Abstract
Despite its recent invasion into the marine realm, the sea otter (Enhydra lutris) has evolved a suite of adaptations for life in cold coastal waters, including limb modifications and dense insulating fur. This uniquely dense coat led to the near-extinction of sea otters during the 18th–20th century fur trade and an extreme population bottleneck. We used the de novo genome of the southern sea otter (E. l. nereis) to reconstruct its evolutionary history, identify genes influencing aquatic adaptation, and detect signals of population bottlenecks. We compared the genome of the southern sea otter with the tropical freshwater-living giant otter (Pteronura brasiliensis) to assess common and divergent genomic trends between otter species, and with the closely related northern sea otter (E. l. kenyoni) to uncover population-level trends. We found signals of positive selection in genes related to aquatic adaptations, particularly limb development and polygenic selection on genes related to hair follicle development. We found extensive pseudogenization of olfactory receptor genes in both the sea otter and giant otter lineages, consistent with patterns of sensory gene loss in other aquatic mammals. At the population level, the southern sea otter and the northern sea otter showed extremely low genomic diversity, signals of recent inbreeding, and demographic histories marked by population declines. These declines may predate the fur trade and appear to have resulted in an increase in putatively deleterious variants that could impact the future recovery of the sea otter.
Keywords: sea otter, giant otter, genomics, population genetics, adaptation, olfaction, demography, deleterious variation, pseudogenes
Background
Within the weasel family (Mustelidae), otters (Lutrinae) are a recent radiation that originated from terrestrial weasel-like ancestors and evolved into semiaquatic hunters that thrive in freshwater and marine habitats. Among the 13 living species of otters, the sea otter (Enhydra lutris) and the freshwater-living giant otter (Pteronura brasiliensis) are the largest (up to 45 kg) and longest (up to 1.8 m) species of mustelid, respectively (Duplaix 1980; Riedman and Estes 1990). The two otter species live in vastly different environments and face different evolutionary pressures. The sea otter is almost entirely aquatic, living in cold coastal marine environments of the North Pacific Ocean, from the northern Japanese archipelago and Kuril Islands to California and Mexico (Kenyon 1969; Riedman and Estes 1990) (supplementary fig. S1A, Supplementary Material online), whereas the semiaquatic giant otter lives along the freshwater streams, rivers, and lakes of South America (supplementary fig. S1B, Supplementary Material online).
The sea otter is a recently derived marine species compared with the three other marine mammal lineages (cetaceans, sirenians, and pinnipeds) which first entered the aquatic environment between 30 and 55 Ma (Berta 2012). Fossil evidence suggests Enhydra only entered the marine realm <2–3 Ma (Riedman and Estes 1990; Boessenecker 2018). The three ancient marine mammal lineages have highly modified body plans and their genomes show dramatic adaptations to life in the sea (Yim et al. 2014; Foote et al. 2015). The sea otter has its own suite of unique marine adaptations, including webbed hind feet, large highly efficient kidneys for osmoregulation, increased lung and blood volumes for flotation and oxygen storage, increased metabolic rate, unique eye structure, high tactile sensitivity, dense bones, and distinct behaviors, including tool use (Murphy et al. 1990; Fish and Stein 1991; Fujii et al. 2015; Ralls et al. 2017; Tinker et al. 2017; Strobel et al. 2018). Other marine mammals have evolved a thick layer of blubber as insulation, but sea otters instead rely on a pelt made up of dense interlocking underhairs that trap air for insulation, forming the densest fur of any mammal (Williams et al. 1992; Kuhn et al. 2010; Kuhn and Meyer 2010a, 2010b; Liwanag et al. 2012).
In contrast to the sea otter, freshwater otter species such as the giant otter do not have to confront the extreme challenges of marine life and instead are adept in both aquatic and terrestrial habitats. For example, freshwater otter species locomote relatively well on land and are able to move several kilometers across terrestrial habitats (Williams et al. 2002). Freshwater otters also have distinct morphological and behavioral adaptations to aquatic habitats. The giant otter has four webbed feet, a paddle-like tail, and an insulating pelt that has similar interlocking hair structure as the sea otter, but the fur is shorter and less dense (Duplaix 1980; Kuhn 2009; Kuhn and Meyer 2009). Given that the sea otter diverged from its freshwater relatives about 5 Ma (Koepfli et al. 2008), the rapid evolution of its suite of adaptations to marine life is remarkable. Comparative analysis of whole-genome sequences of the sea otter and giant otter will enable understanding of the genetic mechanisms underlying the morphological and physiological adaptations that are shared by or are unique to these two species.
In addition to adaptation, genes may also degenerate into pseudogenes during the transition to an aquatic environment as certain sensory functions become unnecessary. The loss of sensory genes, particularly those that encode proteins related to taste and olfaction, has been observed in multiple aquatic mammal lineages (Kishida et al. 2007, 2015; Hayden et al. 2010; Jiang et al. 2012; Sato and Wolsan 2012; Feng et al. 2014; Li and Zhang 2014; Hughes et al. 2018). We therefore predict increased pseudogenization of sensory genes in the sea otter and giant otter compared with terrestrial mammals. As sea otters spend more of their time in the water than the giant otter (Kenyon 1969; Estes 1989), we predict more pseudogenization in the sea otter lineage than the giant otter. However, because both otters are part of a relatively recent semiaquatic radiation (<10 Ma; Willemsen 1992; Wang et al. 2018), we also predict that fewer sensory genes will be pseudogenized in the otter lineages than in more ancient and fully aquatic marine mammals, such as pinnipeds, sirenians, and cetaceans (Kishida et al. 2007, 2015; Hayden et al. 2010; Jiang et al. 2012; Sato and Wolsan 2012; Feng et al. 2014; Li and Zhang 2014; Hughes et al. 2018).
On a more recent timescale, the sea otter is marked by its near-extinction as a result of the fur trade that began in the mid-18th century and lasted until the early 20th century (Kenyon 1969; Riedman and Estes 1990). Specifically, all three recently diverged sea otter subspecies (Asian, northern, and southern sea otters—see supplementary fig. S1A, Supplementary Material online) were devastated by the fur trade, with only a handful of remnant populations surviving. The southern sea otter (E. l. nereis) was extirpated from Baja California and Oregon, and the population in California was devastated, with only 50 individuals estimated to have survived into the 20th century from an ancestral population of 16–20,000 individuals (Riedman and Estes 1990). The population has been slowly recovering over the past century, reaching a current size of ∼3,000 individuals (Tinker and Hatfield 2017), well below the estimated ancestral population size (Riedman and Estes 1990) and occupying only a fraction of the historical range (Tinker and Hatfield 2017). The northern sea otter, which ranged from the Aleutian Islands to Washington state (supplementary fig. S1A, Supplementary Material online), was extirpated from much of its range during the fur trade, with remnant populations surviving in the Aleutians and central Alaska. In the 1960s–1970s, translocations were used to repopulate the sea otter populations of southeast Alaska and the Pacific northwest (Jameson et al. 1982). Translocated sea otters were drawn from two distinct northern sea otter populations from the Aleutian Islands and Prince William Sound (supplementary fig. S1A, Supplementary Material online).
Due to their history of extreme population decline, the genetic diversity and genomic health of remnant sea otter populations has been a concern for decades. Studies based on mitochondrial DNA and microsatellites found low diversity (Cronin et al. 1996; Bodkin et al. 1999; Larson, Jameson, Etnier, et al. 2002; Larson, Jameson, Bodkin, et al. 2002; Aguilar et al. 2008; Larson et al. 2012; Gagne et al. 2018). We predict that this loss of diversity extends genome-wide, and the extreme population bottleneck of the California southern sea otter population may also have resulted in an increase in deleterious genetic variants (genetic load) due to the increased strength of genetic drift in small populations (Ohta 1973; Lynch et al. 1995; Kohn et al. 2006; Akashi et al. 2012). The giant otter has also experienced population fragmentation and decline due to habitat loss during more recent times (Carter and Rosas 1997; Pickles et al. 2012), but not to the same extent as the sea otter. Therefore, we expect the giant otter to harbor higher genome-wide diversity and a lower genetic load compared with the sea otter.
Here, we provide an in-depth genomic comparison of the sea otter and giant otter. We first explore long-term evolutionary trends of positive selection and gene loss within an evolutionary framework. In both otter species, we found evidence for positive polygenic selection on genes related to hair follicle development and a substantial loss of genes underlying sensory systems. We found that the southern and northern sea otter populations both have extremely low levels of genomic diversity, demographic histories marked by multiple potential periods of population disruption, and elevated levels of deleterious variation.
Results
Genome Sequencing, Assembly, and Annotation
We sequenced and assembled a 2.4-Gb genome of a southern sea otter from the wild California population that was maintained at the Monterey Bay Aquarium. The genome was sequenced using high-coverage Illumina short read sequencing and Dovetail Genomics HiRise Scaffolding to join scaffolds (Putnam et al. 2016) (table 1). The 2.6-Gb giant otter genome was sequenced from a single PCR-free library and assembled using the DISCOVAR de novo assembly pipeline at the Broad Institute (table 1). The southern sea otter genome assembly was much more contiguous than the giant otter genome due to the use of HiRise scaffolding, with a scaffold N50 value of 6.6 Mb compared with a contig N50 of 0.12 Mb for the giant otter genome (DISCOVAR yields a contig-only assembly) (table 1). We annotated both genomes using the MAKER2 pipeline (Holt and Yandell 2011) (supplementary information 1, Supplementary Material online) and detected 21,909 and 23,665 protein-coding gene models for the southern sea otter and giant otter, respectively. The higher number of genes in the giant otter is likely due to the more fragmented genome, which can lead to multiple gene models being called for a single gene. We compared the genome assembly and annotation statistics of our southern sea otter and giant otter genomes to the domestic ferret (Mustela putorius furo) reference genome (Peng et al. 2014) and a northern sea otter genome (E. l. kenyoni) from an individual from the translocated wild southeast Alaska population that was maintained at the Vancouver Aquarium (Jones et al. 2017) (table 1). The northern sea otter genome had high contiguity (scaffold N50: 38.7 Mb) due to linked-read 10× Genomics sequencing (table 1). Our southern sea otter genome was used as the primary sea otter genome for the molecular evolution analyses, and the northern sea otter genome was included to investigate population-level differences in sea otter diversity and demographic history.
Table 1.
Comparison of Genome Assembly Statistics.
| Genome Comparison | Southern Sea Otter | Giant Otter | Northern Sea Otter | Domestic Ferret | |
|---|---|---|---|---|---|
| Species | Enhydra lutris nereis | Pteronura brasiliensis | Enhydra lutris kenyoni | Mustela putorius furo | |
| Reference | This paper/UCLA | This paper/Broad Institute | Jones et al. (2017) | Peng et al. (2014) | |
| Assembly Accession | QQQE00000000 | GCA_004024605.1 | GCF_002288905.1 | GCF_000215625.1 | |
| Group | UCLA | Broad Institute | BC Cancer Agency | Ferret Genome Sequencing Consortium | |
| Sequencing/assembly methods | Illumina + Meraculous + Dovetail | Illumina + DISCOVAR | Illumina+10× Genomics/ABySS Supernova Hybrid | Illumina/ALLPATHS-LG | |
| Raw coverage | 81× | 44× | 110× | 162× | |
| Contig N50 | 18.4 kb | 99.9 kb | 244.5 kb | 44.8 kb | |
| Scaffold N50 | 6.6 Mb | N/A (contig-only assembly) | 38.7 Mb | 9.3 Mb | |
| Number of scaffolds | 55,496 | 649,335 | 6,771 | 7,783 | |
| Total length | 2,425.5 Mb | 2,603.5 Mb | 2,455.2 Mb | 2,405.5 Mb | |
| BUSCO v2 Assembly Completeness Statistics (based on 4,104 mammalian core BUSCOs) | |||||
| Complete genes | 93.3% | 85.2% | 96.2% | 95.3% | |
| Complete + partial | 96.9% | 95.5% | 98.0% | 97.8% | |
| Missing | 3.0% | 4.5% | 2.0% | 2.2% | |
| Duplicated | 0.99% | 0.66% | 1.09% | 0.67% | |
| Annotation method | MAKER2 | MAKER2 | NCBI Eukaryotic Genome Annotation Pipeline | NCBI Eukaryotic Genome Annotation Pipeline | |
| Protein-coding gene models | 21,909 | 23,665 | 19,458 | 20,062 | |
Phylogeny and Divergence
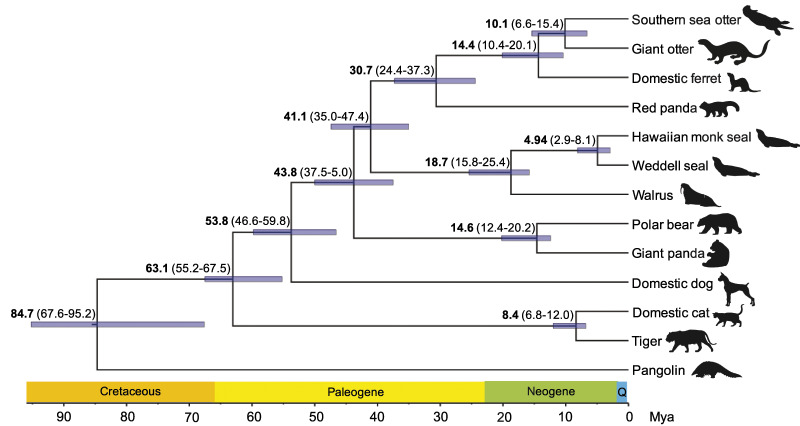
To place our comparative analyses within a well-grounded evolutionary framework, we first reconstructed a phylogeny of the southern sea otter, giant otter, and ten other species of the order Carnivora (supplementary table S1, Supplementary Material online) with RAxML 8.0 (Stamatakis 2014) under the GTRGAMMA model using 4-fold degenerate sites from 784 single-copy orthologous genes which was rooted with the Malayan pangolin (Manis javanica). The reduced number of 1:1 orthologs used in our phylogenomic analysis was due to the lower quality annotations of the giant panda (Ailuropoda melanoleuca) and tiger (Panthera tigris) genomes, resulting in more fragmented alignments relative to the other species. Therefore, the 784 1:1 orthologs we obtained represented the most complete and reliable gene alignments across the 12 carnivoran species and pangolin outgroup. The topology of the inferred phylogeny (fig. 1) is consistent with previous findings based on <10 kb (Eizirik et al. 2010) or <36 kb of DNA sequences (Meredith et al. 2011). We found that the cumulative branch length for the sea otter, giant otter, and domestic ferret (Mustelidae) was significantly greater than for other species, suggesting a higher substitution rate (supplementary fig. S2, Supplementary Material online). Consequently, we estimated divergence times among the 12 carnivoran species using MCMCTree (Yang 2007) under the independent clock model with six fossil and molecular calibrations (supplementary table S2, Supplementary Material online). We found that the southern sea otter and giant otter diverged 10.1 Ma (6.6–15.4 Ma 95% credibility interval [CI]), and that the otters diverged from the domestic ferret and red panda 14.4 (95% CI: 10.4–20.1) and 30.7 (95% CI: 24.4–37.3) Ma, respectively (fig. 1), a result consistent with previous studies (Koepfli et al. 2008; Sato et al. 2009).
Fig. 1.
Phylogenetic tree and divergence times of southern sea otter, giant otter, and ten other carnivoran species. The phylogeny and divergence time estimation was based on 4-fold degenerate sites across 784 1:1 orthologs and was time-calibrated using fossil- and molecular-derived priors. The Malayan pangolin served as the outgroup. Branch lengths represent time before present (Ma). The mean age of each node is shown, with 95% CIs in parentheses and depicted as purple bars around each node. Geological periods are shown above the time axis.
We used a single sea otter sequence (the southern sea otter) as representative of the sea otter lineage in the phylogeny because the split time between the northern and southern sea otter subspecies is too recent (<100,000 years) to reliably estimate divergence time using phylogenetic methods and ancient fossil calibrations. The topology of (domestic ferret (giant otter (northern sea otter, southern sea otter))) was confirmed using the ABBA–BABA test (Green et al. 2010; Durand et al. 2011) which, as expected based on their divergent evolutionary history and widely separated geographic range, showed no significant evidence for any gene flow from the giant otter lineage into either of the two sea otter lineages (Patterson’s D-statistic: −0.00066, Z-test P-value: 0.9).
