Abstract
Atherosclerosis underlies the predominant number of cardiovascular diseases and remains a leading cause of morbidity and mortality worldwide. The development, progression and formation of clinically relevant atherosclerotic plaques involves the interaction of distinct and over-lapping mechanisms which dictate the roles and actions of multiple resident and recruited cell types including endothelial cells, vascular smooth muscle cells, and monocyte/macrophages. The discovery of non-coding RNAs (ncRNAs) including microRNAs, long non-coding RNAs, and circular RNAs, and their identification as key mechanistic regulators of mRNA and protein expression has piqued interest in their potential contribution to atherosclerosis. Accruing evidence has revealed ncRNAs regulate pivotal cellular and molecular processes during all stages of atherosclerosis including cell invasion, growth, and survival; cellular uptake and efflux of lipids, expression and release of pro- and anti-inflammatory intermediaries, and proteolytic balance. The expression profile of ncRNAs within atherosclerotic lesions and the circulation have been determined with the aim of identifying individual or clusters of ncRNAs which may be viable therapeutic targets alongside deployment as biomarkers of atherosclerotic plaque progression. Consequently, numerous in vivo studies have been convened to determine the effects of moderating the function or expression of select ncRNAs in well-characterized animal models of atherosclerosis. Together, clinicopathological findings and studies in animal models have elucidated the multifaceted and frequently divergent effects ncRNAs impose both directly and indirectly on the formation and progression of atherosclerosis. From these findings’ potential novel therapeutic targets and strategies have been discovered which may pave the way for further translational studies and possibly taken forward for clinical application.
Keywords: Atherosclerosis, Non-coding RNA, microRNA, Vascular smooth muscle cells, Endothelial cells, Macrophages
1. General introduction to atherosclerosis
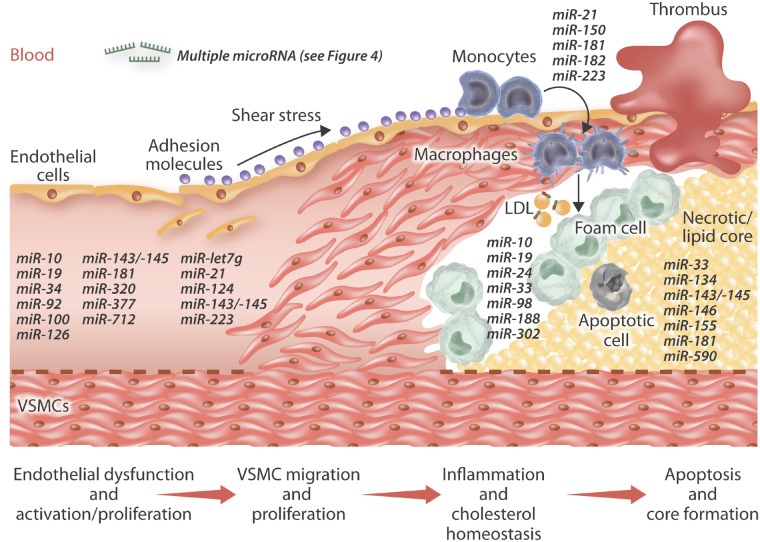
Atherogenesis is initially characterized by substantial alterations in the inner arterial surface. A normal artery consists of three tissues layers: the inner layer (endothelium), a middle layer (intima and media), and the outer layer (adventitia). The permeation, trapping and physicochemical modification of circulating lipoprotein particles in the sub-endothelial space represents the earliest detectable change towards the formation of an atherosclerotic lesion.1 However, although this may be the case in animal models of atherosclerosis, in humans, the accumulation and subsequent modification of lipoproteins is thought to occur where adaptive intimal thickenings have previously developed.2 Adaptive intimal thickenings are primarily located at atheroprone areas in response to disturbed blood flow (such as bifurcations and curved arterial regions) and are distinguished by intimal accrual of vascular smooth muscle cells (VSMCs) embedded within specific extracellular matrix (ECM) proteins such as the proteoglycans decorin and biglycan, which contribute to the accumulation, retention, and subsequent modification of lipoproteins.2
In both humans and animal models, intimal lipid accumulation is associated with changes in endothelial permeability in response to endothelial cell (EC) activation.3 Activated ECs undergo phenotypic changes including abnormal migration, proliferation, and altered expression of adhesion molecules and chemokines. These, in turn, stimulate the adhesion, transmigration, and accretion of inflammatory white blood cells such as monocytes, within the subendothelial space and developing intima. Once within the intima, monocytes differentiate into macrophages and express an array of scavenger receptors and Toll-like receptors, which have been proposed to contribute to the formation of foam cell macrophages.4 In particular, scavenger receptors facilitate the uptake of modified low-density lipoproteins (LDLs) by macrophages in the artery wall, which triggers local inflammation and ultimately leads to the development of the atherosclerotic lesion.4
As part of plaque development within animal models, stimuli released from inflammatory cells induce VSMC translocation from the medial layer of the arterial wall into the intima. Migrating VSMCs lose their characteristic contractile phenotype, start to proliferate and synthesize ECM proteins, thus actively contributing to plaque formation through establishment of a fibrous cap.5 While cellular proliferation is common during the early stages of the atherosclerotic lesion formation, advanced plaques are characterized by significant levels of VSMC and foam cell macrophage apoptosis. Hence, dead cells and lipids build up within the plaque resulting in the development of the lipid-rich necrotic core.6 In the absence of expansive remodelling, plaques generally can cause marked stenosis which limits blood flow and can ultimately result in tissue ischaemia.7 Concurrently, VSMC death alongside focal accrual of protease-rich foam cell macrophages, increases the risk of plaque rupture as the ECM is essential for maintaining the integrity of the fibrous cap and accompanying preservation of plaque stability. If the fibrous cap of a plaque ruptures, blood coagulation components encounter the thrombogenic plaque core resulting in thrombus formation, which if large enough within a coronary plaque will induce a myocardial infarction and possibly death.
2. General introduction to non-coding RNAs
The development of full genome sequencing techniques has made it possible to survey the transcriptomes of multiple organisms to an unprecedented level. In this context, large genomic projects such as FANTOM8,9 and ENCODE10,11 have marked the beginning of the ‘post-genomic era’. These extensive studies have provided the scientific community with the knowledge that although the majority (70–80%) of the mammalian genome is transcribed, only a tiny part (1–2%) of the transcriptionally active regions correspond to protein-coding genes. Pervasive transcription produces a vast repertoire of non-coding RNAs (ncRNAs) of all sizes and shapes, including short ncRNAs (such as microRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (cRNAs). Collectively, ncRNAs have been proposed to play pivotal roles in modulating a previously underestimated complexity in gene regulatory networks.
3. An introduction to lncRNAs
Among ncRNAs, lncRNAs represent the widest and most heterogeneous class. These are transcripts exceeding 200 nucleotides (nt) in length, with no significant protein coding capacity.12 The majority of lncRNAs are transcribed by RNA polymerase II, they undergo splicing, present 5′ caps and are polyadenylated.13 Based on their genomic location relative to their neighbouring protein-coding genes, lncRNAs can be distinguished into intergenic (lincRNAs), exonic, intronic, or fully overlapping. Antisense (AS) lncRNAs are transcribed from the opposite DNA strand overlapping with exons of a protein-coding gene; these have been often shown to contribute to canonical regulation of cognate sense genes.14 LncRNAs contribute to gene expression regulation at different levels by cis or trans-acting. The extensive diversity in their mechanisms of action and functional outputs is strictly linked to their anatomical properties, subcellular localization and interactions with molecular partners. LncRNA may act as a ‘molecular sink’ sequestering different factors from their site of action; they can work as scaffolds assembling molecular effectors; they can guide the localization of ribonucleoprotein complexes to specific target genes and they can function as molecular signals to indicate gene regulation in space and time.15 Examples of lncRNAs intervening in transcriptional and translational regulation, cellular trafficking, nuclear organization, and compartmentalization have been shown.16 Other studies showed that the structure of lncRNAs is more highly conserved across different species than their primary sequence, suggesting a strict link between structural and functional features.17,18 Since their recognition, lncRNAs have been reported to be involved in normal organism development and physiology, as well as in the pathogenesis of multiple diseases.19–21 Additionally, recent years has seen an abundance of studies examining lncRNA expression and modulation in clinical samples, animal models and cell systems mimicking atherosclerosis, and these are discussed below and summarized in Table 1. Their proposed mechanisms of action are also summarized within Figure 1.
Table 1.
Long non-coding and circular RNAs in atherosclerosis-related research
| Non-coding RNA name | Model | Atherogenic/ atheroprotective | Experimentally validated function | References |
|---|---|---|---|---|
| ANRIL | Human vascular tissue and peripheral blood | Atherogenic | Associated to the cardiovascular disease locus 9p21.3 | 22–24 |
| MALAT1 | HUVECs | Atherogenic | Up-regulated upon endothelial dysfunction; induces proliferation | 25 |
| MEG3 | HUVECs | Atherogenic | Up-regulated upon endothelial dysfunction | 25 |
| sONE | ECs | Atherogenic | Inhibits eNOS expression; up-regulated upon hypoxia | 26 , 27 |
| SENCR | Human ECs and VSMCs; HUVECs | Atheroprotective (in early stages) | Flow-responsive, favours endothelial integrity; maintenance of contractile phenotype; increases proliferation | 28–30 |
| MIAT | ECs | Not assessed | Regulates ECs function via control of VEGF expression | 31 |
| Dll4-AS | Human and mouse ECs | Not assessed | Knock-down decreases proliferation and migration and enhances sprouts formation | 32 , 33 |
| MeXis | Human and mouse macrophages | Atheroprotective | Controls cholesterol efflux via regulation of ABCA1 transcription | 34 |
| lnc-Ang362 | Rat VSMCs | Atherogenic | Induced upon AngII stimulation; knock-down impairs proliferation | 35 |
| lincRNA-p21 | Human and mouse; ECs, VSMCs and macrophages | Atheroprotective (in early stages)/upon stenosis | Represses proliferation and induces apoptosis; down-regulated in atherosclerosis models and patients | 36 , 37 |
| HIF-AS1 | VSMCs and ECs | Not assessed | Represses proliferation and induces apoptosis | 38 , 39 |
| cANRIL | Human vascular tissue and peripheral blood | Atheroprotective (in early stages) | Inhibits proliferation via interference with ribosomal RNA maturation | 40 |
| hsa_circ_0124644 | Human peripheral blood | Atherogenic | Biomarker for CAD | 41 |
| hsa_circ_0003575 | HUVECs | Atherogenic | Up-regulated upon oxLDL treatment | 42 |
| hsa_circ_000595 | Human aortic VSMCs | Atherogenic (in advanced lesions) | Up-regulated upon hypoxia; induces apoptosis | 43 , 44 |
| Circ_Lrp6 | Human and mouse VSMCs | Not assessed | Knock-down decreases VSMCs proliferation and migration and reduces stented carotid intima hyperplasia in mouse | 45 |
| cZNF292 | ECs | Not assessed | Up-regulated upon hypoxia; induces proliferation and sprout formation | 46 |
AngII, angiotensin II; CAD, coronary artery disease; ECs, endothelial cells; HUVECs, human umbilical vein endothelial cells; VSMCs, vascular smooth muscle cells.
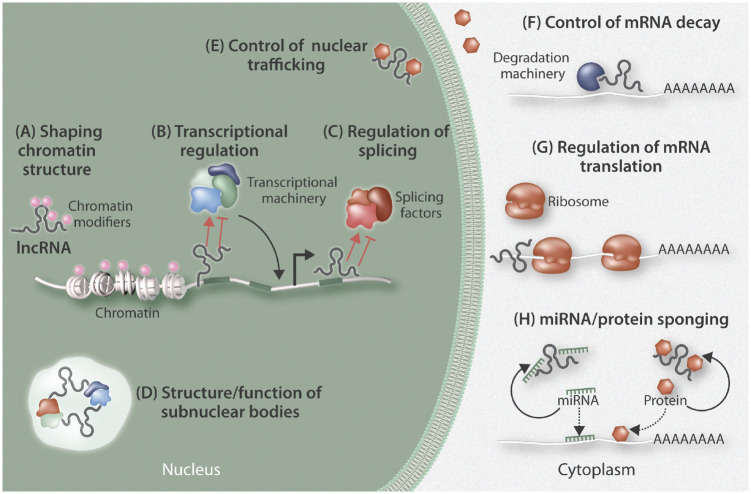
Figure 1.
Overview of the cellular regulation of long non-coding RNAs (lncRNAs). LncRNA may act both within nuclear and cytoplasmic compartments. Within the nucleus, they contribute to shaping chromatin structure and accessibility via recruitment of chromatin modifiers (A); they can regulate transcription rate by modulating transcription factor availability at transcription start sites (B); they can control RNA splicing by directing the splicing machinery (C); they can also work as scaffolding structures through provision of components to aid the formation of specific subnuclear bodies (D); they regulate the shuttling of proteins between cellular compartments (E). In the cytoplasm, lncRNAs can regulate mRNA turnover by guiding the degradation machinery to specific transcripts (F); they can also dictate translational regulation through actions such as blocking ribosome binding to RNA (G), or as ‘molecular sinks’ to sequester different factors (microRNAs and proteins) from their site of action (H).
3.1 LncRNAs in atherosclerosis
In the last decade, genome-wide association studies (GWAS) unveiled an increasing number of genetic loci linked to coronary artery disease (CAD) risk inheritance. Among these, the Chr9p21 locus has been extensively studied, with a special focus on a cluster of five genes which include the 3.8 kb long ANRIL ncRNA and the tumour suppressors cyclin dependent kinase inhibitor CDKN2A/p16INK4A, CDKN2A/p14ARF, CDKN2B/p15INK4B, and methylthioadenosine phosphorylase (MTAP).47 Interestingly, single-nucleotide polymorphisms (SNPs) conferring cardiovascular risk do not span the protein-coding regions of the locus (i.e. CDKN2A/p16INK4A, CDKN2A/p14ARF, CDKN2B/p15INK4B and MTAP), but rather fall within the lncRNA ANRIL introns.48
ANRIL overlaps in antisense orientation the entire CDKN2B/p15INK4B gene and was therefore referred to as CDKN2B antisense RNA (CDKN2B-AS1). More than 20 linear ANRIL isoforms, as well as multiple circular isoforms have been reported (www.ensembl.org). Interestingly, Jarinova et al.49 showed that ANRIL expression was induced by the CAD risk SNP rs1333049 in peripheral blood monocytes (PBMCs), with no significant effects on expression of CDKN2A or CDKN2B. Transcriptional profiling of these genes was later carried out in diverse tissues, primary cells and cell lines relevant to atherosclerosis. Most of the studies investigating ANRIL expression found an association with the Chr9p21 genotype (reviewed in ref.50). In particular, patients carrying the CAD-risk allele were found to predominantly express linear ANRIL isoforms containing the proximal and distal exons; moreover, ANRIL expression in plaques, circulating PBMCs or whole blood correlated with atherosclerosis severity.22–24 Conversely, circular ANRIL (circANRIL) isoforms were down-regulated in patients with the Chr9p21 risk haplotype and inversely correlated with atherosclerotic severity.40 Interestingly, when the effects of Chr9p21 were simultaneously investigated on both ANRIL and CDKN2B in large cohorts, a stronger genotype/expression correlation was identified for ANRIL compared to CDKN2B.22,40,51 Overall, the scenario sees a general trend towards an inverse correlation between circANRIL or CDKN2B, which are down-regulated in patients with the CAD-risk genotype, and linear ANRIL isoforms, which are on the contrary up-regulated. Recent data provided evidence of ANRIL acting also in trans on non-overlapping genes.51 Holdt et al. reported that the functional modules responsible for ANRIL trans-regulation consists of Alu repeats contained in the transcript sequence. These would facilitate recruitment of Polycomb group proteins to Alu-containing promoters of target genes, most likely through RNA: DNA interactions enabled by the presence of highly homologous Alu elements.
ANRIL is expressed in ECs, VSMCs, inflammatory cells and tissues that are affected by atherosclerosis.52 Silencing of ANRIL in human aortic VSMCs by siRNA, alternatively targeting exon1 or exon19, has been shown to differentially modulate the expression of genes controlling apoptosis, proliferation, inflammation, and ECM remodelling; namely BCL2-related protein A1 (BCL2A1), baculoviral IAP repeat containing 3 (BIRC3), cadherin 5 (CDH5), and heparin-binding EGF-like growth factor, thus suggesting isoform-specific regulatory properties.53 Recently Lo Sardo et al.54 generated induced pluripotent stem cell-derived VSMCs from CAD risk and non-risk individuals and deleted the region corresponding to the ∼60 kb risk haplotype (which is depleted of coding genes) by taking advantage of TALEN technology.54 Transcriptional profiling revealed that VSMCs from CAD risk individuals displayed altered gene expression patterns, resembling those previously identified in CAD risk individuals. Furthermore, they exhibited aberrant adhesion, contraction, and proliferation. Deletion of the risk haplotype rescued VSMC normal phenotype and, conversely, forced expression of the lncRNA ANRIL induced risk phenotypes in non-risk VSMCs.
3.1.1 Endothelial cells
It is acknowledged that atherosclerosis is a chronic inflammatory disease which develops at specific regions within the arterial wall such as branch points and prominent curvatures where disturbed blood flow prevails.55 The altered shear stress at such sites can exert profound effects on the ECs including altered migratory and proliferative responses alongside modulating their susceptibility to apoptosis and permeability,55 permitting the insudation of lipoproteins within adaptive intimal thickenings which form at such sites.2 Accordingly, EC-derived ncRNA expression and their contributory roles on cell behaviour have been explored in response to haemodynamic alterations and exposure to pro-atherosclerosis risk factors. Deep sequencing of polyA-RNA from human umbilical vein endothelial cells (HUVECs) showed that the expression levels of some lncRNAs, including the metastasis associated lung adenocarcinoma transcript 1 (MALAT1), MEG3, TUG1, linc00493, and linc00657, were comparable with the ones observed for endothelial coding genes, such as vascular endothelial growth factor (VEGF) receptor 2.25 Upon hypoxic stimuli MALAT1, MEG3, TUG1, and linc00657 were significantly up-regulated, suggesting a link between these lncRNAs and endothelial dysfunction characterizing the initial process of atherogenesis.
