SUMMARY
Intervention trials of vector control methods often require community level randomization with appropriate inferential methods. For many interventions, the possibility of confounding due to the effects of health-care seeking behavior on disease ascertainment remains a concern. The test-negative design, a variant of the case-control method, was introduced to mitigate this issue in the assessment of the efficacy of influenza vaccination (measured at an individual level) on influenza infection. Here, we introduce a cluster-randomized test-negative design that includes randomization of the intervention at a group level. We propose several methods for estimation and inference regarding the relative risk (RR). The inferential methods considered are based on the randomization distribution induced by permuting intervention assignment across two sets of randomly selected clusters. The motivating example is a current study of the efficacy of randomized releases of Wolbachia-infected Aedes aegypti mosquitoes to reduce the incidence of dengue in Yogyakarta City, Indonesia. Estimation and inference techniques are assessed through a simulation study.
Keywords: Case-control, Cluster-randomized trials, Odds ratio, Test-negative design
1. Introduction: the test-negative design
The test-negative design (TND) is a modification of a case-control study that allows for the use of surveillance systems in assessing the impact of an intervention in reducing disease. The design has been widely used to assess the effectiveness of seasonal influenza vaccines since 2005 (Skowronski and others, 2005). The original intent of the TND design was, in large part, to deal with confounding associated with health-care-seeking behavior that can occur in case-control or cohort designs. However, it can also be seen as providing a viable approach to disease ascertainment in cases where longitudinal studies are likely to be ethically difficult or cost prohibitive.
In the TND, individuals seeking care for symptoms consistent with the disease of interest (but not unique to this disease) are recruited and formally tested for the presence of the specific disease. Those testing positive are then compared with those testing negative with regard to a pre-specified exposure or intervention. As noted, the design is popular for evaluating the effectiveness of seasonal influenza vaccination (Sullivan and others, 2014). Here, patients seeking health care for an acute respiratory illness are recruited into the study and tested for influenza—those confirmed as incident cases of influenza are referred to as test-positives, whereas the others are the test-negatives. In addition, the patient’s recent vaccination status is ascertained. In general, assuming that influenza ascertainment was complete, one could estimate influenza incidence proportions for both the vaccinated and unvaccinated population subgroups by dividing the number of influenza cases by the sizes of the vaccinated and unvaccinated, susceptible, and care-seeking populations, respectively, to yield estimates of incidence proportions. However, these population sizes can be extremely difficult to determine accurately. The TND uses the relative frequency of the test-negatives in the two exposure groups as a proxy for the ratio of these population sizes. This yields an estimated RR based on the ratio of the odds of vaccination in patients testing positive for influenza to the equivalent odds in patients testing negative (see Jackson and Nelson, 2013, and below, for further details). No rare disease assumption is required.
Because cases and controls are recruited from the same patient population, and restricted to those seeking care at participating clinics, the design was assumed to eliminate bias caused by health-care-seeking behavior (Jackson and Nelson, 2013; Haber and others, 2015); however, it has recently been demonstrated that this bias is more likely reduced than entirely removed (Sullivan and others, 2016). Several authors have explored the statistical rationale and underlying assumptions of the TND, showing that the odds ratio (OR)—i.e., the odds of vaccination in influenza cases versus that for test-negative controls—is directly equivalent to the RR of influenza comparing vaccinated with unvaccinated individuals, providing underlying assumptions are met (Jackson and Nelson, 2013; Haber and others, 2015). In addition, the causal structure underlying this approach has recently been studied in detail using directed acyclic graphs and causal inference ideas (Sullivan and others, 2016). For the traditional TND examining influenza, vaccination is applied at the individual level, not always possible for some interventions. This article discusses TNDs for interventions randomly applied at a group level.
2. Cluster-randomized trials and application to TNDs
Randomized controlled trials are considered the gold standard for evaluating the efficacy of health interventions, providing the basis of non-model dependent inference. When an intervention is delivered to groups of individuals, e.g., in neighborhoods, or is expected to have a community-wide impact, randomization of the intervention necessarily occurs at the group, rather than individual, level. Such a trial is termed a cluster-randomized trial (CRT)—see Hayes and Moulton (2009).
Investigators implement a CRT by recruiting a cohort of participants, randomly assigning the intervention to groups of individuals, and following the cohort over time to measure the endpoint in groups assigned to each study arm. The RR comparing intervention and non-intervention arms is used to quantify the intervention’s efficacy, equal to one minus RR. The non-independence of individuals within each group in CRTs causes statistical inefficiency, termed the ‘design effect’, and inferential methods must appropriately account for the clustering induced by the design (Hayes and Moulton, 2009, Part C).
Although these statistical challenges have been effectively addressed, prospective CRTs frequently require very large cohorts of individuals to generate a sufficient number of events for hypothesis testing, particularly when the outcome is relatively rare (see e.g., Mortimer and others, 2017). This has significant cost, time, ethical, and logistical implications. Trials of preventive interventions against infectious diseases with acute and transient presentations or narrow diagnostic windows face further challenges due to difficulties in obtaining complete and unbiased ascertainment of outcomes within a cohort. Where blinding to intervention status is not possible due to the nature of the intervention or other reasons, this introduces the potential for detection or performance bias (Wilson and others, 2015) if care-seeking or testing behavior (and therefore case ascertainment) is differential by study arm due to participant’s and/or healthcare providers’ perceptions of the intervention’s efficacy. Estimation of disease incidence in treated and untreated populations through simple clinical-based surveillance of the disease of interest, without employing the TND, also requires knowledge of the size of the source population from which cases arise. In most settings this cannot be estimated with any accuracy, given that case surveillance is likely to occur in only a subset of the total available healthcare providers and most (potentially care-seeking) populations will have the choice of a number of providers, including in the private sector, both within and outside their local residential area.
Here, we propose a method to assess the endpoints in CRTs using the TND that offers the advantage of being more efficient, cost effective, and logistically simpler to achieve than a large prospective cohort, and does not require knowledge of the sizes of populations at risk. We refer to our proposal as a cluster-randomized test-negative design (CR-TND)—see Anders and others (2017). The CR-TND fundamentally alters the standard TND in two key ways: (i) randomization of the exposure and (ii) clustering of participants’ exposure status due to randomized delivery of the intervention at a group-level.
The next section describes the motivating application, a preventive intervention against dengue. Section 4 introduces estimation and inferential methods to assess an intervention’s efficacy using data arising from the new design, based on summary measures at the cluster level (As compared with individual level data). Section 5 provides exploratory simulation results to assess the power of these approaches along with additional properties of the estimation and inferential methods. Inference is examined in terms of the permutation distribution induced by randomization; some comparisons are made with model-based methods—including generalized estimating equations (GEEs) and mixed effects logistic regression—techniques intended to account for within group correlation. Section 6 provides a brief discussion and points towards further research topics.
3. Application
The World Mosquito Program is an international research collaboration that is delivering a paradigm shift in the control of arboviral diseases transmitted by Aedes aegypti mosquitoes. The method utilizes Wolbachia, obligate intracellular endosymbionts that are common in insect species (see e.g., Hilgenboecker and others, 2017) but were not present in A. aegypti mosquitoes until they were stably transinfected in the laboratory. In insects, Wolbachia is maternally transmitted via the egg and manipulates insect reproduction to favor its own population dissemination via cytoplasmic incompatibility. The result is that Wolbachia rapidly enters into naive mosquito populations in a self-sustaining, durable manner. Strikingly, the presence of Wolbachia in A. aegypti mosquitoes renders them more resistant to disseminated arbovirus infection, including dengue, Zika, chikungunya, and yellow fever (Dutra and others, 2016; Johnson, 2015; Rainey and others, 2014). Thus the critical and signature effect of Wolbachia as an intervention is to severely reduce the vectorial capacity of mosquito populations to transmit arboviral infections between humans. For field implementation, the approach seeds wild mosquito populations with Wolbachia through controlled releases of small numbers of Wolbachia-infected mosquitoes.
