Abstract
Rheumatoid arthritis (RA) is a common systemic, inflammatory and autoimmune disorder. MicroRNAs (miRs) are strongly associated with the initiation and progression of RA. However, the functions and mechanisms underlying miR-23 in RA are not completely understood. Therefore, the present study aimed to investigate the molecular mechanisms underlying miR-23 in RA. A bioinformatics tool (StarBase) and a wide range of experimental assays, including reverse transcription-quantitative PCR, western blotting, luciferase reporter assays and ELISAs, were performed to investigate the biological role of miR-23 in RA. The results indicated that miR-23 was downregulated and chemokine C-X-C motif ligand 12 (CXCL12) was upregulated in RA samples compared with healthy samples. Furthermore, miR-23 overexpression suppressed inflammation via reducing TNF-α, IL-1β and IL-8 expression levels compared with the NC mimic group. Regarding the underlying mechanism, compared with NC mimic, miR-23 mimic decreased CXCL12 mRNA expression by binding to its 3'-untranslated region. Additionally, CXCL12 overexpression reversed miR-23 mimic-mediated effects on inflammation. NF-κB signaling is associated with inflammation. Therefore, the present study indicated that CXCL12 promoted inflammation by activating NF-κB signaling. In conclusion, miR-23 inhibited inflammation to alleviate RA by regulating CXCL12 via the NF-κB signaling pathway, which may serve as a potential target for the diagnosis and treatment of RA.
Keywords: microRNA-23, rheumatoid arthritis, chemokine C-X-C motif ligand 12, NF-κB signaling
Introduction
Rheumatoid arthritis (RA), a common systemic autoimmune disease, is primarily characterized by chronic inflammation and cell infiltration in synovial tissues, which contribute to the destruction or loss of cartilage and bone (1-4). Although several risk factors, including genetic factors, viral infection and sex hormones, have been reported to be strongly associated with the occurrence of RA, the etiology of RA is not completely understood (5,6). During the progression of RA, various inflammatory cells are activated by a series of cytokines and chemokines or via cell-cell contact (7). Fibroblast-like synoviocytes (FLSs), a type of inflammatory cell, are highly enriched in the synovial membrane (8). Additionally, FLSs have been widely reported to serve a critical role in the initiation and development of RA (9). Moreover, FLSs secrete a number of proinflammatory cytokines, such as TNF-α, IL-1β and IL-6, resulting in aggravation of chronic inflammation (10). Therefore, further studies investigating the molecular mechanism underlying inflammation in RA are required.
MicroRNAs (miRNAs/miRs), a subgroup of non-coding RNAs, are short and single-stranded RNAs (11), which have been reported to exert an important impact on a broad spectrum of cellular processes, including cell proliferation, differentiation, inflammation and fibrosis in various diseases (12,13). For example, hematopoietic miR-126 inhibits fibrosis in renal ischemia/reperfusion injury via regulating C-X-C motif chemokine receptor 4(14). In addition, it has been reported that miR-21 genetic ablation promotes fibrosis and inflammation to aggravate cardiac remodeling and dysfunction in hypertension (15). Furthermore, the methylation of miR-495 promoter activates NOD-like receptor protein 3 inflammasome to facilitate acute lung injury progression via promoting inflammation (16). It has been reported that miR-23 overexpression induces trophoblast cell apoptosis by binding to X-linked inhibitor of apoptosis (17). In addition, miR-23 promotes myelination in the central nervous system (18). Moreover, it has been demonstrated that miR-23 is downregulated in patients with RA who are treated with anti-TNF-α, indicating that miR-23 may serve as a potential biomarker for RA therapy (19). However, the biological functions and regulatory network of miR-23 in RA are not completely understood.
Furthermore, the regulation of inflammation in diverse diseases via chemokine C-X-C motif ligand 12 (CXCL12) has been widely reported (20,21). For example, CXCL12 promotes α-synuclein-triggered neuroinflammation in Parkinson's disease (22). Additionally, tissue-infiltrating inflammatory cells highly express CXCL12, serving a significant role in chronic periaortitis (23). A previous study demonstrated that CXCL12 downregulation in synovial fibroblasts via DNA demethylation inhibited RA development (24). Nevertheless, the mechanism underlying the miR-23/CXCR12 axis in RA is not completely understood.
The NF-κB signaling pathway is involved in the regulation of inflammation (25-27). However, the association between miR-23 or CXCL12 and NF-κB signaling requires further investigation.
In conclusion, the present study investigated the function and mechanism underlying miR-23 in RA, with the aim to develop a novel diagnostic and therapeutic strategy for the treatment of RA.