The Importance of Reducing False Positives in Analyses of Positive Selection
To detect nonsynonymous changes under positive selection in the southern sea otter and giant otter lineages, we identified 15,317 single-copy orthologs in our two de novo genome assemblies and 13 additional mammal species (see supplementary table S1, Supplementary Material online, for species information). With these sequences, we performed the branch-site test using phylogenetic analysis by maximum likelihood (PAML)’s codeml program (Yang 2007) with the southern sea otter, giant otter, and the joint lineage leading to both otter species as foreground branches.
The branch-site test is highly sensitive to errors in sequence alignment (Wong et al. 2008; Mallick et al. 2009; Schneider et al. 2009; Fletcher and Yang 2010; Markova-Raina and Petrov 2011; Jordan and Goldman 2012; Privman et al. 2012; Harrison et al. 2014) and therefore alignments have the potential to be highly enriched for false positives. We therefore carried out extensive filtering of alignments using multiple filtering techniques to mitigate false positives. First, we aligned orthologous genes with the PRANK algorithm (Löytynoja 2014) and inconsistent residues were masked using GUIDANCE2 (Sela et al. 2015). The alignments were then further masked for inconsistent regions using two different schemes. Specifically, Gblocks (Castresana 2000) was used to select conserved blocks of sequence or a sliding window approach (sliding window alignment masker for PAML [SWAMP]) (Harrison et al. 2014) was used to mask regions of the alignment with excessive amino acid changes. We then visually inspected the alignments of genes identified as being under positive selection (P-value ≤ 0.01) by the branch-site test in each data set, manually evaluating over 600 gene alignments. Genes were flagged as likely containing spurious signals of selection if the regions of the alignment containing the significant amino acid change were close to insertions or deletions, surrounded by large gaps in the sequence due to fragmented annotations or differences in exon structure across species, or near the start or end of the alignment where de novo annotations can sometimes include small amounts of nongenic sequence.
We found that no filtering strategy could fully mitigate the false positives caused by alignment errors. SWAMP-filtered alignments led to a lower number of significant genes, only 13 prior to visual inspection, compared with 325 identified in the Gblocks-filtered alignments. However, we found that 75–88% of alignments with a P-value ≤0.01 from the Gblocks-filtered alignments were likely alignment artifacts, and 38–76% were artifacts under the SWAMP filtering scheme (supplementary table S3, Supplementary Material online; see supplementary table S4A–C, Supplementary Material online, for all genes with P-value ≤0.01 passing visual inspection). The false-positive rate was most extreme for the outlier alignments that passed genome-wide significance with a false discovery rate (q-value) of 10%. Of these outlier alignments, 88–98% failed visual inspection under the Gblocks filtering scheme, and 75–100% failed under SWAMP. Gblocks overall had a higher number of putatively real genes passing inspection, with 18 genes passing inspection compared with only one gene for SWAMP (which was also significant under Gblocks) (table 2 and supplementary table S3, Supplementary Material online). Thus, though the Gblocks filter is too liberal, yielding 307 false positives, the SWAMP filter may be too conservative, possibly missing 17 true positives found with Gblocks.
Table 2.
Genes Identified as Putatively under Positive Selection.
| HGCN Symbol | Foreground Branch | (1) SWAMP q-Value | (1) SWAMP P-Value | (2) Gblocks q-Value | (2) Gblocks P-Value | Gene Name |
|---|---|---|---|---|---|---|
| BEND7 a | S. sea otter | 2.36E-02 | 5.68E-06 | 1.80E-03 | 2.74E-06 | BEN domain containing 7 |
| MAST2 | S. sea otter | 1.00E+00 | 4.32E-02 | 7.14E-03 | 2.53E-05 | Microtubule-associated serine/threonine kinase 2 |
| RELN | S. sea otter | x | x | 1.30E-02 | 6.02E-05 | Reelin |
| FAM111A | S. sea otter | x | x | 1.91E-02 | 9.85E-05 | Family with sequence similarity 111, member A |
| SLC18A3 | S. sea otter | 1.00E+00 | 1.00E+00 | 2.83E-02 | 1.69E-04 | Solute carrier family 18 (vesicular monoamine), member 3 |
| CEP350 | S. sea otter | 1.00E+00 | 3.98E-03 | 2.90E-02 | 1.76E-04 | Centrosomal protein 350 |
| SWAP70 | S. sea otter | 3.75E-01 | 2.33E-04 | 3.73E-02 | 2.40E-04 | SWA-70 protein |
| SLC7A4 | S. sea otter | 3.42E-01 | 1.92E-04 | 4.02E-02 | 2.66E-04 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| ZNF200 | S. sea otter | 1.00E+00 | 3.46E-03 | 4.55E-02 | 3.11E-04 | Zinc finger protein 200 |
| ISG20 | S. sea otter | 1.00E+00 | 6.85E-02 | 6.15E-02 | 4.49E-04 | Interferon-stimulated protein |
| PGR | S. sea otter | x | x | 7.04E-02 | 5.27E-04 | Progesterone receptor |
| TPGS1 | S. sea otter | x | x | 7.88E-02 | 6.14E-04 | Tubulin polyglutamylase complex subunit 1 |
| HADHA | S. sea otter | 6.26E-01 | 4.81E-04 | 7.90E-02 | 6.25E-04 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit |
| SLAMF7 | S. sea otter | 1.00E+00 | 8.75E-03 | 9.76E-02 | 8.20E-04 | SLAM family member 7 |
| PRSS35 | Giant otter | x | x | 4.31E-02 | 2.91E-04 | Protease, serine 35 |
| WBP1 | Giant otter | 4.07E-01 | 2.64E-04 | 7.90E-02 | 6.25E-04 | WW domain-binding protein 1 |
| SIK2 | Ottersb | 4.97E-01 | 3.52E-04 | 7.60E-02 | 5.83E-04 | Salt inducible kinase 2 |
| IQCD | Otters | 1.00E+00 | 1.33E-03 | 8.56E-02 | 6.93E-04 | IQ motif containing D |
Note.—Eighteen genes under positive selection were detected using the branch-site test (Yang 2007) on the branches leading to the southern sea otter, giant otter, and the ancestral otter internal branch. Two different alignment filtering schemes were used: (1) GUIDANCE2 (Sela et al. 2015) plus sliding window filtering (SWAMP; Harrison et al. 2014); or (2) GUIDANCE2 plus conserved block filtering (Gblocks; Castresana 2000). Genes with q ≤ 0.1 under either filtering scheme are shown. Additional details on these genes available in supplementary table S4D, Supplementary Material online. SWAMP: p and q-values based on codeml run on gene alignments filtered first using GUIDANCE2 then stringent SWAMP sliding window masking, with columns of the alignments containing any gaps removed. Genes with length <120 bp after filtering were excluded. Gblocks: p and q-values based from PAML’s codeml run on gene alignments filtered first using GUIDANCE2 then moderately stringent Gblocks conserved block selection filtering, with columns of the alignment with >50% of the sequences containing a gap removed. Genes with length <120 bp after filtering were excluded. “x” denotes a gene that was excluded under one of the filtering schemes if its length postfiltering was <120 bp.
BEND7 is the only gene with q-value ≤ 0.1 under both Gblocks and SWAMP filtering schemes.
The branch leading to both otter lineages.
By applying both filters independently, we found that the genes that had moderate likelihood ratio test (LRT) statistic scores under both filtering schemes tended to pass the visual inspection, whereas those with high LRT scores under only one filtering scheme tended to be artifacts (supplementary figs. S3–S5, Supplementary Material online). Further, all significant outlier genes passing visual inspection contained multinucleotide changes in the codons identified as being under positive selection, which can inflate the signal of selection (Schrider et al. 2011; Venkat et al. 2018). We retained these gene alignments, but noted the presence of the multinucleotide changes (supplementary table S4A–D, Supplementary Material online). Finally, every southern sea otter amino acid change identified as possibly being under selection in the outlier genes (q ≤ 0.1) was confirmed to be present in the northern sea otter sequence as well (supplementary table S4D, Supplementary Material online). Overall, our rigorous examination yielded a relatively small set of 18 significant outlier genes (q ≤ 0.1) passing inspection that may represent real signals of positive selection (table 2).
Positive Selection on Single Genes
We used literature searches, gene ontology enrichment tests (Reimand et al. 2016), and the Entrez Gene (Maglott et al. 2011), Uniprot (UniProt Consortium 2016), International Mouse Phenotyping Consortium (IMPC) (Dickinson et al. 2016), RefSeq (O’Leary et al. 2016) and Online Mendelian Inheritance in Man (OMIM) (Hamosh et al. 2004) databases to explore potential functions of the 18 genes we identified above as being under positive selection (table 2). As the set of significant genes is relatively small, tests of gene ontology enrichment yielded no significant results. Individually, several of these genes have interesting phenotypic associations. On the joint branch leading to both otter lineages, there were two significant genes. One of these (IQCD) could not be directly linked to a particular phenotype, whereas SIK2, which is thought to be involved in insulin regulation (Horike et al. 2003), is associated with the “enlarged heart” phenotype in mice (Dickinson et al. 2016). We found that the giant otter lineage had two genes showing significant signals of positive selection, PRSS35, a member of the serine protease family uniquely expressed in mouse ovaries, and WPB1, a transferase involved in protein glycosylation in yeast, but which could not be linked to a particular phenotype. The southern sea otter foreground branch showed the most intriguing phenotypic associations. There were 14 genes putatively under positive selection, six of which could not be linked to phenotypes (BEND7, HADHA, SLC18A3, CEP350, SWAP70, and SLC7A4) and eight of which had interesting functional associations: FAM111A is related to Kenny Caffey Syndrome and Gracile Bone Dysplasia in humans, disorders marked by short stature and shortening and cortical thickening of limb bones (Unger et al. 2013) (fig. 2a). RELN may be related to brain development and synaptic plasticity (Weeber et al. 2002; Tissir and Goffinet 2003). Two genes are related to immune function (ISG20 and SLAMF7) and four are related to reproduction (MAST2, PGR, TPGS1 and possibly ZNF200). MAST2 is also associated with the following phenotypes in mice: “increased bone mineral content,” “increased mature B-cell number,” “decreased total body fat amount,” and increased and decreased circulating iron levels (Dickinson et al. 2016).
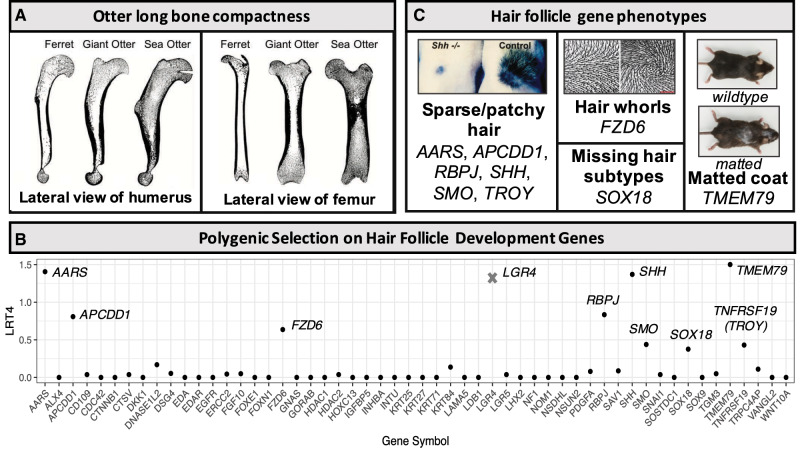
Fig. 2.
Positive selection on single and many genes. (A) Differences in long bones between terrestrial mustelids (domestic ferret), freshwater otters (giant otter), and the sea otter (modified from Houssaye and Botton-Divet [2018] with permission of Oxford University Press and the Linnean Society of London). Lateral views of the forelimb humerus and hindlimb femur based on high-resolution tomography are shown (Houssaye and Botton-Divet 2018). Differences in long bone shape and compactness, including increased thickness of outer cortical bone and increased spongy bone throughout the bone shaft, are observed in both otter species, but are particularly extreme in the sea otter. The positive selection we detected on genes FAM111A and MAST2 may contribute to these skeletal differences (table 2). (B) The gene category identified as being under significant polygenic selection in the ancestral otter lineage. The y axis shows the fourth root of the likelihood ratio test statistic score (LRT4) that is used to generate a cumulative score for the gene set. Genes in the category with the highest LRT4 scores are labeled. Upon visual inspection, LGR4 (marked by gray x) had a fragmented alignment that yielded a false signal of moderate selection, and so the cumulative score was recalculated without it. (C) Mouse and/or human phenotypes associated with mutations or knockouts in the highest LRT4 score genes in the “hair follicle development” category. For the sparse/missing hair phenotype, the image shown is an SHH mouse knockout skin graft onto a wildtype mouse back showing limited hair growth compared with a control (adapted from figure 3 in Chiang et al. [1999] with permission from Elsevier). For the hair whorls phenotype, the image depicts the difference between wildtype (left) and FZD6 knockout (right) hair orientation patterns in mice (adapted from figure 1E and F in Wang et al. [2006]). For the matted phenotype, wildtype and TMEM79 mutant mice with a patchy, matted coat are shown (adapted from figure 3 in Saunders et al. [2013]).
Polygenic Selection
The complex aquatic adaptations of otters may be controlled by variants of small effect in many genes rather than a single gene. Under this scenario, no single gene may show significant evidence of positive selection using the branch-site test described above. A typical gene ontology enrichment approach such as g:Profiler (Reimand et al. 2016) tests for an enrichment of gene ontology categories in the set of significant genes. To detect polygenic selection, we instead employed a different approach, polysel (Daub et al. 2013, 2017), which tested particular gene ontology categories for enrichment of groups of genes showing a slight increase in nonsynonymous divergence based on the branch-site test. To reduce the impact of multiple testing on our results, we a priori chose a set of gene categories to test that are related to complex traits possibly functioning in the unique aquatic transition that the otter lineage experienced (hypoxia resistance, thermoregulation, diet, sensory perception, osmoregulation, and hair growth), rather than testing every known gene ontology category for enrichment of genes under selection. Specifically: 1) hypoxia resistance which may enable otters to dive longer without being harmed by a lack of oxygen, and has been observed under positive selection in marine and high-altitude mammals (Zhang et al. 2014; Tian et al. 2016); 2) thermoregulatory changes to withstand cold temperatures in aquatic environments; 3) a dietary shift to fish and invertebrates from small mammals and birds; 4) altered tactile senses, hearing and vision for underwater behavior (Murphy et al. 1990; Strobel et al. 2018); 5) osmoregulation associated changes to kidney function to concentrate salt content as an adaptation for using sea water as a source of fresh water (Ortiz 2001); and 6) genetic changes to hair development underlying the remarkable hair density of otter species (Kuhn and Meyer 2010a).
Because polysel has the most power to detect selection on the internal foreground branches of a phylogeny (Daub et al. 2013, 2017), we tested for polygenic selection on the lineage leading to the southern sea otter and giant otter. The branch-site LRT scores were from the Gblocks-filtered gene alignments. The top-scoring and only significantly enriched gene ontology category was “hair follicle development” (fig. 2B and supplementary table S5A and B, Supplementary Material online), a gene ontology category which contained 55 genes that were found in our data set, ten of which had moderate-to-low (P-values ranging from 0.02 to 0.89) LRT scores from the branch-site test (supplementary table S5B, Supplementary Material online). The fact that we only found a single GO term under putative polygenic selection could indicate a lack of power to detect subtle amino acid divergence in the other categories, or a lack of polygenic selection. Additional data from multiple individuals could improve power to detect sites under selection by comparing polymorphism and divergence.
To determine whether the signal of positive selection on hair follicle development might be influenced by false positives due to poor sequence alignments, we subjected the sequence alignments of the four top scoring genes in the category (AARS [alanyl-tRNA synthetase], TMEM79 [transmembrane protein 79], SHH [sonic hedgehog], and LGR4 [leucine-rich repeat containing G-protein-coupled receptor 4]) to the same visual inspection (described above) that we did for the genes passing genome-wide significance. The LGR4 gene alignment was highly fragmented, with large gaps in the southern sea otter sequence. The site identified as under positive selection was in a poorly aligned region of the sequence that was surrounded by large gaps. Removal of LGR4 increased the P-value on the “hair follicle development category” from 0.0048 to 0.019, and the q-value from 0.099 to 0.33. However, it remained the top-scoring and only significant gene category in the analysis. The fact that the signal persisted despite the removal of LGR4 is due to the fact that several genes contribute to the category’s signal, and so the removal of LGR4 does not dramatically alter the result.
