas).
3. RESULTS
More than half of the analytic study population was women and African Americans (Table 2). More than 40% of the participants were college graduates and had hypertension. Baseline characteristics were similar by DASH diet and control diet, except for income. Participants assigned to the DASH diet were more likely to have an income ≥$60,000. This variable was included in the multivariable linear regression models as a covariate.
Table 2.
| DASH diet (N=202) | Control diet (n=193) | |
|---|---|---|
| Age category, n (%) | ||
| 18-<30y | 6 (3.0) | 6 (3.1) |
| 31–55y | 158 (78.2) | 136 (70.4) |
| ≥56y | 38 (18.8) | 51 (26.4) |
| Women, n (%) | 119 (58.9) | 104 (53.9) |
| African Americans, n (%) | 115 (56.9) | 111 (57.5) |
| Income, n (%) | ||
| <$29,999 | 61 (30.8) | 64 (34.0) |
| $30,000-$59,999 | 66 (33.3) | 78 (41.5) |
| ≥$60,000 | 71 (35.9) | 46 (24.5) |
| Education, n (%) | ||
| High school graduate or less | 26 (12.9) | 38 (19.7) |
| Some college | 83 (41.3) | 62 (32.1) |
| College graduate | 47 (23.4) | 46 (23.8) |
| Post-graduate | 45 (22.4) | 47 (24.4) |
| Current smoker, n (%) | 21 (31.0) | 21 (27.0) |
| Total energy intake, kcal | 2614.7 (476.3) | 2634.3 (449.0) |
| BMI, kg/m2 | 28.9 (4.6) | 29.5 (4.9) |
| Weight, kg | 82.8 (14.6) | 85.6 (15.8) |
| SBP, mm Hg | 134.2 (9.6) | 135.3 (9.3) |
| DBP, mm Hg | 85.6 (4.8) | 85.7 (4.1) |
| Hypertension status2 | 92 (45.5) | 86 (44.6) |
| 24-hour urinary creatinine, g | 1.4 (0.5) | 1.5 (0.5) |
We compared baseline characteristics of participants according to randomly assigned dietary pattern (DASH diet vs. control diet). Values are n (%) for categorical variables and mean (standard deviation) for continuous variables.
P-value was >0.05 for all characteristics except for income.
SBP ≥140 mm Hg and/or DBP≥ 90 mmHg
BMI, body mass index; DASH, Dietary Approaches to Stop Hypertension; DBP, diastolic blood pressure; SBP, systolic blood pressure.
Detailed information on significant metabolites are provided in Supporting Information Table S1. Out of 938 metabolites, urine levels of 153 metabolites (excluding partially characterized molecules) were significantly different between participants assigned to the DASH-high sodium diet compared to those assigned to the control-high sodium diet (Supporting Information Table S2), after adjusting for participant characteristics. Of 153 metabolites, 40 were in the food component/plant pathway. Urine levels of 157 metabolites were significantly different for participants assigned to the DASH-low sodium diet compared to those assigned to the control-high sodium diet (Supporting Information Table S3). Of the 157 metabolites, 31 were in the food component/plant pathway. With the exception of methyl glucopyranoside (α + β) and β-cryptoxanthin, which were not detected in our sample, the top 10 candidate biomarkers identified in serum samples from the original DASH trial replicated in urine samples from the DASH-Sodium trial: N-methylproline, stachydrine, tryptophan betaine, theobromine, 7-methylurate, chiro-inositol, 3-methylxanthine, and 7-methylxanthine. Of the 97 serum metabolites significantly different for participants assigned to the DASH diet and those assigned to the control diet in the original DASH feeding study, 17 (out of 153 metabolites) and 18 (out of 157 metabolites) in the DASH-high sodium diet and DASH-low sodium diet, respectively, were significant in urine and were similar in terms of direction and magnitude in the DASH-Sodium trial (see Supporting Information Tables S2 and S3 for overlapping metabolites).
The number of significant metabolites in each metabolite category was similar for the DASH-low sodium diet and DASH-high sodium diets relative to the control-high sodium diet (Figure 2). The most common metabolite category was xenobiotics, which includes food components and plants (60 metabolites for the DASH-high sodium diet and 52 metabolites for the DASH-low sodium diet). Several xenobiotics and amino acids were positively associated with DASH-high sodium and DASH-low sodium diets and had small P-values. For instance, urine levels of stachydrine (a xenobiotic) and N-methylglutamate (an amino acid) were higher among participants assigned to the DASH-high sodium diet or the DASH-low sodium diet compared to those assigned to the control-high sodium diet and these two metabolites had the smallest P-values (P-values <1.16 × 10−65 for both). The majority of lipids, specifically those involved in carnitine metabolism or acyl carnitine metabolism, were inversely associated with the DASH diets (28 out of 36 lipids in the DASH-high sodium diet; and 26 out of 32 lipids in the DASH-low-sodium diet) (Figure 3). In general, slightly stronger associations were observed for the DASH-low sodium diet than DASH-high sodium diet. Thirty-four and 42 unique metabolites were observed for the DASH-high sodium diet and DASH-low sodium diet, respectively. For example, 4 peptides in the γ-glutamyl amino acid pathway were unique to the DASH-low sodium diet, whereas no peptides were unique to the DASH-high sodium diet.
Figure 2.