3.1.1.1 sONE
Nitric oxide (NO) plays a vital role in vascular homeostasis and is involved in dysfunction and damage of the vasculature during atherosclerosis. NO is mainly synthesized by three NO synthase (NOS) enzymes, with endothelial NOS (eNOS or NOS3) representing the vascular EC-restricted isoform. Altered eNOS expression results in abnormalities of blood pressure, platelet function, and vessel wall remodelling. In particular, advanced human atherosclerotic plaques are characterized by decreased expression of steady-state eNOS mRNA due to exposure of ECs to diverse injurious stimuli.56 Recently, sONE has been identified as a tail-to-tail overlapping AS lncRNA transcribed from the opposite strand of eNOS in VSMCs, but not within ECs. The knock-down of sONE was associated with augmented levels of eNOS in VSMCs, while sONE overexpression unusually reduced EC eNOS levels in a post-transcriptional manner.26 The expression of sONE is induced by hypoxia, resulting in negative regulation of eNOS expression in ECs.27 Together these experiments suggest that not only does sONE regulate cell-specific eNOS expression but also its expression can be modulated upon atherosclerotic stimuli such as hypoxia. Whether other stimuli involved in the development of atherosclerotic lesions, such as oxidized LDL or inflammation, can affect sONE or eNOS expression remains an open question.
3.1.1.2 SENCR
Recently, the smooth muscle and EC-enriched migrational differentiation-associated lncRNA (SENCR) was shown to be a flow-responsive lncRNA favouring endothelial integrity, suggesting that lncRNA deregulation may provide the interface between shear stress and endothelial damage, ultimately leading to atherosclerosis.28 SENCR levels were shown to be increased in several differentiated human EC lineages exposed to laminar shear stress. This was confirmed also in vivo by taking advantage of humanized SENCR expressing mice; furthermore, this lncRNA was not induced in disturbed shear stress regions. SENCR has a role in preserving EC membrane integrity, as shown by loss-of-function experiments, which highlighted increased EC permeability upon SENCR knock-down. Pull-down and mass spectrometry illustrated the interaction with cytoskeletal-associated protein 4 (CKAP4) through a non-canonical RNA-binding domain. SENCR silencing facilitated the interaction between CKAP4 and cadherin 5 (CDH5 or VE-cadherin), resulting in damaging the structure of adherens junctions through destabilization of the CDH5/CTNND1 complex and augmenting CDH5 internalization.28
3.1.1.3 MALAT1
Recent investigations have demonstrated that MALAT1 can control both epigenetic gene regulation and splicing, and changes in its expression were shown to be associated with metastasis of lung tumours.57 MALAT1 was shown to interact with polycomb 2 (CBX4) and thereby regulate histone modifications to control cellular proliferation.58 The Dimmeler lab showed that MALAT1 expression affects the balance between proliferative and migratory EC phenotype in vitro, and its genetic deletion in vivo impairs vascular growth. Silencing of MALAT1 inhibits proliferation in HUVECs by modulating the expression of cell cycle regulators, and promotes a switch towards a migratory phenotype characterized by increased basal sprouting upon pro-angiogenic conditions.25 MALAT1 expression in ECs is induced under high-glucose conditions or oxidative stress, and its knock-down results in decreased cell viability.59 According to a recent study, in high-glucose cultured ECs, MALAT1 up-regulation initiates an inflammatory cascade ultimately inducing the expression of inflammatory serum amyloid antigen (SAA3).60
3.1.1.4 MIAT
Serum levels of the lncRNA myocardial infarction-associated transcript (MIAT) are increased in patients with coronary atherosclerotic disease compared with healthy subjects, and the increased levels positively correlates with IL-6 and TNFα serum levels.61 Moreover, patients with symptomatic carotid atherosclerosis exhibit increased intra-plaque MIAT expression than individuals with asymptomatic disease or healthy controls.62 A similar pattern was also reported both within plaques and serum of mice with advanced atherosclerosis in comparison to early disease.62 With regards to ECs, MIAT can regulate their function by acting as a competing endogenous RNA, thus preventing miR-150-5p from reaching its target VEGF, an action commonly referred to as a microRNA sponge.31 MIAT knock-down in Apoe-deficient mice achieved through systemic delivery of a MIAT shRNA adenoviral vector decreased aortic atherosclerosis, supporting a pro-atherosclerotic role for this lncRNA.62 Mechanistically, the beneficial effects of MIAT knock-down were attributed to its role as a miR-149 sponge, preventing miR-149 from targeting CD47 within foam cell macrophages and subsequent loss of efficient efferocytosis, a process involved in plaque progression.62 Indeed, plaques from MIAT knock-down mice were deemed more stable than those from control animals due to observed increased collagen and VSMCs content against decreased necrotic core size and macrophage positive area.62
3.1.1.5 Dll4-AS
An antisense lncRNA transcribed from the Delta-like 4 gene, named Dll4-AS, has been shown to affect proliferation, migration, and sprouting in human and mouse ECs through modulating Dll4 expression, which is a specific ligand for the Notch1 receptor on arterial endothelium. The expression of Dll4 and Dll4-AS is driven by the same promoter and transcripts are co-regulated upon Notch-activating or inhibiting stimuli. In particular, silencing of Dll4-AS led to decreased Dll4 mRNA level and resulted in enhanced sprout formation, impaired EC proliferation and migration.32,33
3.1.1.6 ASncmtRNA-2
Vascular cell senescence has been ascribed a role in age-associated cardiovascular diseases. Replicative senescence (RS) and stress-induced premature senescence are provoked respectively by endogenous (telomere erosion) and exogenous (H2O2, UV) stimuli, resulting in cell cycle arrest in Gl and G2 phases. In both scenarios, mitochondria-derived ROS are important players in senescence initiation. In this context, ASncmtRNA-2 is a mitochondrial DNA-transcribed lncRNA whose expression was found to be increased in mouse aged aortas.63 According to in vitro experiments, ASncmtRNA-2 is induced in RS in ECs rather than in VSMCs. The authors proposed that this lncRNA may exert its action through up-regulation of miR-1973 and miR-4485, as both microRNAs were up-regulated by ASncmtRNA-2 over-expression and upon RS, eventually leading to cell cycle arrest.63
3.1.1.7 FLJ11812
Autophagy has been considered to play a protective role in atherosclerosis mainly through degrading long-lived proteins and dysfunctional organelles, as well as by facilitating removal of cholesterol from foam cell macrophages. At the same time, EC autophagy may also destroy the structural stability of the plaque and aggravate thrombosis, potentially triggering acute clinical events.64 In this setting, Ge et al.65 investigated novel factors downstream of the mTOR signalling pathway which would inhibit autophagy in HUVECs. After treatment with 3-benzyl-5-((2-nitrophenoxy) methyl)-dihydrofuran-2(3H)-one (3BDO), which stimulates mTOR, they found that a lncRNA transcribed from the TGFB2 gene and named FLJ11812 was significantly down-regulated in treated cells. This was accompanied by a strong decrease of autophagy-related 13 (ATG13) protein levels. Although the mechanism through which FLJ11812 exerts its regulatory action needs further investigation, it has been proposed that it could be via sequestering of a specific miRNA (miR-4459) targeting ATG13.65
3.1.2 Vascular smooth muscle cells
A main feature of VSMCs is their high level of plasticity which they retain even after differentiation. In normal conditions, VSMCs reside within the media, where they are primarily quiescent and typically contractile. In response to a variety of stimuli (inflammation, cyclic strain, oxLDL, etc.), VSMCs may undergo phenotypic modulation, permitting their proliferation and migration towards the intimal layer alongside taking on a synthetic phenotype, thus actively contributing to the formation of the atherosclerotic plaque.5 VSMC phenotypic modulation is a crucial process for the formation of atherosclerotic lesions, vascular remodelling, and injury repair/stabilization. As such, although VSMC phenotype switching and associated behavioural changes may be deemed detrimental during atherogenesis (particularly in humans), this process is fundamentally beneficial in advanced plaques to ensure maintenance of the protective fibrous cap.
3.1.2.1 SENCR
The aforementioned and proposed AS lncRNA SENCR is expressed in both ECs and VSMCs29 and is transcribed from the upstream of the friend leukaemia virus integration 1 (FLI1) gene locus, overlaps the FLI1 gene, presents transcriptional variants and is mainly localized within the cytosol. In particular, the transcriptional variant specific to VSMCs is explicitly detected within cells displaying a contractile phenotype. Indeed, SENCR knock-down was associated with VSMC de-differentiation and induction of migration through a yet unidentified mechanism. A similar study in ECs showed that SENCR overexpression promoted their proliferation, migration, and angiogenic function.30 Although AS lncRNAs often participate in regulation of sense neighbouring transcripts, no regulatory action has been shown for SENCR on FLI1 to date.
3.1.2.2 HAS2-AS1
VSMCs are responsible for the majority of ECM synthesis within the vessel wall. Hyaluronic acid (HA) is a multifunctional matrix protein and its accumulation can result in vessel wall thickening, thus contributing to vascular injury and atherogenesis.66 Furthermore, HA can affect VSMC function through the accumulation of adhesion molecules involved in the initiation of the immune cascade. Mammalian HA is synthesized at the cell membrane by three HA synthases (HAS): HAS1, HAS2, and HAS3. The expression of a natural antisense RNA to the HAS2 isoform (HAS2-AS1), was detected in osteosarcoma cells67 and in renal proximal tubular epithelial cells.68 In the latter, HAS2-AS1 forms a duplex with HAS2 mRNA, resulting in sense transcript stabilization and increased expression levels upon stimulation with IL-1β or TGF-β1.68 AS-mediated regulation of HA synthesis in VSMCs remains unexplored and it would be interesting to investigate whether a similar mechanism is involved during atherogenesis.
3.1.2.3 Lnc-Ang362
Dysregulated proliferation and hypertrophy of VSMCs can be induced by angiotensin II (Ang II), which can also promote inflammation, fibrosis, and cell growth. Moreover, increased endogenous or exogenous levels of Ang II can promote atherosclerotic plaque formation and progression. Accordingly, Leung et al.35 conducted transcriptome and epigenome profiling of rat VSMCs in response to Ang II treatment. They discovered that an Ang II-regulated lncRNA (lnc-Ang362) functions as the host transcript for miR-221 and miR-222, which are proposed mediators of VSMC function. Indeed, lnc-Ang362 knock-down reduced miR-221 and miR-222 expression and suppressed VSMC proliferation. Taken together the results argue for the possibility of using Ang II-regulated ncRNAs as potential novel therapeutic targets for Ang II-associated cardiovascular diseases such as atherosclerosis.
3.1.2.4 LincRNA-p21
Apoptosis of VSMCs can contribute to weakening of the plaque fibrous cap, and consequently impinge on the stability of atherosclerosis plaque. Similarly, EC loss may promote plaque erosion and encourage thrombus formation and subsequent myocardial infarction, particularly over highly stenotic plaques.69 LincRNA-p21 has been recently shown to repress proliferation and induce apoptosis in VSMCs and mouse macrophages in vitro, potentially through enhancement of p53 transcriptional activity.36 LincRNA-p21 appears to function as a component of the p53 pathway, at least in part, by physically interacting with a p53 repressive complex to down-regulate many p53 target genes.37 Interestingly, LincRNA-p21 was found to be down-regulated in both the Apoe-deficient mouse model of atherosclerosis and patients with CAD.36 Moreover, lincRNA-p21 lentiviral knock-down in the mouse carotid artery injury model resulted in marked neointimal hyperplasia.36 These findings have relevance to the VSMC hyperproliferative response observed during atherogenesis and after surgical interventions of advanced plaques where (re)stenosis can result in further vessel occlusion. In this context, the above experiments raise the possibility that manipulation of lincRNA-p21 expression could be beneficial to treat restenosis and prevent atherogenesis, but unwanted plaque destabilization effects may be encountered in advanced plaques unless localized interventions were deployed.
3.1.2.5 HIF1a-AS1
The brahma-related gene 1 (BRG1) is highly expressed by VSMCs during thoracic aortic aneurysms, where it has the effect of triggering apoptosis and reducing cell proliferation. Similar changes in expression level of the lncRNA HIF 1 alpha-antisense RNA 1 (HIF1a-AS1) were observed as those of BRG1.38 Furthermore, HIF1a-AS1 knock-down markedly promoted VSMC proliferation and reduced susceptibility to apoptosis through increasing Bcl2 expression and decreasing the expression of caspase3 and caspase8 in VSMCs and caspase9 in ECs.39 As such, HIFa-AS1 may also contribute to the development and progression of atherosclerosis through controlling VSMC and EC apoptosis.
3.1.3 Inflammatory cells
The progression and destabilization of atherosclerotic plaques is largely responsible for the majority of cardiovascular related deaths.70 Histopathological findings from human atherosclerotic plaques have illuminated our understanding of how atherosclerotic lesions progress and revealed that increasing vulnerability to rupture is related to perpetual recruitment and accumulation of monocyte/macrophages, their transformation into lipid-laden foam cells, expansion of the lipid/necrotic core, loss of VSMC content alongside decreased collagen deposition.71 Therefore, many of the deleterious characteristics of plaque progression are related to inflammation, particularly monocyte/macrophages, which is supported by the recent results of the CANTOS trial which confirmed a pivotal role for inflammation in the progression and clinical complications of atherosclerosis.72 With regards to the function and behaviour of monocyte/macrophages, ncRNAs have been proposed as harnessing important modulatory roles, such as directing the adhesion, invasion and proliferation of monocytes, affecting macrophage uptake and efflux of modified lipoproteins, macrophage phenotypic polarization, alongside the regulation and secretion of inflammatory mediators and proteases. Collectively, such findings have elucidated the novel mechanistic functions ncRNAs may exert on the inflammatory response during atherosclerosis and identified specific ncRNAs as latent therapeutic targets, concurrent with their assessment within the circulation as prognostic biomarkers of atherosclerotic disease progression.
3.1.3.1 MeXis
Based on mouse studies, MeXis is a lncRNA attributed a crucial role in atherogenesis via regulation of cholesterol metabolism.34 Highly expressed in macrophages, MeXis is up-regulated in response to cholesterol overload. MeXis and the neighbouring cholesterol-efflux gene ABCA1 are co-regulated at the transcriptional level via liver X receptor (LXR)β, which belongs to the sterol-activated nuclear receptor family controlling the expression of genes pivotal for cholesterol homeostasis. Interestingly, MeXis potentiates LXR-dependent transcription of ABCA1, which is defective in MeXis-deficient mice in a tissue-selective manner.34 Mechanistic studies revealed that MeXis exerts its action through mediating binding of the transcriptional co-activator DDX17 to the ABCA1 promoter.34 Interestingly, the LXR-MeXis-ABCA1 axis is conserved in humans, with the MeXis homologue referred to as TCONS00016111. A GWAS from the CARDIoGRAMplus consortium73 identified an association between a SNP overlapping the TCONS00016111 transcript and human CAD, highlighting the potential relevance of this lncRNA to human atherosclerosis.
4. An introduction to circular RNAs
Initially considered as aberrant splicing products, circRNAs are now known to be essential players in the regulation of physiological and pathological processes.74 Most circRNAs derive from precursor mRNA (pre-mRNA) back-splicing events, in which a downstream 5′splice site (ss) is joined and ligated with an upstream 3′ss (reviewed by ref.33). At the basis of RNA circularization, the formation of back-splicing junctions is catalyzed by the canonical spliceosomal machinery and fine-tuned by cis as well as trans elements. Cis-acting regulatory modules include intronic complementary sequences flanking the back-splicing junction, which often consist of repetitive elements, such as Alus in primates.75,76 RNA binding proteins may contribute to circRNA regulation in trans by either facilitating or destabilizing intronic RNA pairing, thus promoting or inhibiting circRNA biogenesis, respectively.77,78 cRNAs are modestly expressed and in most cases less abundant than linear transcripts.75,76,79,80 Interestingly, the expression of circRNAs is tightly regulated both spatially and temporally. A given circular transcript may display high tissue-specificity79 and expression patterns can be characteristic of a certain biological process, developmental stage or disease condition.80,81 CircRNAs are located within both the nuclear and cytoplasmic compartments and can accordingly regulate gene expression through multiple mechanisms. Evidence suggests they can participate to splicing regulation, may act as miRNA or protein ‘sponges’ and can interfere with pre-mRNA processing (reviewed in ref.82). Furthermore, there are novel indications that some endogenous circRNAs are translatable.83 Finally, circRNAs can be secreted in exosomes and body fluids including saliva and serum.84,85 In this context, their increased stability compared to linear transcripts make them potentially ideal biomarkers in clinical practice. The role of circRNA in atherosclerotic disease initiation and progression has been investigated in the last few years. Moreover, clinical and experimental studies have highlighted the potential diagnostic value of these particular transcripts in atherosclerosis prevention and treatment (see Table 1). Their proposed mechanisms of action are also summarized within Figure 2.
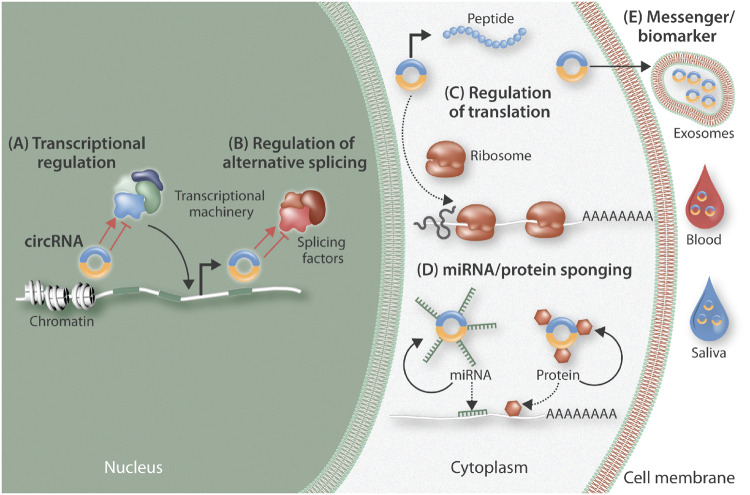
Figure 2.
Overview of the cellular regulation of circular RNAs (circRNAs). CircRNAs may can exert actions within the nucleus, the cytoplasm, or as secreted molecules. Within the nucleus, they can contribute to transcriptional (A) and splicing (B) regulation. Within the cytoplasm, circRNAs can affect translational regulation through actions such as blocking ribosome binding to RNA or alternatively, they can be translated into small pepdtides (C); they can also serve as ‘molecular sinks’ to sequester different factors (microRNAs and proteins) from their site of action (D). CircRNAs can be secreted within exosomes and therefore participate in intercellular communication, while their presence within bodily fluids suggests they may be able to be potentially exploited as biomarkers.
