Abstract
Cervical cancer is the most common gynecological cancer in women worldwide. Human papillomavirus (HPV) is required but not sufficient for developing cervical cancer. HPV E6 and E7 proteins are able to directly interact with certain nuclear receptors; however, whether steroid hormone receptors mediate cervical carcinogenesis is not completely understood. The present study demonstrated via immunohistochemistry that estrogen receptor α (ERα) and androgen receptor (AR) expression were decreased in a sequential manner from healthy cervical tissues to cervical intraepithelial neoplasia tissues and further to cervical cancer (CC) tissues, whereas microRNA (miR)-130a-3p expression levels were higher in CC tissues compared with healthy tissues. Both ERα and AR were direct targets of miR-130a-3p, as determined by performing luciferase reporter assays and western blotting. Functionally, compared with the corresponding control groups, miR-130a-3p knockdown, ERα overexpression and AR overexpression significantly inhibited CC cell proliferation and invasion, as demonstrated by the results obtained from the Cell Counting Kit-8 and Transwell assays in vitro. In addition, antagomiR-130a decreased tumor size and weight in vivo compared with control antagomiR as determined via the xenograft tumor growth assay. Therefore, the results suggested that miR-130a-3p might contribute to tumor progression by suppressing ERα and AR, and serve as a promising candidate target for the treatment of patients with CC.
Keywords: cervical cancer, microRNA, estrogen receptor α, androgen receptor, neoplasm, invasion
Introduction
Cervical cancer (CC) is the most common gynecological cancer in women worldwide (1). In 2019, 13,170 new cases were diagnosed and 4,250 disease-related deaths were predicted in the USA (1). Furthermore, 530,000 new cases are diagnosed yearly worldwide and 20-25% of cases are reported in China (2). Although CC also occurs during perimenopause, unlike endometrial cancer or ovarian cancer, there are few reports on the association between steroid hormone receptors and CC (3). Several in vitro studies have investigated the interactions between estrogen/progesterone and human papillomavirus (HPV), but the results are controversial (4,5).
Estrogen serves a major role in several hormone-dependent malignancies (6). Estrogen and its analogs can activate estrogen-responsive genes by binding to estrogen receptor α (ERα) (7). Previous genomic studies have identified frequent mutations of the ERα gene (ESR1) in CC (8,9). Additionally, it has been demonstrated that viral-codified E6 and E7 proteins could directly interact with androgen receptor (AR) (10). However, to the best of our knowledge, the role of ERα and AR in cervical carcinogenesis is not completely understood.
MircoRNAs (miRNAs/miRs) are a class of short, highly conserved, non-coding RNAs that regulate gene expression by inhibiting translation or inducing mRNA degradation at the post-transcriptional level (11). The aberrant expression of miRNAs has been reported in various tumors (12), such as endometrial cancer (13), ovarian cancer (14), and CC (15). Our previous study demonstrated that miR-107-5p could promote tumor proliferation and invasion by targeting ERα in endometrial carcinoma (16); therefore, it was hypothesized that specific miRNAs may control ERα and AR expression post-transcriptionally during CC progression.
The aim of the present study was to investigate the expression of miR-130a-3p, ERα and AR in healthy cervical and CC tissues, and to explore whether miR-130a-3p could contribute to tumor progression by suppressing ERα and AR.
Materials and methods
Clinical samples
The present study was approved (approval no. GKLW 2017-125) by the Human Investigation Ethical Committee of the International Peace Maternity & Child Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Shanghai, China). Written informed consent was obtained from all patients. A total of 60 female patients (45-68 years old) who underwent radical hysterectomy with lymph node dissection for CC at the International Peace Maternity & Child Health Hospital Affiliated to Shanghai Jiao Tong University School of Medicine between August 2014 and April 2017 were included in the present study. The stages and histological grades of the tumors were determined according to the criteria of the Federation International of Gynecology and Obstetrics Surgical staging system (2018) (17). A total of 20 healthy cervical tissue samples (female subjects; 44-56 years old) were obtained from patients who underwent a hysterectomy to treat other diseases between April 2015 and April 2017 such as myoma or adenomyosis. In addition, 20 cervical intraepithelial neoplasia (CIN) I, 20 CIN II and 30 CIN III tissue samples (female subjects; 34-53 years old) were collected from patients who underwent a cervical biopsy or loop electrosurgical excision procedure between October 2015 and April 2017. All the tissues were collected from each patient or healthy control at the International Peace Maternity & Child Health Hospital Affiliated to Shanghai Jiao Tong University School of Medicine and immediately snap-frozen in liquid nitrogen until further use. All the tissues were pathologically reviewed by two pathologists before use.
Immunohistochemistry (IHC)
All tissue sections (4-µm thick) were processed for IHC as previously described (18,19). The following primary antibodies were used for IHC: Anti-ERα (cat. no. ab75635; 1:200; Abcam) and AR (cat. no. ab74272; 1:250; Abcam).
The expression of ERα and AR was evaluated in terms of staining intensity: (Negative), 1 (weak), 2 (medium) or 3 (strong). The extent of staining was scored as 0 (0%), 1 (1-25%), 2 (26-50%), 3 (51-75%) or 4 (76-100%), according to the percentage of positively stained areas in relation to the whole tumor area. The final staining score (0-12) was calculated by multiplying the intensity score and the extent score (19). The final staining score was used to evaluate the expression of ERα and AR.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from frozen tissues using TRI Reagent (Molecular Research Center). Total RNA was reverse transcribed into mature miRNA using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), using the following RT temperature protocol: 42˚C for 15 min; 85˚C for 5 sec. Subsequently, qPCR was performed using TaqMan MicroRNA assay primers (Applied Biosystems; Thermo Fisher Scientific, Inc.) with TaqMan Universal PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and an ABI Prism 7000 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95˚C for 3 min; 95˚C for 30 sec and 60˚C for 30 sec for 35 cycles; 72˚C for 5 min and maintenance at 4˚C for further use. The sequences of the primers used for qPCR are provided in Table I. miRNA expression levels were normalized to the internal reference gene U6. Relative gene expression was quantified using the 2-ΔΔCq method (20).
