Abstract
Numerous studies have reported the critical roles of long non-coding RNAs (lncRNAs) in the regulation of osteoarthritis (OA) development. The present study aimed to assess the function and regulatory mechanism of a lncRNA, KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1), in OA in vitro. C28/I2 cells were treated with lipopolysaccharide (LPS) to generate an in vitro OA model. The relative expression levels of KCNQ1OT1, microRNA (miR)-211-5p and transcription factor 4 (TCF4) were determined via reverse transcription-quantitative polymerase chain reaction. The associations between KCNQ1OT1, miR-211-5p and TCF4 were confirmed using a dual-luciferase reporter assay. Furthermore, cell viability was assessed using the MTT assay. Inflammatory cytokine levels were measured using ELISA. The protein expression levels of matrix metalloproteinase-3/13, collagen II/X and TCF4 were detected by western blotting. KCNQ1OT1 and TCF4 were highly expressed in the cartilage tissues of patients with OA and C28/I2 cells treated with LPS (OA cells), whereas miR-211-5p was downregulated concomitantly in OA tissues and cells. Knockdown of KCNQ1OT1 stimulated cell viability, and suppressed the inflammation and degradation of the extracellular matrix (ECM) in OA cells. In addition, overexpression of miR-211-5p stimulated cell viability, and inhibited inflammation and degradation of the ECM in OA cells. Notably, miR-211-5p was revealed to be the target of, and was negatively regulated by, KCNQ1OT1. TCF4 was targeted and negatively modulated by miR-211-5p. Transfection of cells with the miR-211-5p inhibitor or pcDNA-TCF4 reversed the suppressive effects of short hairpin RNA (sh)-KCNQ1OT1 on inflammation and ECM degradation, as well as the promotive effect of sh-KCNQ1OT1 on viability in OA in vitro. Therefore, KCNQ1OT1 may regulate the miR-211-5p/TCF4 axis to ameliorate OA in vitro.
Keywords: osteoarthritis, long non-coding RNA KCNQ1OT1, microRNAs, miR-211-5p, transcription factor 4
Introduction
As the most common musculoskeletal disease in the elderly population, osteoarthritis (OA) is characterised by joint pain, tenderness, stiffness, crepitus and limitation of movement (1). Histologically, the major characteristics of OA are early fragmentation of the cartilage surface, vertical clefts of the cartilage, cloning of chondrocytes, variable crystal deposition and violation of the tidemark by blood vessels (2,3). Worldwide disease estimates have demonstrated that 18% of women and 9.6% of men aged 60 and older have symptomatic OA (4). Although there are numerous treatments for OA, including acupuncture therapy, drug treatment and surgical therapy (5,6), OA remains difficult to cure. Therefore, it is imperative to assess the molecular mechanisms underlying OA in order to identify novel therapeutic targets.
Long non-coding RNAs (lncRNAs) are a class of nucleotide sequences >200 nucleotides in length that lack protein-coding potential (7). A previous study reported that lncRNAs may function as important participants in regulating the progression of OA (8). Liu et al (9) reported that the silencing of lncRNA XIST may restrain the development of OA by inhibiting cell apoptosis and extracellular matrix (ECM) degradation (9). Chen et al (10) demonstrated that the lncRNA HOTAIR participates in OA progression by stimulating ECM degradation and chondrocyte apoptosis. Zhang et al (11) revealed that lncRNA SNHG15 suppresses OA progression by acting as a sponge of microRNA (miRNA/miR)-141-3p. Notably, a recent study indicated that lncRNA KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) may serve a critical role in OA progression by sponging hsa-miR-1202/ETS1 interactions (12). However, research on the underlying mechanism of KCNQ1OT1 in OA remains in the exploratory stage.
miRNAs are highly conserved non-coding RNAs with a length of 19-25 nucleotides, which have been reported to participate in the process of OA (13,14). Luo et al (15) reported that miR-34a induces OA synovial cell apoptosis by regulating transforming growth factor-β-induced factor homeobox 2. Zhong et al (16) confirmed that miR-335-5p may markedly alleviate inflammation in OA chondrocytes. Huang et al (17) revealed the essential role of miR-211 in maintaining joint homeostasis and counteracting the progression of OA. Notably, previous studies on the expression and function of miR-211-5p in OA have received significant attention (18,19). It has been reported that miR-211-5p manifests a downregulation in sclerotic bone compared with non-sclerotic OA bone samples, and miR-211-5p has been confirmed to decrease the mineralisation capacity of ossified osteoblasts in OA (18). Furthermore, miR-211-5p has been reported to be markedly diminished in the articular cartilage tissues and functions to restrain ECM degradation and the production of inflammatory cytokines in OA (19). Despite these findings, the interaction between miR-211-5p and KCNQ1OT1 in OA has not been fully understood.
The aim of the present study was to investigate the role and molecular mechanism of KCNQ1OT1 in OA, and to identify the associations between KCNQ1OT1, miR-211-5p and transcription factor 4 (TCF4). Through this work, the present study aimed to develop a solid foundation for targeted OA therapy.
Materials and methods
Cartilage tissue samples
From January 2018 to December 2019, OA cartilage tissues were obtained from the knee joints of 25 patients (12 males and 13 females; age range, 52-70 years; mean age, 60.5±6.4) who underwent artificial total knee replacement surgery. Normal cartilage tissues were derived from the knee joints of 25 subjects (12 males and 13 females; age range, 48-66 years; mean age, 56.7±6.4) who had femoral neck fractures without OA or rheumatoid arthritis. The diagnostic criteria for OA were based on the American College of Rheumatology standards (20). The diagnostic criteria included: i) Pain in the knee, hip and interphalangeal joints, ii) presence of osteophytes detected through X-ray examination, iii) synovial fluid laboratory examination consistent with OA, iv) morning stiffness for <30 min, v) age of at least 40 years, and vi) presence of crepitus. Patients who met i and ii; i, iii, v and vi; or i, iv, v and vi were diagnosed with OA. The clinicopathological data of these patients were as follows: Age range, 53-75 years; mean age, 63.3±5.9 years; sex, 9 men and 16 women; and OA grade I, 2; grade II, 10; and grade III, 13. The present study was approved by the Ethics Committee of The Second Xiangya Hospital of Central South University. Written informed consent was obtained from each participant.
