Obashi et al. review the regulation of molecular diffusion by dendritic spine structures and discuss its role in synaptic functions and plasticity.
Abstract
Spines are tiny nanoscale protrusions from dendrites of neurons. In the cortex and hippocampus, most of the excitatory postsynaptic sites reside in spines. The bulbous spine head is connected to the dendritic shaft by a thin membranous neck. Because the neck is narrow, spine heads are thought to function as biochemically independent signaling compartments. Thus, dynamic changes in the composition, distribution, mobility, conformations, and signaling properties of molecules contained within spines can account for much of the molecular basis of postsynaptic function and regulation. A major factor in controlling these changes is the diffusional properties of proteins within this small compartment. Advances in measurement techniques using fluorescence microscopy now make it possible to measure molecular diffusion within single dendritic spines directly. Here, we review the regulatory mechanisms of diffusion in spines by local intra-spine architecture and discuss their implications for neuronal signaling and synaptic plasticity.
Introduction
Neurons communicate with each other through synapses that organize to create functional circuits. Most excitatory synapses in the central nervous system are formed on dendritic spines, tiny protrusions that extend from dendrites (Bourne and Harris, 2008; Fig. 1 a). The spine typically has a head of 200–1,000-nm diameter, which is connected to the dendritic shaft via a neck of 100–200-nm width (Arellano et al., 2007; Fig. 1 b). The head contains postsynaptic density (PSD) proteins, the actin cytoskeleton, membrane structures, and organelles (Sheng and Hoogenraad, 2007; Fig. 1 c). The molecular composition of spine heads is different from that of the shaft. Because of its characteristic morphology, spines are thought to function as biochemically independent compartments by limiting molecular movement between the spine head and the rest of the dendrite (Adrian et al., 2014; Tønnesen and Nägerl, 2016). Clarifying this regulation is key to understanding how this unitary site of synaptic transmission is controlled. This is particularly crucial to our understanding about how changes in the postsynaptic site lead to synaptic plasticity.
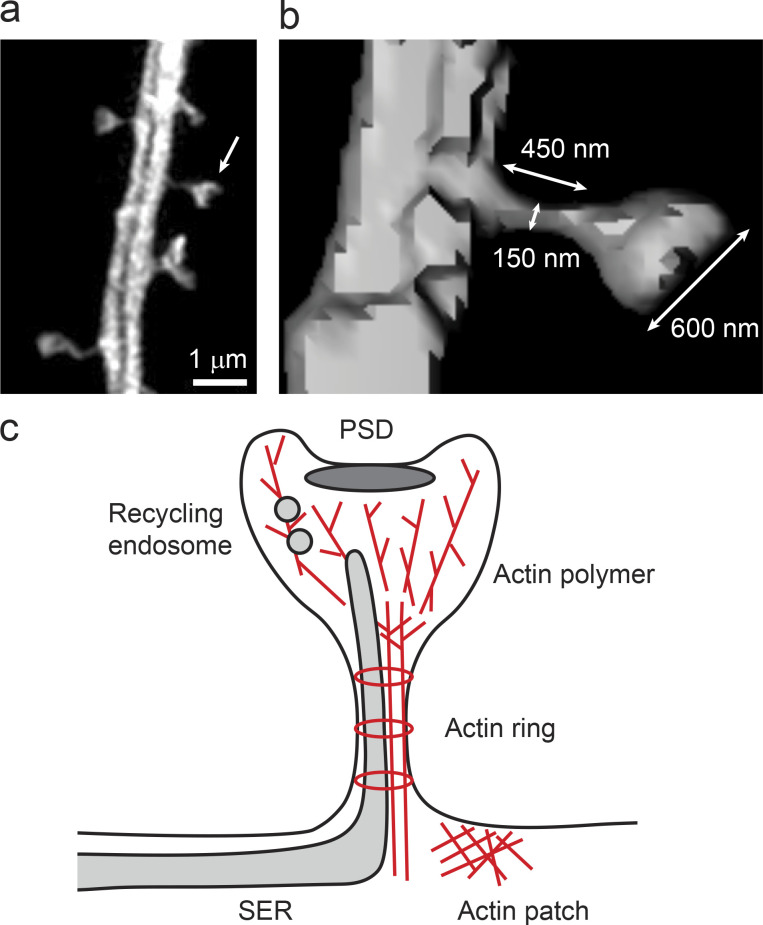
Figure 1.
The shape and internal architecture of dendritic spines. (a) A super-resolution SIM image of a hippocampal neuron dendrite expressing GFP. (b) A surface image of a spine (arrow in a) reconstructed from a SIM image. (c) Schematic representation of a spine containing the PSD, actin cytoskeleton, recycling endosome, and SER.
The control of spine architecture is critical at excitatory synapses in the brain (Alvarez and Sabatini, 2007; Forrest et al., 2018). Excitatory synapses exhibit synaptic plasticity, which changes the strength of synaptic transmission through mechanisms at both pre- and postsynaptic sides (Citri and Malenka, 2008). This process is generally thought to be a basis for changes in neural circuits controlled by experiences—i.e., learning and memory (Humeau and Choquet, 2019; Magee and Grienberger, 2020). Here, the size and shape of spines are strongly correlated with the strength of synaptic transmission (Kasai et al., 2010). Spine volume is proportional to PSD area (Harris and Stevens, 1989) and the number of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate–type glutamate receptors (AMPARs; Nusser et al., 1998; Matsuzaki et al., 2001). Recently, combinational analysis of electrophysiology and correlative light and EM (CLEM) revealed the linear relationship between PSD area and synaptic strength (Holler et al., 2021). Also, longer spine necks attenuate somatic potentials to a greater degree (Araya et al., 2006). Thus, structural and functional plasticity of spines is tightly regulated. Specifically, when synaptic transmission is strengthened (e.g., long-term potentiation [LTP]), spines grow (Matsuzaki et al., 2004). In turn, when synaptic transmission weakens (e.g., long-term depression), spines shrink (Zhou et al., 2004; Oh et al., 2013).
While many molecules involved in the plasticity of spine synapses have been identified (Sala and Segal, 2014), their mechanisms and regulations can only be discovered by monitoring the regulated changes in the composition and signaling properties of these factors within the confined space of the spine’s cytoplasm. To this end, the development of local photolysis of caged-glutamate played an important role (Matsuzaki et al., 2001). This method made it possible to induce structural plasticity locally at a single spine. Spine enlargement is induced by uncaging of caged-glutamate in the absence of Mg2+ or with postsynaptic depolarization in the presence of Mg2+ to activate N-methyl-D-aspartate–type glutamate receptors (NMDARs; Matsuzaki et al., 2004). Conversely, spine shrinkage is induced by low-frequency uncaging of caged-glutamate in the absence of Mg2+ or with postsynaptic depolarization (Oh et al., 2013). Shrinkage can also be induced by glutamate uncaging temporally coupled with back propagation action potential and uncaging of caged–γ-aminobutyric acid (GABA; Hayama et al., 2013). Glutamate uncaging–induced structural plasticity has also been seen to occur in vivo (Noguchi et al., 2019).
