Abstract
Soy products contain isoflavones (genistein, daidzein, and glycitein) that display biological effects when ingested by humans and animals, these effects are species, dose and age dependent. Therefore, the content and quality of isoflavones in soybeans is a key to their biological effect. Our objective was to identify loci that underlie isoflavone content in soybean seeds. The study involved 100 recombinant inbred lines (RIL) from the cross of ‘Essex' by ‘Forrest,' two cultivars that contrast for isoflavone content. Isoflavone content of seeds from each RIL was determined by high performance liquid chromatography (HPLC). The distribution of isoflavone content was continuous and unimodal. The heritability estimates on a line mean basis were 79% for daidzein, 22% for genistein, and 88% for glycitein. Isoflavone content of soybean seeds was compared against 150 polymorphic DNA markers in a one-way analysis of variance. Four genomic regions were found to be significantly associated with the isoflavone content of soybean seeds across both locations and years. Molecular linkage group B1 contained a major QTL underlying glycitein content (P = 0.0001, R2 = 50.2%), linkage group N contained a QTL for glycitein (P = 0.0033, R2 = 11.1%) and a QTL for daidzein (P = 0.0023, R2 = 10.3%) and linkage group A1 contained a QTL for daidzein (P = 0.0081, R2 = 9.6%). Selection for these chromosomal regions in a marker assisted selection program will allow for the manipulation of amounts and profiles of isoflavones (genistein, daidzein, and glycitein) content of soybean seeds. In addition, tightly linked markers can be used in map based cloning of genes associated with isoflavone content.
INTRODUCTION
Foods containing bioactive components are receiving increased attention due to their functionality in disease prevention and treatment. Soybean products contain a plethora of bioactive phytochemicals (e.g., isoflavones, saponins, phytic acids, phytosterols, trypsin inhibitors, phenolic acids, peptides,...). Moreover, soybeans contain plant estrogens commonly called phytoestrogens or more specifically isoflavones. The three major groups of isoflavones found in soybeans are genistein, daidzein, and glycitein. The numerous health benefits of soy phytoestrogens appear to be expanding. Soy phytoestrogens are being implicated in a reduced risk of breast and prostate cancer, cardiovascular disease, and osteoporosis (see [1, 2, 3, 4, 5]). Additionally, phytoestrogens have been shown to have positive and negative effects on animal reproduction and growth (see [6, 7]). The biological effects of these isoflavones appear to be species, dose, and age dependent (Knight and Eden [8]).
The amount of isoflavones in soybean seed can vary up to five fold (Eldridge and Kwolek [9]). Isoflavone content and profile can vary by year, environment, and genotype. Genetic markers closely linked to genes controlling isoflavone content may be used to indirectly select for favorable alleles moreefficiently than direct phenotype selection, as has been the case with other traits of agronomic importance (Lande and Thompson [10]).
In the past decade, much effort has been put into the genetic and genomic analysis of soybeans. DNA markers allowed the construction of genetic linkage maps of soybean. Maps from RFLP (Lark et al. [11], Shoemaker and Specht [12]), RAPD (Chang et al. [13]), AFLP (Keim et al. [14]), and microsatellite markers (Cregan et al. [15]) are available. Gene maps have been very useful for the soybean genome analysis and for the detection of quantitative trait loci (QTL). These maps allowed the identification of many economically important soybean genes conditioning quantitative traits loci (see [16, 17, 13, 18, 19, 20, 21]).
A key enzyme (P450) known as Isoflavone synthase involved in the genistein and daidezien biosynthesis pathways was cloned and studied by (Akashi et al. [22]). In soybean, two genes encoding isoflavone synthase were identified, used in expression studies in Arabidopsis and to isolate their homologous genes from other leguminous (Jung et al. [23]).
The purpose of this study was to determine the heritabilities of daidzein, genistein, and glycitein and to identify genetic markers that are linked to loci conditioning variation in isoflavone production by different soybean varieties. This genetic information would then be used in plant breeding to develop a soybean variety that has a high and consistent concentration of a beneficial isoflavone.
