Abstract
In vivo reflectance confocal microscopy (RCM) enables clinicians to examine lesions’ morphological and cytological information in epidermal and dermal layers while reducing the need for biopsies. As RCM is being adopted more widely, the workflow is expanding from real-time diagnosis at the bedside to include a capture, store, and forward model with image interpretation and diagnosis occurring offsite, similar to radiology. As the patient may no longer be present at the time of image interpretation, quality assurance is key during image acquisition. Herein, we introduce a quality assurance process by means of automatically quantifying diagnostically uninformative areas within the lesional area by using RCM and coregistered dermoscopy images together. We trained and validated a pixel-level segmentation model on 117 RCM mosaics collected by international collaborators. The model delineates diagnostically uninformative areas with 82% sensitivity and 93% specificity. We further tested the model on a separate set of 372 coregistered RCM-dermoscopic image pairs and illustrate how the results of the RCM-only model can be improved via a multimodal (RCM + dermoscopy) approach, which can help quantify the uninformative regions within the lesional area. Our data suggest that machine learning–based automatic quantification offers a feasible objective quality control measure for RCM imaging.
INTRODUCTION
Reflectance confocal microscopy (RCM) offers dermatologists a noninvasive alternative to traditional invasive histopathology for cutaneous diagnostics (Gill et al., 2014; Kelsey et al., 2019; Rajadhyaksha et al., 2017). Providing cellular resolution (0.5–1 μm) on par with histology, in vivo RCM is 92–100% sensitive and 85–97% specific for basal cell carcinomas and 88–92% sensitive and 70–84% specific for melanomas (Guitera et al., 2012; Nori et al., 2004). RCM combined with dermoscopy increases diagnostic specificity by about two times compared with dermoscopy alone and correspondingly reduces the benign-to-malignant biopsy ratio by about two times (Alarcon et al., 2014; Borsari et al., 2016; Pellacani et al., 2016, 2014). RCM is advancing into clinical practice for noninvasively guiding diagnosis and treatment of cancer (Rajadhyaksha et al., 2017). In the clinical setting, layers of quality assurance, both at the time of capture to ensure a quality image set and at the time of interpretation to relay the likelihood of representative sampling, is imperative. We propose that machine learning (ML)-based techniques could be incorporated into the RCM standard operating procedure to add an objective quality control measure as part of a larger quality assurance protocol.
As in vivo RCM is being adopted more widely, the workflow is expanding from real-time diagnosis at the bedside to include a capture, store, and forward model with image interpretation and diagnosis occurring offsite when the patient is no longer in the imaging suite, similar to that in radiology. In this setting, additional imaging because of lack of quality assurance requires the patient to return to the clinic. Therefore, real-time quantitative assessment of image quality has a key role in image capture. The image capture capabilities of clinical RCM devices, including mosaics and image stacks, have been described previously (Scope et al., 2019). Mosaics are captured at several layers in the epidermis and superficial dermis as part of a standard image set, followed by acquisition of image stacks. RCM mosaics typically include the entire lesional area and at least 1–2 mm of surrounding nonlesional skin whenever possible. Thus, each mosaic usually has both lesional and nonlesional regions, with the lesional area containing the diagnostically relevant information and the nonlesional area being diagnostically noncontributory. Inadequate image quality occurs when (i) features used to locate the appropriate skin level for mosaics are not recognized by the technician and (ii) artifacts occur because of motion (patient or device) or obscuring structures (bubbles, particulates, or hair) (Curchin et al., 2011, Gill et al., 2019a, 2019b). Artifacts (e.g., air bubbles, particulates, underillumination, and pixel saturation) and areas where the window is not in contact with skin because of lesional surface contour or voids within wrinkles (Gill et al., 2019a), collectively referred to herein as uninformative areas, represent the main causes of quality degradation in RCM images. Such areas almost always exist in RCM images; although some are clearly visible, others may be subtle, but both can significantly impact the interpretation of the image. Objective aids that could quantify these artifacts at the time of capture could potentially decrease the need for reimaging or subsequent patient visits required because of an inadequate image set. We hypothesize that ML-based techniques could quantify automatically the percentage of uninformative areas in the entire RCM mosaic and, by displaying the location of the artifact on the registered dermoscopic image, allow the user to localize the uninformative areas to within or outside of the lesion and envisage the potential effects of the uninformative areas on diagnosis.