These methods have vastly improved our understanding of the molecular mechanisms of structural plasticity (Nishiyama and Yasuda, 2015). In particular, stimulus-dependent increases in spine size (structural LTP [sLTP]), which are thought to be associated with functional LTP, have been studied extensively as a model of LTP (Nakahata and Yasuda, 2018; Fig. 2). Strong synaptic input causes an influx of Ca2+ through NMDARs that activates Ca2+/calmodulin-dependent protein kinase II (CaMKII) and the downstream signaling cascades. This modification of signaling cascades can affect cytoskeletal organization and membrane trafficking, which are responsible for two subsequent cellular events. First, spine morphology is modulated through cytoskeletal changes (Borovac et al., 2018). Second, synaptic transmission is enhanced by increased AMPAR insertion into the plasma membrane and movement to the PSD (Huganir and Nicoll, 2013). However, the molecular mechanisms that link these two phenomena are not fully understood (Herring and Nicoll, 2016). As sLTP progresses, the molecular composition within spines changes (Bosch et al., 2014; Meyer et al., 2014). Specifically, immediately after sLTP induction, actin-related molecules such as cofilin and actin-related protein 2/3 (Arp2/3) complex accumulate within a stimulated spine. On the other hand, scaffold proteins such as PSD-95 slowly accumulate over tens of minutes. SynGAP, which is localized at the PSD through interaction with PSD-95, escapes from spines immediately after stimulation and contributes to the expression of sLTP (Araki et al., 2015). These changes in molecular compositions immediately after stimulation may be due not only to molecule-specific binding but also to the physical regulation of diffusion (Obashi et al., 2019).
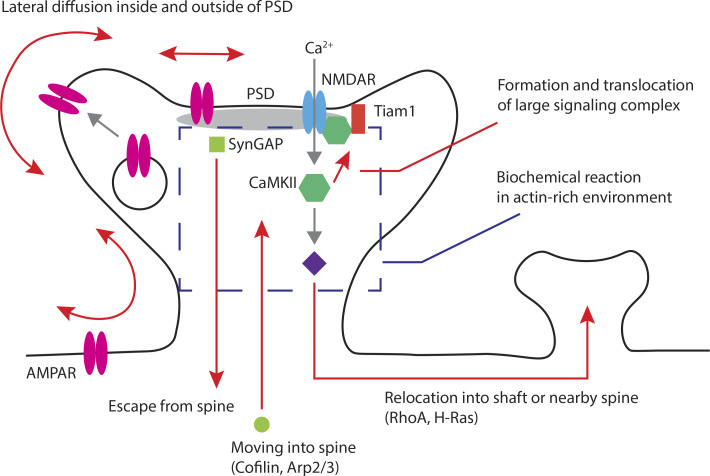
Figure 2.
Molecular motion important in sLTP. Strong synaptic input causes an influx of Ca2+ through NMDARs that activates CaMKII and the downstream signaling cascades. This modification of signaling cascades can affect cytoskeletal organization and membrane trafficking, which regulate spine morphology. Spine morphology affects the molecular exchange between the spine head and the dendritic shaft and lateral diffusion of membrane proteins including AMPARs. Regulation of molecular movements through the spine neck affects the molecular composition within spines. This change affects signal propagation into nearby spines. For example, cofilin and Arp2/3 complex accumulate within spines. SynGAP and activated RhoA escape from spines. Reorganization of the actin cytoskeleton affects movement of large molecules and the formation of a large signaling complex containing CaMKII and Tiam1. Also, the structure of the PSD affects membrane protein diffusion and alters the synaptic trafficking of AMPARs.
In addition to molecular localization, fluorescence lifetime imaging of FRET-based biosensors has made it possible to measure spatiotemporal changes in the activity of signaling molecules involved in sLTP (Yasuda, 2012). These studies have demonstrated a critical relationship between the time that a molecule spends within a spine and the rate of signal inactivation. This relationship determines whether an activated signaling molecule is confined within a single spine or escapes from the spine and interacts with effectors present in the adjacent dendritic shaft or nearby spines (Yasuda, 2017). The signal propagation into nearby spines is most likely related to heterosynaptic plasticity, where activated synapses influence neighbor synapses within the same dendritic segments (Oh et al., 2015; Colgan et al., 2018; Chater and Goda, 2021). Thus, diffusion is a central feature of the regulation of spine structural plasticity. However, because the size of spines is small relative to the spatial resolution of diffraction-limited fluorescence microscopy and measuring methods are limited, elucidation of the mechanism regulating diffusion within spines has been challenging.
To address this gap in understanding, researchers advanced fluorescence microscopy techniques, which enabled us to measure changes in the nanoscale localization, diffusion, and signal activities inside spines. These studies allow us to directly understand how spine structures physically limit molecular diffusion and reveal fundamental mechanisms that control the localization and biochemical signal transduction pathways in neurons. Here, we summarize recent findings that have revealed physical barriers within spines using super-resolution microscopy (Sigal et al., 2018; Table 1) and molecular dynamics measurements (Fig. 3 and Table 2), and we discuss how these barriers serve as a fundamental feature controlling neuronal signaling and synaptic plasticity.
Table 1. List of super-resolution microscopy techniques.
| Technique | Principle | Resolution | Comments | Applications in spines and synapses | |
|---|---|---|---|---|---|
| Lateral (XY) | Axial (Z) | ||||
| CLSM | 250 nm | 500 nm | |||
| TPLSM | Two-photon excitation | 350 nm | 700 nm | Deeper tissue penetration; adaptive optics further improve | Helmchen and Denk, 2005; Ji, 2017 |
| STED | Stimulated emission (Vicidomini et al., 2018) | 20–70 nm | 500 nm | 3-D STED increases axial resolution; chronic in vivo imaging is possible | Nägerl et al., 2008; Berning et al., 2012; Pfeiffer et al., 2018 |
| SIM | Moiré effect with structured illumination (Wu and Shroff, 2018) | 100 nm | 250 nm | No need for special fluorophores; limited resolution improvement | Kashiwagi et al., 2019; Li et al., 2020 |
| SMLM (PALM, STORM) | Photoactivation, photoconversion (Baddeley and Bewersdorf, 2018) | 10–30 nm | 30–60 nm | High spatial resolution; temporal resolution is relatively worse | Dani et al., 2010; Tang et al., 2016 |
| ExM | Physical expansion of sample (Wassie et al., 2019) | 4–20-fold improvement | 4–20-fold improvement | Capable of combining with other imaging techniques, only for fixed samples | Gao et al., 2019; Sarkar et al., 2020 Preprint |
CLSM, confocal laser scanning microscopy; SMLM, single-molecule localization microscopy; STORM, stochastic optical reconstruction microscopy; TPLSM, two-photon laser scanning microscopy.
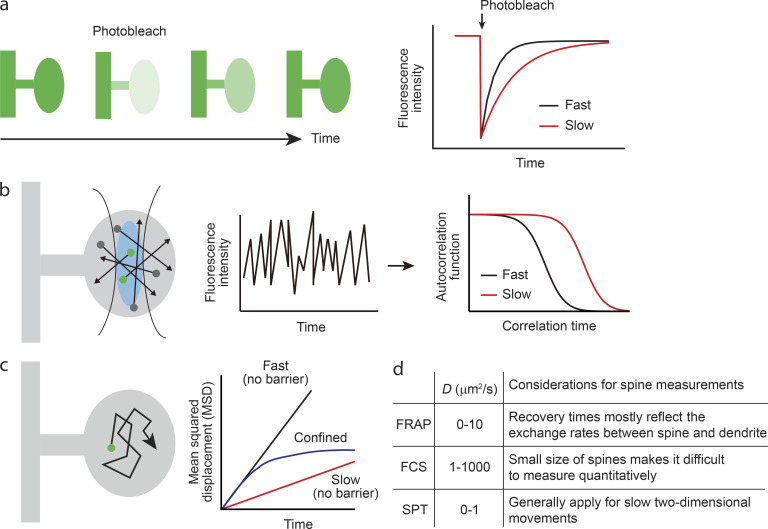
Figure 3.