MATERIALS AND METHODS
Plant material
The study involved 100 recombinant inbred lines (RIL) from Essex × Forrest; two soybean cultivars that contrast for disease resistance, water deficit tolerance, yield potentials, and isoflavone content (see [24, 25, 13, 26, 27]). Soybean seeds were planted in two different environments in southern Illinois, over two years. In 1996 the RILs were planted at the Agronomic Research Center, Carbondale, IL in Stoy soil, Fine-silty, mixed, mesic, Aquic, Hapludalfs where some drought stress occurred in late July and August. In 1997 the RIL were planted at Myer's farm, Desoto, IL in Camden soil, Fine-silty, mixed, mesic, Typic Hapludalfs. Both locations were free from visible disease problems, had low cyst nematode counts and did not show significant soybean sudden death syndrome (SDS) although infestation by F. solani is likely. In 1997 the RILs were relatively free from abiotic stress and rainfall patterns were normal. Seed were harvested from 4.3 m rows trimmed to 3 m, cleaned and stored at 12% (w/w) moisture content.
Two subpopulations from the 100 E × F recombinant inbred lines were analyzed. A total of 40 RILs were used for the first analysis, 10 g of seeds per RIL harvested one year apart at different locations were analyzed separately. The isoflavone content of each sample was analyzed twice for a total of 4 replications. The second subpopulation involved 60 RILs, two samples per RIL harvested at different location one year a part were combined for one single extraction. Each extraction was analyzed twice for isoflavone content. High performance liquid chromatography analysis with photodiode array detection was performed as described by Wang and Murphy [28].
Isoflavone extraction
Two grams of raw soybean seeds with seed coat were ground, mixed with 2 mL of 0.1 N HCL and 10 mL of acetonitrile, stirred for 2 h at room temperature, and filtered through Whatman No. 42 filter paper. The filtrate was taken to dryness under vacuum and temperature below 30°C.
The dry material was redissolved in 10 mL of 80% methanol and then filtered through a 0.45 μm filter unit (Alltech Associates, Deerfield, IL). Twenty microliters of filtrate was applied in the HPLC analysis.
A linear HPLC gradient was employed: Solvent A was 0.1% glacial acetic acid in H2O, and solvent B was 0.1% acetic acid in acetonitrile; following injection of 20 μL of sample, solvent B was increased from 15% to 35% over 50 min and then held at 35% for 10 min. The solvent flow rate was 1 mL/min. A Waters 991 series photodiode array detector monitored from 200 to 350 nm. UV spectra were recorded, and area responses were integrated by Waters software.
Microsatellite markers
Soybean genomic DNA was extracted and purified using the Qiagen Plant Easy DNA Extraction Kit (Qiagen, Hilden, Germany). The microsatellites primers were labeled by phosphorylating the 5' end with 5 μl [γ-32P] ATP (3000 Ci/mmol) for 30 min at 37°C with 10 units of T4 Kinase (Pharmacia, Piscataway, NJ). Radioactive PCR reactions (Meksem et al. [29]) were performed with genomic DNA from our mapping population (F5:13 recombinant inbred lines that segregate for isoflavone content). The PCR products were separated by electrophoresis on a 5% (w/v) polyacrylamide denaturing gel.
Data analysis
Polymorphic DNA markers were compared against all traits by a one-way analysis of variance (SAS Institute Inc., Cary, NC; Wang et al. [28]). The probability of association of each marker with each trait was determined and a significant association was declared if P < 0.009. MapMaker-EXP 3.0 (Lander et al. [30]) was used to estimate map distances (cM, Haldane units) between linked markers. The log10 of the odd ratio (LOD) for grouping markers was set at 2.0. maximum distance was 30 cM. Trait data and marker map were simultaneously analyzed with Mapmaker/QTL 1.1 (Paterson et al. [31]) using the F2-backcross genetic model for trait segregation (Webb et al. [17]), Hnetkovsky et al. [32], Chang et al. [13]) to identify the approximate position of QTL within intervals. Putative QTL were inferred when the LOD score exceeded 2.5 at some point in each interval. The position of the QTL was inferred from the LOD peaks at individual loci detected by maximum likelihood test at positions every 2 cM between adjacent linked markers.
