Abstract
Glioma therapeutic resistance to alkylating chemotherapy is mediated via O6-methylguanine-DNA methyltransferase (MGMT). We hypothesized that a CD45/HAM56/MGMT double-stained cocktail would improve MGMT discrimination in tumor cells versus inflammatory and endothelial cells (IEC). Total MGMT protein was quantified by IHC on 982 glioblastomas (GBM) and 199 anaplastic astrocytomas. Correcting for IEC was done by a CD45/HAM56/MGMT 2-color cocktail. Lowest IEC infiltrates (IEC “cold spots”) were identified to quantitate MGMT as well as the percentage of IEC% in the IEC cold spots. MGMT promoter methylation (PM) was also determined. Among the GBM biopsies, mean uncorrected and corrected MGMT% were 19.87 (range 0–90) and 16.67; mean IEC% was 18.65 (range 1–80). Four hundred and fifty one (45.9%) GBM biopsies were positive MGMT PM. Both uncorrected and corrected MGMT% positivity correlated with PM. All 3 MGMT scores correlated with overall survival (OS) in GBM’s. Cold spot IEC% was also positively associated with OS. These effects remained in a multivariate model after adjusting for age and disease status. Prognosis determined by correcting MGMT% score for IEC% is not improved in this analysis. However, IEC COLD SPOT score does provide additional prognostic information that can be gained from this correction method.
Keywords: Glioblastoma, Glioma, Immunohistochemistry, Inflammatory cell, MGMT, Promoter methylation
INTRODUCTION
From the first therapeutic report of temozolomide to treat human gliomas (1), O6-methylguanine-DNA methyltransferase (MGMT) has been shown to be an important mechanism underlying resistance to treatment (2). MGMT expression is largely, but not exclusively (3), controlled via promoter methylation (PM) (4). The specific measurement of MGMT PM is generally predictive of response and overall survival (OS) in patients with these tumors (5–9). A range of molecular genetic techniques are available for determining MGMT PM status in tumors (10). However, these methods are protected by patent (11), are generally expensive, and are destructive of often limited tissues. We, and others, have previously shown that immunohistochemical localization of MGMT protein is useful in detecting the nuclear enzyme (8, 12). One antiMGMT antibody, MAb 3.1, was developed at Duke (13–15) and its immunohistochemical detection correlated with MGMT protein detection in brain tumors by Western blot (6). Other studies have confirmed the correlation between MGMT PM and protein expression by Western blot (4). Immunohistochemical detection of MGMT has a number of reported confounding factors, including interobserver reliability (10, 16), differences in expression between recurrent versus primary disease, differences between infiltrating edge and center of tumor (17) and variances between methods of detection (6). Not surprisingly, the literature has warnings against the use of MGMT immunostaining as a clinical biomarker in brain tumors (16, 18–20). A number of papers have suggested that benign, MGMT-expressing cells found in tumors, such as endothelial cells, lymphocytes, microglia, and macrophages, may pose a problem in the interpretation of MGMT immunostaining of brain tumors (21–23). Watanabe (24), Nakasu (22, 25), and Sorensen (26) measured MGMT immunohistochemically using serial sections to detect nontumor, inflammatory cell markers using various markers including CD3, CD45, Iba1, CD31, and CD68. In this way, they identified regions with low populations of nonneoplastic cells in which to count MGMT in tumor cells. They found that MGMT percent positivity in tumor cells correlated with outcomes in glioma patients, a finding supported by Sasai (27). It is not clear whether an inflammatory cell infiltrate in a brain tumor is a biological phenomenon without its own confounding effect. Recent papers by Yuan (28) suggested that glioblastomas (GBM) with dense infiltrates of Iba-1-positive microglia exhibit a prolonged survival, a finding contradicted by Sorensen et al (29).
Burke et al has shown in a pilot study the feasibility of double-labelling immunohistochemistry for nuclear MGMT and nontumorous cytoplasmic and membranous elements via CD34, CD45, and CD68 for a corrected MGMT status in GBM (21). Double-labelling immunohistochemistry is an established technique that is now available on some automated immunohistochemistry platforms (e.g. Leica Biosystems, Richmond, IL). Such a technique allows the distinction and simultaneous measurement of MGMT among neoplastic cells and benign indigenous immune and endothelial cells.
For the past 5 years, we have tested for MGMT status in 2 ways, by MGMT PM and by immunohistochemistry. For the immunohistochemistry method, we have prospectively applied a triple-antibody (MGMT, CD45, HAM56), double-labelling (MGMT-brown nuclei; CD45 plus HAM56 cocktail-red cytoplasm) protocol to identify MGMT in both tumor cells and inflammatory and endothelial cells (IEC). In order to focus on tumor cell populations, we sought areas of tumor with least IEC infiltrates. Tumor MGMT% was counted in nonred-cytoplasmic staining cells in areas with fewest IEC infiltrates, that is, “IEC cold spots.” Using this approach, we measured 3 indices: Uncorrected MGMT% (single color, single antibody immunohistochemistry, number of MGMT brown nuclear-stained cells divided by total cells in area), an IEC cold spot percentage (IEC%; number of single color CD45/HAM56 cocktail cytoplasmic-stained cells divided by total cells in area) and a corrected MGMT% (among the nonred-stained cells, the number of cells with MGMT brown-positive nuclei divided by the total number of nonred-stained cells). This technique allowed correction for IEC MGMT positive nuclei in the IEC cold spots. We compared these findings to MGMT PM status. Our analysis on 982 patients with GBM collected over 5 years indicate that both the uncorrected MGMT% score and a complicated double-staining technique to derive a corrected MGMT% tumor nuclear score are both significantly associated with MGMT PM status and with OS. Our results also indicate that the cold spot IEC% found in GBM is associated with a strong positive effect on OS. Similar investigations into anaplastic astrocytomas (AA) did not yield significant results.