Materials and methods
Samples of synovial tissue and serum
Synovial (n=22) and healthy (n=22) tissue samples were obtained from patients with RA (10 males and 12 females; average age, 51 years old) and healthy volunteers (9 males and 13 females; average age, 47 years old) at The Second Changzhou People's Hospital Affiliated to Nanjing Medical University from March 2016 to April 2018. Following surgery, samples were immediately stored at -80˚C. In addition, blood samples were collected from patients with RA (n=22) and healthy volunteers (n=22). Written informed consent was obtained from patients and volunteers prior to surgery. The present study was approved by the Ethics Committee of The Second Changzhou People's Hospital Affiliated to Nanjing Medical University.
FLSs
FLSs were isolated from synovial tissues. Briefly, the synovial tissues were sectioned and digested with 0.25% trypsin to isolate the synoviocytes at 37˚C for 30 min. Following culturing overnight in DMEM supplemented with 10% FBS as well as penicillin (100 IU/ml) and streptomycin (100 µg/ml) at 37˚C, the non-adherent cells were removed and the adherent cells were cultured in DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS and 1% mixture of penicillin and streptomycin at 37˚C in a humidified atmosphere containing 5% CO2. Cells were collected and used for subsequent assays at passage 4-8. FLSs were stimulated with 10 µg/ml lipopolysaccaride (LPS; Sigma-Aldrich; Merck KGaA) for 24 h at 37˚C to establish the RA model.
Cell transfection
miR-23 mimic (5'-AAACCGUUAGGGGUUC-3'), miR-23 inhibitor (5'-GCUGUCAUUCGUUAUC-3') and their corresponding negative controls (NCs; NC mimic; 5'-AAGUGGCAACGAACCG-3'; NC inhibitor; 5'-AUGCACUUAGUAAUGA-3') were synthesized by Shanghai GenePharma Co., Ltd. Small interfering (si)RNAs targeting CXCL12 (siCXCL12; 5'-CGAGGGCGAGCAUGCGUGUUGAUUG-3') and inhibitor κBα (IκBα; siIκBα; 5'-GCUAGGUAAAUCGGUUGGGUCGUGA-3') were obtained from Shanghai GenePharma Co., Ltd., were used to knockdown CXCL12 and IκBα, respectively, and siNC (5'-UCAAGUCCAGCACGACAUUG-3') was used as the negative control. The full length CXCL12 gene was inserted into the pcDNA3.1 vector (Shanghai GenePharma Co., Ltd.) to overexpress CXCL12, whereas the empty pcDNA3.1 vector served as a control. FLSs (5x104) were transfected with 50 nM vectors using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h prior to subsequent assays.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cultured FLSs, serum or synovial tissues using an Eastep Super Total RNA Extraction kit (Promega Corporation). Total RNA was reverse transcribed into cDNA using the Perfect Real Time RT Reagent kit (Takara Bio, Inc.) at 37˚C for 15 min for the measurement of CXCL12 and IκBα expression levels according to the manufacturer's instructions. Additionally, total RNA was reverse transcribed into cDNA using the TransScript Green miRNA RT SuperMix (Beijing Transgen Biotech Co., Ltd.) for the measurement of miR-23 expression levels. Subsequently, qPCR was performed using the SYBR-Green PCR Master mix kit or the TaqMan miRNA assay kit on a 7900HT Fast Real-Time system (all from Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation at 95˚C for 3 min, followed by 40 cycles of denaturation at 95˚C for 30 sec, annealing at 60˚C for 30 sec and extension at 72˚C for 20 sec, followed by a final extension at 72˚C for 5 min. miRNA and mRNA expression levels were quantified using the 2-ΔΔCq method (28) and normalized to the internal reference genes U6 and GAPDH, respectively. Primer sequences used for RT-qPCR analysis were as follows: miR-23 forward, 5'-CCTACTGTCGTCCCAAGACCT-3' and reverse, 5'-GGGGCTCGTGCAGAAGAAT-3'; and CXCL12 forward, 5'-TCCAAACTGTGCCCTTCA-3' and reverse, 5'-CTCTTCTTCTGTCGCTTCTT-3'; and IκBα forward, 5'-TGGCCAGTGTAGCAGTCTTG-3' and reverse, 5'-GACATCAGCACCCAAAGTCA-3'; and GAPDH forward, 5'-GCTGGCGCTGAGTACGTCGT-3' and reverse, 5'-ACGTTGGCAGTGGGGACACG-3'; and U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse 5'-AACGCTTCACGAATTTGCGT-3'.