The remaining nine genes driving the signal of selection include genes involved in hair follicle organogenesis (SHH, SMO [smoothened frizzled class receptor], SOX18 [SRY-box 18], TNFRSF19 [TROY] [TNF receptor superfamily member 19], and RBPJ [recombination signal binding protein for immunoglobulin kappa J region]) and/or changes to hair or fur phenotype in humans or mice (AARS, APCDD1 [APC downregulated 1], FZD6 [frizzled class receptor 6], RBPJ, and TMEM79) (fig. 2B and C). Although none shows a strong selective signal individually, the cumulative effect of small changes across these genes may be responsible for the remarkable pelts of otter species.
Pseudogenization in Otter Genomes
We identified genes that may no longer be functional after the transition to a marine or aquatic environment in the southern sea otter and giant otter. We labeled genes as putative pseudogenes if they passed both of two criteria: 1) The protein sequence in the domestic ferret was not annotated in the southern sea otter and/or giant otter de novo genome assemblies due to insertions or deletions, premature stop codons, frameshifts or retrotransposition, identified using PseudoPipe (Zhang et al. 2006); and 2) the gene was also identified by Ensembl’s Variant Effect Predictor (VEP) (McLaren et al. 2016) as containing high-impact variants (gain/loss of stop codon, loss of splice donor/acceptor, loss of start codon, frameshift) when southern sea otter and giant otter reads were aligned to the domestic ferret genome (supplementary table S6A and B, Supplementary Material online). Using this approach, 113 putative pseudogenes were found in the southern sea otter, 133 in the giant otter, and 45 in both otter species (supplementary table S6A and B, Supplementary Material online). The putative pseudogenes in each lineage were tested for enrichment of gene ontology terms. For each species, we found that all the significant (P < 0.005) categories were related to sensory perception of chemical stimuli (supplementary table S6C, Supplementary Material online). Notably, there were several olfactory receptor genes (ORGs) identified as pseudogenized in each otter lineage, as well as two bitter taste receptors (TAS2R38 [taste receptor 2 group 38] pseudogenized in giant otter and TAS2R40 [ taste receptor 2 group 40] in both otter species), as well as a gene involved in inflammatory response to pathogens (NLRC4 [NLR family CARD domain containing 4]) pseudogenized in both otter species, and a gene related to neuronal development of sensory organs (NAV2) pseudogenized in the southern sea otter (supplementary table S6C, Supplementary Material online). In sum, this pattern of pseudogenization suggests reduced selection on gustatory and olfactory genes in both otter lineages likely related to their evolution in aquatic environments.
Loss of ORGs
To further explore potential gene loss associated with sensory perception in the otter lineage, we characterized the functional and pseudogenized ORG repertoires using our de novo southern sea otter and giant otter genome assemblies and the domestic ferret reference genome. To account for difficulties with ORG detection and differences in genome assembly quality, we used the same procedures on the more contiguous northern sea otter genome (Jones et al. 2017) as an independent reference. We then combined this data set with a larger ORG data set that included the same set of species used to reconstruct the phylogenetic tree in order to generate a gene tree (fig. 1).
We found that the sea otter and giant otter lineages had separately lost hundreds of ORGs since their split from the domestic ferret, their terrestrial relative (fig. 3 and supplementary fig. S6, Supplementary Material online), but showed similar functional ORG repertoires between the otter lineages. The domestic ferret’s ORG repertoire of 816 functional genes and 377 pseudogenes had significantly more functional ORGs than the southern sea otter (524 functional ORGs, 538 pseudogenes), the northern sea otter (569 functional ORGs, 589 pseudogenes), and the giant otter (584 functional ORGs and 671 pseudogenes; chi-square test P-values for all comparisons <2.2 × 10−16; supplementary table S7A, Supplementary Material online). Functional ORGs were classified as Class I and Class II based on their phylogenetic positions (Niimura 2013), and there were no significant differences between the domestic ferret and the otter species in the relative proportions of functional ORGs in the two classes, with Class I ORGs making up 16–18% of the functional repertoire in each species (chi-square test; supplementary table S7A, Supplementary Material online).
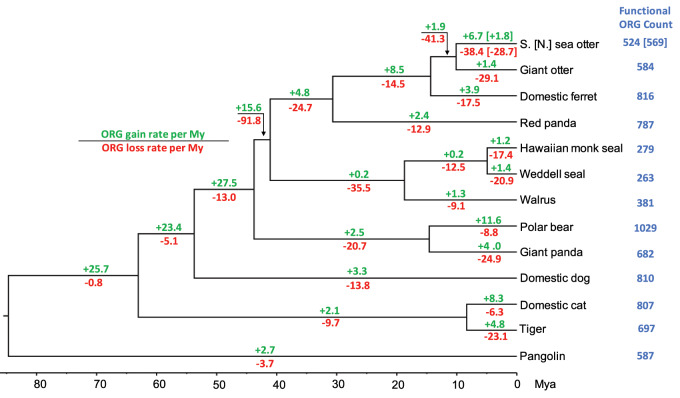
Fig. 3.
Rates of gain and loss of functional ORGs. Rates of ORG gain and loss per million years are shown for each branch of the phylogeny. These rates are calculated by dividing the counts of functional ORG gained or lost (supplementary fig. S6, Supplementary Material online) by the branch lengths calculated from the mean node ages (fig. 1). Rates are shown for both the southern (“S.”) and northern (“N.”) sea otter genomes, with the latter in brackets. The total number of functional ORGs in each species is shown to the right of the phylogeny.
We compared the functional ORG repertoires of other mammalian species (Hughes et al. 2018) with otters (supplementary fig. S7 and table S7B, Supplementary Material online). The two otter species have intermediate-sized ORG repertoires that are smaller than the polar bear, a species that more recently became aquatic (524–584 functional ORGs in otters vs. 1,029 in polar bear), but larger than those of pinnipeds and cetaceans which represent much older radiations and have very low numbers of functional ORGs (263–371 in pinnipeds and 58 in bottlenose dolphin) (Hughes et al. 2018).
When scaled by the branch lengths of the phylogenetic tree, the southern/northern sea otter branch lost 29–38 ORGs per My and the giant otter lost 29 per My (fig. 3). Although the limited taxonomic sampling makes direct rate comparisons difficult, these rates are elevated relative to most of the terrestrial species surveyed, particularly the related domestic ferret (18 genes lost per My), and the ancestral lineage leading to mustelids (15 genes lost per My) (fig. 3). Interestingly, the branch ancestral to the sea otter and giant otter has a high rate of ancestral ORG loss (losing 175 ORGs at a rate of 41 genes lost per My; fig. 3), which is comparable to the rate of loss in the ancestor of pinnipeds (36 genes lost per My).
The moderate difference in the number of functional ORGs between the northern and southern sea otter (524 vs. 569) was not significant (chi-square test P-value: 0.98; supplementary table S7A, Supplementary Material online) and is likely due to differences in the completeness and contiguity of the two genomes (southern sea otter scaffold N50: 6.6 Mb, contig N50: 18.4 kb, genome size: 2,425.5 Mb; northern sea otter scaffold N50: 38.7 Mb, contig N50: 244.5 kb, genome size: 2,455.2 Mb; table 1). The high contiguity of the northern sea otter genome may enable better detection of functional ORGs. For example, all Class I olfactory receptors are generally found on the same chromosome (chromosome 11 in humans), whereas Class II are spread throughout the genome (Niimura and Nei 2003). In the northern sea otter genome assembly, 99% of Class I ORGs were found on a single scaffold with only one on a different scaffold (supplementary table S6, Supplementary Material online), indicating that that region of the chromosome is fully assembled. In the southern sea otter assembly, 74% of Class I ORGs were also found on a single scaffold, but the rest were scattered across 25 other small scaffolds (supplementary table S6, Supplementary Material online). Some functional or pseudogenized Class I and Class II ORG sequences may therefore be missing due to incomplete assembly in the southern sea otter, likely making the northern sea otter ORG count the more accurate one for the sea otter lineage.
Genomic Diversity
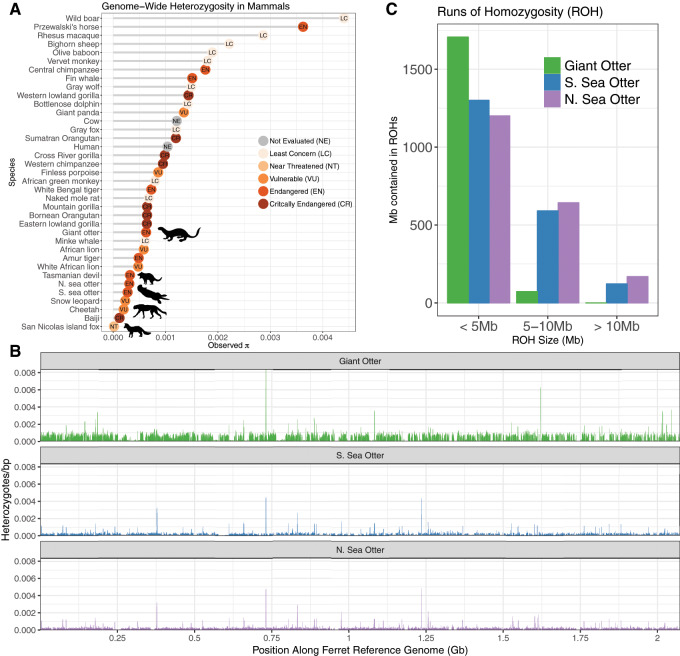
We next assessed patterns of genome-wide variation in the otter genomes. Specifically, we used the domestic ferret genome as an outgroup genome to avoid reference genome bias in our analyses. After mapping the southern sea otter, northern sea otter, and giant otter sequencing reads to the domestic ferret reference genome, we found that all three taxa have very low genome-wide heterozygosity. The southern sea otter and northern sea otter had the same level of genome-wide heterozygosity (0.0003 heterozygous sites/basepair), which is 2× lower heterozygosity than the giant otter (0.0006 heterozygous sites/bp) (fig. 4A). Compared with values of genome-wide heterozygosity for other organisms gathered from the literature and modified from Robinson et al. (2016) (see supplementary table S8, Supplementary Material online, for DOIs and species information), the sea otter genomes have extremely low heterozygosity. Their low levels of heterozygosity are comparable to hallmark low-diversity endangered species such as the cheetah (Dobrynin et al. 2015) and Tasmanian devil (Miller et al. 2011) (fig. 4A). We found that heterozygosity in sliding windows across the genome is generally lower for both sea otters than it is for the giant otter (fig. 4B).
Fig. 4.
Genomic diversity in the southern sea otter and giant otter. (A) Comparison of genome-wide heterozygosity in sea otter, giant otter, and other mammals drawn from the literature, based on Robinson et al. (2016). Dots are colored by the endangered status according to the International Union for Conservation of Nature (IUCN) Red List for Threatened Species. A full table of values with references can be found in supplementary table S8, Supplementary Material online. (B) Sliding window heterozygosity of the giant otter (green), southern sea otter (blue), northern sea otter (purple) sequencing reads mapped to the domestic ferret reference genome (220 scaffolds >3 Mb). (C) ROH across the largest 220 scaffolds (>3 Mb) of the domestic ferret reference genome. The ROH are binned by size of the run, and the y axis depicts the amount of sequence contained in ROH of that size.
The sea otters and giant otter also show differences in the distribution of runs of homozygosity (ROH), calculated across all domestic ferret reference scaffolds >3 Mb. Importantly, ROH were assessed using reads from both species aligned to the domestic ferret reference genome, so the relative lack of contiguity in the giant otter de novo genome is not a factor explaining the different distributions. We found that the southern and northern sea otters have considerably more sequence contained in long ROH 5–10 Mb in length than the giant otter (591 Mb in southern sea otter and 643 Mb in northern sea otter vs. 73 Mb in giant otter), and in extremely long ROH >10 Mb (122 Mb in southern sea otter and 169 Mb in northern sea otter vs. 0 Mb in giant otter). This pattern is an indication of more recent inbreeding in the bottlenecked sea otter populations than in the nonbottlenecked giant otter (fig. 4C).
Historical Demography
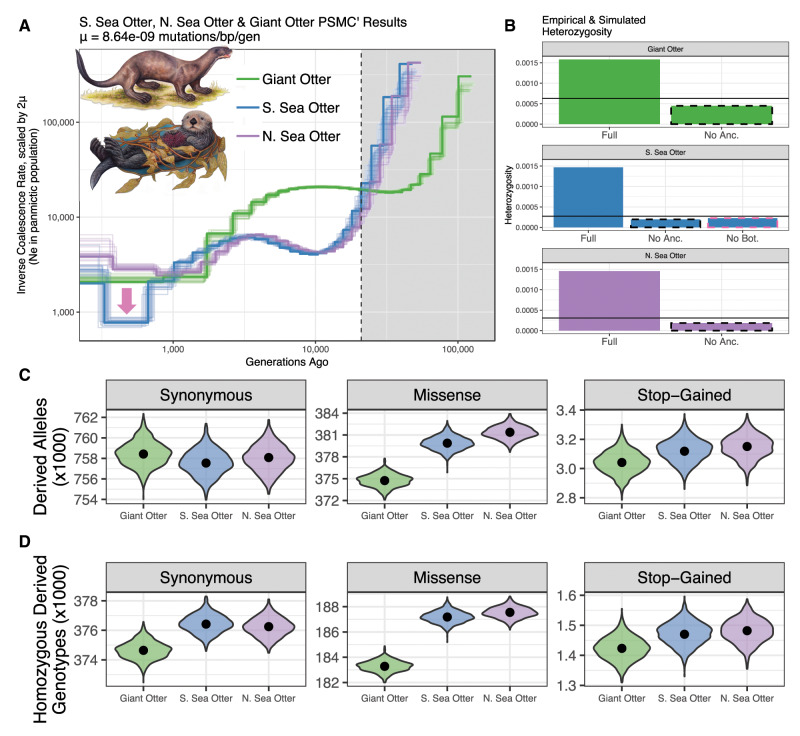
We used the multiple sequentially Markovian coalescent (MSMC) (also known as PSMCʹ [pairwise sequentially Markovian coalescent using the SMCʹ algorithm (the other name for MSMC when it is run on a single genome)]) (Schiffels and Durbin 2014) to infer the history of coalescent rate changes for the southern sea otter, northern sea otter, and giant otter. MSMC calculates the instantaneous inverse coalescence rate (IICR) (Schiffels and Durbin 2014; Mazet et al. 2016) over a series of time intervals which when scaled by two times the mutation rate (2 μ) is equivalent to the effective population size (Ne) in a panmictic population over time (Schiffels and Durbin 2014) (fig. 5A). We scaled each MSMC trajectory by three sets of mutation rates and generation times to assess the range of plausible timings for each part of the trajectory (supplementary fig. S8 and tables S9 and S10A–C, Supplementary Material online). Beichman et al. (2017) showed that trimming off ancient events from MSMC curves can improve the fit to other summaries of the data. We therefore simulated data under the inferred demographic models with and without ancient events of >∼21,000 generations in the past (dotted line in fig. 5A) and compared the heterozygosity predicted by our simulations with empirical heterozygosity. As expected, removing the period of ancient high population size improved the fit to empirical heterozygosity in both species (fig. 5B).
Fig. 5.
Demographic inference and the burden of deleterious variants. (A) MSMC (Schiffels and Durbin 2014) inference of inverse coalescent rate (a proxy for effective size) through time for giant otter (green), southern sea otter (blue), and northern sea otter (purple). Fine lines denote bootstrap replicates. (B) Empirical genome-wide heterozygosity (horizontal black line) for each individual compared with heterozygosity simulated under the full MSMC model and under the “No Ancient (Anc.)” model which trimmed away the ancient shaded area in (A). Southern sea otter heterozygosity was also simulated under the “No Bottleneck (Bot.)” model which excluded ancient events as in the “No Anc.” model and additionally excluded the bottleneck marked by the pink arrow in (A). (C) The count of derived alleles (scaled down by 1000) that are annotated as synonymous (non-amino acid changing), missense (amino acid changing), or stop-gained (resulting in a premature stop codon) relative to domestic ferret. Black dots indicate the empirical count of derived alleles rescaled by the average number of genotyped sites in coding regions between giant otter, southern sea otter, and northern sea otter. The distribution around each point represents 1,000 bootstraps in which the average number of genotyped coding sites was sampled with replacement from each individual. The southern sea otter and northern sea otter had significantly more missense-derived alleles than the giant otter (P-values: giant otter vs. southern sea otter: 4.85 × 10−5, giant otter vs. northern sea otter: 6.74 × 10−8). (D) The count of genotypes (scaled down by 1000) in coding regions that are in the homozygous-derived state relative to domestic ferret, rescaled and bootstrapped as described in (C).