MATERIALS AND METHODS
The prospective collection of the data used in this study and the retrospective analysis were granted waivers of consent by the Duke Institutional Review board (PRO#00007434, PRO#00067457). Protected Health Information was stripped from each patient and replaced with an assigned number that was linked via a separate key kept by a nonparticipating laboratory technician.
The materials for this study were from 1181 patients with primary high-grade astrocytomas (WHO Grades III and IV) analyzed over a 7-year period (2007–2014) who had had their neurosurgeries performed at Duke University Hospital of which there were 982 GBM and 199 AA. Patients’ disease status was labeled “newly diagnosed” (ND) if the time between date of diagnosis and date of surgical procedure was <30 days; otherwise, patients were considered to have “recurrent disease” (RD). None of our patients was tested twice; of the patients who received multiple testing, only the primary result was included for analysis. At the time of diagnosis, the cases were prospectively tested for MGMT% and IEC infiltrates by single stains as well as by a double-stain for IEC MGMT% by immunohistochemistry using a triple-antibody, double-stain procedure to correct for infiltrating inflammatory cells and indigenous endothelial cells as described below. Each case was also independently tested for MGMT PM by a Clinical Laboratories Improvement Act (CLIA) certified outside reference laboratory (University of Pittsburgh Reference Laboratory, Pittsburgh, PA).
The immunohistochemical staining procedure was performed as follows: A hematoxylin and eosin (H&E)-stained slide (Fig. 1A) and 6 serial 5-µm-thick sections were cut for each immunohistochemical analysis (Figs. 1, 2) and considered slides #1–#6. For the triple-stained slides (slide #1), MGMT monoclonal antibody (MAb) application followed CD45/HAM56 dual-stain immunohistochemistry using a sequential staining method on the Bond Autostainer (Leica Biosystems) with the Bond Polymer Refine Detection system with the following antibodies: MGMT (clone 3.1, Santa Cruz; 1:20 dilution), CD45 (clone NCL-LCA, Leica, 1:50 dilution), and HAM56 (clone HAM-56, Cell Marque; 1:50 dilution). The MGMT antibody was visualized with diaminobenzidine ([DAB]; brown nuclei) and the CD45/HAM-56 cocktail was visualized with Texas red (red cytoplasmic and membranous stain). A negative control (slide #2) consisting of a mouse IgG MAb against an irrelevant antigen was performed with each patient’s case. External controls for CD45 and MGMT were performed with each assay. Single immunohistochemistry assays (with negative isotype matched controls) for CD45 (slides #3, #4 respectively) and MGMT (slides #5 and #6, respectively) were run alongside the dual-stained slides (slide #1) for each case. The single MGMT immunohistochemistry stain result was analyzed to determine the “uncorrected MGMT% score.”
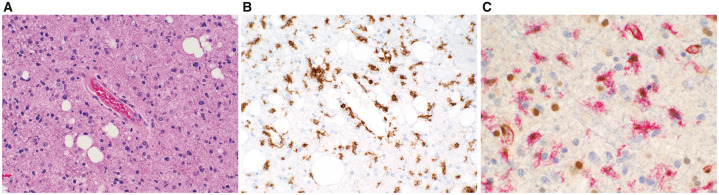
FIGURE 1.
(A) Anaplastic astrocytoma, NOS, WHO Grade III with nuclear pleomorphism and modestly increased cellular density. (B) CD45 immunohistochemistry reveals ∼20% of the cells to be microglia. (C) CD45/HAM56 cocktail (Texas red chromogen) confirms the IEC shown above while the DAB-stained nuclei reflect MGMT enzyme localization. (A: H&E; B: CD45 with DAB chromogen; C: CD45/HAM56 with Texas red chromogen and MGMT with DAB chromogen).
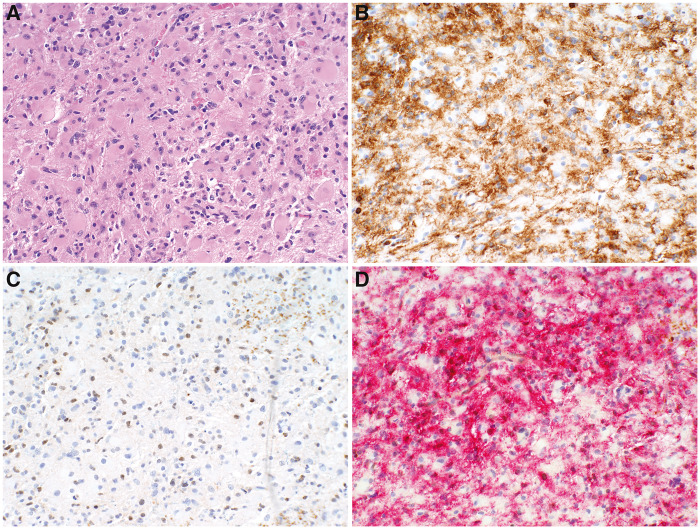
FIGURE 2.
(A) Densely cellular infiltrate of an anaplastic astrocytoma, NOS, WHO Grade III. (B) Serial section with the CD45 individual stain revealing a dense infiltrate of microglia comprising ∼70% of cells. (C) Serial section with the antiMGMT showing ∼20% of the cells to exhibit brown nuclei. (D) CD45/HAM56 cocktail reveals the vast majority of the brown-stained nuclei to be red IEC. (A: CD45 with DAB chromogen; B: CD45 with DAB chromogen; C: CD45/HAM56 cocktail with Texas red chromogen; MGMT with DAB chromogen).