Table I.
Oligo sequences used in the present study.
| Identifier | Sequence (5'→3') |
|---|---|
| miR-130a | F: TTGCGATTCTGTTTTGTGCT |
| R: GTGGGGTCCTCAGTGGG | |
| U6 | F: AGAGCCTGTGGTGTCCG |
| R: CATCTTCAAAGCACTTCCCT | |
| miR-130a inhibitor | F: AUGCCCUUUUAACAUUGCACUG |
| R: UAGUGCAAUAGUAUCGUCGAGAC | |
| miR-130a inhibitor NC | F: CAGUACUUUUGUGUAGUACAA |
| R: GUCCUGAGAAGGCUAGCAUAGAU | |
| miR-130a mimic | F: CAGUGCAAUGUUAAAAGGGCAU |
| R: AUAGCCCUGUACAAUGCUGCUUU | |
| miR-130a mimic NC | F: UUCUCCGAACGUGUCACGUTT |
| R: ACUUUGACAAUACUAUAGUGGAA | |
| ESR1-3'UTR-WT | F: TAAACGCGTGTAACGTGAATACCAC |
| R: TCGATGCGTAACGATGTTCCTCAGTG | |
| ESR1-3'UTR-MT | F: TATTACCGATACGGAAAAGCAATGT |
| R: TGAAGCAACGGAAATGCATAGA | |
| AR-3'UTR-WT | F: TACGAAAAACTTGAATGACAATAC |
| R: GAAAATGAACTTGAGACAAATG | |
| AR-3'UTR-MT | F: GCGATGACACTGCTCCTATAGCGAAT |
| R: TGGGATCCCACTGCTCCCATGGCTTA |
miR, microRNA; NC, negative control; UTR, untranslated region; WT, wild-type; MT, mutant; F, forward; R, reverse.
Western blotting
Cells were harvested and proteins were extracted using Mem-PER Eukaryotic Membrane Protein Extraction Reagent (Pierce; Thermo Fisher Scientific, Inc.) containing complete mini cocktail, NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce; Thermo Fisher Scientific, Inc.) and protease inhibitor cocktail. A total of 20 µg proteins (determined using the BCA method) were separated via 8% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked for 1 h at room temperature with 5% skimmed milk in TBS. Subsequently, the membranes were incubated with rabbit polyclonal primary antibodies targeted against: ERα (cat. no. ab75635; 1:1,000; Abcam), AR (cat. no. ab74272; 1:1,000; Abcam) and β-actin (cat. no. 4970; 1:2,000; Cell Signaling Technology, Inc.) in 10 ml of 5% skimmed milk and incubated at 4˚C overnight. After washing, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat. no. 7074; 1:5,000; Cell Signaling Technology, Inc.) for 1 h at 37˚C. The results were visualized using an enhanced chemiluminescence kit (ECL kit; Pierce; Thermo Fisher Scientific, Inc.) using Kodak XAR-5 film (Sigma-Aldrich; Merck KGaA). β-actin was used as the loading control.
Cell culture and transfections
Human HeLa and SiHa CC cell lines, and 293T cells were obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were cultured in DMEM/F12 (cat. no. 11030; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (cat. no. 16000-44; Gibco; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2.
miR-130a inhibitor (miR-130ai), miR-130a inhibitor negative control (NC; miR-130ai NC), miR-130a mimic (miR-130am) and miR-130a mimic negative control (miR-130am NC) were synthesized by Shanghai GenePharma Co., Ltd. The oligo sequences are provided in Table I. For transfection, cells were seeded into 6-well plates at 70-80% confluence and grown overnight. Subsequently, HeLa or SiHa cells were transfected with 100 nM miR-130ai, miR-130am, miR-130ai NC or miR-130am NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Transfection efficiency of miR-130ai and miR-130am was assessed via RT-qPCR after 24 h (Fig. S1A and B). For vector transduction, ERα-overexpression vector (Ubi-MCS-3FLAG-ESR1), AR-overexpression vector (Ubi-MCS-3FLAG-AR) and empty vector were purchased from Shanghai GeneChem Co., Ltd. HeLa and SiHa cells were transfected with 1 µg/ml Ubi-MCS-3FLAG-ESR1, Ubi-MCS-3FLAG-AR or empty vector using Lipofectamine® 2000 according to the manufacturer's instructions. Transfection efficiency of Ubi-MCS-3FLAG-ESR1 and Ubi-MCS-3FLAG-AR were confirmed via western blotting after 48 h (Fig. S1C and D).
Cell Counting Kit-8 (CCK-8) and cell clonogenic assays
To assess cell proliferation using the CCK-8 assay at 70-80% confluence, transfected cells were serum-starved for 24 h at 5% CO2, 37˚C and then cultured in 96-well plates (1x103 cells/well). Subsequently, at selected time points (24, 48, 72, 96 and 120 h), 20 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.) were added in each well, and incubated at 37˚C for 2 h to measure the rate of cell proliferation. Absorbance was measured at a wavelength of 490 nm using a SpectraMax 190 microplate reader (Bio-Rad Laboratories, Inc.). For the cell clonogenic survival assays, transfected cells were seeded (2x103 cells/plate) into 6-well plates. Following culture for 2 weeks at 5% CO2, 37˚C, cell colonies (-50 cells are visible from one cell) were fixed in 2% formaldehyde in PBS for 2 min and the stained with 0.5% crystal violet for 30 min (both at room temperature). Colonies were visually counted.