Western blotting
Total protein was extracted from FLSs using RIPA buffer (Beyotime Institute of Biotechnology) and protein concentrations were measured using the BCA protein assay kit (Beyotime Institute of Biotechnology). Equal quantities (15 µg/lane) of protein were separated via 10% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore). Following blocking with 5% skimmed milk for 2 h at room temperature, membranes were incubated overnight at 4˚C with the following primary antibodies: Anti-phosphorylated (p)-p65 (1:1,000; cat. no. 3033; Cell Signaling Technology, Inc.), anti-IκBα (1:1,000; cat. no. 4812; Cell Signaling Technology, Inc.) and anti-GAPDH (1:1,000; cat. no. 5174; Cell Signaling Technology, Inc.). Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit (1:3,000; cat. no. GB23303; Wuhan Servicebio Technology Co., Ltd.) secondary antibody in the dark at room temperature for 1 h. Protein bands were visualized using an ECL reagent (EMD Millipore). GAPDH was used as the loading control. Protein expression was quantified using Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Luciferase reporter assay
StarBase V2.0 (http://starbase.sysu.edu.cn/index.php) was used to validate the putative bindings between miR-23 and CXCL12 3'-untranslated region (3'-UTR). The wild-type (WT) or mutant (Mut) binding sequences of CXCL12 3'-UTR were cloned into the pmirGLO vector (Promega Corporation). 293T cells (ATCC; 5x104) were co-transfected with 50 nM miR-23 mimic, miR-23 inhibitor, NC mimic or NC inhibitor and CXCL12-WT or CXCL12-Mut using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to manufacturer's instructions. At 48 h post-transfection, luciferase activities were detected using the Dual-Luciferase Reporter assay system (Promega Corporation) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla (Promega Corporation).
ELISA
Cell culture media from cultured FLSs was harvested and stored at -80˚C. Subsequently, The supernatants were collected by centrifugation at 1,000 x g for 5 min at 4˚C, and the secretory levels of TNF-α, IL-1β and IL-8 in the supernatants of cultured FLSs were measured using a Duoset ELISA kit (cat. no. dy206; R&D Systems, Inc.) according to the manufacturer's instructions.
Statistical analysis
Data are presented as the mean ± standard deviation of three independent experimental repeats. Statistical analyses were performed using SPSS 17.0 (SPSS, Inc.) and GraphPad Prism (version 5; GraphPad Software, Inc.) software. Comparisons between two groups were analyzed using the paired or unpaired Student's t-test. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-23 is downregulated and CXCL12 is upregulated in RA
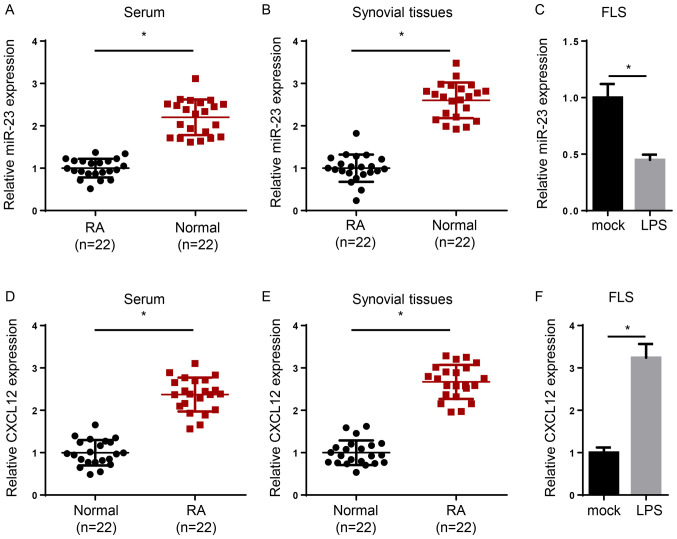
To determine the expression levels of miR-23 and CXCL12 in RA, serum and synovial tissues were collected from patients with RA and healthy volunteers, and RT-qPCR was performed. The expression levels of miR-23 were significantly reduced in the serum and synovial tissues of patients with RA compared with healthy samples (Fig. 1A and B). In addition, induction of FLSs with lipopolysaccharide (LPS) significantly decreased miR-23 expression levels compared with the mock group (Fig. 1C). By contrast, CXCL12 expression levels were significantly increased in the serum or synovial tissues of patients with RA compared with healthy controls (Fig. 1D and E). Moreover, CXCL12 expression levels were significantly increased in LPS-treated FLSs compared with the mock group (Fig. 1F). Overall, the results indicated that miR-23 was downregulated and CXCL12 was upregulated in RA.