The motivation for the proposed CR-TND arises from a planned 2-year trial to evaluate the efficacy of Wolbachia-infected mosquitoes in reducing dengue transmission in Yogyakarta City, Indonesia. The administrative area of the city, with a population in 2015 of 408 000, has a generally higher dengue incidence than surrounding districts. A parallel two-arm non-blinded CR-TND will be conducted. The study site was subdivided into 24 contiguous clusters, each approximately one 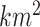 in size, to allow for effective deployment while minimizing cluster interference. Clusters were randomly allocated in a 1 to 1 ratio to receive either Wolbachia-infected mosquito deployments or no intervention. Eligible febrile participants will be recruited from across the study area through primary healthcare clinics, and subsequently classified as virologically confirmed dengue cases (test-positives) or arbovirus-negative controls on the basis of laboratory testing (i.e., those suffering from other febrile illnesses [OFIs]). The Wolbachia exposure distribution in test-positive cases (i.e., whether or not the individual lives in an intervention cluster) will be compared with that for test-negative controls, in order to estimate the efficacy of the intervention.
in size, to allow for effective deployment while minimizing cluster interference. Clusters were randomly allocated in a 1 to 1 ratio to receive either Wolbachia-infected mosquito deployments or no intervention. Eligible febrile participants will be recruited from across the study area through primary healthcare clinics, and subsequently classified as virologically confirmed dengue cases (test-positives) or arbovirus-negative controls on the basis of laboratory testing (i.e., those suffering from other febrile illnesses [OFIs]). The Wolbachia exposure distribution in test-positive cases (i.e., whether or not the individual lives in an intervention cluster) will be compared with that for test-negative controls, in order to estimate the efficacy of the intervention.
The CR-TND approach has been developed for the Yogyakarta trial in preference to a traditional cohort design, in which absolute and relative dengue incidence in treated and untreated populations is measured directly, because of challenges in reliably ascertaining dengue illness episodes prospectively in a large population and in quantifying true exposure to Wolbachia prior to any presenting infection. Passive ascertainment of dengue cases through existing routine disease surveillance systems would provide incomplete and potentially biased, estimates of the population dengue incidence. The surveillance system in Indonesia, and many endemic settings, captures only hospital admissions, and completeness of reporting may be spatially and temporally variable. Specificity of the surveillance data is also imperfect and variable, as case notification is commonly based on a clinical diagnosis of dengue, with only a subset of cases confirmed by laboratory diagnostics. The alternative approach of actively ascertaining dengue cases within a cohort of individuals recruited from treated and untreated areas is challenging both operationally and ethically, due to the large size of the cohorts required, the intensity of contact required to detect and diagnose acute dengue infections, and the potential for biases arising from under-ascertainment of illness events or loss to follow-up. Further, it would be hard to measure individual mobility and Wolbachia levels at the time of any presenting infection without almost continuous monitoring.
4. Cluster-level analyses of CR-TND data
4.1. Test-positive fraction of all tests by cluster
Suppose there are 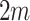 clusters with
clusters with 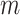 randomly assigned to the intervention and the remainder untreated. All test-positive (cases) and test-negative (controls) individuals are selected, numbering
randomly assigned to the intervention and the remainder untreated. All test-positive (cases) and test-negative (controls) individuals are selected, numbering 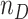 , and
, and 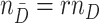 , respectively. The RR associated with the intervention is denoted by
, respectively. The RR associated with the intervention is denoted by 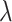 . The simplest data are cluster counts of confirmed test-positives and test-negatives. As our primary approach is to develop inference based on a permutation approach, we take these numbers as fixed, at this point allowing randomness only to enter through the allocation of intervention status to each cluster. Further,
. The simplest data are cluster counts of confirmed test-positives and test-negatives. As our primary approach is to develop inference based on a permutation approach, we take these numbers as fixed, at this point allowing randomness only to enter through the allocation of intervention status to each cluster. Further, 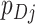 and
and 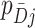 represent the fraction of cases and controls, respectively, in the
represent the fraction of cases and controls, respectively, in the 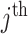 cluster. Let
cluster. Let 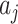 denote the ratio of the number of test-positives (
denote the ratio of the number of test-positives (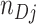 ) to the total number of tests in the
) to the total number of tests in the 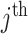 cluster (
cluster (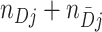 ). Note that
). Note that 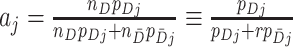 . A proposed test statistic to assess the effect of the intervention is then
. A proposed test statistic to assess the effect of the intervention is then 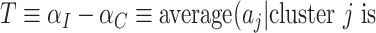
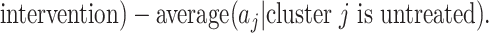
Under the null hypothesis of no intervention effect, for a randomly selected cluster (from either arm) 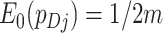 since the
since the 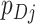 s sum to 1; here the expectation is over the permutation distribution of all possible random allocations and the subscript 0 reinforces that this expectation is under the null. Similarly,
s sum to 1; here the expectation is over the permutation distribution of all possible random allocations and the subscript 0 reinforces that this expectation is under the null. Similarly, 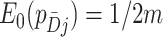 . Thus,
. Thus, 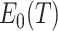 is approximately zero using the delta method. In fact, by the symmetry of its definition,
is approximately zero using the delta method. In fact, by the symmetry of its definition, 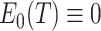 .
.
We now approximate 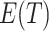 when the intervention affects case counts, changing the relative distribution of cluster test-positives to test-negatives, and how this depends on
when the intervention affects case counts, changing the relative distribution of cluster test-positives to test-negatives, and how this depends on 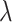 . For a large number of test-positives in the intervention arm, this total is reduced by
. For a large number of test-positives in the intervention arm, this total is reduced by 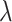 (for
(for 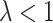 ). Thus, the fraction of the total number of test-positives that occur in the intervention clusters is approximately
). Thus, the fraction of the total number of test-positives that occur in the intervention clusters is approximately 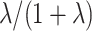 . Hence, for a random cluster,
. Hence, for a random cluster, 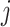 , in the intervention arm,
, in the intervention arm, 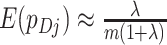 . Similarly, for a random cluster,
. Similarly, for a random cluster, 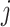 , in the untreated arm,
, in the untreated arm, 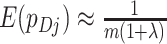 . The
. The 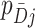 are unaffected by the intervention so that by the delta method,
are unaffected by the intervention so that by the delta method,
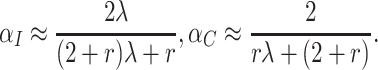 |
(4.1) |
Thus,
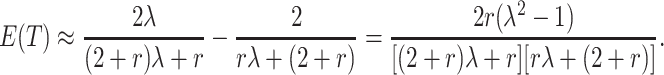 |
(4.2) |
The RHS of (4.2) is zero if and only 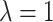 , as noted above. When
, as noted above. When 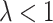 ,
, 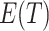
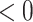 , as expected. Further, (4.2) is quadratic in
, as expected. Further, (4.2) is quadratic in 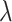 for any given
for any given 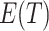 . Thus, we can substitute an estimate of
. Thus, we can substitute an estimate of 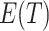 , obtained from differencing the average observed ratios
, obtained from differencing the average observed ratios 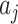 in each arm, into (4.2) and solve for
in each arm, into (4.2) and solve for 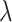 , yielding an approximate estimate of RR. This approach assumes a cluster specific interpretation of the RR when compared with a marginal version; we return to this point later.
, yielding an approximate estimate of RR. This approach assumes a cluster specific interpretation of the RR when compared with a marginal version; we return to this point later.
We illustrate with a simple example, with 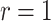 and
and 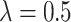 . Here (4.2) yields
. Here (4.2) yields 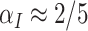 and
and 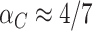 , so that
, so that 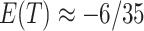 . In reverse, substituting
. In reverse, substituting 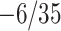 into the LHS of (4.3) yields
into the LHS of (4.3) yields 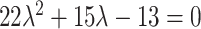 with only one positive solution, namely
with only one positive solution, namely 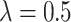 .
.
Turning to inferential methods based on 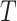 , the null hypothesis can be tested via the permutation distribution of
, the null hypothesis can be tested via the permutation distribution of 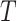 , calculated by examining the estimated
, calculated by examining the estimated 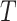 s for all possible randomized intervention allocations, holding the observed data fixed. This permutation distribution can then be used to assess the significance of the observed value of
s for all possible randomized intervention allocations, holding the observed data fixed. This permutation distribution can then be used to assess the significance of the observed value of 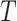 .
.