RESULTS
Distribution and heritability
The frequency distributions of the first subpopulation (n = 40) of recombinant inbred lines for genistein, glycitein, and daidzein content shows no significant departure from normality (P > 0.05).
The frequency distribution of the second subpopulation (n = 60) recombinant inbred lines for daidzein content was peaked and skewed toward Forrest, however this distribution did not significantly depart from normality (P > 0.05). The frequency distribution of genistein was skewed toward Forrest, and therefore the distribution significantly departed from normality. The distribution of glycitein was peaked, not skewed, but significantly departed from normality.
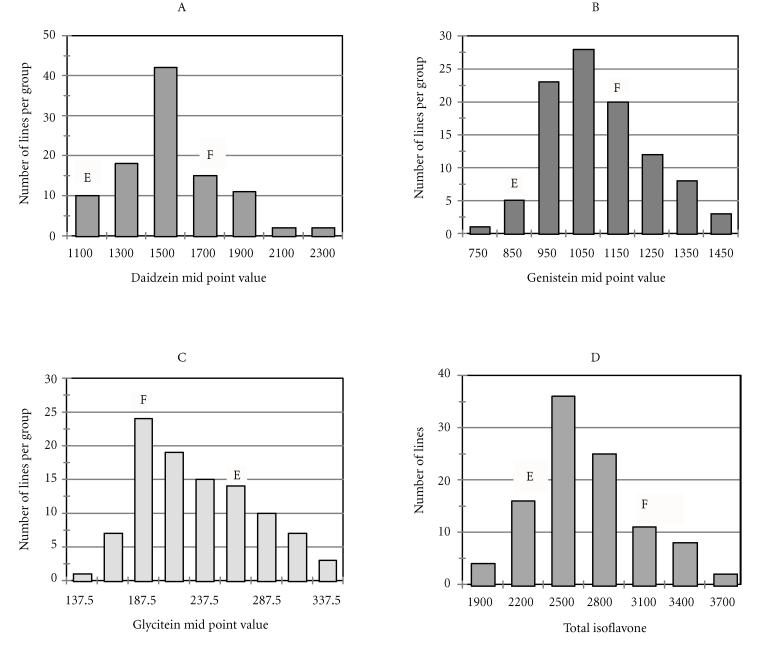
When all 100 RILs were pooled, the frequency distribution of daidzein, genistein, and glycitein all significantly departed from normality (Figure 1). Daidzein showed a peaked distribution that was also skewed toward Essex, genistein had a peaked distribution that was not significantly skewed, glycitein showed a flattened distribution that was not significantly skewed. All distributions were continuous and uni-model. There was transgressive segregation for each isoflavone and total isoflavone content. Recombinant inbred lines showed transgressive segregation for high daidzein and genistein contents and for both low and high glycitein content. The group of RILs with higher total isoflavone content than Forrest were significantly greater than Forrest when considered as a group.
Figure 1.
Frequency distributions of the mean isoflavone content across two locations among 96 recombinant inbred lines from a cross between cultivars Essex and Forrest. The mean isoflavone content for each parent is shown. A, daidzein content. B, genistein content. C, glycitein content. D, Total isoflavone. The isoflavone content is calculated in μg/g of dry weight of soybean seed.
Broad sense line mean heritability estimates from 40 recombinant lines were 79% for daidzein, 22% for genistein and 88% for glycitein. Forrest accumulated more daidzein and genistein but Essex accumulated more glycitein. Heritability was not calculated for the whole population due to the pooling of replicates during isoflavone analysis to reduce cost.
