Abstract
Carotid angioplasty and stenting have developed into reliable options for patients with carotid stenosis. However, postoperative restenosis remains a serious and unresolved problem. Restenosis is partly caused by the proliferation of vascular smooth muscle cells. As certain long non-coding RNAs (lncRNAs) affect cell proliferation and migration, the present study aimed to investigate them as novel biomarkers for restenosis development and to further reveal the potential underlying mechanisms. The expression of lncRNA small nucleolar RNA host gene 1 (SNHG1) and microRNA145 (miR-145) in human carotid artery smooth muscle cells (hHCtASMCs) was analyzed using reverse transcription-quantitative PCR. In addition, a luciferase reporter assay was performed to investigate the interaction between SNHG1 and miR-145. The effects of the SNHG1/miR-145 axis on the proliferation and migration of hHCtASMCs were evaluated by Cell Counting Kit-8 and Transwell assays. Serum SNHG1 and miR-145 expression levels were increased and decreased, respectively, in patients with restenosis (all P<0.001). High SNHG1 and low miR-145 were identified as risk factors for restenosis onset (all P<0.01). Furthermore, decreasing SNHG1 expression levels in hHCtASMCs inhibited cell proliferation and migration. The luciferase reporter assay and expression results demonstrated that miR-145 may be a target of SNHG1 and mediated the effects of SNHG1 on hHCtASMC proliferation and migration. The results obtained suggested that abnormal expression of SNHG1 and miR-145 may be risk factors for restenosis. The present study revealed that the SNHG1/miR-145 axis regulates hHCtASMC proliferation and migration, indicating its potential for restenosis prevention and treatment.
Keywords: carotid stenosis, restenosis, small nucleolar RNA host gene 1, microRNA-145, vascular smooth muscle cells, proliferation, migration
Introduction
Carotid stenosis is the narrowing or contraction of the internal surface of the carotid artery (1). In patients with severe carotid stenosis, the incidence of cerebral ischemic events within 2 years is as high as 26% (2). Carotid stenosis is a major risk factor for stroke that leads to brain damage (3), and it is a common cause of ischaemic stroke and transient ischaemic attack (4). In >60% of cases of cerebral infarction, the cause is carotid stenosis, and severe cerebral infarction may lead to disability or even death of patients (5). Carotid stenosis seriously threatens the health of affected individuals. Over the past decade, carotid angioplasty and stenting (CAS) has been recognized as a safe and effective treatment for patients with carotid stenosis (6). However, restenosis remains a serious and unresolved problem following CAS treatment (7). Therefore, exploring the molecules related to the occurrence and development of restenosis is of great significance to prevent restenosis and improve the efficacy of CAS in the treatment of carotid stenosis. Vascular smooth muscle cells (VSMCs) are the major cell type in blood vessels. Unlike numerous other mature cell types in the adult body, VSMCs cannot terminally differentiate and therefore retain a significant plasticity (8). VSMC proliferation is an important feature of restenosis and one of the important pathological mechanisms of restenosis (9). Thus, exploring the regulatory mechanisms of VSMC proliferation and testing methods of interfering with VSMC proliferation are expected to provide novel ideas and targets for preventing restenosis.
Several studies have indicated that circulating microRNA (miRNA/miR)-145 is significantly associated with vascular restenosis after stent implantation (10,11). Targeted regulation of miR-145 by long-noncoding (lnc)RNA small nucleolar RNA host gene 1 (SNHG1) (12-14). As a potential marker of restenosis, miR-145 has the ability to inhibit VSMC proliferation and migration (15). Yang and Zi (16) reported that SNHG1 participates in the cerebrovascular pathological process of stroke. SNHG1 also has the function of regulating endothelial cell function (17). However, the regulatory effect of SNHG1 on VSMC function, an important mechanism in the pathological process of restenosis, has not been reported. Therefore, the aim of the present study was to analyze the expression level of SNHG1 in patients with restenosis after carotid artery stenosis stenting, and to explore the regulatory effect of SNHG1 on the proliferation and migration of VSMCs.
Materials and methods
Patient inclusion and sample collection
The present study included 20 patients with restenosis and 30 patients without restenosis underwent CAS for carotid stenosis at Cangzhou Central Hospital (Cangzhou, China) between February 2014 and October 2016. The degree of stenosis was determined using the criteria of the North American Symptomatic Carotid Endarterectomy trial (18). Patients were excluded if any of the following applied: i) Acute infection, cough, fever or diarrhea; ii) chronic inflammatory diseases; iii) malignant tumors; iv) severe kidney or liver diseases; v) poor adherence. Blood samples were collected from subjects after CAS, the blood samples were all centrifuged for serum isolation immediately after collection to avoid hemolysis in this experiment and stored at -80˚C for future use. The demographic and clinicopathological characteristics of the patients were recorded and listed in Table I, including age, sex, body mass index, smoking status, drinking status, diabetes, hypertension, hyperlipidemia, total cholesterol (TC), triglyceride, high-density lipoprotein cholesterol and low-density lipoprotein cholesterol (LDL-C). Written informed consent was obtained from patients or their relatives for the use of blood samples and clinical data. Experimental procedures were approved by the guidelines of the Ethics Committee of Cangzhou Central Hospital (Cangzhou, China; approval no. CZCH14h0280).