Cell culture
The normal human cartilage cell line (C28/I2) was purchased from the American Type Culture Collection. C28/I2 cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)-Nutrient Mixture F12 supplemented with 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). All cells were maintained in an incubator with 5% CO2 at 37˚C. An OA model was established in vitro in accordance with previous studies (21,22). In brief, C28/I2 cells were treated with lipopolysaccharide (LPS; 5 µg/ml; Sigma-Aldrich; Merck KGaA) for 12 h at 37˚C, and untreated C28/I2 cells were used as controls.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was synthesised using a high-capacity RT kit at 42˚C for 45 min (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed using SYBR Green PCR Master mix (Takara Biotechnology Co., Ltd.). The thermocycling conditions were as follows: 95˚C for 5 min, 40 cycles of denaturation at 95˚C for 10 sec, annealing at 50˚C for 1 min and extension at 72˚C for 30 sec. All primers were purchased from Invitrogen (Thermo Fisher Scientific, Inc.), and their sequences are presented in Table I. Experimental results were calculated using the 2-ΔΔCq method (23). KCNQ1OT1 and TCF4 expression levels were normalised to GAPDH, and miR-211-5p expression levels were normalised to U6.
Table I.
Primers for reverse transcription-quantitative polymerase chain reaction.
| Gene | Forward | Reverse |
|---|---|---|
| KCNQ1OT1 | 5'-TTGGTAGGATTTTGTTGAGG-3' | 5'-CAACCTTCCCCTACTACC-3' |
| miR-211-5p | 5'-TCGGCAGGTCCCTTTGTCATCC-3' | 5'-TGCAGGTCAACTGGTGTCGT-3' |
| TCF4 | 5'-CCTGGCTATGCAGGAATGTT-3' | 5'-CAGGAGGCGTACAGGAAGAG-3' |
| GAPDH | 5'-CCAGGTGGTCTCCTCTGA-3' | 5'-GCTGTAGCCAAATCGTTGT-3' |
| U6 | 5'-CTCGCTTCGGCAGCACA-3' | 5'-AACGCTTCACGAATTTGCGT-3' |
miR, microRNA; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; TCF4, transcription factor 4.
Cell transfection
When C28/I2 cells reached 80% confluence, they were seeded onto 6-well cell culture plates (5x103 cells/well). The short hairpin (sh) RNA targeting KCNQ1OT1 (sh-KCNQ1OT1; 5'-GGUUCAGAUUCAUAAACUAGA-3'), sh-negative control (sh-NC; 5'-CAUAGUCGAAUUUCGCUAGUGAGUU-3'), miR-NC (5'-ACCGCUAAUCAUACGAAUACAC-3'), miR-211-5p mimics (5'-UUCCCUUUGUCAUCCUUCGCCU-3'), miR-211-5p inhibitor (5'-AGGCAAAGGAUGACAAGGGAA-3'), inhibitor NC (5'-CAGUACUUUUGUAGUACAAA-3'), overexpression plasmids (pcDNA-KCNQ1OT1 and pcDNA-TCF4) and empty vector (pcDNA-NC) were purchased from Guangzhou RiboBio Co., Ltd. C28/I2 cells (6x105 cells/well) were transfected with these factors (all, 50 nM) for 48 h at 37˚C with Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols.
Dual-luciferase reporter (DLR) assay
The targeting relationship between KCNQ1OT1 and miR-211-5p was analysed using the lncBase Predicted database (version 2.0; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted). Additionally, the targeting relationship between miR-211-5p and TCF4 was predicted using TargetScan (release 7.2; http://www.targetscan.org/vert_72/) and miRDB (version 4.0; http://mirdb.org/). The 3'-untranslated region (UTR) of KCNQ1OT1 or TCF4 harbouring the predicted binding sites for miR-211-5p was introduced into the pGL3 vector (Promega Corporation) to construct the KCNQ1OT1 wild-type (wt)/mutant type (mut) or TCF4 wt/mut. Next, C28/I2 cells (1x105 cells/well) were seeded onto a 24-well plate and co-transfected with one of the aforementioned plasmids (80 ng) and miR-211-5p mimics or miR-NC (50 nM) using Lipofectamine 3000. After 48 h at 37˚C, the relative luciferase activity was determined using the DLR Assay kit (Promega Corporation). The activity of firefly luciferase was normalized to that of Renilla luciferase.
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (EMD Millipore) in accordance with the manufacturer's protocol. In brief, C28/I2 cells were transfected with miR-211-5p mimics or miR-NC. After 48 h, the cells were collected and lysed using RIP lysis buffer (Beyotime Institute of Biotechnology). An anti-argonaute2 (Ago2) antibody (1:5,000; cat. no. 03-110; EMD Millipore) was conjugated to magnetic beads and incubated at 55˚C for 30 min with the whole cell extract. Samples were then centrifuged (at 450 x g for 20 min at 20˚C) and washed three times with Hank's Balanced Salt Solution (Sigma-Aldrich; Merck KGaA). The immunoprecipitated RNAs were isolated, and an aforementioned RT-qPCR assay was used to detect the expression levels of KCNQ1OT1 and TCF4.
MTT assay
Transfected cells were seeded onto 96-well plates (5x103 cells/well). After 48 h of culture, the cells were incubated with 10 µl MTT reagent (Beyotime Institute of Biotechnology) for another 4 h. Thereafter, 100 µl dimethyl sulfoxide was added to dissolve the formazan. The optical density was measured at 490 nm using a spectrophotometer (Thermo Fisher Scientific, Inc.).
ELISA
The levels of IL-6 (cat. no. PD6050; R&D Systems, Inc.), IL-1β (cat. no. PDLB50; R&D Systems, Inc.) and TNF-α (cat. no. PDTA00D; R&D Systems, Inc.) in culture media were measured using Quantikine ELISA kits according to the manufacturer's protocols. The optical density was determined at 450 nm using a microplate reader (Molecular Devices, LLC).