4.1 CircularRNAs in atherosclerosis
As atherosclerosis underlies the bulk of cardiovascular disorders, new and highly sensitive/convenient diagnostic biomarkers patterning atherosclerosis development are required which may aid monitoring disease progression, and therefore be highly valuable in terms of human health, as well as social economics. Their structural properties alongside identification of circRNA presence in body fluids such as plasma and saliva, pave the way for their application as biomarkers. In the field of cancer, hsa_circ_002059, whose expression is significantly higher in gastric cancer tissue compared to healthy adjacent tissue, has been proposed as a potential biomarker for diagnosis of gastric cancer.86 Further examples are provided by circ-ITCH 2087 and hsa_circ_0005075 21,88 in oesophageal cancer and hepatocellular carcinoma, respectively. In this context, by using microarray technology, Zhao et al.41 profiled peripheral blood circRNA expression in CAD patients and matched healthy controls and revealed hsa_circ_0124644 was a sensitive and specific disease biomarker. Arrays examining circRNA expression have proved to be valid tools for differential expression analysis in pathologies of interest, with some advantages compared to RNA-sequencing. Indeed, low count numbers associated to such transcripts often impair accuracy in quantification, making analysis prone to an increased rate of error,89 unless extremely high sequencing depth is adopted. Furthermore, although computational approaches for circRNA detection are seeing continual improvement, annotation and analysis pipelines are rather complex and generally not widely available.90 Conversely, microarray technology is characterized by high sensitivity and is relatively unaffected by lowered transcript levels specific to circRNAs.91
4.1.1 Circ-000595
In a study investigating differential expression of circRNAs in abdominal aortic aneurysms, heightened expression of the circRNA hsa_circ_000595 was associated with disease progression through regulating VSMCs apoptosis under hypoxic conditions.43 Upon cobalt chloride (CoCl2)-induced hypoxia in vitro, circ_000595 was up-regulated and subsequent siRNA-directed silencing decreased hypoxia-induced apoptosis rates in VSMCs.43 Furthermore, circ_000595 knock-down was shown to be associated with increased expression of miR-19a, which is known to confer atheroprotection via flow-regulated control of endothelial proliferation.44 As hypoxia is a characteristic feature of atherosclerotic lesions,92 it would be interesting to further explore the role of circ_000595 in the broader context of atherogenesis and disease progression.
4.1.2 cZNF292
RNA-seq analysis of ECs cultured in 0.2% O2 or normoxic conditions revealed cZNF292 as another example of a hypoxia-induced circRNA.46In vitro experiments revealed that cZNF292 could stimulate proliferation.46 Subsequent silencing of cZNF292 (and not its linear counterpart) in HUVECs impaired sprouting and tube formation in matrigel assays and reduced proliferation rates. Of notice, levels of the ZNF292 pre-mRNA or mRNA host-gene remained unaltered.
4.1.3 Circ_Lrp6
A well-elucidated function of cRNAs is miRNA-sponging. Due to their miRNA-complementary binding sites, circRNA can ‘capture’ these and prevent them from reaching their sites of action. Recently Hall et al.45 discovered that a circRNA alternatively spliced from the lipoprotein receptor 6 (Lrp6) gene locus, serves as a natural miR-145 sponge. Circ_Lrp6 modulates the action of miR-145 by sequestering the latter in P-bodies, ultimately regulating VSMC migration, proliferation, and differentiation. In this context, the ratio between circ_Lrp6-bound/unbound miR-145 has been shown to be crucial in vascular disease pathology, in both human and mouse.
4.1.4 circANRIL
Probably, the most exhaustively characterized circRNA in atherosclerosis is circANRIL, which represents an example of disease-linked circularized transcript whose function and mechanism of action have been recently partially unveiled. Burd et al.93 initially found that besides the aforementioned linear ANRIL, a circRNA variant of the latter was transcribed and back-spliced from the atherosclerotic vascular disease risk region on chromoseome 9p21.3, in proximity to the INK4/ARF (CDKN2a/b) locus. Interestingly, they proposed SNPs characterizing this region would ultimately lead to vascular disease susceptibility by regulating ANRIL splicing and circANRIL production. A few years later, Holdt et al.40 demonstrated that circANRIL was involved in ribosomal RNA (rRNA) maturation in VSMCs and macrophages. In detail, pre-rRNA processing and ribosome biogenesis is impaired by binding of circANRIL to Pescadillo homologue 1 (PES1), an essential 60S-preribosomal assembly factor, resulting in nucleolar stress, activation of p53, and a subsequent increased apoptosis and decreased proliferative rate. Accordingly, the authors propose an atheroprotective role of circANRIL involving suppression of cellular proliferating during the early stages of atherosclerotic plaque development. In concert, linear ANRIL would promote while circANRIL would protect from excessive proliferation, suggesting that the genotype of Chr9p21 is crucial in regulating the balance of linear and circANRIL levels in VSMCs and macrophages. As such, a shift in the ratio towards the linear isoform of ANRIL would favour atherogenesis.40 Indeed, exogenous circANRIL expression was shown to be beneficial in a rat model of coronary atherogenesis.94 In this study, the effects of low or high exogenous circANRIL expression were evaluated by monitoring circulating levels of total cholesterol, triglycerides, LDL, and matrix metalloproteinase-9 (MMP-9), alongside pro-inflammatory and pro-apoptotic markers in ECs. All were found to be decreased in the low-expressed circANRIL group, while high-density lipoprotein (HDL) levels alongside mRNA and protein expression levels of anti-apoptotic bcl-2 were increased.94 Curiously, opposing effects were observed in the other group, that is upon elevated levels circANRIL. Taken together, the results confirm the protective role of circANRIL in atherosclerosis but adds an essential piece of information: protective effects are reverted when doses are beyond a certain threshold.
A well-described function of circRNAs, especially if residing within the cytosolic compartment, is microRNA-binding and trapping.79,95 Thus, a crucial point is the investigation of the presence of miRNA binding sites within circRNAs sequences. Although network analysis revealed the presence of miRNA target sequences in many disease-relevant circular transcripts detected in vascular cells,42,43 the molecular mechanisms and the cellular pathways underlying circRNA contribution to atherosclerosis remain vastly unexplored. However, there are a large number of circRNAs lacking sequences for interaction with miRNAs,46 thus raising the point that circRNA modulation of miRNA activity may represent only the tip of the iceberg of a wider array of modes of action. It is clear molecular investigation and the discovery of novel circRNA ‘functional prototypes’ is required to permit further research within this relatively new area in the context of atherosclerosis.45
5. An introduction to microRNAs
MicroRNAs (miRNAs, miRs) are short ncRNAs usually between 18 and 22 nucleotides long which harbour the ability to post-transcriptionally control mRNA/protein expression through either inhibition of translation or promotion of target messenger (m)RNA degradation. Within the nucleus, polymerase II positively regulates production of primary microRNAs (pri-miRs) which are then processed into smaller precursor forms (pre-miRs) by the Class 3 Ribonucleasese III Drosha in order to permit their export into the cytoplasm. Once within the cytoplasm, pre-miRs are further processed by a Class 4 ribonuclease III family member, Dicer, resulting in the formation of a mature and biologically functional microRNA which can bind the 3′ untranslated regions (3′-UTR) of target mRNA and therefore control their expression. Due to their small size, microRNAs have been predicted to yield the capacity to modulate approximately 90% of mammalian genes and hence proposed to exert an essential role in regulating key cellular functions.96 Predictive algorithms have identified that individual microRNAs can bind and regulate a large number of divergent mRNAs, accounting for the discrepancy in the ratio of microRNAs and mRNAs, although more recent evidence has shown that multiple mRNA targets of a single microRNA may cluster within a given functional network. Furthermore, due to the hairpin structure of precursor microRNA their processing results in the generation of -3p and -5p strands, which can bind complimentary and distinct mRNAs. Owing to these unique characteristics, microRNAs have been put forward as pivotal regulators of mRNA and protein expression throughout all stages of atherosclerosis supported by human clinical and pathological studies which have analysed the expression of individual microRNAs alongside their predicted targets, in addition to similar investigations in animal models of atherosclerosis. Built upon such findings, over 45 studies have assessed the effects of modulating microRNA expression and function on the pathogenesis of atherosclerosis in multiple mouse models. Differing strategies have been deployed to moderate individual microRNA function in vivo including the use of miR mimics (also referred to as agomirs) or viral vectors (including adeno- or lenti-viruses) to over-express/restore levels of specific microRNA. Similarly, reduction or complete deficiency in expression of a select microRNA can be achieved through deployment of microRNA inhibitors (also referred to as antagomirs) or with genetically-modified mice.
5.1 MicroRNAs in atherosclerosis
5.1.1 Human studies
In humans, the pre-cursors of mature coronary and carotid atherosclerotic plaques are adaptive and pathological intimal thickenings, which are characterized by intimal accumulation of VSMCs and distinct ECM proteins at regions of disturbed shear stress (such as bifurcations and curved arterial regions), and subsequent deposition and modification of lipoproteins alongside accrual of monocyte/macrophages.2 MicroRNA profiling of non-disease coronary arteries and those with early plaques revealed expression of miR-29, miR-100, miR-155, miR-199, miR-221, miR-363, miR-497, and miR-508 were up-regulated in early lesions while miR-490, miR-1273, and miR-1284 levels were down-regulated.97 A comparison of healthy thoracic arteries and atherosclerotic lesions from aortic, carotid and femoral arteries demonstrated miR-21, miR-34, miR-146, and miR-210 levels were increased in atherosclerotic arteries.98 Analysis of carotid lesions and healthy mammary arteries revealed miR-520 and miR-105 expression to be down-regulated and miR-15, miR-26, miR-30, miR-98, miR-125, miR-152, miR-181, miR-185, and miR-422 levels increased within atherosclerotic plaques.99 Furthermore, symptomatic carotid plaques (deemed unstable) exhibited increased expression of miR-100, miR-127, miR-133, and miR-145 when compared with symptomatic lesions (classed as stable).100 Lastly, evaluation of coronary atherosclerotic plaques demonstrated elevated miR-181 expression and concomitant lowered miR-24 levels in plaques categorized as unstable when matched to stable plaques.101,102
The expression of circulating microRNAs has also been assessed, particularly with the consideration that changes in blood levels of select microRNAs could represent valid biomarkers of atherosclerosis and importantly its stage of progress. Indeed, circulating levels of miR-29, miR-126, miR-145, and miR-155 were increased in patients with optical coherence tomography-defined thin-capped fibroatheromas, inferring these microRNAs as causal in plaque stability alongside their potential as biomarkers of rupture-prone plaques.103 Comparison of patients with stable CAD and healthy control subjects revealed decreased circulating miR-155, miR-145, and let-7c levels in the patients with CAD.104 Likewise, blood levels of miR-17, miR-19, miR-29, miR-30, miR-92, miR-126, miR-145, miR-150, miR-155, miR-181, miR-222, miR342, miR-378, and miR-484 were diminished in patients with stable disease in comparison to non-diseased individuals.105,106 Comparing patients with stable and unstable CAD, circulating miR-155 plasma levels were reduced in patients presenting with clinical events such as unstable angina or myocardial infarction.107 Similarly, circulating miR-1, miR-122, miR-126, miR-133, miR-199, miR-433, and miR-485 levels were elevated in angina patients, whilst increased miR-337 levels characterized stable angina patients and increased miR-145 delineated unstable angina patients.108 Lastly, plasma levels of miR-132, miR-150, and miR-186 were collectively predictive of unstable angina in comparison to healthy subjects.109
Additionally, microRNA levels within peripheral blood mononuclear cells (PBMCs) have also been considered predictive for atherosclerosis-related clinical events. Indeed, microRNA profiling within peripheral blood cell samples from acute myocardial infarction patients revealed 121 significantly dysregulated microRNAs when compared with healthy individuals, and identified miR-663 up-regulation as a strong indicator of acute myocardial infarction—although the authors did not identify if the dysregulated microRNAs are as a result of plaque rupture or the myocardial infarction itself.110 Expression of miR-155 was lower in PBMCs from patients with clinically-relevant coronary artery atherosclerosis and inversely associated with the atherogenic risk factors age, hypertension, LDL cholesterol level, and smoking.107 Two separate studies have shown elevated PBMC expression of miR-146 is associated with CAD risk,111,112 while the miR-135a/miR147 ratio within PBMCs has also shown promise as an atherosclerotic disease risk predictor.113 Meanwhile, assessment of dysregulated microRNAs in obese and lean individuals and restricted to CD14 positive monocytes demonstrated reduced levels of miR-181a, miR-181b, and miR-181d were related to obesity, but only diminished miR-181a levels correlated with angiography-defined CAD in obese individuals.114 Lastly, expression profiles within lymphocytes have also been examined, revealing miR-122 expression is increased within CD14-ve lymphocytes of unstable angina and acute MI patients compared to stable angina and healthy control individuals.115 While miR-155 levels are elevated in CD4+ T lymphocytes of unstable angina patients with marked coronary artery stenosis compared with subjects with mild stenosis or no stenosis.116
Taken together, the assessment of microRNA expression with plaques can assist in the identification of candidate causal microRNAs while evaluation of circulating and blood cell-derived microRNAs may provide the identification of potential predictive biomarkers of disease progression (see Figure 4). Although, the baseline characteristics of patients, their existing medical therapies, and the manifestation of contraindicative diseases need to be considered when drawing conclusions from microRNA profiling studies, and such confounding issues may explain why there are discrepancies in between clinical studies.
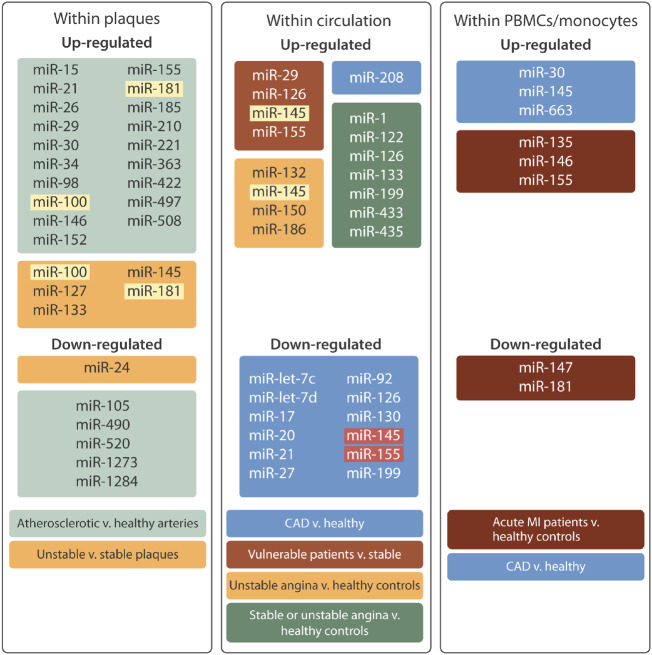
Figure 4.
MicroRNA expression in human atherosclerotic plaques and circulating blood. This diagram illustrates the dysregulated microRNAs identified through profiling approaches within atherosclerotic plaques, circulating plasma samples, and peripheral blood mononuclear cells (PBMCs). Coloured boxes indicate the patient cohorts from within which the dysregulated microRNAs were identified. MicroRNA depicted by yellow highlighting have been verified in two independent studies, while microRNA with red highlighting have been independently reported to be up- and down-regulated.
5.1.2 Animal studies
The differential expression of microRNAs has also been assessed in hypercholesterolaemic mice using a carotid artery double ligation model to generate lesions characterized as stable and unstable.117 Microarray analysis demonstrated increased expression of miR-138, miR-142, miR-322, miR-335, and miR-450 in plaques deemed unstable (due to the presence of intraplaque haemorrhage), compared to stable lesions, implying a role for these microRNA in plaque progression.117 While there have been scores of in vitro studies using vascular and inflammatory cells to determine the expression and function of microRNAs, these are too numerous to include within this review. Accordingly, only studies which have directly ascertained the influential roles of select microRNA to the development and progression of atherosclerosis are discussed in detail. Most such studies rely on the use of genetically modified mouse models of atherosclerosis (such as Apoe or LDL receptor (Ldlr)-deficient mice) and two distinct pharmacological approaches to moderate the activity of individual microRNA in vivo. Individual microRNAs can be over-expressed or restored using either synthetic double-stranded RNA molecules (commonly termed mimics or agomirs), or with viral expression constructs. Conversely, the action of microRNAs can be suppressed/inhibited with chemically modified anti-miR oligonucleotides (commonly termed antagomirs). Deploying such approaches, there has been a rapid growth in the number of publications assessing microRNA modulation in mouse models of atherosclerosis, and these are discussed below and summarized in Table 2. In particular, the cellular origin of the modulated microRNA and its potential target mRNA are highlighted, and therefore the studies have been delineated by their proposed cellular source and modulation by the assessed microRNA.
Table 2.