For simplicity in carrying out simulations and for additional insight, we can analytically evaluate the null permutation variance of 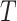 . As noted, we consider the observed values of
. As noted, we consider the observed values of 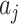 for
for 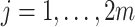 to be fixed. Under the null hypothesis the
to be fixed. Under the null hypothesis the 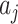 s for the
s for the 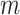 intervention clusters are simply a random sample of
intervention clusters are simply a random sample of 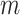 of these values, also true for the untreated
of these values, also true for the untreated 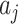 s. The variance of the
s. The variance of the 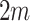 fixed values of
fixed values of 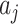 across all clusters depends on: (i) the variability of both the
across all clusters depends on: (i) the variability of both the 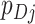 s and
s and 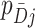 s, i.e., the distribution of cluster test-positives and test-negatives, respectively, (ii) the covariance of the
s, i.e., the distribution of cluster test-positives and test-negatives, respectively, (ii) the covariance of the 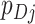 s and the
s and the 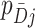 s, i.e., how test-positives and test-negatives covary across the clusters, and (iii) the value of
s, i.e., how test-positives and test-negatives covary across the clusters, and (iii) the value of 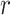 . However, we do not need to analytically derive this variance as we will empirically estimate it in due course. We refer to the variance of the
. However, we do not need to analytically derive this variance as we will empirically estimate it in due course. We refer to the variance of the 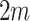
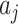 s by
s by 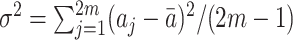 where
where 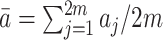 .
.
In the 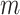 intervention clusters, the permutation variance of
intervention clusters, the permutation variance of 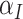 is
is 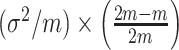 where the second term is the finite population correction factor. The variance of
where the second term is the finite population correction factor. The variance of 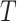 must accommodate that
must accommodate that 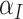 and
and 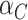 are correlated due to the finite number of clusters, and the fact that we are conditioning on the observed data and computing expectations and variances according to the permutation distribution. In fact, under the null hypothesis,
are correlated due to the finite number of clusters, and the fact that we are conditioning on the observed data and computing expectations and variances according to the permutation distribution. In fact, under the null hypothesis, 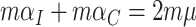 (where
(where 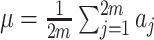 ), so that
), so that 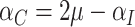 . Hence, under the null hypothesis,
. Hence, under the null hypothesis, 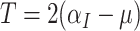 . Finally, therefore,
. Finally, therefore,
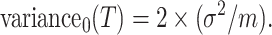 |
(4.3) |
Note that, when 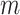 is sufficiently large, the RHS of (4.3) is just what you would obtain by naively treating
is sufficiently large, the RHS of (4.3) is just what you would obtain by naively treating 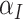 and
and 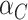 as independent averages with a common variance
as independent averages with a common variance 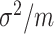 .
.
The variance, 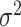 , can be estimated by the variance of the
, can be estimated by the variance of the 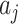 s in either the intervention or untreated clusters. Since these arms both contain
s in either the intervention or untreated clusters. Since these arms both contain 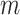 clusters, an average of these two estimates suffices–the pooled variance estimator of the two-sample t-test. With this estimate,
clusters, an average of these two estimates suffices–the pooled variance estimator of the two-sample t-test. With this estimate, 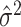 , used in (4.3), the standardized test statistic is thus
, used in (4.3), the standardized test statistic is thus 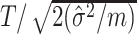 , equivalent to the two-sample t-test statistic comparing the observed
, equivalent to the two-sample t-test statistic comparing the observed 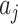 s across the two arms. Of course, if the null hypothesis is true, we would know the variability of the
s across the two arms. Of course, if the null hypothesis is true, we would know the variability of the 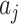 s exactly since all would be observed but this variance would overestimate the variance of
s exactly since all would be observed but this variance would overestimate the variance of 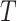 away from the null. We can then compare the standardized test statistic to a
away from the null. We can then compare the standardized test statistic to a 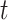 distribution with
distribution with 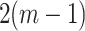 degrees of freedom to assess significance, and this provides a close approximation to the permutation distribution result so long as
degrees of freedom to assess significance, and this provides a close approximation to the permutation distribution result so long as 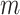 is large.
is large.
For any 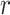 , the difference in the average cluster means of the
, the difference in the average cluster means of the 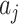 s across intervention arms estimates
s across intervention arms estimates 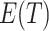 and thus directly relates to an estimate of
and thus directly relates to an estimate of 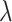 , through (4.2). As
, through (4.2). As 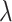 moves away from the null value, the absolute size of
moves away from the null value, the absolute size of 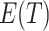 monotonically increases. Hence, in addition to point estimation, a confidence interval for
monotonically increases. Hence, in addition to point estimation, a confidence interval for 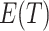 directly yields a corresponding confidence interval for
directly yields a corresponding confidence interval for 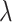 . Note that, in the scenario with an intervention effect, it is not straightforward to approximate the permutation variance of
. Note that, in the scenario with an intervention effect, it is not straightforward to approximate the permutation variance of 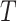 . Nevertheless, we recommend treating the average
. Nevertheless, we recommend treating the average 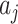 ’s as if they come from independent samples since this is correct at the null. Away from the null, the variances of
’s as if they come from independent samples since this is correct at the null. Away from the null, the variances of 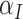 and
and 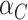 are not equal so that it might be better to base confidence intervals on the Welch version of the t-test, using separate estimates of the variances in the two arms and modifying the number of degrees of freedom appropriately using the Welch–Satterthwaite formula (Welch, 1938; Satterthwaite, 1946; Welch, 1947). We examine the empirical coverage properties of this approach in Section 5. Note that the differentiation between the intervention arms—given by
are not equal so that it might be better to base confidence intervals on the Welch version of the t-test, using separate estimates of the variances in the two arms and modifying the number of degrees of freedom appropriately using the Welch–Satterthwaite formula (Welch, 1938; Satterthwaite, 1946; Welch, 1947). We examine the empirical coverage properties of this approach in Section 5. Note that the differentiation between the intervention arms—given by 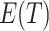 —gets smaller as
—gets smaller as 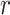 increases from 1. Perversely, this approach is then optimal when
increases from 1. Perversely, this approach is then optimal when 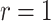 and loses power as
and loses power as 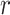 increases. However, this effect is small as examined quantitatively in simulations.
increases. However, this effect is small as examined quantitatively in simulations.
4.2. ORs from collated cluster data
Consider now an alternative method to both assess the intervention’s efficacy and provide an estimate of 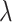 . Following Jackson and Nelson (2013), Table 1 is the 2 x 3 table that classifies health-care-seeking individuals by their intervention status and outcomes, with
. Following Jackson and Nelson (2013), Table 1 is the 2 x 3 table that classifies health-care-seeking individuals by their intervention status and outcomes, with 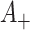 the total number of individuals who both experience the intervention, are detected by the surveillance system, and test positive for the outcome of interest with similar definitions for other entries. There is an analogous classification of (unobserved) individuals who do, or would, not seek care when experiencing such infections; the generalizability of any intervention efficacy estimate from observed data in Table 1 to the entire population depends on an untestable assumption that efficacy is not modified by health-care-seeking behavior—see Jackson and Nelson (2013) and Sullivan and others (2016).
the total number of individuals who both experience the intervention, are detected by the surveillance system, and test positive for the outcome of interest with similar definitions for other entries. There is an analogous classification of (unobserved) individuals who do, or would, not seek care when experiencing such infections; the generalizability of any intervention efficacy estimate from observed data in Table 1 to the entire population depends on an untestable assumption that efficacy is not modified by health-care-seeking behavior—see Jackson and Nelson (2013) and Sullivan and others (2016).
Table 1.
Stratification of population based on intervention status, infection, and health-care-seeking behavior
| Seek care | ||||
|---|---|---|---|---|
| Infected with dengue | Infected with OFI | Not Infected | Total | |
| Intervention |
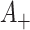
|
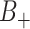
|
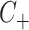
|
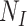
|
| Control |
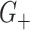
|
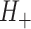
|
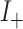
|
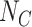
|
OFI refers to other febrile illnesses with similar presenting conditions to the infection of interest that can be discriminated on the basis of a specific laboratory test. Adapted from Figure 1 of Jackson and Nelson (2013).
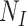 is the total number of exposed susceptible individuals in the population who would seek care if they experience symptoms; an analogous definition describes
is the total number of exposed susceptible individuals in the population who would seek care if they experience symptoms; an analogous definition describes 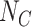 for controls. In principle, the incidence of the disease outcome in the exposed health-care-seeking population can then be estimated by
for controls. In principle, the incidence of the disease outcome in the exposed health-care-seeking population can then be estimated by 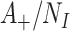 , and RR by
, and RR by 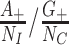 . Unfortunately, it is often difficult to obtain accurate details for the susceptible care-seeking population sizes
. Unfortunately, it is often difficult to obtain accurate details for the susceptible care-seeking population sizes 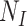 and
and 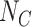 . The TND therefore exploits the incidence of OFIs in the population as a basis for assessing the relative population sizes
. The TND therefore exploits the incidence of OFIs in the population as a basis for assessing the relative population sizes 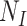 and
and 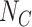 , under the assumption that incidence of OFIs is independent of the intervention. The latter assumption means that
, under the assumption that incidence of OFIs is independent of the intervention. The latter assumption means that 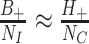 so that
so that 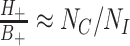 . Thus
. Thus 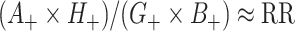 . In other words, the empirical OR from the data of Table 1 provides a direct estimate of RR. Note that this is a marginal OR with no reference to within cluster characteristics.