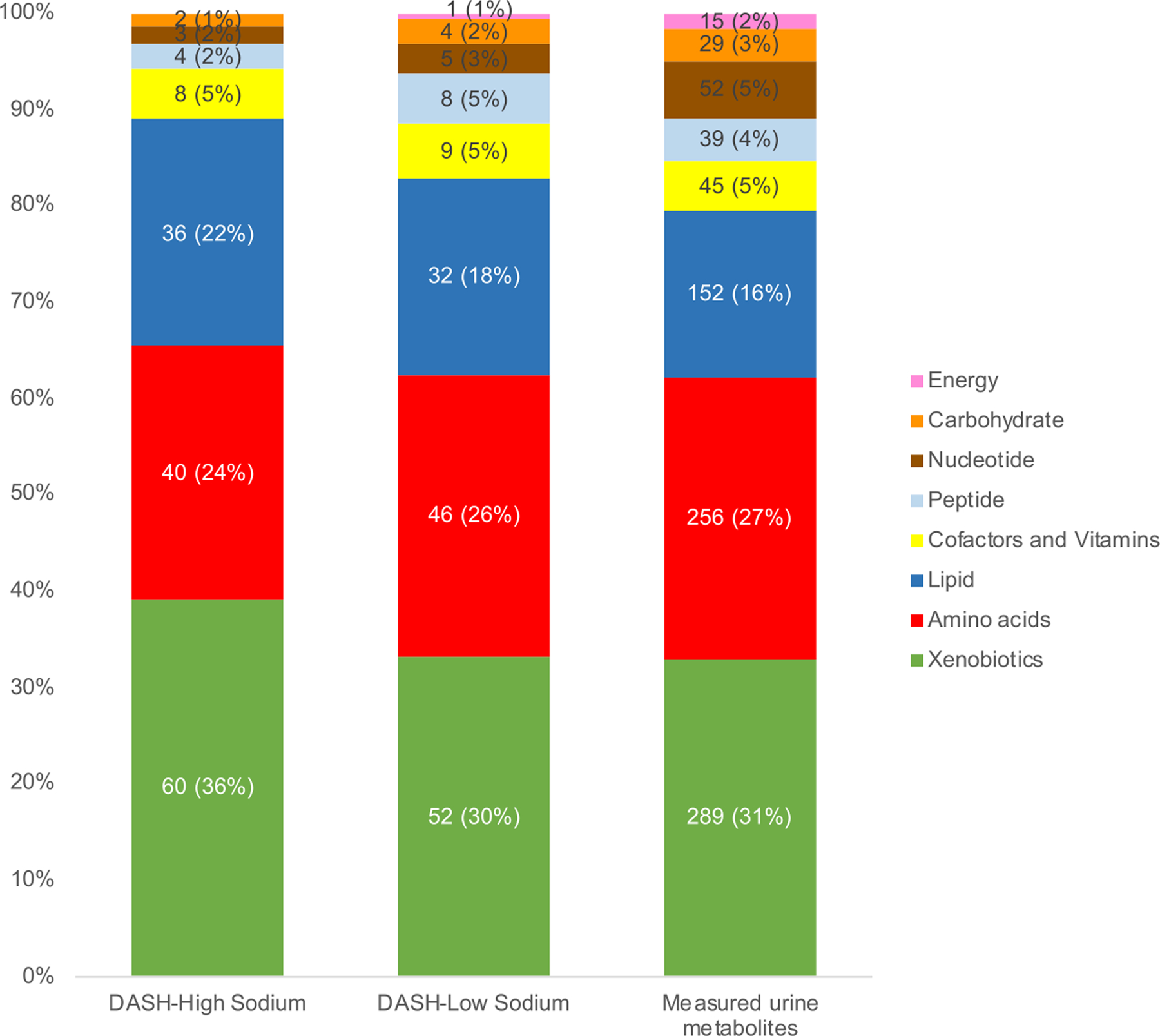
Number and percentages of metabolites by metabolite categories significantly different between the DASH diets and the control-high sodium diet
Numbers within the graph represents number of metabolites (%). “Measured urine metabolites” represents the distribution of all measured metabolites in our study.
DASH, Dietary Approaches to Stop Hypertension
Figure 3.
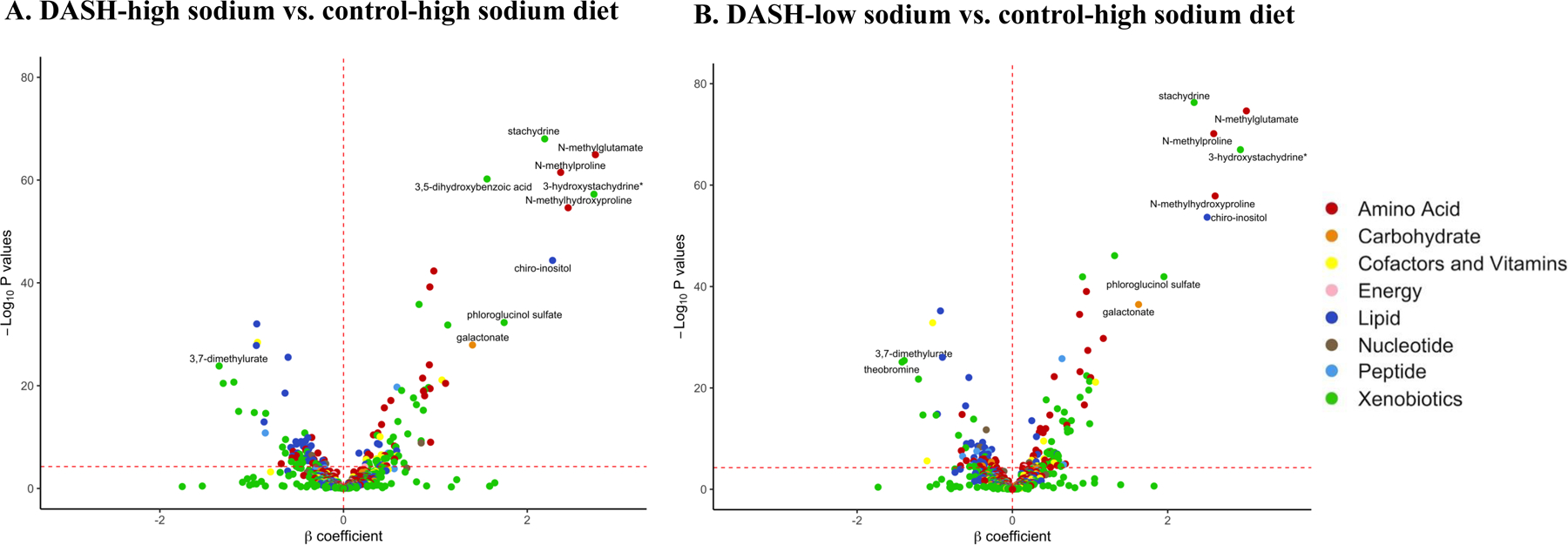
Volcano plots of P-values and β coefficients for the association between individual metabolites and DASH diets
The red horizontal dashed line represents the Bonferroni-adjusted threshold (5.33 × 10−5) and the red vertical dashed line is set at β coefficient=0. Positive β coefficients (to the right of the red vertical dashed line) indicate that the level of metabolites was higher in individuals randomly assigned to DASH diet relative those assigned to the control diet. Negative β coefficients (to the left of the red vertical dashed line) indicate that the level of metabolites was lower in individuals randomly assigned to DASH diet relative those assigned to the control diet. The top 10 metabolites representative of the DASH-high sodium diet and DASH-low sodium diet relative to the control-high sodium diet are labeled.