MicroRNAs in atherosclerosis-related research
| MicroRNA name | Model | Atherogenic/Athero-protective | Main cellular origin & function | Target mRNA | References |
|---|---|---|---|---|---|
| miR-let-7g | Apoe KO + miR mimic | Atherogenic | VSMC; proliferation/migration | LOX1 | 118 |
| miR-10a | Apoe KO | Atheroprotective | Mac; foam cell formation | LCOR/NCOR2 | 119 |
| Apoe KO | Atheroprotective | EC; inflammatory activation | GATA6 | 120 | |
| miR-10b | Apoe KO | Atherogenic | Mac; foam cell apoptosis | ABCA1 | 121 |
| miR-19 | Apoe KO + miR mimic or antagomir | Atherogenic | Mac; foam cell formation | ABCA1 | 122 |
| miR-19b | Apoe KO collar model + mimic-rich microparticles | Atherogenic | EC; inflammatory activation | SOCS3 | 123 |
| miR-21 | Ldlr KO + miR KO | Atheroprotective | Mac; foam cell apoptosis | MAP2K3 | 124 |
| Apoe KO + miR KO or over-expression | Atheroprotective | VSMC; proliferation | REST/PTEN | 125 | |
| miR-23a | Apoe KO + antagomir | Atherogenic | Mac; foam cell formation | ABCA1/ABCG1 | 126 |
| miR-24 | Apoe KO + miR antagomir | Atheroprotective | Mac; proteolysis & invasion | MMP14 | 101 |
| Apoe KO + miR mimic | Atherogenic | Mac/Hepat; lipid metabolism | SCARB1 | 127 | |
| miR-30c | Apoe KO + miR lentiviral over-expression or inhibition | Atheroprotective | Hepat; lipid metabolism | MTTP | 128 |
| Apoe KO + miR mimic | Atheroprotective | Hepat; lipid metabolism | 129 | ||
| miR-33 | Reversa + miR antagomir | Atherogenic | Mac; foam cell formation | ABCA1/ABCG1 | 130 |
| Apoe KO ± miR KO BMT | Atherogenic/no effect | Mac/Hepat; lipid metabolism | 131 | ||
| Ldlr KO + miR antagomir | No effect | 132 | |||
| Ldlr KO + miR antagomir | Atherogenic | 133 | |||
| Ldlr KO + miR antagomir | Atherogenic | 134 , 135 | |||
| Ldlr KO ± miR KO BMT | No effect/atherogenic | 136 | |||
| Ldlr KO + miR antagomir | Atherogenic | 137 | |||
| Ldlr KO + miR antagomir | Atherogenic | 138 | |||
| miR-33b | Apoe KO + miR-knockin mouse | Atherogenic | Mac; foam cell formation | ABCA1/ABCG1 | 139 |
| miR-34a | Apoe KO + miR agomir | Atherogenic | EC; apoptosis | BCL2 | 140 |
| miR-92a | Ldlr KO + miR antagomir | Atherogenic | EC; inflammatory activation | SOCS5 | 141 |
| miR-98 | Apoe KO + miR agomir or antagomir | Atheroprotective | Mac; foam cell formation | LOX1 | 142 |
| miR-100 | Apoe KO + miR mimic or antagomir | Atheroprotective | EC; anti-inflammatory | MTOR | 143 |
| miR-124 | Apoe KO + miR mimic or inhibitor | Atherogenic | VSMC; collagen synthesis | P4HA1 | 144 |
| miR-126 | Apoe KO + miR KO or mimic | Atheroprotective | EC; proliferation | DLK1 | 145 |
| miR-134 | Apoe KO + miR agomir or antagomir | Atherogenic | Mac; foam cell formation | ANGPTL4 | 146 |
| miR-143/-145 | Apoe KO + microparticles | Atheroprotective | EC; microvesicle production | 147 | |
| Apoe KO miR lentiviral SMC-specific over-expression | Atheroprotective Atherogenic | VSMC; phenotypic modulation | MYOCD/KLF4 | 148 | |
| Ldlr KO + double miR KO | Mac; foam cell formation | ABCA1/SCARB1 | 149 | ||
| miR-146a | Ldlr KO ± miR KO BMT | Atheroprotective/genic atheroprotective | Mac/Hepat; lipid metabolism Mac; pro-inflammatory | SORT | 150 |
| Apoe: Ldlr KO or Ldlr KO + miR mimic | No effect | Mac; cholesterol metabolism | IRAK1/TRAF6 | 151 | |
| Ldlr KO ± miR KO BMT | Atheroprotective | EC; inflammatory activation | CCL2/CCL5 | 152 | |
| Apoe KO + E-selectin targeting miR+ve microparticles | 153 | ||||
| miR-150 | Apoe KO + miR KO | Atherogenic | Mac; pro-inflammatory | PDLIMI | 154 |
| miR-155 | Ldlr KO mouse ± miR KO BMT | Atheroprotective Atherogenic | Mac; pro-inflammatory | 155 | |
| Apoe KO mouse ± miR KO BMT | Atherogenic | Mac; foam cell formation | BCL6 | 156 | |
| Apoe KO mouse ± miR KO BMT | 157 | ||||
| miR-181b | Apoe KO or Ldlr KO mouse + miR antagomir | Atherogenic | Mac/VSMC; proteolysis/ECM | TIMP3/ELN | 102 |
| Apoe KO + miR mimic | Atheroprotective | Mac; anti-inflammatory | NOTCH1 | 158 | |
| Apoe KO + miR mimic | Atheroprotective | EC; inhibits NFκB activation | KPNA4 | 159 | |
| miR-182 | Apoe KO + miR mimic or antagomir | Atherogenic | Mac; pro-inflammatory | HDAC9 | 160 |
| miR-188 | Apoe KO + miR mimic or inhibitor | Atheroprotective | Mac; anti-inflammatory | OLR1 | 161 |
| miR-223 | Apoe KO + miR KO or antagomir | Atheroprotective | VSMC; growth & apoptosis | IGF1R | 162 |
| miR-302 | Ldlr KO + miR antagomir | Atherogenic | Mac; foam cell formation | ABCA1 | 163 |
| Hepat; cholesterol clearance | |||||
| miR-320 | Apoe KO + miR mimic or antagomir | Atherogenic | EC; pro-inflammatory | SRF | 164 |
| miR-590 | Apoe KO + miR mimic or antagomir | Atheroprotective | Mac; lipid metabolism | LPL | 165 |
| miR-712 | Apoe KO ± carotid ligation + miR mimic or antagomir | Atherogenic | EC; pro-inflammatory | TIMP3 | 166 |
Target mRNA which have not been validated are presented in italics.
Apoe KO, apolipoprotein E-deficient mice; BMT, bone-marrow transplantation; EC, endothelial cell; ECM, extracellular matrix; Hepat, hepatocytes; Ldlr KO, Ldlr-deficient mice; Mac, macrophages; Mono, monocyte; VSMC, vascular smooth muscle cell.
5.1.2.1 Endothelial cells
miR-10
An atheroprotective role has been proposed for miR-10a as expression of this microRNA is reduced within the athero-susceptible inner curvature of the aortic arch in healthy rats and hypercholesterolaemic mice where disturbed flow is prevalent.120 Supporting findings have also been demonstrated within a swine model and miR-10a suggested to retard a pro-inflammatory switch in ECs.167 Based upon previous cancer studies, it was shown that co-administration of RARα/RXRα-selective agonists restored EC miR-10a expression and was associated with inhibition of atherosclerosis development at the aortic arch inner curvature, which could be prevented by systemic delivery of a miR-10a antagomir.120 Moreover, the atheroprotective effects seen with RARα/RXRα-selective agonists mirrored those achieved through administration of a miR-10a mimic, and the beneficial effects were attributed to repression of GATA6/VCAM1 signalling within ECs.120
miR-19
Circulating levels of miR-19b are elevated within patients with angiographically identified CAD when compared with those with negative identification, and the circulating miR-19b is predominantly located within endothelial microparticles,168 although the mechanism for their release is unclear. Nonetheless, administration of endothelial microparticles (derived from miR-19b mimic transfected HUVECs) accelerated atherosclerosis development in the collar-induced Apoe-deficient mouse model associated with increased macrophage and lipid content, although VSMC content was also augmented.123 It was proposed that miR-19b microparticles accumulate within the peri-vascular adipose tissue around the arteries and target SOCS3 expression to subsequently promote the expression of pro-inflammatory molecules such as TNF-α and IL-6 thus encouraging atherosclerosis, as this effect was lost when the peri-vascular adipose tissue was removed before delivery of miR-19b containing microparticles.123
miR-34
In vitro studies revealed HUVEC miR-34a expression is down-regulated in response to atheroprotective high shear stress and conversely up-regulated under atheroprone oscillatory shear stress when compared with static conditions, promoting a pro-inflammatory EC phenotype potentially through targeting of SIRT1 although this was not directly confirmed.169 Moreover, miR-34a expression is increased within human carotid and femoral atherosclerotic plaques when compared with non-diseased thoracic arteries,98 while plasma levels of miR-34a are elevated in patients with CAD or hypercholesterolaemic Apoe-deficient mice related to healthy controls and wild-type mice respectively.170 Further studies in Apoe-deficient mice demonstrated miR-34 inhibition reduced aortic root atherosclerosis, in part through direct targeting of BCL2 and associated suppression of EC apoptosis (induced by oxLDL within in vitro experiments).140 However, it has also been shown that miR-34 inhibition prevented oxLDL-induced EC apoptosis through directly targeting HDAC1, although elevated Bcl2 protein expression was also reported in support of the above.171 Heightened miR-34a expression has also been recently associated with promoting VSMC senescence and subsequent vascular calcification (a complication of atherosclerosis) through targeting of SIRT1,172 as also proposed within ECs,169 and may therefore represent an additional mechanism through which miR-34 levels may affect atherosclerotic plaque development.
miR-92
Studies of human carotid plaques and the aortic arch of hypercholesterolaemic Ldlr-deficient mice revealed miR-92a expression is up-regulated in response to pro-atherogenic flow conditions alongside raised plasma cholesterol levels, and specifically by ECs.141 As such, inhibition of miR-92 through systemic administration of a specific antagomir reduced atherosclerotic plaque size within the aortic root of Ldlr-deficient mice which was associated with diminished macrophage number and increased collagen content.141 These beneficial effects were attributed to re-established EC expression of the negative regulator of cytokine signalling, SOCS5.141
miR-100
Evidence from a murine ischaemia–reperfusion model identified miR-100 as an endothelial-enriched microRNA which exerts anti-angiogenic properties through suppression of mTOR,173 suggesting protective role for this microRNA in cardiovascular diseases. Assessment of human carotid plaques revealed that while miR-100 expression does not differ between stable plaques and non-diseased mammary arteries, levels were markedly decreased in unstable atherosclerotic lesions.143 Concordantly, intravenous administration of a miR-100 antagomir accelerated atherogenesis in Ldlr-deficient mice, while over-expression achieved through systemic delivery of a miR-100 mimic protected from aortic plaque formation.143 Mechanistic studies revealed miR-100 imparts an anti-inflammatory effect on the vasculature by dampening leucocyte–endothelial interactions through direct targeting of mTOR and Raptor, which permits EC autophagy and subsequent inhibition of NFκB activity.143
miR-126
Studies in humans and mice have shown that miR-126-3p and miR-126-5p (miR-126*) are consistently the most abundant microRNAs expressed in resting ECs and protect from vascular inflammation.174,175 Interestingly, depressed expression of miR-126-5p, but not miR-126-3p, has been reported in ECs at sites of disturbed shear stress and therefore considered atheroprone.145 Mechanistic studies revealed loss of miR-126-5p suppresses EC proliferation through up-regulation of the Notch1 signalling pathway inhibitor DLK1.145 Further in vivo investigation demonstrated that miR-126-deficeint mice exhibit exacerbated atherogenesis within the aortic root and the carotid artery (ligation-induced) of Apoe-deficient mice, which could be rescued through administration of a miR-126 mimic and was associated with restored EC proliferative capacity.145
miR-143/145
miR-143 and miR-145 are closely related microRNAs and commonly co-transcribed, and as such are regularly studied in unison. Findings from studies appraising plasma and atherosclerotic plaque microRNA expression in patients with symptomatic atherosclerosis have provided conflicting results on the association between expression of miR-143/miR-145 and atherosclerosis. While circulating levels of miR-145 are inversely related with the extent of coronary fibroatheroma and macrophage plaque content in humans, trans-coronary plasma levels of miR-145 were positively associated with the presence of thin-cap fibroatheromas, as identified through optimal coherence tomography (OCT).103 In agreement, intra-plaque miR-145 levels were heightened in patients with symptomatic carotid disease compared to asymptomatic plaques.100,103 In line with these findings, Ldlr-deficient mice harbouring miR-143 and miR-145 deletion exhibit reduced aortic atherosclerosis compared to miR-143/145 expressing Ldlr-deficient mice.149 However, a focussed array of human advanced coronary plaques alongside non-atherosclerotic mammary arteries revealed that miR-143 levels were decreased in atherosclerotic lesions.176 Similarly, miR-145 expression was attenuated within aortic plaques of Apoe-deficient mice when compared with non-diseased animals, and in human carotid plaques in contrast to plaque-free arteries.148 Additionally, plasma levels of miR-145 are reduced in patients with angiographically identified CAD compared to healthy controls.105 Suggesting a beneficial role for miR-143 and miR-145. In relation, it is now well-accepted that KLF2 plays a central role in mediating the atheroprotective endothelial phenotype generated by shear stress.177 Accordingly, profiling of microRNA changes in KLF2 overexpressing HUVECs in order to mimic levels observed in HUVECs exposed to prolonged laminar flow, revealed miR-143 and miR-145 as two of the most highly up-regulated microRNAs.147 Furthermore, atheroprotective shear stress and statin administration up-regulated EC miR-143/145 expression in a KLF2-dependent manner.147 Additionally, KLF2 signalling encouraged the generation of EC-derived extracellular vesicles enriched in miR-143/145 which can be transferred to VSMCs to maintain an atheroprotective smooth muscle cell phenotype.147 Accordingly, systemic delivery of extracellular vesicles derived from KLF2-overexpressing ECs reduced aortic atherosclerotic lesion size in Apoe-deficient mice.147 In agreement, lentiviral VSMC-restricted over-expression of miR-145 reduced atherosclerotic burden at multiple vascular beds within Apoe-deficient mice which was associated with promoting a contractile VSMC phenotype.148 Interestingly, it has also been suggested that VSMC miR-145 can be transported to macrophages under atherogenic stimuli, targeting ABCA1 and subsequently perturbing cholesterol efflux and enhanced foam cell formation.149 The contradictory results reported above reveal the need for future studies to clarify the therapeutic and diagnostic potential of miR-143/145.
miR-320
Circulating levels of miR-320a are elevated in patients with CAD compared to non-diseased individuals,164 suggesting a pro-atherogenic role for this microRNA. Indeed, intravenous delivery of a miR-320a over-expression plasmid induced aortic atherogenesis in Apoe-deficient mice, which was related with promoting a pro-inflammatory EC phenotype, characterized by reduced NO production and increased expression of inflammatory cytokines (including IL-6 and MCP-1) alongside a significant increase in plasma total cholesterol, triglyceride, and LDL levels.164 Interestingly, miR-320a over-expression in wild-type mice also induced aortic atherosclerotic plaque development. Conversely, administration of miR-320 anti-sense retarded aortic atherosclerosis.164 Mechanistic in vitro studies revealed miR-320 directly targets and decreases EC expression of SRF, retarding cellular proliferation and promoting their susceptibility to apoptosis,164 characteristics associated with atherosclerotic plaque progression.
miR-377
A recent study in rats reported that hepatic miR-377 expression was modulated by the consumption of distinct dietary lipids,178 suggesting that altered miR-377 levels may affect the development of atherosclerosis. Supportingly, patients with aberrant elevated plasma levels of triglyceride, a risk factor for atherosclerosis, concomitantly display reduced circulating levels of miR-377.179 Studies in Apoe-deficient mice demonstrated that exogenous addition of miR-377 suppressed plasma triglyceride levels in response to high-fat feeding and reduced aortic root atherogenesis, while conversely miR-377 antagomir administration accelerated lesion development.179 Mechanistic insight gained from studies in ECs proposed enhanced miR-377 levels suppress DNMT1 expression which permits lipoprotein lipase (LPL) binding to ECs and subsequent hydrolysis of triglycerides and a reduction in their circulating levels.179
miR-712
Analytical comparisons of microRNA expression in mouse ECs subjected to atheroprone disturbed flow in vitro or in vivo alongside cells under atheroprotective laminar shear stress identified miR-712 as a flow-sensitive microRNA up-regulated under disturbed flow conditions.166 Further in vitro studies established TIMP-3 within the endothelium as a miR-712 target under disturbed flow, inducing endothelial inflammation and increased permeability.166 Accordingly, in Apoe-deficient mice with either spontaneous atherosclerosis or induced through partial left carotid ligation, systemic delivery of a miR-712 antagomir blunted atherogenesis and was linked with restored TIMP-3 expression and reduced proteolytic activity within the vessel wall, mirroring findings achieved through adenoviral over-expression of TIMP-3 in the partial carotid ligation model.166 Positive findings in human ECs confirmed miR-205 as a potential homologue of murine miR-712 and demonstrated miR-205 down-regulated EC TIMP-3 expression, and showed human EC miR-205 expression is flow sensitive.166
5.1.2.2 Vascular smooth muscle cells
miR-let-7g
Pertinent to atherosclerosis, miR-let-7g has been shown to modulate oxLDL-induced apoptosis and proliferation of VSMCs, associated with changes in the expression of LOX1.180 Confirmatory findings demonstrated over-expression of LOX-1 induced VSMC proliferation and migration were both attenuated by miR-let-7g over-expression, and confirmed LOX1 as a direct target of miR-let-7g.118 In line with the effects observed in vitro, systemic administration of a miR-let-7g specific mimic reduced atherosclerotic lesion size within the aortae of high-fat fed Apoe-deficient mice, which was associated with reduced intra-plaque expression of LOX1 although cellular differences were not examined.118
miR-21
Relevant to atherogenesis, miR-21 has been shown to promote the growth of VSMCs and subsequent neointimal formation which underlies restenosis after surgical interventions in patients with CAD.181–183 Dysregulated VSMC growth and neointimal formation are shared characteristics of adaptive intimal thickenings, the precursors of atherosclerotic plaque sin humans.2 Accordingly, VSMC proliferation and migration, and by analogy increased miR-21 levels, can be considered detrimental during atherogenesis and conversely beneficial in advanced lesions by maintaining plaque stability through preservation of the fibrous cap. Indeed, mature carotid plaques deemed unstable in humans and within Apoe-deficient mice express reduced miR-21 levels, predominantly lost from fibrous cap VSMCs.184 Using the carotid ligation/cast model in Apoe-deficient mice to induce unstable plaques as evidence by the presence of intra-plaque haemorrhage,125 Jin et al.184 demonstrated that systemic loss of miR-21 resulted in the generation of plaques with unstable characteristics, which was associated with miR-21-dependent regulation of the VSMC anti-proliferative transcription factor REST. Furthermore, using ultrasound-targeted microbubble destruction to achieve local delivery and accumulation of a miR-21 mimic within established unstable plaques (generated through carotid ligation/cast model) improved plaque composition and stability as indicated by increased VSMC proliferation and number, attributed to reduced expression of the miR-21 targets PTEN and REST.184
Macrophages also express miR-21 levels where targeting of PTEN and PDCD4 is proposed to modulate efferocytosis-induced macrophage polarization185 and foam cell formation,184 suggesting miR-21 may also regulate intra-plaque inflammation. Moreover, advanced human plaques (which contain macrophages) exhibit increased miR-21 levels when compared with non-diseased arteries (which contain limited numbers of macrophages).98 However, bone-marrow transplantation of miR-21 deficient cells aggravated aortic atherosclerosis in Ldlr-deficient mice,124 which was associated with increased foam cell formation and associated apoptosis as a suggested result of restored MAP2K3 expression (a miR-21 target) which can negatively regulate ABCA1 and therefore cholesterol efflux capacity.124 Similarly, miR-21/Apoe double-deficient mice exhibited accelerated atherogenesis associated with heightened macrophage accumulation and foam cell formation.184 Finally, it has been postulated that the dual effects of miR-21 on macrophages and VSMCs are through cross-talk between these two cell types, as it has been shown that macrophages from miR-21/Apoe double-deficient mice release factors which exert anti-proliferative effects on VSMCs.184
miR-124
The expression of miR-124 is up-regulated in the monocytes of smokers compared to former and non-smokers and is elevated levels of miR-124 in whole blood was associated with an increased risk of sub-clinical atherosclerosis.186 Fluorescent in situ hybridization of Apoe-deficient mouse aortic plaques revealed miR-124 was predominantly localized to VSMCs.144 Further in vitro studies have identified the miR-124 regulates VSMC fibrillar collagen metabolism through targeting P4HA1.144 Although administration of a miR-124 mimic or inhibitor had no effect on aortic plaque size or macrophage accumulation, in line with the in vitro observations effects on VSMC and collagen content were detected, with miR-124 mimic delivery exerting an adverse effect, whereas miR-124 inhibition was beneficial.144
miR-223
Increased circulating levels of miR-223 have been reported within acute myocardial infarction patients compared with healthy controls.187 Furthermore, analysis of serum samples from patients with confirmed angiographically-defined coronary atherosclerosis demonstrated elevated miRNA-223 levels served as a positive predictor of adverse cardiovascular events including death.188 A further study confirmed serum levels of miR-223 are elevated within patients or mice with atherosclerosis when compared with non-diseased controls, which was associated with increased expression of miR-223 within atherosclerotic plaques of both species.162 The primary cellular sources of miR-223 were identified as leukocytes and platelets, and in vitro studies revealed miR-223 from these cells could be transported via microparticles into the vessel wall where they accumulate within VSMCs and down-regulate IGF-1R expression to suppress cell growth and promote apoptosis.162In vivo studies established that systemic delivery of a miR-223 inhibitor to Apoe-deficient mice limited atherogenesis as observed through a decrease in plaque size at the aortic root.162 Yet subjection of miR-223 deficient mice to carotid artery ligation injury resulted in accelerated neointimal formation when compared with wild-type mice,162 which could translate to a deleterious effect on advanced atherosclerotic lesions as VSMC growth and survival are essential for maintenance of the beneficial fibrous cap and subsequent protection from plaque destabilization. Accordingly, although miR-223 inhibition may harbour therapeutic potential for retarding atherogenesis and restenosis, it may exert adverse effects on plaque stability and preclude its use in atherosclerotic patients. Indeed, it is plausible the detected increases in circulating levels of miR-223 after myocardial infarction187 may be due in part to plaque VSMCs regenerating the fibrous cap after a rupture, a phenomenon known to occur in human coronary events.189
5.1.2.3 Macrophages
miR-10
As mentioned earlier, miR-10a has been proposed to exert an atheroprotective role through preventing the transformation of ECs into a pro-inflammatory phenotype. Further studies have also indicated miR-10a may also afford beneficial effects on atherosclerosis through direct targeting of LCOR and NCOR2 within macrophages, thus promoting fatty acid degradation subsequently limiting foam cell formation.119 Supporting, miR-10a expression was inversely associated with plaque progression in mice and humans, especially lipid/necrotic core size.119 Furthermore, blocking the interaction between miR-10a and LCOR through administration of target site blockers, heightened atherosclerosis development in Apoe-deficient mice.119 Conversely, miR-10b appears to play a deleterious role in advanced atherosclerosis as human atherosclerotic plaques express higher levels of miR-10b compared with healthy arteries without atherosclerosis.99 Moreover, inhibition of miR-10b suppressed progression of established aortic and brachiocephalic plaques in Apoe-deficient mice which was associated with increased intra-plaque macrophage ABCA1 expression (and by inference improved cholesterol efflux) and diminished macrophage apoptosis, resulting in plaques with more stable characteristics, however, no beneficial effects of miR-10b silencing were observed on atherogenesis within the same model.121
miR-19
Similarly to miR-10b, miR-19b has been shown to specifically target and down-regulate ABCA1 expression within macrophages and therefore retard cholesterol efflux and drive foam cell formation.122 Consequently, systemic delivery of a miR-19b mimic to Apoe-deficient mice lowered plasma HDL levels and alongside increased LDL levels, and subsequently increased aortic plaque size and deleteriously altered lesion composition and ABCA1 expression.122 Whereas administration of miR-19b antisense oligonucleotides exerted opposite effects.122 Although not assessed, given that EC-derived microparticles rich in miR-19b promote atherosclerosis and can be transferred to macrophages,123 it is plausible that this mechanism may be in part be responsible for the above observed effects of miR-19b on atherogenesis.