Quantitative and objective analysis of RCM images via ML has been investigated in several studies (Bozkurt et al., 2017a,b, 2018; Ghanta et al., 2017; Hames et al., 2016; Kose et al., 2020; Kurugol et al., 2015). As most of the diagnostic information within the RCM images lies in the architectural and morphological patterns (Bozkurt et al., 2018; Gill et al., 2014), texture analysis–based methods have been our focus. Initial efforts concentrated on automated delineation of skin layers within RCM stacks (Bozkurt et al., 2017b; Ghanta et al., 2017; Hames et al., 2016; Kurugol et al., 2015) using hand-crafted mathematical relations between the pixels (intensity profile [Kurugol et al., 2015], textons [Julesz, 1981], log-Gabor [Field, 1985], or wavelet transform features [Laine and Fan, 1993; Randen and Husoy, 1999]). Similar textural analysis methods were also used by Koller et al. (2011) to detect malignancy in RCM images. These initial studies relied on applying a classification model over a set of textural features that are hand-picked by a human expert. As such, these approaches were limited to the interpretation of human readers and not necessarily optimal for tasks such as classification, detection, or segmentation. Deep neural networks or deep learning alleviate this problem by learning how task-relevant, highly distinguishable, and potentially unique mathematical relations within signal samples (e.g., image pixels) can be extracted to accomplish the target task (e.g., classification or segmentation). With recent advances in computational resources (e.g., graphical processing units and cloud computing) and availability of large digital datasets (e.g., microscopy images, scanned pathology slides, and radiology images), a specific version of deep neural networks called convolutional neural networks have been shown to perform on par with humans at image recognition and segmentation applications (Esteva et al., 2017; Lecun et al., 2015; Litjens et al., 2017; Han et al., 2018).
Initial application of convolutional neural networks on RCM images was shown by Bozkurt et al. (2017a and 2017b) for segmentation of skin layers and by Bozkurt et al. (2018) and Kose et al. (2020) for a pixel-level (semantic) segmentation of diagnostically significant morphological patterns at the dermal-epidermal junction (DEJ) in RCM mosaics of melanocytic lesions (Gill et al., 2014). Similar to analysis of histology slides, clinicians typically examine morphological patterns in RCM mosaics starting from low magnification (low resolution and large field of view), followed by close inspection of suspicious areas with higher magnification (higher resolution and smaller field of view). Likewise, Multiscale Encoder Decoder Network (MED-Net) (Bozkurt et al., 2018; Kose et al., 2020) pixel-wise labels the RCM mosaics by analyzing them at multiple scales, mimicking the clinicians’ way of examining lesions and incorporating the segmentation results at consecutive scales. RCM imaging artifacts and topographical voids without window contact (uninformative areas), such as morphological patterns, have a distinct textural appearance (Gill et al., 2019a) and are suitable for ML-based segmentation. Moreover, they typically obscure the textural and morphological details in the acquired images, resulting in decreased diagnostic value of the data. Hence, identifying and quantifying these areas can serve as a form of quality control. In the studies by Bozkurt et al. (2018) and Kose et al. (2020), we trained and tested a segmentation model on 56 mosaics and achieved 79% sensitivity and 97% specificity in identifying uninformative areas in RCM mosaics captured at the DEJ of melanocytic neoplasms.
In this article, we explore applying ML-based techniques to automatically identify and quantify uninformative areas within RCM mosaics captured at the DEJ of atypical pigmented lesions. Our first aim was to train our MED-Net model (Bozkurt et al., 2018; Kose et al., 2020) on a dataset of 117 RCM mosaics from seven clinical sites, which were pixel-wise labeled by two RCM experts (CAF and MG) in consensus to identify uninformative areas and calculate our model’s sensitivity and specificity on this larger dataset. Our second aim was to utilize MED-Net segmentation results to better understand how frequently diagnostically uninformative areas in the RCM mosaic fall within the lesional area (the area of diagnostic interest) and whether MED-Net can be utilized to detect image quality trends across sites. For this experiment, we tested MED-Net on a dataset collected by multiple imaging technicians at five clinics consisting of 372 coregistered RCM DEJ mosaic and dermoscopic image pairs. If a lesion did not occupy the entire field of view, the dermoscopic and mosaic (referred to hereafter as entire mosaic area) images contained areas with lesion (referred to hereafter as lesional area) and areas that are lesion-free and comprised of surrounding skin. The same readers manually labeled, by consensus, the lesional area (the portion of the image that contains the lesion only) on the dermoscopic images for this part of the study. As the images are coregistered, the lesional area segmented in the dermoscopic image essentially delineates the lesional area in the paired RCM mosaic, allowing for calculation of uninformative areas within the lesion. Finally, we describe how MED-Net could be implemented to detect images that require operator attention or reimaging and provide an objective assessment of image quality to both the imaging technician and the clinician interpreting the RCM images.