* Metabolites that are not officially confirmed based on a standard.
In the biplot, there was a clear differentiation in the urine metabolome between the DASH diets and the control diet in the overall study population (Figure 4). The first two components explained 36.7% of the variance for the DASH-high sodium diet and 36.8% of variance for the DASH-low diet. The differentiation was also clear when we examined the urine metabolome by sex (Supporting Information Figure S1) and race (Supporting Information Figure S2). The first two components explained a similar proportion of the variance for the DASH dietary patterns in sex and race subgroups relative to the overall study population.
Figure 4.
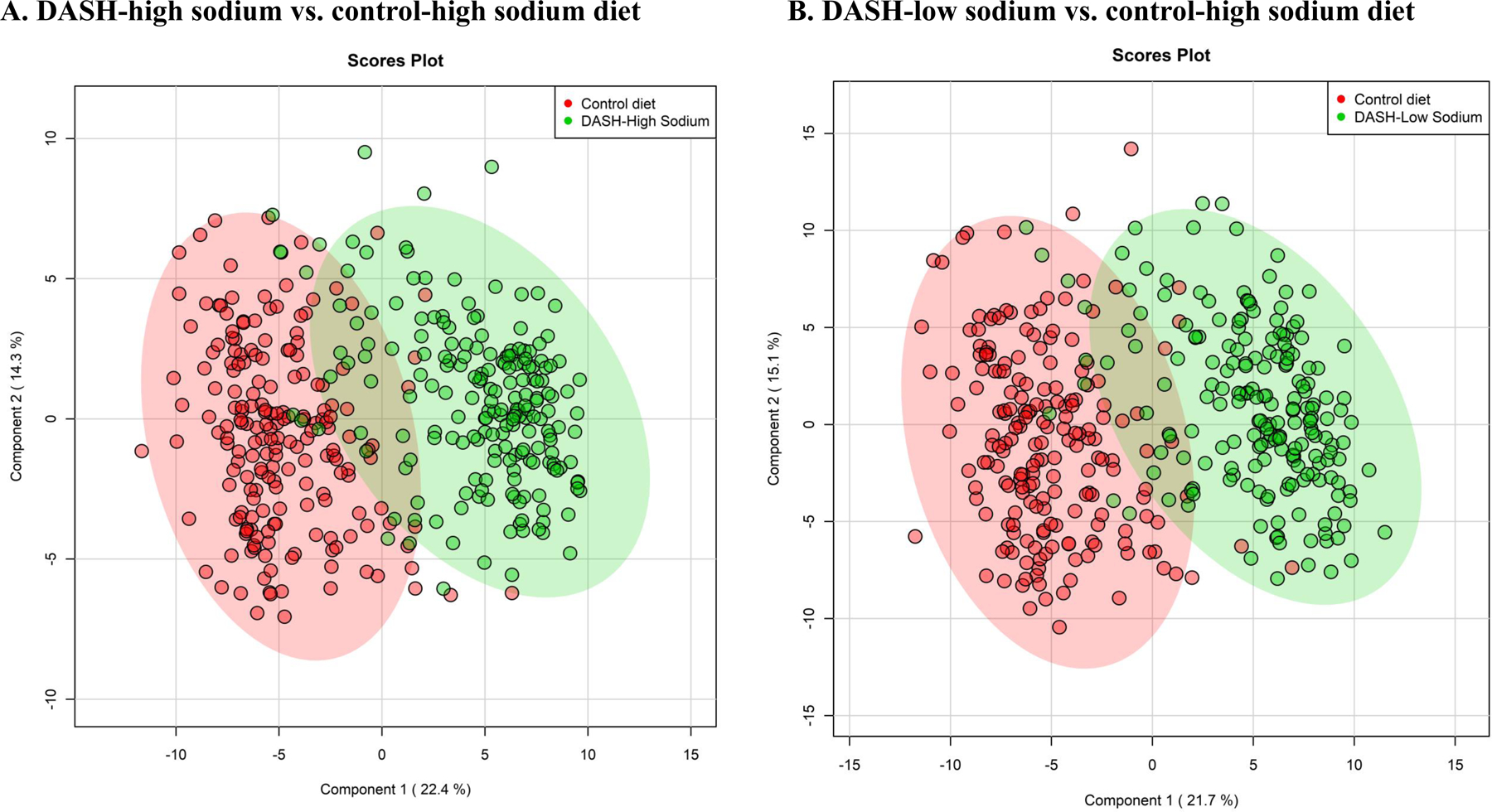
Scores plot derived from principal component analysis of significant metabolites for the DASH diets and control-high sodium diet
Plots were created from partial least squares-discriminant analysis.