Polymorphism and linkage
A total of 400 microsatellite markers were tested and 133 were found to be polymorphic within the E × F population. The present report summarizes the data from the 133 SSR loci. One hundred and seven markers were mapped to 18 linkage groups encompassing about 2823 cM with an average distance of 26 cM between markers plus 26 markers that were not linked but map to known locations in the soybean genome (see [15]). The recombination distances and orders of markers in linkage groups agree with values reported for the soybean genome of about 3000 cM within 20 linkage groups (see [12, 33, 15, 34, 29]). There was an average of six to seven markers per linkage group. The actual number of markers per linkage group ranged from three on linkage group A1 to 16 on linkage group G. The large number of markers on linkage group G was the result of marker saturation around the Rhg1 gene for the soybean cyst nematode (H. glycines1) resistance. The F5:10 lines were heterogeneous at 12% of loci scored by codominant markers.
GENOMIC REGIONS ASSOCIATED WITH ISOFLAVONE CONTENT
Subpopulation of 40
Using subpopulation of 40 RILs with data from each plot replicate, three chromosomal region on two different molecular linkage groups were found to contain quantitative trait loci (QTL) for seed isoflavone content (Table 1). A region on linkage group B1 identified by the microsatellite marker Satt251 was significantly (P = 0.0001, R2 = 49%) associated with glycitein content (Figure 2). The linked markers Satt197 and Satt415 were also significantly associated with glycitein content. The interval containing the QTL spanning about 10 cM had LOD score of 10.6 and explained about 50% of total variation in glycitein content. The region derived the beneficial allele from Essex.
Table 1.
Chromosomal regions associated with Isoflavone content of the soybean seed in Essex × Forrest recombinant inbred lines.
| Maker | L.Gc | Traitd | P value | R2 | LODa | QTL var.b | Isoflavone means (μg/g) | |
| Essex ± SEM | Forrest ± SEM | |||||||
| 40 RILs | ||||||||
| BARC-Satt251 | B1 | Gly | 0.0001 | 50.0 | 5.5 | 55 | 236 ± 124 | 171 ± 694 |
| BARC-Satt197 | B1 | Gly | 0.0001 | 49.0 | 4.4 | 46 | 234 ± 12 | 165 ± 7 |
| BARC-Sat116 | K | Gly | 0.001 | 27.0 | H | 218 ± 3.0 | 171 ± 4.0 | |
| Gen | 0.005 | 21.0 | H | 1156 ± 4.0 | 918 ± 6.0 | |||
| BARC-Satt337 | K | Daid | 0.008 | 21.3 | H | 1460 ± 81 | 1197 ± 53 | |
| 60 RILs | ||||||||
| BARC-Satt251 | B1 | Gly | 0.0001 | 50.0 | 7.0 | 49 | 244.6 ± 8.1 | 179.5 ± 5.7 |
| BARC-Satt197 | B1 | Gly | 0.0001 | 35.4 | 5.0 | 37 | 239.8 ± 9.8 | 184.7 ± 8.1 |
| BARC-Satt302 | H | Gly | 0.003 | 19.5 | 2.0 | 19 | 236 ± 12.3 | 194 ± 7.1 |
| ALL RILs | ||||||||
| BARC-Satt251 | B1 | Gly | 0.0001 | 50.2 | 10.6 | 51 | 242 ± 6.6 | 175 ± 4.3 |
| BARC-Satt197 | B1 | Gly | 0.0001 | 41.2 | 8.3 | 41 | 238 ± 7.3 | 176 ± 5.7 |
| BARC-Satt237 | N | Gly | 0.0033 | 11.1 | 2.3 | 20 | 211 ± 6.6 | 182 ± 5.7 |
| BARC-Satt080 | N | Daid | 0.0023 | 10.3 | 3.2 | 26 | 1230 ± 29 | 1381 ± 40 |
| BARC-Satt276 | A1 | Daid | 0.0081 | 9.6 | 2.7 | 27 | 1258 ± 42 | 1280 ± 35 |
aLOD indicate how much more probable the data are to have arisen assuming the presence of a locus than assuming its absence; LOD threshold = 2.0; H QTL LOD score was less than 2.0.
bAmount of variability in the trait explained by the marker loci based on MapMakerQTL1.1.
cL.G: Linkage group.
dGly: Glycitein; Daid: Daidzein; Gen: Genistein.