RESULTS
We trained and tested the MED-Net model on a set of 117 RCM mosaics with 5-fold stratified cross-validation, where one fifth of the data is reserved for testing and the remaining is used for training and validation for five different trials. MED-Net–detected diagnostically uninformative areas with 82 ± 9.8% sensitivity, 93 ± 2.4% specificity, and a Dice coefficient of 0.78 ± 5.9. Figure 1 illustrates comparative inference results obtained using the MED-Net model versus the expert segmentation. Areas overlaid in green correspond to uninformative areas over which the algorithm and expert consensus concur. Yellow areas were labeled as uninformative by MED-Net only and magenta areas were labeled as uninformative by experts only. As illustrated, areas with too little information for interpretation (underillumination or saturation) or which are physically obscured because of the presence of extensive particulates, bubbles, and/or hairs were deemed uninformative by the model. These initial findings suggest that MED-Net is capable of detecting uninformative areas in RCM mosaics. The model that achieved the best sensitivity and specificity was used in the second phase of the study.
Figure 1. Mosaics with color overlays highlighting diagnostically uninformative areas.
(a–c) Top row: The experts’ manual segmentation and the MED-Net segmentation results are color-coded in the overlays as follows: green, areas where MED-Net and experts concur; yellow, areas labeled as diagnostically uninformative by MED-Net only; and magenta, areas labeled as diagnostically uninformative by experts only. Bottom row: Respective mosaics without the overlays are shown. MED-Net, Multiscale Encoder Decoder Network.
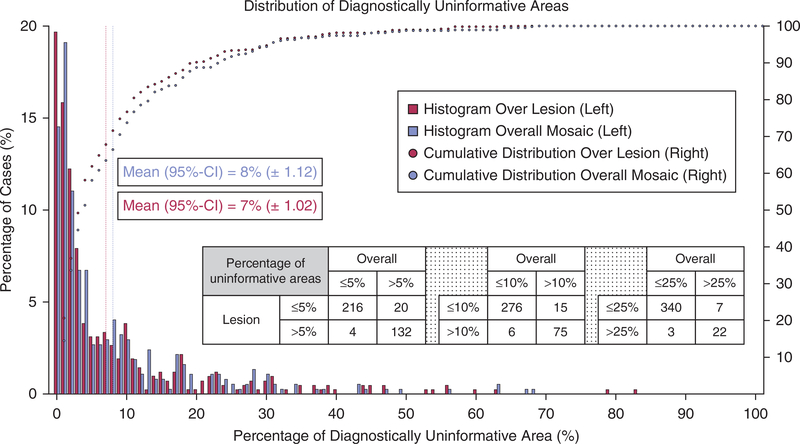
To understand the frequency with which uninformative areas occur in routine imaging with the future goal of identifying a quantitative metric that could be used to identify RCM images of low quality, we applied our MED-Net model over a separate, larger dataset of 372 samples. Each RCM image in the dataset has a coregistered dermoscopy counterpart that was manually labeled in consensus (by CAF and MG) for lesional areas. In this dataset, we labeled only the dermoscopy images because our aim was to illustrate how the distribution of MED-Net–detected uninformative areas varied between the entire mosaic versus the lesional area within each mosaic. The results presented in Figure 2 show, on average, that uninformative areas covered 8% (95% confidence interval, ±1.12) of the entire mosaicked area and 7% (95% confidence interval, ±1.02) of the lesional area within this dataset. Conducting a paired Wilcoxon signed rank test resulted in a P-value < 0.001, showing that the difference between the two distributions is significant. As shown in the table in Figure 2, the MED-Net–estimated percentage of uninformative areas within the entire mosaic and the lesional area are 94–97% concordant.
Figure 2. Distribution of MED-Net–detected uninformative areas over the lesional area versus the entire mosaic.
Bar chart shows histogram of lesional areas (red) and the entire RCM mosaic area (blue) with a given percentage of uninformative areas. On average, uninformative areas cover 7% (95% CI, ±1.02) of the lesional area and 8% (95% CI, ±1.12) of the entire mosaic area (y-axis on the left side). Dotted curves show the percentage of dataset (y-axis on right) containing less than × percentage of uninformative area (x-axis). The red dotted line shows analysis over the lesional area and blue dotted line over the entire mosaic area. The graph shows that 80% of the lesional areas have <10% diagnostically uninformative area, whereas 75% of the mosaics have <10% diagnostically uninformative area. The inset table shows the concordance between the entire mosaic measure and only lesional area measure in terms of the number of cases. CI, confidence interval; MED-Net, Multiscale Encoder Decoder Network; RCM, reflectance confocal microscopy.