DASH, Dietary Approaches to Stop Hypertension
Nine out of the top 10 metabolites representing each of the DASH diets were identical (N-methylglutamate, N-methylhydroxyproline, 3-hydroxystachydrine, N-methylproline, chiro-inositol, stachydrine, phloroglucinol sulfate, galactonate, and 3,7-dimethylurate). and the order of these influential metabolites determined by VIP scores were similar between the two DASH diets (Table 3). Urine levels of the majority of metabolites were higher among participants assigned to the DASH diets relative to the control-high sodium diet, whereas levels of 3,7-dimethylurate and theobromine were lower among participants assigned to the DASH diets relative to the control-high sodium diet (Figure 3).
Table 3.
Top 10 influential metabolites that distinguish the DASH diets relative to the control diet1
| DASH-high sodium diet relative to control-high sodium diet | DASH-low sodium diet relative to control-high sodium diet | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Superpathway | Metabolite | β | P-value | VIP | C-statistics in the testing sample | C-statistics in the validation sample | β | P-value | VIP | C-statistics in the testing sample | C-statistics in the validation sample |
| Amino Acid | N-methylglutamate | 2.74282 | 1.15 × 10−65 | 3.65 | 0.918 | 0.943 | 3.01342 | 2.39 × 10−75 | 3.87 | 0.946 | 0.952 |
| Amino Acid | N-methylhydroxyproline | 2.44555 | 2.43 × 10−55 | 3.26 | 0.898 | 0.910 | 2.61079 | 1.40 × 10−58 | 3.36 | 0.911 | 0.912 |
| Xenobiotics | 3-hydroxystachydrine* | 2.72683 | 5.67 × 10−58 | 3.18 | 0.897 | 0.925 | 2.93629 | 1.05 × 10−67 | 3.35 | 0.940 | 0.929 |
| Amino Acid | N-methylproline† | 2.36431 | 3.18 × 10−62 | 3.15 | 0.912 | 0.933 | 2.59277 | 7.35 × 10−71 | 3.35 | 0.939 | 0.945 |
| Lipid | chiro-inositol†2 | 2.27567 | 4.26 × 10−45 | 2.98 | 0.864 | 0.879 | 2.50780 | 2.20 × 10−54 | 3.19 | 0.891 | 0.905 |
| Xenobiotics | stachydrine† | 2.19013 | 9.55 × 10−69 | 2.92 | 0.933 | 0.944 | 2.34008 | 5.19 × 10−77 | 3.01 | 0.953 | 0.954 |
| Xenobiotics | 3,5-dihydroxybenzoic acid3 | 1.56259 | 6.09 × 10−61 | 1.97 | 0.920 | 0.898 | — | — | — | — | — |
| Xenobiotics | phloroglucinol sulfate | 1.74828 | 5.26 × 10−33 | 1.86 | 0.830 | 0.882 | 1.95133 | 1.15 × 10−42 | 2.13 | 0.903 | 0.866 |
| Carbohydrate | galactonate | 1.40436 | 1.24 × 10−28 | 1.82 | 0.808 | 0.849 | 1.62469 | 3.54 × 10−37 | 2.04 | 0.878 | 0.843 |
| Xenobiotics | 3,7-dimethylurate | −1.35530 | 1.46 × 10−24 | 1.72 | 0.795 | 0.824 | −1.39313 | 4.06 × 10−26 | 1.64 | 0.809 | 0.867 |
| Xenobiotics | theobromine†3 | — | — | — | — | — | −1.42144 | 8.04 × 10−26 | 1.62 | 0.805 | 0.875 |
| Overall C-statistics | 0.981 | 0.977 | 0.975 | 0.968 | |||||||
β coefficient modeling loge(metabolite) and P-values were derived from multivariable linear regression models. Variable Importance in Projection (VIP) scores were derived from partial least-squares discriminant analysis (PLS-DA). C-statistics were calculated by building logistic regression models with incremental addition of each of the individual metabolites and diet interventions as the outcome. The overall C-statistic was calculated after adding the top 10 influential metabolites to the model.
Chiro-inositol may be classified as belonging to a different class of metabolites (superpathway) in studies not using the Metabolon platform (e.g., cyclohexanol / polyol of sugar).
3,5-dihydroxybenzoic acid was not one of the top 10 influential metabolites that distinguished the DASH-low sodium diet relative to control-high sodium diet. Theobromine was not one of the top 10 influential metabolites that distinguished the DASH-high sodium diet relative to control-high sodium diet.
Metabolites that are not officially confirmed based on a standard.
Metabolites that were reported as the top 10 highly influential metabolites in discriminating between the DASH and control diets in the original DASH trial [7].
DASH, Dietary Approaches to Stop Hypertension
Among the top 10 influential metabolites, N-methylproline, chiro-inositol, stachydrine, and theobromine were also among the top 10 metabolites in the original DASH feeding study. Other influential urine metabolites (N-methylglutamate, 3,5-dihydroxybenzoic acid, phloroglucinol sulfate, galactonate) were identified as novel biomarkers of the DASH diet in the present study. Two additional candidate biomarkers from the original DASH trial (3-methylxanthine, tryptophan betaine) replicated in our study in the top 15 influential metabolites (Supporting Information Table S4). L-urobilin, one of the top 15 influential metabolites for the DASH-low sodium diet which was unique to this diet, had a negative coefficient, meaning that L-urobilin levels were lower on the DASH-low sodium diet vs. the control-high sodium diet.