miR-23
Circulating levels of miR-23a are elevated within the plasma of atherosclerotic Apoe-deficient mice compared to wild-type controls,126,170 and within patients with advanced coronary170 or carotid126 atherosclerosis related to non-diseased individuals. Silencing of miR-23a in vivo through administration of a selective antagomir to Apoe-deficient mice decreased aortic root atherosclerotic plaque size and was associated with increased intra-plaque macrophage expression of both ABCA1 and ABCG1 alongside favourable effects on plaque composition.126 Mechanistic in vitro findings confirmed ABCA1 and ABCG1 as direct targets of miR-23a and revealed that oxLDL increases macrophage miR-23a expression while miR-23a inhibition increased macrophage cholesterol efflux and suppressed foam cell formation, potentially underlying the favourable effects of miR-23a silencing in vivo.126
miR-24
Polarization of human macrophages with GM-CSF is associated with down-regulation of miR-24 alongside a concomitant increase in MMP-14 protein levels and subsequent heightened invasive capacity, when compared with M-CSF matured macrophages.101 Increased macrophage expression of MMP-14 in conjunction with reduced miR-24 levels are also observed in unstable human coronary plaques whilst the opposite pattern is observed in stable lesions.101 Accordingly, systemic delivery of a miR-24 inhibitor to Apoe-deficient mice with established brachiocephalic artery atherosclerosis enhanced lesion progression which was associated with elevated intra-plaque macrophage MMP-14 expression and a deleterious shift in plaque composition.101 Despite the previous study showing the favourable effects of miR-24 on plaque progression were independent of changes in plasma cholesterol levels, a similar study in Apoe-deficient mice proposed miR-24 promotes atherogenesis through direct targeting of SCARB1 (SRB1) within hepatocytes which diminishes HDL-cholesterol ester clearance and subsequently elevates plasma cholesterol levels.127 Such disparate effects of miR-24 modulation may therefore represent the opposing effects of miR-24 on atherogenesis and the progression of established atherosclerotic lesions.
miR-98
LOX-1, a receptor for ox-LDL is a predicted target of miR-98 and divergent LOX1 mRNA and protein expression compared to mIR-98 is observed in macrophages after exposure to oxLDL.142 Further findings confirmed LOX1 as a direct target of miR-98 and exposure of oxLDL-treated macrophage to a miR-98 mimic lowered LOX-1 levels and retarded foam cell formation.142 Similarly, administration of a miR-98 agomir retarded intimal LOX-1 expression and associated lipid accumulation within the aortae of high-fat fed Apoe-deficient mice, while enhanced aortic LOX-1 expression alongside increased lipid content was observed with miR-98 antagomir delivery.142 However, effects on plaque size and composition were not reported, limiting the further extrapolation of the above findings.
miR-134
PBMCs from patients with unstable CAD exhibited higher levels of miR-134 when compared with those from patients with stable disease.113 Moreover, miR-134 has been shown to directly bind the 3′ UTR of ANGPTL4 and supress its expression within macrophages which inadvertently permits enhanced lipoprotein lipase activity and subsequent foam cell formation alongside heightened pro-inflammatory cytokine release.190 Accordingly, systemic administration of a miR-134 agomir increased aortic atherosclerotic plaque size in Apoe-deficient mice which was associated with decreased ANGPTL4 levels and concomitant increased expression and activity of lipoprotein lipase and lipid content within plaques, whilst opposing effects were observed in mice which received a miR-134 antagomir.146
miR-146
Elevated levels of miR-146 have been detected within human aortic and femoral artery atherosclerotic plaques,98 and a SNP in the miR146a gene which alters miR-146a expression has been proposed as an indicator of CAD susceptibility.191 Whole body deficiency of miR-146a in Ldlr-deficient mice resulted in decreased aortic arch plaque size in conjunction with lowered plasma LDL cholesterol levels, which collectively indicates a pro-atherosclerotic role for miR-146a.150 Furthermore, through deployment of a bone-marrow transplantation approach, monocyte/macrophage-derived miR-146a was proposed as the central effector of atherogenesis within the Ldlr-deficient model, through targeting of SORT1 and subsequent modulation of plasma LDL levels.150 However, it should be noted that the pro-atherogenic effects of monocyte/macrophage-restricted miR-146a were only observed with prolonged hypercholesterolaemia and within the aortic arch as opposed to short-term feeding and other vascular beds including the aortic root.150 A similar study also demonstrated miR-146a deficiency exclusively in haematopoietic cells regulates circulating cholesterol levels in Ldlr-deficient mice, but does not affect atherogenesis after either short- or long-term high-fat feeding.152 Paradoxically, an atheroprotective effect for miR-146a has been proposed as systemic delivery of a miR-146 mimic to Apoe/Ldlr double-deficient mice or Ldlr-deficient suppressed atherogenesis within the aortic root, which was associated with decreased intra-plaque macrophage content but without affecting plasma cholesterol levels.151 Accompanying mechanistic studies revealed that Apoe favourably regulated macrophage miR-146a levels through the transcription factor PU.1, with heightened miR-146a levels retarding the expression of IRAK1 and TRAF6 to subsequently diminish NFκβ-driven pro-inflammatory responses.151 Equally in ECs miR-146a has been shown to down-regulate TRAF6 levels, thus suppressing NFκβ activation and preventing pro-inflammatory stimulation of ECs,192 inferring endothelial miR‐146 expression may play a protective role against the development of atherosclerosis. In accordance with these findings, implied endothelium-directed delivery of miR-146a-loaded E-selectin-targeting synthetic microparticles decreased aortic atherosclerosis in Apoe-deficient mice which was associated with reduced intra-plaque macrophage content.153 Collectively, these studies reveal that a more nuanced approach is required if miR-146a is pursued as a therapeutic target for atherosclerosis prevention, exemplified by the fact that mice deficient for miR-146a except in bone marrow-derived cells display increased atherosclerosis compared to mice lacking miR-146a in all cells (including ECs).150 To further complicate matters, neutrophil derived miR-146a has been associated with the increased frequency of future adverse cardiovascular events in patients with overt cardiovascular disease.193
miR-150
Elevated circulating levels of miR-150 delineate patients with a diagnosis of unstable angina compared to patients with non-coronary chest pain (exclusion of coronary stenosis during angiogram) or healthy subjects.109 Moreover, human coronary arteries harbouring advanced plaques demonstrated increased miR-150 expression when related to non-diseased vessels, with a similar pattern observed in high-fat fed Apoe-deficient mice.154 In line with these observations, aortic root plaque size was reduced in Apoe-deficient mice also lacking miR-150 when compared with Apoe-deficient mice alone.154 Additionally, plaques from miR-150 deficient mice were deemed more stable due to increased content of collagen and smooth muscle cells alongside decreased lipid and macrophage accumulation.154 A similar atheroprotective effect was detected after bone-marrow transplantation from miR-150 deficient mice into Apoe-deficient animals, implying macrophage-derived miR-150 drives atherosclerosis in this model.154In vitro findings indicated miR-150 facilitates inflammation through targeting of PDLIM1 in macrophages, resulting in heightened NFκβ activation.154
miR-155
Multiple lines of evidence have demonstrated elevated expression of miR-155 within human plaques at various vascular beds,98,156,157 and in plaques from hypercholesteraemic mice,156,157 suggesting a deleterious role for miR-155 in atherosclerosis. Elevated circulating levels of miR-155 have also been reported in patients with OCT-defined advanced coronary plaques.103 However, diminished circulating levels of miR-155 were detected in patients with either a previous history of CAD or angiogram-defined coronary stenosis when compared with control subjects.104–106 Nonetheless, mechanistic in vitro studies revealed macrophage miR-155 expression is associated with foam cell formation156 and pro-inflammatory macrophage polarization, effects regulated by miR-155 targeting of BCL6 and subsequent NFκβ-activation.157 Consequently, Apoe-deficient mice with either whole-body156 or haematopoietic-restricted deletion156,157 of miR-155 exhibited reduced atherosclerosis at the aortic root, which was reversed upon BCL6 silencing.157 Conversely, haematopoietic-restricted miR-155 loss in Ldlr-deficient mice aggravated atherogenesis through proposed anti-inflammatory effects on intra-plaque macrophages.155 Such opposing effects on atherogenesis could be a result of the differing models deployed to study atherosclerosis as the degree of hypercholesterolaemia is markedly diverse between high fat-fed Apoe- and Ldlr-deficient mice, and ensuing macrophage foam cell formation is known to regulate miR-155 expression.155
miR-181
Analysis of human circulating monocyte subsets identified members of the miR-181 family were increased within non-classical (CD14+CD16++) monocytes compared to their classical (CD14++CD16−) counterparts and elevated within atherosclerotic carotid arteries in comparison to healthy vessels.99 Specifically, human unstable coronary plaques exhibited increased miR-181b levels compared to stable lesions, with expression largely restricted to pro-inflammatory foam cell macrophages.102 Similar studies in Apoe-deficient mice and Ldlr-deficient mice demonstrated systemic delivery of a miR-181b inhibitor mutually diminished atherosclerotic plaque formation and progression of established atherosclerotic plaques.102 A dual beneficial effect of miR-181b inhibition was identified and attributed to restoration of foam cell macrophage TIMP-3 protein expression resulting in associated diminished intra-plaque proteolysis, alongside increased VSMC elastin production, actions expected to favour plaque stability.102 On the contrary, plasma miR-181b levels were shown to be lower in patients with angiogram-defined obstructive CAD159 or after suffering from acute stroke.158 Moreover, two independent in vivo studies demonstrated miR-181b systemic over-expression through administration of specific mimics reduced aortic plaque formation in Apoe-deficient mice.158,159 The favourable actions of miR-181b elevation were ascribed in one study to repressed EC KPNA4 (also known as IPOA3) expression and associated dulling of NFκβ-activity,159 whilst suppression of NOTCH1 levels/signalling which permitted anti-inflammatory macrophage polarization was put forward by An et al.158 The divergent reported effects of miR-181b modulation on atherosclerosis may highlight the differing effectiveness of agomir/mimics and miR inhibitors deploying locked nucleic acid (LNA)-modification to target and accumulate within atherosclerotic plaques, as LNA-miR inhibitors display increased sensitivity and specificity alongside superior stability, therefore facilitating their accrual within lesions and ability to target intra-plaque cells.194
miR-182
Profiling of whole blood samples demonstrated elevated miR-182 levels in patients undergoing elective coronary artery bypass grafting compared with healthy controls, highlighting miR-182 as a likely biomarker and regulator of progressive atherosclerosis.195 Indeed, miR-182 agomir administration to Apoe-deficient mice induced the development of larger aortic plaques against control mice whereas systemic delivery of an antagomir blunted plaque formation.160 Further in vitro an in vivo analysis confirmed miR-182 targets the histone deacetylase HDAC9, which upon miR-182-dependent down-regulation within macrophages facilitates augmented LPL expression which sequentially permits lipid accumulation and pro-inflammatory foam cell macrophage formation.160
miR-188
In a mouse model of myocardial infarction, decreased levels of miR-188 were reported and restoration of miR-188 expression attenuated myocardial infarction size through targeting of ATG7 and associated inhibition of autophagy and autophagic cell death within the heart.196 A similar experimental approach was deployed in atherosclerotic Apoe-deficient mice and revealed systemic delivery of a miR-188 mimic suppressed the development of aortic atherosclerosis while a miR-188 inhibitor increased the size of aortic plaques.161 Effects on ATG7 expression and autophagy were not examined in the atherosclerosis study and the anti-atherosclerotic actions of elevated miR-188 were attributed to reduced foam cell macrophage formation alongside related decreased expression and release of pro-inflammatory-connected factors including IL-6, IL-1β, and TNFα, although no direct targets were identified or validated.161
miR-302
A microarray study assessing the effect of modified LDL exposure on macrophage microRNA expression demonstrated miR-302a levels were down-regulated upon contact with either acetylated- or oxidized-LDL and was associated with a concomitant increase in expression of the cholesterol efflux genes ABCA1 and ABCG1.163 Validation studies confirmed ABCA1 as a miR-302a target and supporting in vitro and in vivo findings established miR-302 over-expression suppresses cholesterol efflux and facilitates both foam cell macrophage formation and dysregulated hepatic cholesterol clearance.163 Accordingly, administration of a miR-302a inhibitor to Ldlr-deficient mice resulted in the formation of smaller aortic atherosclerotic plaques with increased VSMC and macrophage content but reduced necrotic core size and was also associated with increased circulating HDL levels,163 suggesting that targeting of miR-302a may have therapeutic potential through dual anti-atherosclerotic effects on foam cell formation and circulating lipoprotein metabolism.
miR-590
While a previous study has shown that increased LPL expression in macrophages through a miR-182/HDAC9 axis promotes a pro-inflammatory macrophage phenotype and is subsequently pro-atherosclerotic,160 miR-590 directly targets and supresses LPL levels and facilitates anti-inflammatory macrophage polarization,165,197 and would therefore be expected to exert an anti-atherosclerotic role. Accordingly, miR-590 agomir delivery to Apoe-deficient mice decreased aortic atherosclerotic plaque formation, while miR-590 antagomir administration accelerated atherogenesis.165 The effects on atherosclerosis were associated with reciprocal changes in plaque macrophage LPL expression and circulating LDL-cholesterol levels.165
5.1.2.1 Hepatocytes and lipid metabolism
An important contributory role for microRNA regulation of lipid metabolism has been recently highlighted,198 and due to the strong links between circulating lipoprotein profiles and CAD, have been associated to indirectly promote atherosclerosis. Although the focus of this review is on microRNAs which have been directly demonstrated to influence atherosclerosis, some examples of microRNAs which modulate lipid metabolism and therefore by association potentially atherosclerosis, include miR-21, miR-27a/b, and miR-122. Indeed, miR-122 has been identified as a liver-enriched and liver-specific microRNA which can regulate total serum cholesterol and triglyceride levels,199 in part through modulation of PPAR signalling family members such as PPARα and PPARβ/δ.200 Relatedly, miR-21 expression was attenuated within livers of high-fat fed mice compared to chow fed mice and was therefore attributed a role in lipid metabolism,201 also associated with regulation of PPARα.202 Finally, miR-27a/b may regulate lipid metabolism through effects on lipid synthesis and secretion from cells, again by targeting members of the PPAR family including PPARα and PPARγ, alongside other direct cholesterol efflux mRNAs such as ABCA1.203,204 Additionally, a recent study performed in baboons identified a novel molecular mechanism whereby LDL-C levels influence monocyte microRNA expression and may therefore affect atherosclerosis initiation through an additional pathway.205 Nonetheless, some microRNAs such as miR-30 and miR-33 have been shown to directly modulate atherosclerosis.