Western blotting
Total proteins were extracted using radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology). Total protein was quantified using a BCA Protein assay kit (Invitrogen; Thermo Fisher Scientific, Inc.). Protein samples (20 µg per lane) were fractionated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes. After blocking with 5% skimmed milk-Tris-buffered saline with TBST (Tween-20, 0.05%) for 2 h at 25˚C, the membranes were incubated with primary antibodies (Abcam) against matrix metalloproteinase (MMP)-3 (dilution, 1:1,000; cat. no. ab53015), MMP-13 (dilution, 1:1,000; cat. no. ab51072), collagen II (dilution, 1:2,000; cat. no. ab34712), collagen X (dilution, 1:1,000; cat. no. ab182563), TCF4 (dilution, 1:1,000; cat. no. ab130014) and β-actin (dilution, 1:5,000; cat. no. ab6276) at 4˚C overnight. After the membranes were washed with TBST three times, the horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000; cat. no. 14709; Cell Signaling Technology, Inc.) was added and incubated at 37˚C for 1 h. β-actin was selected as the internal reference. The immunoreactive bands were visualised using an enhanced chemiluminescent detection kit (Thermo Fisher Scientific, Inc.), and the relative protein expression levels were semiquantified using the ChemiDoc XRS system (Bio-Rad Laboratories, Inc.).
Statistical analysis
All data derived from independent experiments performed in triplicate are presented as the mean ± standard deviation. A paired Student's t-test was employed for comparisons between two groups. Differences between multiple groups were compared using a one-way analysis of variance followed by Tukey's multiple comparisons test. P<0.05 was considered to indicate a statistically significant difference.
Results
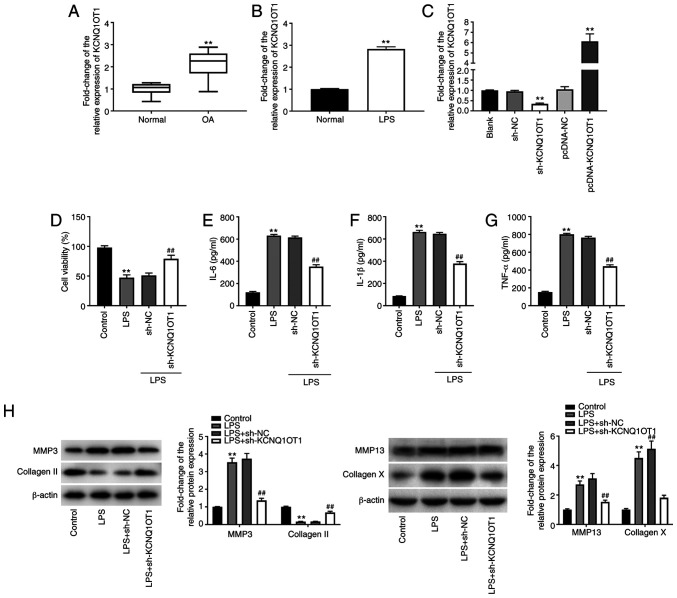
Silencing of lncRNA KCNQ1OT1 alleviates LPS-induced chondrocyte injury in C28/I2 cells. To assess the function of lncRNA KCNQ1OT1 in OA, the expression levels of KCNQ1OT1 were detected through RT-qPCR. KCNQ1OT1 was significantly upregulated in OA cartilage tissues compared with that in normal cartilage tissues (P<0.01; Fig. 1A). KCNQ1OT1 expression levels were also increased in C28/I2 cells treated with LPS (P<0.01; Fig. 1B). KCNQ1OT1 was then silenced or overexpressed to investigate its role in OA. As shown in Fig. 1C, KCNQ1OT1 expression was markedly diminished post-transfection with sh-KCNQ1OT1, whereas it was increased following transfection with pcDNA-KCNQ1OT1 in C28/I2 cells (P<0.01). The MTT assay demonstrated that cell viability was suppressed by LPS in C28/I2 cells, and sh-KCNQ1OT1 enhanced cell viability in LPS-induced C28/I2 cells (all P<0.01; Fig. 1D). In addition, the ELISA results indicated that IL-6, IL-1β and TNF-α levels in the culture medium were enhanced by LPS, whereas the levels of IL-6, IL-1β and TNF-α were decreased by sh-KCNQ1OT1 in C28/I2 cells treated with LPS (all P<0.01; Fig. 1E-G). Furthermore, the expression levels of ECM degradation-related proteins (MMP-3/-13 and collagen II) and the hypertrophy marker collagen X were determined. Western blot analysis revealed that the expression levels of MMP-3/-13 and collagen X were enhanced by LPS in C28/I2 cells, whereas collagen II expression levels were decreased by LPS in C28/I2 cells (all P<0.01; Fig. 1H). MMP-3/-13 and collagen X were diminished, whereas collagen II was increased following transfection of LPS-treated C28/I2 cells with sh-KCNQ1OT1 (all P<0.01; Fig. 1H).
Figure 1.
Silencing of KCNQ1OT1 alleviates chondrocyte injury caused by LPS in C28/I2 cells. (A) Relative expression level of KCNQ1OT1 in OA cartilage tissues and normal cartilage tissues was determined by RT-qPCR. **P<0.01 vs. normal. (B) Relative expression level of KCNQ1OT1 in C28/I2 cells was determined by RT-qPCR. **P<0.01 vs. normal. (C) Relative expression level of KCNQ1OT1 in C28/I2 cells was determined by RT-qPCR post-transfection with sh-KCNQ1OT1/NC or pcDNA-KCNQ1OT1/NC. **P<0.01 vs. blank. (D) Cell viability was assessed by MTT assay. **P<0.01 vs. control; ##P<0.01 vs. LPS + sh-NC. Levels of (E) IL-6 (F) IL-1β and (G) TNF-α in culture medium were measured by ELISA. **P<0.01 vs. control; ##P<0.01 vs. LPS + sh-NC. (H) Protein expression levels of MMP-3, MMP-13, collagen X and collagen II were detected by western blotting. **P<0.01 vs. control; ##P<0.01 vs. LPS + sh-NC. LPS, lipopolysaccharide; OA, osteoarthritis; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; sh, short hairpin RNA; NC, negative control; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; MMP, matrix metalloproteinase.