As detailed in the Materials and Methods section, the dataset contains images from four different institutes and five clinical sites (two separate clinical sites from one institute). Analyzing the data coming from each site individually, we found that the mean (standard deviation) percentage of uninformative areas within the entire mosaic ranged between 4% and 17% (6–23%) across sites and the mean (standard deviation) percentage of uninformative areas in the lesional area ranged from 3% to 15% (4–18%) across sites. Only one out of five clinics showed statistics deviating from the overall distribution given in Figure 2, with a mean (standard deviation) percentage of uninformative areas of 17% (23%) and 15% (18%) within the entire mosaic and the lesional area, respectively. Repeating the analysis without this outlier, we found a mean (standard deviation) percentage of uninformative areas of 7% (8%) and 6% (8%) within the entire mosaic and the lesional area, respectively, across the four remaining sites. If we quantize the results presented in Figure 2 to show regions of percent uninformative area within the entire mosaic as 0–5, 5–10, 10–15, 15–25, and 25–100, then 220, 62, 28, 33, and 29 of the mosaics fall into the given regions, respectively. Conversely, 236, 55, 28, 28, and 25 mosaics fall into the given regions of percent uninformative area within the lesional area, respectively.
In Figure 3, we present exemplar mosaics that contain a high percentage of uninformative areas within the lesion highlighted by our MED-Net algorithm. In Figure 3a, the algorithm highlights a bubble (within the index-matching oil used on the skin) that obscures the lesion both in the dermoscopic and RCM images (Gill et al., 2019a). In Figure 3b, the dermoscopic image appears artifact-free; however, the algorithm detects a shadow obscuring the lesion, which is caused by a bubble in the objective lens immersion gel (Gill et al., 2019a). In Figure 3c and d, the dermoscopic image again appears artifact-free, but the algorithm detects a large uninformative area over the entire lesion caused by insufficient contact between the RCM window and portions of the skin surface. In all of these cases, the uninformative areas represent more than 20% (average, 42.8%) of the lesional area. By contrast, some cases harbored significant uninformative areas, but only a small portion of the uninformative area fell within the lesional area and, therefore, adequate lesional sampling was likely still achieved (Figure 4).
Figure 3. Coregistered dermoscopic and DEJ mosaic image pairs with color overlays showing the extent of uninformative areas obscuring the mostly diagnostic areas.
Uninformative areas cover (a) 31.3%, (b) 20.8%, (c) 62.7%, and (d) 56.4%, of the lesional area. MED-Net segmentation results are color-overlaid on RCM mosaics (row 3) and coregistered dermoscopic images (row 2). Raw images without overlays are shown in rows 1 and 4. In (a) and (b), the algorithm detected the bubbles within the oil and gel, respectively, that obscure the lesion. In (c) and (d), the algorithm detected the black areas indicating where the imaging window was not in contact with the skin because of (c) the curved surface of the body at that site or (d) irregular lesional surface contour. DEJ, dermal-epidermal junction; MED-Net, Multiscale Encoder Decoder Network; RCM, reflectance confocal microscopy.
Figure 4. Coregistered dermoscopic and DEJ mosaic image pairs with color overlays showing the extent of uninformative areas obscuring the perilesional skin.
Uninformative areas cover (a) 27.3%, (b) 32.0%, (c) 13.0%, and (d) 34% of the entire mosaic with limited lesional involvement. MED-Net segmentation results are color-overlaid on RCM mosaics (row 3) and coregistered dermoscopic images (row 2). Raw images without overlays are shown in rows 1 and 4. In (a), poor skin-to-window contact on the left is due to curved body contour. In (b) and (d), the papular lesion contour results in similar poor window contact in perilesional skin. In (c), alternating areas of saturation and underillumination (black) are due to patient motion. In (c), the lesional area is free of uninformative areas and reimaging is unnecessary. In (a), (b), and (d), reimaging is unnecessary, but collecting additional mosaics to capture needed anatomic skin levels in all portions of the lesion is recommended. DEJ, dermal-epidermal junction; MED-Net, Multiscale Encoder Decoder Network; RCM, reflectance confocal microscopy.