For analysis, the single-stained CD45 slide (slide #3; Figs. 1B, 2A, B) was examined microscopically to determine the area of tumor with the least infiltrates of CD45-positive cells (IEC cold spots). In these cold spots, the percentage of CD45-positive cells among 100 cells was recorded (cold spot IEC%). The area on the slide was marked and the matching area on the serial slide similarly identified. A single-stained MGMT slide (slide #5) was also examined to determine uncorrected MGMT% score in the IEC cold spot using the Automated Cellular Imaging System (ACIS; DAKO, San Juan Capistrano, CA). The ACIS system measured the percentage of nuclei staining positively with MGMT MT3.1 within the field of interest. Slide #1 (triple antibody, dural chromogen-stained slide) was then analyzed microscopically (Figs. 1C, 2C) in the IEC cold spot by the same technician who determined the corrected MGMT% score of MGMT positive nuclei found in cells negative for IEC red staining and recorded the value. The corrected results were then compared with the ACIS determined uncorrected MGMT% score (slide #5) minus the CD45-positive cell percentage (slide #3), to compare against the manual count. In order to overcome interobserver variation, all of the results were quantitated by the same 2 individuals, initially by a technologist (A.A.) then by a neuropathologist (R.E.M.). A final consensus quantitation of results was determined at this time along with determination of sufficiency of neoplastic tissue and discussion of technical issues. Data analysts (A.C. and T.D.) recorded results in a REDCapTM database. Survival data were abstracted and entered into a REDCap database (E.L.). In cases in which <50 neoplastic cells were identifiable (for example, in a needle biopsy), the case was reported as “Insufficient for analysis.” Over the 5 years of the present study, these rare insufficient cases were excluded from the database and were not available for this analysis.
According to the literature provided with each test result, PM was performed by MethyLight PCR, confirmed with methylation-specific PCR at the University of Pittsburgh Medical Center (Pittsburgh, PA) referral laboratory at the time of the pathologic analysis as part of the patient’s clinical care (30) using the following method described in their common methodology report: Manual microdissection was used to separate neoplastic tissue and normal adjacent tissue. In order for a tissue target to be accepted for analysis, microdissection of a minimum of 50% of tumor cells was required. DNA was isolation was per standard laboratory procedure. Optical density readings provided DNA concentration results. The DNA samples underwent sodium bisulfite treatment; bisulfite-converted DNA was recovered using EpiTect Bisulfite KitTM (Qiagen, Germantown, MD). Bisulfite-treated DNA was amplified per described method (31–33). The results of MethyLight PCR were confirmed by methylation-specific PCR and agarose gel electrophoresis (34). The specimen was considered negative for MGMT PM when the methylation Index was 0 and no amplification was detected by methylation-specific PCR (32).
Statistical Methods
OS was computed from date of procedure from which the tissue tested was procured until date of death or last follow-up if censored. The effects of both MGMT% and IC% on survival were assessed using Cox proportional hazard models in both the univariate and multivariate settings and demonstrated visually using a Kaplan Meier graph. Youden’s J statistic, calculated as the sum of the sensitivity and specificity less 1, was used to identify an optimal labeling index cut-off within ND and RD patients.
RESULTS
Demographics
Diagnosis, gender distribution, recurrence status, MGMT PM results, and IDH mutational status are shown in Table 1. There were 1,181 patients evaluated in total, including 473 females and 708 males with a mean age of 55 (SD: 14.24) years (Table 2). There were 982 patients with GBM, of which 647 were ND and 335 were RD. Of the 199 diagnosed with AA, 103 were ND and 96 were RD (Table 1). The survival of GBM patients was significantly worse among those with RD than among ND patients, with median OS of 13.1 months (95% CI: 11.4–13.9) and 16.0 months (95% CI: 14.6–17.1), respectively. Among patients with AA, the difference between ND and RD subgroups was not significant, with OS median of 22.2 months (95% CI: 18.4–33.1) and 19.5 months (95% CI: 16.2–29.7), respectively (Table 3). Over two-thirds of the patient and tumor data were collected prior to the discovery of the significance of IDH status and survival. We instituted prospective clinical testing of all gliomas in December 2011. Of the samples available, 17/51 AA patients and 18/329 of GBM patients had tumors with mutated IDH1 (Table 2).
TABLE 1.
Gender Diagnosis Status, MGMT and IDH1 by Diagnosis
| Diagnosis |
||||
|---|---|---|---|---|
| AA |
GBM |
|||
| N | % | N | % | |
| Total # patients | 199 | 16.85 | 982 | 83.15 |
| Gender | ||||
| Female | 75 | 6.35 | 398 | 33.70 |
| Male | 124 | 10.50 | 584 | 49.45 |
| Diagnosis status | ||||
| Newly diagnosed | 103 | 8.72 | 647 | 54.78 |
| Recurrent | 96 | 8.13 | 335 | 28.37 |
| MGMT promoter methylation status | ||||
| Identified | 115 | 9.74 | 451 | 38.19 |
| Not identified | 84 | 7.11 | 531 | 44.96 |
| IDH1 diagnostics | ||||
| Missing | 148 | 12.53 | 653 | 55.29 |
| Intact | 34 | 2.88 | 311 | 26.33 |
| Mutation | 17 | 1.44 | 18 | 1.52 |
AA, anaplastic astrocytoma; GBM, glioblastoma; MGMT, O6-methylguanine-DNA methyltransferase.
TABLE 2.