. In other words, the empirical OR from the data of Table 1 provides a direct estimate of RR. Note that this is a marginal OR with no reference to within cluster characteristics.
In the CR-TND, the two rows of Table 1 correspond to data from different clusters since every individual in a given cluster is either exposed to the intervention or not. The observed log OR from Table 1 is 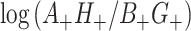 . We first assess the properties of this random variable (induced by randomization and keeping the data fixed) under the null hypothesis, i.e., under the permutation distribution. Under random sampling,
. We first assess the properties of this random variable (induced by randomization and keeping the data fixed) under the null hypothesis, i.e., under the permutation distribution. Under random sampling, 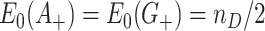 where
where 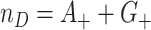 . Analogously,
. Analogously, 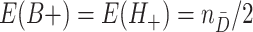 . Then, by the delta method,
. Then, by the delta method, 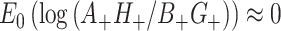 ; of course, this expectation is exactly 0 because of the symmetry between the counts caused by randomization. Thus, this log(OR) is centered at the correct value assuming the null. For testing we can again revert to the full permutation distribution. Again, we can analytically approximate the variance of the cumulative log(OR) estimate at the null hypothesis for use in simulations and to provide approximate permutation inference.
; of course, this expectation is exactly 0 because of the symmetry between the counts caused by randomization. Thus, this log(OR) is centered at the correct value assuming the null. For testing we can again revert to the full permutation distribution. Again, we can analytically approximate the variance of the cumulative log(OR) estimate at the null hypothesis for use in simulations and to provide approximate permutation inference.
As before, under the null, the permutation variance of 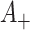 is simply
is simply 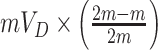 where
where 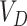 is the variance of the test-positive counts
is the variance of the test-positive counts 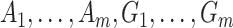 across both intervention and untreated clusters (again using
across both intervention and untreated clusters (again using 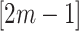 in the denominator of the definition of
in the denominator of the definition of 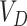 ). Here, we use
). Here, we use 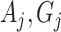 , etc. to refer to the entries of Table 1 specific to the
, etc. to refer to the entries of Table 1 specific to the 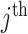 cluster. Conditional on the observed data,
cluster. Conditional on the observed data, 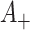 and
and 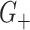 are not independent under the randomization distribution since
are not independent under the randomization distribution since 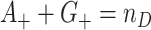 . Thus the null variance of
. Thus the null variance of 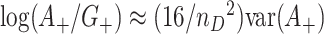 using the delta method and the fact that
using the delta method and the fact that 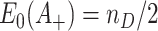 . Finally, putting these two observations together yields
. Finally, putting these two observations together yields
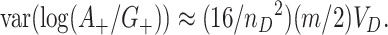 |
(4.4) |
Similarly,
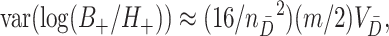 |
(4.5) |
where 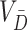 is the variance of all
is the variance of all 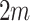 cluster test-negative counts.
cluster test-negative counts.
For approximate estimation of the variance of the log(OR), note that 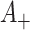 and
and 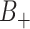 may be correlated because of characteristics of the clusters that may induce test-positives and test-negatives to tend to be high together (e.g., the population size and density within a given cluster) or possibly negatively associated. We can approximate the covariance of the two terms
may be correlated because of characteristics of the clusters that may induce test-positives and test-negatives to tend to be high together (e.g., the population size and density within a given cluster) or possibly negatively associated. We can approximate the covariance of the two terms 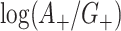 and
and 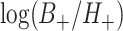 by exploiting the delta method, so that
by exploiting the delta method, so that
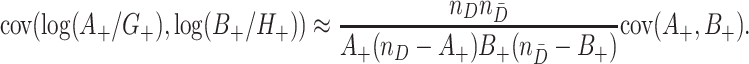 |
(4.6) |
Putting (4.4), (4.5) and (4.6) together then yields
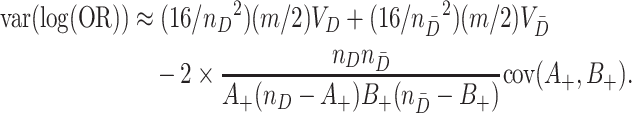 |
(4.7) |
It remains to estimate 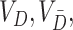 and
and 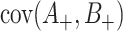 . As before, under the null,
. As before, under the null, 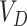 could be estimated by using a variance estimator of the test-positive counts
could be estimated by using a variance estimator of the test-positive counts 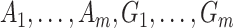 from both intervention and untreated clusters. However, this yields a poor estimate with an intervention effect. We thus use a pooled estimate, the average of the variances of the
from both intervention and untreated clusters. However, this yields a poor estimate with an intervention effect. We thus use a pooled estimate, the average of the variances of the 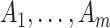 and
and 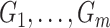 , separately estimated (using
, separately estimated (using 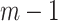 in the denominator for the estimated variances).
in the denominator for the estimated variances). 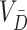 can be estimated analogously. Finally,
can be estimated analogously. Finally, 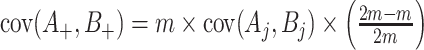 using finite population sampling methods—see Tam (1985) for the less familiar use for covariances. The term
using finite population sampling methods—see Tam (1985) for the less familiar use for covariances. The term 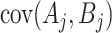 can be estimated from the covariance of the observed
can be estimated from the covariance of the observed 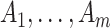 and
and 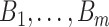 in the intervention clusters, again using
in the intervention clusters, again using 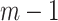 in the denominator of the covariance estimate.
in the denominator of the covariance estimate.
A simple example illustrates the effectiveness of a Gaussian approximation to the permutation distribution (at the null) based on the proposed sample estimate of (4.7). Table 1 in supplementary material available at Biostatistics online shows an assumed distribution of dengue and OFI cases for 10 clusters (mimicking more complete data used in later simulations). From these distributions, a random set of 100 cases and 100 controls were selected once. There are 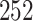 =
= 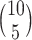 possible cluster intervention allocations. The exact standard deviation of the permutation distribution of
possible cluster intervention allocations. The exact standard deviation of the permutation distribution of 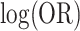 , with no intervention effect, is 0.1566. Over the 252 possible intervention allocations the average estimated standard deviation of
, with no intervention effect, is 0.1566. Over the 252 possible intervention allocations the average estimated standard deviation of 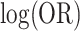 based on (4.7) is 0.1529, a close approximation. For comparison, for a simple random intercept logistic regression, the average model-based standard deviation estimate is 0.2843. The average estimated robust standard deviation based on a standard GEE (assuming an exchangeable correlation structure - see below) is unreliable at this small of a sample size. However, when 16 of the 252 permutation distributions with unreliably large estimates are removed, the average estimated standard deviation is 0.1407. Further discussion of these two approaches is provided below. For
based on (4.7) is 0.1529, a close approximation. For comparison, for a simple random intercept logistic regression, the average model-based standard deviation estimate is 0.2843. The average estimated robust standard deviation based on a standard GEE (assuming an exchangeable correlation structure - see below) is unreliable at this small of a sample size. However, when 16 of the 252 permutation distributions with unreliably large estimates are removed, the average estimated standard deviation is 0.1407. Further discussion of these two approaches is provided below. For 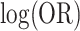 , using the 252 possible intervention allocations yields lower and upper 2.5% percentile thresholds of
, using the 252 possible intervention allocations yields lower and upper 2.5% percentile thresholds of 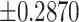 for the exact permutation distribution; the lower and upper thresholds, induced by
for the exact permutation distribution; the lower and upper thresholds, induced by 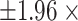 the average standard deviation estimate based on (4.7) are
the average standard deviation estimate based on (4.7) are 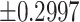 .The performance of GEE improved as sample size increased, resulting in a stable underestimate of the permutation variance the performance of the mixed effects estimator was more variable and not always as poor as this specific case and also improves as the sample sizes of cases and controls increase. Further simulation evidence of Type 1 error rates and power associated with using this approximate OR inference approach is given in the next section.