Imaging techniques to measure diffusion inside dendritic spines. (a) FRAP. Fluorescence intensity change is measured after photobleaching fluorescent molecules in a spine head. Fluorescence recovery rate is mostly determined by the exchange rate between spine and dendrite. (b) FCS. The fluctuation of fluorescence intensity from the detection volume fixed inside a spine head (blue region in left panel) is recorded as a function of time (center panel). Since the fluorescence intensity fluctuates as the molecules enter and leave the fixed detection volume, the characteristics of intensity fluctuation essentially contain information about local diffusion speed. To estimate the diffusion coefficient, the autocorrelation function of fluorescence intensity fluctuation is calculated (right panel). (c) SPT. In SPT, molecular trajectory is directly measured with video microscopy. To analyze the speed and pattern of molecular motion, mean squared displacement (MSD) is calculated. For diffusion without barrier, MSD increases linearly against time. On the other hand, for diffusion within the compartment, MSD converges to a certain value, which corresponds to compartment size. (d) Comparison of three measurement techniques.
Table 2. List of fluorescence molecular dynamic measurement techniques.
| Technique | Principle | Applications in spines and synapses |
|---|---|---|
| FRAP | Fluorescent molecules in a small region are photobleached, and subsequent movement of surrounding nonbleached fluorescent molecules into the photobleached area is monitored (Lippincott-Schwartz et al., 2018). | Svoboda et al., 1996; Bloodgood and Sabatini, 2005 |
| FCS | Fluctuation of fluorescence intensity from the detection volume fixed at a specific position is recorded, and a temporal correlation is analyzed (Elson, 2011). | Chen et al., 2015; Obashi et al., 2019 |
| RICS | Spatial correlation is analyzed from raster-scanned images (Digman and Gratton, 2011). | Obashi et al., 2019 |
| SPT | The movement of a single particle is tracked using time-lapse imaging, and a trajectory is made and analyzed. To detect single particles, the density of fluorescence particles should be kept low (Shen et al., 2017). | Borgdorff and Choquet, 2002; Varela et al., 2016 |
| SPT-PALM | Only a small number of photoactivatable fluorescent proteins in the field of view are activated and tracked until they are bleached (Manley et al., 2010). | Frost et al., 2010b; Nair et al., 2013 |
Spine structures and diffusion
The complex physical structures of spines can impact the diffusion of molecules inside the spine cytoplasm and between spines and their parental dendritic shafts (Fig. 1 c and Fig. 2). For example, consider the diffusional translocation of molecules between the PSD and dendritic shaft. For cytoplasmic proteins, because a spine is connected to the dendritic shaft through a narrow neck, proteins must pass through the neck by diffusion or slow active transport. A spine neck functions as a diffusion barrier because of its narrow width (Svoboda et al., 1996). Molecular complexes with actin filaments and related proteins, such as synaptopodin and ankyrin-G, that maintain this characteristic neck morphology may also affect diffusion. Furthermore, the cytoplasm within spines is likely to be not homogeneous but organized with multiple nanoscale domains with different biophysical properties (Frost et al., 2010a; MacGillavry and Hoogenraad, 2015). Thus, these locally dense cytoskeletal and membranous structures can limit the molecular path of diffusion within a spine by specific binding interactions or nonspecific local steric effects. These factors will change the residence time of proteins within spines.
Besides cytosolic proteins, membrane protein diffusion can be regulated by structures on and near the plasma membrane. For example, the cortical cytoskeleton affects the movement of membrane proteins (Kusumi et al., 2012). Furthermore, specialized membrane domains with a high density of membrane-associated structures, such as synaptic contact sites, accumulate many relatively immobile molecules and limit membrane protein diffusion (Trimble and Grinstein, 2015). Lastly, spines are not simply spherical. Boundaries between the spine shaft and neck—and also the spine head and neck—can contain high curvatures. Also, large spine heads contain a concave surface (Kashiwagi et al., 2019). Thus, local concavities, undulations, and convexities may affect the possible path a molecule can take (Simon et al., 2014; Klaus et al., 2016). From all these factors, the shape and internal architecture of spines can have strong effects on diffusion for both cytosolic and membranous proteins.
Influences of spine morphology on diffusional coupling between spines and dendrites
Although the cytoplasm of spines is directly connected to the cytoplasm of dendritic shafts, a narrow neck is thought to limit diffusion of both cytosolic and membrane molecules between two compartments (Holcman and Schuss, 2011; Kusters et al., 2013; Ramirez et al., 2015). FRAP is a method that can be used to measure the diffusional speeds from an exchange rate between nonbleached and bleached molecules after bleaching fluorescent molecules in a small region (Lippincott-Schwartz et al., 2018; Fig. 3, a and d). Local photoactivation or photoconversion and subsequent measurements of fluorescence intensity is another technique comparable to FRAP (Bancaud et al., 2010). When fluorescence bleaching is performed in a spine head, the speed of fluorescence recovery mostly reflects the rate of molecular exchange between the head and the connected dendrite. Because the diffusion of small molecules in a head is faster than the rate of molecular exchange between spines and dendrites, the fast component of intra-spine diffusion is more difficult to detect in FRAP recovery curves (Svoboda et al., 1996). FRAP or photoactivation experiments of cytoplasmic and membrane-anchored fluorescent proteins showed that diffusional coupling between spines and dendrites varies between spines (Bloodgood and Sabatini, 2005; Ashby et al., 2006). Since the shape of spines is diverse, it has been proposed that this diversity underlies variability in spine–dendrite coupling. However, because the details of spine morphology cannot be analyzed with the spatial resolution of diffraction-limited fluorescence microscopy, a relationship between the shape of spines and diffusional coupling had not been directly demonstrated.
Recently, however, super-resolution microscopy has made it possible to analyze spine shape in living neurons with a spatial resolution of ∼50 nm (Nägerl et al., 2008). Influences of spine morphology on diffusional coupling were verified experimentally for the first time by directly comparing the morphological features of spines and diffusional coupling. This comparison was achieved by stimulated emission depletion (STED) microscopy of spines combined with FRAP of YFP or Alexa dyes applied to the same spine (Takasaki and Sabatini, 2014; Tønnesen et al., 2014). These direct comparisons indicated that the diversity in diffusional couplings could be explained solely by the diversity of spine shapes for more than half of the measured spines. In other words, it was shown that for many spines, the exchange rate (τ) of small molecules within spines could be explained by a single-compartment model (Svoboda et al., 1996) described by the shape of the spine:
where V is the head volume, L is the length of the neck, A is the cross-sectional area of the neck, and D is the diffusion coefficient of molecules. Also, sLTP induction made a spine neck thicker and shorter (Tønnesen et al., 2014). This change in the spine neck complements the decrease in the coupling rate associated with the increase in the spine head volume. This coordinated morphological change appears to maintain molecular concentration in a spine.