Age, MGMT, and IEC by Diagnosis
| Diagnosis |
||||
|---|---|---|---|---|
| AA |
GBM |
|||
| n | Mean, SD | n | Mean, SD | |
| Age at procedure | 199 | 47.27, 15.12 | 982 | 56.70, 13.52 |
| Uncorrected MGMT% | 193 | 22.01, 18.54 | 934 | 19.87, 16.41 |
| Corrected MGMT% | 194 | 19.14, 16.80 | 933 | 16.67, 15.22 |
| IEC% | 182 | 17.68, 15.37 | 910 | 18.65, 13.73 |
AA, anaplastic astrocytoma; GBM, glioblastoma; IEC, inflammatory and endothelial cells; MGMT, O6-methylguanine-DNA methyltransferase.
TABLE 3.
GBM and AA Deaths Stratified by ND and RD Along With Various Survival Estimates
| Diagnosis, Status | Total | # Failed | Median Survival in Months (95% CI) | 1 Year Survival (95% CI) | 2 Year Survival (95% CI) | 3 Year Survival (95% CI) | 5 Year Survival (95% CI) |
|---|---|---|---|---|---|---|---|
| AA, ND | 103 | 67 | 22.2 (18.4, 33.1) | 76.4% (66.6%, 83.6%) | 44.6% (33.9%, 54.7%) | 37% (26.5%, 47.5%) | 19.4% (10.8%, 29.9%) |
| AA, RD | 96 | 54 | 19.5 (16.2, 29.7) | 80.4% (70.3%, 87.3%) | 42.6% (31.2%, 53.4%) | 35.9% (25%, 47%) | 28% (17.6%, 39.4%) |
| GBM, ND | 647 | 534 | 16 (14.6, 17.1) | 63.5% (59.5%, 67.2%) | 28.1% (24.4%, 31.9%) | 14.2% (11.3%, 17.4%) | 3.6% (2.1%, 5.8%) |
| GBM, RD | 335 | 267 | 13.1 (11.4, 13.9) | 52.7% (47%, 58.1%) | 20.7% (16.1%, 25.7%) | 12.5% (8.7%, 17%) | 5.9% (3.2%, 9.7%) |
AA, anaplastic astrocytoma; GBM, glioblastoma; ND, newly diagnosed; RD, recurrent disease.
MGMT PM Status Is Associated With Prolonged OS
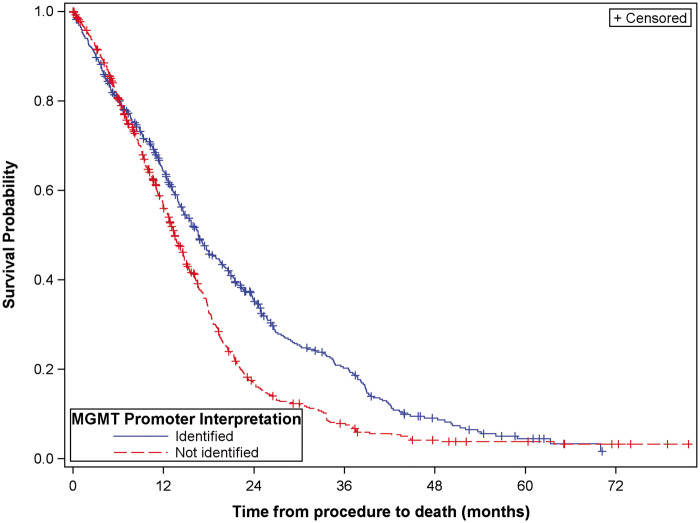
Mean ages at procedure as well as mean uncorrected and corrected MGMT% scores for both AA and GBMs are shown in Table 2. MGMT PM status (present vs absent) was significantly associated with prolonged OS in GBM’s (HR 0.718; 95% CI: 0.624, 0.827; Fig. 3). There was no evidence that PM had a similar effect on OS in AA’s.
FIGURE 3.
Kaplan Meier curve demonstrating association between MGMT PM and overall survival.
Uncorrected MGMT% Immunohistochemistry Scores in the IEC Cold Spots Are Associated With MGMT PM Status and Prolonged OS
Among the GBM tumor samples, mean uncorrected MGMT% was 19.87 (range 0–90) and MGMT PM was found in 54%. MGMT PM score and uncorrected MGMT% score by immunohistochemistry were inversely associated in GBM (Wilcoxon signed rank p < 0.0001). The raw MGMT% score was compared with the “gold standard” of PM positive or negative by PCR. This analysis does show a significant relationship between the positive MGMT PM testing and the uncorrected IHC results among the GBM. Interestingly, this analysis also demonstrated a significant relationship between MGMT positivity and the uncorrected IHC results among AA. As a result of this analysis, we were able to calculate sensitivity and specificity with the MGMT IHC score versus the MGMT PM score (gold standard). For the ND gliomas, the specificity was 0.602 and the sensitivity was 0.582 while for the recurrent tumors, the specificity was 0.591 and sensitivity was 0.577. OS was highly correlated with uncorrected MGMT% based on an analysis of combining AA and GBM patient groups (Pearson’s r = 0.8625, p < 0.0001). Uncorrected MGMT% score had a significant association with OS in GBM’s (HR for 5% increase in MGMT%: 1.030; 95% CI: 1.007, 1.054), but there was not similar evidence in AA patients (Table 4).
TABLE 4.