DISCUSSION
Quality standards are critical in diagnostic medicine, and ensuring adequate representative sampling is necessary to achieve an accurate diagnosis. Minimizing the amount of diagnostically uninformative areas therefore is critical in RCM imaging. During the first aim of this study, MED-Net continued to identify uninformative areas with very good accuracy when we increased our dataset from 56 mosaics (79% sensitivity and 97% specificity) (Bozkurt et al., 2018; Kose et al., 2020) to 117 mosaics (82% sensitivity and 93% specificity) in this study. These findings support our hypothesis that ML-based techniques could quantify automatically the percentage of uninformative areas in RCM mosaics. Further, as a lesion may not fill the entire mosaicked image (mean ± standard deviation, 74 ± 24% of the entire mosaic is lesional area in our dataset) and the area of clinical and/or dermoscopic concern may be focal, the location of uninformative areas is as important a quality measure as the overall amount. In certain lesions, as little as 10% uninformative area, if located within a focus of diagnostic concern, could preclude accurate diagnosis. Therefore, the overlay of the algorithm-detected uninformative areas on the coregistered dermoscopic image is critical in determining whether an obscured area may be an area of diagnostic importance. Providing an automated objective assessment of image quality as well as the location of uninformative areas at the time of imaging could prompt an imaging technician to reimage a lesion while the patient is still in the imaging suite, avoiding patient recalls, if the uninformative areas are obscuring a significant portion of the lesion. The information also provides the clinician interpreting the RCM image set an objective image quality measure that can be referenced and incorporated into the report as a confidence measure regarding representative sampling. Also, as part of the quality assurance program, an automated measurement of image quality provides a useful objective data point for tracking a technician’s performance both during a single encounter and over time to determine if remediation is necessary.
In the second aim of our study, we applied our MED-Net algorithm to 372 DEJ mosaics of pigmented lesions from five different clinical sites as a pilot study to explore how commonly diagnostically uninformative areas are encountered across the entire mosaic and within the lesional area. In this dataset, approximately 90% of the mosaics contained <25% uninformative area and approximately 75% contained <10% uninformative area. However, the amount of uninformative area that falls within the lesional area is more important to quantify. Our results show that for a given percentage of uninformative area within the overall mosaic, the amount of uninformative area within the lesional area only is overestimated. Results in the concordance table in Figure 2 show that using the two modalities together increases the specificity of uninformative area quantification within diagnostically relevant regions (e.g., lesional area) by 13–24%. Similarly, as shown in Figure 2, among the 343 mosaics that contained <25%, 282 mosaics that contained <10%, and 220 mosaics that contained <5% uninformative areas in the entire mosaic, 24 had >25% uninformative area, 6 had >10% uninformative area, and 4 had >5% uninformative area within the lesional area. The results show that using two modalities together increases the sensitivity of detection of uninformative areas within the lesion by only 1–2%. These findings suggest that combining the visual overlay of the diagnostically uninformative areas with the percentage of diagnostically uninformative area over the entire mosaic may provide sufficient objective information and avoid the cumbersome task of requiring the clinician to delineate the lesional area on the coregistered dermoscopic image. In the future, a separate algorithm that delineates lesional area on dermoscopic images could be incorporated to completely automate calculating the percentage of uninformative area within the lesion.
The results of our pilot study also suggest that incorporating our MED-Net algorithm could identify technicians that require additional training or remediation and provide an objective assessment tool that could decrease the number of patient recalls because of inadequate image quality. Four of the five clinical sites showed very similar percentages of diagnostically uninformative areas (on average, 4–9% in the entire mosaic and 3–8% in the lesional area, whereas the outlier site had on average 17% in the entire mosaic and 15% in the lesional area). Interestingly, this same site was flagged during the original study by the clinical coordinator, and the imaging technician was remediated. Because in clinical practice a site may have only one imaging technician, a larger prospective study would be helpful to delineate standards for imaging quality assessment guidelines. For our dataset, hypothetically setting the threshold for reimaging between 5% and 25% uninformative area within the lesion (rather than entire mosaic) would lead to a 13–24% decrease in patient recalls for reimaging (Figure 2). Furthermore, imaging quality guidelines may help technicians in day-to-day decision making and also allow a clinic to determine whether an imaging technician has received sufficient training or requires remediation, as there would be an accepted standard for average uninformative area.