Once the ancient events are trimmed, the giant otter’s MSMC trajectory shows a relatively constant IICR, followed by a steady decline in population size to the present day (fig. 5A). The southern and northern sea otters’ MSMC trajectories are much more dynamic, with a more ancient period of decline in IICR detected in both sea otters, followed by a period of increasing IICR, and a more recent dip in IICR in the southern sea otter population. These declines could represent population bottlenecks, which is the usual interpretation of dips in PSMC or MSMC curves. However, the fluctuations could also be caused by changes in population structure, admixture or migration (Mazet et al. 2016; Beichman et al. 2017; Chikhi et al. 2018). The more ancient dip in IICR observed in both northern and southern sea otters occurred ∼9,000–10,000 generations ago, or ∼35–45 ka assuming a 4-year generation time (see supplementary fig. S8 and table S10A and B, Supplementary Material online, for additional mutation rate and generation time scaling). The more recent dip observed only the southern sea otter trajectory occurred ∼300–700 generations ago or ∼1–3 ka assuming a 4-year generation time (and therefore long before the 18th–20th century fur trade) and would correspond to a population effective size of ∼800 individuals if the IICR is used as a proxy for Ne (see supplementary fig. S8 and table S10A, Supplementary Material online, for other scalings).
We used coalescent simulations to test whether the presence of the recent dip in IICR was consistent with a bottleneck in the southern sea otter population in California prior to the fur trade (marked by the pink arrow in fig. 5A). We found that simulations under the model with and without the bottleneck did not dramatically alter the fit of simulated heterozygosity to empirical heterozygosity. The simulated distributions of heterozygosity across 100-kb windows were highly similar, mainly differing in the number of windows with zero heterozygous sites (supplementary fig. S9, Supplementary Material online). This finding indicates that there is not sufficient information contained in a single genome to distinguish among models based on one, two, or three recent bottlenecks. Nonetheless, the fluctuations in the sea otter MSMC trajectories compared with the relatively stable trajectory of the giant otter point to a more dynamic population history, potentially influenced by multiple changes in population size or structure in the southern sea otter.
Deleterious Variation
We used VEP (McLaren et al. 2016) to classify the impact of coding variants in the southern sea otter, northern sea otter, and giant otter relative to the domestic ferret reference genome. We classified variants as synonymous (non-amino acid changing), missense (amino acid changing), or stop-gained (introducing a premature stop codon) and rescaled the counts by the number of called sites in coding regions for each species. We found that the southern sea otter, northern sea otter, and giant otter showed similar numbers of synonymous (putatively neutral) derived alleles relative to the domestic ferret (fig. 5C and supplementary table S11, Supplementary Material online), which is expected under neutral models (Simons et al. 2014) (Z-test P-values for all comparisons >0.6; supplementary table S11, Supplementary Material online). However, the southern and northern sea otters have significantly higher counts of missense-derived alleles, 1.4–1.8% more than what is observed in the giant otter, indicating a potentially higher additive genetic load in small populations (Z-test P-values: giant otter vs. southern sea otter, 4.8 × 10−5; and giant otter vs. northern sea otter, 6.7 × 10−8; fig. 5C and supplementary table S11, Supplementary Material online). The number of missense-derived alleles did not differ between the northern and southern sea otter populations (supplementary table S11, Supplementary Material online). We did not find significant differences in the number of stop-gained variants between the southern sea otter, northern sea otter, and giant otter suggesting that potentially highly deleterious variants are eliminated in sea otter populations despite past population bottleneck(s) (Z-test P-values for all comparisons > 0.3; fig. 5C and supplementary table S11, Supplementary Material online).
We also found that the southern and northern sea otters had more derived alleles in the homozygous state compared with the giant otter (fig. 5D and supplementary table S11, Supplementary Material online). For synonymous homozygous-derived genotypes, the difference was not significant after correction for multiple testing (supplementary table S11, Supplementary Material online). However, the sea otters had 2.1–2.3% more missense homozygous-derived genotypes than the giant otter, which was highly significant (Z-test P-values: giant otter and southern sea otter: 5.8 × 10−10, giant otter and northern sea otter: 2.7 × 10−12; supplementary table S11, Supplementary Material online). This difference is notable because recessive deleterious alleles that appear in the homozygous state would have a higher impact on fitness (Peischl and Excoffier 2015). However, we did not observe a significant difference between the southern sea otter, northern sea otter, and giant otter in the stop-gained homozygous-derived genotype category, implying that potentially highly deleterious variants are eliminated when they appear in the homozygous state regardless of differences in demography among these populations (Z-test P-values > 0.2 for all comparisons; fig. 5D and supplementary table S11, Supplementary Material online).
Discussion
We report a large-scale comparative genomic analysis of the marine-adapted sea otter from California and Alaska and the freshwater giant otter from South America, two highly diverged otter species. We found that these large, semiaquatic mustelids diverged from each other 10.1 Ma and from the domestic ferret 14.4 Ma (fig. 1) and that the southern sea otter showed intriguing signals of positive selection on genes related to reproduction, immune function, memory, and limb development (table 2). Both species showed a signal of polygenic positive selection acting on genes related to hair follicle development (fig. 2). Each species also experienced considerable loss of sensory genes, particularly olfactory receptors, consistent with patterns observed in other aquatic mammals (supplementary fig. S7A, Supplementary Material online). At the population level, we compared the giant otter to the southern sea otter from California and its Alaskan relative, the northern sea otter, both of which experienced extreme population bottlenecks due to the fur trade. We found that the southern and northern sea otters had extremely low genetic diversity (fig. 4A and B), with long ROH indicative of recent inbreeding (fig. 4C). The sea otters and the giant otter had differing demographic histories (fig. 5A) possibly reflecting the different climatic histories of the North Pacific and tropical South America and the impacts of indigenous hunting prior to the European fur trade. Due to this likely history of population declines, southern and northern sea otters showed elevated levels of putatively deleterious variants relative to the giant otter (fig. 5C and D) which could impact the capacity for the populations to recover.
Phylogeny and Divergence Times
Our phylogeny and associated divergence times (fig. 1) are largely consistent with previous phylogenetic analyses of carnivoran (Eizirik et al. 2010; Meredith et al. 2011; Nyakatura and Bininda-Emonds 2012) and, specifically, musteloid evolution (Koepfli et al. 2008; Sato et al. 2009, 2012; Harding and Smith 2009; Yu et al. 2011; Waku et al. 2016). Our estimate of sea otter and giant otter divergence of 10.1 Ma (fig. 1) is slightly higher than earlier estimates based on a much smaller sampling of orthologous genes (Koepfli et al. 2008; Nyakatura and Bininda-Emonds 2012), which place the divergence at 7–9 Ma, and lower than another estimate of 15 Ma based on the mitochondrial genome (Waku et al. 2016). However, the previous estimates fall into our 95% CI of 6.6–15.4 Ma (fig. 1). Furthermore, our estimate of the age of divergence between the otter lineage and domestic ferret of 14.4 (10.4–20.1) Ma (fig. 1) is concordant with past estimates of a divergence time of 10–17 Ma (Harding and Smith 2009; Sato et al. 2009, 2012; Yu et al. 2011; Nyakatura and Bininda-Emonds 2012; Waku et al. 2016) but higher than estimates which place the divergence around 9 Ma (Koepfli et al. 2008; Eizirik et al. 2010). In general, our results confirm the findings from previous studies that lutrines and other mustelids began to diversify during the Miocene epoch.
The Importance of Reducing False Positives in Analyses of Positive Selection
A growing body of literature highlights the impact of alignment errors on false-positive rates in the branch-site test for positive selection (Wong et al. 2008; Mallick et al. 2009; Schneider et al. 2009; Fletcher and Yang 2010; Markova-Raina and Petrov 2011; Jordan and Goldman 2012; Privman et al. 2012; Harrison et al. 2014). Alignment errors can be exacerbated by evolutionary distance between species (Rosenberg 2005) and fragmented or incorrect gene annotations due to assembly errors (Schneider et al. 2009; Markova-Raina and Petrov 2011). Our results serve as an empirical case study of the impact of alignment errors and different filtering methods on the results of the branch-site test for positive selection. Alignment with PRANK (Löytynoja 2014) and residue filtering using GUIDANCE2 (Sela et al. 2015) followed by conserved sequence selection using the more lenient Gblocks (Castresana 2000) or a sliding window mask using the highly stringent SWAMP (Harrison et al. 2014) were each imperfect solutions to the problem of alignment error. Even after filtering, 75–100% of significant gene alignments failed visual inspection (supplementary table S3, Supplementary Material online). Though it is labor intensive, we recommend comparing multiple filtering schemes of varying levels of stringency through rigorous visual inspection of significant alignments. This approach can largely mitigate the impact of the extremely high false-positive rate of the branch-site test, while still retaining some genes that may be incorrectly excluded by the most stringent automated filters.
Our results, and those of others, are also likely influenced by the presence of multinucleotide changes within a codon as the codeml branch-site test implemented in codeml assumes that nucleotide changes are successive not simultaneous which can inflate a signal of selection (Venkat et al. 2018). Every significant gene in our data set showed the presence of at least one codon with multinucleotide changes. We still explored the functions of these genes, as they may be biologically relevant, but we caution that the presence of these changes may not represent successive nucleotide changes to the protein and could therefore be inflating the signal of positive selection. Nonetheless, our resulting set of 18 significant genes (table 2, described below), although relatively small, is not inflated by spurious signals of selection caused by major alignment errors. Therefore, these genes are likely to more accurately represent the role of positive selection during the evolutionary histories of the southern sea otter and giant otter.
Positively Selected Genes
The first evidence of aquatic adaptations in the otter lineage are found in lutrine remains from Middle Miocene deposits (Willemsen 1992; Wang et al. 2018) and further fossils indicate that the sea otter (E. lutris) entered the North Pacific Ocean in the Pliocene–Pleistocene, 1–3 Ma (Riedman and Estes 1990; Boessenecker 2018). We predicted that there would be detectable genomic signals of the transition to semiaquatic life, both in the ancestral lineage leading to sea otter and giant otter and in the sea otter lineage as it rapidly adapted to marine life. Using the southern sea otter genome assembly as a representative sequence for the sea otter lineage, we found several genes that may be related to morphological modifications and behavioral patterns that make them unique among the Lutrinae. In particular, sea otters have divergent forelimb and hindlimb bone shape, density, and length compared with freshwater otters and terrestrial mustelids (Tarasoff 1972; Fish and Stein 1991; Willemsen 1992; Mori et al. 2015; Botton-Divet et al. 2016, 2018; Houssaye and Botton-Divet 2018). Changes in long-bone shape and compactness are observed between all otter species and terrestrial mustelids, with sea otters being the most divergent (Botton-Divet et al. 2016, 2018; Houssaye and Botton-Divet 2018). Sea otters have the shortest relative femur length, which is thought to reduce drag when swimming by bringing the limb closer to the body (Samuels et al. 2013; Mori et al. 2015). They have distinct forelimb bone shapes, as their forelimb is not used for locomotion, but primarily for manipulation of resources (Botton-Divet et al. 2018). Their long bones are denser relative to body size, possibly to provide ballast that enables diving (Tarasoff 1972; Fish and Stein 1991; Willemsen 1992; Mori et al. 2015). Freshwater otters show a trend toward compact long bones as well with thickened outer (cortical) bone and some spongy (trabecular) bone in the shaft of the long bone relative to terrestrial mustelids (fig. 2A) (Houssaye and Botton-Divet 2018). In sea otters this pattern is more extreme, with thick and compact bones that have trabecular bone spread throughout the entire shaft of the long bone (fig. 2A) (Houssaye and Botton-Divet 2018). Positive selection on FAM111A and MAST2 may be involved in these changes to sea otter limbs. FAM111A is related to Kenny Caffey Syndrome and Gracile Bone Dysplasia in humans, both of which involve the shortening and thinning of limbs and higher bone density, among other phenotypes (Unger et al. 2013). In particular, a clinical feature of Kenny Caffey Syndrome is cortical thickening of tubular bones (Unger et al. 2013). FAM111A may therefore play a role in the increased cortical thickness observed in sea otter long bones (Houssaye and Botton-Divet 2018) (fig. 2A). MAST2 is a kinase that functions as part of spermiogenesis (Lumeng et al. 1999) and may therefore be related to sea otter reproduction. However, it is also statistically associated with increased bone mineralization in mice (Dickinson et al. 2016) which may implicate it in the increased bone compactness and expansion of the trabecular network observed in sea otters (Houssaye and Botton-Divet 2018) (fig. 2A).
We also found genes under selection in the southern sea otter that may be involved in brain development (RELN), reproduction (MAST2, PGR, TPGS1, and ZNF200) and immune function (ISG20 and SLAMF7) (table 2). Positive selection on RELN, which encodes the reelin protein, is particularly interesting, as it is involved in the regulation of synaptic plasticity in response to experience (Weeber et al. 2002; Tissir and Goffinet 2003). Sea otters display interindividual variation in prey preferences and tool use that are transmitted along matrilines (Estes et al. 2003; Fujii et al. 2015) which may involve genetic adaptations for increased memory and learning abilities.
In addition to amino acid changes in single genes, adaptations in the southern sea otter and giant otter may be due to more evolutionarily rapid genetic changes, such as changes to gene regulation, copy number variants, or transposable elements which were not examined in this study. Mutations with individually weak effects spread across dozens of genes working in concert could also be responsible for complex adaptive aquatic traits in otters. Traditional gene ontology enrichment analysis that only considers genes showing a genome-wide statistically significant effect may miss cases of polygenic selection (Daub et al. 2017). We found that the “hair follicle development” gene ontology category was enriched for genes under moderate positive selection in the internal branch leading to the sea otter and giant otter. No single hair-related gene had a strong enough signal of selection to rise to genome-wide significance, demonstrating the increase in power using a polygenic approach on the full data set.
Several of the genes in the category that drive the signal of selection are directly involved in hair folliculogenesis (TROY, SHH, SMO, SOX18, and RBPJ). TROY acts early in the ectodysplasin A pathway to initiate follicle development and is expressed in early development of the hair follicle (Millar 2002; Pispa et al. 2008). SMO and SHH, a gene vital to embryonic development of everything from the brain to limbs, appear to be essential in the middle stages of hair follicle development, regulating the growth and proliferation of epithelial cells (St-Jacques et al. 1998; Chiang et al. 1999; Karlsson et al. 1999; Reddy et al. 2001; Millar 2002; Gritli-Linde et al. 2007). Transcription factor SOX18 also may participate in the further development of the hair follicle (Pennisi, Bowles, et al. 2000; Pennisi, Gardner, et al. 2000; Millar 2002), and RBPJ, a regulator of the Notch signaling pathway, is associated with the differentiation of hair follicle cells (Yamamoto et al. 2003; Blanpain et al. 2006). Interestingly, most of these genes (SHH, SMO, SOX18, and RBPJ) appear to be active in the later stages of hair follicle development, rather than the initial dermal signal (Millar 2002). Modifications that result in the remarkable underhairs of otters would be expected to occur during these later stages under von Baer’s laws of embryonic development, which predict that special characteristics of a species appear later in development and are less likely to have downstream effects on other structures that might reduce fitness (von Baer 1828; Abzhanov 2013).
Knockouts or mutations in these genes can have dramatic impacts on hair phenotypes in mice and humans (fig. 2C). SHH knockout mice have a much lower density of hair follicles (St-Jacques et al. 1998; Chiang et al. 1999) (fig. 2C). Knockouts and mutations in AARS (Lee et al. 2006), RBPJ (Blanpain et al. 2006), APCDD1 (Shimomura et al. 2010), and TROY (Pispa et al. 2008) can lead to sparse or patchy hair. TMEM79 is associated with the matted coat phenotype in mice (Saunders et al. 2013) (fig. 2C), FZD6 is associated with changes in hair orientation (Guo et al. 2004; Wang et al. 2006, 2010) (fig. 2C) and SOX18 is associated with missing hair subtypes (Pennisi, Bowles, et al. 2000; Pennisi, Gardner, et al. 2000).