KCNQ1OT1 may serve as a competitive endogenous RNA of miR-211-5p
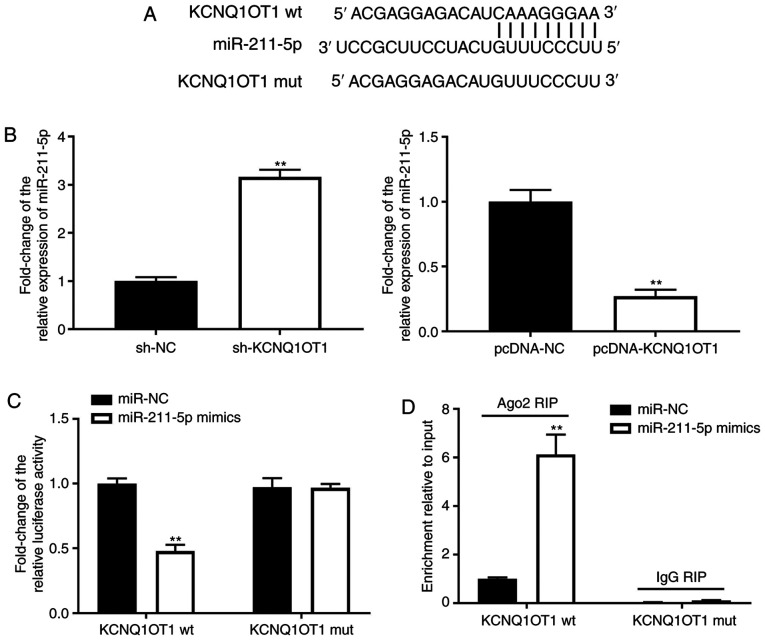
To further investigate the mechanism of KCNQ1OT1 in OA, the binding sites of KCNQ1OT1 were predicted using lncBase Predicted v.2. miR-211-5p was predicted to be the target of KCNQ1OT1 (Fig. 2A). The expression levels of miR-211-5p were increased by sh-KCNQ1OT1 but inhibited by pcDNA-KCNQ1OT1 in C28/I2 cells (P<0.01; Fig. 2B-C). Furthermore, the DLR and RIP assays were utilised to verify the interaction between KCNQ1OT1 and miR-211-5p. As shown in Fig. 2D, the relative luciferase activity of C28/I2 cells was decreased post-transfection with the KCNQ1OT1 wt vector and miR-211-5p, compared with C28/I2 cells transfected with the KCNQ1OT1 wt vector and miR-NC (P<0.01). As shown in Fig. 2E, KCNQ1OT1 immunoprecipitates obtained from C28/I2 cells were notably enriched in the miR-211-5p mimics group, compared with the miR-NC group (P<0.01).
Figure 2.
KCNQ1OT1 may serve as a competitive endogenous RNA of miR-211-5p. (A) Binding sequence between KCNQ1OT1 and miR-211-5p was predicted by lncBase Predicted v.2. (B) Relative expression level of miR-211-5p after transfection with sh-KCNQ1OT1/NC or pcDNA-KCNQ1OT1/NC into C28/I2 cells was detected by RT-qPCR. **P<0.01 vs. sh-NC. (C) Targeting association between KCNQ1OT1 and miR-211-5p was validated by dual-luciferase reporter assay. (D) Targeting association between KCNQ1OT1 and miR-211-5p was validated by RIP assay. **P<0.01 vs. miR-NC. miR, microRNA; sh, short hairpin RNA; NC, negative control; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; wt, wild type; mut, mutant; RIP, RNA immunoprecipitation.
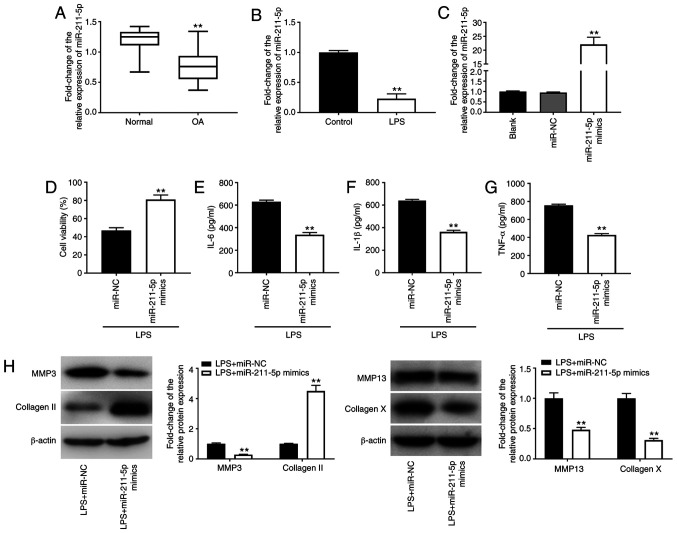
Overexpression of miR-211-5p reduces LPS-induced chondrocyte injury in C28/I2 cells
Through investigating the role of miR-211-5p in OA, it was revealed that the expression levels of miR-211-5p in OA cartilage tissues were downregulated compared with those in normal cartilage tissues (P<0.01; Fig. 3A). miR-211-5p expression levels were also decreased in LPS-induced C28/I2 cells compared with those in C28/I2 cells not treated with LPS (P<0.01; Fig. 3B). Following transfection with miR-211-5p mimics, miR-211-5p expression levels exhibited a significant increase in C28/I2 cells (P<0.01; Fig. 3C), indicating that overexpression of miR-211-5p was successful in subsequent functional studies. As shown in Fig. 3D, cell viability was increased by transfection of LPS-treated C28/I2 cells with miR-211-5p mimics (P<0.01). Overexpression of miR-211-5p also suppressed the inflammatory response by decreasing the levels of IL-6, IL-1β and TNF-α in C28/I2 cells treated with LPS (all P<0.01; Fig. 3E-G). In addition, the upregulation of collagen II and downregulation of MMP-3/-13/collagen X implied that overexpression of miR-211-5p may suppress ECM degradation and chondrocyte hypertrophy (all P<0.01; Fig. 3H).
Figure 3.
Overexpression of miR-211-5p reduces chondrocyte injury caused by LPS in C28/I2 cells. (A) Relative expression level of miR-211-5p in OA cartilage tissues and normal cartilage tissues was determined by RT-qPCR. **P<0.01 vs. normal. (B) Relative expression level of miR-211-5p in C28/I2 cells was determined by RT-qPCR. **P<0.01 vs. control. (C) Relative expression level of miR-211-5p in C28/I2 cells was determined by RT-qPCR following transfection with miR-211-5p mimics. **P<0.01 vs. blank. (D) Cell viability was assessed by MTT assay. **P<0.01 vs. LPS + miR-NC. Levels of (E) IL-6, (F) IL-1β and (G) TNF-α in culture medium were measured by ELISA. **P<0.01 vs. LPS + miR-NC. (H) Protein expression levels of MMP-3 MMP-13, collagen X and collagen II were detected by western blotting. **P<0.01 vs. LPS + miR-NC. miR, microRNA; LPS, lipopolysaccharide; OA, osteoarthritis; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control; MMP, matrix metalloproteinase.