C-statistics for the models which included the top 10 influential metabolites were highly predictive of DASH-high sodium diet (0.981 in the testing sample; 0.977 in the validation sample) and DASH-low sodium diet (0.975 in the testing sample; 0.968 in the validation sample) (Table 3). For individual metabolites, C-statistics were the highest for stachydrine for the two DASH diets (range=0.944–0.945) and lowest for 3,7-dimethylurate (C-statistic=0.824) for the DASH-high sodium diet and galactonate for the DASH-low sodium diet (C-statistic=0.843) in the validation sample. When we used candidate biomarkers from the original DASH feeding study instead of our top 10 metabolites, C-statistics in the testing sample were 0.956 for the DASH-high sodium diet and 0.966 for the DASH-low sodium diet; and were 0.958 in the validation sample for the DASH-high-sodium diet, and 0.980 for the DASH-low sodium diet.
4. DISCUSSION
In the DASH-Sodium trial, a feeding study of adults with similar characteristics to the original DASH trial, we replicated 8 urine metabolites which were the most influential in distinguishing the DASH diet and control diet in the original DASH feeding study, after adjusting for participant characteristics and accounting for multiple comparisons. Additionally, we identified several novel biomarkers of the DASH diet through PLS-DA (N-methylglutamate, 3,5-dihydroxybenzoic acid, phloroglucinol sulfate, and galactonate), and found that nine out of the top 10 urine metabolites which represented the DASH-high sodium diet and the DASH-low sodium diet were identical. N-methylproline, chiro-inositol, stachydrine, theobromine, 3-methylxanthine, and tryptophan betaine which were identified as the most predictive serum metabolites of the DASH diet in the original DASH feeding study replicated as influential urine metabolites of DASH diets in the present study. Considering the similarities we observed in these metabolites, our results reveal that serum and urinary samples may be interchangeable for characterizing the nutritional metabolome.[25]
Comparison of findings from the DASH-Sodium trial to the original DASH feeding trial
The DASH-Sodium trial was an ideal dataset to conduct a replication study of the original DASH trial given the similarities in study design. The original DASH study recruited participants who were prehypertensive or had stage 1 hypertension, randomly assigned participants to either the DASH diet or control diet, and participants followed their assigned diets for 8 weeks.[26] This prior study reported that 97 serum metabolites were significantly different between the DASH diet and control diet,[7] and our study found 153 urine metabolites that were significantly different between the DASH-high sodium diet and control-high sodium diet and 157 urine metabolites that were significantly different between the DASH-low sodium diet and control-high sodium diet. Using multivariate methods, 6 of the top 10 serum metabolites of the DASH diet replicated as influential urine metabolites in the DASH-high sodium diet and the DASH-low sodium diet in the DASH-Sodium trial. These results suggest that these 6 compounds (representing amino acids, food components and other xenobiotics, a lipid) are highly likely to be objective biomarkers of the DASH dietary pattern in both serum and urine, and may be used to improve dietary assessment. The direction of the association was the same in serum and urine specimens (e.g., high in serum is high in urine), suggesting higher intake and excretion of metabolites.
Comparison of findings from the DASH-Sodium trial to other studies
Several metabolites (stachydrine, N-methylproline, and carnitine), which have been associated with the DASH dietary patterns in our study, were reported as potential biomarkers of healthy dietary patterns in prior studies. Serum stachydrine (also known as proline betaine) and N-methylproline were positively associated with greater adherence to healthy dietary patterns (Healthy Eating Index, DASH diet, alternate Mediterranean diet index, lower dietary acid load) in observational studies.[12,27–29] In a controlled feeding setting, urine levels of stachydrine increased after 19 healthy individuals consumed a diet with the highest concordance to the World Health Organization (WHO) healthy eating guidelines (higher intakes of whole grains, fruits, vegetables, dietary fiber, and lower intakes of total fat, sugar, and salt) for 72 hours.[30] In contrast, urine levels of carnitine, which has been linked to red meat intake, decreased when these individuals consumed a diet with the least concordance to the WHO guidelines.[30] However, other than these 3 metabolites, we did not find much overlap with other urine metabolomics studies focused on dietary patterns.[31–33] This may be due to differences in platforms utilized for metabolomic profiling [Metabolon (LC/MS) in our study vs. proton nuclear magnetic resonance spectroscopic profiling in other studies], differences in study populations (individuals with elevated blood pressure vs. healthy individuals), differences in dietary patterns (DASH vs. WHO guidelines), or simply due to a paucity of metabolomics research on dietary patterns.
Novel biomarkers of the DASH diet
We identified additional urine metabolites that may be novel biomarkers of the DASH diet. For example, N-methylglutamate, 3,5-dihydroxybenzoic acid, phloroglucinol sulfate, and galactonate were influential metabolites that were predictive of the DASH-high sodium diet and DASH-low sodium diet relative to the control-high sodium diet. N-methylglutamate is classified as glutamic acid or derivatives of glutamic acids, a common dietary amino acid that is rich in plant protein.[34,35] Glutamic acid has been shown to serve as a substrate for glutathione and arginine, both of which can improve bioavailability of nitric oxide, a vasodilator.[36,37] This suggests that glutamate metabolism may be a pathway through which the DASH diet decreases blood pressure. In the Navy Colon Adenoma Study, serum N-methylglutamate was associated with self-reported citrus and juice intake.[25] 3,5-dihydroxybenzoic acid is a metabolite of alkylresorcinols, phenolic compounds found in cereals or whole grain breads, and has been reported to result from microbial transformation of dietary polyphenols.[38,39] Feeding studies have found that urine levels of 3,5-dihydroxybenzoic acid increase after whole wheat or rye consumption, and this metabolite has been proposed as a biomarker of whole grain intake.[40] One study suggested that diets which include 3,5-dihydroxybenzoic acid may be helpful for controlling dyslipidemia.[41] Phloroglucinol sulfate is a metabolite of phlorotannin, a polyphenolic compound, and, similar to 3,5-dihydroxybenzoic acid, is hypothesized to be produced from microbial degradation of polyphenols.[42] Phloroglucinol sulfate in urine has been most frequently detected in biological samples after seaweed intake, but has also been detected after consuming other foods such as grapes, beans, and lentils.[42–44] Galactonate is derived from galactose, a monosaccharide that is found in dairy, syrups, and small amounts in pulses and seeds.[45] Serum concentration of galactonate was positively associated with the DASH diet in the original DASH feeding study and was identified as one of the top 5 important metabolites which distinguished higher vs. lower adherence to the DASH diet using food frequency questionnaire data from the Cancer Prevention Study-II Nutrition Cohort.[7,11] Although these metabolites were associated with consumption of individual foods, to our knowledge, our study is the first to identify N-methylglutamate, 3,5-dihydroxybenzoic acid, and phloroglucinol sulfate as potential urine biomarkers of the DASH diet. It would be useful to determine if these compounds are representative of the DASH diet in other study populations.
