Abstract
We investigated 113 adult Brazilian patients with glioblastoma (GBM) for comparison with patients from distinct geographical areas and evaluation of suitability for novel targeted therapies. Patients were assessed for clinical features and tumor genomic characteristics such as ROS1 and NTRK1 rearrangements, KIT, PDGFRA, and KDR amplification, and RB1 deletion using multicolor fluorescence in situ hybridization. The majority of patients were male (53%), over 40 years (94%), with tumor located in single site (64%), in the right cerebral hemisphere (60%), and underwent partial resection (71%); 14% presented complications after surgery. The main clinical sign at diagnosis was focal abnormality (57%); frontal (31%); and temporal (20%) regions were most commonly affected. Median hospitalization time was 20 days, median survival was 175 days. One tumor was positive for rearrangement in NTRK1 and another in ROS1 (0.9% each). PDGFRA was amplified in 20% of cases, often co-amplified with KDR (>90%) and KIT (>60%). RB1 was deleted in 16% of patients. There was no association between these molecular abnormalities and patient survival. However, older age, complications after surgery, and right-sided tumors were independent variables associated with patient survival. This study contributes information on the molecular profile of glioblastomas in Latin America possibly supporting new target therapies.
Keywords: Fluorescence in situ hybridization, Glioblastoma, KDR, KIT, NTRK1, PDGFRA, ROS1
INTRODUCTION
Malignant central nervous system (CNS) neoplasms are among the most feared human cancers, mainly because patients have a very poor prognosis and the disease directly impacts the quality of life and cognitive function. Glioblastoma (GBM) is a high-grade glioma with predominantly astrocytic differentiation with nuclear atypia, cellular pleomorphism, mitotic activity, microvascular proliferation, and necrosis (1), which despite current therapies usually causes patient’s death within 1 year after diagnosis (2). GBM is the most frequent primary CNS tumor, with an annual incidence of 3.2 per 100 000 in the United States (3). The frequency of GBM in Brazil has not been reported, but incidence of CNS tumors in 2012 was 5.7 per 100 000, the highest rate in comparison to other countries in South America and similar to Canada and Australia (4).
Genetic profile of solid tumors may differ according to geographic areas. In non-small cell lung carcinomas, for instance, frequencies of the most common activating EGFR mutations are twice as high in patients of Asian origin (>30%) compared to their Caucasian (10%–15%) and African-American (<5%) counterparts (5, 6). Importantly, numerous molecular tumor markers such as EGFR mutations are common. These studies aim not only to improve the quality and precision of therapies, but also to guide the implementation of new assays in molecular diagnostic laboratories, and the regulation and approval of new cancer drugs, to determine objective response to specific targeted therapies, and to provide a significantly better overall survival (OS). There is indication that molecular diversity also occurs among GBMs (Supplementary DataTables S1 and S2), which justifies studies in geographical regions such as Brazil and other Latin America countries.
Current treatment for GBM combines surgery, radiation therapy, and chemotherapy, but these interventions only provide a small survival benefit. Patients who underwent chemoradiation followed by maintenance treatment with temozolomide and alternating electric fields via transducer arrays applied to the scalp showed significantly longer, although still poor, OS (7). Recently, genomic studies have been performed in GBMs aiming to identify oncogenic pathways that could be potentially inhibited by new targeted therapies. Genomic alterations activating oncogenes (mainly receptor tyrosine kinases, RTKs) and inactivating tumor suppressor genes have been reported in GBMs. RTKs are activated by point mutations, gene amplifications (8), and gene fusions (9). Among the clinically relevant oncogenic pathways in GBM are the ones including the genes NTRK1 and ROS1, for which numerous specific inhibitors are already approved or under development (10). Several molecular fusions involving these genes have been found in GBM, such as BCAN-NTRK1, NFASC-NTRK (9, 11), ARHGEF2-NTRK1, CHTOP-NTRK1 (12), GOPC-ROS1 (13), and CEP85L-ROS1 (9). Amplifications of the KIT, PDGFRA, and KDR genes have also been reported (14), and there are drugs in development for brain tumors with such molecular profiles (15). Loss of tumor suppressor genes may also be therapeutically targeted and there are studies evaluating the response of antitumor drug acting on the RB1 pathway, thus supporting stratification of patients based on the GBM’s RB1 suppressor gene status (16). Moreover, heterogeneity in genetic abnormalities is an outstanding feature of GBM that may contribute to response to treatment (17).
This study aimed to examine for the first time the frequency and clinical significance of rearrangements in ROS1 and NTRK1, amplification of KIT, PDGFRA, and KDR, and deletion of RB1 in a Brazilian cohort of adult patients with GBM, using fluorescence in situ hybridization (FISH).