Univariate Associations of MGMT and IEC% With OS
| Diagnosis | Parameter | HR (95% CI) | p value |
|---|---|---|---|
| AA | Uncorrected MGMT% | 1.007 (0.997, 1.017) | 0.1947 |
| Corrected MGMT% | 1.005 (0.995, 1.016) | 0.3388 | |
| IEC% | 1.002 (0.990, 1.014) | 0.7245 | |
| GBM | Uncorrected MGMT% | 1.006 (1.001, 1.011) | 0.0118 |
| Corrected MGMT% | 1.006 (1.002, 1.011) | 0.0096 | |
| IEC% | 0.993 (0.988, 0.999) | 0.0163 |
AA, anaplastic astrocytoma; GBM, glioblastoma; IEC, inflammatory and endothelial cells; MGMT, O6-methylguanine-DNA methyltransferase, OS, overall survival.
The maximum value of Youden’s statistic was used to select an optimal cut-off point for uncorrected MGMT% immunohistochemistry as a predictor of MGMT PM status. The optimal point for ND patients was 19.19; whereas the cut-off for recurrent patients was 19.38. These cut-points corroborate previous studies indicating an immunohistochemical percent positive range of 10%–20% as cut-points (1, 17, 24).
Corrected MGMT% in the IEC Cold Spots Is Similar to Uncorrected MGMT%
Among the GBM patients, mean corrected MGMT% was 19.87 (range 0–90). Lymphocytes and microglia express MGMT protein (21, 22); an effect that could potentially interfere with the immunohistochemical detection of MGMT in gliomas. Variable survival results have been reported between inflammatory cell infiltrates and OS (18, 35–38). Both effects were therefore evaluated. To correct for the contribution of inflammatory cell MGMT immunohistochemical positivity on uncorrected MGMT% score, we analyzed the MGMT% score in IEC cold spots, that is, areas of tumor with lowest density of red-staining CD45-positive/HAM56-positive cells by microscopic examination of the stained slide and confirmed by correlation with the CD45-stained slide. Corrected MGMT% was only determined from nonred-staining (CD45-negative/HAM56-negative) cells. The resultant corrected MGMT% was highly correlated with the uncorrected MGMT% based on an analysis of combining AA and GBM patient groups (Pearson’s r = 0.8625, p < 0.0001). Similar to uncorrected MGMT%, corrected MGMT% was significantly associated with OS in GBM’s, but this association was not observed in AA’s (Table 4).
Cold Spot IEC% Is Associated With OS
We then separately examined the distribution of the IEC in AAs and GBMs and its association with OS. The mean cold spot IEC% in GBMs was 18.65 (SD 13.73); increasing cold spot IEC% had a significant association on prolonged OS in GBM’s (HR: 0.993; 95% CI: 0.988, 0.999). There was no similar evidence of an association in AA’s (Table 4).
Uncorrected MGMT% and Cold Spot IEC% Are Associated With OS in a Multivariate Setting
In a multivariate Cox model (Table 5) including both uncorrected MGMT% and cold spot IEC% adjusted for age at procedure, diagnosis, and diagnosis status, there was evidence that the risk of death increased with MGMT% (HR: 1.008, 95% CI: 1.003, 1.012) and decreased as cold spot IEC% increased (HR: 0.992, 95% CI: 0.987, 0.997). As a secondary analysis, IDH1 mutational status was also included in this multivariate setting, and the adjusted associations of MGMT% and IEC% remained (Table 6).
TABLE 5.
Hazard Ratios From a Multivariate Cox Model Predicting OS
| Parameter | p Value | HR | 95% Confidence Limits | |
|---|---|---|---|---|
| Age at procedure | <.0001 | 1.024 | 1.018, 1.030 | |
| Diagnosis | AA vs GBM | <.0001 | 0.586 | 0.476, 0.721 |
| Diagnosis status | ND vs RD | 0.0002 | 0.755 | 0.652, 0.874 |
| MGMT% | 0.0005 | 1.008 | 1.003, 1.012 | |
| IEC% | 0.0021 | 0.992 | 0.987, 0.997 |
AA, anaplastic astrocytoma; IEC, inflammatory and endothelial cells; GBM, glioblastoma; MGMT, O6-methylguanine-DNA methyltransferase; ND, newly diagnosed; OS, overall survival, RD, recurrent disease.
TABLE 6.
Hazard Ratios From a Multivariate Cox Model Predicting OS That Also Includes IDH1 Mutational Status
| Parameter | p Value | HR | 95% Confidence Limits | |
|---|---|---|---|---|
| Age at procedure | <.0001 | 1.024 | 1.018, 1.030 | |
| Diagnosis | AA vs GBM | <0.0001 | 0.591 | 0.480, 0.727 |
| Diagnosis status | ND vs RD | 0.0001 | 0.752 | 0.649, 0.871 |
| IDH1 | Mutant vs intact | 0.4877 | 0.763 | 0.356, 1.637 |
| IDH1 | NOS vs intact | 0.2322 | 0.905 | 0.769, 1.066 |
| MGMT% | 0.0018 | 1.007 | 1.003, 1.012 | |
| LCA% | 0.0037 | 0.992 | 0.987, 0.998 |
AA, anaplastic astrocytoma; IEC, inflammatory and endothelial cells; GBM, glioblastoma; MGMT, O6-methylguanine-DNA methyltransferase; ND, newly diagnosed; OS, overall survival; RD, recurrent disease.
DISCUSSION
Previous studies have debated the utility of MGMT immunohistochemistry detection in high-grade gliomas (4). The method has a number of reported confounding factors, including interobserver reliability (10, 16), infiltrating microglia and indigenous endothelial cells (23), differences in expression between recurrent versus primary disease (39), and variances between methods of detection (6). In order to overcome the issue of interobserver variation (16), these cases were reviewed by the same 2 individuals, a technologist (A.A.) and a neuropathologist (R.E.M.). Further, the uncorrected MGMT% was determined using a digital imaging system, as highlighted by Araki et al (40).