Table I.
Baseline characteristics of the study subjects.
| Variable | non-ISR (n=30) | ISR (n=20) | P-value |
|---|---|---|---|
| Age (years) | 64.5±7.2 | 67.5±8.5 | 0.199 |
| Male sex | 26 | 17 | 0.868 |
| BMI (kg/m2) | 24.7±2.5 | 25.1±3.1 | 0.640 |
| Smoking status | 12 | 9 | 0.726 |
| Alcohol consumption | 12 | 10 | 0.166 |
| Diabetes | 8 | 10 | 0.018 |
| Hypertension | 20 | 16 | 0.304 |
| Hyperlipidemia | 13 | 11 | 0.419 |
| TC (mg/dl) | 149.0±36.3 | 169.88±36.9 | 0.046 |
| TG (mg/dl) | 143.0±48.5 | 134.6±48.0 | 0.548 |
| HDL-C (mg/dl) | 40.1±6.7 | 41.7±7.1 | 0.433 |
| LDL-C (mg/dl) | 96.6±19.9 | 84.2±18.5 | 0.031 |
BMI, body mass index; TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ISR, in-stent restenosis.
Cell culture and transfection
The human carotid artery smooth muscle cell (hHCtASMC) line was purchased from the Cell Applications, Inc. and was cultured with Medium 231 and Smooth Muscle Growth Supplement (Thermo Fisher Scientific, Inc.) in an incubator containing air with 5% CO2 at 37˚C.
Cell transfection was used to achieve the in vitro regulation of SNHG1 and miR-145 in the present study. Short interfering (si)RNA targeting SNHG1 (si-SNHG1), negative control siRNA (si-NC), mimics NC, miR-145 mimics, inhibitor NC and miR-145 inhibitor were synthesized from GenePharma and were transfected into hHCtASMCs using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers' protocol . Cells treated with only transfection reagen ts were set as a mock group. After 48 h of transfection, the cells were used for further analyses. The transfected sequences were as follows (from 5' to 3'): si-SNHG1, 5'-CUUAAAGUGUUAGCAGACATT-3'; si-NC, 5'-UUCUCCGAACGUGUCACGUTT-3'; miR-145 mimic, 5'-GUCCAGUUUUCCCAGGAAUCCCU-3'; miR-145 inhibitor, 5'-AGGGAUUCCUGGGAAAACUGGAC-3'; mimics NC, 5'-UUCUCCGAACGUGUCACGU-3'; inhibitor NC, 5'-CAGUACUUUUGUGUAGUACAA-3'.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was obtained from fresh serum samples and cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the RNA was reversely transcribed into single-stranded complementary DNA with a PrimeScript reverse transcriptase reagent kit (Takara Bio, Inc.) following the manufacturer's protocol. The expression of SNHG1 and miR-145 was measured by real-time qPCR, which was performed using a SYBR-Green I Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.) on a 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used: Initial denaturation at 95˚C for 10 min; followed by 40 cycles of 95˚C for 20 sec, 58˚C for 15 sec and 72˚C for 20 sec. U6 was used as an endogenous control for miR-145 and GAPDH was used as an endogenous control for SNHG1. The oligonucleotide primer sequences were as follows: SNHG1 forward, 5'-CCCCATGATGGTTCCTCAGTT-3' and reverse, 5'-GGAAAGCAAGTGCAGGTTAGTC-3'; GAPDH forward, 5'-TGCACCACCAACTGCTTAGC-3' and reverse, 5'-GGCATGCACTGTGGTCATGAG-3'; miR-145 forward, 5'-GCCGAGGUCCAGUUUUCCCC-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'. The final expression value was calculated using the 2-ΔΔCq method (19).
Cell proliferation analysis
A Cell Counting Kit 8 (CCK-8) assay was used to analyze the cell proliferation. Cells were seeded into a 96-well plate at 5x103 cells/well and incubated for 0, 24, 48 or 72 h. Subsequently, as per the manufacturer's protocol, 10 µl CCK-8 reagent (Beyotime Institute of Biotechnology) was added to each well, followed by further incubation for 2 h. The optical density at 450 nm was determined using a microplate reader.
Transwell assay
Transwell chambers (Corning, Inc.) 8.0 µm pore size filter membranes (Corning, Inc.) without Matrigel coating (for migration assay) were used to analyze the migration ability of hHCtASMCs. Cells (2x105 cells/well) in serum-free culture medium were seeded into the upper chambers, while the lower chambers were filled with medium containing 2% FBS (Gibco; Thermo Fisher Scientific, Inc.). After incubation at 37˚C for 48 h, the cells that had migrated to the lower chambers were stained with 0.1% crystal violet for 10 min at room temperature and the cell number in five random fields was counted under an inverted microscope (Olympus Corp.).