MATERIALS AND METHODS
Sample
A total of 113 patients subjected to resection of GBM in 2 Southern Brazilian hospitals (Porto Alegre, RS) from January 2009 to May 2015 were included in the study after approval by the Institutions’ Research Review Boards. Formalin-fixed paraffin-embedded (FFPE) tumor sections were centrally revised by a pathologist (V.O.P.), and GBM diagnosis was confirmed according to the 2007 World Health Organization (WHO) Classification of Tumors of the CNS (18). Since these tumors were not tested for IDH mutations, according to the 2016 WHO Classification (1) they are designated as GBM NOS. Demographic profile of patients was noted, including data about clinical profile, tumor characteristics, and OS. For survival evaluation, patients were divided based on age distribution as <40, 40–59, and >60 years old (19). For clinical signs and symptoms, groups were created for focal disturbance (i.e. aphasias and motor deficit), increased intracranial pressure (i.e. headache, vomiting, and nausea), behavioral change (i.e. mental confusion and disorientation), seizure and decreased consciousness (i.e. sensory changes and somnolence) (20, 21). The anatomical site of tumor was defined as frontal, temporal, parietal, or other (i.e. corpus callosum, occipital region, or more than a single site) (19, 22). The extension of the resection was classified as “partial” (10%–90% resected) or “gross-total resection” (more than 90% resected) (22). Cases with a previous low-grade tumor were considered CNS recurrences. Complications after surgery were noted until hospital discharge. The median follow-up of the study was 175 days (minimum of 3 days, maximum of 7 years).
FISH Assays
FISH assays were performed on 4-μm FFPE sections using commercial (NTRK1, KIT, CEP4, RB1, and LSI 13q34 from Abbott Molecular, Abbott Park, IL, USA; and ROS1 from Cytocell Ltd, Cambridge, UK) or laboratory developed DNA probes (PDGFR and KDR) diluted in tDenHyb-2 hybridization buffer (Insitus Biotechnologies, Albuquerque, NM) as described in Supplementary DataTable S3.
In the FISH break-apart assays, a specimen was considered positive for NTRK1 and ROS1 rearrangement when ≥15% of tumor cells showed single 3′ or single 5′ gene signals, or 5′-3′ gene signals split by more than 1 signal diameter (23). In the FISH enumeration assays, gene amplification was accepted when the gene: control ratio was ≥2, or there was ≥15 copies of gene in ≥10% of tumor cells (24). In the RB1 FISH assay, deletion was considered when ratio gene: control RB1:13q34 < 0.8 (25) or there was a single copy of gene signal in >50% of tumor nuclei (16).
Statistical Analysis
The quantitative variables were described as mean (and standard deviation) or median (and interquartile range). Absolute and relative frequencies were used for categorical variables, as well as Chi-square test with Yates correction and Fisher exact test. To evaluate survival, Kaplan-Meier method was applied and the curves were compared using the log-rank test. The confounding factors were controlled by multivariate Cox proportional hazards model when the variables presented p < 0.25 in the univariate analysis. The effect measure used was the hazard ratio in conjunction with a confidence interval of 95%. The significance level adopted was 5% (p < 0.05) and the analyses were performed in the SPSS program version 21.0.
RESULTS
Clinical findings of the 113 patients are summarized (Table 1) and presented in detail (Supplementary DataTable S4). Approximately half of patients (53.1%) were male, and their age ranged from 18 to 83 years (mean 56.9, SD 12.3; 93.8% were over 40 years). The main clinical sign at diagnosis was focal disturbance (57.1%), and seizure was described in 16.1% of patients. Frontal (31%) and temporal (20.4%) regions were the most affected regions, with the tumors mainly located on the right cerebral hemisphere (59.5%). Most patients had a single site affected (63.7%) and underwent partial tumor resection (70.6%). Cases that presented previously a low-grade tumor are indicated in Supplementary DataTable S4. Fourteen patients (13.7%) presented complications after surgery. The median hospitalization time was 20 days (IQR: 13.5; 28) and the median OS was 175 days (95% CI: 126.1–223.9) for a follow-up of 21–60 months. At the end of the study, only 4 patients (3.5%) were still alive. The observed survival rates were 23% at 1 year and 3.4% at 5 years.
TABLE 1.
Clinical and Molecular Characteristics of the Patients (n = 113)
| Variables | N (%) |
|---|---|
| Sex (n = 113) | |
| Male | 60 (53.1) |
| Female | 53 (46.9) |
| Age (n = 113) mean 56.9 years; SD 12.3 years | |
| <40 years | 7 (6.2) |
| 40–59 years | 54 (47.8) |
| ≥60 years | 52 (46.0) |
| Clinical feature groups (n = 112) | |
| Focal disturbance | 64 (57.1) |
| Increase ICP | 58 (51.8) |
| Behavioral change | 27 (24.1) |
| Seizure | 18 (16.1) |
| Decreased consciousness | 14 (12.5) |
| Tumor location (n = 113) | |
| Frontal | 35 (31.0) |
| Temporal | 23 (20.4) |
| Parietal | 11 (9.7) |
| Other | 44 (38.9) |
| Tumor side (n = 111) | |
| Right | 66 (59.5) |
| Left | 43 (38.7) |
| Bilateral | 2 (1.8) |
| Number of involved regions in brain (n = 113) | |
| One region | 72 (63.7) |
| Two or more regions | 41 (36.3) |
| Prior CNS tumor (n = 102) | |
| No | 95 (93.1) |
| Yes | 7 (6.9) |
| Resection (n = 109) | |
| Partial | 77 (70.6) |
| Gross total | 32 (29.4) |
| Complications after surgery (n = 102) | |
| No | 88 (86.3) |
| Yes | 14 (13.7) |
| NTRK1 rearrangement (n = 113) | |
| No | 112 (99.1) |
| Yes | 1 (0.9) |
| ROS1 rearrangement (n = 113) | |
| No | 112 (99.1) |
| Yes | 1 (0.9) |
| KIT amplification (n = 113) | |
| No | 99 (87.6) |
| Yes | 14 (12.4) |
| PDGFRA amplification (n = 113) | |
| No | 91 (80.5) |
| Yes | 22 (19.5) |
| KDR amplification (n = 113) | |
| No | 93 (82.3) |
| Yes | 20 (17.7) |
| RB1 deletion (n = 113) | |
| No | 95 (84.1) |
| Yes | 18 (15.9) |
N, number of patients; %, percentage; ICP, intracranial pressure; CNS, central nervous system.