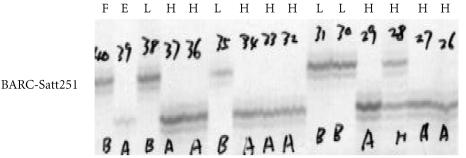
Figure 2.
Autoradiograph showing PCR amplification of genomic DNA from the low glycitein Forrest (F), the high glycitein parent (E) and the F5:13 selected RIL segregating for glycitein soybean seed content with the BARC-Sat251 primers. The labeled PCR products were electrophoresed on an 5% polyacrylamide denaturing gel (H: High content, L: Low content of glycitein seed content.
A region on linkage group K identified by Sat_116 was significantly associated (P = 0.001 and 0.005, R2 = 27% and 21%) with glycitein and genistein, respectively. The linked markers Satt326 was also significantly associated with glycitein and genistein content. The region derived the beneficial allele for both glycitein and genistein seed content from Essex. Another region on linkage group K identified by Satt337 was significantly associated (P = 0.008, R2 = 21%) with daidzein content. The region derived the beneficial allele from Essex.
Subpopulation of 60 RILs
Using the subpopulation of 60 RILs, with data from pooled plot replicates, two chromosomal regions on two different molecular linkage groups were found to contain QTL for seed isoflavone content (Table 1). A region on linkage group B1 identified by the microsatellite marker Satt251 was significantly (P = 0.0001, R2 = 50%) associated with glycitein content. The linked markers Satt197 and Satt415 were also significantly associated with glycitein content. The interval containing the QTL spanned about 10 cM and had a peak LOD score of 7.0 and explained about 49% of total variation in glycitein content. The region derived the beneficial allele from Essex. A region on linkage group H identified by Satt302 was significantly associated (P = 0.003, R2 = 19.5%) with glycitein content. There were no other linked polymorphic markers that were significant associated with the trait. The region derived the beneficial allele from Essex.
The pooled population of 100 RILs
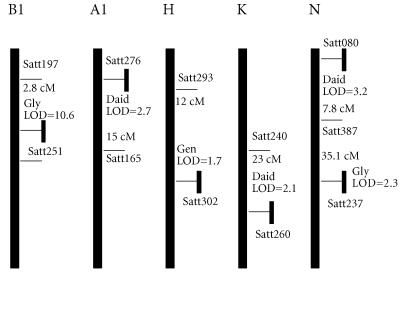
Using the whole population of 100 RILs, with data from pooled plot replicates, four chromosomal regions on three different molecular linkage groups were found to contain QTL for seed isoflavone content (Table 1 and Figure 3). A region on linkage group B1 identified previously in the subpopulations of 40 and 60 RILs by the microsatellite marker Satt251 was again significantly (P = 0.0001, R2 = 49%) associated with glycitein content. The linked markers Satt197 and Satt415 were also significantly associated with glycitein content. The interval containing the QTL spanned about 10 cM and had a peaked LOD score of 10.6 and explained about 51% of total variation in glycitein content. The region derived the beneficial allele from Essex.
Figure 3.
Location of microsatellite markers and QTL that condition phytoestrogen content. END indicates the likely position of the telomere on that linkage group, the disjunct bar represents the rest of the linkage group. Marker names and distances and peak LOD score for the interval are given. LOD scores are from single locus analyses of additive gene effects using Mapmaker/QTL 1.1. Genetic distances were from the recombinant inbred line function of Mapmaker/EXP 3.0.
A region on linkage group N identified by Satt080 was significantly associated (P = 0.002, R2 = 10.3%) with daidzein content. The interval containing the QTL spanned about 7.8 cM between Satt080 and Satt387, had a peak LOD score of 3.2 and explained bout 26% of total variation in soybean seed daidzein content. The region derived the beneficial allele from Forrest. Another region on linkage group N identified by Satt237 was significantly associated (P = 0.0003, R2 = 11.1%) with glycitein content. The interval had a peak LOD score of 2.3 and explained about 20% of the total variation in soybean seed glycitein content. The region derived the beneficial allele from Essex. A region on linkage group A1 identified by Satt276 was significantly associated (P = 0.008, R2 = 9.6%) with soybean seed daidzein content. The interval (15 cM) had a peak LOD score of 2.7 and explained about 27% of total variation in seed glycitein content. The region derived the beneficial allele from Forrest.