miR-30
Marked expression of miR-30c is observed within the liver in comparison to other tissues, where it is proposed to regulate lipoprotein production (such as ApoB) through targeting of the microsomal triglyceride transfer protein (MTTP) alongside decreasing lipid synthesis independent of modulation of MTTP.128In vivo, lentiviral hepatic-directed over-expression of miR-30c suppressed plasma cholesterol levels in high-fat fed C57Bl/6 mice which was associated with reduced hepatic expression of MTTP.128 Similar effects were observed in Apoe-deficient mice alongside reduced atherogenesis within the aorta whereas liver-directed delivery of a miR-30c inhibitor elevated circulating ApoB and cholesterol levels alongside increasing aortic plaque size compared with control animals.128 Further studies in Apoe-deficient mice deploying systemic delivery of a miR-30c mimic showed similar beneficial effects on lowering plasma cholesterol levels and mitigating aortic plaque development, even in animals with pre-existing hypercholesterolaemia.129 Collectively, these studies support therapeutic strategies to increase liver miR-30c expression to prevent atherosclerosis progression, especially in patients who respond poorly to other lipid-lowering treatments. Moreover, the mouse studies demonstrated that hepatosteatosis (a common side effect of conventional MTTP inhibitors) was avoided with miR-30c over-expression presumably through reducing hepatic lipid synthesis, further supporting elevation of hepatic miR-30c levels as a pharmacological approach to mitigate hypercholesterolaemia and atherosclerosis.128,129
miR-33
There has been intense interest in miR-33a and miR-33b with regards to their possible deleterious role in atherosclerosis, driven by their ability to target cholesterol efflux related mRNAs such as ABCA1, and the identification of their intergenic location within SREBF2 and SREBF1 respectively, transcription factors with prominent roles in lipid metabolism regulation.206 However, analysis of human carotid endarterectomy samples revealed miR-33a levels were decreased in plaques compared with adjacent plaque edge regions deemed relatively healthy, whereas miR-33b expression was not significantly altered,139 inferring a more prominent role for miR-33a in atherosclerosis as opposed to miR-33b. Though, findings utilizing a miR-33b transgenic ‘knock-in’ mouse (deployed as mice express negligible miR-33b transcript levels), demonstrated miR-33b and SREBF1 expression is elevated within the livers of hypercholesterolaemic mice, while miR-33a and SREBF2 levels are decreased139 suggesting the actions of these two miR-33 members may be tissue or cell specific. Direct assessment of miR-33 perturbation on atherosclerosis have been predominantly carried out in the Ldlr-deficient mouse model and support pro-atherosclerotic roles for miR-33a and miR-33b associated with regulatory roles in lipid metabolism.133,137,139 Moreover, mice double deficient for Apoe and miR-33 (presumably both isoforms) exhibit elevated HDL-cholesterol plasma levels and suppressed aortic atherogenesis.131 Conversely, bone-marrow transplantation from miR-33 donor mice into Apoe-deficient recipient had no effect on plaque size or macrophage content, although lipid accumulation within lesions was lowered despite no effect on circulating HDL-cholesterol levels.131 In both experiments the authors proposed that loss of miR-33 reduced intra-plaque lipid accumulation through restoration of macrophage ABCA1 and ABCG1 expression subsequently enabling enhanced cholesterol efflux from foam cell macrophages.131 Equally, global deficiency of miR-33 in Ldlr-deficient raised cholesterol levels but did not affect aortic atherosclerosis, while miR-33 haematopoietic-restricted deficiency did not affect plasma cholesterol levels but retarded plaque development, an effect which was lost when mice were reconstituted with bone-marrow cells from miR-33b over-expressing mice.136 Of note, systemic miR-33 loss induced obesity and insulin resistance in Ldlr-deficient mice, which was absent in mice reconstituted with miR-33-deficeint bone-marrow cells.136 These findings imply the positive therapeutic effects of miR-33 inhibition would require targeting of intra-plaque macrophages (to enhance reverse cholesterol transport) whilst averting deleterious systemic effects related to heightened metabolic disease. However, numerous studies have shown systemic miR-33 inhibition attenuates atherosclerosis in Ldlr-deficient mice134,135,137,138 or the diabetic REVERSA mouse model,130 largely independent of effects on lipid metabolism. Although long-term (14 weeks) miR-33 inhibition exerted no beneficial change on aortic atherosclerosis in hypercholesteraemic Ldlr-deficient mice.132 Furthermore, although miR-33 antagonism appears anti-atherosclerotic harmful elevations in circulating triglyceride levels alongside development of hepatosteatosis have been reported.131,132 Accordingly, if miR-33 inhibition is to be pursued therapeutically, macrophage-specific targeting approaches will be essential to limit unwanted off-target effects.
6. Conclusions
The above findings demonstrate the wealth of studies investigating the expression and roles of ncRNAs pertinent to atherosclerosis. Mechanistic studies have revealed how ncRNAs can acutely function and behaviour of vascular and inflammatory cells, such as ECs, VSMCs, and macrophages. Clinico-pathological and animal studies (see Figures 3–5) have further elucidated the contribution of ncRNAs to atherosclerotic plaque formation and progression and highlighted processes which are modulated, including lipid metabolism, EC activation, modulation of VSMC phenotype, inflammatory cell recruitment, macrophage polarization and foam cell formation, and aberrant proteolysis. However, there are incidence where the action of a ncRNA is beneficial during atherogenesis but potentially detrimental in advanced plaques, hampering their therapeutic potential. For instance, miR-21 and miR-145 are markedly up-regulated in human plaques and within vessels displaying restenosis (after stent deployment for example) implying a detrimental role for these microRNAs in both pathologies. Yet modulating miR-21 or miR-145 levels in animal models exerts divergent effects, as both are considered atheroprotective where VSMC growth is required for maintain fibrous cap integrity, but associated with restenosis where VSMC growth is detrimental,181,183,207 resulting in intimal formation analogous to atherogenesis in humans.
Figure 3.
Proposed roles of long non-coding and circular RNAs in atherosclerotic plaque development, progression and stability. The association of long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) are shown during the different stages of atherosclerotic plaque development. Atherogenesis is initially characterized by substantial alterations in the inner arterial surface: stress stimuli (nitric oxide, hypoxia, oxidative stress, shear stress…) trigger endothelial cell (EC) activation. The activated endothelium express numerous adhesion molecules (such as VCAM-1 and depicted as green circles) promoting the recruitment of monocytes (in green) from the blood stream and, at the same time, stimulating vascular smooth muscle cell (VSMC) migration and proliferation. VSMCs acquire a synthetic phenotype and contribute to the formation of the protective fibrous cap (in pink) by secreting differing extracellular matrix (ECM) proteins. However, perpetual monocyte recruitment, their differentiation into macrophages and their associated accrual of lipids (such as modified LDL) results in macrophage foam cell formation. During atherosclerotic plaque progression, foam cell macrophages undergo apoptosis and drive the formation of the necrotic/lipid-rich core (depicted in yellow), while VSMC apoptosis and dysregulated proteolysis drives thinning of the protective fibrous cap, both of which characterizes advanced unstable plaques.
Figure 5.
Proposed roles microRNAs in atherosclerotic plaque development, progression, and stability. The association of microRNAs are shown during the different stages of atherosclerotic plaque development. Atherogenesis is initially characterized by substantial alterations in the inner arterial surface: stress stimuli (nitric oxide, hypoxia, oxidative stress, shear stress…) trigger endothelial cell (EC) activation. The activated endothelium express numerous adhesion molecules (such as VCAM-1 and depicted as green circles) promoting the recruitment of monocytes (in green) from the blood stream and, at the same time, stimulating vascular smooth muscle cell (VSMC) migration and proliferation. VSMCs acquire a synthetic phenotype and contribute to the formation of the protective fibrous cap (in pink) by secreting differing extracellular matrix (ECM) proteins. However, perpetual monocyte recruitment, their differentiation into macrophages and their associated accrual of lipids (such as modified LDL) results in macrophage foam cell formation. During atherosclerotic plaque progression, foam cell macrophages undergo apoptosis and drive the formation of the necrotic/lipid-rich core (depicted in yellow), while VSMC apoptosis and dysregulated proteolysis drives thinning of the protective fibrous cap, both of which characterizes advanced unstable plaques.
Members of all the ncRNA families can target multiple genes, pathways and processes alongside controlling other non-coding classes, such as lncRNAs acting as microRNA sponges, adding further complexity in attempt to elucidate the roles of ncRNAs in atherosclerosis. However, there is also devil in the details as microRNAs can target mRNAs within common regulatory networks, suggesting that modulating select microRNAs may be a means to effect specific biological mechanisms and signalling pathways within atherosclerotic arteries alongside other organs associated with atherosclerotic risk including the liver. Furthermore, there is an expanding armamentarium of tools available for researchers and clinicians to modulate the expression and function of ncRNAs which may offer attractive therapeutic strategies to manage all stages of atherosclerosis. Any such therapeutics will have to take into consideration the wide-range of potential substrates (and therefore possible off-target effects) ncRNAs harbour, necessitating the need for sophisticated delivery and targeting approaches. These will include cell type-specific delivery, as recently demonstrated using microRNA-containing microparticles enriched with miR-146a and miR-181b to selectively target ECs.153 Similarly, deploying ultrasound-targeted microbubbles to permit local delivery of miR-21 to carotid plaques184 represents another novel stratagem. Such targeting strategies are essential to ensure the therapeutic potential of anti-atherosclerotic treatments to control select ncRNA are fully exploited; for example, achieving macrophage-specific perturbation of miR-33 to spare detrimental off-target effects on the liver. A precedent has been set for ncRNA therapies as recently evidenced in clinical trials for several diseases.208 Especially, anti-sense oligonucleotides have been deployed to suppress miR-122 in hepatitis C patients, and chemically-modified mimics to exogenously increase levels of miR-16, miR-29, or miR-155 as treatments for various forms of cancer are currently under assessment.208 Such developments should foster new clinical studies exploiting ncRNA therapeutics in atherosclerosis but will necessitate robust identification and validation of significant candidate ncRNAs for atherosclerotic plaque development, progression and rupture. Moreover, nuanced and sophisticated delivery and targeting approaches will be necessary to circumvent likely off-target effects and toxicities, and to enable the deployment of ncRNA preventative and treatment therapeutics in patients with all stages of atherosclerosis.
Conflict of interest: none declared.
Funding
J.L.J. is the recipient and funded through a British Heart Foundation Senior Basic Science Research Fellowship (FS/18/1/33234). Research in Lars Maegdefessel’s labs is funded through the German Research Council (DFG Heisenberg Professorship, Collaborative Research Center, CRC1123), the German Center for Cardiovascular Research (DZHK, Junior Research Group), the European Research Council (ERC Starting Grant NORVAS), the Swedish Research Council (VR, 2015-03140), the Swedish Heart-Lung Foundation (HLF, 20180680), and a Ragnar Söderberg Foundation Fellowship (M14-55).
References
- 1. Skalen K, Gustafsson M, Rydberg EK, Hulten LM, Wiklund O, Innerarity TL, Boren J. Subendothelial retention of atherogenic lipoproteins in early atherosclerosis. Nature 2002;417:750–754. [DOI] [PubMed] [Google Scholar]
- 2. Kolodgie FD, Burke AP, Nakazawa G, Virmani R. Is pathologic intimal thickening the key to understanding early plaque progression in human atherosclerotic disease? Arterioscler Thromb Vasc Biol 2007;27:986–989. [DOI] [PubMed] [Google Scholar]
- 3. Leitinger N. Oxidized phospholipids as modulators of inflammation in atherosclerosis. Curr Opin Lipidol 2003;14:421–430. [DOI] [PubMed] [Google Scholar]
- 4. Hansson GK. Inflammation, atherosclerosis, and coronary artery disease—reply. N Engl J Med 2005;353:429–430. [DOI] [PubMed] [Google Scholar]
- 5. Allahverdian S, Chaabane C, Boukais K, Francis GA, Bochaton-Piallat ML. Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc Res 2018;114:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tabas I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat Rev Immunol 2010;10:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glagov S, Weisenberg E, Zarins CK, Stankunavicius R, Kolettis G. Compensatory enlargement of human atherosclerotic coronary plaques. N Engl J Med 1987;316:1371–1375. [DOI] [PubMed] [Google Scholar]
- 8. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest ARR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato X, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SPT, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Babu MM, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CAM, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science 2005;309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 9.FANTOM Consortium and the RIKEN PMI and CLST (DGT)Forrest ARR, Kawaji H, Rehli M, Baillie JK, MJL de Hoon, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple CA, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JAC, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drablos F, Edge ASB, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furuno M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Sui SJH, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov A, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JFJ, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, Van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Nozaki T, Ogishima S, Ohkura N, Ohmiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JGD, Rackham OJL, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Satoh H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng GJ, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, 't Hoen PAC, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyoda H, Toyoda T, Valen E, van de Wetering M, van den Berg LM, Verardo R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells CA, Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zhao XB, Zucchelli S, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume DA, Carninci P, Hayashizaki Y.. A promoter-level mammalian expression atlas. Nature 2014;507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feingold EA, Good PJ, Guyer MS, Kamholz S, Liefer L, Wetterstrand K, Collins FS, Gingeras TR, Kampa D, Sekinger EA, Cheng J, Hirsch H, Ghosh S, Zhu Z, Pate S, Piccolboni A, Yang A, Tammana H, Bekiranov S, Kapranov P, Harrison R, Church G, Struhl K, Ren B, Kim TH, Barrera LO, Qu C, Van Calcar S, Luna R, Glass CK, Rosenfeld MG, Guigo R, Antonarakis SE, Birney E, Brent M, Pachter L, Reymond A, Dermitzakis ET, Dewey C, Keefe D, Denoeud F, Lagarde J, Ashurst J, Hubbard T, Wesselink JJ, Castelo R, Eyras E, Myers RM, Sidow A, Batzoglou S, Trinklein ND, Hartman SJ, Aldred SF, Anton E, Schroeder DI, Marticke SS, Nguyen L, Schmutz J, Grimwood J, Dickson M, Cooper GM, Stone EA, Asimenos G, Brudno M, Dutta A, Karnani N, Taylor CM, Kim HK, Robins G, Stamatoyannopoulos G, Stamatoyannopoulos JA, Dorschner M, Sabo P, Hawrytycz M, Humbert R, Wallace J, Yu M, Navas PA, McArthur M, Noble WS, Dunham I, Koch CM, Andrews RM, Clelland GK, Wilcox S, Fowler JC, James KD, Groth P, Dovey OM, Ellis PD, Wraight VL, Mungall AJ, Dhami P, Fiegler H, Langford CF, Carter NP, Vetrie D, Snyder M, Euskirchen G, Urban AE, Nagalakshmi U, Rinn J, Popescu G, Bertone P, Hartman S, Rozowsky J, Emanuelsson O, Royce T, Chung S, Gerstein M, Lian Z, Lian J, Nakayama Y, Weissman S, Stoic V, Tongprasit W, Sethi H, Jones S, Marra M, Shin H, Schein J, Clamp M, Lindblad-Toh K, Chang J, Jaffe DB, Kamal ES, Lander ES, Mikkelsen TS, Vinson J, Zody MC, de Jong PJ, Osoegawa K, Nefedov M, Zhu B, Baxevanis AD, Wolfsberg TG, Collins FS, Crawford GE, Holt E, Vasicek TJ, Zhou D, Luo S, Green ED, Bouffard GG, Margulies EH, Portnoy ME, Hansen NF, Thomas PJ, Mcdowell JC, Maskeri B, Young AC, Idol JR, Blakesley RW, Schuler G, Miller W, Hardison R, Elnitski L, Shah P, Salzberg SL, Pertea M, Majoros WH, Haussler D, Thomas D, Rosenbloom KR, Clawson H, Siepe A, Kent WJ, Weng Z, Jin S, Halees A, Burden H, Karaoz U, Fu Y, Yu Y, Ding C, Cantor CR, Kingston RE, Dennis J, Green RD, Singer MA, Richmond TA, Norton JE, Farnham PJ, Oberley MJ, Inman DR, McCormick MR, Kim H, Middle CL, Pirrung MC, Fu XD, Kwon YS, Ye Z, Dekker J, Tabuchi TM, Gheldof N, Dostie J, Harvey SC ENCODE Project Consortium. The ENCODE (ENCyclopedia of DNA elements) project. Science 2004;306:636–640. [DOI] [PubMed] [Google Scholar]
- 11. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan XA, Ruan YJ, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pelechano V, Steinmetz LN, Coding RNA. Gene regulation by antisense transcription. Nat Rev Genet 2013;14:880–893. [DOI] [PubMed] [Google Scholar]
- 15. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marín-Béjar O, Huarte M. Long noncoding RNAs: from identification to functions and mechanisms. Adv Genomics Genet 2015;5:257–274. [Google Scholar]
- 17. Li R, Zhu H, Luo Y. Understanding the functions of long non-coding RNAs through their higher-order structures. Int J Mol Sci 2016;17:E702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith MA, Gesell T, Stadler PF, Mattick JS. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res 2013;41:8220–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7–21. [DOI] [PubMed] [Google Scholar]
- 20. Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther 2013;23:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform 2017;18:558–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J, Teupser D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol 2010;30:620–627. [DOI] [PubMed] [Google Scholar]
- 23. Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, Yamamoto K, Onishi M, Sugimoto K, Katsuya T, Awata N, Ikebe K, Gondo Y, Oike Y, Ohishi M, Rakugi H. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012;220:449–455. [DOI] [PubMed] [Google Scholar]