Luciferase reporter assay
The bioinformatics database, StarBase v.2.0 (http://starbase.sysu.edu.cn/starbase2/), was used to decipher miRNA-target interactions. After determining that miR-145 contained a binding site of SNHG1, a luciferase reporter assay was used to confirm the interaction between miR-145 and SNHG1. The wild-type (WT) and mutant-type (MUT) of the SNHG1 sequence that contained the binding site for miR-145 were individually combined into the pGL3-luciferase basic reporter vector (Promega Corp.). The MUT sequence was obtained using a QuickMutation kit (Beyotime Institute of Biotechnology). The respective combined vectors were then co-transfected into hHCtASMCs with miR-145 mimics or mimics NC using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). Changes in luciferase activity were analyzed by Dual Luciferase Reporter Assay System (Promega Corp.) and normalized to Renilla luciferase activity.
Statistical analysis
Values are expressed as the mean ± standard deviation and analyzed with SPSS 21.0 (IBM Corp.) and GraphPad 7.0 (GraphPad Software, Inc.). Differences between groups were analyzed with Student's t-test or one-way ANOVA with Tukey's multiple-comparison test. Pearson correlation analysis was used to analyze the correlation between the expression of SNHG1 and miR-145. Logistic regression analysis was used to analyze the correlation between SNHG1, miR-145 and the occurrence of restenosis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of SNHG1 and miR-145 in patients with restenosis
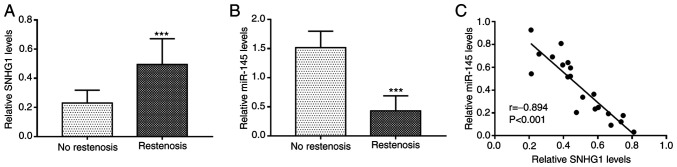
The relative expression levels of SNHG1 and miR-145 in the serum of patients with and without restenosis was determined by RT-qPCR. The results revealed that the expression of SNHG1 was significantly upregulated (P<0.001; Fig. 1A), while that of miR-145 was downregulated (P<0.001; Fig. 1B) in the serum of patients with restenosis compared with the patients without restenosis. There was a significant negative correlation between the expression levels of SNHG1 and miR-145 in serum of patients with restenosis (r=-0.894; P<0.001; Fig. 1C).
Figure 1.
Expression of SNHG1 and miR-145 in patients with restenosis. (A) The expression of SNGH1 was higher in patients with restenosis than that in patients without restenosis. (B) miR-145 levels were lower in patients with restenosis. (C) Serum SNHG1 and miR-145 levels were significantly negatively correlated in patients with restenosis. ***P<0.001 vs. non-restenosis group. miR, microRNA; SNHG1, small nucleolar RNA host gene 1.
Association of SNHG1 and miR-145 with restenosis
The clinical data of all patients, including age, sex, BMI, smoking and drinking status, hypertension, hyperlipidemia, TC, TG, HDL-C, LDL-C and the expression levels of SNHG1 and miR-145 were included in the univariate and multivariate logistic regression analysis. The continuous data, including age, TC, TG, HDL-C, LDL-C, SNHG1 and miR-145, were classified based on median value, and 24 kg/m2 was used as a threshold for BMI. The results indicated that SNHG1 (P=0.008), miR-145 (P=0.002), diabetes (P=0.049), TC (P=0.044) and LDL-C (P=0.032) were independently associated with the occurrence of restenosis and are thus potential risk factors for the occurrence of restenosis in patients following CAS (Table II).
Table II.
Results of the logistic analysis for patients with restenosis.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | OR | 95% CI | P-value | OR | 95% CI | P-value |
| Age (≥65 years vs. <65 years) | 1.531 | 0.862-2.371 | 0.268 | 1.325 | 0.856-2.369 | 0.286 |
| Gender (male vs. female) | 1.226 | 0.701-1.894 | 0.794 | 1.123 | 0.952-1.236 | 0.758 |
| BMI (≥24 kg/m2 vs. <24 kg/m2) | 1.112 | 0.891-2.555 | 0.121 | 1.523 | 0.758-2.569 | 0.162 |
| Smoking status (yes vs. no) | 1.336 | 0.740-2.397 | 0.549 | 1.156 | 0.652-3.296 | 0.521 |
| Alcohol consumption (yes vs. no) | 1.401 | 0.812-2.306 | 0.521 | 2.212 | 0.785-4.621 | 0.596 |
| Diabetes (yes vs. no) | 1.505 | 1.038-2.377 | 0.041 | 1.526 | 1.068-2.768 | 0.049 |
| Hypertension (yes vs. no) | 1.520 | 1.013-1.886 | 0.046 | 2.425 | 0.856-3.965 | 0.132 |
| Hyperlipidemia (yes vs. no) | 1.131 | 0.801-2.089 | 0.407 | 1.132 | 0.925-1.995 | 0.856 |
| TC (high vs. low) | 1.721 | 1.124-2.579 | 0.031 | 2.569 | 1.451-3.874 | 0.044 |
| TG (high vs. low) | 1.524 | 0.951-2.185 | 0.082 | 1.589 | 0.802-3.552 | 0.356 |
| HDL-C (high vs. low) | 1.208 | 0.712-1.941 | 0.507 | 1.124 | 0.936-1.696 | 0.784 |
| LDL-C (low vs. high) | 1.896 | 1.375-3.752 | 0.016 | 2.232 | 1.521-4.132 | 0.032 |
| miR-145 (low vs. high) | 2.597 | 1.812-3.997 | <0.001 | 2.896 | 1.812-4.712 | 0.002 |
| SNHG1 (high vs. low) | 2.366 | 1.672-3.413 | 0.001 | 2.751 | 1.685-4.556 | 0.008 |
miR, microRNA; SNHG1, small nucleolar RNA host gene 1; OR, odds ratio; BMI, body mass index; TC, total cholesterol; TG, triglycerides; H/LDL-C, high/low-density lipoprotein cholesterol.