Metabolomic markers of sodium intake
The top 10 influential metabolites representative of the DASH-high sodium and DASH-low sodium diets were nearly identical, and the first 6 influential metabolites were the same. This observation was not unexpected because, in contrast to the control-high sodium diet, the DASH-high sodium and DASH-low sodium diets were similar in terms of most nutrients and foods, with the exception of sodium.[2] Metabolites in the γ-glutamyl amino acid pathway were different, and lower in the DASH-low sodium diet relative to the control-high sodium diet. This finding is consistent with a prior report that investigated differences in the serum metabolomic profile between the high- and intermediate-sodium phase and between the high- and low-sodium phase among those randomly assigned to the DASH diet.[19] Lower levels of metabolites in the γ-glutamyl amino acid pathway were identified during the DASH-low sodium phase. Based on these results, it was hypothesized that lower sodium intake may reduce γ-glutamyltransferase (a protein which plays an important role in glutathione metabolism) by lowering oxidative stress, and this may be a potential mechanism by which sodium reduction resulted in lower blood pressure. This previous study also hypothesized that sodium intake may have an effect on the gut microbiome given that serum levels of several metabolites (e.g.,4-ethylphenylsulfate, indole-related metabolites) produced by the gut microflora changed with reduced sodium intake.[19]
L-urobilin was one of the top 15 influential metabolites that was unique to the DASH-low sodium diet. Urine levels of L-urobilin were lower among individuals during the DASH-low sodium phase compared to the control diet-high sodium diet phase, consistent with a prior report.[19] Sodium intake may affect L-urobilin, because L-urobilin levels increase with lack of water intake.[34] A study using data from the DASH-Sodium trial found that sodium reduction decreased thirst and urine volume (a proxy for fluid intake) in individuals assigned to the control diet intervention whereas no change was observed for those assigned to the DASH diet intervention.[46] This suggests that less fluid intake may have increased L-urobilin among those in the control diet relative to the DASH diet. However, it is unclear if this metabolite can be considered a biomarker of sodium reduction, because water or fluid intake was not assessed in the DASH-Sodium trial, and other factors such as liver dysfunction and gut microorganisms can influence levels of urobilin.[47,48]
Strengths and limitations
The strengths of our study come from the study design. We used stored urine samples from a well-designed and carefully conducted feeding study. In the DASH-Sodium trial, participants received all meals and ate one of their meals onsite on weekdays. Thus, our data improves upon a prior metabolomics study which used self-reported data to identify biomarkers of the DASH dietary pattern.[11] Next, all identified metabolites were confirmed using authentic standards. In addition, participants were randomly assigned to either the DASH diet or control diet, which minimized confounding. We adjusted for socio-demographic and clinical factors in multivariable linear regressions models to be consistent with the original DASH trial, address residual confounding, and improve precision. Lastly, the DASH-Sodium trial was designed to be generalizable to US adults, considering that approximately half of the study population consisted of individuals who were women and minority race.
Several limitations should be considered. The sample size was modest, with approximately 400 participants. Degradation of metabolites is possible given that biospecimens were stored at −80°C for over 20 years. However, it is likely any degradation of metabolites that did occur was similar for the diet interventions. In the DASH-Sodium trial, the intervention period was relatively short (30 days for each intervention period); thus, we were unable to address issues related to dietary intake over a longer period of time. However, on the basis of the metabolites identified, there is no basis to expect that the intervention period was inadequate to assess the metabolomic fingerprint of the DASH diet.
In conclusion, most candidate biomarkers of the DASH diet identified in serum in the original DASH feeding study replicated in urine in the present study, and several metabolites (N-methylproline, chiro-inositol, stachydrine, theobromine, 3-methylxanthine, tryptophan betaine) replicated as predictive metabolites of the DASH diet. These results highlight the possibility that serum and urine metabolites may offer similar information in nutritional metabolomics. Further, our results suggest that these replicated metabolites are highly likely to be biomarkers of the DASH diet. Next, we must capitalize on these consistent associations to improve the measurement of diet in more generalizable settings and understand the physiologic implications of metabolomic alterations associated with the DASH diet which mediate the salutary effects of the DASH diet.
Supplementary Material
ACKNOWLEDGEMENTS / FUNDING
Dr. Rebholz was supported by a Mentored Research Scientist Development Award from the National Institute of Diabetes and Digestive and Kidney Diseases (K01 DK107782). This research was funded by grants from the National Heart, Lung, and Blood Institute (R21 HL143089, R56 HL153178). The funding agencies had no role in study design, data collection, analysis, drafting of the manuscript, and the decision to submit this manuscript for publication.