The independent variables associated with patient survival in multivariate analyses were age (p = 0.009), tumor side (p = 0.022), and complications after surgery (p = 0.003) (Table 2). Patients older than 60 years presented a risk 3.38 times higher for death when compared with those younger than 40 years. Patients with tumor located on the left side had 42% lower chance of death when compared with those with tumor on the right side. Clinical complications after surgery were significantly associated with decreased survival (p = 0.003), with a risk 2.69 times higher for death.
TABLE 2.
Multivariate Analysis (Cox Regression) of Variables Associated With Survival
| Variable | Number of Patients | Median Survival Days (95% CI) | 1-Year Survival | 5-Year Survival | HR (95% CI)* | p Value |
|---|---|---|---|---|---|---|
| Age | ||||||
| <40 years | 7 | 619 (0–1422) | 57.1% | 14.3% | 1.00 | — |
| 40–59 years | 54 | 220 (123–317) | 27.8% | 4.6% | 1.74 (0.72–4.21) | 0.223 |
| ≥60 years | 52 | 123 (75.9–170) | 13.5% | 0 | 3.38 (1.35–8.84) | 0.009 |
| Tumor side | ||||||
| Right | 66 | 164 (130–197) | 21.2% | 0 | 1.00 | — |
| Left | 43 | 220 (111–329) | 25.6% | 7% | 0.58 (0.37–0.92) | 0.022 |
| Complications after surgery | ||||||
| No | 88 | 216 (160–271) | 26.1% | 3.4% | 1.00 | — |
| Yes | 14 | 77 (0–161) | 0 | 0 | 2.69 (1.40–5.18) | 0.003 |
Adjusted by age, increase ICP (intracranial pressure), frontal location, tumor side, number of involved regions, resection, complication.
HR, hazard ratio; %, percentage; CI, confidence interval.
The molecular cytogenetic results are summarized (Table 1) and presented in detail (Supplementary DataTable S5). Molecular fusions were rare: case 61 was positive for NTRK1 rearrangement (Fig. 1A) with a single 5′NTRK1 signal pattern, and case 31 was positive for ROS1 gene rearrangement (Fig. 1B) with a single 3′ROS1 signal pattern. Neither of these 2 patients had remarkable clinical features. Attempts to identify the fusion partners in these rearrangements were unsuccessful due to inadequate quality of DNA preservation.
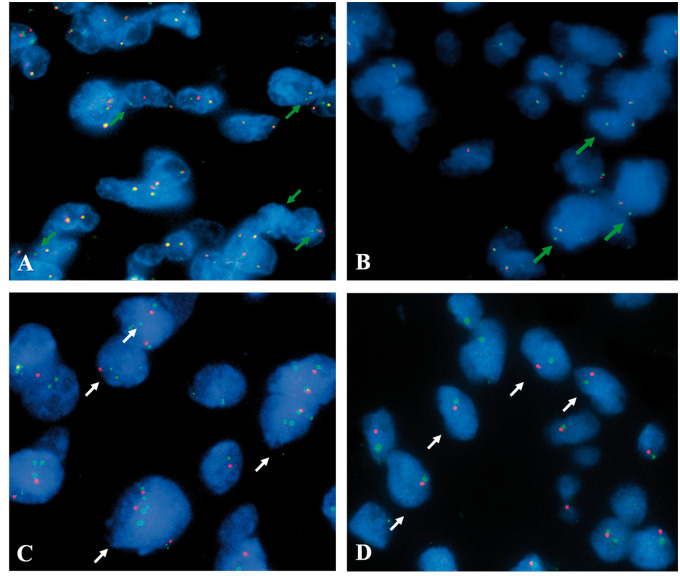
FIGURE 1.
Microscope fields of tumors showing positive patterns identified by the presence of single green signals (green arrows) for NTRK1(A) in GMB case 61, and ROS1 rearrangements (B) in GBM case 31. (C) Microscope fields showing tumor cells with loss of the RB1 gene, with unbalanced loss of RB1 (red signal) in comparison with the control probe 13q34 (green signal) in GBM case 41. (D) Case 96 with a pattern suggestive of monosomy 13 in GBM case 96. Representative nuclei are indicated by white arrows.