.The performance of GEE improved as sample size increased, resulting in a stable underestimate of the permutation variance the performance of the mixed effects estimator was more variable and not always as poor as this specific case and also improves as the sample sizes of cases and controls increase. Further simulation evidence of Type 1 error rates and power associated with using this approximate OR inference approach is given in the next section.
For confidence interval calculations (away from the null) we need to evaluate the randomization distribution of the log(OR) estimate assuming an intervention effect. Following Jackson and Nelson (2013), note that the intervention only affects the counts 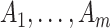 by assumption. These are each replaced in turn by
by assumption. These are each replaced in turn by 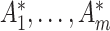 which reflect altered test-positive counts in the intervention clusters. For large populations,
which reflect altered test-positive counts in the intervention clusters. For large populations, 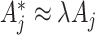 for the intervention clusters. Note that this specifically uses the assumption that the intervention effect is the same for all clusters. An alternative approach might model the intervention effect that allows variation with cluster characteristics, or, for example, with the size of
for the intervention clusters. Note that this specifically uses the assumption that the intervention effect is the same for all clusters. An alternative approach might model the intervention effect that allows variation with cluster characteristics, or, for example, with the size of 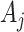 itself. We ignore this “second order” phenomenon and leave this for future analysis. The common reduction of the
itself. We ignore this “second order” phenomenon and leave this for future analysis. The common reduction of the 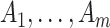 has two immediate implications: first, under the randomization distribution,
has two immediate implications: first, under the randomization distribution, 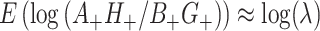 ; second, there is no change to the variance formula (4.7) since all the ORs for different permutations are simply shifted by approximately
; second, there is no change to the variance formula (4.7) since all the ORs for different permutations are simply shifted by approximately 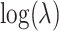 . Despite the remaining usefulness of (4.7) , we do now have to modify the estimates of
. Despite the remaining usefulness of (4.7) , we do now have to modify the estimates of 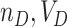 , and cov
, and cov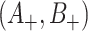 due to the replacement of each
due to the replacement of each 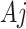 with
with 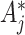 . The necessary adjustment is achieved by simply increasing the observed
. The necessary adjustment is achieved by simply increasing the observed 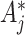 s by the common factor
s by the common factor 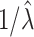 to obtain an estimate of
to obtain an estimate of 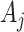 (in the
(in the 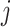 intervention clusters), en route to an estimate of
intervention clusters), en route to an estimate of 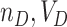 , and cov
, and cov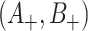 as before.
as before.
The above approach ignores any random variation of 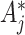 around
around 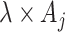 . Two potential sampling models might be employed here to account for this additional variation when the counts
. Two potential sampling models might be employed here to account for this additional variation when the counts 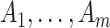 are smaller due to an intervention effect. The first is to assume that for the intervention clusters, and given
are smaller due to an intervention effect. The first is to assume that for the intervention clusters, and given 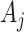 ,
, 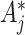 is Binomial with probability
is Binomial with probability 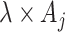 . The alternative is to assume that
. The alternative is to assume that 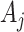 is Poisson with rate parameter
is Poisson with rate parameter 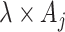 . Essentially, we are assuming here that given all the test-positive and test-negative counts in all the clusters, the
. Essentially, we are assuming here that given all the test-positive and test-negative counts in all the clusters, the 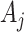 s in the intervention clusters are ‘filtered’ by an additional layer of randomness to generate the observed
s in the intervention clusters are ‘filtered’ by an additional layer of randomness to generate the observed 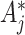 s. The Binomial approach essentially assumes that
s. The Binomial approach essentially assumes that 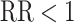 but this can achieved without loss of generality by switching the intervention label. By first conditioning on all counts (including the unobserved
but this can achieved without loss of generality by switching the intervention label. By first conditioning on all counts (including the unobserved 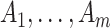 in the intervention clusters), we can see that the variance in (4.7) is subsequently increased approximately by either
in the intervention clusters), we can see that the variance in (4.7) is subsequently increased approximately by either 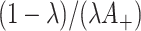 for the Binomial case, or
for the Binomial case, or 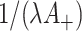 for the Poisson. (The latter is more conservative and therefore recommended.)
for the Poisson. (The latter is more conservative and therefore recommended.)
4.3. OR estimates via GEE and random effects logistic regression models
The CR-TND design yields clustered binary outcome data where interest may focus on estimation of the OR. It is natural, therefore, to consider applying GEEs or random intercept logistic regression methods. For GEE, we focus on use of a working exchangeable correlation structure within groups. Both of these methods are well known and easily implementable using standard software, and account for clustering through use of a robust variance method (GEE) or via an appropriate assumed random effects distribution. However, both methods are also known to suffer in performance in situations with small number of clusters, each containing a large number of observations. Also, at the outset, it is important to note that these two approaches are designed to estimate different parameters: GEE focuses on the marginal, or population averaged, OR whereas the random effects model targets the cluster-specific OR (see, Hayes and Moulton, 2009, Chapter 9.3). As defined in Section 2, the OR used to estimate intervention efficacy is a marginal OR.
In the simple example above with 10 clusters when the sample size is small, GEE is unreliable in estimating the variability of the log OR; the random effects model also overestimates the relevant standard deviation although significantly less so. As the sample size increases, both methods tend to underestimate the standard deviation. In general, with small numbers of clusters, the GEE technique suffers from inflated Type 1 error rates (Bellamy and others, 2000). Pan and Wall (2002) describe approaches to correcting for this through use of a 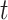 distribution as reference as opposed to the standard Normal. Morel and others (2003) suggest an alternative modification for making inferential statements. As pointed out by Hayes and Moulton (2009), however, this tactic requires addition calculations to determine the relevant degrees of freedom.
distribution as reference as opposed to the standard Normal. Morel and others (2003) suggest an alternative modification for making inferential statements. As pointed out by Hayes and Moulton (2009), however, this tactic requires addition calculations to determine the relevant degrees of freedom.
Recent research has examined the impact of small numbers of clusters on point and interval estimation of a fixed effect that covers both GEE and random effects logistic regression. See McNeish and Stapleton (2016) for an overview. While comparisons differ depending on the context, the general consensus is that GEE performs poorly with small numbers of clusters whereas random effects models provide reasonable inference. We consider both methods in the simulations.
5. Simulations
We used simulations that exploited historical information on the incidence of dengue and OFIs in Yogyakarta City. As discussed above, the city was divided into 24 non-overlapping contiguous clusters. The design randomly allocates 12 of these clusters to the intervention, with the remainder left untreated. Dengue incident cases, along with relevant comparative OFIs will be collected over a 2-year period. Table 2 of supplementary material available at Biostatistics online contains the frequency of recorded (hospitalized) dengue cases in the 24 clusters for each of nine distinct 2-year periods covering 2003–2014. During this period, there was no available data for 2004 and 2009, so that the first 2-year interval was for 2003 and 2005; similarly, the 2008–2010 interval included data for 2008 and 2010. Otherwise each 2-year period covered consecutive years. Data for the distribution for OFIs (Table 3 of supplementary material available at Biostatistics online) is only available for a single 2-year period from 2014 to 2015.
Table 2.
The proportion of simulations that returned significant results for each intervention effect of interest (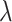 )
)
Relative risk (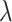 ) ) |
Test-positive fraction | Odds ratio | GEE | Random effects |
|---|---|---|---|---|
| 1 | 0.0497 | 0.0749 | 0.0779 | 0.0743 |
| 0.6 | 0.4916 | 0.5795 | 0.5936 | 0.6143 |
| 0.5 | 0.7498 | 0.8238 | 0.8266 | 0.8445 |
| 0.4 | 0.9298 | 0.9620 | 0.9603 | 0.9670 |
| 0.3 | 0.9951 | 0.9985 | 0.9983 | 0.9988 |
The GEE assumed an exchangeable correlation matrix. Each approach was applied to the results of the 10 000 random intervention allocations with 1000 cases and 4000 controls (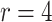 ).
).
Table 3.