Abbreviations:
- DBP
diastolic blood pressure
- DASH
Dietary Approaches to Stop Hypertension
- PLS-DA
partial least-squares discriminant analysis
- SBP
systolic blood pressure
- VIP
variable importance in projection
Footnotes
Clinical Trial Registration: https://clinicaltrials.gov/ct2/show/NCT00000608; Unique identifier: NCT00000608
DATA SHARING AND DATA ACCESSIBILITY
The DASH-Sodium trial data are available through the National Heart, Lung and Blood Institute (NHLBI) Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC): https://biolincc.nhlbi.nih.gov/studies/dashsodium/. The remaining metabolomics data that support the results of the present study are available from the corresponding author upon reasonable request.
CONFLICT OF INTEREST
The authors have no conflicts of interests to disclose.
6. REFERENCES
- [1].US Department of Health and Human Services, US Department of Agriculture, Dietary Guidelines for Americans, 2015–2020, US Government Printing Office, Washington, DC: 2015. [Google Scholar]
- [2].Appel LJ, Moore TJ, Obarzanek E, Vollmer WM, Svetkey LP, Sacks FM, Bray GA, Vogt TM, Cutler JA, Windhauser MM, Lin P-H, Karanja N, Simons-Morton D, McCullough M, Swain J, Steele P, Evans MA, Miller ER, Harsha DW, N Engl J Med 1997, 336, 1117–1124. [DOI] [PubMed] [Google Scholar]
- [3].Fung TT, Chiuve SE, McCullough ML, Rexrode KM, Logroscino G, Hu FB, Arch. Intern. Med 2008, 168, 713–720. [DOI] [PubMed] [Google Scholar]
- [4].Rebholz CM, Crews DC, Grams ME, Steffen LM, Levey AS, Miller III ER, Appel LJ, Coresh J, Am J Kidney Dis 2016, 68, 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schwingshackl L, Bogensberger B, Hoffmann G, J Acad Nutr Diet 2018, 118, 74–100.e11. [DOI] [PubMed] [Google Scholar]
- [6].Soltani S, Arablou T, Jayedi A, Salehi-Abargouei A, Nutrition Journal 2020, 19, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rebholz CM, Lichtenstein AH, Zheng Z, Appel LJ, Coresh J, Am J Clin Nutr 2018, 108, 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Freedman LS, Schatzkin A, Midthune D, Kipnis V, J Natl Cancer Inst 2011, 103, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tzoulaki I, Ebbels TMD, Valdes A, Elliott P, Ioannidis JPA, Am J Epidemiol 2014, 180, 129–139. [DOI] [PubMed] [Google Scholar]
- [10].Hernández‐Alonso P, Papandreou C, Bulló M, Ruiz‐Canela M, Dennis C, Deik A, Wang DD, Guasch‐Ferré M, Yu E, Toledo E, Razquin C, Corella D, Estruch R, Ros E, Fitó M, Arós F, Fiol M, Serra‐Majem L, Liang L, Clish CB, Martínez‐González MA, Hu FB, Salas‐Salvadó J, Molecular Nutrition & Food Research 2019, 63, 1900140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McCullough ML, Maliniak ML, Stevens VL, Carter BD, Hodge RA, Wang Y, Am J Clin Nutr 2019, 109, 1439–1451. [DOI] [PubMed] [Google Scholar]
- [12].Playdon MC, Ziegler RG, Sampson JN, Stolzenberg-Solomon R, Thompson HJ, Irwin ML, Mayne ST, Hoover RN, Moore SC, Am J Clin Nutr 2017, 106, 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mazzilli KM, McClain KM, Lipworth L, Playdon MC, Sampson JN, Clish CB, Gerszten RE, Freedman ND, Moore SC, J Nutr 2019, DOI 10.1093/jn/nxz300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guasch-Ferré M, Zheng Y, Ruiz-Canela M, Hruby A, Martínez-González MA, Clish CB, Corella D, Estruch R, Ros E, Fitó M, Dennis C, Morales-Gil IM, Arós F, Fiol M, Lapetra J, Serra-Majem L, Hu FB, Salas-Salvadó J, Am J Clin Nutr 2016, 103, 1408–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jenab M, Slimani N, Bictash M, Ferrari P, Bingham SA, Hum Genet 2009, 125, 507–525. [DOI] [PubMed] [Google Scholar]
- [16].Sacks FM, Svetkey LP, Vollmer WM, Appel LJ, Bray GA, Harsha D, Obarzanek E, Conlin PR, Miller ER, Simons-Morton DG, Karanja N, Lin PH, DASH-Sodium Collaborative Research Group, N Engl J Med 2001, 344, 3–10. [DOI] [PubMed] [Google Scholar]
- [17].Svetkey LP, Sacks FM, Obarzanek E, Vollmer WM, Appel LJ, Lin PH, Karanja NM, Harsha DW, Bray GA, Aickin M, Proschan MA, Windhauser MM, Swain JF, McCarron PB, Rhodes DG, Laws RL, J Am Diet Assoc 1999, 99, S96–104. [DOI] [PubMed] [Google Scholar]
- [18].Giffen CA, Carroll LE, Adams JT, Brennan SP, Coady SA, Wagner EL, Biopreserv Biobank 2015, 13, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Derkach A, Sampson J, Joseph J, Playdon MC, Stolzenberg-Solomon RZ, Am J Clin Nutr 2017, 106, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E, Anal. Chem 2009, 81, 6656–6667. [DOI] [PubMed] [Google Scholar]
- [21].Ford L, Kennedy AD, Goodman KD, Pappan KL, Evans AM, Miller LAD, Wulff JE, Wiggs BR, Lennon JJ, Elsea S, Toal DR, J Appl Lab Med 2020, 5, 342–356. [DOI] [PubMed] [Google Scholar]