Our findings may help to explain the extreme density of otter fur, as many phenotypes associated with the genes we found under moderate selection are related to hair density in mice and humans (Liwanag et al. 2012). There may also be a strong role for gene regulatory changes during folliculogenesis which could be further explored with studies of gene expression in skin cells of the sea otter and other mustelids. Finally, research on genetic markers and genes associated with pelt quality in mink (a related semiaquatic mustelid) may benefit from our approach used to detect polygenic selection (Thirstrup et al. 2014; Cai et al. 2017).
Patterns of Pseudogenization in Otter Genomes
The sense of smell, used to detect prey, predators, and interact with conspecifics, and the sense of taste, used to forage and avoid toxins, are less critical for aquatic existence (Thewissen and Nummela 2008; Thewissen 2009). Aquatic mammals have reduced olfactory gene repertoires across independent evolutionary lineages (Hayden et al. 2010; Hughes et al. 2018). They have also been shown to have lost some or all their taste receptor genes, particularly sweet and bitter taste receptors, likely due to swallowing food whole (in the case of cetaceans) and a reduced need to avoid bitter plant toxins when foraging (Jiang et al. 2012; Sato and Wolsan 2012; Feng et al. 2014; Li and Zhang 2014). We found that the sea otter and giant otter genomes both had significant enrichment of pseudogenized genes related to sensing stimuli and development of the sensory apparatus (supplementary table S6C, Supplementary Material online) including pseudogenization of two bitter taste receptors and hundreds of ORGs. The loss of one bitter taste receptor in the southern sea otter and two in giant otter suggest the pressure to avoid plant toxins may be reduced in semiaquatic carnivores. However, the sea otter is under strong pressure to avoid toxins which can induce paralytic shellfish poisoning and has been shown to be able to avoid contaminated shellfish (Kvitek et al. 1991).
We observed a more profound loss of ORGs in the sea otter and giant otter lineages, which suggests reduced selection for olfactory acuity in otters as a consequence of aquatic or semiaquatic life. Otter species may have reduced olfactory requirements compared with terrestrial mustelids due to their increased time underwater. Terrestrial mustelids use scent extensively, with complex scent profiles exuded from anal scent glands that can be used to distinguish between individuals, mark territory, and assess reproductive status (Burger 2005). Freshwater otters have retained anal scent glands and engage in territory marking and mate assessment using scent (Duplaix 1980; Kean et al. 2011). Sea otters have been observed sniffing the air, and are thought to rely on their sense of smell for interacting with conspecifics, assessing female reproductive status, and detecting threats. However, they lack anal scent glands, do not engage in territory marking (Riedman and Estes 1990; Thewissen 2009), and like pinnipeds, have reduced olfactory bulbs (Radinsky 1968; Gittleman 1991; Pihlström 2008) and olfactory turbinate areas (Van Valkenburgh et al. 2011) compared with terrestrial mammals. These physiological changes indicate that olfactory sensing and signaling is reduced in sea otters relative to other mustelids as the marine environment makes olfactory acuity less advantageous, and other senses, such as sea otter’s highly acute tactile perception (Strobel et al. 2018), more important. We predicted a concomitant loss of ORGs in sea otter, and possibly the giant otter, associated with their ancestral aquatic radiation, and more recently, the invasion of the sea otter lineage into the marine environment. However, as both species are derived from semiaquatic ancestors and are still observed using olfaction for some purposes, we also predicted that the reduction in ORGs would not be as extreme as that observed in ancient, fully aquatic, marine mammal lineages (Hayden et al. 2010; Jiang et al. 2012; Sato and Wolsan 2012; Feng et al. 2014; Li and Zhang 2014; Hughes et al. 2018).
Hayden et al. (2010) and Hughes et al. (2018) compared the size of functional ORG repertoires across a large array of terrestrial and aquatic species. The number of functional ORGs roughly corresponds to the length of time a species has been aquatically adapted and how fully aquatic it is (supplementary fig. S7, Supplementary Material online). The polar bear is the most recently adapted marine mammal (<1.5 Ma) (Berta 2012). As it spends a large amount of time foraging on ice, it retains an acute sense of smell (Thewissen 2009) and a correspondingly large functional ORG repertoire of ∼1,029 functional genes (Hughes et al. 2018) (fig. 3 andsupplementary fig. S7, Supplementary Material online). In contrast, cetaceans, one of the most ancient (∼50 Ma) and highly modified marine mammal lineages, have the smallest set of functional ORGs (58 functional genes) (Kishida et al. 2007; Hayden et al. 2010; Hughes et al. 2018) (supplementary fig. S7, Supplementary Material online). Cetaceans are entirely aquatic and have partially or entirely lost the olfactory bulb apparatus (Thewissen 2009). Pinnipeds are a more recent (30 Ma) marine mammal lineage that hunts underwater but interacts with conspecifics out of the water (Thewissen 2009). The pinnipeds have intermediate-sized ORG repertoires (263–381 functional genes, fig. 3) that is reduced relative to terrestrial carnivores such as the domestic dog, but larger than that of cetaceans (fig. 3 and supplementary fig. S7, Supplementary Material online).
We found that the southern sea otter, northern sea otter, and giant otter have moderately sized ORG repertoires that are reduced compared with their terrestrial relative, the domestic ferret, but larger than that of the pinnipeds and cetaceans (fig. 3 and supplementary figs. S6 and S7, Supplementary Material online). The rate of ORG loss in the ancestral otter lineage of 41 genes lost per My was high relative to the rate in other terrestrial carnivorans and was comparable to that observed in the ancestral pinniped branch of 36 genes lost per Ma (fig. 3). Our analyses revealed that ∼175 ORGs were lost in the ancestor of the sea otter and the giant otter but that a greater number of genes were lost in the sea otter and giant otter lineages independently (>275 ORGs lost in each lineage) (fig. 3). This intriguing pattern indicates that there was an independent loss of functional ORGs in the sea otter and giant otter lineages over a timescale of ∼10 Ma (fig. 3). These independent losses nevertheless yielded functional ORG repertoires of similar size in each species. This finding was surprising as we expected that the sea otter would have a smaller ORG repertoire than the giant otter given that the former is more fully aquatic. We did not observe any significant differences between the relative proportions of Class I (fish-like receptors) or Class II (tetrapod receptors) ORGs among the domestic ferret, sea otter, or giant otter, indicating that although their functional repertoires are different in size they have similar distributions between classes in mustelid species.
The reduced functional ORG repertoire we observed in otter species may reflect their relatively recent aquatic evolution. Although otters use olfaction to interact with conspecifics and sense threats (Duplaix 1980; Riedman and Estes 1990), selection to maintain a large ORG repertoire may be reduced once these behaviors occur underwater. Otter sensory genes may therefore be under relaxed selection leading to gene loss without an impact on fitness. The range or sensitivity of otter olfaction is likely reduced compared with terrestrial mustelids.
Genomic Diversity
At the population level, patterns of genetic diversity, demographic history, and deleterious variation differed between the sea otter and giant otter, both of which are endangered species. Both the southern and northern sea otters appear to be at risk from extremely low levels of genetic diversity and an elevated burden of putatively deleterious genetic variants. Sea otters once numbered from 150,000 to 300,000 individuals worldwide prior to the harvest for the fur trade that began in the mid-18th century (Johnson 1982). Following the fur trade, sea otter numbers are thought to have been reduced to 1,000–2,000 individuals worldwide and scattered across a handful of small remnant populations. Some of these populations have now recovered to sizes of tens of thousands of individuals (Johnson 1982; Riedman and Estes 1990). The southern sea otter population in California has partially recovered from a population size estimated at <100 individuals to a current size of ∼3,000 individuals but well below its historical abundance (Riedman and Estes 1990; Tinker and Hatfield 2017). Northern sea otter populations in Alaska have recovered in many areas to historical abundance levels (Riedman and Estes 1990).
Population bottlenecks can have a long-lasting imprint on overall genomic diversity (Nei et al. 1975). The extremely low genomic diversity we observe in two sea otter genomes corresponds with findings of low mitochondrial haplotype (Cronin et al. 1996; Bodkin et al. 1999; Larson, Jameson, Etnier, et al. 2002; Larson, Jameson, Bodkin, et al. 2002) and microsatellite diversity (Larson, Jameson, Etnier, et al. 2002; Larson, Jameson, Bodkin, et al. 2002; Aguilar et al. 2008; Larson et al. 2012) in sea otter populations. In fact, previous studies have suggested the California southern sea otter population has the lowest diversity of all sea otter populations (Aguilar et al. 2008; Larson et al. 2012). We found that both the southern and northern sea otters show extremely low levels of genome-wide heterozygosity comparable to that of the classic examples of low-diversity mammals such as the Tasmanian devil and the cheetah (fig. 4A). However, these comparisons need to be accepted with qualification as the choice of individual and differences in genotype calling and filtering could impact relative levels of diversity between studies. Although low heterozygosity does not always imply an obvious decrease in fitness (DeWoody and DeWoody 2005; Robinson et al. 2016, 2018), low levels of genetic diversity and inbreeding depression have been associated with low sperm quality, mortality, and increased disease susceptibility in the cheetah, which experienced at least two bottlenecks ∼100,000 and ∼12,000 years ago (Dobrynin et al. 2015). Tasmanian devils are at risk of extinction due to a contagious cancer, which may be caused in part by their very low levels of genetic diversity in immune genes (Miller et al. 2011; Murchison et al. 2012).
In addition to low heterozygosity, we also find greater evidence for recent inbreeding in sea otters based on the presence of long ROH in the two genomes sequenced (fig. 4C). This result may reflect the impact of recent small population sizes due to exploitation in the fur trade. The distribution of ROH has been shown to be skewed toward higher ROH in small, isolated populations that have experienced population bottlenecks and recent inbreeding (Brüniche-Olsen et al. 2018; Kardos et al. 2018).
Historical Demography
Although long ROH are indicative of recent inbreeding events, genome-wide heterozygosity is generally driven by more ancient demography (Tajima 1983). Therefore, the extremely low heterozygosity in sea otters may not only be the result of the relatively brief bottleneck induced by the 18th–20th century fur harvest. As suggested by others for the California population, ancient bottlenecks may also have contributed to low diversity (Aguilar et al. 2008; Larson et al. 2012). Our inferred demographic trajectory in MSMC, although not a literal representation of changes in population size (Mazet et al. 2016; Beichman et al. 2017; Chikhi et al. 2018), also suggested past events of population instability occurred, particularly in southern sea otter (fig. 5A). The fluctuations in the trajectory could be due to changes in population size or population structure, a combination of both, or other more complex factors such as linked selection (Nadachowska-Brzyska et al. 2013; Mazet et al. 2016; Schrider et al. 2016; Beichman et al. 2017; Chikhi et al. 2018).
Our best estimates for the more ancient demographic events seen in sea otter populations suggest they occurred ∼30–40 ka, but conceivably they could have occurred as recently as 26 ka or deeper in time (>50 ka) if the demographic history is scaled using different mutation rates and generation times (see supplementary fig. S8 and table S10A–C, Supplementary Material online, for alternate scalings). The last glacial maximum was ∼20 ka and global sea levels were ∼140 m lower than today, impacting coastal habitats in California and Alaska. In California, San Francisco Bay was dry and large amounts of estuarine habitat were eliminated (Hewitt 2000; Jacobs et al. 2004; Dolby et al. 2016, 2018). In Alaska, changes in glaciation levels altered coastlines dramatically (Mann and Peteet 1994). Although it would require extensive habitat modeling to understand how changes to sea level might have impacted sea otter population dynamics (Pyenson and Lindberg 2011; Dolby et al. 2018), it is an intriguing finding that there is disturbance in both sea otter MSMC trajectories spanning this time period of climate change (fig. 5A and supplementary fig. S8, Supplementary Material online). The fact that these fluctuations are not observed in the giant otter population during this time period lends credence to the impacts of the last glacial maximum, which, though it caused drying and restriction of the rainforest (Hewitt 2000), likely had less impact on the tropical habitat of the giant otter than on the coastal habitat of the sea otter.
The more recent putative decline in the MSMC trajectory (∼1–3 ka) of the southern sea otter is much older than any bottleneck that might be caused by the fur trade. The decline could possibly relate to population depletion or fragmentation and gene flow limitation caused by indigenous Californians as previously suggested (Simenstad et al. 1978; Aguilar et al. 2008; Larson et al. 2012). Archeological evidence of hunting of marine mammals begins ∼9 ka on the coast of California (Hildebrandt and Jones 1992; Braje and Rick 2011) and a high level of exploitation of sea otters ∼750–2,500 ya is consistent with the estimated timing of this decrease in population size (Hildebrandt and Jones 1992; Braje and Rick 2011). We found that information from a single genome lacks power to distinguish between a model with a recent bottleneck from one without it (fig. 5B and supplementary fig. S9, Supplementary Material online). However, the MSMC results are still useful as a preliminary reconstruction of the demographic history of the California southern sea otter population. Sequencing 10–20 additional individuals will allow multiple bottleneck models to be explicitly tested using a site frequency spectrum or approximate Bayesian computation approach (Beaumont 2010; Excoffier et al. 2013; Beichman et al. 2018). If the California southern sea otter population experienced multiple bottlenecks prior to the fur trade, it will be important to incorporate that demographic history into any studies of adaptation or deleterious variation at the population level.
Deleterious Variation
The extremely low heterozygosity and known history of at least one fur-trade-related bottleneck in southern and northern sea otters suggests they may have suffered fitness consequences from deleterious variation and inbreeding. Purifying selection is less effective at removing deleterious variants from a population during periods of small population size (Ohta 1973; Akashi et al. 2012) which can lead to an increase in putatively deleterious missense (nonsynonymous)-derived variants (Lohmueller et al. 2008; Henn et al. 2015; Marsden et al. 2016; Robinson et al. 2016, 2018). The increase of deleterious alleles can lower the overall fitness of the population, impeding recovery (Kohn et al. 2006; Harrisson et al. 2014). The fact that we observed similar numbers of synonymous-derived alleles that are putatively neutral in the sea otter and giant otter but a dramatic increase in the number of possibly deleterious missense-derived alleles suggests an increased burden of deleterious variants due to one or more historical bottlenecks (fig. 5C). The sea otters also had an increase of derived mutations in the homozygous state relative to the giant otter (fig. 5D). If some of these mutations are recessive and deleterious, the sea otter may be exposed to fitness impacts due to elevated homozygosity (Peischl and Excoffier 2015). Interestingly, we do not observe a significant difference between the numbers of homozygous loss of function mutations between the sea otters and giant otter, consistent with purging of highly deleterious recessive variants once they are exposed to selection in the homozygous state (Henn et al. 2016; Robinson et al. 2018).
These findings are relevant to conservation of rare and endangered species. The primary barriers to population recovery in the sea otter are population fragmentation, resource limitation, and predation by sharks and killer whales (Estes et al. 1998; U.S. Fish and Wildlife Service 2003; Tinker et al. 2016). However, the low genetic diversity and burden of putatively deleterious alleles have the potential to impact population recovery goals as well. The increased burden of deleterious variants coupled with extremely low genetic diversity may have long-term fitness impacts for sea otter populations, slowing recovery and limiting their genetic response to environmental challenges such as disease or climate change (Kohn et al. 2006; Harrisson et al. 2014). Although logistically challenging, long-term recovery goals of increasing genetic diversity and counteracting elevated deleterious variation through translocation and/or captive breeding programs may help to bolster sea otters’ persistence given future environmental threats.
Materials and Methods
See Supplementary Material online for additional details on all analyses.
Genome Sequencing, Assembly, and Annotation
We sequenced the southern sea otter genome from genomic DNA extracted using the Qiagen DNEasy kit from a blood sample taken from a female resident sea otter named Gidget at the Monterey Bay Aquarium, Monterey, California, during a standard veterinary examination (Permit #MA186914-2). We initially sequenced a standard Illumina TruSeq library on the HiSeq4000 (150 bp paired-end reads). We then generated a preliminary contig assembly using Meraculous (Chapman et al. 2011). Next, sequencing of a single Chicago library (Dovetail Genomics, Santa Cruz, CA) was carried out on an Illumina HiSeq4000 (100 bp PE), followed by HiRise scaffolding (Putnam et al. 2016). The de novo giant otter genome was sequenced at the Broad Institute (Cambridge, MA) from a tissue sample of a male giant otter from the Frankfurt Zoological Garden born and collected in 1994 (ID #1384). The genome was sequenced from a single PCR-free library and assembled into contigs using the DISCOVAR de novo assembly pipeline (https://software.broadinstitute.org/software/discovar/blog/, last accessed March 2019). Genome completeness was assessed with BUSCO v2 (Simão et al. 2015), using the gVolante web interface (Nishimura et al. 2017).