Figure 1.
miR-23 is downregulated and CXCL12 is upregulated in RA. miR-23 expression levels in (A) serum, (B) synovial tissues and (C) FLS cells. CXCL12 expression levels in (D) serum, (E) synovial tissues and (F) FLS cells. *P<0.05. miR, microRNA; CXCL12, chemokine C-X-C motif ligand 12; RA, rheumatoid arthritis; LPS, lipopolysaccharide; FLS, fibroblast-like synoviocytes.
miR-23 suppresses inflammation and binds to the 3'-UTR of CXCL12
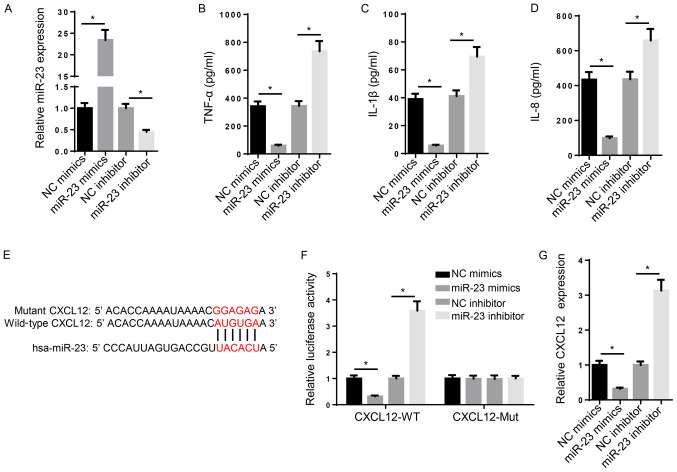
Several miRNAs have been associated with the inhibition of RA progression (29). In addition, miR-23 is upregulated following anti-TNF-α/disease-modifying antirheumatic drugs combination therapy, thus suggesting that miR-23 may serve as a potential biomarker for RA treatment (19). However, the biological functions and molecular mechanism underlying miR-23 in RA are not completely understood. In the present study, a series of experiments were designed to investigate the role of miR-23 in RA. Compared with NC mimic and NC inhibitor, miR-23 mimic and miR-23 inhibitor significantly increased and decreased the expression levels of miR-23, respectively (Fig. 2A). Subsequently, the effect of miR-23 on inflammation was investigated. TNF-α, IL-1β and IL-8 are typical markers of inflammation (30). The ELISA results suggested that the levels of TNF-α, IL-1β and IL-8 were significantly increased in miR-23 inhibitor-transfected FLSs compared with the NC inhibitor group, whereas miR-23 mimic significantly decreased the secretory levels of inflammatory markers compared with the NC mimic group (Fig. 2B-D). Furthermore, the mechanism underlying the effects of miR-23 on RA progression was investigated. Increasing evidence has suggested that miRNAs regulate mRNA expression at the post-transcriptional level via interacting with the 3'-UTR of the target mRNA (31). The binding sequence between miR-23 and CXCL12 3'-UTR (3280-3287 bp) was predicted using StarBase (http://starbase.sysu.edu.cn/) (Fig. 2E). Furthermore, the luciferase reporter assay indicated that miR-23 mimic significantly decreased the luciferase activity of CXCL12-WT compared with NC mimic, whereas miR-23 inhibitor displayed the opposite effect compared with NC inhibitor. By contrast, the luciferase activity of CXCL12-Mut was not significantly altered by miR-23 mimic or inhibitor compared with NC mimic and inhibitor, respectively (Fig. 2F). Similarly, the RT-qPCR results suggested that miR-23 mimic and miR-23 inhibitor significantly decreased and increased CXCL12 mRNA expression levels compared with NC mimic and NC inhibitor, respectively (Fig. 2G). Collectively, the aforementioned results suggested that miR-23 suppressed inflammation and bound to the 3'-UTR of CXCL12.
Figure 2.
miR-23 suppresses inflammation and binds to the 3'-UTR of CXCL12. (A) Transfection efficacy of miR-23 mimic and miR-23 inhibitor. ELISAs were performed to assess the effects of miR-23 overexpression and knockdown on the secretory levels of (B) TNF-α, (C) IL-1β and (D) IL-8. (E) The binding sequences between miR-23 and CXCL12 were predicted using StarBase. (F) The luciferase reporter assay was performed to verify the interaction between miR-23 and CXCL12. (G) CXCL12 expression levels were negatively regulated by miR-23 in FLS cells. *P<0.05. miR, microRNA; UTR, untranslated region; CXCL12, chemokine C-X-C motif ligand 12; WT, wild-type; Mut, mutant; NC, negative control.