- [22].Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR, Metabolomics 2007, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA, J Am Soc Mass Spectrom 2016, 27, 1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barr DB, Wilder LC, Caudill SP, Gonzalez AJ, Needham LL, Pirkle JL, Environ. Health Perspect 2005, 113, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Playdon MC, Sampson JN, Cross AJ, Sinha R, Guertin KA, Moy KA, Rothman N, Irwin ML, Mayne ST, Stolzenberg-Solomon R, Moore SC, Am J Clin Nutr 2016, 104, 776–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sacks FM, Obarzanek E, Windhauser MM, Svetkey LP, Vollmer WM, McCullough M, Karanja N, Lin PH, Steele P, Proschan MA, Ann Epidemiol 1995, 5, 108–118. [DOI] [PubMed] [Google Scholar]
- [27].Playdon MC, Moore SC, Derkach A, Reedy J, Subar AF, Sampson JN, Albanes D, Gu F, Kontto J, Lassale C, Liao LM, Männistö S, Mondul AM, Weinstein SJ, Irwin ML, Mayne ST, Stolzenberg-Solomon R, Am J Clin Nutr 2017, 105, 450–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rebholz CM, Surapaneni A, Levey AS, Sarnak MJ, Inker LA, Appel LJ, Coresh J, Grams ME, J Nutr 2019, 149, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim H, Hu EA, Wong K, Yu B, Steffen LM, Seidelmann SB, Boerwinkle E, Coresh J, Rebholz CM, J Nutr in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Garcia-Perez I, Posma JM, Gibson R, Chambers ES, Hansen TH, Vestergaard H, Hansen T, Beckmann M, Pedersen O, Elliott P, Stamler J, Nicholson JK, Draper J, Mathers JC, Holmes E, Frost G, The Lancet Diabetes & Endocrinology 2017, 5, 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].O’Sullivan A, Gibney MJ, Brennan L, Am J Clin Nutr 2011, 93, 314–321. [DOI] [PubMed] [Google Scholar]
- [32].Lindqvist HM, Rådjursöga M, Torstensson T, Jansson L, Ellegård L, Winkvist A, J Nutr n.d., DOI 10.1093/jn/nxaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wellington N, Shanmuganathan M, de Souza RJ, Zulyniak MA, Azab S, Bloomfield J, Mell A, Ly R, Desai D, Anand SS, Britz-McKibbin P, Nutrients 2019, 11, 2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A, Nucleic Acids Res. 2018, 46, D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stamler J, Brown IJ, Daviglus ML, Chan Q, Kesteloot H, Ueshima H, Zhao L, Elliott P, INTERMAP Research Group, Circulation 2009, 120, 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Robaczewska J, Kedziora-Kornatowska K, Kozakiewicz M, Zary-Sikorska E, Pawluk H, Pawliszak W, Kedziora J, J. Physiol. Pharmacol 2016, 67, 331–337. [PubMed] [Google Scholar]
- [37].Ligthart-Melis GC, van de Poll MC, Boelens PG, Dejong CH, Deutz NE, van Leeuwen PA, Am J Clin Nutr 2008, 87, 1282–1289. [DOI] [PubMed] [Google Scholar]
- [38].Zhu Y, Shurlknight KL, Chen X, Sang S, J Nutr 2014, 144, 114–122. [DOI] [PubMed] [Google Scholar]
- [39].Zamora-Ros R, Achaintre D, Rothwell JA, Rinaldi S, Assi N, Ferrari P, Leitzmann M, Boutron-Ruault M-C, Fagherazzi G, Auffret A, Kühn T, Katzke V, Boeing H, Trichopoulou A, Naska A, Vasilopoulou E, Palli D, Grioni S, Mattiello A, Tumino R, Ricceri F, Slimani N, Romieu I, Scalbert A, Sci Rep 2016, 6, DOI 10.1038/srep26905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Söderholm PP, Koskela AH, Lundin JE, Tikkanen MJ, Adlercreutz HC, Am J Clin Nutr 2009, 90, 1167–1171. [DOI] [PubMed] [Google Scholar]
- [41].Juurlink BH, Azouz HJ, Aldalati AM, AlTinawi BM, Ganguly P, Nutrition Journal 2014, 13, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Garcia-Aloy M, Ulaszewska M, Franceschi P, Estruel‐Amades S, Weinert CH, Tor‐Roca A, Urpi‐Sarda M, Mattivi F, Andres-Lacueva C, Molecular Nutrition & Food Research 2020, 64, 1901137. [DOI] [PubMed] [Google Scholar]
- [43].Baldrick FR, McFadden K, Ibars M, Sung C, Moffatt T, Megarry K, Thomas K, Mitchell P, Wallace JMW, Pourshahidi LK, Ternan NG, Corona G, Spencer J, Yaqoob P, Hotchkiss S, Campbell R, Moreno-Rojas JM, Cuevas FJ, Pereira-Caro G, Rowland I, Gill CIR, Am J Clin Nutr 2018, 108, 688–700. [DOI] [PubMed] [Google Scholar]
- [44].Xi M, Dragsted LO, Genes & Nutrition 2019, 14, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Caballero B, Trugo LC, Finglas PM, Encyclopedia of Food Sciences and Nutrition, Academic, San Diego, CA: 2003. [Google Scholar]
- [46].Juraschek SP, Miller ER, Chang AR, Anderson CAM, Hall JE, Appel LJ, Hypertension 2020, 75, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stenemo M, Ganna A, Salihovic S, Nowak C, Sundström J, Giedraitis V, Broeckling CD, Prenni JE, Svensson P, Magnusson PKE, Lind L, Ingelsson E, Ärnlöv J, Fall T, ESC Heart Failure 2019, 6, 764–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, Ibba I, Lai MA, Orrù S, Blois S, Loizedda AL, Griffin JL, Usai P, Caboni P, Atzori L, Manzin A, Sci Rep 2017, 7, DOI 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