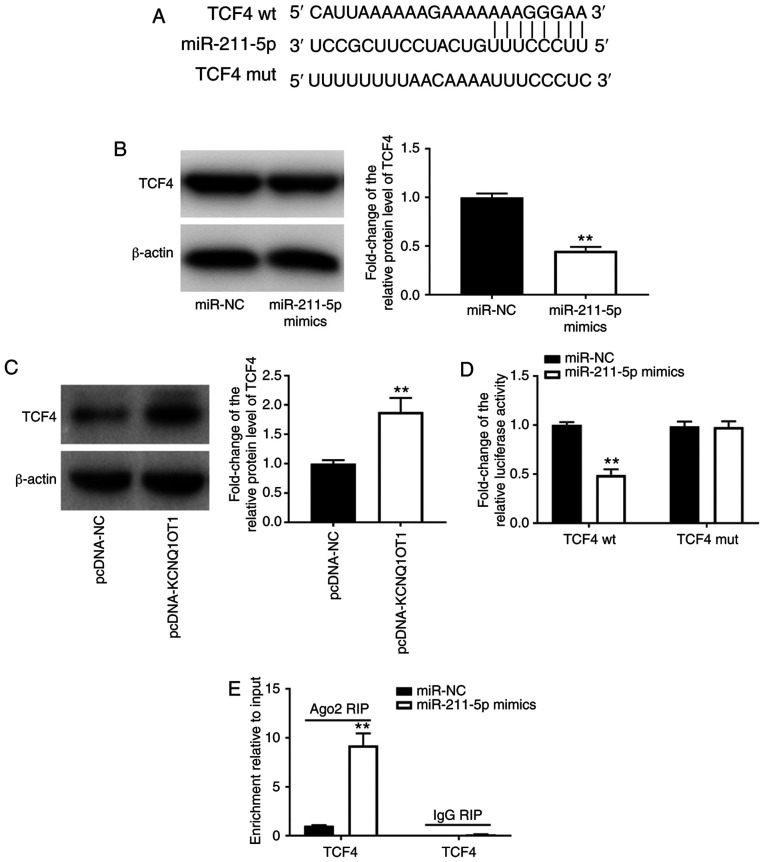
TCF4 is a target gene of miR-211-5p
TargetScan and miRDB software were employed to search for the target gene of miR-211-5p. It was discovered that miR-211-5p had certain complementary bases with the sequence of TCF4 3'-UTR, indicating that TCF4 may be the target gene of miR-211-5p (Fig. 4A). Western blot analysis revealed that overexpression of miR-211-5p markedly decreased the protein expression levels of TCF4 (P<0.01; Fig. 4B), whereas KCNQ1OT1 overexpression increased the protein expression levels of TCF4 (P<0.01; Fig. 4C). The results of DLR and RIP assays verified the interaction between miR-211-5p and TCF4. It was revealed that the relative luciferase activity was markedly decreased by the introduction of miR-211-5p mimics in C28/I2 cells transfected with the TCF4 wt vector (P<0.01; Fig. 4D) but did not differ following the introduction of miR-211-5p mimics in C28/I2 cells transfected with the TCF4 mut vector (Fig. 4D). The RIP assay demonstrated that endogenous TCF4 was significantly enriched in the miR-211-5p mimic group compared with that in the miR-NC group (P<0.01; Fig. 4E), revealing the direct binding between miR-211-5p and TCF4.
Figure 4.
TCF4 is a target gene of miR-211-5p. (A) Binding sequence between miR-211-5p and TCF4 was predicted by TargetScan and miRDB. (B) Relative protein expression level of TCF4 was detected by western blotting. **P<0.01 vs. miR-NC. (C) Relative protein expression level of TCF4 was detected by western blotting following transfection of C28/I2 cells with pcDNA-KCNQ1OT1. **P<0.01 vs. pcDNA-NC. (D) Targeting association between miR-211-5p and TCF4 was validated by dual-luciferase reporter assay. **P<0.01, vs. miR-NC. (E) Targeting association between miR-211-5p and TCF4 was validated by RIP assay. **P<0.01, vs. miR-NC. miR, microRNA; NC, negative control; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; TCF4, transcription factor 4; wt, wild type; mut, mutant; RIP, RNA immunoprecipitation.