In the gene amplification analyses (Table 1), PDGFRA was most commonly amplified (19.5%), followed by KDR (17.7%) and KIT (12.4%), and most often the 3 genes were co-amplified. Among the 19.5% of cases harboring gene amplification in the 4q12 locus, 63.6% (14 cases) had co-amplification of the 3 genes, 27.3% (6 cases) had co-amplification of PDGFRA and KDR and 9.1% (2 cases) had only PDGFRA amplification. An amplification-positive case for all 3 genes is illustrated in Figure 2A, and examples of co-amplification of PDGFRA and KDR, and of amplification of PDGFRA alone are shown in Figures 2B and 3, respectively. We confirmed that the selected criteria for definition of amplification had not excluded specimens harboring high gene copy numbers by verifying that the few borderline negative specimens had on average less than 5 gene copies per cell. One interesting observation was the intra-tumoral heterogeneity regarding the distribution of the amplification within the tumors. Out of the 22 amplified cases, the amplification was diffusely distributed in 16 cases while it was confined to specific tumor areas (focal distribution) in 6 cases.
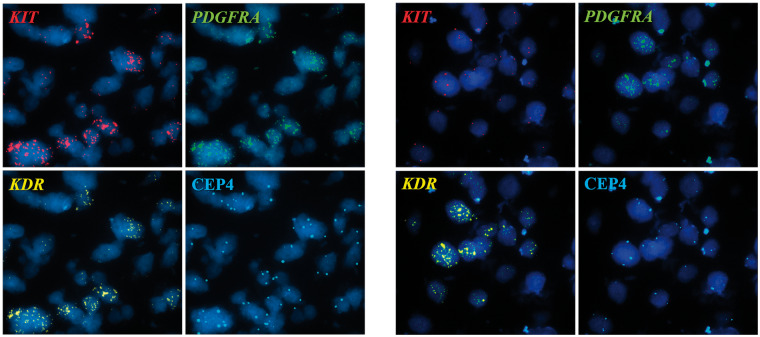
FIGURE 2.
Segmented images of microscope fields showing co-amplification of genes. KIT gene shown in red signal; PDGFRA gene shown in green signal, and KDR gene shown in yellow signal; CEP4 (aqua signal) corresponded to a control probe recognizing the centromere of chromosome 4. (A) GBM case 70 showing co-amplification of KIT, PDGFRA, and KDR. (B) GBM case 69 showing co-amplification of the PDGFRA and KDR genes, and KIT gene with similar copy number as the control CEP4 (aqua signal).
FIGURE 3.
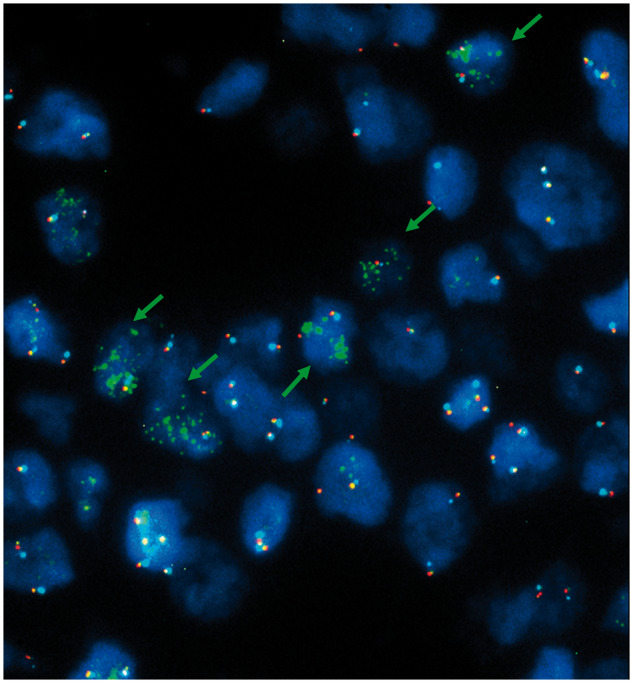
Microscope field illustrating amplification only for the PDGFRA gene (green signals) in case 85 (green arrows).
In the RB1 deletion analyses, 18 cases (15.9%) presented loss of the gene and this phenotype had no association with survival (p = 0.167). Half of cases met both criteria for deletion, whereas 4 tumors were positive only by the ratio gene: control RB1:13q34 < 0.8 (25) criterion, comprising cases with gene deletion followed by chromosome duplication (Fig. 1C), and 5 tumors qualified for deletion based only on the criterion of >50% of tumor nuclei with a single copy of gene signal (16), comprising cases carrying loss of both gene and control likely due to chromosome 13 monosomy (Fig. 1D).
A subset of this cohort (n = 40) was previously investigated (26) for the frequency and clinical significance of EGFR amplification, aneuploidy of chromosomes 7 (gain) and 10 (loss), and deletions in PTEN, TP53, and 1p/19q. Supplementary DataTable S6 combines all molecular data for that patient subset and a summary is presented in Table 3. None of these 40 patients carried 1p/19q co-deletion or rearrangements in ROS1 or NTRK1 thus these 3 markers were excluded. Overlap of genetic alterations detected in each of the studies was only seen in 11 cases (27.5%). EGFR amplification and monosomy of chromosome 10, 2 of the most common abnormalities observed by Koshiyama et al (26), were rarely found in tumors harboring amplification of KIT, PDGFRA, or KDR, the most common abnormalities in this study. However, amplifications in the later genes were found in about 25% of patients with PTEN or TP53 deletions, while these deletions were only rarely concurrent with RB1 deletions.