In summary, our results confirm that ML-based image assessment can accurately detect diagnostically uninformative areas and suggest that incorporating ML-based image quality assessment may be useful both for establishing quality standards and as an objective quality assessment tool, enabling quantitative quality control at two key steps in the workflow process, (i) at the time of capture, to reduce patient recall for reimaging because of poor image quality and (ii) at the time of image interpretation, to assist in assessing the likelihood of nonrepresentative sampling. As in all cases, the clinician interpreting the images must still apply a qualitative assessment of whether sampling is representative or whether the small amount of uninformative area is present in the portion of the lesion with the dermoscopic feature of concern (i.e., a dermoscopic island suggestive of melanoma arising in a nevus). The limitations of our study include small sample size and its retrospective nature. The datasets used in our experiments were collected for diagnostic reading studies and were captured on a narrower range of diagnoses than a typical clinical practice. A large multicenter prospective study on a wider subset of lesions is needed to confirm our findings. Finally, based on our findings, we hypothesize that ML-based image quality assessment may achieve similar results in other in vivo imaging modalities where the diagnostic information lies in the morphological (or textural) patterns.
MATERIALS AND METHODS
At six clinical sites, five from a National Institutes of Health-funded clinical study conducted in the U.S. (Memorial Sloan Kettering Cancer Center, New York, New York; Memorial Sloan Kettering Cancer Center, Hauppauge, New York; University of Rochester, Rochester, New York; Loma Linda University, Loma Linda, California; and Skin and Cancer Associates, Plantation, Florida) and one from Italy (University of Modena and Reggio Emilia), 489 (117 + 372) mosaics were collected. Cases submitted in the U.S. were captured using a skin-to-window (tissue ring) tissue coupling device (VivaScope 1500, Caliber Imaging and Diagnostics, Andover, MA), a through-the-window macroscopic imager that captures a dermoscopic image registered to the RCM mosaics. For these cases, the surface of the skin is found by imaging up and down within the skin layers during real-time live RCM imaging, and the starting imaging location (zero level) is set at the stratum corneum, or skin surface. Through a semi-automated imaging protocol assistant, four 6 mm × 6 mm mosaics were captured at predefined depths approximately corresponding to the stratum granulosum, suprabasal layer, DEJ, and papillary dermis. These image sets were available in their entirety; however, only the dermoscopic image and the mosaic captured at the DEJ were used in this study. Only mosaics of the DEJ were provided by the Italian group; we did not have corresponding dermoscopic images, and therefore, this portion of the dataset was only used for MED-Net model training. All 372 dermoscopic images used in the study were coregistered to the RCM images. The images were manually labeled by our expert readers (CAF and MG) for lesional areas. The labeling was conducted in Seg3D (Seg3D, 2016) using the paintbrush tool, which enables the experts to interact with images at pixel-level detail. The experts examined the cases together and annotated the images in consensus.
MED-Net (Bozkurt et al., 2018; Kose et al., 2020) is a semantic segmentation network that can analyze input images at different scales and output a pixel-wise segmentation map. It is composed of multiple subnetworks nested in a hierarchical manner. In an N-level network, the first subnetwork takes the (2N−1)-times subsampled version of the image as input and predicts a low-resolution segmentation. The next subnetwork that follows takes this output and the (2N−2)-times subsampled version of the image to generate a higher resolution segmentation. The process is repeated until the final subnetwork (Level N) that processes the image at its input resolution. In this way, each subnetwork analyzes the image at a different scale and passes its results to the next subnetwork to increase the detail level of the segmentation gradually. This multiscale analysis capability is crucial for the detection of the uninformative areas within RCM mosaics, as they appear in different sizes and shapes.
The MED-Net model was originally developed for the segmentation of diagnostically significant patterns (meshwork, ring, nested, and aspecific) including nonlesional and diagnostically uninformative areas in RCM mosaics at the DEJ of melanocytic lesions. Full technical details as well as the training parameters can be found in our previous publications (Bozkurt et al., 2018; Kose et al., 2020). The model was trained and tested on 117 mosaics (5-fold cross-validation), which were pixel-wise labeled for the previously mentioned six classes by two RCM experts (CAF and MG). The model was trained end-to-end using the Keras library (Chollet 2015) on a single graphical processing unit with 12-gigabyte memory (NVIDIA P100, TitanX, and TitanV for different instances, all from Nvidia, Santa Clara, CA). The training takes between 12 and 24 hours depending on the graphical processing unit used.