DISCUSSION
The phenotypic characterization of isoflavone content among the RILs showed that distributions were typical of polygenic traits with little evidence for genes of large effect on the trait. There were transgressive segregants for all isoflavones and total content suggesting that it will be possible to advance isoflavone content above 5–6 mg/g by breeding. The existence of these RILs showed it is possible to combine high glycitein (Essex trait) with high daidzein and genistein (Forrest traits) in a single cultivar. The heritabilities of daidzein, genistein, and glycitein are estimated in the subpopulation of 40 RILs but not the larger populations since replication were maintained separately in the extraction and analysis of isoflavones. However, the heritabilities are in agreement with previous reports and reflect a significant G × E interaction (Wang and Murphy [28]). Molecular markers provide effective breeding tools for selecting traits with moderate heritabilities (Paterson et al. [31]).
Seven different chromosomal regions on five different linkage groups were found to contain loci that influenced isoflavone content in soybean seeds across two environments in two consecutive years. Three of the seven regions were found to control soybean seed glycitein content only, three controlled seed daidzein content and one controlled both seed genistein and glycitein content. Only one of the seven regions controlling seed isoflavones content was detected in both sub populations and the pooled population. This region, located between BARC-Satt251 and BARC-Satt197 on linkage group B1 of the soybean map had a consistently strong effect on soybean seed glycitein content and could be a candidate for positional gene isolation.
Three QTL for glycitein content were detected in this population suggesting glycitein content of soybean is controlled by a few major genes. In contrast, daidzein and genistein may be under the control of a larger number of polygenes that would be below the resolving power of this experiment (Paterson et al. [31]). The detection of QTL with large effect for glycitein content and fewer QTL with smaller effects for genestein and daidzein content of soybean may reflect the environmental stability of glycitein content compared to daidzein and genistein contents (Wang and Murphy, 1993) or the number of loci controlling the traits. Further experiments that reduce genotype by environment interaction (G × E) or larger opulations are needed to resolve these hypotheses.
Verification of QTL is important, due to the expectation of common errors in their assignment. The QTL for glycitein content on linkage group B1 that appeared in all subsamples of the study population is likely to be real. However, the QTL presence and effectiveness in new environments, near isogeneic lines and cultivars of diverse genetic background remain to be determined (Meksem et al. [29], Prabhu et al. [35]).
There were no QTL that were associated with total isoflavone content suggesting that the cultivars do not contrast for loci that control overall pathway flux. However, one of the loci controlling the isoflavone content of the soybean seed (on linkage group K) was in coupling with a region previously found to be associated with high soybean yield (Njiti et al. [26]). While the loci are unlikely to be closely linked this association suggests that increased isoflavones contents of soybean seed can be achieved without compromising yield.
In conclusion, the identification of loci controlling the accumulation of specific soybean isoflavones suggests that both the isoflavone content and profiles of soybean can be manipulated. The ability to manipulate both profile and total isoflavone content is important because the effect of each isoflavone may be species and dose dependent. Additionally, specific isoflavone profiles may confer different results depending on the target population, so reducing unwanted isoflavones while enhancing beneficial isoflavones could be a key-breeding target. Manipulation of isoflavone contents and profiles will result in the creation of special purpose value added soybeans. Future research needs to focus on the production of a cultivar that consistently produces 5–6 mg/g of total isoflavone, with a white hilum and non-GMO herbicide resistance for the international soy protein isolate market.
ABBREVIATIONS
E × F Essex × Forrest
LG linkage group;
QTL quantitative trait loci;
HPLChigh performance liquid chromatography
AFLP amplified fragments length polymorphism;
RFLP restriction fragments length polymorphism;
RIL recombinant inbred lines;
SDS sudden death syndrome.