To address the issue of infiltrating microglia and indigenous endothelial cells (17, 21, 22, 27, 29), previous studies have used a combination of antibodies including CD45 (21), CD68 (macrophage/microglia) (23), Iba1 (microglia) (26), CD31 (endothelial cells) (26), and/or CD34 (endothelial cells) (21, 22, 24). In our diagnostic neuropathology service, we have routinely used HAM56 to identify both macrophages and endothelial cells in tumors for years and found its biphenotypic affinity provides simplicity and efficiency in this application (41, 42). Furthermore, other studies have found that CD34 can be found in dysplastic neurons, gangliogliomas, hemangiopericytomas, and other gliomas (43). Similarly, CD31 has been used to identify transdifferentiation in GBM (44), thus indicating that no endothelial marker is perfect. Five years ago, we incorporated this MAb into a 3-antibody/dual-color technique that allowed us to correct for CD45-positive/HAM56-positive red cytoplasmic staining inflammatory cells and endothelial cells while also counting “naked” MGMT-positive DAB-brown nuclei.
Molecular pathology has also been used to investigate MGMT status and has exploited the fact that MGMT expression is largely controlled via methylation of the promoter region of the gene, an epiphenomenon that is inheritable through the cell lineage (45). The advantage that immunohistochemistry brings is its ease and speed of application relative to molecular techniques and familiarity to most diagnostic laboratories (24). In the present study, we confirmed what has been variably reported by others (39, 46), that the immunohistochemical detection of MGMT protein was inversely associated with MGMT PM in GBM.
Isocitrate dehydrogenase mutation appears to result in widespread PM, and may actually represent a biomarker for more widespread methylation-inhibition of other drug resistance mechanisms (47). Therefore, we investigated IDH mutational status in these cases. In the present study, over two-thirds of the patient and tumor data were collected prior to the discovery of the significance of IDH status and survival. Of those samples with IDH1 testing, only 9.2% (35/380) of tumors (33% of AA and 5% of GBM) demonstrated IDH1 mutations (Table 1) suggesting a minor role in controlling the MGMT PM status (48). Given the scope of this study and the small number of events and limited follow-up within this subset, a formal analysis of IDH1 is not appropriate. However, for the 329 patients for whom IDH status was known, IDH1 mutational status was not found to be associated with survival in GBMs, though there is a trend towards a beneficial effect for IDH mutated tumors, especially noted at 2 years (Table 7). Additionally, the effects of MGMT% and IEC% remained when adjusted for IDH1 in a multivariate setting (Table 6). In addition, the survival data can be compared with the previous studies on the effect of MGMT on survival (16, 17, 34) that were performed on IDH status naïve tumors. It may not be appropriate to compare the current study to previous studies where tumors were not stratified by IDH status, as, in hindsight, those studies suffer from the drawback of IDH status being a potentially unmeasured confounding variable. This supports the need for further studies to adequately understand the effect of IDH. The immunohistochemical method described in this paper was not found to be predictive of survival in lower-grade astrocytomas (WHO Grade III) nor, as previously reported from our laboratory (49), in oligodendrogliomas. In reviewing these cases, our results confirm those of Yuan et al that lower-grade gliomas have more obviously infiltrative features (leading edge effect) in which benign glia become incorporated into the tumors (36). These benign glial elements are not identified by either the CD45 or the HAM56 antibodies and are thus included in the neoplastic cell counts. These benign cells are strong expressers of MGMT (3). Because PM of MGMT is strongly associated with the malignant genotype, molecular detection of this epiphenomenon is recommended over immunohistochemical detection in diffuse gliomas (WHO Grades II–IV).
TABLE 7.
Univariate Analysis of Survival Stratified by IDH Status in 329 GBM Patients
| IDH1 Diagnostics | Total | # Failed | Median Survival in Months (95% CI) | 1 Year Survival (95% CI) | 2 Year Survival (95% CI) | 3 Year Survival (95% CI) | 5 Year Survival (95% CI) |
|---|---|---|---|---|---|---|---|
| Intact | 311 | 202 | 13.7 (12.5, 15.3) | 57.1% (51%, 62.8%) | 22% (16.4%, 28.1%) | 10.1% (5.5%, 16.3%) | 7.1% (3%, 13.5%) |
| Mutation | 18 | 7 | 29.7 (7, 29.7) | 70.5% (42.8%, 86.6%) | 58.8% (27.4%, 80.4%) | 0% | 0% |
Another finding of this analysis was that the cold spot IEC% was independently predictive of a survival advantage to the GBM patient with a high cold spot IEC%. The data were collected prospectively with a standardized procedure, and the results were not found to be significant in AA. The data were collected in regions identified with lowest IEC% without regard to MGMT immunohistochemistry status. Previous studies have noted a survival advantage for dense IEC infiltrates in the center of a glioma (18, 35, 36) though none was analyzed by cold spot identification, a measure that may index the extent to which infiltrating IEC have overcome inhibitory signals and permeated the tumor. Other studies have not identified a relationship among tumor infiltrating lymphocytes, microglia, and OS in gliomas (37, 38). The inverse relationship between the effect on survival of MGMT immunohistochemistry scores and cold spot IEC% lends credence to a previous hypothesis that tumor infiltrating lymphocyte counts may be associated with methylation status of GBMs (50).
Previous authors have recommended using both MGMT PM quantitative results and MGMT IHC quantitative results. The present study supports that contention in that, independently, patients with either a negative IHC result or a positive methylation result survive longer relative to patients with a negative methylation test or a positive IHC result (51).