The bias for the test-positive fraction quadratic estimates of the relative risk and the standard deviation of the difference in arm-specific averages (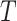 ) from 10 000 unconstrained intervention allocations
) from 10 000 unconstrained intervention allocations
| Ratio = 1 | Ratio = 4 | |||||
|---|---|---|---|---|---|---|
Relative risk (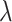 ) ) |
Bias | Average estimated standard deviation of 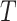
|
True standard deviation of 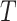
|
Bias | Average estimated standard deviation of 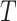
|
True standard deviation of 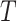
|
| 1 | 0.0264 | 0.0559 | 0.0564 | 0.0340 | 0.0397 | 0.0401 |
| 0.6 | 0.0386 | 0.0552 | 0.0557 | 0.0180 | 0.0392 | 0.0389 |
| 0.5 | 0.0390 | 0.0546 | 0.0551 | 0.0140 | 0.0389 | 0.0379 |
| 0.4 | 0.0380 | 0.0536 | 0.0540 | 0.0099 | 0.0385 | 0.0365 |
| 0.3 | 0.0351 | 0.0520 | 0.0521 | 0.0053 | 0.0379 | 0.0344 |
We carried out simulations assuming that the dengue distribution was exactly as identified for a given time period, but with a consistent OFI distribution across all time periods (the 2014–2015 distribution). For each simulation, 1000 dengue cases were assigned to the 24 clusters according to the dengue distribution, and 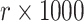 OFIs were assigned according to the OFI distribution. These allocations provided the base data from which we subsequently applied intervention assignment labels to 12 of the 24 clusters at random (according to a permutation distribution) for each distinct simulation. The cluster intervention assignment was permuted 10 000 times at random.
OFIs were assigned according to the OFI distribution. These allocations provided the base data from which we subsequently applied intervention assignment labels to 12 of the 24 clusters at random (according to a permutation distribution) for each distinct simulation. The cluster intervention assignment was permuted 10 000 times at random.
At the null, no further data modifications were required in computing various estimates and test statistics. Away from the null, we applied various values of 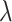 to deterministically reduce the dengue cases in the 12 intervention clusters before selecting cases (while maintaining the total number of cases at 1000). While this reduction could be applied stochastically, this was not considered necessary given that the permutation approach conditions on the true fixed number of cases in each cluster and bases inference on simply permuting the intervention assignment. Note that this applies a cluster-specific reduction in cases corresponding to the chosen
to deterministically reduce the dengue cases in the 12 intervention clusters before selecting cases (while maintaining the total number of cases at 1000). While this reduction could be applied stochastically, this was not considered necessary given that the permutation approach conditions on the true fixed number of cases in each cluster and bases inference on simply permuting the intervention assignment. Note that this applies a cluster-specific reduction in cases corresponding to the chosen 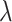 .
.
We first examine the power of several approaches to testing for an intervention effect: (i) comparison of the average test-positive fraction across intervention arms as outlined in Section 4.1, using the t-test statistic (assuming variance homogeneity across the two arms), (ii) using a test statistic based on an estimated OR from aggregated cluster data (by arm) as described in Section 4.2, using the variance estimate (4.7) (on the log scale), (iii) GEE (with a working exchangeable correlation structure) OR from aggregated OR data that employs a robust variance calculation, and (iv) a mixed effects logistic regression model with random intercept terms by cluster, the latter two approaches mentioned in Section 4.3. For each method, the same 10 000 random permutations of intervention assignment were used to generate results. Subsequently simulation results were averaged across the nine distinct two-year time periods.
Given the relatively small number of clusters in the Yogyakarta trial, constrained randomization was used to ensure balance between study arms for some key cluster covariates including historical dengue incidence, OFI incidence, demographics, population, and area. Constrained randomization restricts the number of permuted intervention assignments that are allowed in the random selection. After filtering 100 000 random allocations by these balancing criteria, 247 balanced allocations were retained, i.e., 494 potential allocations of the intervention to either arm. Computational considerations make it difficult to examine the exact permutation distribution of the average test-positive fraction difference or the aggregate OR test statistic over all simulations and so we focus here on the approximations derived above.
Table 2 shows the power of the various methods for testing the null hypothesis of no intervention effect for values of 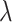 ranging from 1.0 down to 0.3 with
ranging from 1.0 down to 0.3 with 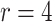 (with results averaged across the nine historical dengue distributions discussed above). The average Type 1 error control is extremely close to 5% for the test-positive fraction approach, and very slightly anti-conservative for the OR method. Decomposing the results for the nine specific dengue cluster distributions used (not shown here) exhibits very little variation where the range of Type 1 errors is from 4.6% to 5.2% for the test-positive fraction method and from 6.7% to 8.2% for the OR test. The extremes do not necessarily occur for the same dengue distribution across clusters for the two techniques. GEE and the random effects model perform similarly to the direct OR technique. This suggests that while 10 clusters were insufficient to reliably use such models, 24 may be enough.
(with results averaged across the nine historical dengue distributions discussed above). The average Type 1 error control is extremely close to 5% for the test-positive fraction approach, and very slightly anti-conservative for the OR method. Decomposing the results for the nine specific dengue cluster distributions used (not shown here) exhibits very little variation where the range of Type 1 errors is from 4.6% to 5.2% for the test-positive fraction method and from 6.7% to 8.2% for the OR test. The extremes do not necessarily occur for the same dengue distribution across clusters for the two techniques. GEE and the random effects model perform similarly to the direct OR technique. This suggests that while 10 clusters were insufficient to reliably use such models, 24 may be enough.
With regards to power, both the test-positive fraction and direct OR methods exhibit excellent power, on average, for values of the true RR equal or lower than 0.5, as exhibited in Table 2. The OR method exhibits slightly improved power, when compared with the test-positive fraction method, somewhat more than can be explained by its slight anti-conservativeness at the null. The random effects model is in turn very slightly more powerful than the direct OR technique. There is considerably more variation in the power of both methods over the nine varying dengue distribution scenarios. For example, with the RR, 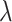 at 0.5, the power of the test-positive fraction approach ranges from 52% to 94% (with average of 75%); at the same
at 0.5, the power of the test-positive fraction approach ranges from 52% to 94% (with average of 75%); at the same 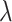 , the power ranges from 61% to 98% for the OR strategy. Again, the extremes occur at differing assumed dengue distributions for the two methods. Table 4 of supplementary material available at Biostatistics online provides similar results for
, the power ranges from 61% to 98% for the OR strategy. Again, the extremes occur at differing assumed dengue distributions for the two methods. Table 4 of supplementary material available at Biostatistics online provides similar results for 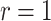 . Given the way the simulations were performed, it is immediately apparent why the results are identical for the direct OR approach. Results for the other three techniques are very similar with an incremental increase in power for the test-positive fraction method as previously noted (the results for the GEE method only appear the same–differences occur beyond the fourth decimal place).
. Given the way the simulations were performed, it is immediately apparent why the results are identical for the direct OR approach. Results for the other three techniques are very similar with an incremental increase in power for the test-positive fraction method as previously noted (the results for the GEE method only appear the same–differences occur beyond the fourth decimal place).
Table 4.
The bias and standard deviation for the random effects odds ratio estimates of the relative risk from 10 000 unconstrained intervention allocations
| Ratio = 1 | Ratio = 4 | |||||
|---|---|---|---|---|---|---|
Relative risk (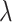 ) ) |
Bias | Average estimated standard deviation of log(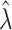 ) ) |
True standard deviation of log(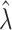 ) ) |
Bias | Average estimated standard deviation of log(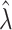 ) ) |
True standard deviation of log(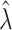 ) ) |
| 1 | 0.0284 | 0.2239 | 0.2366 | 0.0293 | 0.2262 | 0.2390 |
| 0.6 | 0.0169 | 0.2240 | 0.2364 | 0.0171 | 0.2263 | 0.2388 |
| 0.5 | 0.0141 | 0.2240 | 0.2365 | 0.0142 | 0.2263 | 0.2390 |
| 0.4 | 0.0113 | 0.2242 | 0.2364 | 0.0113 | 0.2264 | 0.2388 |
| 0.3 | 0.0083 | 0.2248 | 0.2438 | 0.0081 | 0.2270 | 0.2367 |
Tables 5 and 6 of supplementary material available at Biostatistics online present analogous results for the situation where assignment of the intervention labels is constrained as described above. Now, on average, the test-positive fraction and OR methods are both quite conservative in this situation (as is the random effects model), although the OR test remains considerably less so. For this case, at least, it appears that the test thresholds should be relaxed (thereby gaining additional power) to produce a 5% Type I error; this is, of course, most easily achieved by using the exact permutation distribution. The constrained randomization power is only modestly greater than for unconstrained randomization although this would likely be improved by using the exact permutation distribution in each case.