Transwell invasion assay
Cells were plated (2x105 cells/well) in serum-free medium in Transwell chambers (8-µm pore size; BD Biosciences) pre-coated with Matrigel (4˚C for 2 h). Complete DMEM/F12 medium containing 10% FBS was added to 24-well plates as a chemoattractant (lower chamber). After 24 h of incubation at 5% CO2, 37˚C, the cells were fixed with 4% paraformaldehyde for 1 h at room temperature. The cells on the apical side of each insert were removed by mechanical scraping. The cells that migrated to the basal side of the membrane were stained with 0.1% crystal violet at room temperature and visualized under a Leica DMI 3000B light microscope (Leica Microsystems, Inc.; magnification, x200).
Luciferase assays
By searching for potential miRNAs, the miR-130a-3p targeting site was identified within the 3'untranslated region (UTR) of ESR1 and AR, as detected by three software algorithms (TargetScan v3.1, targetscan.org; Pictar, pictar.mdc-berlin.de; and MiRanda, miranda.org). To examine whether miR-130a-3p could modulate ERα and AR expression, a DNA fragment comprising a partial wild-type (WT) 3'UTR of ESR1or AR was constructed, as well as a corresponding mutant (MT) 3'UTR of ESR1 or AR. The plasmids were synthesized and cloned into the pGL3-REPORT luciferase vector (Promega Corporation) containing the luciferase gene to generate pGL3-ESR1/AR-3'UTR-WT and pGL3-ESR1/AR-3'UTR-MT. 293T cells (5x104/well) were seeded into 24-well plates and transfected with 0.2 µg of either pGL3-ESR1/AR-3'UTR-WT or pGL3-ESR1/AR-3'UTR-MT. The luciferase reporter assay was performed as previously described (16,21,22).
Xenograft tumor growth assays
The animal experiments were performed in strict accordance with the Guideline for the Care and Use of Laboratory Animals of China. The protocol was approved by the Committee on the Ethics of Animal Experiments of Shanghai Jiaotong University [approval no. SCXK (hu) 2018-0007]. All efforts were made to minimize animal suffering. A total of 10 female BALB/c nude mice (age, 5 weeks; weight, 15 g) were obtained from the Chinese Academy of Sciences. Animals were maintained at standard controlled conditions (room temperature, 22±1˚C; relative humidity, 50-60%) on a 12 h light/dark cycle and free access to food and water. HeLa cells were harvested and resuspended (5x106 cells/200 µl) in sterile saline. Mice (n=5 per group; miR-130a-targeting antagomiR group and the control antagomiR group) were subcutaneously injected with 200 µl HeLa cells in the subdermal space on the medial side of the neck. After ~1 week, when tumors reached an average volume of ~20 mm3, each tumor was directly injected with antagomiR-130a (cat. no. B05001) or control antagomiR (cat. no. B04007) (both from Cytiva; 40 ml PBS containing 1 µg antagomiR-130a or control antagomiR) on day 0 (when the tumors reached an average volume of 20 mm3), 5 and 9(23). Tumor volume was measured every 7 days until the end of the experiment and was calculated using the following formula: Volume=(largest diameter x smallest diameter2) x0.5. At the end of the xenograft experiment (day 21), mice were sacrificed by CO2 inhalation (30% volume displacement per minute). Following sacrifice, tumor weight was determined.
In silico analysis of ESR1, AR and disease-free survival (DFS) of patients with CC
In silico analysis of the association between ESR1 or AR and DFS of patients with cervical cancer was performed using online published data obtained from Gene Expression Profiling Interactive Analysis (GEPIA; gepia.cancer-pku.cn) and The Cancer Genome Atlas (TCGA) network (cBioPortal for Cancer Genomics; cbioportal.org) (24-26). Overall survival (OS), DFS, disease-specific survival (DSS) and progression-free survival (PFS) were analyzed using the cBioPortal for Cancer Genomics database (25,26).
Statistical analysis
Comparisons between two groups were analyzed using an unpaired Student's t-test. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. DFS was assessed by using the Kaplan-Meier method with the log-rank test Data are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using SPSS software (version 16.0; SPSS, Inc.). All experiments were carried out in triplicate and repeated at least three times.
Results
In silico analysis of ESR1 and AR expression, and association with survival of patients with CC
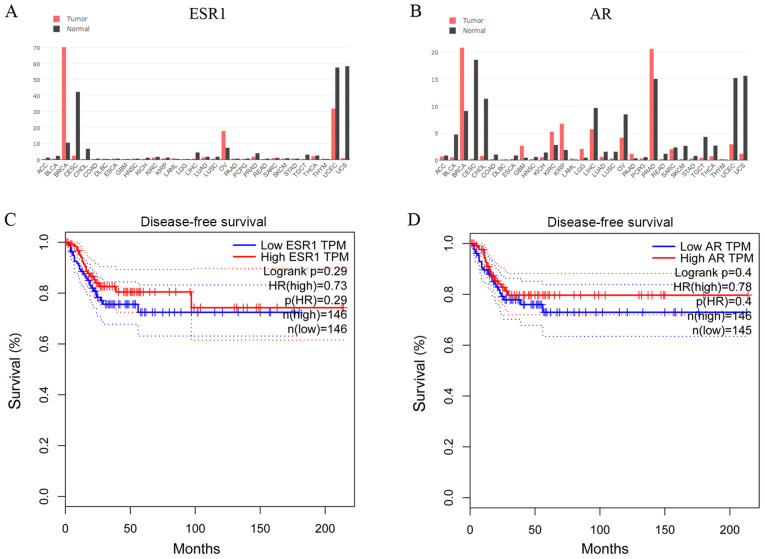
To establish whether steroid hormone receptors affected CC progression, the GEPIA database was used to examine cross-cancer alteration summaries of ESR1 and AR. Patients with CC displayed notably lower ESR1 and AR expression levels compared with healthy individuals (Fig. 1A and B). The associations between ESR1 and AR expression and DFS of patients with CC (n=292) were assessed in an independent dataset obtained from the GEPIA database (24). The DFS of the high-ESR1 group [hazard ratio (HR), 0.73; 95% confidence interval (CI), 0.61-0.90] and the low-ESR1 group (95% CI, 0.62-0.82) are presented in Fig. 1C. The DFS of the high-AR group (HR, 0.78; 95% CI, 0.72-0.88) and the low-AR group (95% CI, 0.63-0.83) are presented in Fig. 1D. In another TCGA network (cBioPortal for Cancer Genomics) (25,26), the OS, DFS, DSS and PFS of patients with CC from the cBioPortal database were also analyzed (data not shown). Although not significant, patients with CC and lower ESR1 and AR expression displayed poorer survival rates compared with patients with CC and higher ESR1 and AR expression.