The southern sea otter genome was annotated using five iterative rounds of the MAKER2 pipeline (Holt and Yandell 2011). Whole-blood RNA-Seq data from the southern sea otter and NCBI (National Center for Biotechnology Information) protein data from domestic ferret, domestic dog, and domestic cat were used for training the AUGUSTUS gene predictor (Stanke and Waack 2003). The giant otter genome was then annotated using AUGUSTUS trained on the sea otter data. The sea otter and giant otter annotations yielded 21,909 and 23,665 gene models, respectively (table 1). Dovetail Hi-Rise scaffolding, although increasing scaffold contiguity, does not add additional sequence to the assembly, resulting in long scaffolds containing numerous gaps (0.073% of genome contained in gaps prior to Hi-Rise scaffolding; 0.411% after). These sequence gaps were disruptive to the gene predictor. For example, when they occur in the middle of the gene, the gene predictor cannot complete the gene model which results in the fragmentation of gene models even if present on long scaffolds. This finding suggests future assemblies make use of long-read technologies in addition to Dovetail HiRise scaffolding to increase contig size and fill in gaps (additional details in supplementary information 1, Supplementary Material online).
Mapping to the Domestic Ferret Reference Genome
To directly compare genetic diversity between the southern sea otter and giant otter, the raw sequencing reads used for de novo genome assembly were also aligned to the domestic ferret (Mustela putorius furo) genome (Assembly Accession #GCF_000215625.1). To investigate population-level differences between sea otter populations, we also mapped sequencing reads from the northern sea otter genome (Jones et al. 2017) downloaded from Sequence Read Archive (SRA; Library SRX2967283) to the domestic ferret reference genome. All otter species are equidistant from the domestic ferret. We chose this approach for several reasons. First, we wished to avoid potential reference bias that would come from aligning the reads used to generate the sea otter and giant otter assemblies against those same genomes. Second, we wished to avoid the confounding influence of variable de novo genome assembly quality (Lehri et al. 2017). Third, we wanted to assess diversity between the otters at the same regions of the genome. Lastly, using an established reference genome enabled access to additional resources only available for such genomes such as the Ensembl VEP (McLaren et al. 2016) (additional details in supplementary information 2, Supplementary Material online).
Phylogeny and Divergence Time Estimation
We identified 784 high-confidence “one-to-one” (1:1) gene orthologs from the gene annotations of 12 carnivore species (southern sea otter, giant otter, domestic ferret, red panda, polar bear, giant panda, Hawaiian monk seal, Weddell seal, walrus, domestic dog, tiger, and domestic cat; see supplementary table S1, Supplementary Material online, for accessions) and the Malayan pangolin as an outgroup using ProteinOrtho (Lechner et al. 2011). All gene groups identified as single-copy orthologs were then aligned in a codon-aware manner in PRANK (Löytynoja 2014) and conserved blocks were selected using Gblocks (Castresana 2000). The phylogeny was reconstructed using RAxML 8.0 (Stamatakis 2014) under the GTRGAMMA model and divergence times were estimated with the MCMCTree tool from the PAML software package (Yang 2007) (additional details in supplementary information 3, Supplementary Material online). The topology of the domestic ferret, giant otter, southern sea otter, and northern sea otter was confirmed using the number of shared versus private derived alleles and the ABBA–BABA test. (See supplementary information 4, Supplementary Material online, for details.)
Positive Selection on Single Genes
To find orthologous genes for positive selection analyses, we compared genome annotations from a wide range of taxa in the Laurasiatheria (southern sea otter, giant otter, domestic ferret, giant panda, polar bear, Weddell seal, walrus, domestic dog, domestic cat, microbat, megabat, dolphin, domestic cow, and domestic horse) and the human (Euarchontoglires) (supplementary table S1, Supplementary Material online, for accessions and Latin species names) using ProteinOrtho (Lechner et al. 2011).
The nucleotide sequences corresponding to the orthologous protein sequences were aligned for each gene using GUIDANCE2 (Sela et al. 2015). We found that using genome assemblies and annotations that vary in quality in our alignments led to many spurious alignments, particularly due to assembly and annotation artifacts. We used two filtering schemes in addition to masking of inconsistent residues with GUIDANCE2 to attempt to remedy these problems. First, we used the common alignment filtering approach Gblocks (Castresana 2000) with moderately stringent parameters to select conserved blocks of sequence. Others have noted that Gblocks can still lead to many false-positive results (Jordan and Goldman 2012; Harrison et al. 2014). Therefore, we separately employed a sliding window masking approach using SWAMP (Harrison et al. 2014) with stringent parameters to mask regions of the southern sea otter and giant otter sequences that have too many amino acid changes within a window.
The codeml branch-site test for positive selection was run on every gene alignment under each filtering scheme for three foreground branches: southern sea otter, giant otter, and the internal branch leading to both otter species. Significance was calculated using the LRT, and for each filter-type, a false-discovery rate correction applied across all branch-genes combinations (all genes surveyed, across all three foreground branches).
To assess the success of the different filters, the alignments of all 638 genes that had an uncorrected P-value ≤0.01 in any test were visually inspected and those with annotation and alignment artifacts that could yield false positives were excluded. Genes with high-impact VEP mutations when aligned to the ferret were also excluded as they could be pseudogenized. Genes containing multinucleotide changes were noted, but not excluded. The genes passing inspection with P-values ≤0.01 were tested for gene ontology enrichment with g:Profiler (Reimand et al. 2016). For the top outlier genes (q ≤ 0.1), every amino acid change identified by the Bayes empirical Bayes method as putatively under selection in the southern sea otter or shared otter lineages, regardless of confidence level, was confirmed by BLASTing to the northern sea otter sequence (supplementary table S4D, Supplementary Material online). The only site that failed to match the northern sea otter sequence was a codon in CEP350, which in the southern sea otter was AGA (Arginine), whereas in the northern sea otter and domestic ferret was GGA (Glycine) (supplementary table S4D, Supplementary Material online). The remaining six Bayes empirical Bayes sites in CEP350 matched the northern sea otter sequence (additional details in supplementary information 5, Supplementary Material online).
Polygenic Selection
We used a method to detect low levels of polygenic selection on the otters foreground branch (Daub et al. 2017). This method, called polysel (Daub et al. 2017), uses information from all genes that went through the codeml branch-site test to detect subtle signals of polygenic selection in genes within a pathway. The user provides input gene pathways or categories to test and the method adds together the fourth root of the LRT statistic (“LRT4,” used to down-weight outliers) of all genes in that pathway that are present in the data set to generate a “SUMSTAT” score for each category. The method then generates a null distribution for each pathway by randomly sampling the user’s gene set to generate 1 million “pathways” of the same size as the pathway being tested but composed of a random selection of genes whose association has no biological relevance and no expectation of being under selection. By examining how often the SUMSTAT statistic of the null pathways exceeds that of the true pathway, the method generates a P-value which is then corrected for multiple testing. This statistical approach avoids the subjectivity of trying to choose gene sets that might not be under selection to act as a null set and instead generates the null sets through random sampling.
Our input pathways were chosen to test broad a priori hypotheses about aquatic adaptation. GO pathways or categories containing the following terms in their description: “hair follicle,” “osmoregulation,” “thermoregulation,” “diet,” “hypoxia,” and “sensory perception” were selected. Our input gene set was filtered using the more moderate Gblocks filtering scheme. For the significantly enriched hair follicle development pathway, we tested the robustness of the result by removing a fragmented gene alignment (LGR4) from the input data set and carrying out the full analysis without it, which raised the category’s P-value from 0.0048 to 0.019, and the q-value from 0.099 to 0.33. We also performed the same polysel analysis on the codeml results for the genes filtered using the highly conservative SWAMP filtering scheme and did not observe any significantly enriched pathways. This finding suggests that alignment artifacts may have influenced the results or that SWAMP’s filtering is overly conservative (additional details in supplementary information 6, Supplementary Material online).
Detection of Pseudogenes
To find genes that have been pseudogenized, we employed the PseudoPipe program (Zhang et al. 2006) which uses query protein sequences from the species itself or a relative to detect pseudogenized versions of the query proteins in the genome. We used domestic ferret protein sequences as a query to search for pseudogenized genes in the southern sea otter and giant otter genomes. We also used a second approach to identify loss-of-function genes that is independent of the quality of de novo genome assembly and annotation. Specifically, we used our alignments of southern sea otter and giant otter reads to the domestic ferret reference genome as a secondary measure of gene loss. We extracted all variants annotated by VEP as “HIGH” impact that occurred in a known protein domain in a canonical transcript, as these variants are the most likely to disrupt protein function (additional details in supplementary information 7, Supplementary Material online).
PseudoPipe yielded 1,060 (southern sea otter) and 1,132 (giant otter) genes that were classified as pseudogenized in the de novo genome assemblies. These counts included 649 genes which were classified as pseudogenized in both species. Our second approach using VEP yielded 1,619 (southern sea otter) and 1,503 (giant otter) genes that were characterized in Uniprot and potentially disrupted by a high impact SNP or indel in a protein-coding domain relative to the domestic ferret sequence, 900 of which were putatively disrupted in both southern sea otter and giant otter. Most intriguingly, there was very little overlap between our two approaches, with only 113 putative pseudogenes found in common between PseudoPipe and VEP for southern sea otter, 133 for giant otter, and 45 genes found in both species by both methods (supplementary table S6A and B, Supplementary Material online). The genes held in common were searched for enrichment of particular functions using g:Profiler (Reimand et al. 2016) using all domestic ferret genes as a background set. We also reviewed the literature to uncover associations between genes and phenotypes in mice and humans.
Loss of ORGs
We followed the protocols of Niimura (2013) and Montague et al. (2014) to detect putatively functional, truncated and pseudogenized ORGs in the domestic ferret, southern sea otter, northern sea otter, and giant otter genomes. ORGs were classified as Class I and Class II by phylogenetic position. To generate the gene tree, functional ORG sequences were aligned to a data set of ORG sequences from six additional Caniformia species (Montague et al. 2014): red panda, walrus, Hawaiian monk seal, Weddell seal, polar bear, giant panda, and domestic dog with domestic cat, tiger and Malayan pangolin as outgroup species (see supplementary table S1, Supplementary Material online, for accessions). The gene tree was built using SATé (Liu et al. 2009, 2012) and Notung 2.6 (Durand et al. 2006; Vernot et al. 2008) was used for the reconciliation of ORG gene trees and the species tree. The per-branch counts of gained and lost ORGs from the reconciled gene tree and species tree (supplementary fig. S6, Supplementary Material online) were scaled by the branch lengths of the phylogeny (fig. 1) to estimate rates of gain and loss per million years for each branch (fig. 3). For comparison, functional ORG counts based on genomic data of additional mammal species were gathered from the recent study by Hughes et al. (2018) (see table S1 in Hughes et al. [2018] and supplementary table S7B, Supplementary Material online) (additional details in supplementary information 8, Supplementary Material online).
Genomic Diversity
Genome-wide heterozygosity was calculated based on the reads mapped to the domestic ferret genome. Sliding window heterozygosity was calculated using a python script modified from Robinson et al. (2016). We calculated the distribution of ROH using PLINK (Purcell et al. 2007) (additional details in supplementary information 9, Supplementary Material online).
Mutation Rate Inference
See supplementary information 10, Supplementary Material online, for mutation rate calculations the range of mutation rates used for scaling demographic inference.
Historical Demography
The MSMC (Schiffels and Durbin 2014), also known as PSMC′ when used on a single genome, was used to infer the distribution of coalescence rates (inversely proportional to effective population size under certain assumptions). Coalescent rates were scaled to estimates of population size and time intervals to generations and years using a range of mutation rates and generation times (supplementary fig. S8 and table S10A–C, Supplementary Material online).
To test whether trimming away ancient events, including the high inferred ancient population size, would improve the fit to empirical heterozygosity, we simulated data using MaCS (Chen et al. 2009) under the full demographic model inferred using MSMC and under a trimmed model that removed events >16–17,000 generations in the past (time index 33 for southern sea otter, 31 for northern sea otter, and index 20 for giant otter). To test whether a recent southern sea otter bottleneck was consistent with observed genome-wide heterozygosity, we carried out further simulations without the recent bottleneck indicated by the pink arrow in figure 5A. The simulated distributions of heterozygosity with and without the bottleneck were highly similar, suggesting a lack of power to obtain greater resolution on that event from a single genome (supplementary fig. S9, Supplementary Material online) (additional details in supplementary information 11, Supplementary Material online).
Deleterious Variation
To assess a proxy for the load of deleterious mutations, we examined the number of derived alleles in coding sequence between southern sea otter, northern sea otter, and giant otter. Coding sequence was defined using the annotation from the domestic ferret reference genome (MusPutFur1.0, Ensembl release 91), and variants in coding regions were annotated using Ensembl’s VEP (McLaren et al. 2016) against the 90_MusPutFur1.0 domestic ferret database. We counted the number of changes per species annotated as “missense_variant,” “synonymous_variant,” and “stop_gained” in canonical transcripts as representing nonsynonymous, synonymous, and putative loss of function changes, respectively. As the southern sea otter and giant otter had differing numbers of sites passing all filters across the genome, we rescaled these numbers by the total number of coding sites called in each species, then multiplied by the average number of coding sites called between species for each category of site (synonymous, missense, and stop-gained). We also compared the total number of homozygous-derived genotypes in each species across the three categories which were rescaled in a similar manner.
Bootstrapping of these rescaled estimates of the number of derived alleles per category and the number of homozygous-derived genotypes per category were carried out by randomly sampling with replacement of the average number of called coding sites from each species and recording the number of synonymous, nonsynonymous, and stop-gained changes in homozygous or heterozygous states in each of 1,000 bootstrap replicates. We tested for differences in the number of derived alleles and homozygous-derived genotypes between southern sea otter and giant otter using a two-tailed Z-test. P-values were then calculated based on the z-scores from a standard normal distribution (additional details in supplementary information 12, Supplementary Material online).
Ethics Approval and Consent to Participate
Sea otter samples were taken from “Gidget” under the Monterey Bay Aquarium’s scientific research permit #MA186914-2 and from wild individuals under MTT’s permit #MA672624-18, with the sampling protocol approved by UC Santa Cruz’s Institutional Animal Care and Use Committee (IUCAC) in December, 2015 (protocol reference ID TINKT1510).
Supplementary Material
Acknowledgments
The authors are deeply grateful for the advice, help, and knowledge provided by the following collaborators. James Estes (UC Santa Cruz), Michelle Staedler, and Andy Johnson (Monterey Bay Aquarium) for invaluable advice, aid acquiring samples, and comments on the manuscript. Christine DeAngelo, Marissa Young, Athena Copenhaver (Monterey Bay Aquarium), and Lilian Carswell (U.S. Fish and Wildlife Service) for coordinating the sea otter sampling and giving aid and guidance; The U.S. Geological Survey, Western Ecological Research Center for supporting fieldwork and wild sea otter sampling; Christof Schenck & Elke Schenck and the Giant Otter Project at the Frankfurt Zoological Society for the providing the giant otter sample; and Diane Genereux (Broad Institute of MIT and Harvard) for very helpful comments on the manuscript. They also thank the editor and anonymous reviewers for suggestions that significantly improved the manuscript. The authors would also like to thank Eduardo Eizirik (Pontifícia Universidade Católica do Rio Grande do Sul) for advice; Joann Shih (UCLA undergraduate) for creating the paintings and silhouettes used in the figures; Rena Schweizer (University of Montana), Jacqueline Robinson (UC San Francisco), Daniel Chavez, Tanya Phung, Jazlyn Mooney, and Clare Marsden (UCLA) for advice and scripting assistance; Teia Schweizer, Colin Shew, and Amber deVries (UCLA) for assistance with laboratory work; Beth Shapiro (UC Santa Cruz) for advice on genome sequencing and assembly; David Jacobs (UCLA) for information on the history of the California coast; Blaire Van Valkenburgh, Justin Keller, and Brandon Hupka (UCLA) for information on otter skeletal anatomy; Ken Peterson, Daniel Potter, and the Monterey Bay Aquarium for helping us tell the public this story. “Gidget” the sea otter for her genomic material. Additional animal silhouettes are from PhyloPic [phylopic.org] (Public Domain Mark 1.0 license). The work was supported by the Monterey Bay Aquarium, UCLA Academic Senate, the Catalyst grant program of the UC President's office, and National Science Foundation (DEB Small Grant #1556705). A.C.B. was supported by the National Institutes of Health (NIH) Training Grant in Genomic Analysis and Interpretation (T32 HG-002536) and the NSF Graduate Research Fellowship Program. K.P.K. and S.K. were partially supported by funding from St. Petersburg State University, Russia (Genome Russia Grant no. 1.52.1647.2016). S.K. was also supported by a grant from the Russian Foundation for Basic Research (RFBR) (Grant No. 17-00-00144). This work used the Extreme Science and Engineering Discovery Environment (XSEDE) Data Oasis and Comet systems at the San Diego Supercomputer Center (allocation number DEB160007), which is supported by National Science Foundation grant number ACI-1548562. WJM and GL were supported by NSF grant IOS-1456506. The authors have no competing interests.