Table 5.
The coverage averaged across the 10 000 intervention allocations and nine time periods for the proposed odds ratio method, the random effects odds ratio estimates, and the test-positive fraction method using pooled variance estimation
Any value of 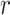
|
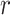 = 1 = 1
|
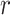 = 4 = 4
|
|||
|---|---|---|---|---|---|
Relative risk (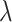 ) ) |
Direct odds ratio method | Random effects method | Test-positive method | Random effects method | Test-positive method |
| 1 | 0.9251 | 0.9258 | 0.9494 | 0.9257 | 0.9507 |
| 0.6 | 0.9628 | 0.9635 | 0.9478 | 0.9635 | 0.9544 |
| 0.5 | 0.9629 | 0.9636 | 0.9463 | 0.9636 | 0.9575 |
| 0.4 | 0.9629 | 0.9638 | 0.9445 | 0.9638 | 0.9626 |
| 0.3 | 0.9629 | 0.9642 | 0.9426 | 0.9640 | 0.9713 |
The proposed odds ratio method is invariant to 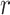 .
.
We also examined point and interval estimation of the RR based on the test-positive fraction (using both the homogeneous variance assumption and the Welch adjusted method), OR, and random effects logistic regression techniques. We examined the identical 10 000 random permutations used for our power calculations above for both 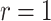 and
and 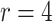 . Table 3 shows the bias of point estimates based on the test-positive fraction quadratic estimation procedure, with a comparison of the average estimated standard deviation of the differences of the test-positive fractions (i.e.,
. Table 3 shows the bias of point estimates based on the test-positive fraction quadratic estimation procedure, with a comparison of the average estimated standard deviation of the differences of the test-positive fractions (i.e., 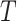 ), based on the estimator given in (4.3) with pooled variance, to the true standard deviation of
), based on the estimator given in (4.3) with pooled variance, to the true standard deviation of 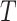 across the 10 000 simulations. We show the bias on the scale of
across the 10 000 simulations. We show the bias on the scale of 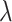 and the standard deviation comparison on the (symmetric) scale for
and the standard deviation comparison on the (symmetric) scale for 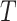 on which confidence intervals are first calculated. In practice, these confidence intervals are subsequently transformed back to
on which confidence intervals are first calculated. In practice, these confidence intervals are subsequently transformed back to 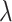 . The table shows very little bias in the estimation strategy (with very slight overestimation, i.e., closer to the null here) and that (4.3) provides a very good approximation to the variance of
. The table shows very little bias in the estimation strategy (with very slight overestimation, i.e., closer to the null here) and that (4.3) provides a very good approximation to the variance of 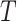 , even away from the null where the common variance assumption is not exactly satisfied. This suggests that there will be little value in turning to the more complex Welch version of the t-test. This is examined further below when we consider confidence interval coverage.
, even away from the null where the common variance assumption is not exactly satisfied. This suggests that there will be little value in turning to the more complex Welch version of the t-test. This is examined further below when we consider confidence interval coverage.
A similar analysis was implemented for the direct OR estimator where bias is assessed on the OR scale but standard deviations are compared on the log scale (where confidence intervals are calculated). Note that here, the term “bias” is a misnomer as the OR estimator targets a marginal effect whereas, in the simulated data, 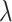 denotes a cluster-specific effect. As noted above, the OR estimate for a specific sample has zero bias as a single random draw from the permutation distribution of the OR estimators. Thus, here the bias term refers to the difference between the population-averaged, or marginal OR, and the true cluster-specific OR. This difference moves the OR estimate slightly towards the null as would be expected. Specifically, the bias is 0.0287, 0.0172, 0.0143, 0.0115, and 0.0086 for
denotes a cluster-specific effect. As noted above, the OR estimate for a specific sample has zero bias as a single random draw from the permutation distribution of the OR estimators. Thus, here the bias term refers to the difference between the population-averaged, or marginal OR, and the true cluster-specific OR. This difference moves the OR estimate slightly towards the null as would be expected. Specifically, the bias is 0.0287, 0.0172, 0.0143, 0.0115, and 0.0086 for 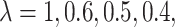 and
and 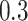 , respectively. The average estimated standard deviation of
, respectively. The average estimated standard deviation of 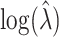 is 0.2348 whereas the true standard deviation is 0.2435, these values not depending on
is 0.2348 whereas the true standard deviation is 0.2435, these values not depending on 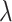 ; there is no variation of the true standard deviation of
; there is no variation of the true standard deviation of 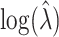 as
as 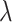 changes since the simulations at differing
changes since the simulations at differing 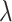 do not allow for stochastic variation around the reduced
do not allow for stochastic variation around the reduced 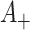 counts as previously noted. Thus, for any given permutation labeling, the estimator
counts as previously noted. Thus, for any given permutation labeling, the estimator 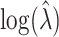 is simply shifted by the fixed amount
is simply shifted by the fixed amount 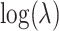 . When
. When 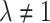 , the simulation average of the estimated standard deviation of
, the simulation average of the estimated standard deviation of 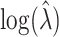 over permuted intervention assignments also does not depend on the true value of
over permuted intervention assignments also does not depend on the true value of 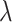 when the variant of (4.7) is used in estimation after changing the observed
when the variant of (4.7) is used in estimation after changing the observed 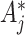 counts. This is because the
counts. This is because the 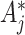 s are deterministically obtained by multiplying the (fixed but unobserved)
s are deterministically obtained by multiplying the (fixed but unobserved) 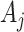 s by
s by 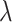 . For variance estimation, Section 4.2 then suggests inflating
. For variance estimation, Section 4.2 then suggests inflating 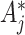 by
by 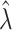 to estimate the original
to estimate the original 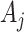 . But, as noted,
. But, as noted, 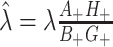 , so that
, so that 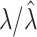 does not depend on the assumed
does not depend on the assumed 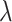 . By the same token, the estimated
. By the same token, the estimated 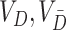 , and cov
, and cov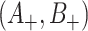 in (4.7) using the modified
in (4.7) using the modified 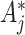 s also do not depend on
s also do not depend on 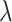 . Note that, at the null, no modification of the observed
. Note that, at the null, no modification of the observed 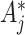 s is necessary if one assumes
s is necessary if one assumes 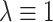 : in this case, without modification, the average estimated standard deviation of
: in this case, without modification, the average estimated standard deviation of 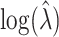 is 0.2363, very similar to that recorded when adjustments are made even at the null. These simulations indicate that (4.7) provides a good approximation to the true permutation variation of the estimator.
is 0.2363, very similar to that recorded when adjustments are made even at the null. These simulations indicate that (4.7) provides a good approximation to the true permutation variation of the estimator.
Table 4 provides analogous average results based on a simple random effects logistic regression model. The bias is acceptably small (and here the random effects model indeed targets the appropriate cluster-specific effect), and the model-based standard deviation estimator (on the log scale) adequately estimates the true standard deviation with this number of clusters.
Finally, Table 5 provides coverage properties of approximate confidence interval methods associated with the three methods. The OR method works on the log scale and uses (4.7) to provide the relevant estimated standard deviation, before subsequently transforming back by exponentiating; the test-positive fraction method is based on the scale of 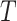 , using (4.3) for standard deviation estimates and then transforms back to the scale of the RR (noting that this assumes two independent samples of
, using (4.3) for standard deviation estimates and then transforms back to the scale of the RR (noting that this assumes two independent samples of 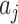 s in the two arms as discussed in Section 4.1); and the simple random effects model also works on the log scale using model-based variance estimation before transforming back to the original scale. The coverage estimates are again averaged across the simulated permuted intervention assignment labels for a specific dengue and OFI distribution and then averaged across the nine possible scenarios. All of the methods provide reasonable coverage for each case considered with little to choose between them. The Welch–Satterthwaite modification to the test-positive fraction method makes very little difference to coverage, very slightly improving the performance when
s in the two arms as discussed in Section 4.1); and the simple random effects model also works on the log scale using model-based variance estimation before transforming back to the original scale. The coverage estimates are again averaged across the simulated permuted intervention assignment labels for a specific dengue and OFI distribution and then averaged across the nine possible scenarios. All of the methods provide reasonable coverage for each case considered with little to choose between them. The Welch–Satterthwaite modification to the test-positive fraction method makes very little difference to coverage, very slightly improving the performance when 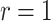 (e.g., in the best case considered, increasing coverage to 0.9434 when
(e.g., in the best case considered, increasing coverage to 0.9434 when 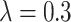 ) but working in the opposite direction when
) but working in the opposite direction when 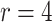 (e.g., in the worst case considered, increasing coverage to 0.9651 when
(e.g., in the worst case considered, increasing coverage to 0.9651 when 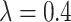 ).