Acknowledgments
ACKNOWLEDGEMENT
The authors acknowledge Dr. Michael Schmidt, Dr. Patricia Murphy and all the student workers and graduate students in the geonomic laboratory. Research funded in part by C-FAR
Contributor Information
K. Meksem, Email: Ga4082@siu.edu.
V. N Njiti, Email: Ga4082@siu.edu.
W. J Banz, Email: Tw3a@siu.edu.
M. J Iqbal, Email: Ga4082@siu.edu.
My. M Kassem, Email: Ga4082@siu.edu.
D. L Hyten, Email: Ga4082@siu.edu.
J. Yuang, Email: Ga4082@siu.edu.
T. A Winters, Email: Tw3a@siu.edu.
D. A Lightfoot, Email: Ga4082@siu.edu.
References
- Banz WJ, Williams MP, Lightfoot DA, Winters TA. The effects of soy protein and soy phytoestrogens on symptoms associated with cardiovascular disease in rats. J Med Food. 1999. In press. [DOI] [PubMed]
- Cassidy A, Bingham S, Setchell K. Biological effects of a diet of soy protein rich in isoflavones on the menstrual cycle of premenopausal women. Am J Clin Nutr. 1994;60:333–340. doi: 10.1093/ajcn/60.3.333. [DOI] [PubMed] [Google Scholar]
- Anderson JW, Johnstone BM, Cook-Newell MA. Meta-analysis of the effects of soy protein intake on serum lipids. N Engl J Med. 1995;333:276–282. doi: 10.1056/NEJM199508033330502. [DOI] [PubMed] [Google Scholar]
- Messina M, Barnes S. The role of soy products in reducing risk of cancer. J Natl Cancer Inst. 1991;17:541–6. doi: 10.1093/jnci/83.8.541. [DOI] [PubMed] [Google Scholar]
- Anthony MS, Clarkson TB, Hughes CL, Jr, Morgan TM, Burke GL. Soybean isoflavones improve cardiovascular risk factors without affecting the reproductive system of peripubertal rhesus monkeys. J Nutr. 1996;126:43–50. doi: 10.1093/jn/126.1.43. [DOI] [PubMed] [Google Scholar]
- Greer-Baney JA, Banz WJ, Lightfoot DA, Winters TA. Dietary soy protein and soy isoflavones: A histological examination of reproductive tissues in rats. J Med Food. 1999. In press. [DOI] [PubMed]
- Adams NR. Detection of the effects of phytoestrogens on sheep and cattle. J Anim Sci. 1995;73:1509–1515. doi: 10.2527/1995.7351509x. [DOI] [PubMed] [Google Scholar]
- Knight DC, Eden JA. A review of the clinical effects of phytoestrogens. Obsted Gynecol. 1996;87:897–904. [PubMed] [Google Scholar]
- Eldridge CE, Kwolek WF. Soybean isoflavones: Effects of the environment and variety on the composition. J Agric Food Chem. 1983;31:394–396. doi: 10.1021/jf00116a052. [DOI] [PubMed] [Google Scholar]
- Lande R, Thompson R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics. 1990;124:743–756. doi: 10.1093/genetics/124.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark KG, Weisemann JM, Matthews BF, Palmer R, Chase K, Macalma T. A genetic map of soybean (Glycine max L.) using an intraspecific cross of two cultivars: ‘Minsoy' and ‘Noir1'. Theor Appl Genet. 1993;86:901–906. doi: 10.1007/BF00211039. [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Specht JE. Integration of the soybean molecular and classical genetic linkage groups. Crop Science. 1995;35:436–446. [Google Scholar]
- Chang SJC, Doubler TW, Kilo V, et al. Two additional loci underlying durable field resistance to soybean sudden death syndrome (SDS) Crop Science. 1996;36:1624–1628. [Google Scholar]
- Keim P, Shoemaker RC, Palmer RG. A high density soybean genetic map based on AFLP. Crop Science. 1996;36:786–792. [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, et al. An intergrated genetic linkage map of the soybean genome. Crop Science. 1999;39:1464–1490. [Google Scholar]
- Mansur LM, Orf JH, Chase K, Jarvick T, Cregan PB, Lark KG. Genetic mapping of agronomic traits using recombinant inbred lines. Crop Science. 1996;36:1327–1336. [Google Scholar]