CXCL12 overexpression reverses the inhibitory effect of miR-23 on inflammatory cytokine expression
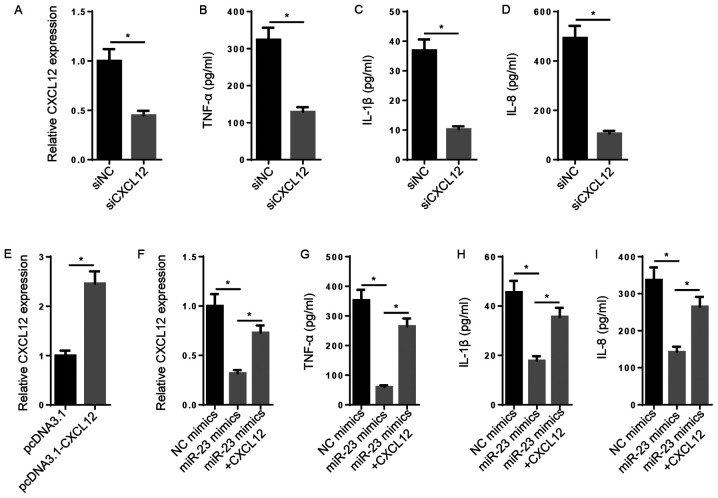
Subsequently, the biological function of CXCL12 in RA was explored. The results indicated that the levels of CXCL12, TNF-α, IL-1β and IL-8 were significantly decreased in the siCXCL12 group compared with the siNC group (Fig. 3A-D). CXCL12 expression levels were significantly increased in pcDNA3.1-CXCL12-transfected FLSs compared with pcDNA3.1-transfected FLSs (Fig. 3E). Furthermore, miR-23 mimic-mediated decreased levels of CXCL12 mRNA were significantly reversed following CXCL12 overexpression (Fig. 3F). Likewise, the inhibitory effect of miR-23 overexpression on TNF-α, IL-1β and IL-8 levels was significantly reversed by CXCL12 overexpression (Fig. 3G-I). The results demonstrated that miR-23 inhibited inflammation via regulating CXCL12 expression.
Figure 3.
CXCL12 overexpression reverses the inhibitory effect of miR-23 on inflammatory cytokine expression. (A) Transfection efficiency of siCXCL12. ELISAs were performed to assess the effects of CXCL12 knockdown on the secretory levels of (B) TNF-α, (C) IL-1β and (D) IL-8. (E) Transfection efficiency of pcDNA3.1-CXCL12. (F) The effects of miR-23 and CXCL12 co-overexpression on CXCL12 expression. ELISAs were performed to assess the effects of miR-23 and CXCL12 co-overexpression on the secretory levels of (G) TNF-α, (H) IL-1β and (I) IL-8. *P<0.05. CXCL12, chemokine C-X-C motif ligand 12; miR, microRNA; si, small interfering RNA; NC, negative control.
NF-κB signaling is crucial for miR-23-mediated regulation of inflammatory cytokine expression
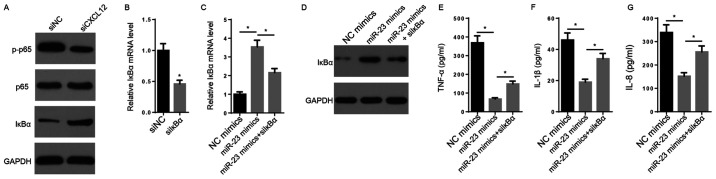
It has been reported that NF-κB signaling, an inflammation-associated signaling pathway, may be activated by CXCL12 via the CXCL12/CXCR7 axis (32). CXCL12 knockdown decreased the protein expression levels of p-p65 and increased the protein expression levels of IκBα compared with the siNC group (Fig. 4A). The transfection efficacy assay indicated that the expression of IκBα was significantly decreased in siIκBα-transfected FLSs compared with siNC-transfected FLSs (Fig. 4B). The RT-qPCR and western blotting results indicated that the mRNA and protein expression levels of IκBα were markedly increased by miR-23 mimic compared with NC mimic, but decreased following co-transfection with siIκBα (Fig. 4C and D). Furthermore, the suppressive effects of miR-23 mimic on TNF-α, IL-1β and IL-8 secretory levels were significantly reversed by co-transfection with siIκBα (Fig. 4E-G). In conclusion, the results suggested that CXCL12 promoted inflammation by activating NF-κB signaling.
Figure 4.
NF-κB signaling is crucial for miR-23-mediated regulation of inflammatory cytokine expression. (A) The effects of CXCL12 knockdown on the protein expression levels of p65, p-p65 and IκBα were measured via western blotting. (B) IκBα expression levels following transfection with siIκBα and siNC were measured via RT-qPCR. IκBα (C) mRNA and (D) protein expression levels were measured via RT-qPCR and western blotting, respectively. ELISAs were performed to measure the secretory levels of (E) TNF-α, (F) IL-1β and (G) IL-8. *P<0.05. miR, microRNA; CXCL12, chemokine C-X-C motif ligand 12; p, phosphorylated; IκBα, inhibitor κBα; si, small interfering RNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR.
Discussion
RA impacts the quality of life of patients (33). Modern advanced therapies, including surgical resection, medical treatment and other complementary therapies, have been applied for the treatment of RA (34). However, the treatment of RA is challenging due to inflammatory cell infiltration and bone destruction (35).