Besides the work focusing on cytoplasmic proteins, the influence of spine shape on the diffusional coupling of membrane molecules has also been investigated (Adrian et al., 2017). The spine–dendrite diffusional coupling was tested by photoactivated localization microscopy (PALM) and photoconversion experiments using membrane-anchored mEos3.2 as a probe. This study showed that even if spines have the same surface area and neck width, the diffusional coupling varies between different spine shapes. Therefore, a model spine was created based on the experimentally measured spine shape parameters, and a simulation was conducted on the model spine and compared with the experiment. As a result, although experimental results tended to provide slower diffusion kinetics than simulation values, experiments showed a good correlation with simulations based on the spine shape parameters alone.
Experiments have confirmed that spine morphology is a major factor determining the diffusional coupling for both cytoplasmic and membrane-bound molecules in dendrites. However, for some spines, the simulated and experimental results diverge. One possibility is that the effects of local intra-spine architectures on molecular diffusion vary for each spine. Another possibility is that there was insufficient spatial resolution for reconstructing the spine morphology. Although the above studies used rotationally symmetric shapes as model spines, actual spines are not rotationally symmetrical structures and generally have a more complicated morphology and surface features (Nägerl et al., 2008; Berning et al., 2012; Kashiwagi et al., 2019; Zaccard et al., 2020; Fig. 1, a and b). Thus, it is possible that estimations of spine shape were insufficient or that the fine structure of spines affects diffusion. In this regard, developing an analysis method for spine morphology from both the experimental and computational sides is key (Okabe, 2020a; Tamada et al., 2020). Recently, Kashiwagi et al. (2019) developed a 3-D structured illumination microscopy (SIM)–based nanoscale analysis of spine morphology. Direct comparison of SIM images and serial-section EM images revealed that the basic morphological features were highly correlated among these images. This indicates the high precision of SIM-based nanoscale spine analysis. To analyze spines computationally, SIM images were converted into a computational geometry, and morphological features were calculated. Then, these features were analyzed by principal component analysis. By mapping the temporal changes of spine morphology obtained by live-cell SIM imaging in the dimension-reduced feature space, the authors revealed that the spine population can be categorized based on different simplified morphological dynamics.
Also, expansion microscopy (ExM) is another new and important imaging technique for spine structural analysis (Wassie et al., 2019). However, it can only be applied to fixed samples. Since ExM samples are transparent, 3-D super-resolution imaging is available for thick samples with large volumes (Gao et al., 2019). With recent developments in sample preparation technology, ExM has the potential to investigate spine morphology and localization of multiple biomolecules and organelles within a single sample (Chozinski et al., 2016; Tillberg et al., 2016; Karagiannis et al., 2019 Preprint; Sun et al., 2021). Minimizing the distortion of isotropy during expansion will be important for nanoscale morphological analysis. In the future, combining dynamic fluorescence measurements and structural measurements gained from EM (CLEM) will be a powerful approach to evaluate the effects of spine ultrastructure on molecular diffusion in greater nanoscale detail (Maco et al., 2013; Taraska, 2015; Luckner et al., 2018).
Along with biochemical compartmentalization, dendritic spines have been proposed to be important for electrical compartmentalization (Yuste, 2013; Araya, 2014; Tønnesen and Nägerl, 2016). Spine morphology, particularly spine neck morphology, is thought to be critical for this effect (Cartailler et al., 2018). Several studies have sought to measure neck resistance based on morphological analysis using EM (Harris and Stevens, 1989; Tamada et al., 2020), super-resolution microscopy (Tønnesen et al., 2014), FRAP of small molecules (Svoboda et al., 1996; Tønnesen et al., 2014), glutamate uncaging (Araya et al., 2006; Takasaki and Sabatini, 2014), calcium imaging (Grunditz et al., 2008; Harnett et al., 2012), voltage imaging (Popovic et al., 2015; Acker et al., 2016; Kwon et al., 2017), and intracellular recordings directly from spine heads (Jayant et al., 2017). However, results were not completely consistent, and the degree of electrical compartmentalization is still unclear. Thus, the relationship between spine morphology and electrical signaling of the synapse is still an open question. Likewise, how morphological changes in the neck induced by LTP affect dendritic computation will be an important area of future study (Araya et al., 2014; Tazerart et al., 2020).
Actin cytoskeleton
The cytoskeleton in spines is primarily composed of actin (Hotulainen and Hoogenraad, 2010; Okabe, 2020b). Actin is present in high densities in both the head and neck regions (Korobova and Svitkina, 2010). Actin polymers are essential in controlling the localization of PSD molecules and in changing and maintaining spine morphology (Frost et al., 2010a; Bertling and Hotulainen, 2017). In addition to these functions, dense actin polymers in spines may regulate synaptic functions by controlling diffusion because the intracellular cytoskeleton and membrane structures influence diffusion (Novak et al., 2009). If this regulation occurs in spines, variations in the distribution of intra-spine structures can be a factor in the large deviations between the measured values of diffusional coupling and the value predicted from models. A ratio of the spine FRAP recovery time of Alexa dyes to that of YFP was comparable to that of hydrodynamic radii (Tønnesen et al., 2014). This suggests that the suppression of diffusion by actin polymers is weak for molecules with the size of GFP. However, suppressive effects on molecular diffusion by the cytoskeleton, such as actin polymers, is dependent on the size of molecules (Baum et al., 2014; Katrukha et al., 2017). Thus, diffusion of larger molecules may be influenced to a greater degree by actin polymers.
Because the shape of spines affects the recovery time of FRAP measurement, it is difficult to investigate the effects of intra-spine structure on molecular diffusion using FRAP alone. Therefore, there is a need for a method capable of measuring diffusion directly in confined spaces. Lu et al. (2014) measured the motion of mEOS2-fused CaMKIIα in spines by single-particle tracking (SPT)–PALM. SPT can directly evaluate diffusional speed in spines because it analyzes the molecular movement trajectory of single molecules (Fig. 3, c and d). The SPT measurement showed that CaMKIIα exhibited at least three different diffusion modes within spines: (1) a free diffusion component, (2) a component bound to immobile molecules, and (3) a component moving at an intermediate velocity. Depolymerization of actin polymers by latrunculin A reduced the proportion of molecules with intermediate velocities in spines while concomitantly increasing the free diffusion component. Also, diffusional speeds of CaMKIIα were slower and the ratio of the intermediate component was larger in spines than in dendrites. Because the transition between free and bound states would occur rarely during the measurement period due to the slow unbinding rate of CaMKII from actin polymers, transient binding alone does not explain the mechanism for the intermediate velocity. Although the details are unclear, CaMKII motion is restricted by actin polymers through a mechanism distinct from direct binding, including a molecular sieve effect or transient binding to actin-associated molecules.