Figure 1.
In silico analysis of ESR1 and AR expression, and the association with patient survival in CC. Genomics of cross-cancer alteration summary of (A) ESR1 and (B) AR genes in tumor and healthy tissues. (C) DFS of the high-ESR1 (HR, 0.73; 95% CI, 0.61-0.90) and low-ESR1 (95% CI, 0.62-0.82) groups. (D) DFS of the high-AR (HR, 0.78; 95% CI, 0.72 to 0.88) and low-AR (95% CI, 0.63-0.83) groups. ESR1, estrogen receptor α gene; AR, androgen receptor; DFS, disease-free survival; CI, confidence interval; HR, hazard ratio.
ERα and AR expression is significantly decreased in CC
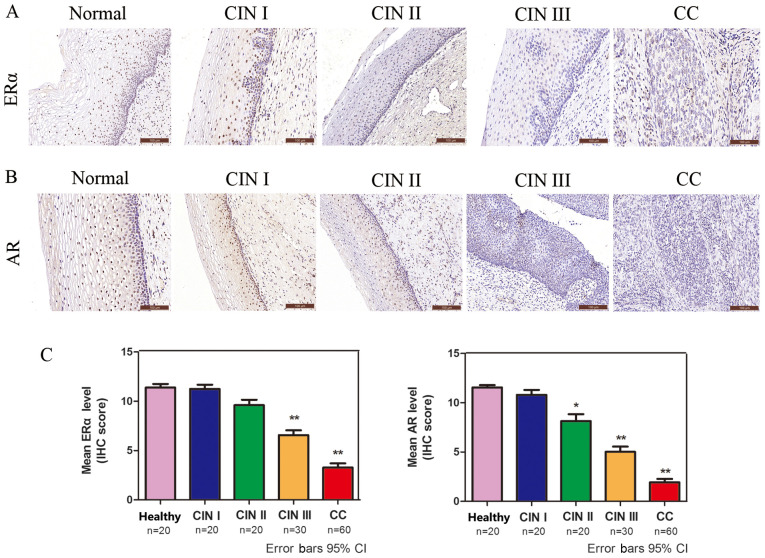
The protein expression levels of ERα and AR in CC were analyzed via IHC. ERα- and AR-positive immunostaining was observed in the cell nucleus (Fig. 2A and B). The protein expression levels of ERα and AR were decreased in a sequential manner from healthy cervical to CIN and further to CC tissues (Fig. 2C), suggesting that low ERα and AR expression was associated with high-grade lesions.
Figure 2.
ERα and AR are significantly decreased in CC. Immunohistochemical analysis of (A) ERα and (B) AR in healthy cervical, CIN and squamous cell cervical carcinoma tissues (magnification, x200). (C) Quantification of immunohistochemical staining in healthy cervical epithelium (n=20), CIN I (n=20), CIN II (n=20), CIN III (n=30) and CC (n=60) tissues. *P<0.05 and **P<0.01. ERα, estrogen receptor α; AR, androgen receptor; CC, cervical cancer; CIN, cervical intraepithelial neoplasia; IHC, immunohistochemistry; CI, confidence interval.
ERα and AR are targets of miR-130a-3p
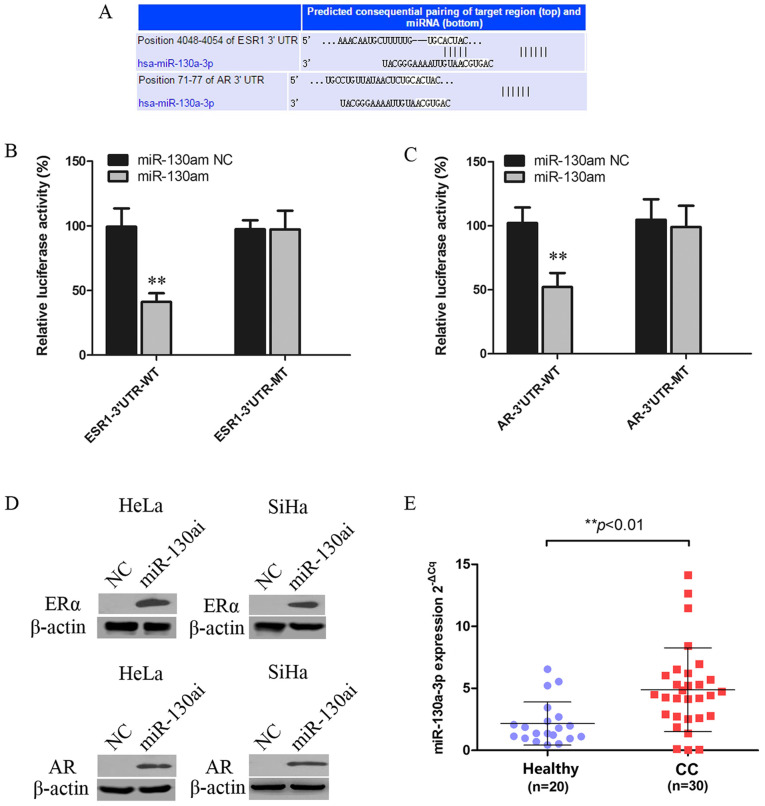
miRNAs can regulate gene expression by inhibiting translation or inducing mRNA degradation at the post-transcriptional level (27). The miR-130a-3p targeting site was identified within the 3'UTRs of ESR1 and AR (Fig. 3A). 293T cells were co-transfected with WT or MT vector and miR-130am or miR-130am NC. miR-130am significantly decreased the relative luciferase activity of ESR1-3'UTR-WT and AR-3'UTR-WT compared with miR-130am NC. By contrast, miR-130am did not significantly alter the luciferase activity of ESR1-3'UTR-MT or AR-3'UTR-MT compared with miR-130am NC (Fig. 3B and C).