TABLE 3.
Frequency of Concurrent Positive Biomarkers in the Subset of 40 Adult GBM Cases Combining Data From Previous (Koshiyama et al [26]) and Current Study
| Concurrently Positive | Biomarker Positive for Koshiyama et al (26) With KIT Amplification |
Biomarker Positive for Koshiyama et al (26) With PDGFRA Amplification |
Biomarker Positive for Koshiyama et al (26) With KDR Amplification |
Biomarker Positive for Koshiyama et al (26) With RB Deletion |
||||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | |
| EGFR | 1 | 5.9 | 3 | 17.6 | 1 | 5.9 | 2 | 11.8 |
| Polysomy 7 | 2 | 10.0 | 4 | 20.0 | 3 | 15.0 | 2 | 10.0 |
| Monosomy 10 | 1 | 4.8 | 3 | 14.3 | 1 | 4.8 | 2 | 9.5 |
| PTEN | 4 | 28.6 | 5 | 35.7 | 4 | 28.6 | 1 | 7.1 |
| TP53 | 2 | 22.2 | 3 | 33.3 | 3 | 33.3 | 1 | 11.1 |
DISCUSSION
Few studies have examined adult onset GBM in Brazil for molecular profile, clinical findings, and survival (26–29). Two studies (26, 29) also applied molecular cytogenetic technology (as in our study) but focused on different genes, which highlights the relevance of this study. The prevalence in the cohort of males (M:F = 1.13:1) and older adults (55–85 years) agrees with data from the United States, Switzerland (1) and Brazil (26–28). Our study was intentionally designed to target adult population and showed that older age seemed to associate with poor prognosis, in agreement with the literature (26, 27, 29–31). The worse prognosis of elderly patients may be explained by their higher comorbidities and lower resistance to medical procedures and treatment (2, 32), but in developing countries the delay in disease diagnosis is also an important negative factor.
GBMs develop rapidly and the symptoms depend especially on the tumor location and size. Common symptoms include focal neurological signs, seizures, mood, and personality changes, or symptoms of increased intracranial pressure (21). The majority of patients in this cohort presented 1 type of focal disturbance (57.1%) and increased intracranial pressure (51.8%) (Table 1). Occurrence of seizures has been reported in 20%–76% of patients and we observed it in 16.1%. Seizure can correlate with a better survival, since it may be linked to reduced tumor aggressiveness and it could be a factor for earlier diagnosis (30).
Few studies have evaluated the impact of the brain side in which the tumor developed. Connon et al (32) observed more right-side tumors, in agreement with our study (59.5%), while Mariniello et al verified more cases with left-side tumors (33). To the best of our knowledge, there was no previous report of tumor-side association with survival, whereas in our study the patients with left-side tumors presented a lower risk of death. This observation may be due to the location on the left side of the Wernicke and Broca's areas, responsible for speech, comprehension, or understanding of written and spoken language, which generate symptoms leading to earlier diagnosis (31). The majority of cases with 1 type of aphasia (73.7%) had the tumor located on the left side. In this Brazilian cohort, most tumors were located on frontal and temporal lobes and no association was found with survival. However, there are reports that frontal and temporal tumors are typically associated with fewer symptoms or vague effects (i.e. behavioral changes and hearing or speech difficulties), which could be easily confounded with typical aging changes, resulting in late diagnosis, and consequently worse prognosis (32).
The extent of the brain tumor resection is highly dependent on its location, size, and difficulty in accessing. The majority of our cases underwent partial resection (70.6%), in agreement with other studies (19, 31). Cases that underwent diagnosis through biopsy were not included, mainly to ensure enough tissue to perform the molecular analysis, since necrotic areas unsuitable for analysis are common in GBM. There was no association between type of resection (gross total vs partial) and survival, as also found by Korshunov et al (34) in high-grade gliomas. In the present investigation, the occurrence of clinical complications was significantly associated with decreased survival in agreement with a German study (35), and there was no statistically significant difference in the median time of hospitalization between patients with (21 days, CI: 16–45.5) and without complications (19 days, CI: 13–26) (p = 0.070).
The overall median survival rate (OS) for these Brazilian GBM patients was 175 days (5.8 months), which is somewhat similar to reports from other regions, such as Australia (OS = 4.7 months in patients over 80 years) (32) and Switzerland (OS = 4.9 months in patients at different ages) (2). Some other European cohorts had higher OS (9.5–12.4 months) (30, 35, 36). The rates were lower for 1-year survival and similar for 5-year survival in our cohort compared to cohorts from other regions (28%–68% and 2.9%–14%, respectively) (2, 30, 36). It is noteworthy that Brazilian investigators have associated low socio-economic level with poor prognosis for tumors such as breast and uterine cancers as consequence of late diagnosis in public health care services (37, 38). Similar rationale could explain why the GBM patients in our sample had a poorer prognosis when compared with the patients from developed regions.