In conclusion, a MGMT% score corrected by using a complicated 3-antibody/2-color immunohistochemical method does not provide significantly improved prognostic data versus an uncorrected score; both are inversely correlated with MGMT PM status and both are associated with a shortened survival. However, an IEC% derived from a tumor cold spot does provide additional prognostic information that can be gained from this correction method.
ACKNOWLEDGMENTS
The authors greatly appreciate the technical advice and assistance provided by the technicians of the Cell Image Laboratory of Duke Department of Pathology as well as the support of the members of the Duke Preston Robert Tisch Neuro-oncology laboratory.
This study was supported by a grant from the Pediatric Brain Tumor Foundation and the Kyrie Foundation.
The authors have no duality or conflicts of interest to declare.
REFERENCES
- 1. Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol 1998;16:3851–7 [DOI] [PubMed] [Google Scholar]
- 2. Friedman HS, Johnson SP, Dong Q, et al. Methylator resistance mediated by mismatch repair deficiency in a glioblastoma multiforme xenograft. Cancer Res 1997;57:2933–6 [PubMed] [Google Scholar]
- 3. Sasai K, Akagi T, Aoyanagi E, et al. O6-methylguanine-DNA methyltransferase is downregulated in transformed astrocyte cells: Implications for anti-glioma therapies. Mol Cancer 2007;6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spiegl-Kreinecker S, Pirker C, Filipits M, et al. O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro Oncol 2010;12:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 2004;10:1871–4 [DOI] [PubMed] [Google Scholar]
- 6. Maxwell JA, Johnson SP, Quinn JA, et al. Quantitative analysis of O6-alkylguanine-DNA alkyltransferase in malignant glioma. (Comparative Study Research Support, N.I.H., Extramural). Mol Cancer Ther 2006;5:2531–9 [DOI] [PubMed] [Google Scholar]
- 7. Quinn JA, Desjardins A, Weingart J, et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J Clin Oncol 2005;23:7178–87 [DOI] [PubMed] [Google Scholar]
- 8. Chinot OL, Barrie M, Fuentes S, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol 2007;25:1470–5 [DOI] [PubMed] [Google Scholar]
- 9. Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. (Multicenter Study). J Neurooncol 2010;97:311–22 [DOI] [PubMed] [Google Scholar]
- 10. Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, methylight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 2012;118:4201–11 [DOI] [PubMed] [Google Scholar]
- 11. Vlassenbroeck IB, Biero K, Methylation detection of MGMT. In: USPTO, ed. WO2009/037441, Vol. US9050280 B2. USA: MDxHealth SA, 2008 [Google Scholar]
- 12. McLendon RE, Cleveland L, Pegram C, et al. Immunohistochemical detection of the DNA repair enzyme O6-methylguanine-DNA methyltransferase in formalin-fixed, paraffin-embedded astrocytomas. Lab Invest 1998;78:643–4 [PubMed] [Google Scholar]
- 13. Ostrowski LE, von Wronski MA, Bigner SH, et al. Expression of O6-methylguanine-DNA methyltransferase in malignant human glioma cell lines. Carcinogenesis 1991;12:1739–44 [DOI] [PubMed] [Google Scholar]
- 14. Brent TP, von Wronski MA, Edwards CC, et al. Identification of nitrosourea-resistant human rhabdomyosarcomas by in situ immunostaining of O6-methylguanine-DNA methyltransferase. Oncol Res 1993;5:83–6 [PubMed] [Google Scholar]
- 15. Brent TP, von Wronski M, Pegram CN, et al. Immunoaffinity purification of human O6-alkylguanine-DNA alkyltransferase using newly developed monoclonal antibodies. Cancer Res 1990;50:58–61 [PubMed] [Google Scholar]
- 16. Preusser M, Charles Janzer R, Felsberg J, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: Observer variability and lack of association with patient survival impede its use as clinical biomarker. (Multicenter Study) Brain Pathol 2008;18:520–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Capper D, Mittelbronn M, Meyermann R, et al. Pitfalls in the assessment of MGMT expression and in its correlation with survival in diffuse astrocytomas: Proposal of a feasible immunohistochemical approach. Acta Neuropathol 2008;115:249–59 [DOI] [PubMed] [Google Scholar]
- 18. Brell M, Ibanez J, Tortosa A. O6-Methylguanine-DNA methyltransferase protein expression by immunohistochemistry in brain and non-brain systemic tumours: Systematic review and meta-analysis of correlation with methylation-specific polymerase chain reaction. BMC Cancer 2011;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brell M, Tortosa A, Verger E, et al. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res 2005;11:5167–74 [DOI] [PubMed] [Google Scholar]
- 20. Mason S, McDonald K. MGMT testing for glioma in clinical laboratories: Discordance with methylation analyses prevents the implementation of routine immunohistochemistry. J Cancer Res Clin Oncol 2012;138:1789–97 [DOI] [PubMed] [Google Scholar]
- 21. Burke E, Grobler M, Elderfield K, et al. Double-labelling immunohistochemistry for MGMT and a “cocktail” of non-tumourous elements is a reliable, quick and easy technique for inferring methylation status in glioblastomas and other primary brain tumours. Acta Neuropathol Commun 2013;1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakasu S, Fukami T, Baba K, et al. Immunohistochemical study for O6-methylguanine-DNA methyltransferase in the non-neoplastic and neoplastic components of gliomas. J Neurooncol 2004;70:333–40 [DOI] [PubMed] [Google Scholar]