- Webb DM, Baltazar BM, Rao-Arelli AP, et al. Genetic mapping of soybean cyst-nematode race-3 resistance loci in soybean PI 437.654. Theor Appl Genet. 1995;91:574–581. doi: 10.1007/BF00223282. [DOI] [PubMed] [Google Scholar]
- Lee SH, Bailey MA, Mian MAR, et al. RFLP loci associated with soybean seed protein and oil content across populations and locations. Theor Appl Genet. 1996;93:649–657. doi: 10.1007/BF00224058. [DOI] [PubMed] [Google Scholar]
- Vierling RA, Faghihi J, Ferris VR, Ferris JM. Association of RFLP markers conferring broad-based resistance to the soybean cyst nematode (Heterodera glycines) Theor Appl Genet. 1996;92:83–86. doi: 10.1007/BF00222955. [DOI] [PubMed] [Google Scholar]
- Concibido V, Boutin S, Denny R, Hautea R, Orf J, Young ND. Genome mapping of a soybean cyst nematode resistance genes in ‘Peking', PI 91763, and PI 88788 using DNA markers. Crop Science. 1997;37:258–264. [Google Scholar]
- Mian MAR, Bailey MA, Ashley DA, et al. Molecular markers associated with water use efficiency and leaf ash in soybean. Crop Science. 1996;36:1252–1257. [Google Scholar]
- Akashi T, Aoki T, Ayabe Si. Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 1999;121:821–828. doi: 10.1104/pp.121.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau SM, et al. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol. 2000;18:208–212. doi: 10.1038/72671. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Camper HM. Registration of Essex soybeans. Crop Science. 1973;13:495. [Google Scholar]
- Hartwig EE, Epps JM. Registration of forrest soybeans. Crop Science. 1973;13:287. [Google Scholar]
- Njiti VN, Schmidt CA, Schmidt ME, Lightfoot DA. Mapping loci underlying soybean yield in Illinois. Soybean Genet Newsl. 1997;24:136–138. [Google Scholar]
- Cho Y, Lightfoot DA, Wood AJ. Survey of trigonelline concentrations in salt-stressed leaves of cultivated glycine max. Phytochemistry. 1999;52:1235–1238. [Google Scholar]
- Wang H, Murphy PA. Isoflavones composition of American and Japanese soybeans in Iowa: Effects of variety, crop year and location. J Agric Chem. 1994;42:1674–1677. [Google Scholar]
- Meksem K, Doubler TW, Chancharoenchai K, et al. Clustering among loci underlying soybean resistance to Fusarium solani, SDS and SCN in near-isogenic lines. Theor Appl Genet. 1999;99:1131–1142. [Google Scholar]
- Lander E, Green P, Abrahamson J, et al. MAPMAKER: AN interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Paterson A, Lander E, Lincoln S, Hewitt J, Paterson S, Tanksley S. Resolution of quantitative traits into mendelian factors using a complete RFLP linkage map. Nature. 1988;335:721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- Hnetkovsky N, Chang SC, Doubler TW, Gibson PT, Lightfoot DA. Genetic mapping of loci underlying field resistance to sudden death syndrome. Crop Science. 1996;36:392–400. [Google Scholar]
- Chang SJC, Doubler TW, Kilo V, et al. Association of field resistance to soybean sudden death syndrome (SDS) and cyst nematode (SCN) Crop Science. 1997;37:965–971. [Google Scholar]
- Mudge J, Cregan PB, Kenworthy JP, Orf JH, Young WD. Two microsatellite markers that flank the major soybean cyst nematode resistance locus. Crop Science. 1997;37:1611–1615. [Google Scholar]
- Prabhu RR, Njiti VN, Bell-Johnson B, et al. Selecting soybean cultivars for dual resistance to soybean cyst nematode and sudden death syndrome using two DNA markers. Crop Science. 1999;39:982–987. [Google Scholar]