Obashi et al. (2019) used fluorescence correlation spectroscopy (FCS) and raster image correlation spectroscopy (RICS) to measure the diffusion of biologically inert probes within spines. FCS is a method for estimating diffusion speed from the time taken for fluorescent molecules to pass through the detection volume excited by a high numerical aperture objective and is capable of measuring fast diffusion within a small cellular compartment (Elson, 2011; Fig. 3, b and d). RICS is another method for estimating diffusion speed from the spatial similarity of fluorescence intensity in a scanned image (Digman and Gratton, 2011). Since FCS and RICS are affected by the small size of spines due to the boundary effect, it is not possible to measure the diffusion coefficient accurately (Jiang et al., 2020). Still, by averaging, values proportional to the actual values can be obtained. Diffusion of GFP and GFP tandem pentamer (GFP5) were compared, and only diffusion of GFP5 within spines was enhanced by depolymerizing actin with latrunculin A treatment. Molecular dynamics simulation confirmed that the diffusion of molecules over the size of GFP5 was suppressed by actin polymers with a density (380 µM) estimated from the values in the literature and experiments.
Together, these experiments support the idea that a meshwork of dense actin polymers in spines acts as a physical barrier to the diffusion of larger (>100 kD) molecules (Fig. 4 a). Photoactivation experiments of intra-spine photoactivatable GFP (PA-GFP)–actin showed that there are at least three groups of actin polymers with different reorganization rates (Honkura et al., 2008). In addition, experiments with SPT-PALM of PA-GFP–actin showed that a rate of actin filament polymerization increased near PSDs (Frost et al., 2010b). PALM analysis also revealed that actin-related molecules within spines are arranged in a manner specific for each molecule (Chazeau et al., 2014). These results suggest that the diffusional control by actin polymers in spines may differ between each subcompartment.
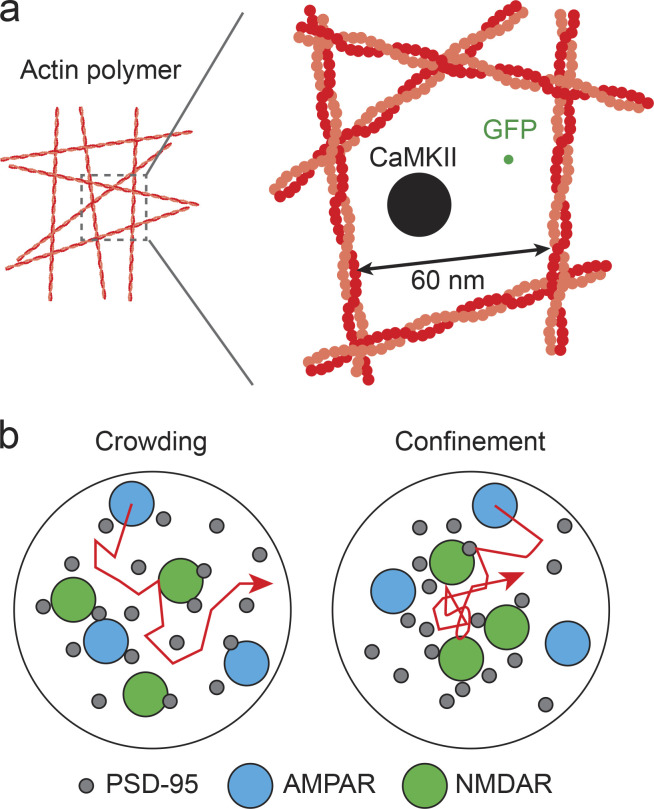
Figure 4.
Diffusion within network of actin polymers and PSD. (a) Comparison of the size of actin polymer network and diffusion molecules. Average distance between actin polymers is estimated for actin polymers with 380 µM (Obashi et al., 2019). GFP is represented as a diameter of 3 nm and CaMKII is represented as a diameter of 20 nm (Myers et al., 2017). (b) A schematic model of AMPAR diffusion within a crowded PSD. Different localization patterns of molecules cause different diffusion patterns. Such mechanisms will occur within the PSD. Density and size of molecules are based on the literature (Okabe, 2007; Li et al., 2016).
It was also shown that reorganization of the actin cytoskeleton immediately after sLTP induction (Chazeau and Giannone, 2016; Mikhaylova et al., 2018) enhanced the diffusion of larger molecules within the spine head (Obashi et al., 2019). Further, FRAP experiments showed that diffusional coupling and synaptic translocation of large synaptic molecules, such as CaMKII and T cell lymphoma invasion and metastasis-inducing protein 1 (Tiam1), were facilitated at the initial phase of sLTP. Thus, the reorganization of actin polymers regulates molecular translocation between dendrites and the PSD in coordination with morphological changes of the spine neck (Tønnesen et al., 2014). The enhancement of molecular diffusion by actin may also be related to the formation of a large signaling complex containing CaMKII and Tiam1 and may be an important physical mechanism responsible for the initiation of sLTP (Saneyoshi et al., 2019).
Membranous organelles
Along with the cytoskeleton, dendrites also contain many membranous organelles and compartments, and some are present in spines (Bourne and Harris, 2008). Smooth ER (SER) and recycling endosomes are present in <50% of spines. Spine apparatus, which is composed of stacked SER, is present in 10–20% of spines. Localizations of these organelles change after LTP induction, and spines containing SER are larger than those without SER (Chirillo et al., 2019; Kulik et al., 2019; Perez-Alvarez et al., 2020). While mitochondria are abundant in dendritic shafts but rarely present in spines (Wu et al., 2017), synaptic activation relocates mitochondria into spines (Li et al., 2004). Therefore, variations in the diffusional coupling between spines and dendrites could be due to the heterogeneous localization of these organelles (Cugno et al., 2019). Holbro et al. (2009) compared the diffusional coupling of ER-containing spines and ER-free spines by using an ER-targeted GFP probe. FRAP recovery times of RFP were not different among ER-containing and ER-free spines, indicating that the ER does not block cytoplasmic diffusion between spines and dendritic shafts. Understanding both spine morphology and the volume of ER within spines in the future will clarify the effects of excluded volume by the ER and other organelles in more detail.
Structures around spine necks
Molecules present in spine necks may physically control diffusion by forming a complex higher-order structure. Platinum replica EM showed the presence of Arp2/3 complex within necks and a longitudinal network of branched and linear actin filaments (Korobova and Svitkina, 2010). SPT-PALM of PA-GFP–actin also showed that actin polymers in necks are dynamically reorganized and that they are arranged in many orientations (Frost et al., 2010b). Thus, actin polymers in spine necks may affect molecular diffusion. Synaptopodin, for example, is an actin-binding protein located predominantly in spine necks. It is colocalized with the spine apparatus (Vlachos, 2012). Wang et al. (2016) used SPT to measure metabotropic glutamate receptor 5 (mGluR5) diffusion around necks. They compared the diffusion of mGluR5 around the necks of spines containing (or not containing) synaptopodin. The diffusion of mGluR5 decreases around spine necks near synaptopodin clusters. Further, latrunculin A treatment specifically enhanced the diffusion around spine necks near synaptopodin clusters. These results suggest that synaptopodin regulates the actin polymer network around spine necks. This actin complex can act as a diffusion barrier for membrane proteins.