Figure 3.
ERα and AR are targets of miR-130a-3p. (A) Putative miR-130a-3p binding site in the 3'UTRSs of ESR1 and AR, as predicted by TargetScan, Pictar and MiRanda. Luciferase assay of 293T cells co-transfected with (B) pGL3-ESR1-3'UTR-WT, pGL3-ESR1-3'UTR-MT, (C) pGL3-AR-3'UTR-WT or pGL3-AR-3'UTR-MT and miR-130am or NC. (D) ERα and AR protein expression levels in HeLa and SiHa cells following transfected with miR-130ai NC or miR-130ai. (E) miR-130a-3p expression levels in CC (n=30) and healthy cervical (n=20) tissues. **P<0.01 vs. NC. EERα, estrogen receptor α; AR, androgen receptor; miR, microRNA; 3'UTR, 3'untranslated region; WT, wild-type; MT, mutant; miR-130am, miR-130a mimics; miR-130ai, miR-130a inhibitor; NC, negative control; CC, cervical cancer.
To further investigate the functional role of deregulated miR-130a-3p in CC cells, the effects of miR-130ai on the expression of ERα and AR were examined. miR-130ai notably increased the protein expression levels of ERα and AR compared with miR-130ai NC in HeLa and SiHa cells (Fig. 3D), supporting its role as a functional suppressor of ERα and AR. Additionally, the levels of miR-130a-3p in 20 healthy cervical tissues and 30 CC tissues were detected via RT-qPCR. The results indicated that miR-130a-3p expression levels were significantly higher in CC tissues compared with healthy cervical tissues (Fig. 3E).
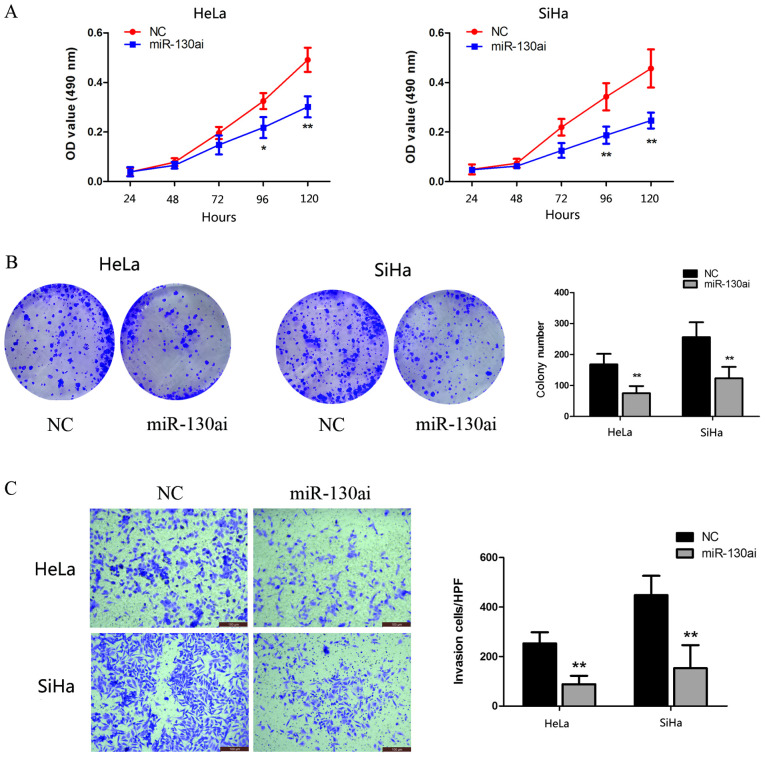
miR-130a-3p knockdown inhibits CC cell proliferation and invasion
To identify the biological function of miR-130a-3p, HeLa and SiHa cells were transfected with miR-130ai. Compared with miR-130ai NC, miR-130ai significantly inhibited HeLa and SiHa cell proliferation, as indicated by the CCK-8 and clonogenic assay results (Fig. 4A and B). Subsequently, the role of miR-130a-3p in the HeLa and SiHa cell invasion was investigated following transfection with miR-130ai. miR-130ai significantly decreased cell invasion compared with miR-130ai NC (Fig. 4C), suggesting that miR-130a-3p enhanced cell invasion.
Figure 4.
miR-130a-3p promotes cervical cancer cell proliferation and invasion. HeLa and SiHa cell proliferation, as determined by performing (A) Cell Counting Kit-8 and (B) clonogenic assays (6-wells). (C) HeLa and SiHa cell invasion (magnification, x200). *P<0.05 and **P<0.01. miR, microRNA; OD, optical density; NC, negative control; HPF, high-power fields; miR-130ai, miR-130a inhibitor.