- 24. Arslan S, Berkan O, Lalem T, Ozbilum N, Goksel S, Korkmaz O, Cetin N, Devaux Y, Network CT. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis 2017;266:176–181. [DOI] [PubMed] [Google Scholar]
- 25. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014;114:1389–1397. [DOI] [PubMed] [Google Scholar]
- 26. Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, Scherer SW, Nakabayashi K, Marsden PA. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem 2004;279:37982–37996. [DOI] [PubMed] [Google Scholar]
- 27. Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem 2007;282:15652–15666. [DOI] [PubMed] [Google Scholar]
- 28. Lyu Q, Xu S, Lyu Y, Choi M, Christie CK, Slivano OJ, Rahman A, Jin Z-G, Long X, Xu Y, Miano JM. SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc Natl Acad Sci USA 2019;116:546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol 2014;34:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, Brittan M, Hunter A, McBride M, McClure J, Miano JM, Emanueli C, Mills NL, Mountford JC, Baker AH. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther 2016;24:978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015;116:1143–1156. [DOI] [PubMed] [Google Scholar]
- 32. Li KG, Chowdhury T, Vakeel P, Koceja C, Sampath V, Ramchandran R. Delta-like 4 mRNA is regulated by adjacent natural antisense transcripts. Vasc Cell 2015;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilusz JE. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA 2018;9:e1478.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, Eskin A, Casero D, Zhang Z, Sandhu J, Salisbury D, Rajbhandari P, Civelek M, Hong C, Ito A, Liu X, Daniel B, Lusis AJ, Whitelegge J, Nagy L, Castrillo A, Smale S, Tontonoz P. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med 2018;24:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res 2013;113:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014;130:1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang S, Zhang X, Yuan Y, Tan M, Zhang L, Xue X, Yan Y, Han L, Xu Z. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surg 2015;47:439–446. [DOI] [PubMed] [Google Scholar]
- 39. He Q, Tan JY, Yu B, Shi WH, Liang K. Long noncoding RNA HIF1A-AS1A reduces apoptosis of vascular smooth muscle cells: implications for the pathogenesis of thoracoabdominal aorta aneurysm. Pharmazie 2015;70:310–315. [PubMed] [Google Scholar]
- 40. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016;7:12429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li CY, Ma L, Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother 2017;95:1514–1519. [DOI] [PubMed] [Google Scholar]
- 43. Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, Wu Z, Li D, Chen X. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep 2015;12:6656–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci USA 2010;107:3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hall IF, Climent M, Quintavalle M, Farina FM, Schorn T, Zani S, Carullo P, Kunderfranco P, Civilini E, Condorelli G, Elia L. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res 2019;124:498–510. [DOI] [PubMed] [Google Scholar]
- 46. Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res 2015;117:884–890. [DOI] [PubMed] [Google Scholar]
- 47. Matarin M, Brown WM, Singleton A, Hardy JA, Meschia JF, Investigators I. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke 2008;39:1586–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harismendy O, Notani D, Song XY, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 2011;470:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jarinova O, Stewart AFR, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, McPherson R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol 2009;29:1671–1677. [DOI] [PubMed] [Google Scholar]
- 50. Holdt LM, Teupser D. Long noncoding RNA ANRIL: lnc-ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of atherosclerosis. Front Cardiovasc Med 2018;5:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gabel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLos Genet 2013;9:e1003588.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol 2012;32:196–206. [DOI] [PubMed] [Google Scholar]
- 53. Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, Ohishi M, Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates pathways in VSMC. Biochem Biophys Res Commun 2012;419:612–616. [DOI] [PubMed] [Google Scholar]
- 54. Lo Sardo V, Chubukov P, Ferguson W, Kumar A, Teng EL, Duran M, Zhang L, Cost G, Engler AJ, Urnov F, Topol EJ, Torkamani A, Baldwin KK. Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell 2018;175:1796–1810.e1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chiu J-J, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol Rev 2011;91:327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilcox JN, Subramanian RR, Sundell CL, Tracey WR, Pollock JS, Harrison DG, Marsden PA. Expression of multiple isoforms of nitric oxide synthase in normal and atherosclerotic vessels. Arterioscler Thromb Vasc Biol 1997;17:2479–2488. [DOI] [PubMed] [Google Scholar]
- 57. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031–8041. [DOI] [PubMed] [Google Scholar]
- 58. Yang LQ, Lin CR, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011;147:773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 2014;5:e1506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med 2015;19:1418–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tan J, Liu S, Jiang Q, Yu T, Huang K. LncRNA-MIAT increased in patients with coronary atherosclerotic heart disease. Cardiol Res Pract 2019;2019:6280194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ye Z-M, Yang S, Xia Y-P, Hu R-T, Chen S, Li B-W, Chen S-L, Luo X-y, Mao L, Li Y, Jin H, Qin C, Hu B. LncRNA MIAT sponges miR-149-5p to inhibit efferocytosis in advanced atherosclerosis through CD47 upregulation. Cell Death Dis 2019;10:138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bianchessi V, Badi I, Bertolotti M, Nigro P, D'Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A, Lauri A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J Mol Cell Cardiol 2015;81:62–70. [DOI] [PubMed] [Google Scholar]
- 64. Martinet W, De Meyer G. Autophagy in atherosclerosis a cell survival and death phenomenon with therapeutic potential. Circ Res 2009;104:304–317. [DOI] [PubMed] [Google Scholar]
- 65. Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, Kung H, Zhao B, Miao J. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 2014;10:957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Viola M, Karousou E, D'Angelo ML, Moretto P, Caon I, De Luca G, Passi A, Vigetti D. Extracellular matrix in atherosclerosis: hyaluronan and proteoglycans insights. Curr Med Chem 2016;23:2958–2971. [DOI] [PubMed] [Google Scholar]
- 67. Chao H, Spicer AP. Natural antisense mRNAs to hyaluronan synthase 2 inhibit hyaluronan biosynthesis and cell proliferation. J Biol Chem 2005;280:27513–27522. [DOI] [PubMed] [Google Scholar]
- 68. Michael DR, Phillips AO, Krupa A, Martin J, Redman JE, Altaher A, Neville RD, Webber J, Kim MY, Bowen T. The Human Hyaluronan Synthase 2 (HAS2) gene and its natural antisense RNA exhibit coordinated expression in the renal proximal tubular epithelial cell. J Biol Chem 2011;286:19523–19532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bentzon JF, Falk E. Plaque erosion: new insights from the road less travelled. Circ Res 2017;121:8–10. [DOI] [PubMed] [Google Scholar]
- 70. Falk E, Nakano M, Bentzon JF, Finn AV, Virmani R. Update on acute coronary syndromes: the pathologists' view. Eur Heart J 2013;34:719–728. [DOI] [PubMed] [Google Scholar]
- 71. Davies MJ. Stability and instability: two faces of coronary atherosclerosis—the Paul Dudley White Lecture 1995. Circulation 1996;94:2013–2020. [DOI] [PubMed] [Google Scholar]
- 72. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 73. Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen SF, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu XF, Lu YC, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu LM, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang XL, Zhang WH, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FUR, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang LY, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu DF, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJF, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O'Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M; Consortium CD. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nature Genet 2015;47:1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol 2017;14:992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014;159:134–147. [DOI] [PubMed] [Google Scholar]
- 77. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 2017;67:214–227 e217. [DOI] [PubMed] [Google Scholar]
- 78. Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017;544:115–119. [DOI] [PubMed] [Google Scholar]
- 79. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 80. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015;18:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell 2018;71:428–442. [DOI] [PubMed] [Google Scholar]
- 83. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 2017;66:22–37 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lin X, Lo HC, Wong DT, Xiao X. Noncoding RNAs in human saliva as potential disease biomarkers. Front Genet 2015;6:175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li PF, Chen SC, Chen HL, Mo XY, Li TW, Shao YF, Xiao BX, Guo JM. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015;444:132–136. [DOI] [PubMed] [Google Scholar]
- 87. Li F, Zhang LY, Li W, Deng JQ, Zheng J, An MX, Lu JC, Zhou YF. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015;6:6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Qin ML, Liu G, Huo XS, Tao XM, Sun XM, Ge ZH, Yang J, Fan J, Liu L, Qin WX. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016;16:161–169. [DOI] [PubMed] [Google Scholar]
- 89. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu W, Seok J, Mindrinos MN, Schweitzer AC, Jiang H, Wilhelmy J, Clark TA, Kapur K, Xing Y, Faham M, Storey JD, Moldawer LL, Maier RV, Tompkins RG, Wong WH, Davis RW, Xiao W; Inflammation, Host Response to Injury Large-Scale Collaborative Research Program. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci USA 2011;108:3707–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hulten LM, Levin M. The role of hypoxia in atherosclerosis. Curr Opin Lipidol 2009;20:409–414. [DOI] [PubMed] [Google Scholar]
- 93. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Song CL, Wang JP, Xue X, Liu N, Zhang XH, Zhao Z, Liu JG, Zhang CP, Piao ZH, Liu Y, Yang YB. Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell Physiol Biochem 2017;42:1202–1212. [DOI] [PubMed] [Google Scholar]
- 95. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 96. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- 97. Wang R, Dong LD, Meng XB, Shi Q, Sun WY. Unique MicroRNA signatures associated with early coronary atherosclerotic plaques. Biochem Biophys Res Commun 2015;464:574–579. [DOI] [PubMed] [Google Scholar]
- 98. Raitoharju E, Lyytikäinen L-P, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kähönen M, Karhunen PJ, Laaksonen R, Lehtimäki T. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011;219:211–217. [DOI] [PubMed] [Google Scholar]
- 99. Bidzhekov K, Gan L, Denecke B, Rostalsky A, Hristov M, Koeppel TA, Zernecke A, Weber C. microRNA expression signatures and parallels between monocyte subsets and atherosclerotic plaque in humans. Thromb Haemost 2012;107:619–625. [DOI] [PubMed] [Google Scholar]
- 100. Cipollone F, Felicioni L, Sarzani R, Ucchino S, Spigonardo F, Mandolini C, Malatesta S, Bucci M, Mammarella C, Santovito D, de Lutiis F, Marchetti A, Mezzetti A, Buttitta F. A unique MicroRNA signature associated with plaque instability in humans. Stroke 2011;42:2556–2563. [DOI] [PubMed] [Google Scholar]
- 101. Di Gregoli K, Jenkins N, Salter R, White S, Newby AC, Johnson JL. MicroRNA-24 regulates macrophage behavior and retards atherosclerosis. Arterioscler Thromb Vasc Biol 2014;34:1990–2000. [DOI] [PubMed] [Google Scholar]
- 102. Di Gregoli K, Mohamad Anuar NN, Bianco R, White SJ, Newby AC, George SJ, Johnson JL. MicroRNA-181b controls atherosclerosis and aneurysms through regulation of TIMP-3 and elastin. Circ Res 2017;120:49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Leistner DM, Boeckel JN, Reis SM, Thome CE, De Rosa R, Keller T, Palapies L, Fichtlscherer S, Dimmeler S, Zeiher AM. Transcoronary gradients of vascular miRNAs and coronary atherosclerotic plaque characteristics. Eur Heart J 2016;37:1738–1749. [DOI] [PubMed] [Google Scholar]
- 104. Faccini J, Ruidavets JB, Cordelier P, Martins F, Maoret JJ, Bongard V, Ferrieres J, Roncalli J, Elbaz M, Vindis C. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci Rep 2017;7:42916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating MicroRNAs in patients with coronary artery disease. Circ Res 2010;107:677–684. [DOI] [PubMed] [Google Scholar]
- 106. Weber M, Baker MB, Patel RS, Quyyumi AA, Bao G, Searles CD. MicroRNA expression profile in CAD patients and the impact of ACEI/ARB. Cardiol Res Pract 2011;2011:532915.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhu G-F, Yang L-X, Guo R-W, Liu H, Shi Y-K, Ye J-S, Yang Z-H. MicroRNA-155 is inversely associated with severity of coronary stenotic lesions calculated by the Gensini score. Coron Artery Dis 2014;25:304–310. [DOI] [PubMed] [Google Scholar]
- 108. D'Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, Rubino M, Marenzi G, Colombo GI, Achilli F, Maggiolini S, Capogrossi MC, Pompilio G. Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS One 2013;8:e80345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zeller T, Keller T, Ojeda F, Reichlin T, Twerenbold R, Tzikas S, Wild PS, Reiter M, Czyz E, Lackner KJ, Munzel T, Mueller C, Blankenberg S. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J 2014;35:2106–2114. [DOI] [PubMed] [Google Scholar]
- 110. Meder B, Keller A, Vogel B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J, Leidinger P, Meese E, Katus HA, Rottbauer W. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol 2011;106:13–23. [DOI] [PubMed] [Google Scholar]
- 111. Xiong X-D, Cho M, Cai X-P, Cheng J, Jing X, Cen J-M, Liu X, Yang X-L, Suh Y. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat Res 2014;761:15–20. [DOI] [PubMed] [Google Scholar]
- 112. Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin–angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci 2010;119:395–405. [DOI] [PubMed] [Google Scholar]
- 113. Hoekstra M, van der Lans CAC, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, van Berkel TJC, Biessen E. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun 2010;394:792–797. [DOI] [PubMed] [Google Scholar]
- 114. Hulsmans M, Sinnaeve P, Van der Schueren B, Mathieu C, Janssens S, Holvoet P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J Clin Endocrinol Metab 2012;97:E1213–E1218. [DOI] [PubMed] [Google Scholar]
- 115. Li X-D, Yang Y-J, Wang L-Y, Qiao S-B, Lu X-F, Wu Y-J, Xu B, Li H-F, Gu D-F. Elevated plasma miRNA-122, -140-3p, -720, -2861, and -3149 during early period of acute coronary syndrome are derived from peripheral blood mononuclear cells. Plos One 2017;12:e0184256.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ye Z-L, Lu H-L, Su Q, Li L. Association between the level of CD4(+) T lymphocyte microRNA-155 and coronary artery disease in patients with unstable angina pectoris. J Geriatr Cardiol 2018;15:611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chen YC, Bui AV, Diesch J, Manasseh R, Hausding C, Rivera J, Haviv I, Agrotis A, Htun NM, Jowett J, Hagemeyer CE, Hannan RD, Bobik A, Peter K. A novel mouse model of atherosclerotic plaque instability for drug testing and mechanistic/therapeutic discoveries using gene and MicroRNA expression profiling. Circ Res 2013;113:252–265. [DOI] [PubMed] [Google Scholar]
- 118. Liu MX, Tao GZ, Liu QF, Liu K, Yang XC. MicroRNA let-7g alleviates atherosclerosis via the targeting of LOX-1 in vitro and in vivo. Int J Mol Med 2017;40:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wei Y, Corbalán-Campos J, Gurung R, Natarelli L, Zhu M, Exner N, Erhard F, Greulich F, Geißler C, Uhlenhaut NH, Zimmer R, Schober A. Dicer in macrophages prevents atherosclerosis by promoting mitochondrial oxidative metabolism. Circulation 2018;138:2007–2020. [DOI] [PubMed] [Google Scholar]
- 120. Lee DY, Yang TL, Huang YH, Lee CI, Chen LJ, Shih YT, Wei SY, Wang WL, Wu CC, Chiu JJ. Induction of microRNA-10a using retinoic acid receptor-alpha and retinoid x receptor-alpha agonists inhibits atherosclerotic lesion formation. Atherosclerosis 2018;271:36–44. [DOI] [PubMed] [Google Scholar]
- 121. Wang DL, Wang WT, Lin WQ, Yang WQ, Zhang PW, Chen M, Ding D, Liu CQ, Zheng JK, Ling WH. Apoptotic cell induction of miR-10b in macrophages contributes to advanced atherosclerosis progression in ApoE(-/-) mice. Cardiovasc Res 2018;114:1794–1805. [DOI] [PubMed] [Google Scholar]
- 122. Lv YC, Tang YY, Peng J, Zhao GJ, Yang J, Yao F, Ouyang XP, He PP, Xie W, Tan YL, Zhang M, Liu D, Tang DP, Cayabyab FS, Zheng XL, Zhang DW, Tian GP, Tang CK. MicroRNA-19b promotes macrophage cholesterol accumulation and aortic atherosclerosis by targeting ATP-binding cassette transporter A1. Atherosclerosis 2014;236:215–226. [DOI] [PubMed] [Google Scholar]
- 123. Li CL, Li SF, Zhang F, Wu MY, Liang HZ, Song JX, Lee CY, Chen H. Endothelial microparticles-mediated transfer of microRNA-19b promotes atherosclerosis via activating perivascular adipose tissue inflammation in apoE(-/-) mice. Biochem Biophys Res Commun 2018;495:1922–1929. [DOI] [PubMed] [Google Scholar]
- 124. Canfran-Duque A, Rotllan N, Zhang XB, Fernandez-Fuertes M, Ramirez-Hidalgo C, Araldi E, Daimiel L, Busto R, Fernandez-Hernando C, Suarez Y. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol Med 2017;9:1244–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sasaki T, Kuzuya M, Nakamura K, Cheng XW, Shibata T, Sato K, Iguchi A. A simple method of plaque rupture induction in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol 2006;26:1304–1309. [DOI] [PubMed] [Google Scholar]
- 126. Yang S, Ye ZM, Chen SC, Luo XY, Chen SL, Mao L, Li YA, Jin HJ, Yu C, Xiang FX, Xie MX, Chang J, Xia YP, Hu B. MicroRNA-23a-5p promotes atherosclerotic plaque progression and vulnerability by repressing ATP-binding cassette transporter A1/G1 in macrophages. J Mol Cell Cardiol 2018;123:139–149. [DOI] [PubMed] [Google Scholar]
- 127. Ren K, Zhu X, Zheng Z, Mo ZC, Peng XS, Zeng YZ, Ou HX, Zhang QH, Qi HZ, Zhao GJ, Yi GH. MicroRNA-24 aggravates atherosclerosis by inhibiting selective lipid uptake from HDL cholesterol via the post-transcriptional repression of scavenger receptor class Btype I. Atherosclerosis 2018;270:57–67. [DOI] [PubMed] [Google Scholar]