Rearrangements involving neurotrophic tropomyosin receptor kinase type 1 (NTRK1) are recurrent in papillary thyroid, lung, and colon cancers (39). The fusions reported seem to contribute to the initiation and maintenance of malignancy (11) and thus are good targets for new drugs in cancer therapy. In-frame fusions involving NTRK1 were found in different studies with GBM samples (9, 11). Zheng et al (12) reported an in-frame fusion involving ARHGEF2 and NTRK1, and 2 in-frame fusions involving CHTOP and NTRK1 on a brain tumor set of 115 samples. The ROS proto-oncogene 1 (ROS1) belongs to a subfamily of insulin RTK genes and is activated by fusion in a variety of tumors, with at least 26 different partners (40). The first fusion protein discovered in GBM involved ROS1 and GOPC (alias FIG), in which an intra-chromosomal deletion of 240 kb led to a constitutively active kinase, suggesting an oncogenic activity (13). In lung cancer, ROS1 rearrangements define a unique molecular subclass that respond to the tyrosine kinase inhibitor crizotinib (41). This study includes the first report of positive GBM case for each of the NTRK1 and ROS1 rearrangements in Brazilian patients, and this low frequency is in accordance with the literature (9, 11, 12).
The gene RB transcriptional corepressor 1 (RB1) encodes the retinoblastoma protein (Rb), a component of regulatory of the cell cycle (16). Brennan et al (8) reported Rb function loss in 7.6% of GBM samples by direct RB1 deletion/mutation, a lower frequency than in our study (15.9%). However, Rodriguez et al (25) found RB1 deletion/chromosome 13 monosomy in 25% of GBM samples, and no association between this alteration and survival was detected. Other authors have demonstrated higher proportions of RB1 deletion in GBM (34%), but they have not identified association between RB1 status and survival (16). Bäcklund et al (42) also found no association between RB1 deletion and survival but there was a significantly shorter median survival when alterations in numerous genes on the Rb pathway (CDKN2A, CDKN2B, RB1, and CDK4) were grouped.
The 4q12 chromosome region harbors 3 genes encoding potentially drug-targeted RTKs: KIT (proto-oncogene RTK), PDGFRA (platelet-derived growth factor receptor alpha) and KDR (kinase insert domain receptor), of which the main mechanism of activation is amplification, reported in frequencies ranging from 4.4% to 21.6%, 8.5% to 36%, and 3.3% to 36%, respectively (8, 14, 43, 44). In our cohort, we verified that amplification frequencies were within the ranges described in the literature, but no significant correlation was found between the amplification and patients’ OS, which is consistent with the reports by Joensu et al (43) and Nobusawa et al (44). However, Burford et al (14) found that amplifications of these genes were individually associated with poor clinical outcome in GBM patients. However, it cannot be ruled out that these discrepant results are due to small sample sizes.
Among the cases classified as positive for amplification in our study, 27% presented heterogeneity in the intra-tumoral distribution of amplifications, similarly to the description by Snuderl et al (17), who also used FISH technology. FISH is an optimal platform to detect tumor subpopulations due to its in situ nature. Intra-tumor heterogeneity was also reported in GBM for expression pattern of the PDGF genes (28), and has been postulated as partially responsible for the failures in clinical trials using targeted therapies against RTKs in GBM (17).
Despite the variety of genetic alterations that may occur in GBM, there are 3 major cellular pathways involved: RTK/PI3K/PTEN and the suppressors p53 and Rb1. Most GBMs present aberrations in all 3 pathways (45), but in the subset of 40 tumors more extensively analyzed, amplification of the EGFR and the PDGFRA/KIT/KDR genes were almost non-overlapped. This complex network of changes plays an important role in the tumor evolution and proliferative advantage and must be considered in adaptive response of new therapies in development (17).
Novel therapeutic agents targeting the oncogenic pathways investigated in this study are in progress. A search on the website ClinicalTrials.gov (46) on July 2, 2018 identified 4 clinical trials in phases 1 and 2 with different NTRK1 inhibitors and tumors, including GBM. No studies with GBM and ROS1 inhibitors were listed, but there were 60 studies in different phases evaluating this gene in other tumors, especially in lung cancer. In trials involving KIT, PDGFRA, and KDR inhibitors, there were respectively 3, 4, and 8 trials in different phases and using distinct drugs, including patients with GBM. Agents targeting the Rb pathway were listed in one phase 1 clinical trial, including GBM.
We did not investigate IDH mutation in this sample mainly because the project was approved and all tumors were collected before the release of the 2016 WHO Classification of CNS tumors. In addition, this document identified the mean age at diagnosis of IDH-mutant GBM as close to 44 years and the median as 48 years. The mean age of our GBM population was 56.9 years (SD 12.3, median 58 years); 94% of patients were ≥40 years and 62% were ≥55 years old. Therefore, based on this parameter, there is a reduced likelihood of impact of IDH mutations in our results.