sh-KCNQ1OT1 may regulate the miR-211-5p/TCF4 axis to inhibit OA progression
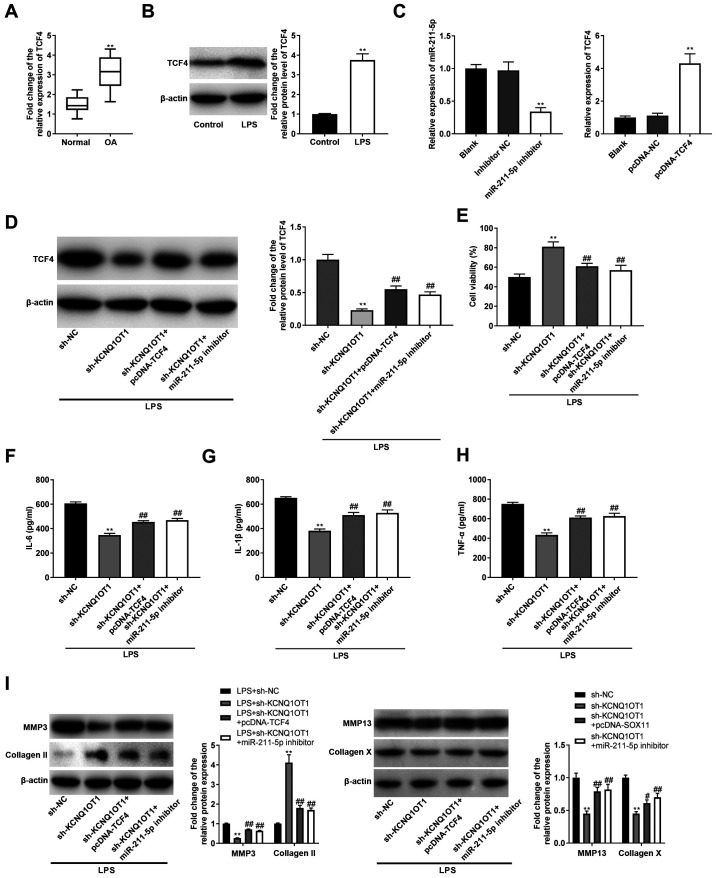
To confirm the associations between KCNQ1OT1, miR-211-5p and TCF4 in OA, the expression levels of TCF4 in OA were determined. It was revealed that the mRNA expression levels of TCF4 were upregulated in the cartilage tissues of patients with OA, and the protein expression levels of TCF4 were also upregulated in LPS-induced C28/I2 cells, compared with their respective controls (all P<0.01; Fig. 5A and B). In addition, the expression levels of miR-211-5p were decreased by transfection with miR-211-5p inhibitor, and TCF4 expression levels were increased post-transfection with pcDNA-TCF4 (P<0.01; Fig. 5C). Subsequently, rescue experiments were implemented, and it was revealed that sh-KCNQ1OT1 led to a significant decrease in TCF4 in LPS-treated C28/I2 cells (P<0.01; Fig. 5D). The suppressive effect of sh-KCNQ1OT1 on TCF4 was reversed by pcDNA-TCF4 or the miR-211-5p inhibitor in LPS-treated C28/I2 cells (all P<0.01; Fig. 5D). Cell viability was promoted by sh-KCNQ1OT1 in LPS-treated C28/I2 cells (P<0.01; Fig. 5E), whereas the effect of sh-KCNQ1OT1 on cell viability was reversed by pcDNA-TCF4 or the miR-211-5p inhibitor (all P<0.01; Fig. 5E). Furthermore, inflammation was suppressed by sh-KCNQ1OT1 in LPS-treated C28/I2 cells (all P<0.01; Fig. 5F-H). The suppressive effects of sh-KCNQ1OT1 on IL-6, IL-1β and TNF-α were reversed by pcDNA-TCF4 or the miR-211-5p inhibitor in LPS-induced C28/I2 cells (all P<0.01; Fig. 5F-H). As illustrated in Fig. 5I, the decrease in MMP-3/-13/collagen X and increase in collagen II induced by transfection of LPS-induced C28/I2 cells with sh-KCNQ1OT1 (all P<0.01) was reversed by transfection with pcDNA-TCF4 or the miR-211-5p inhibitor (P<0.05).
Figure 5.
KCNQ1OT1 indirectly regulates expression of TCF4 by competitively binding to miR-211-5p. (A) Relative expression level of TCF4 in OA cartilage tissues and normal cartilage tissues was determined by RT-qPCR. **P<0.01 vs. normal. (B) Relative protein expression level of TCF4 in C28/I2 cells was determined by western blotting. **P<0.01 vs. control. (C) Expression levels of miR-211-5p and TCF4 were detected by RT-qPCR. **P<0.01 vs. inhibitor NC or pcDNA-NC. (D) Relative protein expression level of TCF4 in transfected C28/I2 cells were determined by western blotting. **P<0.01 vs. LPS + sh-NC; ##P<0.01 vs. LPS + sh-KCNQ1OT1. (E) Cell viability was assessed by MTT assay. **P<0.01 vs. LPS + sh-NC; ##P<0.01 vs. LPS + sh-KCNQ1OT1. Levels of (F) IL-6, (G) IL-1β and (H) TNF-α in culture medium were measured by ELISA. **P<0.01 vs. LPS + sh-NC; ##P<0.01 vs. LPS + sh-KCNQ1OT1. (I) Protein expression levels of MMP-3, MMP-13, collagen X and collagen II were detected by western blotting. **P<0.01 vs. LPS + sh-NC; ##P<0.01 vs. LPS + sh-KCNQ1OT1. miR, microRNA; OA, osteoarthritis; LPS, lipopolysaccharide; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; sh, short hairpin RNA; NC, negative control; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; TCF4, transcription factor 4; MMP, matrix metalloproteinase.
Discussion
OA is characterised as a degenerative joint disease that poses a serious threat to public health (24). Previous studies on lncRNA profiles have demonstrated upregulation of various lncRNAs is involved in the pathogenesis of OA, including lncRNA ANRIL (25), lncRNA CTBP1-AS2(26) and lncRNA FAS-AS1(27). Consistent with these findings, the present study demonstrated that lncRNA KCNQ1OT1 was markedly upregulated in OA cartilage tissues and LPS-induced C28/I2 cells. Therefore, it was hypothesized that KCNQ1OT1 may be a therapeutic target for OA in the clinic. Furthermore, an increasing number of studies have reported that KCNQ1OT1 may serve vital roles in cell proliferation and inflammation (28-30). Li et al (29) revealed that silencing KCNQ1OT1 promoted cell viability, and decreased the production of TNF-α, IL-6 and IL-1β in H9c2 cells induced by oxygen and glucose deprivation/reoxygenation (29). Ye et al (30) revealed that KCNQ1OT1 suppressed the proliferation of vascular smooth muscle cells and secretion of inflammatory cytokines in intimal hyperplasia (30). Furthermore, KCNQ1OT1 has been demonstrated to have a crucial impact on OA progression by sponging the hsa-miR-1202/ETS1 axis (12). The present study reported that knockdown of KCNQ1OT1 promoted cell viability, suppressed inflammation and decreased the degradation of ECM in OA cells. These results validated the crucial impact of KCNQ1OT1 on the progression of OA in vitro. Given these results, measurements of cell viability, inflammatory factor levels and ECM degradation in OA may determine whether KCNQ1OT1 is involved in OA, thereby highlighting a potential targeted treatment for OA in the clinic.