- 128. Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med 2013;19:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Irani S, Pan XY, Peck BCE, Iqbal J, Sethupathy P, Hussain MM. MicroRNA-30c mimic mitigates hypercholesterolemia and atherosclerosis in mice. J Biol Chem 2016;291:18397–18409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Distel E, Barrett TJ, Chung K, Girgis NM, Parathath S, Essau CC, Murphy AJ, Moore KJ, Fisher EA. miR33 inhibition overcomes deleterious effects of diabetes mellitus on atherosclerosis plaque regression in mice. Circ Res 2014;115:759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− Mice. JAHA 2012;1:e003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Marquart TJ, Wu J, Lusis AJ, Baldán Á. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol 2013;33:455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, Mattison JA, de Cabo R, Suarez Y, Fernandez-Hernando C. A regulatory role for MicroRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol 2013;33:2339–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, Rinehold K, van Solingen C, Fullerton MD, Cecchini K, Rayner KJ, Steinberg GR, Zamore PD, Fisher EA, Loke P, Moore KJ. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest 2015;125:4334–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ouimet M, Ediriweera H, Afonso MS, Ramkhelawon B, Singaravelu R, Liao XH, Bandler RC, Rahman K, Fisher EA, Rayner KJ, Pezacki JP, Tabas I, Moore KJ. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler Thromb Vasc Biol 2017;37:1058–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Price NL, Rotllan N, Canfrán-Duque A, Zhang X, Pati P, Arias N, Moen J, Mayr M, Ford DA, Baldán Á, Suárez Y, Fernández-Hernando C. Genetic dissection of the impact of miR-33a and miR-33b during the progression of atherosclerosis. Cell Rep 2017;21:1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 2011;121:2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Rotllan N, Ramírez CM, Aryal B, Esau CC, Fernández-Hernando C. Therapeutic silencing of MicroRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice—brief report. Arterioscler Thromb Vasc Biol 2013;33:1973–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Nishino T, Horie T, Baba O, Sowa N, Hanada R, Kuwabara Y, Nakao T, Nishiga M, Nishi H, Nakashima Y, Nakazeki F, Ide Y, Koyama S, Kimura M, Nagata M, Yoshida K, Takagi Y, Nakamura T, Hasegawa K, Miyamoto S, Kimura T, Ono K. SREBF1/MicroRNA-33b axis exhibits potent effect on unstable atherosclerotic plaque formation in vivo. Arterioscler Thromb Vasc Biol 2018;38:2460–2473. [DOI] [PubMed] [Google Scholar]
- 140. Su G, Sun G, Liu H, Shu L, Liang Z. Downregulation of miR-34a promotes endothelial cell growth and suppresses apoptosis in atherosclerosis by regulating Bcl-2. Heart Vessels 2018;33:1185–1194. [DOI] [PubMed] [Google Scholar]
- 141. Loyer X, Potteaux S, Vion A-C, Guerin CL, Boulkroun S, Rautou P-E, Ramkhelawon B, Esposito B, Dalloz M, Paul J-L, Julia PL, Maccario J, Boulanger CM, Mallat Z, Tedgui A. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res 2014;114:434–443. [DOI] [PubMed] [Google Scholar]
- 142. Dai Y, Wu XQ, Dai DS, Li J, Mehta JL. MicroRNA-98 regulates foam cell formation and lipid accumulation through repression of LOX-1. Redox Biol 2018;16:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pankratz F, Hohnloser C, Bemtgen X, Jaenich C, Kreuzaler S, Hoefer I, Pasterkamp G, Mastroianni J, Zeiser R, Smolka C, Schneider L, Martin J, Juschkat M, Helbing T, Moser M, Bode C, Grundmann S. MicroRNA-100 suppresses chronic vascular inflammation by stimulation of endothelial autophagy. Circ Res 2018;122:417–432. [DOI] [PubMed] [Google Scholar]
- 144. Chen WJ, Yu FP, Di MX, Li MM, Chen YF, Zhang Y, Liu XL, Huang XZ, Zhang M. MicroRNA-124-3p inhibits collagen synthesis in atherosclerotic plaques by targeting prolyl 4-hydroxylase subunit alpha-1 (P4HA1) in vascular smooth muscle cells. Atherosclerosis 2018;277:98–107. [DOI] [PubMed] [Google Scholar]
- 145. Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RTA, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med 2014;20:368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ye Q, Tian GP, Cheng HP, Zhang X, Ou X, Yu XH, Tan RQ, Yang FY, Gong D, Huang C, Pan YJ, Zhang J, Chen LY, Zhao ZW, Xie W, Li L, Zhang M, Xia XD, Zheng XL, Tang CK. MicroRNA-134 promotes the development of atherosclerosis via the ANGPTL4/LPL Pathway in apolipoprotein E knockout mice. J Atheroscler Thromb 2018;25:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147. Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJG, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol 2012;14:249–256. [DOI] [PubMed] [Google Scholar]
- 148. Lovren F, Pan Y, Quan A, Singh KK, Shukla PC, Gupta N, Steer BM, Ingram AJ, Gupta M, Al-Omran M, Teoh H, Marsden PA, Verma S. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation 2012;126:S81–S90. [DOI] [PubMed] [Google Scholar]
- 149. Sala F, Aranda JF, Rotllan N, Ramírez CM, Aryal B, Elia L, Condorelli G, Catapano AL, Fernández-Hernando C, Norata GD. MiR-143/145 deficiency protects against progression of atherosclerosis in Ldlr(−/−) mice. Thromb Haemost 2014;112:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Cheng HS, Besla R, Li A, Chen ZQ, Shikatani EA, Nazari-Jahantigh M, Hammoutene A, Nguyen MA, Geoffrion M, Cai L, Khyzha N, Li T, MacParland SA, Husain M, Cybulsky MI, Boulanger CM, Temel RE, Schober A, Rayner KJ, Robbins CS, Fish JE. Paradoxical suppression of atherosclerosis in the absence of microRNA-146a. Circ Res 2017;121:354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Li K, Ching D, Luk FS, Raffai RL. Apolipoprotein E enhances MicroRNA-146a in monocytes and macrophages to suppress nuclear factor-kappa B-driven inflammation and atherosclerosis. Circ Res 2015;117:e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. del Monte A, Arroyo AB, Andrés-Manzano MJ, García-Barberá N, Caleprico MS, Vicente V, Roldán V, González-Conejero R, Martínez C, Andrés V. miR-146a deficiency in hematopoietic cells is not involved in the development of atherosclerosis. PLoS One 2018;13:e0198932.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ma ST, Tian XY, Zhang YR, Mu CF, Shen HF, Bismuth J, Pownall HJ, Huang Y, Wong WT. E-selectin-targeting delivery of microRNAs by microparticles ameliorates endothelial inflammation and atherosclerosis. Sci Rep 2016;6:22910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Gong FH, Cheng WL, Wang HP, Gao MM, Qin JJ, Zhang Y, Li X, Zhu XY, Xia H, She ZG. Reduced atherosclerosis lesion size, inflammatory response in miR-150 knockout mice via macrophage effects. J Lipid Res 2018;59:658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Donners M, Wolfs IMJ, Stöger LJ, van der Vorst EPC, Pöttgens CCH, Heymans S, Schroen B, Gijbels MJJ, de Winther M. Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice. PLoS One 2012;7:e35877.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Du F, Yu F, Wang YZ, Hui Y, Carnevale K, Fu MG, Lu H, Fan DP. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol 2014;34:759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, Weber C, Schober A. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest 2012;122:4190–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. An TH, He QW, Xia YP, Chen SC, Baral S, Mao L, Jin HJ, Li YN, Wang MD, Chen JG, Zhu LQ, Hu B. MiR-181b antagonizes atherosclerotic plaque vulnerability through modulating macrophage polarization by directly targeting Notch1. Mol Neurobiol 2017;54:6329–6341. [DOI] [PubMed] [Google Scholar]
- 159. Sun XH, He SL, Wara AKM, Icli B, Shvartz E, Tesmenitsky Y, Belkin N, Li DZ, Blackwell TS, Sukhova GK, Croce K, Feinberg MW. Systemic delivery of MicroRNA-181b inhibits nuclear factor-kappa B activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ Res 2014;114:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Cheng HP, Gong D, Zhao ZW, He PP, Yu XH, Ye Q, Huang C, Zhang X, Chen LY, Xie W, Zhang M, Li L, Xia XD, Ouyang XP, Tan YL, Wang ZB, Tian GP, Zheng XL, Yin WD, Tang CK. MicroRNA-182 promotes lipoprotein lipase expression and atherogenesis by targeting histone deacetylase 9 in apolipoprotein E-knockout mice. Circ J 2017;82:28–38. [DOI] [PubMed] [Google Scholar]
- 161. Zhang XF, Yang Y, Yang XY, Tong Q. MiR-188-3p upregulation results in the inhibition of macrophage proinflammatory activities and atherosclerosis in ApoE-deficient mice. Thromb Res 2018;171:55–61. [DOI] [PubMed] [Google Scholar]
- 162. Shan Z, Qin SS, Li W, Wu WB, Yang J, Chu MP, Li XK, Huo YQ, Schaer GL, Wang SM, Zhang CX. An endocrine genetic signal between blood cells and vascular smooth muscle cells role of MicroRNA-223 in smooth muscle function and atherogenesis. J Am Coll Cardiol 2015;65:2526–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Meiler S, Baumer Y, Toulmin E, Seng K, Boisvert WA. MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arterioscler Thromb Vasc Biol 2015;35:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Chen C, Wang Y, Yang SL, Li HP, Zhao G, Wang F, Yang L, Wang DW. MiR-320a contributes to atherogenesis by augmenting multiple risk factors and down-regulating SRF. J Cell Mol Med 2015;19:970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. He PP, OuYang XP, Li Y, Lv YC, Wang ZB, Yao F, Xie W, Tan YL, Li L, Zhang M, Lan G, Gong D, Cheng HP, Zhong HJ, Liu D, Huang C, Li ZX, Zheng XL, Yin WD, Tang CK. MicroRNA-590 inhibits lipoprotein lipase expression and prevents atherosclerosis in apoE knockout mice. Plos One 2015;10: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Son DJ, Kumar S, Takabe W, Woo Kim C, Ni C-W, Alberts-Grill N, Jang I-H, Kim S, Kim W, Won Kang S, Baker AH, Woong Seo J, Ferrara KW, Jo H. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat Commun 2013;4:3000.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. PNAS 2010;107:13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Li S, Ren J, Xu N, Zhang J, Geng Q, Cao C, Lee C, Song J, Li J, Chen H. MicroRNA-19b functions as potential anti-thrombotic protector in patients with unstable angina by targeting tissue factor. J Mol Cell Cardiol 2014;75:49–57. [DOI] [PubMed] [Google Scholar]
- 169. Fan W, Fang R, Wu X, Liu J, Feng M, Dai G, Chen G, Wu G. Shear-sensitive microRNA-34a modulates flow-dependent regulation of endothelial inflammation. J Cell Sci 2015;128:70–80. [DOI] [PubMed] [Google Scholar]
- 170. Han H, Qu GJ, Han CH, Wang YH, Sun TT, Li FQ, Wang JX, Luo SS. MiR-34a, miR-21 and miR-23a as potential biomarkers for coronary artery disease: a pilot microarray study and confirmation in a 32 patient cohort. Exp Mol Med 2015;47:e138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Li Y, Zhang K, Mao W. Inhibition of miR34a prevents endothelial cell apoptosis by directly targeting HDAC1 in the setting of atherosclerosis. Mol Med Rep 2018;17:4645–4650. [DOI] [PubMed] [Google Scholar]
- 172. Badi I, Mancinelli L, Polizzotto A, Ferri D, Zeni F, Burba I, Milano G, Brambilla F, Saccu C, Bianchi ME, Pompilio G, Capogrossi MC, Raucci A. miR-34a promotes vascular smooth muscle cell calcification by downregulating SIRT1 (Sirtuin 1) and Axl (AXL Receptor Tyrosine Kinase). Arterioscler Thromb Vasc Biol 2018;38:2079–2090. [DOI] [PubMed] [Google Scholar]
- 173. Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, Sluijter JPG, Hoefer I, Pasterkamp G, Bode C, Moser M. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 2011;123:999–1009. [DOI] [PubMed] [Google Scholar]
- 174. Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A 2008; 105:1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Fish JE, Santoro MM, Morton SU, Yu S, Yeh R-F, Wythe JD, Ivey KN, Bruneau BG, Stainier DYR, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 2008;15:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Parahuleva MS, Lipps C, Parviz B, Holschermann H, Schieffer B, Schulz R, Euler G. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci Rep 2018;8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. van Thienen JV, Fledderus JO, Dekker RJ, Rohlena J, van Ijzendoorn GA, Kootstra NA, Pannekoek H, Horrevoets A. Shear stress sustains atheroprotective endothelial KLF2 expression more potently than statins through mRNA stabilization. Cardiovasc Res 2006;72:231–240. [DOI] [PubMed] [Google Scholar]
- 178. Casas-Agustench P, Fernandes FS, Tavares do Carmo MG, Visioli F, Herrera E, Dávalos A. Consumption of distinct dietary lipids during early pregnancy differentially modulates the expression of microRNAs in mothers and offspring. PLoS One 2015;10:e0117858.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Chen LY, Xia XD, Zhao ZW, Gong D, Ma XF, Yu XH, Zhang Q, Wang SQ, Dai XY, Zheng XL, Zhang DW, Yin WD, Tang CK. MicroRNA-377 inhibits atherosclerosis by regulating triglyceride metabolism through the DNA methyltransferase 1 in apolipoprotein E-knockout mice. Circ J 2018;82:2861.. [DOI] [PubMed] [Google Scholar]
- 180. Ding Z, Wang X, Khaidakov M, Liu S, Mehta JL. MicroRNA hsa-let-7g targets lectin-like oxidized low-density lipoprotein receptor-1 expression and inhibits apoptosis in human smooth muscle cells. Exp Biol Med (Maywood) 2012;237:1093–1100. [DOI] [PubMed] [Google Scholar]
- 181. Wang D, Deuse T, Stubbendorff M, Chernogubova E, Erben RG, Eken SM, Jin H, Li Y, Busch A, Heeger C-H, Behnisch B, Reichenspurner H, Robbins RC, Spin JM, Tsao PS, Schrepfer S, Maegdefessel L. Local MicroRNA modulation using a novel Anti-miR-21–eluting stent effectively prevents experimental in-stent restenosis. Arterioscler Thromb Vasc Biol 2015;35:1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. McDonald RA, Halliday CA, Miller AM, Diver LA, Dakin RS, Montgomery J, McBride MW, Kennedy S, McClure JD, Robertson KE, Douglas G, Channon KM, Oldroyd KG, Baker AH. Reducing in-stent restenosis: therapeutic manipulation of miRNA in vascular remodeling and inflammation. J Am Coll Cardiol 2015;65:2314–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. McDonald RA, White KM, Wu J, Cooley BC, Robertson KE, Halliday CA, McClure JD, Francis S, Lu R, Kennedy S, George SJ, Wan S, van Rooij E, Baker AH. miRNA-21 is dysregulated in response to vein grafting in multiple models and genetic ablation in mice attenuates neointima formation. Eur Heart J 2013;34:1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Jin H, Li DY, Chernogubova E, Sun CY, Busch A, Eken SM, Saliba-Gustafsson P, Winter H, Winski G, Raaz U, Schellinger IN, Simon N, Hegenloh R, Matic LP, Jagodic M, Ehrenborg E, Pelisek J, Eckstein HH, Hedin U, Backlund A, Maegdefessel L. Local delivery of miR-21 stabilizes fibrous caps in vulnerable atherosclerotic lesions. Mol Ther 2018;26:1040–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Das A, Ganesh K, Khanna S, Sen CK, Roy S. Engulfment of apoptotic cells by macrophages: a role of MicroRNA-21 in the resolution of wound inflammation. J Immunol 2014;192:1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. de Ronde MWJ, Kok MGM, Moerland PD, Van den Bossche J, Neele AE, Halliani A, van der Made I, de Winther MPJ, Meijers JCM, Creemers EE, Pinto-Sietsma S-J. High miR-124-3p expression identifies smoking individuals susceptible to atherosclerosis. Atherosclerosis 2017;263:377–384. [DOI] [PubMed] [Google Scholar]
- 187. Li C, Fang Z, Jiang T, Zhang Q, Liu C, Zhang C, Xiang Y. Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med Genomics 2013;6:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Schulte C, Molz S, Appelbaum S, Karakas M, Ojeda F, Lau DM, Hartmann T, Lackner KJ, Westermann D, Schnabel RB, Blankenberg S, Zeller T. miRNA-197 and miRNA-223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS One 2015;10:e0145930.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Burke AP, Kolodgie FD, Farb A, Weber DK, Malcom GT, Smialek J, Virmani R. Healed plaque ruptures and sudden coronary death—evidence that subclinical rupture has a role in plaque progression. Circulation 2001;103:934–940. [DOI] [PubMed] [Google Scholar]
- 190. Lan G, Xie W, Li L, Zhang M, Liu D, Tan Y-L, Cheng H-P, Gong D, Huang C, Zheng X-L, Yin W-D, Tang C-K. MicroRNA-134 actives lipoprotein lipase-mediated lipid accumulation and inflammatory response by targeting angiopoietin-like 4 in THP-1 macrophages. Biochem Biophys Res Commun 2016;472:410–417. [DOI] [PubMed] [Google Scholar]
- 191. Bao M-H, Xiao Y, Zhang Q-S, Luo H-Q, Luo J, Zhao J, Li G-Y, Zeng J, Li J-M. Meta-analysis of miR-146a polymorphisms association with coronary artery diseases and ischemic stroke. IJMS 2015;16:14305–14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI, Fish JE. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med 2013;5:1017–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Arroyo AB, Reyes-García A, Rivera-Caravaca JM, Valledor P, García-Barberá N, Roldán V, Vicente V, Martínez C, González-Conejero R. MiR-146a regulates neutrophil extracellular trap formation that predicts adverse cardiovascular events in patients with atrial fibrillation. Arterioscler Thromb Vasc Biol 2018;38:892–902. [DOI] [PubMed] [Google Scholar]
- 194. Sluijter JPG, Pasterkamp G. MicroRNAs: the Swing voters in vascular disease waiting for a program. Circ Res 2017;120:5–7. [DOI] [PubMed] [Google Scholar]
- 195. Taurino C, Miller William H, McBride Martin W, McClure John D, Khanin R, Moreno María U, Dymott Jane A, Delles C, Dominiczak Anna F. Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci 2010;119:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Wang K, Liu C-Y, Zhou L-Y, Wang J-X, Wang M, Zhao B, Zhao W-K, Xu S-J, Fan L-H, Zhang X-J, Feng C, Wang C-Q, Zhao Y-F, Li P-F. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015;6:6779.. [DOI] [PubMed] [Google Scholar]
- 197. He P-P, Ouyang X-P, Tang Y-Y, Liao L, Wang Z-B, Lv Y-C, Tian G-P, Zhao G-J, Huang L, Yao F, Xie W, Tang YL, Chen W-J, Zhang M, Li Y, Wu J-F, Peng J, Liu X-Y, Zheng X-L, Yin W-D, Tang C-K. MicroRNA-590 attenuates lipid accumulation and pro-inflammatory cytokine secretion by targeting lipoprotein lipase gene in human THP-1 macrophages. Biochimie 2014;106:81–90. [DOI] [PubMed] [Google Scholar]
- 198. Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol 2011;22:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87–98. [DOI] [PubMed] [Google Scholar]
- 200. Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepää AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 2009;23:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res 2012;56:1665–1674. [DOI] [PubMed] [Google Scholar]
- 202. Kida K, Nakajima M, Mohri T, Oda Y, Takagi S, Fukami T, Yokoi T. PPARα is regulated by miR-21 and miR-27b in human liver. Pharm Res 2011;28:2467–2476. [DOI] [PubMed] [Google Scholar]
- 203. Jennewein C, von Knethen A, Schmid T, Brüne B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J Biol Chem 2010;285:11846–11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Wang T, Li M, Guan J, Li P, Wang H, Guo Y, Shuai S, Li X. MicroRNAs miR-27a and miR-143 regulate porcine adipocyte lipid metabolism. IJMS 2011;12:7950–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Karere GM, Glenn JP, Birnbaum S, Garcia R, VandeBerg JL, Cox LA. Identification of coordinately regulated microRNA-gene networks that differ in baboons discordant for LDL-cholesterol. PLoS One 2019;14:e0213494.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Hussain MM, Goldberg IJ. Human MicroRNA-33b promotes atherosclerosis in Apoe-/- mice. Arterioscler Thromb Vasc Biol 2018;38:2272–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Cheng Y, Liu X, Yang J, Lin Y, Xu D-Z, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res 2009;105:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203.. [DOI] [PubMed] [Google Scholar]