Another protein that has been implicated in diffusional control of membrane proteins is ankyrin-G. Ankyrin-G forms nanodomain at perisynaptic membranes and in spine necks (Smith et al., 2014). AMPARs accumulated in spines with ankyrin-G clusters and showed slower spine–dendrite coupling. Ankyrin-G is the major cytoskeletal scaffold of the axon initial segment (AIS; Leterrier, 2018). Ankyrin-G and actin scaffolds densely accumulate at the AIS and inhibit diffusion of the membrane and cytoplasmic molecules (Winckler et al., 1999; Nakada et al., 2003; Song et al., 2009). Also, super-resolution microscopy recently revealed the presence of membrane-associated periodic skeleton composed of actin rings, spectrin, and accompanying proteins in the axon including the AIS (Xu et al., 2013; Zhong et al., 2014). At the AIS, the actin rings and associated structures act as a diffusion barrier to membrane proteins (Albrecht et al., 2016). Adding to the axon, membrane-associated periodic skeleton was also observed in the dendrites and spine necks (Bär et al., 2016; Sidenstein et al., 2016). Therefore, it is interesting to postulate that a molecular complex with actin filaments similar to the AIS is also present in spine necks and could regulate molecular diffusion in this small compartment.
Another cytoskeletal component, septin 7, localizes to the base of spines and acts as a diffusion barrier for membrane-bound molecules (Ewers et al., 2014). Recently, actin patches were found at the base of spines and were shown to be remodeled by synaptic activity. These structures modulate microtubule entry into spines and the transport of lysosomes (Schätzle et al., 2018; van Bommel et al., 2019). It is interesting to ask whether actin patches at spine bases affect molecular diffusion. There are still many unknown features at the spine neck, and how these structures limit the diffusion of cytoplasmic and membrane molecules to control neuronal functions remains unclarified.
Molecular crowding in the PSD
The PSD is a membrane-associated structure containing densely packed postsynaptic molecules (Sheng and Hoogenraad, 2007). It was originally identified as an electron-dense structure in EM (Okabe, 2007). The number and location of receptors and adhesion molecules in PSDs are directly related to synaptic function (Chen et al., 2018). SPT studies indicate that AMPARs diffuse laterally into and out of PSDs and regulate synaptic function by controlling the number and location of AMPARs (Choquet and Hosy, 2020). Because there are many scaffold proteins in PSDs, membrane proteins including AMPARs accumulate in PSDs due to intermolecular binding. Furthermore, because the molecular density in PSDs is high, the accumulation of membrane proteins may be regulated by the suppression of mobility within the PSD and molecular exchange at the boundary of PSDs (Gerrow and Triller, 2010; Kokolaki et al., 2020).
To check this possibility, Li et al. (2016) combined FRAP, SPT, and Monte Carlo simulation to investigate the effect of molecular crowding of PSDs on the lateral diffusion of membrane molecules. When the intracellular domain size of membrane proteins was large, diffusion within the PSD and the exchange rate between the inside and outside of the PSD decreased. Super-resolution microscopy showed that the distribution of PSD-95, a major scaffolding protein of the PSD, within PSDs is not uniform (Fukata et al., 2013; MacGillavry et al., 2013; Nair et al., 2013; Broadhead et al., 2016; Gwosch et al., 2020). Interestingly, the simulation showed that the residence time of membrane proteins within PSDs was longer in the condition of experimentally measured PSD-95 distribution, while the residence time decreased with a random distribution of PSD-95 (Li et al., 2016).
Recently, the shape of PSDs inside spines induced by sLTP was analyzed by CLEM (Sun et al., 2019 Preprint). It was shown that rearrangements of PSD shape occurred immediately after induction of sLTP (<3 min), and the PSD took more complex morphology. This increased structural complexity persisted in the late phase (120 min). PSD size and the accumulation of PSD-95 increased slowly over several tens of minutes after sLTP induction (Meyer et al., 2014), whereas synaptic transmission efficiency increased immediately (Matsuzaki et al., 2004). This difference in time may be explained by a mechanism in which the acute ultrastructural changes of the PSD without net growth of the molecular assembly alter the mobility of AMPARs by changing the distribution of a physical barrier, leading to alternations in the number and localization of AMPARs (Fig. 4 b). In future studies, it will be necessary to clarify how coordination between intermolecular binding and physical diffusion barriers in PSDs supports both acute accumulation of AMPARs and their subsequent stabilization in stimulated spines. Further, 3-D SIM imaging revealed that the concave surface of the spine head, which interacts with presynaptic membranes, is enlarged and stabilized by sLTP induction (Kashiwagi et al., 2019). In the future, it will be interesting to determine the relationships between concave membrane surfaces, PSD morphologies, and the dynamics of receptors and adhesion molecules at single spines.
In addition, although AMPAR has been thought to be present as a tetramer (Greger et al., 2007), recent observations of SPT have shown that the majority of diffusive AMPARs are monomers or dimers (Morise et al., 2019). Molecular diffusion in the monomer form increases an exchange rate between the inside and the outside of PSDs, making it possible to efficiently change the AMPAR composition within synapses. It remains to be seen whether other molecular complexes, such as NMDARs and cell adhesion molecules, also modulate their diffusion within the molecularly dense PSD by changing their oligomeric state.
Conclusion and outlook
Here, we have highlighted key recent findings on the relationship between molecular diffusion and physical barriers within spines. The regulation of molecular diffusion is important for sLTP expression (Fig. 2). Spine structural changes during sLTP will affect synaptic function in a coordinated manner (Fig. 5). For example, after sLTP induction, the actin network is reorganized and diffusion of large molecules is enhanced (Obashi et al., 2019). This facilitates the formation of large signaling complexes and the rearrangement of protein complexes within spines. At the same time, spine necks become wider and shorter, and spine heads enlarge (Tønnesen et al., 2014). Changes in actin and spine morphology enhance the molecular movement between the PSD and the shaft and are important for the relocation of proteins (Fig. 2). These structural changes occur in the early phase of sLTP. Thus, the cooperative regulation of diffusion might act as a precise temporal switch of sLTP induction. Also, this enhancement of molecular exchange affects the relocation of activated signaling molecules into the shaft or nearby synapses, which leads to heterosynaptic plasticity (Yasuda, 2017). Potentiation of synaptic transmission requires synaptic trafficking of AMPARs (Choquet and Hosy, 2020). Although both spine morphology (Adrian et al., 2017) and PSD structure (Li et al., 2016) affect membrane protein diffusion, how structural changes associated with sLTP induction affect diffusion will be clarified in the future. Furthermore, the effects of transient SER visits (Perez-Alvarez et al., 2020) and structural changes around spine necks are an important area for future work. Although the relationship between structure and diffusion in sLTP is critical, the difficulty of measurements with a small single spine has made a comprehensive view difficult to obtain. Thus, future work will be necessary to clarify how structural changes affect diffusion and how this physical change to dendritic spines cooperatively modulates synaptic functions.
Figure 5.
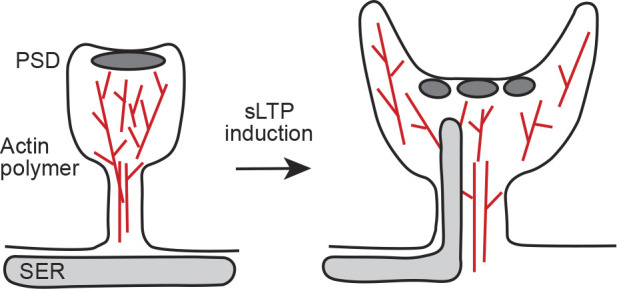
Changes in the shape and internal architecture of spines after induction of sLTP. At the initial phase of sLTP, a spine head expands. In addition, the spine neck becomes wider and shorter (Tønnesen et al., 2014), and a concave surface area of spine head is increased (Kashiwagi et al., 2019). The actin polymer network is reorganized (Obashi et al., 2019), and the SER visits within a spine transiently (Perez-Alvarez et al., 2020). Also, PSD shape becomes more complex (Sun et al., 2019 Preprint). These physical changes should occur in concert and will affect molecular composition and biochemical signaling through diffusional regulation. These physical changes will act as a precise temporal switch of sLTP induction.