Several studies have reported that miR-211-5p is decreased in OA tissues and serves a regulatory role in OA (18,19). Prasadam et al (18) reported that miR-211-5p was downregulated in the OA meniscectomy model and was confirmed to participate in decreasing the mineralisation capacity of ossified osteoblasts in OA. Liu and Luo (19) determined that miR-211-5p was markedly diminished in OA articular cartilage tissues, and was able to restrain ECM degradation and the production of inflammatory cytokines in OA. Similar to the aforementioned outcomes, miR-211-5p was downregulated in OA tissues and cells in the present study, and overexpression of miR-211-5p was shown to facilitate cell viability, suppress inflammation and decrease ECM degradation, implying that miR-211-5p may alleviate OA in vitro. Furthermore, previous studies have reported that lncRNAs exert their regulatory functions by interacting with miRNAs in OA (31,32). More importantly, the crucial role of PVT1/miR-211-3p axis (33) and XIST/miR-211(34) axis in OA have been verified. In the present study, miR-211-5p was identified as a target of KCNQ1OT1 and was inversely modulated by it. Furthermore, inhibition of miR-211-5p reversed the suppressive effects of sh-KCNQ1OT1 on inflammation and ECM degradation, as well as the promotive effect of sh-KCNQ1OT1 on viability in OA cells. Given these outcomes, it was deduced that knockdown of KCNQ1OT1 attenuated OA by regulating miR-211-5p in vitro.
TCF4 is a crucial risk gene on human chromosome 18 that has been reported to be associated with OA (35,36). Previous studies have confirmed the high expression of TCF4 in OA (22,36), and revealed that TCF4 may induce chondrocyte apoptosis and cartilage degradation to facilitate OA development (37). In agreement with previous studies, the present study reported that TCF4 expression was increased in OA tissues and cells, implying that TCF4 contributed toward the development of OA. Furthermore, a previous study revealed that TCF4 may serve as the target gene of numerous miRNAs in OA, including miR-137(38), miR-130a-3p (22) and miR-93-5p (37). In the present study, TCF4 was targeted and inversely regulated by miR-211-5p. Based on the aforementioned results, it was concluded that miR-211-5p exerted an inhibitory effect on the progression of OA by targeting TCF4. Furthermore, it was revealed that TCF4 was positively regulated by KCNQ1OT1, and overexpression of TCF4 reversed the suppressive effects of sh-KCNQ1OT1 on inflammation and ECM degradation, and reversed the promotive effect of sh-KCNQ1OT1 on proliferation in OA in vitro. Therefore, it was concluded that sh-KCNQ1OT1 suppressed OA progression by modulating the miR-211-5p/TCF4 axis in vitro.
In conclusion, KCNQ1OT1 and TCF4 expression was increased, whereas miR-211-5p was decreased in OA tissues and cells. It was suggested that KCNQ1OT1 acted as a sponge for miR-211-5p and TCF4 was targeted by miR-211-5p. Knockdown of KCNQ1OT1 facilitated cell viability, but inhibited inflammation and ECM degradation by mediating the miR-211-5p/TCF4 axis in vitro. However, the present study has certain limitations. There may be other possible mediators aside from TCF4. In addition, in vivo experiments were not conducted to elucidate the regulatory mechanism of the KCNQ1OT1/miR-211-5p/TCF4 axis. These limitations should be addressed in future studies. Nonetheless, the present study may provide a solid foundation for treating OA in clinical settings.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DA made substantial contributions to the conception and design of the study. TW, YG, ZC and WW made substantial contributions to the acquisition, analysis and interpretation of data, as well as the drafting and revision of the manuscript. All authors agreed to be accountable for the study, and read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University. Written informed consent was obtained from each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ashford S, Williard J. Osteoarthritis: A review. Nurse Pract. 2014;39:1–8. doi: 10.1097/01.NPR.0000445886.71205.c4. [DOI] [PubMed] [Google Scholar]
- 2.Madry H, Luyten FP, Facchini A. Biological aspects of early osteoarthritis. Knee Surg Sports Traumatol Arthrosc. 2012;20:407–422. doi: 10.1007/s00167-011-1705-8. [DOI] [PubMed] [Google Scholar]
- 3.Xia B, Di C, Zhang J, Hu S, Jin H, Tong P. Osteoarthritis pathogenesis: A review of molecular mechanisms. Calcif Tissue Int. 2014;95:495–505. doi: 10.1007/s00223-014-9917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolf AD, Pfleger B. Burden of major musculoskeletal conditions. Bull World Health Organ. 2003;81:646–656. [PMC free article] [PubMed] [Google Scholar]
- 5.Migliore A, Paoletta M, Moretti A, Liguori S, Iolascon G. The perspectives of intra-articular therapy in the management of osteoarthritis. Expert Opin Drug Deliv. 2020;17:1213–1226. doi: 10.1080/17425247.2020.1783234. [DOI] [PubMed] [Google Scholar]
- 6.Taruc-Uy RL, Lynch SA. Diagnosis and treatment of osteoarthritis. Prim Care. 2013;40:821–836. doi: 10.1016/j.pop.2013.08.003. vii. [DOI] [PubMed] [Google Scholar]
- 7.Fritah S, Niclou SP, Azuaje F. Databases for lncRNAs: A comparative evaluation of emerging tools. RNA. 2014;20:1655–1665. doi: 10.1261/rna.044040.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: Novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(e12313) doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Liu K, Tang C, Shi Z, Jing K, Zheng J. Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomed Pharmacother. 2020;128(110349) doi: 10.1016/j.biopha.2020.110349. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Zhang L, Li E, Zhang G, Hou Y, Yuan W, Qu W, Ding L. Long-chain non-coding RNA HOTAIR promotes the progression of osteoarthritis via sponging miR-20b/PTEN axis. Life Sci. 2020;253(117685) doi: 10.1016/j.lfs.2020.117685. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Huang CR, Pan S, Pang Y, Chen YS, Zha GC, Guo KJ, Zheng X. Long non-coding RNA SNHG15 is a competing endogenous RNA of miR-141-3p that prevents osteoarthritis progression by upregulating BCL2L13 expression. Int Immunopharmacol. 2020;83(106425) doi: 10.1016/j.intimp.2020.106425. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Gao J, Su G, Xiang Y, Wan L. MicroRNA-1202 plays a vital role in osteoarthritis via KCNQ1OT1 has-miR-1202-ETS1 regulatory pathway. J Orthop Surg Res. 2020;15(130) doi: 10.1186/s13018-020-01655-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swingler TE, Niu L, Smith P, Paddy P, Le L, Barter MJ, Young DA, Clark IM. The function of microRNAs in cartilage and osteoarthritis. Clin Exp Rheumatol. 2019;37 (Suppl 120):S40–S47. [PubMed] [Google Scholar]