Emerging evidence has demonstrated that miRNA expression is dysregulated in FLSs or synovial tissues of patients with RA, which is potentially associated with the pathology of RA (36). Although miRNAs do not encode protein products, they may modulate gene expression at the post-transcriptional level via interacting with the 3'-UTR of target mRNAs (29,37). For instance, miR-338-5p promotes cell viability, proliferation and migration of RA-FLSs via targeting nuclear factor of activated T cells 5(38). In addition, miR-20a regulates the secretion of IL-6, IL-1β and TNF-α in FLSs via targeting apoptosis signal-regulating kinase 1(39). Furthermore, it has been previously demonstrated that miR-23 is downregulated in patients with RA who are treated with anti-TNF-α (19). Therefore, the present study further explored the function and mechanism underlying miR-23 in RA. The results suggested that miR-23 negatively regulated the expression of IL-8, IL-1β and TNF-α in FLSs, indicating that miR-23 served as an anti-inflammatory factor in FLSs. On the basis of the underlying mechanism, it was predicted that miR-23 directly targeted CXCL12 3'-UTR.
CXCL12 has been widely reported to regulate inflammatory cell infiltration. For example, high mobility group box 1 induces tissue damage by recruiting inflammatory cells via forming a complex with CXCL12(40). Furthermore, it has been demonstrated that CXCL12 accumulation results in an increase in matrix metalloproteinases, resulting in the perpetuation of RA (41). However, the present study focused on the effect of CXCL12 on inflammation. The results suggested that CXCL12 knockdown reduced inflammation, whereas CXCL12 overexpression reversed the suppressive effect of miR-23 mimic on inflammation.
NF-κB signaling is closely associated with inflammatory cell infiltration (42-44). On the basis of the underlying mechanism, phosphorylation of NF-κB (p65) and downregulation of IκBα may activate NF-κB signaling (45). Therefore, it was hypothesized that CXCL12 induces inflammation via NF-κB signaling. The present study indicated that CXCL12 knockdown increased IκBα and decreased p-p65 expression levels compared with siNC, resulting in inactivation of the NF-κB signaling pathway. Furthermore, rescue assays in FLSs indicated that IκBα knockdown significantly reversed miR-23 mimic-mediated downregulation of the inflammatory process. Overall, the results suggested that miR-23 was downregulated and CXCL12 was upregulated in RA.
Collectively, the present study suggested that miR-23 inhibited inflammation to alleviate RA by regulating CXCL12 via the NF-κB signaling pathway. The novel mechanism identified in the present study may provide a potential target for RA treatment. However, the present study had a number of limitations. Firstly, an animal model was not established, therefore the role of miR-23 in vivo requires further investigation. Secondly, although miR-23 may serve a role in cell proliferation, migration and apoptosis in RA, the present study only focused on inflammation. Finally, as RA pathology is complex, future studies should investigate further miR-23-mediated mechanisms.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by Changzhou Sci&Tech Program (grant no. CJ20200101).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BG and XC designed the present study. GS, YW, YG and LZ performed all the experiments, analyzed the data and prepared the figures. BG drafted the initial manuscript. XC reviewed and revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from patients and volunteers prior to surgery. The present study was approved by the Ethics Committee of The Second Changzhou People's Hospital Affiliated to Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Šenolt L. Rheumatoid arthritis. Vnitr Lek. 2018;64:98–106. (In Czech) [PubMed] [Google Scholar]
- 2.Wasserman A. Rheumatoid arthritis: Common questions about diagnosis and management. Am Fam Physician. 2018;97:455–462. [PubMed] [Google Scholar]
- 3.Katz P. Causes and consequences of fatigue in rheumatoid arthritis. Curr Opin Rheumatol. 2017;29:269–276. doi: 10.1097/BOR.0000000000000376. [DOI] [PubMed] [Google Scholar]
- 4.Baum R, Gravallese EM. Bone as a target organ in rheumatic disease: Impact on osteoclasts and osteoblasts. Clin Rev Allergy Immunol. 2016;51:1–15. doi: 10.1007/s12016-015-8515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burmester GR, Pope JE. Novel treatment strategies in rheumatoid arthritis. Lancet. 2017;389:2338–2348. doi: 10.1016/S0140-6736(17)31491-5. [DOI] [PubMed] [Google Scholar]