ERα and AR are functionally targets of miR-130a-3p, and are involved in CC cell proliferation and invasion
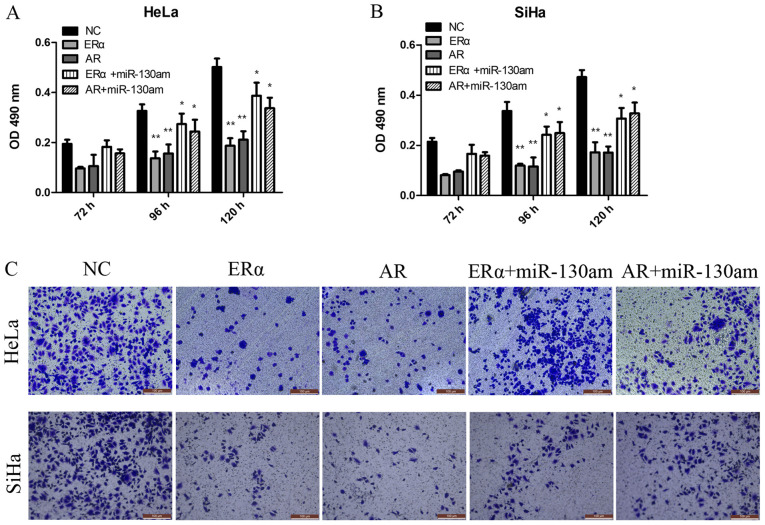
To investigate the biological function of ERα and AR, vector transduction was performed to overexpress ERα and AR in HeLa and SiHa cells. Compared with the NC group, ERα overexpression and AR overexpression significantly inhibited HeLa and SiHa cell proliferation and invasion, as assessed by performing CCK-8 and Transwell assays (Fig. 5).
Figure 5.
ERα and AR are functional targets of miR-130a-3p, and are involved in cervical cancer cell proliferation and invasion. (A) HeLa and (B) SiHa cell proliferation. (C) HeLa and SiHa cell invasion (magnification, x200). *P<0.05 and **P<0.01 vs. NC. ERα, estrogen receptor α; AR, androgen receptor; miR, microRNA; OD, optical density; NC, negative control; miR-130am, miR-130a mimics.
To address whether the biological function effects of ERα and AR expression were predominately due to the regulation of miR-130a-3p, the present study investigated whether miR-130a-3p, ERα and AR functioned via the same signaling pathway to modulate CC cell proliferation and invasion. The results suggested that miR-130am significantly rescued CC cell proliferation and invasion in ERα- and AR-overexpression HeLa and SiHa cells (Fig. 5). The results suggested that ERα and AR were functional targets of miR-130a-3p in CC cells.
Blocking miR-130a-3p inhibits tumorigenicity in vivo
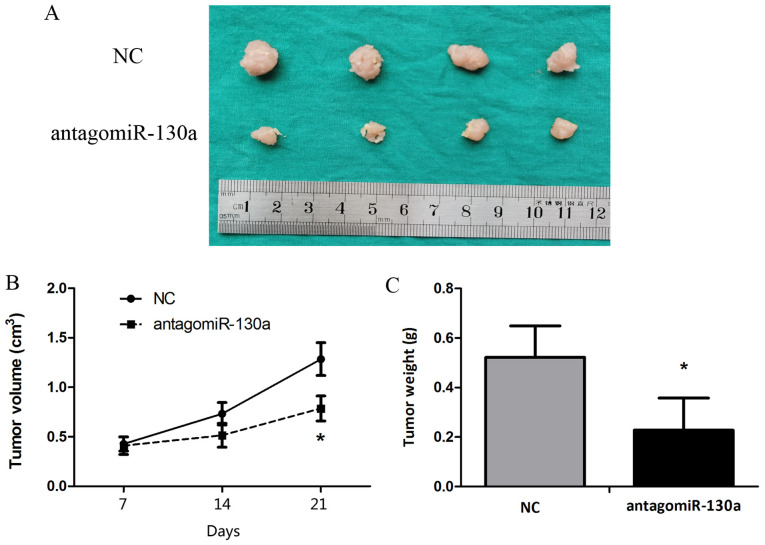
Animal studies were conducted to assess the effect of miR-130a-3p on tumor growth in nude mice. HeLa cells were subcutaneously injected into subdermal space on the medial side of the neck of nude mice to establish tumors, which were treated by direct intratumoral injection when they became clearly palpable. After monitoring tumor growth for 21 days, the antagomiR-130a group displayed significantly decreased tumor size and weight compared with the control antagomiR group (Fig. 6).
Figure 6.
Blocking miR-130a-3p inhibits tumorigenicity in a mouse xenograft model. Tumor cells were injected subcutaneously into mice in the subdermal space on the medial side of the neck. The short and long diameters of the tumors were measured every 7 days, and the tumor volume (cm3) were calculated. (A) Representative image of tumors. Tumor (B) volume and (C) weight. *P<0.05 vs. NC. miR, microRNA; NC, negative control.
Discussion
To the best of our knowledge, the present study suggested for the first time that miR-130a-3p promoted CC cell proliferation and invasion by directly targeting ERα and AR. Despite advances in diagnostic and screening techniques, and the availability of vaccines, CC remains the fourth largest cause of cancer-related deaths in women worldwide (1). Numerous established risk factors of CC have been reported, including exposure to HPV, a high number of sexual partners and young age at onset of sexual activity (2). Risk factors that are associated with progression from HPV infection to pre-cancer include oral contraceptive use and smoking (28). The importance of estrogen and ERα at all carcinogenic steps was strongly supported in certain mouse models (29,30); however, few human studies have been conducted. Therefore, the exact role of hormonal factors in progression to pre-cancer and cancer is not completely understood.
ERα is usually expressed in healthy cervical tissues, but its expression is decreased or absent in invasive CC, which indicated that ERα expression is lost during the development of CC (31). Zhai et al (32) reported that restoration of ESR1 expression in ERα-negative CC reduced cell invasiveness in cell culture, and concluded that loss of ERα expression serves a major role in mediating CC invasion and progression. However, other studies have indicated that estrogenic stimulation can influence cervical tumorigenesis (32,33) in agreement with previous studies, the present study suggested that the expression of ERα and AR was decreased in a sequential manner from healthy cervical tissues to CIN tissues and further to CC tissues, suggesting that the loss of ERα and AR served a major role in mediating CC progression. The role of AR in CC is not completely understood. A previous study reported that long-term androgen treatment of female-to-male transsexuals is associated with morphological alterations of the cervix (34). In another study, Noël et al (10) demonstrated that the loss of AR expression is a frequent and common event in high-grade squamous intraepithelial lesion and invasive squamous CC, resulting from complex interactions between high risk HPVs and AR. However, the underlying regulatory mechanisms and the downstream biological effects of the loss of ERα and AR in CC malignancy are not completely understood.