Knockdown of SNHG1 inhibits the proliferation and migration of hHCtASMCs
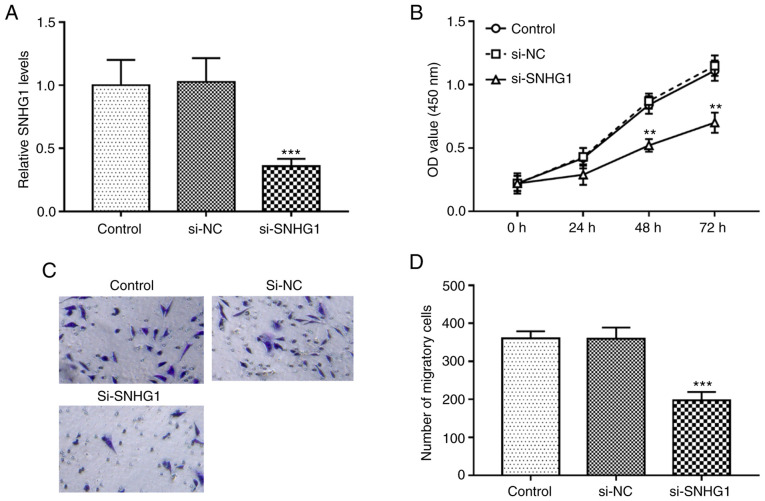
In the present study, the regulatory effect of SNHG1 on the proliferation and migration of hHCtASMCs, which is associated with restenosis, was analyzed. By transfection of si-SNHG1, the expression levels of SNHG1 in hHCtASMCs were successfully downregulated (P<0.001; Fig. 2A). The results of the in vitro assays indicated that cell proliferation and migration were significantly inhibited after silencing of SNHG1 (all P<0.01; Fig. 2B-D).
Figure 2.
Knockdown of SNHG1 inhibits cell proliferation and migration of hHCtASMCs. (A) The expression of SNHG1 was successfully reduced by si-SNHG1 in hHCtASMCs. (B) In hHCtASMCs, knockout of SNHG1 inhibited cell proliferation. (C and D) The migration of hHCtASMCs decreased significantly after transfection with si-SNHG1. (C) Representative images of Transwell membranes (magnification, x100) and (D) quantified numbers of migratory cells in each group. **P<0.01 and ***P<0.001 vs. control. si-SNHG1, short interfering RNA targeting small nucleolar RNA host gene 1; hHCtASMCs, human carotid artery smooth muscle cells; NC, negative control; OD, optical density.
SNHG1 directly inhibits miR-145 expression in hHCtASMCs
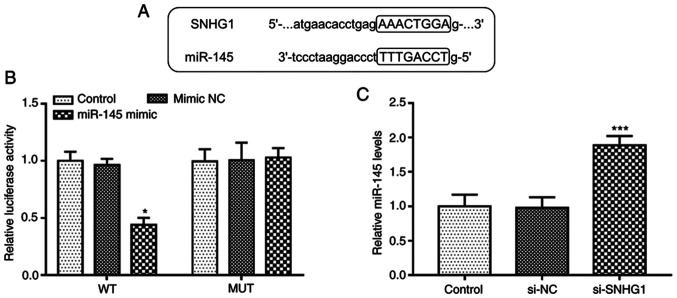
The complementary sequences of miR-145 and SNHG1 at StarBase v.2.0 web are presented in Fig. 3A. To confirm the accuracy of this prediction, a luciferase reporter assay was performed in hHCtASMCs. The experimental results indicated that upregulation of miR-145 suppressed the luciferase activity in the WT-SNHG1 group (P<0.05; Fig. 3B), while it was not affected in the MUT group. Furthermore, RT-qPCR, indicated that silencing of SNHG1 significantly increased the expression levels of miR-145 in hHCtASMCs (P<0.05; Fig. 3C). It was thus suggested that SNHG1 is able to directly interfere with the expression of miR-145 in hHCtASMCs.
Figure 3.