Author Contributions
A.C.B., K.P.K., K.E.L., and R.K.W. conceived the study. K.E.L. and R.K.W. jointly supervised the work. A.C.B. carried out genome sequencing and assembly of the sea otter genome, based on samples from M.J.M. J.J., K.L.T., and E.K.K. carried out sequencing and assembly of the giant otter genome, based on a sample from K.P.K., originally obtained from the Frankfurt Zoo, Frankfurt, Germany. A.C.B. annotated both the southern sea otter and giant otter genomes based on RNA samples from M.J.M. and M.T.T. A.C.B. carried out all analyses and generated all figures, except for detecting olfactory receptors for the nonmustelid carnivorans and generating the olfactory receptor gene tree, which were carried out by G.L. and W.M., and phylogenetic and timetree inference, which were carried out by K.P.K., P.D., and S.K. A.C.B., K.P.K., G.L., W.M., K.E.L., and R.K.W. interpreted the results. A.C.B., K.P.K., K.E.L., and R.K.W. wrote the manuscript, with comments from M.J.M., M.T.T., S.K., K.L.T., P.D., and W.M.
References
- Abzhanov A. 2013. von Baer’s law for the ages: lost and found principles of developmental evolution. Trends Genet. 2912:712–722. [DOI] [PubMed] [Google Scholar]
- Aguilar A, Jessup DA, Estes J, Garza JC. 2008. The distribution of nuclear genetic variation and historical demography of sea otters. Anim Conserv. 111:35–45. [Google Scholar]
- Akashi H, Osada N, Ohta T. 2012. Weak selection and protein evolution. Genetics 1921:15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA. 2010. Approximate Bayesian computation in evolution and ecology. Annu Rev Ecol Evol Syst. 411:379–406. [Google Scholar]
- Beichman AC, Huerta-Sanchez E, Lohmueller KE. 2018. Using genomic data to infer historic population dynamics of nonmodel organisms. Annu Rev Ecol Evol Syst. 49:433–456. [Google Scholar]
- Beichman AC, Phung TN, Lohmueller KE. 2017. Comparison of single genome and allele frequency data reveals discordant demographic histories. G3 7:3605–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta A. 2012. Return to the sea: the life and evolutionary times of marine mammals. Berkeley: University of California Press. [Google Scholar]
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E. 2006. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2021:3022–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodkin JL, Ballachey BE, Cronin MA, Scribner KT. 1999. Population demographics and genetic diversity in remnant and translocated populations of sea otters. Conserv Biol. 136:1378–1385. [Google Scholar]
- Boessenecker RW. 2018. A middle Pleistocene sea otter from northern California and the antiquity of Enhydra in the Pacific Basin. J Mammal Evol. 251:27–35. [Google Scholar]
- Botton-Divet L, Cornette R, Fabre A-C, Herrel A, Houssaye A. 2016. Morphological analysis of long bones in semi-aquatic mustelids and their terrestrial relatives. Integr Comp Biol. 566:1298–1309. [DOI] [PubMed] [Google Scholar]
- Botton-Divet L, Houssaye A, Herrel A, Fabre A-C, Cornette R. 2018. Swimmers, diggers, climbers and more, a study of integration across the mustelids’ locomotor apparatus (Carnivora: Mustelidae). Evol Biol. 452:182–195. [Google Scholar]
- Braje TJ, Rick TC. 2011. Human impacts on seals, sea lions, and sea otters: integrating archaeology and ecology in the northeast Pacific. Berkeley: University of California Press. [Google Scholar]
- Brüniche-Olsen A, Kellner KF, Anderson CJ, Dewoody JA. 2018. Runs of homozygosity have utility in mammalian conservation and evolutionary studies. Conserv Genet. 196:1295–1307. [Google Scholar]
- Burger B.V. 2004. Mammalian Semiochemicals. In: Schulz S, editor. The Chemistry of Pheromones and Other Semiochemicals II. Topics in Current Chemistry, vol 240. Springer, Berlin, Heidelberg. p. 231–278. [Google Scholar]
- Cai Z, Petersen B, Sahana G, Madsen LB, Larsen K, Thomsen B, Bendixen C, Lund MS, Guldbrandtsen B, Panitz F. 2017. The first draft reference genome of the American mink (Neovison vison). Sci Rep. 71:14564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SK, Rosas FC. 1997. Biology and conservation of the giant otter Pteronura brasiliensis. Mamm Rev. 271:1–26. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 174:540–552. [DOI] [PubMed] [Google Scholar]
- Chapman JA, Ho I, Sunkara S, Luo S, Schroth GP, Rokhsar DS. 2011. Meraculous: de novo genome assembly with short paired-end reads. PLoS One 68:e23501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GK, Marjoram P, Wall JD. 2009. Fast and flexible simulation of DNA sequence data. Genome Res. 191:136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Swan RZ, Grachtchouk M, Bolinger M, Litingtung Y, Robertson EK, Cooper MK, Gaffield W, Westphal H, Beachy PA. 1999. Essential role for sonic hedgehog during hair follicle morphogenesis. Dev Biol. 2051:1–9. [DOI] [PubMed] [Google Scholar]
- Chikhi L, Rodríguez W, Grusea S, Santos P, Boitard S, Mazet O. 2018. The IICR (inverse instantaneous coalescence rate) as a summary of genomic diversity: insights into demographic inference and model choice. Heredity (Edinb). 1201:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin MA, Bodkin J, Ballachey B, Estes J, Patton JC. 1996. Mitochondrial-DNA variation among subspecies and populations of sea otters (Enhydra lutris). J Mammal. 772:546–557. [Google Scholar]
- Daub JT, Hofer T, Cutivet E, Dupanloup I, Quintana-Murci L, Robinson-Rechavi M, Excoffier L. 2013. Evidence for polygenic adaptation to pathogens in the human genome. Mol Biol Evol. 307:1544–1558. [DOI] [PubMed] [Google Scholar]
- Daub JT, Moretti S, Davydov II, Excoffier L, Robinson-Rechavi M. 2017. Detection of pathways affected by positive selection in primate lineages ancestral to humans. Mol Biol Evol. 346:1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWoody YD, DeWoody JA. 2005. On the estimation of genome-wide heterozygosity using molecular markers. J Hered. 962:85–88. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. 2016. High-throughput discovery of novel developmental phenotypes. Nature 5377621:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrynin P, Liu S, Tamazian G, Xiong Z, Yurchenko AA, Krasheninnikova K, Kliver S, Schmidt-Küntzel A, Koepfli K-P, Johnson W. 2015. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 16:277.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolby GA, Ellingson RA, Findley LT, Jacobs DK. 2018. How sea level change mediates genetic divergence in coastal species across regions with varying tectonic and sediment processes. Mol Ecol. 274:994–1011. [DOI] [PubMed] [Google Scholar]
- Dolby GA, Hechinger R, Ellingson RA, Findley LT, Lorda J, Jacobs DK. 2016. Sea-level driven glacial-age refugia and post-glacial mixing on subtropical coasts, a palaeohabitat and genetic study. Proc R Soc B. 2831843:20161571.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplaix N. 1980. Observations on the ecology and behavior of the giant river otter Pteronura brasiliensis in Suriname. Rev Décol. 34:495–620. [Google Scholar]
- Durand D, Halldórsson BV, Vernot B. 2006. A hybrid micro–macroevolutionary approach to gene tree reconstruction. J Comput Biol. 132:320–335. [DOI] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 288:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik E, Murphy WJ, Koepfli K-P, Johnson WE, Dragoo JW, Wayne RK, O’Brien SJ. 2010. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 561:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J.A. 1989. Adaptations for Aquatic Living by Carnivores. In: Gittleman J.L., editor. Carnivore Behavior, Ecology, and Evolution. Springer, Boston, MA. p. 242–282. [Google Scholar]
- Estes JA, Riedman ML, Staedler MM, Tinker MT, Lyon BE. 2003. Individual variation in prey selection by sea otters: patterns, causes and implications. J Anim Ecol. 721:144–155. [Google Scholar]
- Estes JA, Tinker MT, Williams TM, Doak DF. 1998. Killer whale predation on sea otters linking oceanic and nearshore ecosystems. Science 2825388:473–476. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 910:e1003905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. 2014. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol. 66:1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish FE, Stein BR. 1991. Functional correlates of differences in bone density among terrestrial and aquatic genera in the family Mustelidae (Mammalia). Zoomorphology 1106:339–345. [Google Scholar]
- Fletcher W, Yang Z. 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2710:2257–2267. [DOI] [PubMed] [Google Scholar]
- Foote AD, Liu Y, Thomas GWC, Vinař T, Alföldi J, Deng J, Dugan S, van Elk CE, Hunter ME, Joshi V, et al. 2015. Convergent evolution of the genomes of marine mammals. Nat Genet. 473:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii JA, Ralls K, Tinker MT. 2015. Ecological drivers of variation in tool-use frequency across sea otter populations. Behav Ecol. 262:519–526. [Google Scholar]
- Gagne RB, Tinker MT, Gustafson KD, Ralls K, Larson S, Tarjan LM, Miller MA, Ernest HB. 2018. Measures of effective population size in sea otters reveal special considerations for wide-ranging species. Evol Appl. 1110:1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittleman JL. 1991. Carnivore olfactory bulb size: allometry, phylogeny and ecology. J Zool. 2252:253–272. [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz M-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 3285979:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A, Hallberg K, Harfe BD, Reyahi A, Kannius-Janson M, Nilsson J, Cobourne MT, Sharpe PT, McMahon AP, Linde A. 2007. Abnormal hair development and apparent follicular transformation to mammary gland in the absence of hedgehog signaling. Dev Cell. 121:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Hawkins C, Nathans J. 2004. Frizzled6 controls hair patterning in mice. Proc Natl Acad Sci U S A. 10125:9277–9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. 2004. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33(Database issue):D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding LE, Smith FA. 2009. Mustela or Vison? Evidence for the taxonomic status of the American mink and a distinct biogeographic radiation of American weasels. Mol Phylogenet Evol. 523:632–642. [DOI] [PubMed] [Google Scholar]
- Harrison PW, Jordan GE, Montgomery SH. 2014. SWAMP: sliding window alignment masker for PAML. Evol Bioinform. 10:197–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisson KA, Pavlova A, Telonis‐Scott M, Sunnucks P. 2014. Using genomics to characterize evolutionary potential for conservation of wild populations. Evol Appl. 79:1008–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 201:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. 2015. Estimating the mutation load in human genomes. Nat Rev Genet. 166:333.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci U S A. 1134:E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 4056789:907.. [DOI] [PubMed] [Google Scholar]
- Hildebrandt WR, Jones TL. 1992. Evolution of marine mammal hunting: a view from the California and Oregon coasts. J Anthropol Archaeol. 114:360–401. [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike N, Takemori H, Katoh Y, Doi J, Min L, Asano T, Sun XJ, Yamamoto H, Kasayama S, Muraoka M, et al. 2003. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J Biol Chem. 27820:18440–18447. [DOI] [PubMed] [Google Scholar]
- Houssaye A, Botton-Divet L. 2018. From land to water: evolutionary changes in long bone microanatomy of otters (Mammalia: Mustelidae). Biol J Linn Soc. 1252:240–249. [Google Scholar]
- Hughes GM, Boston EM, Finarelli JA, Murphy WJ, Higgins DG, Teeling EC. 2018. The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol Biol Evol. msy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DK, Haney TA, Louie KD. 2004. Genes, diversity, and geologic process on the Pacific Coast. Annu Rev Earth Planet Sci. 321:601–652. [Google Scholar]
- Jameson RJ, Kenyon KW, Johnson AM, Wight HM. 1982. History and status of translocated sea otter populations in North America. Wildl Soc Bull (1973–2006). 10:100–107. [Google Scholar]
- Jiang P, Josue J, Li X, Glaser D, Li W, Brand JG, Margolskee RF, Reed DR, Beauchamp GK. 2012. Major taste loss in carnivorous mammals. Proc Natl Acad Sci U S A. 10913:4956–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AM. 1982. Status of Alaska sea otter populations and developing conflicts with fisheries. Transactions of the North American Wildlife and Natural Resources Conference. 47:293–299. [Google Scholar]
- Jones S, Haulena M, Taylor G, Chan S, Bilobram S, Warren R, Hammond S, Mungall K, Choo C, Kirk H, et al. 2017. The genome of the northern sea otter (Enhydra lutris kenyoni). Genes 812:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan G, Goldman N. 2012. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol Biol Evol. 294:1125–1139. [DOI] [PubMed] [Google Scholar]
- Kardos M, Åkesson M, Fountain T, Flagstad Ø, Liberg O, Olason P, Sand H, Wabakken P, Wikenros C, Ellegren H. 2018. Genomic consequences of intensive inbreeding in an isolated wolf population. Nat Ecol Evol. 21:124–131. [DOI] [PubMed] [Google Scholar]
- Karlsson L, Bondjers C, Betsholtz C. 1999. Roles for PDGF-A and sonic hedgehog in development of mesenchymal components of the hair follicle. Development 12612:2611–2621. [DOI] [PubMed] [Google Scholar]
- Kean EF, Müller CT, Chadwick EA. 2011. Otter scent signals age, sex, and reproductive status. Chem Senses. 366:555–564. [DOI] [PubMed] [Google Scholar]
- Kenyon KW. 1969. The sea otter in the eastern Pacific Ocean. North Am Fauna. 68:1–352. [Google Scholar]
- Kishida T, Kubota S, Shirayama Y, Fukami H. 2007. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol Lett. 34:428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T, Thewissen J, Hayakawa T, Imai H, Agata K. 2015. Aquatic adaptation and the evolution of smell and taste in whales. Zool Lett. 1:9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli K-P, Deere KA, Slater GJ, Begg C, Begg K, Grassman L, Lucherini M, Veron G, Wayne RK. 2008. Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 6:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. 2006. Genomics and conservation genetics. Trends Ecol Evol (Amst). 2111:629–637. [DOI] [PubMed] [Google Scholar]
- Kuhn RA. 2009. Comparative analysis of structural and functional hair coat characteristics, including heat loss regulation, in the Lutrinae (Carnivora: Mustelidae). PhD thesis, University of Hamburg, Hamburg: 1–225.