- 6.Mellado M, Martínez-Muñoz L, Cascio G, Lucas P, Pablos JL, Rodríguez-Frade JG. T cell migration in rheumatoid arthritis. Front Immunol. 2015;6(384) doi: 10.3389/fimmu.2015.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoxha M. A systematic review on the role of eicosanoid pathways in rheumatoid arthritis. Adv Med Sci. 2018;63:22–29. doi: 10.1016/j.advms.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Bustamante MF, Garcia-Carbonell R, Whisenant KD, Guma M. Fibroblast-like synoviocyte metabolism in the pathogenesis of rheumatoid arthritis. Arthritis Res Ther. 2017;19(110) doi: 10.1186/s13075-017-1303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doody KM, Bottini N, Firestein GS. Epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes. Epigenomics. 2017;9:479–492. doi: 10.2217/epi-2016-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Zhang T, Xu W, Ding J, Yin F, Xu J, Sun W, Wang H, Sun M, Cai Z, Hua Y. Sarcoma-targeting peptide-decorated polypeptide nanogel intracellularly delivers shikonin for upregulated osteosarcoma necroptosis and diminished pulmonary metastasis. Theranostics. 2018;8:1361–1375. doi: 10.7150/thno.18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: An overview. Methods Mol Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 13.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 14.Bijkerk R, van Solingen C, de Boer HC, van der Pol P, Khairoun M, de Bruin RG, van Oeveren-Rietdijk AM, Lievers E, Schlagwein N, van Gijlswijk DJ, et al. Hematopoietic microRNA-126 protects against renal ischemia/reperfusion injury by promoting vascular integrity. J Am Soc Nephrol. 2014;25:1710–1722. doi: 10.1681/ASN.2013060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Zhang C, Wang C, Sun J, Wang D, Zhao Y, Xu X. miR-210 promotes human osteosarcoma cell migration and invasion by targeting FGFRL1. Oncol Lett. 2018;16:2229–2236. doi: 10.3892/ol.2018.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying Y, Mao Y, Yao M. NLRP3 inflammasome activation by microRNA-495 promoter methylation may contribute to the progression of acute lung injury. Mol Ther Nucleic Acids. 2019;18:801–814. doi: 10.1016/j.omtn.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Hou A, Gao X, Zhang J, Zhang L, Wang J, Li H, Song Y. Lentivirus-mediated miR-23a overexpression induces trophoblast cell apoptosis through inhibiting X-linked inhibitor of apoptosis. Biomed Pharmacother. 2017;94:412–417. doi: 10.1016/j.biopha.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 18.Lin ST, Huang Y, Zhang L, Heng MY, Ptácek LJ, Fu YH. MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci USA. 2013;110:17468–17473. doi: 10.1073/pnas.1317182110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castro-Villegas C, Pérez-Sánchez C, Escudero A, Filipescu I, Verdu M, Ruiz-Limón P, Aguirre MA, Jiménez-Gomez Y, Font P, Rodriguez-Ariza A, et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFα. Arthritis Res Ther. 2015;17(49) doi: 10.1186/s13075-015-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssens R, Struyf S, Proost P. The unique structural and functional features of CXCL12. Cell Mol Immunol. 2018;15:299–311. doi: 10.1038/cmi.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Wang M, Guo YY, Sun T, Li YJ, Yang Q, Zhang K, Liu SB, Zhao MG, Wu YM. Systemic inflammation induces anxiety disorder through CXCL12/CXCR4 pathway. Brain Behav Immun. 2016;56:352–362. doi: 10.1016/j.bbi.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Wang H, Hu Y, Zhang YS, Wen L, Yin F, Wang Z, Zhang Y, Li S, Miao Y, et al. Inhibition of CaMKIIα activity enhances antitumor effect of fullerene C60 nanocrystals by suppression of autophagic degradation. Adv Sci (Weinh) 2019;6(1801233) doi: 10.1002/advs.201801233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicastro M, Vescovini R, Maritati F, Palmisano A, Urban ML, Incerti M, Fenaroli P, Peyronel F, Benigno GD, Mangieri D, et al. Fibrocytes in chronic periaortitis: A novel mechanism linking inflammation and fibrosis. Arthritis Rheumatol. 2019;71:1913–1922. doi: 10.1002/art.41024. [DOI] [PubMed] [Google Scholar]
- 24.Karouzakis E, Rengel Y, Jüngel A, Kolling C, Gay RE, Michel BA, Tak PP, Gay S, Neidhart M, Ospelt C. DNA methylation regulates the expression of CXCL12 in rheumatoid arthritis synovial fibroblasts. Genes Immun. 2011;12:643–652. doi: 10.1038/gene.2011.45. [DOI] [PubMed] [Google Scholar]