SNHG1 directly inhibits miR-145 expression in hHCtASMCs. (A) The predicted target sequence in the miR-145 for SNHG1 binding. (B) In a hHCtASMC cell line, the luciferase activity of the SNHG1-WT reporter plasmid was decreased by miR-145 overexpression, while that of the SNHG1-MUT plasmid was not affected. (C) Knockdown of SNHG1 increased the expression levels of miR-145 in hHCtASMCs. *P<0.05 and ***P<0.001 vs. control. miR, microRNA; si-SNHG1, short interfering RNA targeting small nucleolar RNA host gene 1; WT, wild-type; MUT, mutant; NC, negative control; hHCtASMCs, human carotid artery smooth muscle cells.
miR-145 mediates the effects of SNHG1 on hHCtASMC proliferation and migration
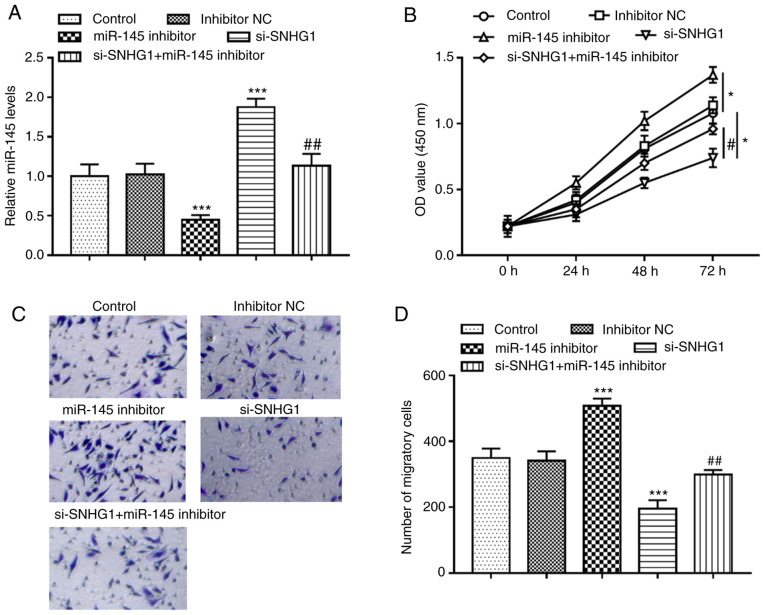
miR-145 inhibitor significantly inhibited the expression of miR-145, but knockout of SNHG1 promoted the expression of miR-145 in hHCtASMCs. When hHCtASMCs were co-transfected with si-SNHG1 and miR-145 inhibitor, it was indicated that miR-145 inhibitor was able to abolish the increase of miR-145 expression caused by knockout of SNHG1 (all P<0.01; Fig. 4A). Analysis of the cells' activities under different treatment conditions revealed that silencing of miR-145 significantly promoted cell proliferation and migration but knockout of SNHG1 suppressed cell proliferation and migration, while in the case of co-regulation of SNHG1 and miR-145, it was observed that miR-145 inhibitor significantly reversed the inhibitory effect of SNHG1 silencing on the proliferation and migration of hHCtASMCs (all P<0.05; Fig. 4B-D), indicating that miR-145 mediated the regulatory effect of SNHG1 on cell functions of hHCtASMCs.
Figure 4.
miR-145 mediates the effects of SNHG1 on hHCtASMC proliferation and migration. (A) The expression level of miR-145 was decreased by miR-145 inhibitor and increased by si-SNHG1, and knockout of SNHG1 reversed the downregulation of miR145 expression levels by miR-145 inhibitor. (B) Effect of miR-145 inhibitor and si-SNHG1 on the proliferation of hHCtASMCs. (C and D) Effect of miR-145 inhibitor and si-SNHG1 on the migration of hHCtASMCs. (C) Representative images of Transwell membranes (magnification, x100) and (D) quantified numbers of migratory cells in each group. The proliferation and migration of hHCtASMCs were significantly increased after transfection with miR-145 inhibitor and significantly decreased after knockout of SNHG1. si-SNHG1 was able to eliminate the promotion effect of miR-145 inhibitor on the proliferation and migration of human colon cancer cells. *P<0.05 and ***P<0.001 vs. control; #P<0.05, ##P<0.01 vs. si-SNHG1. miR, microRNA; si-SNHG1, short interfering RNA targeting small nucleolar RNA host gene 1; hHCtASMCs, human carotid artery smooth muscle cells; NC, negative control; OD, optical density.