- Kuhn RA, Ansorge H, Godynicki S, Meyer W. 2010. Hair density in the Eurasian otter Lutra lutra and the sea otter Enhydra lutris. Acta Theriol. 553:211–222. [Google Scholar]
- Kuhn RA, Meyer W. 2009. Infrared thermography of the body surface in the Eurasian otter Lutra lutra and the giant otter Pteronura brasiliensis. Aquat Biol. 6:143–152. [Google Scholar]
- Kuhn RA, Meyer W. 2010a. Comparative hair structure in the Lutrinae (Carnivora: Mustelidae). Mammalia. 74:291–303. [Google Scholar]
- Kuhn RA, Meyer W. 2010b. A note on the specific cuticle structure of wool hairs in otters (Lutrinae). Zool Sci. 2710:826–829. [DOI] [PubMed] [Google Scholar]
- Kvitek RG, DeGunge AR, Beitler MK. 1991. Paralytic shellfish poisoning toxins mediate feeding behavior of sea otters. Limnol Oceanogr. 362:393–404. [Google Scholar]
- Larson S, Jameson R, Bodkin J, Staedler M, Bentzen P. 2002. Microsatellite DNA and mitochondrial DNA variation in remnant and translocated sea otter (Enhydra lutris) populations. J Mammal. 833:893–906. [Google Scholar]
- Larson S, Jameson R, Etnier M, Fleming M, Bentzen P. 2002. Loss of genetic diversity in sea otters (Enhydra lutris) associated with the fur trade of the 18th and 19th centuries. Mol Ecol. 1110:1899–1903. [DOI] [PubMed] [Google Scholar]
- Larson S, Jameson R, Etnier M, Jones T, Hall R. 2012. Genetic diversity and population parameters of sea otters, Enhydra lutris, before fur trade extirpation from 1741–1911. PLoS One 73:e32205.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. 2011. Proteinortho: detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. 2006. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 4437107:50.. [DOI] [PubMed] [Google Scholar]
- Lehri B, Seddon AM, Karlyshev AV. 2017. The hidden perils of read mapping as a quality assessment tool in genome sequencing. Sci Rep. 7:43149.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 312:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. 2009. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 3245934:1561–1564. [DOI] [PubMed] [Google Scholar]
- Liu K, Warnow TJ, Holder MT, Nelesen SM, Yu J, Stamatakis AP, Linder CR. 2012. SATé-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol. 611:90.. [DOI] [PubMed] [Google Scholar]
- Liwanag HEM, Berta A, Costa DP, Abney M, Williams TM. 2012. Morphological and thermal properties of mammalian insulation: the evolution of fur for aquatic living. Biol J Linn Soc Lond. 1064:926–939. [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 4517181:994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. In: Russell D, editor. Multiple Sequence Alignment Methods. Methods in Molecular Biology (Methods and Protocols), vol 1079. Totowa (NJ): Humana Press. p. 155–170. Available from: https://link.springer.com/protocol/10.1007/978-1-62703-646-7_10. [DOI] [PubMed] [Google Scholar]
- Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, Chamberlain JS. 1999. Interactions between β2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat Neurosci. 27:611–617. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R. 1995. Mutation accumulation and the extinction of small populations. Am Nat. 1464:489–518. [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. 2011. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 39(Database issue):D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Gnerre S, Muller P, Reich D. 2009. The difficulty of avoiding false positives in genome scans for natural selection. Genome Res. 195:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DH, Peteet DM. 1994. Extent and timing of the last glacial maximum in southwestern Alaska. Quat Res. 4202:136–148. [Google Scholar]
- Markova-Raina P, Petrov D. 2011. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res. 216:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, Vilà C, Marques-Bonet T, Schnabel RD, Wayne RK, Lohmueller KE. 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci U S A. 1131:152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet O, Rodriguez W, Grusea S, Boitard S, Chikhi L. 2016. On the importance of being structured: instantaneous coalescence rates and human evolution—lessons for ancestral population size inference? Heredity. 1164:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The ensembl variant effect predictor. Genome Biol. 171:122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, et al. 2011. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334:521–524. [DOI] [PubMed] [Google Scholar]
- Millar SE. 2002. Molecular mechanisms regulating hair follicle development. J Invest Dermatol. 1182:216–225. [DOI] [PubMed] [Google Scholar]
- Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F, et al. 2011. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proc Natl Acad Sci U S A. 10830:12348–12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SMJ, Minx P, Hillier LW, Koboldt DC, Davis BW, et al. 2014. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci U S A. 11148:17230–17235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Suzuki S, Koyabu D, Kimura J, Han S-Y, Endo H. 2015. Comparative functional anatomy of hindlimb muscles and bones with reference to aquatic adaptation of the sea otter. J Vet Med Sci. 775:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, Stewart C, et al. 2012. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 1484:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CJ, Bellhorn RW, Williams T, Burns MS, Schaeffel F, Howland HC. 1990. Refractive state, ocular anatomy, and accommodative range of the sea otter (Enhydra lutris). Vision Res. 301:23–32. [DOI] [PubMed] [Google Scholar]
- Nadachowska-Brzyska K, Burri R, Olason PI, Kawakami T, Smeds L, Ellegren H. 2013. Demographic divergence history of pied flycatcher and collared flycatcher inferred from whole-genome re-sequencing data. PLoS Genet. 911:e1003942.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. 1975. The bottleneck effect and genetic variability in populations. Evolution 291:1–10. [DOI] [PubMed] [Google Scholar]
- Niimura Y. 2013. Identification of Chemosensory Receptor Genes from Vertebrate Genomes. In: Touhara K, editor. Pheromone Signaling. Methods in Molecular Biology (Methods and Protocols), vol 1068. Totowa (NJ): Humana Press. p. 95–105. Available from: https://link.springer.com/protocol/10.1007/978-1-62703-619-1_7. Last accessed on March 2019. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. 2003. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci U S A. 10021:12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura O, Hara Y, Kuraku S. 2017. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 3322:3635–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyakatura K, Bininda-Emonds O. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol. 10:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature 2465428:96–98. [DOI] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44(D1):D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz RM. 2001. Osmoregulation in marine mammals. J Exp Biol. 204(Pt 11):1831–1844. [DOI] [PubMed] [Google Scholar]
- Peischl S, Excoffier L. 2015. Expansion load: recessive mutations and the role of standing genetic variation. Mol Ecol. 249:2084–2094. [DOI] [PubMed] [Google Scholar]
- Peng X, Alföldi J, Gori K, Eisfeld AJ, Tyler SR, Tisoncik-Go J, Brawand D, Law GL, Skunca N, Hatta M, et al. 2014. The draft genome sequence of the ferret (Mustela putorius furo) facilitates study of human respiratory disease. Nat Biotechnol. 3212:1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Bowles J, Nagy A, Muscat G, Koopman P. 2000. Mice null for Sox18 are viable and display a mild coat defect. Mol Cell Biol. 2024:9331–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, Muscat G, Abbott C, Koopman P. 2000. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet. 244:434–437. [DOI] [PubMed] [Google Scholar]
- Pickles RSA, Groombridge JJ, Rojas VDZ, Damme PV, Gottelli D, Ariani CV, Jordan WC. 2012. Genetic diversity and population structure in the endangered giant otter, Pteronura brasiliensis. Conserv Genet. 131:235–245. [Google Scholar]
- Pihlström H. 2008. Comparative anatomy and physiology of chemical senses in aquatic mammals. In: Thewissen JGM, Nummela S, editors. Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates. Berkeley: University of California Press. 95–109. [Google Scholar]
- Pispa J, Pummila M, Barker PA, Thesleff I, Mikkola ML. 2008. Edar and Troy signalling pathways act redundantly to regulate initiation of hair follicle development. Hum Mol Genet. 1721:3380–3391. [DOI] [PubMed] [Google Scholar]
- Privman E, Penn O, Pupko T. 2012. Improving the performance of positive selection inference by filtering unreliable alignment regions. Mol Biol Evol. 291:1–5. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O’Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 263:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyenson ND, Lindberg DR. 2011. What happened to gray whales during the Pleistocene? The ecological impact of sea-level change on benthic feeding areas in the North Pacific Ocean. PLoS One 67:e21295.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radinsky LB. 1968. Evolution of somatic sensory specialization in otter brains. J Comp Neurol. 1344:495–505. [DOI] [PubMed] [Google Scholar]
- Ralls K, McInerney NR, Gagne RB, Ernest HB, Tinker MT, Fujii J, Maldonado J. 2017. Mitogenomes and relatedness do not predict frequency of tool-use by sea otters. Biol Lett. 133:20160880.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. 2001. Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech Dev. 107(1–2):69–82. [DOI] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. 2016. g:profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44(W1):W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedman M, Estes JA. 1990. The sea otter (Enhydra lutris): behavior, ecology, and natural history. US Fish and Wildlife Service Biological Report, 90, I–III, 1–126.
- Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK. 2018. Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr Biol. 2821:3487–3494.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Vecchyo D-D, Fan Z, Kim BY, vonHoldt BM, Marsden CD, Lohmueller KE, Wayne RK. 2016. Genomic flatlining in the endangered island fox. Curr Biol. 269:1183–1189. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS. 2005. Evolutionary distance estimation and fidelity of pair wise sequence alignment. BMC Bioinformatics 6:102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels JX, Meachen JA, Sakai SA. 2013. Postcranial morphology and the locomotor habits of living and extinct carnivorans. J Morphol. 2742:121–146. [DOI] [PubMed] [Google Scholar]
- Sato JJ, Wolsan M. 2012. Loss or major reduction of umami taste sensation in pinnipeds. Naturwissenschaften 998:655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JJ, Wolsan M, Minami S, Hosoda T, Sinaga MH, Hiyama K, Yamaguchi Y, Suzuki H. 2009. Deciphering and dating the red panda’s ancestry and early adaptive radiation of Musteloidea. Mol Phylogenet Evol. 533:907–922. [DOI] [PubMed] [Google Scholar]
- Sato JJ, Wolsan M, Prevosti FJ, D’Elía G, Begg C, Begg K, Hosoda T, Campbell KL, Suzuki H. 2012. Evolutionary and biogeographic history of weasel-like carnivorans (Musteloidea). Mol Phylogenet Evol. 633:745–757. [DOI] [PubMed] [Google Scholar]
- Saunders SP, Goh CSM, Brown SJ, Palmer CNA, Porter RM, Cole C, Campbell LE, Gierlinski M, Barton GJ, Schneider G, et al. 2013. TMEM79/Matt is the matted mouse gene and is a predisposing gene for atopic dermatitis in human subjects. J Allergy Clin Immunol. 1325:1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 468:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Souvorov A, Sabath N, Landan G, Gonnet GH, Graur D. 2009. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol Evol. 1:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hourmozdi JN, Hahn MW. 2011. Pervasive multinucleotide mutational events in eukaryotes. Curr Biol. 2112:1051–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Shanku AG, Kern AD. 2016. Effects of linked selective sweeps on demographic inference and model selection. Genetics 2043:1207–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, Belli S, Petukhova L, Schinzel A, Brivanlou AH, et al. 2010. APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature 4647291:1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simenstad CA, Estes JA, Kenyon KW. 1978. Aleuts, sea otters, and alternate stable-state communities. Science 2004340:403–411. [DOI] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, Sella G. 2014. The deleterious mutation load is insensitive to recent population history. Nat Genet. 463:220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Waack S. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J, Danielian PS, McMahon JA, Lewis PM, Paus R, McMahon AP. 1998. Sonic hedgehog signaling is essential for hair development. Curr Biol. 819:1058–1069. [DOI] [PubMed] [Google Scholar]
- Strobel SM, Sills JM, Tinker MT, Reichmuth CJ. 2018. Active touch in sea otters: in-air and underwater texture discrimination thresholds and behavioral strategies for paws and vibrissae. J Exp Biol. 22118:jeb181347.. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 1052:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasoff FJ. 1972. Comparative aspects of the hind limbs of the river otter, sea otter, and seals. In: Harrison RJ, editor. Functional Anatomy of Marine Mammals. New York: Academic Press. pp 333–359. [Google Scholar]
- Thewissen, J. G. M. 2009. “Sensory biology: overview,” In: Perrin, WF, Würsig, B. and Thewissen, JGM, editors. Encyclopedia of Marine Mammals, 2nd Edn. Burlington, MA: Academic Press, pp. 1003–1005. [Google Scholar]
- Thewissen JGM, Nummela S. 2008. Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates. Berkeley, CA: University of California Press. [Google Scholar]
- Thirstrup JP, Anistoroaei R, Guldbrandtsen B, Christensen K, Fredholm M, Nielsen VH. 2014. Identifying QTL and genetic correlations between fur quality traits in mink (Neovison vison). Anim Genet. 451:105–110. [DOI] [PubMed] [Google Scholar]
- Tian R, Wang Z, Niu X, Zhou K, Xu S, Yang G. 2016. Evolutionary genetics of hypoxia tolerance in cetaceans during diving. Genome Biol Evol. 83:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker MT, Bodkin JL, Ben-David M, Estes JA. 2017. Otters: Enhydra lutris and Lontra felina. In: Wursig, B, Thewissen H, Kovacs K, editors. Encyclopedia of marine mammals. 3rd ed. New York: Elsevier Inc. p. 664–671. [Google Scholar]
- Tinker MT, Hatfield BB. 2017. California sea otter (Enhydra lutris nereis) census results, Spring 2017. Reston (VA: ): U.S. Geological Survey. Available from: http://pubs.er.usgs.gov/publication/ds1067 [Google Scholar]
- Tinker MT, Hatfield BB, Harris MD, Ames JA. 2016. Dramatic increase in sea otter mortality from white sharks in California. Mar Mam Sci. 321:309–326. [Google Scholar]
- Tissir F, Goffinet AM. 2003. Reelin and brain development. Nat Rev Neurosci. 46:496–505. [DOI] [PubMed] [Google Scholar]
- Unger S, Górna MW, Le Béchec A, Do Vale-Pereira S, Bedeschi MF, Geiberger S, Grigelioniene G, Horemuzova E, Lalatta F, Lausch E, et al. 2013. FAM111A mutations result in hypoparathyroidism and impaired skeletal development. Am J Hum Genet. 926:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2016. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Fish and Wildlife Service. 2003. Final revised recovery plan for the southern sea otter (Enhydra lutris nereis). Available from: https://www.cabdirect.org/cabdirect/abstract/20137204961.
- Van Valkenburgh B, Curtis A, Samuels JX, Bird D, Fulkerson B, Meachen‐Samuels J, Slater GJ. 2011. Aquatic adaptations in the nose of carnivorans: evidence from the turbinates. J Anat. 2183:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat A, Hahn MW, Thornton JW. 2018. Multinucleotide mutations cause false inferences of lineage-specific positive selection. Nat Ecol Evol. 2:1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Stolzer M, Goldman A, Durand D. 2008. Reconciliation with non-binary species trees. J Comput Biol. 158:981–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Baer KE. 1828. Über entwickelungsgeschichte der thiere: beobachtung und reflexion. Köningsberg: Bornträger. [Google Scholar]
- Waku D, Segawa T, Yonezawa T, Akiyoshi A, Ishige T, Ueda M, Ogawa H, Sasaki H, Ando M, Kohno N, et al. 2016. Evaluating the phylogenetic status of the extinct Japanese otter on the basis of mitochondrial genome analysis. PLoS One 113:e0149341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grohé C, Su DF, White SC, Ji X, Kelley J, Jablonski NG, Deng T, You Y, Yang X. 2018. A new otter of giant size, Siamogale melilutra sp. nov. (Lutrinae: Mustelidae: Carnivora), from the latest Miocene Shuitangba site in north-eastern Yunnan, south-western China, and a total-evidence phylogeny of lutrines. J Syst Palaeontol. 161:39–65. [Google Scholar]
- Wang Y, Badea T, Nathans J. 2006. Order from disorder: self-organization in mammalian hair patterning. Proc Natl Acad Sci U S A. 10352:19800–19805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chang H, Nathans J. 2010. When whorls collide: the development of hair patterns in frizzled 6 mutant mice. Development 13723:4091–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Förster E, Sweatt JD, Herz J. 2002. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 27742:39944–39952. [DOI] [PubMed] [Google Scholar]
- Willemsen GF. 1992. A revision of the Pliocene and Quaternary Lutrinae from Europe. Leiden (The Netherlands): Nationaal Natuurhistorisch Museum.
- Williams TD, Allen DD, Groff JM, Glass RL. 1992. An analysis of California sea otter (Enhydra lutris) pelage and integument. Mar Mamm Sci. 81:1–18. [Google Scholar]
- Williams TM, Ben-David M, Noren S, Rutishauser M, McDonald K, Heyward W. 2002. Running energetics of the North American river otter: do short legs necessarily reduce efficiency on land? Comp. Biochem Physiol A Mol Integr Physiol. 1332:203–212. [DOI] [PubMed] [Google Scholar]
- Wong KM, Suchard MA, Huelsenbeck JP. 2008. Alignment uncertainty and genomic analysis. Science 3195862:473–476. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Tanigaki K, Han H, Hiai H, Honjo T. 2003. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr Biol. 134:333–338. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yim H-S, Cho YS, Guang X, Kang SG, Jeong J-Y, Cha S-S, Oh H-M, Lee J-H, Yang EC, Kwon KK, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 461:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Peng D, Liu J, Luan P, Liang L, Lee H, Lee M, Ryder OA, Zhang Y. 2011. On the phylogeny of Mustelidae subfamilies: analysis of seventeen nuclear non-coding loci and mitochondrial complete genomes. BMC Evol Biol. 11:92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fan Z, Han E, Hou R, Zhang L, Galaverni M, Huang J, Liu H, Silva P, Li P, et al. 2014. Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 107:e1004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M. 2006. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 2212:1437–1439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.