- 25.Sun SC. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afonina IS, Zhong Z, Karin M, Beyaert R. Limiting inflammation-the negative regulation of NF-κB and the NLRP3 inflammasome. Nat Immunol. 2017;18:861–869. doi: 10.1038/ni.3772. [DOI] [PubMed] [Google Scholar]
- 27.Novack DV. Role of NF-κB in the skeleton. Cell Res. 2011;21:169–182. doi: 10.1038/cr.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Moran-Moguel MC, Petarra-Del Rio S, Mayorquin-Galvan EE, Zavala-Cerna MG. Rheumatoid arthritis and miRNAs: A critical review through a functional view. J Immunol Res. 2018;2018(2474529) doi: 10.1155/2018/2474529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegemann N, Wondimu A, Ullrich K, Schmidt MF. Synovial MMP-3 and TIMP-1 levels and their correlation with cytokine expression in canine rheumatoid arthritis. Vet Immunol Immunopathol. 2003;91:199–204. doi: 10.1016/s0165-2427(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018;22:5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao YX, Zhou CH, Zeng H, Zuo DQ, Wang ZY, Yin F, Hua YQ, Cai ZD. The role of the CXCL12-CXCR4/CXCR7 axis in the progression and metastasis of bone sarcomas (review) Int J Mol Med. 2013;32:1239–1246. doi: 10.3892/ijmm.2013.1521. [DOI] [PubMed] [Google Scholar]
- 33.Coskun Benlidayi I. Sleep impairment: An obstacle to achieve optimal quality of life in rheumatoid arthritis. Rheumatol Int. 2018;38:2183–2192. doi: 10.1007/s00296-018-4155-5. [DOI] [PubMed] [Google Scholar]
- 34.Wasserman AM. Diagnosis and management of rheumatoid arthritis. Am Fam Physician. 2011;84:1245–1252. [PubMed] [Google Scholar]
- 35.2018 Chinese guideline for the diagnosis and treatment of rheumatoid arthritis. Zhonghua Nei Ke Za Zhi. 2018;57:242–251. doi: 10.3760/cma.j.issn.0578-1426.2018.04.004. Chinese Rheumatology Association. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 36.Salehi E, Eftekhari R, Oraei M, Gharib A, Bidad K. MicroRNAs in rheumatoid arthritis. Clin Rheumatol. 2015;34:615–628. doi: 10.1007/s10067-015-2898-x. [DOI] [PubMed] [Google Scholar]
- 37.Song YJ, Li G, He JH, Guo Y, Yang L. Bioinformatics-based identification of microRNA-regulated and rheumatoid arthritis-associated genes. PLoS One. 2015;10(e0137551) doi: 10.1371/journal.pone.0137551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo T, Ding H, Jiang H, Bao N, Zhou L, Zhao J. miR-338-5p regulates the viability, proliferation, apoptosis and migration of rheumatoid arthritis fibroblast-like synoviocytes by targeting NFAT5. Cell Physiol Biochem. 2018;49:899–910. doi: 10.1159/000493222. [DOI] [PubMed] [Google Scholar]
- 39.Philippe L, Alsaleh G, Pichot A, Ostermann E, Zuber G, Frisch B, Sibilia J, Pfeffer S, Bahram S, Wachsmann D, Georgel P. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann Rheum Dis. 2013;72:1071–1079. doi: 10.1136/annrheumdis-2012-201654. [DOI] [PubMed] [Google Scholar]
- 40.Cecchinato V, D'Agostino G, Raeli L, Nerviani A, Schiraldi M, Danelon G, Manzo A, Thelen M, Ciurea A, Bianchi ME, et al. Redox-mediated mechanisms fuel monocyte responses to CXCL12/HMGB1 in active rheumatoid arthritis. Front Immunol. 2018;9(2118) doi: 10.3389/fimmu.2018.02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Zheng Q, Xing X, Dong Y, Wang Y, You Y, Chen R, Hu C, Chen J, Gao D, et al. Matrix stiffness-upregulated LOXL2 promotes fibronectin production, MMP9 and CXCL12 expression and BMDCs recruitment to assist pre-metastatic niche formation. J Exp Clin Cancer Res. 2018;37(99) doi: 10.1186/s13046-018-0761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castejón ML, Rosillo MA, Montoya T, González-Benjumea A, Fernández-Bolaños JG, Alarcón-de-la-Lastra C. Oleuropein down-regulated IL-1β-induced inflammation and oxidative stress in human synovial fibroblast cell line SW982. Food Funct. 2017;8:1890–1898. doi: 10.1039/c7fo00210f. [DOI] [PubMed] [Google Scholar]
- 43.Zhang N, Liu Z, Luo H, Wu W, Nie K, Cai L, Tan S, Chen X, Huang Y, Liu J, et al. FM0807 decelerates experimental arthritis progression by inhibiting inflammatory responses and joint destruction via modulating NF-κB and MAPK pathways. Biosci Rep. 2019;39(BSR20182263) doi: 10.1042/BSR20182263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao Z, Xing L, Boyce BF. NF-κB p100 limits TNF-induced bone resorption in mice by a TRAF3-dependent mechanism. J Clin Invest. 2009;119:3024–3034. doi: 10.1172/JCI38716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noort AR, Tak PP, Tas SW. Non-canonical NF-κB signaling in rheumatoid arthritis: Dr Jekyll and Mr Hyde? Arthritis Res Ther. 2015;17(15) doi: 10.1186/s13075-015-0527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.