The MED-Net segmentation model was then applied to a separate dataset of 372 samples. The mosaics in this test set are distinct from the training set. The lesional area in the RCM mosaics was found by projecting the manually labeled lesional area from the coregistered dermoscopic image over the RCM mosaics. Using this information, we calculated the diagnostically uninformative areas that fell within the lesional area in the RCM images. Inference of the trained model to annotate the uninformative areas over a 6 mm × 6 mm mosaic takes approximately 2 minutes using a graphical processing unit. The statistical analysis between the manually annotated dermoscopy images and the MED-Net segmentations was implemented in MAT-LAB programming environment. The MED-Net framework is in the process of being integrated into the RCM imaging software for real-time clinical testing and validation.
Data availability statement
The deep learning model that was trained to generate the presented results and the code will be made available at https://github.com/kkose/RCM-Artifact upon the publication of the technical paper.
ACKNOWLEDGMENTS
The authors would like to thank Caterina Longo and Giovanni Pellacani for providing RCM mosaics for the training and testing of the MED-Net semantic segmentation model. This project was supported by National Institutes of Health grant R01CA199673 from NCI. This project was also supported in part by MSKCC’s Cancer Center core support National Institutes of Health grant P30CA008748 from NCI. The image capture for U.S. clinical sites was supported by National Institutes of Health grant 5R44CA058054-06. The authors would like to thank NVIDIA Corporation for their donation of the TitanV graphical processing unit used for this project. This graphical processing unit is provided through their GPU Grant Program.
Abbreviations:
- DEJ
dermal-epidermal junction
- MED-Net
Multiscale Encoder Decoder Network
- ML
machine learning
- RCM
reflectance confocal microscopy
Footnotes
CONFLICT OF INTEREST
Christi Alessi Fox is a shareholder and employee of Caliber I.D. Inc, the manufacturer of the VivaScope Systems. Milind Rajadhyaksha is a shareholder of Caliber I.D. Inc, the manufacturer of the VivaScope Systems. Melissa Gill is a consulting investigator on an investigator–initiated study sponsored by DBV Technologies. The remaining authors state no conflict of interest.
REFERENCES
- Alarcon I, Carrera C, Palou J, Alos L, Malvehy J, Puig S. Impact of in vivo reflectance confocal microscopy on the number needed to treat melanoma in doubtful lesions. Br J Dermatol 2014;170:802–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsari S, Pampena R, Lallas A, Kyrgidis A, Moscarella E, Benati E, et al. Clinical indications for use of reflectance confocal microscopy for skin cancer diagnosis. JAMA Dermatol 2016;152:1093–8. [DOI] [PubMed] [Google Scholar]
- Bozkurt A, Kose K, Alessi-Fox C, Dy JG, Brooks DH, Rajadhyaksha M. Unsupervised delineation of stratum corneum using reflectance confocal microscopy and spectral clustering. Skin Res Technol 2017a;23:176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt A, Kose K, Alessi-Fox C, Gill M, Dy J, Brooks D, et al. A Multiresolution Convolutional Neural Network with partial label training for annotating reflectance confocal microscopy images of skin. In: Frangi A, Schnabel J, Davatzikos C, Alberola-López C, Fichtinger G, editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Cham, Switzerland: Springer; 2018. p. 292–9. [Google Scholar]
- Bozkurt A, Kose K, Coll-Font J, Alessi-Fox C, Brooks DH, Dy JG, et al. Delineation of skin strata in reflectance confocal microscopy images using recurrent convolutional networks with Toeplitz attention. arXiv 2017b;arXiv:1712.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet F Keras, https://keras.io; 2015. (accessed 1 August 2018).