Discussion
Carotid stenosis is a major risk factor for stroke that leads to brain damage (3). In the last decade, CAS has been developed into a viable alternative treatment to carotid endarterectomy for patients with symptomatic moderate and high-grade stenosis (20). However, restenosis still remains an unresolved issue following CAS treatment (7). Thus, in order to develop more effective therapeutic approaches for preventing restenosis, an improved understanding of the molecular mechanisms of restenosis is important. One of the important characteristics of restenosis is abnormal proliferation of VSMCs (21). Therefore, it is urgent to understand the molecular mechanisms underlying restenosis and develop specific drugs that target VSMCs. lncRNAs are critical regulatory factors for VSMC function (22). For instance, lncRNA-taurine upregulated gene 1 knockdown attenuated oxidized LDL-induced injury through regulating the proliferation and apoptosis of VSMCs and human umbilical vein endothelial cells via the miR-148b/insulin-like growth factor 2 axis, providing a novel mechanism for the pathogenesis of atherosclerosis (23). In occlusive vascular disease, lncRNA nuclear enriched abundant transcript 1 was discovered as a novel therapeutic target (24). Tao et al (25) indicated that maternally expressed gene 3 may be a negative regulator of spiral artery remodeling via suppressing extravillous trophoblast (EVT) invasion and EVT-mediated VSMC loss. However, the effect of lncRNAs on the proliferation and migration of VSMCs during restenosis has remained largely elusive.
SNHG1 is an lncRNA that is located at 11q12.3 and has 11 exons (26). It has been indicated that SNHG1 is able to participate in cell biological activities, including cell proliferation, migration, invasion and apoptosis (27). SNHG1 has been found to be involved in the pathogenesis of certain diseases. For instance, lncRNA SNHG1 exerted protective effects against oxygen/glucose deprivation-induced injury via sponging miR-338, thus upregulating hypoxia-inducible factor (HIF)-1α/VEGF-A in brain microvascular endothelialscells (16). Zhang et al (28) indicated that SNHG1 exerted a neuroprotective effect mediated by HIF-1α/VEGF signaling through acting as a competing endogenous (ce)RNA for miR-18a. These results revealed a novel function of SNHG1, contributed to a broad understanding of ischemic stroke and provided novel therapeutic options for this disease. However, the results of the present study demonstrated an opposite role of SNHG1 in the progression of restenosis after CAS. It is revealed that the inhibition of SNHG1 served a protective role against restenosis by inhibiting carotid artery smooth muscle cell proliferation and migration. The effect of SNHG1 on the pathologic changes of blood vessels may be dependent on the different pathological states and the types of cells affected by SNHG1, such as endothelial cells or smooth muscle cells. In the present study, SNHG1 was significantly upregulated in serum samples of patients with restenosis. In the present study, serum samples but not plasma samples were used, as plasma contains fibrinogen, which may affect certain proteinsindicators (29). In numerous studies of the roles of ncRNAs in the human circulation, serum samples may better reflect the exact differential expression of these RNAs in various pathologicalsstates (30-32). Furthermore, the blood samples were all centrifuged for serum isolation immediately after collection to avoid hemolysis in this experiment. In addition, the in vitro experimental results indicated that knockout of SNHG1 significantly inhibited hHCtASMC migration and proliferation. The results of the logistic multivariate regression analysis suggested that SNHG1 was independently associated with the occurrence of restenosis and was a potential risk factor for the occurrence of restenosis. Therefore, SNHG1 may have a regulatory role in the development of restenosis.
In recent years, the concept of ceRNA has been proposed as a novel regulatory mechanism in a variety of pathological processes, meaning that lncRNA may function as a ceRNA to sponge miRNA and competitively interact withsmiRNAs (33). In addition, miRNAs have an important role in the diagnosis of various diseases. Diagnosis of human diseases at an early stage contributes to an increased probability of cure and blood tests are easier and faster to perform than tissue tests for early disease screening. Thus, blood tests are considered comparatively more feasible and noninvasive, and may be performed morlyfrequently (34). In the present study, miR-145 binding sites on SNHG1 were discovered. Based on available studies, downregulation of miR-145 in a wide range of diseases, including immune-mediated neuroinflammators diseases (35), colorectal cancer (CC) (36), non-small cell lung cancer (NSC) (37) and breasr cancer (38), may provide novel diagnostic approaches. Previous studies have indicated that the SNHG1/miR-145 axis has an important role in a variety of diseases. For instance Lu et al (12) suggested that downregulation of SNHG1 inhibited NSCLC cell viability, proliferation, migration and invasion, but this inhibition was alleviated by miR-145-5p, which indicated that SNHG1 promoted NSCLC progression by regulating the miR-145-5p/metadherin (MTDH) axis. In CRC, SNHG1 promotes cell proliferation by acting as a sponge of miR-145, suggesting that SNHG1/miR-145 may be a potential target for CRt treatment (13). The present data confirmed that miR-145 was overexpressed in patients with restenosis. Furthermore, inhibition of miR-145 in hHCtASMCs promoted cell migration and proliferation, and the expression level of SNHG1 was negatively correlated with miR-145 in patient serum. Furthermore, a luciferase reporter assay confirmed that SNHG1 was able to specifically associate with miR-145 in hHCtASMCs. SNHG1 affected hHCtASMC proliferation and migration by regulating miR-145. Furthermore, inhibition of miR-145 partially reversed the inhibition of hHCtASMC proliferation and migration by SNHG1 knockout. Thus, based on previous studies and the results of the present experiments, SNHG1 may be involved in the progression of restenosis by directly regulating miR-145.