- 23. Hsu CY, Lin SC, Ho HL, et al. Exclusion of histiocytes/endothelial cells and using endothelial cells as internal reference are crucial for interpretation of MGMT immunohistochemistry in glioblastoma. Am J Surg Pathol 2013;37:264–71 [DOI] [PubMed] [Google Scholar]
- 24. Watanabe R, Nakasu Y, Tashiro H, et al. O6-methylguanine DNA methyltransferase expression in tumor cells predicts outcome of radiotherapy plus concomitant and adjuvant temozolomide therapy in patients with primary glioblastoma. Brain Tumor Pathol 2011;28:127–35 [DOI] [PubMed] [Google Scholar]
- 25. Nakasu S, Fukami T, Jito J, et al. Prognostic significance of loss of O6-methylguanine-DNA methyltransferase expression in supratentorial diffuse low-grade astrocytoma. Surg Neurol 2007;68:603–8;discussion8–9 [DOI] [PubMed] [Google Scholar]
- 26. Dahlrot RH, Dowsett J, Fosmark S, et al. Prognostic value of O-6-methylguanine-DNA methyltransferase (MGMT) protein expression in glioblastoma excluding nontumour cells from the analysis. Neuropathol Appl Neurobiol 2018;44:172–84 [DOI] [PubMed] [Google Scholar]
- 27. Sasai K, Nodagashira M, Nishihara H, et al. Careful exclusion of non-neoplastic brain components is required for an appropriate evaluation of O6-methylguanine-DNA methyltransferase status in glioma: Relationship between immunohistochemistry and methylation analysis. Am J Surg Pathol 2008;32:1220–7 [DOI] [PubMed] [Google Scholar]
- 28. Yuan JX, Bafakih FF, Mandell JW, et al. Quantitative analysis of the cellular microenvironment of glioblastoma to develop predictive statistical models of overall survival. J Neuropathol Exp Neurol 2016;75:1110–23 [DOI] [PubMed] [Google Scholar]
- 29. Sorensen MD, Dahlrot RH, Boldt HB, et al. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol Appl Neurobiol 2018;44:185–206 [DOI] [PubMed] [Google Scholar]
- 30. Christians A, Hartmann C, Benner A, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. (Research Support, Non-U.S. Gov't). PLoS One 2012;7:e33449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vlassenbroeck I, Califice S, Diserens AC, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn 2008;10:332–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn 2006;8:209–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. (Research Support, Non-U.S. Gov't). N Engl J Med 2005;352:997–1003 [DOI] [PubMed] [Google Scholar]
- 35. Boker DK, Kalff R, Gullotta F, et al. Mononuclear infiltrates in human intracranial tumors as a prognostic factor. Influence of preoperative steroid treatment. I. Glioblastoma. Clin Neuropathol 1984;3:143–7 [PubMed] [Google Scholar]
- 36. Yuan Q, Matsumoto K, Nakabeppu Y, et al. A comparative immunohistochemistry of O6-methylguanine-DNA methyltransferase and p53 in diffusely infiltrating astrocytomas. Neuropathology 2003;23:203–9 [DOI] [PubMed] [Google Scholar]
- 37. Rossi ML, Hughes JT, Esiri MM, et al. Immunohistological study of mononuclear cell infiltrate in malignant gliomas. Acta Neuropathol 1987;74:269–77 [DOI] [PubMed] [Google Scholar]
- 38. Rossi ML, Jones NR, Candy E, et al. The mononuclear cell infiltrate compared with survival in high-grade astrocytomas. Acta Neuropathol 1989;78:189–93 [DOI] [PubMed] [Google Scholar]
- 39. Lalezari S, Chou AP, Tran A, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro-Oncol 2013;15:370–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Araki Y, Mizoguchi M, Yoshimoto K, et al. Quantitative digital assessment of MGMT immunohistochemical expression in glioblastoma tissue. Brain Tumor Pathol 2011;28:25–31 [DOI] [PubMed] [Google Scholar]
- 41. Cummings TJ, Hulette CM, Bigner SH, et al. Ham56-immunoreactive macrophages in untreated infiltrating gliomas. Arch Pathol Lab Med 2001;125:637–41 [DOI] [PubMed] [Google Scholar]
- 42. Hulette CM, Downey BT, Burger PC. Macrophage markers in diagnostic neuropathology. Am J Surg Pathol 1992;16:493–9 [DOI] [PubMed] [Google Scholar]
- 43. Perry AB, D. Practical surgical Neuropathology: A diagnsotic Approach. Philadelphia, PA: Churchill, Livingstone, 2010 [Google Scholar]
- 44. El Hallani S, Colin C, El Houfi Y, et al. Tumor and endothelial cell hybrids participate in glioblastoma vasculature. Biomed Res Int 2014;2014:827327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042–54 [DOI] [PubMed] [Google Scholar]
- 46. Tang K, Jin Q, Yan W, et al. Clinical correlation of MGMT protein expression and promoter methylation in Chinese glioblastoma patients. (Research Support, Non-U.S. Gov't). Med Oncol 2012;29:1292–6 [DOI] [PubMed] [Google Scholar]
- 47. Guo C, Pirozzi CJ, Lopez GY, et al. Isocitrate dehydrogenase mutations in gliomas: Mechanisms, biomarkers and therapeutic target. (Review) Curr Opin Neurol 2011;24:648–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McLendon RE, Herndon JE, West B, et al. Survival analysis of presumptive prognostic markers among oligodendrogliomas. Cancer 2005;104:1693–9 [DOI] [PubMed] [Google Scholar]
- 50. Rutledge WC, Kong J, Gao J, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res 2013;19:4951–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kristensen LS, Michaelsen SR, Dyrbye H, et al. Assessment of quantitative and allelic MGMT methylation patterns as a prognostic marker in glioblastoma. J Neuropathol Exp Neurol 2016;75:246–55 [DOI] [PMC free article] [PubMed] [Google Scholar]