There are still too few studies providing molecular tumor data in Latin American and specifically in Brazilian patients and this study aims to contribute toward a larger and accurate regional molecular profiling of adult GBM. NTRK1 and ROS1 gene fusions are rare in GBMs from Southern Brazil and it is uncertain whether they correlate with clinical or demographic characteristics. Deletion of RB1 and amplification of the KIT, PDGFRA, and KDR genes are more frequent events and, although these markers did not appear to impact patient’s survival, they comprise targets for new therapeutic regimens. Importantly, amplifications in PDGFRA/KIT/KDR and in EGFR were largely nonconcurrent. This study has shown similarities to patient populations from other areas and supports the accrual of Brazilian patients with GBM for clinical trials focusing on drugs targeting amplification/overexpression of EGFR and PDGFRA but not for ROS1 and NTRK1 tyrosine kinase inhibitors. Moreover, it is noteworthy that older age, complications after surgery and right-sided tumors were associated with patient survival and these variables may have been impacted by the suboptimal conditions of health care systems in developing countries that lead to late cancer diagnosis and delayed treatment.
Supplementary Material
This work was supported by the Molecular Pathology Shared Resource of the University of Colorado Cancer Center (NCI P30 CA046934), the research productivity fellowship Brazil from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (301834/2016-4), and the scholarship provided by the Brazilian Agency Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) to P.T.
The authors have no duality or conflicts of interest to declare.
Supplementary Data can be found at academic.oup.com/jnen.
REFERENCES
- 1. Louis DN, Ohgaki H, Wiestler OD, et al., eds. World Health Organization Histological Classification of Tumours of the Central Nervous System, Revised 4th ed. Lyon: International Agency for Research on Cancer (IARC; ) 2016:408 [Google Scholar]
- 2. Ohgaki H, Dessen P, Jourde B. Genetic pathways to glioblastoma: A population-based study. Cancer Res 2004;64:6892–9 [DOI] [PubMed] [Google Scholar]
- 3. Ostrom QT, Gittleman H, Xu J, et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol 2016;18:v1–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GloboCan 2012. Cancer Incidence and Mortality Worldwide. International Agency for Research on Cancer (IARC). Online Analysis. Available at: http://globocan.iarc.fr/Pages/burden_sel.aspx. Accessed February 18, 2018
- 5. Zhou W, Christiani DC. East meets West: Ethnic differences in epidemiology and clinical behaviors of lung cancer between East Asians and Caucasians. Chin J Cancer 2011;30:287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leidner RS, Fu P, Clifford B, et al. Genetic abnormalities of the EGFR pathway in African American patients with non-small-cell lung cancer. JCO 2009;27:5620–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA 2017;318:2306–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah N, Lankerovich M, Lee H, et al. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics 2013;14:818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ardini E, Menichincheri M, Banfi P, et al. Entrectinib, a pan-TRK, ROS1, and ALK inhibitor with activity in multiple molecularly defined cancer indications. Mol Cancer Ther 2016;15:628–39 [DOI] [PubMed] [Google Scholar]
- 11. Kim J, Lee Y, Cho HJ, et al. NTRK1 fusion in glioblastoma multiforme. PLoS One 2014;9:e91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479–84 [DOI] [PubMed] [Google Scholar]
- 13. Charest A, Lane K, McMahon K, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer 2003;37:58–71 [DOI] [PubMed] [Google Scholar]
- 14. Burford A, Little SE, Jury A, et al. Distinct phenotypic differences associated with differential amplification of receptor tyrosine kinase genes at 4q12 in glioblastoma. PLoS One 2013;8:e71777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng S, Dhruv H, Armstrong B, et al. Integrated genomic analysis of survival outliers in glioblastoma. Neuro Oncol 2017;19:833–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldhoff P, Clarke J, Smirnov I, et al. Clinical stratification of glioblastoma based on alterations in retinoblastoma tumor suppressor protein (RB1) and association with the proneural subtype. J Neuropathol Exp Neurol 2012;71:83–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 2011;20:810–7 [DOI] [PubMed] [Google Scholar]
- 18. Kleihues P, Burger PC, Aldape KD, et al. Glioblastoma. In: Louis DN, Ohgaki H, Wiestler OD, et al., eds. WHO Classification of Tumours of the Central Nervous System, 4th ed. Lyon: International Agency for Research on Cancer (IARC: ) 2007:33–49 [Google Scholar]
- 19. Simpson JR, Horton J, Scott C, et al. Influence of location and extent of surgical resection on survival of patients with glioblastoma multiforme: Results of three consecutive Radiation Therapy Oncology Group (RTOG) clinical trials. Int J Radiat Oncol Biol Phys 1993;26:239–44 [DOI] [PubMed] [Google Scholar]
- 20. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372:2481–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Preusser M, de Ribaupierre S, Wöhrer A, et al. Current concepts and management of glioblastoma. Ann Neurol 2011;70:9–21 [DOI] [PubMed] [Google Scholar]