However, there are certain limitations to the present study, including small sample size, no deconvolution analysis to reduce cell-type heterogeneity, as well as the lack of cell experiments under pathological conditions, animal experiments and further mechanistic analyses. Therefore, further studies are required to explore the effect of the SNHG1/miR-145 axis on the function of VSMCs under pathological conditions, such as hypoxia, as well as to confirm the role of the SNHG1/miR-145 axis in restenosis in vivo.
Overall, the present study indicated that the SNHG1/miR-145 axis regulates the proliferation and migration of hHCtASMCs, and both SNHG1 and miR-145 are independently associated with and potential risk factors for restenosis. The present results suggested that the SNHG1/miR-145 axis may serve as a meaningful non-invasive biomarker for the prevention and treatment of restenosis and provide novel insight into the pathogenesis of restenosis.
Acknowledgements
Not applicable.
Funding Statement
Funding: The current study was supported by the Cangzhou Science and Technology Plan Project grant no. 151302109).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HM analyzed and interpreted the data. HM and AD performed the experiments and wrote and revised the manuscript. HM and AD checked and confirmed the authenticity of the raw data, and read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from each patient and the experimental procedures were approved by the guidelines of the Ethics Committee of Cangzhou Central Hospital (Cangzhou, China; approval no. CZCH14h0280).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Berman SS, Bernhard VM, Erly WK, McIntyre KE, Erdoes LS, Hunter GC. Critical carotid artery stenosis: Diagnosis, timing of surgery, and outcome. J Vasc Surg. 1994;20:499–508. doi: 10.1016/0741-5214(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 2.Puccinelli F, Roffi M, Murith N, Sztajzel R. Management of carotid artery stenosis. Rev Med Suisse. 2017;13:894–849. (In French) [PubMed] [Google Scholar]
- 3.Kelly R. Selections from current literature: Prevention of stroke in non-rheumatic atrial fibrillation and carotid artery stenosis. Fam Pract. 1992;9:231–236. doi: 10.1093/fampra/9.2.231. [DOI] [PubMed] [Google Scholar]
- 4.Cheng SF, Brown MM, Simister RJ, Richards T. Contemporary prevalence of carotid stenosis in patients presenting with ischaemic stroke. Br J Surg. 2019;106:872–878. doi: 10.1002/bjs.11136. [DOI] [PubMed] [Google Scholar]
- 5.Ijas P, Nuotio K, Vikatmaa P, Soinne L. Carotid artery stenosis as predictor of the risk of cerebral and cardiac infarction. Duodecim. 2014;130:2193–2200. (In Finnish) [PubMed] [Google Scholar]
- 6.Fanelli F, Boatta E, Cannavale A, Corona M, Lucatelli P, Wlderk A, Cirelli C, Salvatori FM. Carotid artery stenting: Analysis of a 12-year single-center experience. J Endovasc Ther. 2012;19:749–756. doi: 10.1583/JEVT-12-3944MR.1. [DOI] [PubMed] [Google Scholar]
- 7.AbuRahma AF, Abu-Halimah S, Hass SM, Nanjundappa A, Stone PA, Mousa A, Lough E, Dean LS. Carotid artery stenting outcomes are equivalent to carotid endarterectomy outcomes for patients with post-carotid endarterectomy stenosis. J Vasc Surg. 2010;52:1180–1187. doi: 10.1016/j.jvs.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 8.Frismantiene A, Philippova M, Erne P, Resink TJ. Smooth muscle cell-driven vascular diseases and molecular mechanisms of VSMC plasticity. Cell Signal. 2018;52:48–64. doi: 10.1016/j.cellsig.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Zhang MJ, Zhou Y, Chen L, Wang YQ, Wang X, Pi Y, Gao CY, Li JC, Zhang LL. An overview of potential molecular mechanisms involved in VSMC phenotypic modulation. Histochem Cell Biol. 2016;145:119–130. doi: 10.1007/s00418-015-1386-3. [DOI] [PubMed] [Google Scholar]
- 10.Yuan Y, Liu X, Hao S, He Q, Shen Z. Plasma levels of miR-143 and miR-145 are associated with coronary in-stent restenosis within 1 year of follow-up after drug-eluting stent implantation. Ann Transl Med. 2020;8(756) doi: 10.21037/atm-20-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He M, Gong Y, Shi J, Pan Z, Zou H, Sun D, Tu X, Tan X, Li J, Li W, et al. Plasma microRNAs as potential noninvasive biomarkers for in-stent restenosis. PLoS One. 2014;9(e112043) doi: 10.1371/journal.pone.0112043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32:3957–3967. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 13.Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget. 2018;9:2128–2139. doi: 10.18632/oncotarget.23255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan X, Liu X. lncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 2019;23:2351–2361. doi: 10.1111/jcmm.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Mao D, Li C, Li M. miR-145-5p inhibits vascular smooth muscle cells (VSMCs) proliferation and migration by dysregulating the transforming growth factor-b signaling cascade. Med Sci Monit. 2018;24:4894–4904. doi: 10.12659/MSM.910986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Zi XH. lncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1α axis. Brain Res. 2019;1714:174–181. doi: 10.1016/j.brainres.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Liang S, Ren K, Li B, Li F, Liang Z, Hu J, Xu B, Zhang A. lncRNA SNHG1 alleviates hypoxia-reoxygenation-induced vascular endothelial cell injury as a competing endogenous RNA through the HIF-1α/VEGF signal pathway. Mol Cell Biochem. 2020;465:1–11. doi: 10.1007/s11010-019-03662-0. [DOI] [PubMed] [Google Scholar]