Although new imaging techniques have demonstrated the connection between diffusion and physical barriers, little is known about how changes in the movement of molecules alter synaptic functions (Reshetniak et al., 2020b). Because of the small volume of the spine, very small molecules with high diffusivity, such as Ca2+, are expected to spread rapidly (∼1 ms) by diffusion (Chen and Sabatini, 2012). For large molecules such as signaling complexes, it remains to be seen whether spatially uniform diffusion takes place or whether local heterogeneity in the spine cytoplasm results in a more complex pattern of diffusion. It is also necessary to clarify whether such changes affect local biochemical signaling events and molecular localizations. The number of molecules per spine could influence the magnitude of functional changes (Okabe, 2007; Ribrault et al., 2011). Furthermore, the changes in diffusion induced by alterations in spine structure will affect the stability of the structure. This will subsequently change the molecule’s diffusivity. Thus, it will be interesting to investigate whether this type of mutual relationship exists within spines.
New imaging techniques will help to answer these questions. By applying fast 3-D SPT to intra-spine measurements, it will be possible to investigate the spatial heterogeneity of diffusion in single spines of living neurons in detail (Hou et al., 2020; Xiang et al., 2020). STED-FCS/fluorescence cross-correlation spectroscopy can also detect changes in intermolecular interactions (Lanzanò et al., 2017). In addition to the development of new measurement techniques, molecular dynamics simulations based on experimental data will become increasingly important in the future (Okabe, 2020a; Reshetniak et al., 2020a; Vasan et al., 2020). Spine morphology and intra-spine structures, which affect diffusion, are closely related. Thus, it is difficult to investigate the effect of one without changing the other experimentally. Molecular dynamics simulation is a useful tool to examine how molecular motion is adjusted by combining elements that are difficult to verify experimentally (Bell et al., 2019). Furthermore, the shape of spines and intra-spine components, such as the actin cytoskeleton, which are the structural basis of spines, differ from spine to spine. Here, a combination of quantitative measurements and simulations based on experimental data will help us to understand molecular events more quantitatively.
Although we reviewed work using fluorescence microscopy, details of spine morphology and intra-spine structures have also been revealed by EM at the nanoscale (Bourne and Harris, 2012; Tao et al., 2018). However, it is difficult to observe specific molecular localizations with EM. On the other hand, super-resolution microscopy is suitable for obtaining a nanoscale picture of molecular positions within spines. Yet, it is still difficult to observe dense structures such as actin polymers (Kommaddi et al., 2018). Therefore, in the future, it will be essential to combine the advantages of each technique, observing internal structures at the nanoscale using EM and measuring molecular localization with super-resolution microscopy (CLEM; Taraska, 2019; Hoffman et al., 2020). Of course, dynamic intracellular structures such as lipid rafts and biomolecular condensates are also likely to affect molecular mobility (Sezgin et al., 2017; Chen et al., 2020). Thus, it will be key to overlay molecular mobilities from living cells over the static structural information of CLEM. We believe that combinations of multiple imaging modalities, along with modeling, will allow for a more in-depth understanding of synapses at the molecular level. These data will reveal how the elaborate architecture, density, and compartmentalization of subcellular components influence the highly tuned, dynamic, and changeable actions of synapses in the brain.
Acknowledgments
Néstor Saiz served as editor.
We thank members of the Taraska laboratory and Okabe laboratory for comments on the manuscript. J.W. Taraska is supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health. S. Okabe is supported by Grants-in-Aid for Scientific Research (20H00481) and the Japan Agency for Medical Research and Development (JP20gm1310003 and JP20gm5010003).
The authors declare no competing financial interests.
Author contributions: K. Obashi, J.W. Taraska, and S. Okabe wrote and edited the paper.
References
- Acker, C.D., Hoyos E., and Loew L.M.. 2016. EPSPs measured in proximal dendritic spines of cortical pyramidal neurons. eNeuro. 3: ENEURO.0050-15.2016. 10.1523/ENEURO.0050-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian, M., Kusters R., Wierenga C.J., Storm C., Hoogenraad C.C., and Kapitein L.C.. 2014. Barriers in the brain: resolving dendritic spine morphology and compartmentalization. Front. Neuroanat. 8:142. 10.3389/fnana.2014.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian, M., Kusters R., Storm C., Hoogenraad C.C., and Kapitein L.C.. 2017. Probing the interplay between dendritic spine morphology and membrane-bound diffusion. Biophys. J. 113:2261–2270. 10.1016/j.bpj.2017.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht, D., Winterflood C.M., Sadeghi M., Tschager T., Noé F., and Ewers H.. 2016. Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. J. Cell Biol. 215:37–46. 10.1083/jcb.201603108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, V.A., and Sabatini B.L.. 2007. Anatomical and physiological plasticity of dendritic spines. Annu. Rev. Neurosci. 30:79–97. 10.1146/annurev.neuro.30.051606.094222 [DOI] [PubMed] [Google Scholar]
- Araki, Y., Zeng M., Zhang M., and Huganir R.L.. 2015. Rapid dispersion of SynGAP from synaptic spines triggers AMPA receptor insertion and spine enlargement during LTP. Neuron. 85:173–189. 10.1016/j.neuron.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya, R. 2014. Input transformation by dendritic spines of pyramidal neurons. Front. Neuroanat. 8:141. 10.3389/fnana.2014.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya, R., Jiang J., Eisenthal K.B., and Yuste R.. 2006. The spine neck filters membrane potentials. Proc. Natl. Acad. Sci. USA. 103:17961–17966. 10.1073/pnas.0608755103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya, R., Vogels T.P., and Yuste R.. 2014. Activity-dependent dendritic spine neck changes are correlated with synaptic strength. Proc. Natl. Acad. Sci. USA. 111:E2895–E2904. 10.1073/pnas.1321869111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano, J.I., Benavides-Piccione R., Defelipe J., and Yuste R.. 2007. Ultrastructure of dendritic spines: correlation between synaptic and spine morphologies. Front. Neurosci. 1:131–143. 10.3389/neuro.01.1.1.010.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby, M.C., Maier S.R., Nishimune A., and Henley J.M.. 2006. Lateral diffusion drives constitutive exchange of AMPA receptors at dendritic spines and is regulated by spine morphology. J. Neurosci. 26:7046–7055. 10.1523/JNEUROSCI.1235-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley, D., and Bewersdorf J.. 2018. Biological insight from super-resolution microscopy: what we can learn from localization-based images. Annu. Rev. Biochem. 87:965–989. 10.1146/annurev-biochem-060815-014801 [DOI] [PubMed] [Google Scholar]
- Bancaud, A., Huet S., Rabut G., and Ellenberg J.. 2010. Fluorescence perturbation techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, photoactivation, photoconversion, and FLIP. Cold Spring Harb. Protoc. 2010:pdb.top90. 10.1101/pdb.top90 [DOI] [PubMed] [Google Scholar]