).
In the simulation scenarios examined, the random effects logistic regression model is reasonable. However, it is premature to speculate that this will remain true in other simulation scenarios or with a different number of clusters. In Section 4.2 we showed that, with 10 clusters, the performance of a random effects logistic regression model is unsatisfactory. Further research will be needed to demonstrate conditions where the latter approach is reliable.
6. Discussion
The CR-TND provides an excellent approach to evaluating the efficacy of an intervention randomly applied to clusters that allows for clinic-based surveillance of disease outcomes. Our simulations are necessarily limited although motivated by the specific application. Even here, the simulations only consider one OFI distribution where, in reality, the observed distribution may differ. Further analysis of the various methods in a wider variety of other settings would be valuable. In particular, evaluation of the methods for data generated by designs other than parallel arm interventions is of immediate interest. Consideration of the CR-TND with a stepped wedge design (Hussey and Hughes, 2007) may provide an appealing alternative design in many contexts.
The methods considered here use cluster summaries of the observed frequencies of dengue and OFI outcomes. The methods also assume no interference across cluster boundaries in terms of the intervention and outcome. In the Yogyakarta trial, data will be collected on mobility of participants in the immediate time window proceeding the relevant clinic visit, information that will account for the percentage of time spent in intervention and untreated clusters. In addition, contemporaneous assessments of Wolbachia prevalence in trapped mosquitoes by cluster will be measured throughout the trial. Both of these will allow construction of an index of “Wolbachia exposure” prior to disease onset precipitating a clinic visit. The ability to capture an exposure assessment immediately prior to the onset of clinical symptoms presents a significant advantage to a cohort design where such measurements would be challenging absent constant surveillance. This kind of exposure measure, and other factors of interest, introduce individual level covariates that may explain some of the variation in dengue, as compared to other OFI, incidence. Such data then demands the extension of the permutation-based inference methods considered here to allow for individual-level explanatory covariates. This requires extension of the methods of Small and others (2008) (and earlier authors) to the CR-TND context. This article summarizes methods to extend exact permutation inference to account for covariance adjustment in cluster-randomized trials with continuous outcomes and sharp null hypotheses. More recent work extends these methods to handle composite null hypotheses with binary outcomes (Fogarty and others, 2017; Keele and others, 2017). Extension of these ideas to the CR-TND design represents an important area of current research.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Christl Donnelly and Immo Kleinschmidt for inspiration and helpful comments. Conflict of Interest: None declared.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at http://biostatistics.oxfordjournals.org.
REFERENCES
- Anders, K. L., Cutcher, Z., Kleinschmidt, I., Donnelly, C. A., Ferguson, N. M., Indriani, C., O’Neill, S. L., Jewell, N. P. and Simmons, C. P. (2018). Cluster randomized test-negative design (CR-TND) trials: A novel and efficient method to assess the efficacy of community level dengue interventions. American Journal Epidemiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy, S. L., Gibberd, R., Hancock, L., Howley, P., Kennedy, B., Klar, N., Lipsitz, S. and Ryan, L. (2000). Analysis of dichotomous outcome data for community intervention studies. Statistical Methods in Medical Research 9, 135–159. [DOI] [PubMed] [Google Scholar]
- Dutra, H. L., Rocha, M. N., Dias, M. N., Mansur, S. B., Caragata, E. P., and Moreira, L. A. (2016). Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aaedes aegypti mosquitoes. Cell Host and Microbe 19, 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty, C. B., Shi, P., Mikkelsen, M. E. and Small, D. S. (2017). Randomization inference and sensitivity analysis for composite null hypotheses with binary outcomes in matched observational studies. Journal of American Statistical Association 112, 321–331. [Google Scholar]
- Haber, M., An, Q., Foppa, I. M., Shay, D. K., Ferdinands, J. M. and Orenstein, W. A. (2015). A probability model for evaluating the bias and precision of influenza vaccine effectiveness estimates from case-control studies. Epidemiology and Infection 143, 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, R. J. and Moulton, L. H. (2009). Cluster Randomised Trials. Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- Hilgenboecker, K., Hammerstein, P., Schlattman, P., Telschow, A. and Werren, J. H. (2008). How many species are infected with Wolbachia?–A statistical analysis of current data. FEMS Microbiology Letters 281, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, M. A. and Hughes, J. P. (2007). Design and analysis of stepped wedge cluster randomized trials. Contemporary Clinical Trials 28, 182–191. [DOI] [PubMed] [Google Scholar]
- Jackson, M. L. and Nelson, J. C. (2013). The test-negative design for estimating influenza vaccine effectiveness. Vaccine 31, 2165–2168. [DOI] [PubMed] [Google Scholar]
- Johnson, K. N. (2015). The impact of Wolbachia on virus infection in mosquitoes. Viruses 7, 5705–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele, L., Small, D. S. and Grieve R. (2017). Randomization-based instrumental variables methods for binary outcomes with an application to the ‘IMPROVE’ trial. Journal of Royal Statistical Society : Series A 180,569–586. [Google Scholar]
- McNeish D. and Stapleton, L. M. (2016). Modeling clustered data with very few clusters. Multivariate Behavioral Research 51, 495–518. [DOI] [PubMed] [Google Scholar]
- Morel, J. G., Bokossa, M. C. and Neerchal, N. K. (2003). Small sample correction for the variance of GEE estimators. Biometrical Journal 45, 395–409. [Google Scholar]
- Mortimer, K., Ndamala, C. B., Naunje, A. W., Malava, J., Katundu, C., Weston, W., Havens, D., Pope, D., Bruce, N. G. and Nyirenda, M. (2017). A cleaner burning biomass-fuelled cookstove intervention to prevent pneumonia in children under 5 years old in rural Malawi (the Cooking and Pneumonia Study): a cluster randomised controlled trial. Lancet 389, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, W. and Wall, M. M. (2002). Small-sample adjustments in using the sandwich variance estimator in generalized estimating equations. Statistics in Medicine 21, 1429–1441. [DOI] [PubMed] [Google Scholar]
- Rainey, S. M., Shah, P., Kohl, A. and Dietrich, J. (2014). Understanding the Wolbachia-mediated inhibition of arboviruses in mosquitoes: progress and challenges. Journal of General Virology 95, 517–530. [DOI] [PubMed] [Google Scholar]
- Satterthwaite, F. E. (1946). An approximate distribution of estimates of variance components. Biometrics Bulletin 2, 110–114. [PubMed] [Google Scholar]
- Skowronski, D. M., Gilbert, M. and Tweed, S. A. (2005). Effectiveness of vaccine against medical consultation due to laboratory-confirmed influenza: results from a sentinel physician pilot project in British Columbia, 2004-2005. Canada Communication Disease Report 18, 181–191. [PubMed] [Google Scholar]
- Small, D. S., Ten Have, T. R. and Rosenbaum, P. R. (2008). Randomization inference in a group-randomized trial of treatments for depression: covariate adjustment, noncompliance, and quantile effects. Journal of American Statistical Association 103, 271–279. [Google Scholar]
- Sullivan, S. G., Feng, S. and Cowling, B. J. (2014). Influenza vaccine effectiveness: potential of the test-negative design. A systematic review. Expert Review of Vaccines 13, 1571–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, S. G., Tchetgen Tchetgen, E. J. and Cowling, B. J. (2016). Theoretical basis of the test-negative design for assessment of influenza vaccine effectiveness. American Journal of Epidemiology 184, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, S. M. (1985). On covariance in finite population sampling. The Statistician 34, 429–433. [Google Scholar]
- Welch, B. L. (1938). The significance of the difference between two means when the population variances are unequal. Biometrika 29, 350–362. [Google Scholar]
- Welch, B. L. (1947). The generalization of “student’s’ problem when several different population variances are involved. Biometrika 34, 28–35. [DOI] [PubMed] [Google Scholar]
- Wilson, A. L., Boelaert, M., Kleinschmidt, I., Pinder, M., Scott, T. W., Tusting, L. S. and Lindsay, S. W. (2015). Evidence-based vector control? Improving the quality of vector control trials. Trends in Parasitology 31, 380–390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