- 22. Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: Recursive partitioning analysis. Neuro Oncol 2004;6:227–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013;19:1469–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 2005;97:643–55 [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez FJ, Scheithauer BW, Giannini C, et al. Epithelial and pseudoepithelial differentiation in glioblastoma and gliosarcoma: A comparative morphologic and molecular genetic study. Cancer 2008;113:2779–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koshiyama DB, Trevisan P, Graziadio C, et al. Frequency and clinical significance of chromosome 7 and 10 aneuploidies, amplification of the EGFR gene, deletion of PTEN and TP53 genes, and 1p/19q deficiency in a sample of adult patients diagnosed with glioblastoma from Southern Brazil. J Neurooncol 2017;135:465–72 [DOI] [PubMed] [Google Scholar]
- 27. Uno M, Oba-Shinjo SM, Silva R, et al. IDH1 mutations in a Brazilian series of glioblastoma. Clinics (Sao Paulo) 2011;66:163–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cantanhede IG, de Oliveira JRM. PDGF family expression in glioblastoma multiforme: Data compilation from Ivy Glioblastoma Atlas Project database. Sci Rep 2017;7:15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinto LW, Araújo MB, Vettore AL, et al. Glioblastomas: Correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Arch 2008;452:481–90 [DOI] [PubMed] [Google Scholar]
- 30. Toledo M, Sarria-Estrada S, Quintana M, et al. Prognostic implications of epilepsy in glioblastomas. Clin Neurol Neurosurg 2015;139:166–71 [DOI] [PubMed] [Google Scholar]
- 31. Liu TT, Achrol AS, Mitchell LA, et al. Computational identification of tumor anatomic location associated with survival in 2 large cohorts of human primary glioblastomas. AJNR Am J Neuroradiol 2016;37:621–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Connon FV, Rosenthal MA, Drummond K. Glioblastoma multiforme in the very elderly. Neurosurg Rev 2016;39:55–61 [DOI] [PubMed] [Google Scholar]
- 33. Mariniello G, Peca C, De Caro MB, et al. Glioblastoma in the elderly: The impact of advanced age on treatment and survival. J Neurol Surg A Cent Eur Neurosurg 2014;75:276–81 [DOI] [PubMed] [Google Scholar]
- 34. Korshunov A, Sycheva R, Golanov A. Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: The utility of fluorescence in situ hybridization. Clin Cancer Res 2004;10:7820–6 [DOI] [PubMed] [Google Scholar]
- 35. Ening G, Osterheld F, Capper D, et al. Risk factors for glioblastoma therapy associated complications. Clin Neurol Neurosurg 2015;134:55–9 [DOI] [PubMed] [Google Scholar]
- 36. Korshunov A, Sycheva R, Golanov A. The prognostic relevance of molecular alterations in glioblastomas for patients age < 50 years. Cancer 2005;104:825–32 [DOI] [PubMed] [Google Scholar]
- 37. Caetano R, Vianna CMM, Thuler LCS, et al. Custo-efetividade no diagnóstico precoce do cancer do colo uterino no Brasil. Rev Saúde Coletiva 2006;16:99–118 [Google Scholar]
- 38. Schneider IJC, d'Orsi E. Sobrevida em cinco anos e fatores prognósticos em mulheres com cancer de mama em Santa Catarina, Brasil. Caderno Saúde Pública 2009;25:1285–96 [DOI] [PubMed] [Google Scholar]
- 39. Park DY, Choi C, Shin E, et al. NTRK1 fusions for the therapeutic intervention of Korean patients with colon cancer. Oncotarget 2016;7:8399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uguen A, De Braekeleer M. ROS1 fusions in cancer: A review. Future Oncol 2016;12:1911–28 [DOI] [PubMed] [Google Scholar]
- 41. Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bäcklund LM, Nilsson BR, Goike HM, et al. Short postoperative survival for glioblastoma patients with a dysfunctional RB1 pathway in combination with no wild-type PTEN. Clin Cancer Res 2003;9:4151–8 [PubMed] [Google Scholar]
- 43. Joensuu H, Puputti M, Sihto H, et al. Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J Pathol 2005;207:224–31 [DOI] [PubMed] [Google Scholar]
- 44. Nobusawa S, Stawski R, Kim YH, et al. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: A population-based study. Neuropathology 2011;31:583–8 [DOI] [PubMed] [Google Scholar]
- 45. Belden CJ, Valdes PA, Ran C, et al. Genetics of glioblastoma: A window into its imaging and histopathologic variability. Radiographics 2011;31:1717–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.US National Library of Medicine: ClinicalTrials.gov. Available at: https://clinicaltrials.gov/. Accessed July 2, 2018
- 47. Nobusawa S, Lachuer J, Wierinckx A, et al. Intratumoral patterns of genomic imbalance in glioblastomas. Brain Pathol 2010;20:936–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Little SE, Popov S, Jury A, et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res 2012;72:1614–20 [DOI] [PubMed] [Google Scholar]
- 50. Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A 2012;109:3041–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phillips JJ, Aranda D, Ellison DW, et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol 2013;23:565–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Becker AP, Scapulatempo-Neto C, Carloni AC, et al. KIAA1549: BRAF gene fusion and FGFR1 hotspot mutations are prognostic factors in pilocytic astrocytomas. J Neuropathol Exp Neurol 2015;74:743–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.