- CIBC. Seg3D: volumetric image segmentation and visualization. Scientific computing and Imaging Institute (SCI). http://www.seg3d.org. 2016. (accessed 15 July 2019). [Google Scholar]
- Curchin CES, Wurm EMT, Lambie DLj, Longo C, Pellacani G, Soyer HP. First experiences using reflectance confocal microscopy on equivocal skin lesions in Queensland. AustralasJ Dermatol 2011;52:89–97. [DOI] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:1 1 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field DJ. Relations between the statistics of natural images and the response properties of cortical cells. J Opt Soc Am A 1985;4:2379–94. [DOI] [PubMed] [Google Scholar]
- Ghanta S, Jordan MI, Kose K, Brooks DH, Rajadhyaksha M, DyJG. A marked poisson process driven latent shape model for 3D segmentation of reflectance confocal microscopy image stacks of human skin. IEEE Trans Image Process 2017;26:172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M, Alessi-Fox C, Kose K. Artifacts and landmarks: pearls and pitfalls for in vivo reflectance confocal microscopy of the skin using the tissue-coupled device. Dermatol Online J 2019a;25. [PubMed] [Google Scholar]
- Gill M, Grant-Kels JM, Fox CA. Absence of lesional features on reflectance confocal microscopy: quality control steps to avoid false-negative results. J Am Acad Dermatol 2019b;81:e71–3. [DOI] [PubMed] [Google Scholar]
- Gill M, Longo C, Farnetani F, Cesinaro AM, González S, Pellacani G. Non-invasive in vivo dermatopathology: identification of reflectance confocal microscopic correlates to specific histological features seen in melanocytic neoplasms. J Eur Acad Dermatol Venereol 2014;28:1069–78. [DOI] [PubMed] [Google Scholar]
- Guitera P, Menzies SW, Longo C, Cesinaro AM, Scolyer RA, Pellacani G. In vivo confocal microscopy for diagnosis of melanoma and basal cell carcinoma using a two-step method: analysis of 710 consecutive clinically equivocal cases. J Invest Dermatol 2012;132:2386–94. [DOI] [PubMed] [Google Scholar]
- Hames SC, Ardigò M, Soyer HP, Bradley AP, Prow TW. Automated segmentation of skin strata in reflectance confocal microscopy depth stacks. PLOS ONE 2016;11:e0153208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol 2018;138:1529–38. [DOI] [PubMed] [Google Scholar]
- Textons Julesz B., the elements of texture perception, and their interactions. Nature 1981;290:91–7. [DOI] [PubMed] [Google Scholar]
- Kelsey A, Grant-Kels JM, Rabinovitz H, Oliviero M, Scope A. Reflectance confocal microscopy can help the dermatopathologist in the diagnosis of challenging skin lesions. Am J Derm Pathol 2019;41:128–34. [DOI] [PubMed] [Google Scholar]
- Koller S, Wiltgen M, Ahlgrimm-Siess V, Weger W, Hofmann-Wellenhof R, Richtig E, et al. In vivo reflectance confocal microscopy: automated diagnostic image analysis of melanocytic skin tumours. J Eur Acad Dermatol Venereol 2011;25:554–8. [DOI] [PubMed] [Google Scholar]
- Kose K, Bozkurt A, Alessi-Fox C, Gill M, Longo C, Pellacani G, et al. Segmentation of cellular patterns in confocal images of melanocytic lesions in vivo via a Multiscale Encoder-Decoder Network (MED-Net). arXiv 2020;arXiv:2001.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurugol S, Kose K, Park B, Dy JG, Brooks DH, Rajadhyaksha M. Automated delineation of dermal-epidermal junction in reflectance confocal microscopy image stacks of human skin. J Invest Dermatol 2015;135: 710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A, Fan J. Texture classification by wavelet packet signatures. IEEE T Pattern Anal Machine Intell 1993;15:1186–91. [Google Scholar]
- Lecun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- Nori S, Rius-Díaz F, Cuevas J, Goldgeier M, Jaen P, Torres A, et al. Sensitivity and specificity of reflectance-mode confocal microscopy for in vivo diagnosis of basal cell carcinoma: a multicenter study. J Am Acad Dermatol 2004;51:923–30. [DOI] [PubMed] [Google Scholar]
- Pellacani G, Pepe P, Casari A, Longo C. Reflectance confocal microscopy as a second-level examination in skin oncology improves diagnostic accuracy and saves unnecessary excisions: a longitudinal prospective study. Br J Dermatol 2014;171:1044–51. [DOI] [PubMed] [Google Scholar]
- Pellacani G, Witkowski A, Cesinaro AM, Losi A, Colombo GL, Campagna A, et al. Cost-benefit of reflectance confocal microscopy in the diagnostic performance of melanoma. J Eur Acad Dermatol Venereol 2016;30: 413–9. [DOI] [PubMed] [Google Scholar]
- Rajadhyaksha M, Marghoob A, Rossi A, Halpern AC, Nehal KS. Reflectance confocal microscopy of skin in vivo: from bench to bedside. Lasers Surg Med 2017;49:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randen T, Husoy JH. Filtering for texture classification: a comparative study. IEEE T Pattern Anal Machine Intell 1999;21:291–310. [Google Scholar]
- Scope A, Dusza SW, Pellacani G, Gill M, Gonzalez S, Marchetti MA, et al. Accuracy of tele-consultation on management decisions of lesions suspect for melanoma using reflectance confocal microscopy as a stand-alone diagnostic tool. J Eur Acad Dermatol Venereol 2019;33:439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The deep learning model that was trained to generate the presented results and the code will be made available at https://github.com/kkose/RCM-Artifact upon the publication of the technical paper.