- 18.Gasecki AP, Hachinski VC, Mendel T, Barnett HT. Endarterectomy for symptomatic carotid stenosis. Review of the European and North American symptomatic carotid surgery trials. Nebr Med J. 1992;77:121–123. [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Moresoli P, Habib B, Reynier P, Secrest MH, Eisenberg MJ, Filion KB. Carotid stenting versus endarterectomy for asymptomatic carotid artery stenosis: A systematic review and meta-analysis. Stroke. 2017;48:2150–2157. doi: 10.1161/STROKEAHA.117.016824. [DOI] [PubMed] [Google Scholar]
- 21.Luo T, Cui S, Bian C, Yu X. Crosstalk between TGF-β/smad3 and BMP/BMPR2 signaling pathways via miR-17-92 cluster in carotid artery restenosis. Mol Cell Biochem. 2014;389:169–176. doi: 10.1007/s11010-013-1938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Dong Y, Zhao Z. lncRNA MEG8 regulates vascular smooth muscle cell proliferation, migration and apoptosis by targeting PPARα. Biochem Biophys Res Commun. 2019;510:171–176. doi: 10.1016/j.bbrc.2019.01.074. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Zheng X, Cheng J, Zhang K, Ma C. lncRNA TUG1 regulates proliferation and apoptosis by regulating miR-148b/IGF2 axis in ox-LDL-stimulated VSMC and HUVEC. Life Sci. 2020;243(117287) doi: 10.1016/j.lfs.2020.117287. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed ASI, Dong K, Liu J, Wen T, Yu L, Xu F, Kang X, Osman I, Hu G, Bunting KM, et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc Natl Acad Sci USA. 2018;115:E8660–E8667. doi: 10.1073/pnas.1803725115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao H, Liu X, Liu X, Liu W, Wu D, Wang R, Lv G. lncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling. Mol Reprod Dev. 2019;86:686–695. doi: 10.1002/mrd.23147. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, Li D, Yang T. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 27.Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta. 2019;494:38–47. doi: 10.1016/j.cca.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Luo X, Chen F, Yuan W, Xiao X, Zhang X, Dong Y, Zhang Y, Liu Y. lncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem. 2018;119:5460–5472. doi: 10.1002/jcb.26705. [DOI] [PubMed] [Google Scholar]
- 29.Zhang A, Sun H, Wang X. Serum metabolomics as a novel diagnostic approach for disease: A systematic review. Anal Bioanal Chem. 2012;404:1239–1245. doi: 10.1007/s00216-012-6117-1. [DOI] [PubMed] [Google Scholar]
- 30.Xie J, Cao Y. Expression of TGF-β1 and miR-99a in serum of patients with early spontaneous abortion and correlation with hormone levels during pregnancy. Exp Ther Med. 2019;17:4593–4597. doi: 10.3892/etm.2019.7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Hu Y, Gong C, Zhang C. Overexpression of miR-518b in non-small cell lung cancer serves as a biomarker and facilitates tumor cell proliferation, migration and invasion. Oncol Lett. 2020;20:1213–1220. doi: 10.3892/ol.2020.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Qiu F, Tian F, Shi X, Gao A, Song L, Liu J. Changes of miR-155 expression in serum of uremic patients before and after treatment and risk factors analysis. Exp Ther Med. 2020;20:3352–3360. doi: 10.3892/etm.2020.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao C, Yang F, Qin Z, Jing X, Shu Y, Shen H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer. 2019;19(1103) doi: 10.1186/s12885-019-6297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharaf-Eldin WE, Kishk NA, Gad YZ, Hassan H, Ali MA, Zaki MS, Mohamed MR, Essawi ML. Extracellular miR-145, miR-223 and miR-326 expression signature allow for differential diagnosis of immune-mediated neuroinflammatory diseases. J Neurol Sci. 2017;383:188–198. doi: 10.1016/j.jns.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Sheng N, Tan G, You W, Chen H, Gong J, Chen D, Gong J, Chen D, Zhang H, Wang Z. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6:1331–1340. doi: 10.1002/cam4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Ding J, He M, Chen Y, Wang R, Han Z, Xing EZ, Zhang C, Yeh S. Estrogen receptor beta promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene. 2019;38:1225–1238. doi: 10.1038/s41388-018-0463-1. [DOI] [PubMed] [Google Scholar]
- 38.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.