Aberrant expression of miR-130a has been reported in several types of cancer, including gastric (35), esophageal (36) and breast cancer (21), as well as in CC (15,37). However, the role of miR-130a in tumor development and progression is contradictory, as it can serve as an oncogene or tumor suppressor gene by regulating various canonical signaling pathways or target genes (38,39). miR-130a is a potential oncomiR candidate in adriamycin-resistant breast cancer and platinum-resistant ovarian cancer, whereas it serves as a suppressive miRNA in cisplatin-resistant hepatoma cells (39). To date, the role of miR-130a in the progression of CC carcinogenesis is not completely understood. Yin et al (40) reported that high expression of miR-130a was significantly associated with lymph node metastasis and an advanced clinical stage of CC. The results of the present study indicated that the expression of miR-130a in CC tissues was obviously higher compared with healthy cervical tissues, and miR-130a knockdown inhibited CC cell proliferation and invasion in vitro and in vivo compared with miR-130ai NC and control antagomiR, respectively. Moreover, the present study indicated that miR-130a could directly bind to the 3'UTR of ESR1 and AR mRNA, suggesting that miR-130a mediated CC progression by functionally regulating ERα and AR.
In conclusion, the present study indicated that miR-130a-3p may contribute to tumor progression by suppressing ERα and AR, and it may serve as a promising candidate target for the treatment of patients with CC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the National Natural Science Foundation of China (grant nos. 81172477, 81572547 and 81402135), the Project of Shanghai Key Clinical Department (grant no. shslczdzk06302), the Project of the Science and Technology Commission of Shanghai Municipality (grant nos. 11ZR1440800 and 13JC1401303), the Project of Outstanding Subject Leaders of the Shanghai Health System (grant no. XBR2013097) and the Shanghai Jiao Tong University Medicine-Engineering Fund (grant no. YG2017MS41).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QF and YW conceived and designed the study. QF, TH, XS, XY, JW, YaL, TN and SG performed the experiments. QF, TH, XS and YuL analyzed the data. QF, TH and YW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved (approval no. GKLW 2017-125) by the Human Investigation Ethical Committee of the International Peace Maternity & Child Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Shanghai, China). The animal experiment protocol was approved by the Committee on the Ethics of Animal Experiments of Shanghai Jiaotong University [approval no. SCXK (hu) 2018-0007].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Zhao F, Qiao Y. Cervical cancer prevention in China: A key to cancer control. Lancet. 2019;393:969–970. doi: 10.1016/S0140-6736(18)32849-6. [DOI] [PubMed] [Google Scholar]
- 3.Brasseur K, Gevry N, Asselin E. Chemoresistance and targeted therapies in ovarian and endometrial cancers. Oncotarget. 2017;8:4008–4042. doi: 10.18632/oncotarget.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu MH, Huang CJ, Liu ST, Liu PY, Ho CL, Huang SM. Physical and functional interactions of human papillomavirus E2 protein with nuclear receptor coactivators. Biochem Biophys Res Commun. 2007;356:523–528. doi: 10.1016/j.bbrc.2007.02.162. [DOI] [PubMed] [Google Scholar]
- 5.Remoue F, Jacobs N, Miot V, Boniver J, Delvenne P. High intraepithelial expression of estrogen and progesterone receptors in the transformation zone of the uterine cervix. Am J Obstet Gynecol. 2003;189:1660–1665. doi: 10.1016/s0002-9378(03)00852-4. [DOI] [PubMed] [Google Scholar]
- 6.Liang J, Shang Y. Estrogen and cancer. Annual Rev Physiol. 2013;75:225–240. doi: 10.1146/annurev-physiol-030212-183708. [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 8.Yang XM, Wu ZM, Huang H, Chu XY, Lou J, Xu LX, Chen YT, Wang LQ, Huang OP. Estrogen receptor 1 mutations in 260 cervical cancer samples from Chinese patients. Oncol Lett. 2019;18:2771–2776. doi: 10.3892/ol.2019.10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sood S, Patel FD, Ghosh S, Arora A, Dhaliwal LK, Srinivasan R. Epigenetic alteration by DNA methylation of ESR1, MYOD1 and hTERT Gene promoters is useful for prediction of response in patients of locally advanced invasive cervical carcinoma treated by chemoradiation. Clin Oncol (R Coll Radiol) 2015;27:720–727. doi: 10.1016/j.clon.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Noël JC, Bucella D, Fayt I, Simonart T, Buxant F, Anaf V, Simon P. Androgen receptor expression in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Int J Gynecol Pathol. 2008;27:437–441. doi: 10.1097/PGP.0b013e318160c599. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, Zhang HJ, Wang LH, Wan XP. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12(155) doi: 10.1186/1476-4598-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv T, Song K, Zhang L, Li W, Chen Y, Diao Y, Yao Q, Liu P. miRNA-34a decreases ovarian cancer cell proliferation and chemoresistance by targeting HDAC1. Biochem Cell Biol. 2018;96:663–671. doi: 10.1139/bcb-2018-0031. [DOI] [PubMed] [Google Scholar]