- 14.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3(e3740) doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo C, Liang JS, Gong J, Zhang HL, Feng ZJ, Yang HT, Zhang HB, Kong QH. The function of microRNA-34a in osteoarthritis. Bratisl Lek Listy. 2019;120:386–391. doi: 10.4149/BLL_2019_063. [DOI] [PubMed] [Google Scholar]
- 16.Zhong G, Long H, Ma S, Shunhan Y, Li J, Yao J. Corrigendum to ‘miRNA-335-5p relieves chondrocyte inflammation by activating autophagy in osteoarthritis’ [Life Sci 226 (2019) 164-172] Life Sci. 2020;240(117135) doi: 10.1016/j.lfs.2019.117135. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Zhao L, Fan Y, Liao L, Ma PX, Xiao G, Chen D. The microRNAs miR-204 and miR-211 maintain joint homeostasis and protect against osteoarthritis progression. Nat Commun. 2019;10(2876) doi: 10.1038/s41467-019-10753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasadam I, Batra J, Perry S, Gu W, Crawford R, Xiao Y. Systematic identification, characterization and target gene analysis of microRNAs involved in osteoarthritis subchondral bone pathogenesis. Calcif Tissue Int. 2016;99:43–55. doi: 10.1007/s00223-016-0125-7. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Luo J. miR-211-5p contributes to chondrocyte differentiation by suppressing Fibulin-4 expression to play a role in osteoarthritis. J Biochem. 2019;166:495–502. doi: 10.1093/jb/mvz065. [DOI] [PubMed] [Google Scholar]
- 20.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Li S, Zou Y. Knockdown of lncRNA H19 relieves LPS-induced damage by modulating miR-130a in osteoarthritis. Yonsei Med J. 2019;60:381–388. doi: 10.3349/ymj.2019.60.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo X, Wang J, Wei X, Wang S, Wang A. Knockdown of lncRNA MFI2-AS1 inhibits lipopolysaccharide-induced osteoarthritis progression by miR-130a-3p/TCF4. Life Sci. 2020;240(117019) doi: 10.1016/j.lfs.2019.117019. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Endo Y, Nwawka OK, Smith S, Burket JC. Tarsometatarsal joint communication during fluoroscopy-guided therapeutic joint injections and relationship with patient age and degree of osteoarthritis. Skeletal Radiol. 2018;47:271–277. doi: 10.1007/s00256-017-2806-3. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Huang TL, Zhang GD, Jiang JT, Guo PY. lncRNA ANRIL impacts the progress of osteoarthritis via regulating proliferation and apoptosis of osteoarthritis synoviocytes. Eur Rev Med Pharmacol Sci. 2019;23:9729–9737. doi: 10.26355/eurrev_201911_19535. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Li J, Shao W, Shen N. lncRNA CTBP1-AS2 is upregulated in osteoarthritis and increases the methylation of miR-130a gene to inhibit chondrocyte proliferation. Clin Rheumatol. 2020;39:3473–3478. doi: 10.1007/s10067-020-05113-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhu JK, He TD, Wei ZX, Wang YM. lncRNA FAS-AS1 promotes the degradation of extracellular matrix of cartilage in osteoarthritis. Eur Rev Med Pharmacol Sci. 2018;22:2966–2972. doi: 10.26355/eurrev_201805_15051. [DOI] [PubMed] [Google Scholar]
- 28.Chen B, Ma J, Li C, Wang Y. Long noncoding RNA KCNQ1OT1 promotes proliferation and epithelialmesenchymal transition by regulation of SMAD4 expression in lens epithelial cells. Mol Med Rep. 2018;18:16–24. doi: 10.3892/mmr.2018.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Dai Y, Yan S, Shi Y, Han B, Li J, Cha L, Mu J. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun. 2017;491:1026–1033. doi: 10.1016/j.bbrc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Ye B, Wu ZH, Tsui TY, Zhang BF, Su X, Qiu YH, Zheng XT. lncRNA KCNQ1OT1 suppresses the inflammation and proliferation of vascular smooth muscle cells through IkappaBa in intimal hyperplasia. Mol Ther Nucleic Acids. 2020;20:62–72. doi: 10.1016/j.omtn.2020.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 2018;17:2411–2422. doi: 10.1080/15384101.2018.1526603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao L, Wang Y, Wang Q, Huang J. lncRNA FOXD2-AS1 regulates chondrocyte proliferation in osteoarthritis by acting as a sponge of miR-206 to modulate CCND1 expression. Biomed Pharmacother. 2018;106:1220–1226. doi: 10.1016/j.biopha.2018.07.048. [DOI] [PubMed] [Google Scholar]
- 33.Xu K, Meng Z, Xian XM, Deng MH, Meng QG, Fang W, Zhang D, Long X. doi: 10.1016/j.mcp.2020.101560. lncRNA PVT1 induces chondrocyte apoptosis through upregulation of TNF-alpha in synoviocytes by sponging miR-211-3p. Mol Cell Probes: 101560, 2020. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Lv G, Wang B, Kuang L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem Biophys Res Commun. 2018;503:2555–2562. doi: 10.1016/j.bbrc.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Wu L, Guo H, Sun K, Zhao X, Ma T, Jin Q. Sclerostin expression in the subchondral bone of patients with knee osteoarthritis. Int J Mol Med. 2016;38:1395–1402. doi: 10.3892/ijmm.2016.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma B, Zhong L, van Blitterswijk CA, Post JN, Karperien M. T cell factor 4 is a pro-catabolic and apoptotic factor in human articular chondrocytes by potentiating nuclear factor kappaB signaling. J Biol Chem. 2013;288:17552–17558. doi: 10.1074/jbc.M113.453985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue H, Tu Y, Ma T, Wen T, Yang T, Xue L, Cai M, Wang F, Guan M. miR-93-5p attenuates IL-1β-induced chondrocyte apoptosis and cartilage degradation in osteoarthritis partially by targeting TCF4. Bone. 2019;123:129–136. doi: 10.1016/j.bone.2019.03.035. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Fang L, Ye L, Ma S, Huang H, Lan X, Ma J. miR-137 targets the inhibition of TCF4 to reverse the progression of osteoarthritis through the AMPK/NF-κB signaling pathway. Biosci Rep. 2020;40(BSR20200466) doi: 10.1042/BSR20200466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.