- 15.He L, Wang HY, Zhang L, Huang L, Li JD, Xiong Y, Zhang MY, Jia WH, Yun JP, Luo RZ, Zheng M. Prognostic significance of low DICER expression regulated by miR-130a in cervical cancer. Cell Death Dis. 2014;5(e1205) doi: 10.1038/cddis.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao W, Zhang Y, Li S, Fan Q, Qiu M, Wang Y, Li Y, Ji X, Yang Y, Sang Z, et al. miR1075p promotes tumor proliferation and invasion by targeting estrogen receptoralpha in endometrial carcinoma. Oncol Rep. 2019;41:1575–1585. doi: 10.3892/or.2018.6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright JD, Matsuo K, Huang Y, Tergas AI, Hou JY, Khoury-Collado F, St Clair CM, Ananth CV, Neugut AI, Hershman DL. Prognostic performance of the 2018 international federation of gynecology and obstetrics cervical cancer staging guidelines. Obstet Gynecol. 2019;134:49–57. doi: 10.1097/AOG.0000000000003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Q, Qiu MT, Zhu Z, Zhou JH, Chen L, Zhou Y, Gu W, Wang LH, Li ZN, Xu Y, et al. Twist induces epithelial-mesenchymal transition in cervical carcinogenesis by regulating the TGF-β/Smad3 signaling pathway. Oncol Rep. 2015;34:1787–1794. doi: 10.3892/or.2015.4143. [DOI] [PubMed] [Google Scholar]
- 19.Bao W, Qiu H, Yang T, Luo X, Zhang H, Wan X. Upregulation of TrkB promotes epithelial-mesenchymal transition and anoikis resistance in endometrial carcinoma. PLoS One. 2013;8(e70616) doi: 10.1371/journal.pone.0070616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Pan Y, Wang R, Zhang F, Chen Y, Lv Q, Long G, Yang K. MicroRNA-130a inhibits cell proliferation, invasion and migration in human breast cancer by targeting the RAB5A. Int J Clin Exp Pathol. 2015;8:384–393. [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng L, Kang Y, Zhang L, Zou W. MiR-133a-5p inhibits androgen receptor (AR)-induced proliferation in prostate cancer cells via targeting FUsed in Sarcoma (FUS) and AR. Cancer Biol Ther. 2020;21:34–42. doi: 10.1080/15384047.2019.1665393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One. 2008;3(e4029) doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(pl1) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical cancer markers: Epigenetics and microRNAs. Lab Med. 2018;49:97–111. doi: 10.1093/labmed/lmx080. [DOI] [PubMed] [Google Scholar]
- 28.Luhn P, Walker J, Schiffman M, Zuna RE, Dunn ST, Gold MA, Smith K, Mathews C, Allen RA, Zhang R, et al. The role of co-factors in the progression from human papillomavirus infection to cervical cancer. Gynecol Oncol. 2013;128:265–270. doi: 10.1016/j.ygyno.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung SH, Wiedmeyer K, Shai A, Korach KS, Lambert PF. Requirement for estrogen receptor alpha in a mouse model for human papillomavirus-associated cervical cancer. Cancer Res. 2008;68:9928–9934. doi: 10.1158/0008-5472.CAN-08-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung SH, Franceschi S, Lambert PF. Estrogen and ERalpha: Culprits in cervical cancer? Trends Endocrinol Metab. 2010;21:504–511. doi: 10.1016/j.tem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Romero R, Garrido-Guerrero E, Rangel-Lopez A, Manuel-Apolinar L, Piña-Sánchez P, Lazos-Ochoa M, Mantilla-Morales A, Bandala C, Salcedo M. The cervical malignant cells display a down regulation of ER-α but retain the ER-β expression. Int J Clin Exp Pathol. 2013;6:1594–1602. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhai Y, Bommer GT, Feng Y, Wiese AB, Fearon ER, Cho KR. Loss of estrogen receptor 1 enhances cervical cancer invasion. The American journal of pathology. 2010;177:884–895. doi: 10.2353/ajpath.2010.091166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deligeoroglou E, Michailidis E, Creatsas G. Oral contraceptives and reproductive system cancer. Annals of the New York Academy of Sciences. 2003;997:199–208. doi: 10.1196/annals.1290.023. [DOI] [PubMed] [Google Scholar]
- 34.Miller N, Bedard YC, Cooter NB, Shaul DL. Histological changes in the genital tract in transsexual women following androgen therapy. Histopathology. 1986;10:661–669. doi: 10.1111/j.1365-2559.1986.tb02520.x. [DOI] [PubMed] [Google Scholar]
- 35.Jiang H, Yu WW, Wang LL, Peng Y. miR-130a acts as a potential diagnostic biomarker and promotes gastric cancer migration, invasion and proliferation by targeting RUNX3. Oncology reports. 2015;34:1153–1161. doi: 10.3892/or.2015.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu SG, Qin XG, Zhao BS, et al. Differential expression of miRNAs in esophageal cancer tissue. Oncology letters. 2013;5:1639–1642. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chi C, Mao M, Shen Z, Chen Y, Chen J, Hou W. HOXD-AS1 Exerts Oncogenic Functions and Promotes Chemoresistance in Cisplatin-Resistant Cervical Cancer Cells. Human gene therapy. 2018;29:1438–1448. doi: 10.1089/hum.2017.256. [DOI] [PubMed] [Google Scholar]
- 38.Hofsjo A, Bohm-Starke N, Bergmark K, Masironi B, Sahlin L. Sex steroid hormone receptor expression in the vaginal wall in cervical cancer survivors after radiotherapy. Acta oncologica. 2019;58:1107–1115. doi: 10.1080/0284186X.2019.1598574. [DOI] [PubMed] [Google Scholar]
- 39.Zhang HD, Jiang LH, Sun DW, Li J, Ji ZL. The role of miR-130a in cancer. Breast cancer. 2017;24:521–527. doi: 10.1007/s12282-017-0776-x. [DOI] [PubMed] [Google Scholar]
- 40.Yin S, Zhang Q, Wang Y, Li S, Hu R. MicroRNA-130a regulated by HPV18 E6 promotes proliferation and invasion of cervical cancer cells by targeting TIMP2. Experimental and therapeutic medicine. 2019;17:2837–2846. doi: 10.3892/etm.2019.7226. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.