- Bär, J., Kobler O., van Bommel B., and Mikhaylova M.. 2016. Periodic F-actin structures shape the neck of dendritic spines. Sci. Rep. 6:37136. 10.1038/srep37136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, M., Erdel F., Wachsmuth M., and Rippe K.. 2014. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat. Commun. 5:4494. 10.1038/ncomms5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, M., Bartol T., Sejnowski T., and Rangamani P.. 2019. Dendritic spine geometry and spine apparatus organization govern the spatiotemporal dynamics of calcium. J. Gen. Physiol. 151:1017–1034. 10.1085/jgp.201812261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berning, S., Willig K.I., Steffens H., Dibaj P., and Hell S.W.. 2012. Nanoscopy in a living mouse brain. Science. 335:551. 10.1126/science.1215369 [DOI] [PubMed] [Google Scholar]
- Bertling, E., and Hotulainen P.. 2017. New waves in dendritic spine actin cytoskeleton: From branches and bundles to rings, from actin binding proteins to post-translational modifications. Mol. Cell. Neurosci. 84:77–84. 10.1016/j.mcn.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Bloodgood, B.L., and Sabatini B.L.. 2005. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 310:866–869. 10.1126/science.1114816 [DOI] [PubMed] [Google Scholar]
- Borgdorff, A.J., and Choquet D.. 2002. Regulation of AMPA receptor lateral movements. Nature. 417:649–653. 10.1038/nature00780 [DOI] [PubMed] [Google Scholar]
- Borovac, J., Bosch M., and Okamoto K.. 2018. Regulation of actin dynamics during structural plasticity of dendritic spines: Signaling messengers and actin-binding proteins. Mol. Cell. Neurosci. 91:122–130. 10.1016/j.mcn.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Bosch, M., Castro J., Saneyoshi T., Matsuno H., Sur M., and Hayashi Y.. 2014. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 82:444–459. 10.1016/j.neuron.2014.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, J.N., and Harris K.M.. 2008. Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 31:47–67. 10.1146/annurev.neuro.31.060407.125646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, J.N., and Harris K.M.. 2012. Nanoscale analysis of structural synaptic plasticity. Curr. Opin. Neurobiol. 22:372–382. 10.1016/j.conb.2011.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhead, M.J., Horrocks M.H., Zhu F., Muresan L., Benavides-Piccione R., DeFelipe J., Fricker D., Kopanitsa M.V., Duncan R.R., Klenerman D., et al. 2016. PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci. Rep. 6:24626. 10.1038/srep24626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartailler, J., Kwon T., Yuste R., and Holcman D.. 2018. Deconvolution of voltage sensor time series and electro-diffusion modeling reveal the role of spine geometry in controlling synaptic strength. Neuron. 97:1126–1136.e10. 10.1016/j.neuron.2018.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater, T.E., and Goda Y.. 2021. My Neighbour Hetero-deconstructing the mechanisms underlying heterosynaptic plasticity. Curr. Opin. Neurobiol. 67:106–114. 10.1016/j.conb.2020.10.007 [DOI] [PubMed] [Google Scholar]
- Chazeau, A., and Giannone G.. 2016. Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell. Mol. Life Sci. 73:3053–3073. 10.1007/s00018-016-2214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazeau, A., Mehidi A., Nair D., Gautier J.J., Leduc C., Chamma I., Kage F., Kechkar A., Thoumine O., Rottner K., et al. 2014. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 33:2745–2764. 10.15252/embj.201488837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and Sabatini B.L.. 2012. Signaling in dendritic spines and spine microdomains. Curr. Opin. Neurobiol. 22:389–396. 10.1016/j.conb.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.H., Kellner Y., Zagrebelsky M., Grunwald M., Korte M., and Walla P.J.. 2015. Two-photon correlation spectroscopy in single dendritic spines reveals fast actin filament reorganization during activity-dependent growth. PLoS One. 10:e0128241. 10.1371/journal.pone.0128241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Tang A.H., and Blanpied T.A.. 2018. Subsynaptic spatial organization as a regulator of synaptic strength and plasticity. Curr. Opin. Neurobiol. 51:147–153. 10.1016/j.conb.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Wu X., Wu H., and Zhang M.. 2020. Phase separation at the synapse. Nat. Neurosci. 23:301–310. 10.1038/s41593-019-0579-9 [DOI] [PubMed] [Google Scholar]
- Chirillo, M.A., Waters M.S., Lindsey L.F., Bourne J.N., and Harris K.M.. 2019. Local resources of polyribosomes and SER promote synapse enlargement and spine clustering after long-term potentiation in adult rat hippocampus. Sci. Rep. 9:3861. 10.1038/s41598-019-40520-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, D., and Hosy E.. 2020. AMPA receptor nanoscale dynamic organization and synaptic plasticities. Curr. Opin. Neurobiol. 63:137–145. 10.1016/j.conb.2020.04.003 [DOI] [PubMed] [Google Scholar]
- Chozinski, T.J., Halpern A.R., Okawa H., Kim H.J., Tremel G.J., Wong R.O., and Vaughan J.C.. 2016. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods. 13:485–488. 10.1038/nmeth.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri, A., and Malenka R.C.. 2008. Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology. 33:18–41. 10.1038/sj.npp.1301559 [DOI] [PubMed] [Google Scholar]
- Colgan, L.A., Hu M., Misler J.A., Parra-Bueno P., Moran C.M., Leitges M., and Yasuda R.. 2018. PKCα integrates spatiotemporally distinct Ca2+ and autocrine BDNF signaling to facilitate synaptic plasticity. Nat. Neurosci. 21:1027–1037. 10.1038/s41593-018-0184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugno, A., Bartol T.M., Sejnowski T.J., Iyengar R., and Rangamani P.. 2019. Geometric principles of second messenger dynamics in dendritic spines. Sci. Rep. 9:11676. 10.1038/s41598-019-48028-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani, A., Huang B., Bergan J., Dulac C., and Zhuang X.. 2010. Superresolution imaging of chemical synapses in the brain. Neuron. 68:843–856. 10.1016/j.neuron.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman, M.A., and Gratton E.. 2011. Lessons in fluctuation correlation spectroscopy. Annu. Rev. Phys. Chem. 62:645–668. 10.1146/annurev-physchem-032210-103424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson, E.L. 2011. Fluorescence correlation spectroscopy: past, present, future. Biophys. J. 101:2855–2870. 10.1016/j.bpj.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers, H., Tada T., Petersen J.D., Racz B., Sheng M., and Choquet D.. 2014. A septin-dependent diffusion barrier at dendritic spine necks. PLoS One. 9:e113916. 10.1371/journal.pone.0113916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, M.P., Parnell E., and Penzes P.. 2018. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 19:215–234. 10.1038/nrn.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, N.A., Kerr J.M., Lu H.E., and Blanpied T.A.. 2010a. A network of networks: cytoskeletal control of compartmentalized function within dendritic spines. Curr. Opin. Neurobiol. 20:578–587. 10.1016/j.conb.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, N.A., Shroff H., Kong H., Betzig E., and Blanpied T.A.. 2010b. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 67:86–99. 10.1016/j.neuron.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata, Y., Dimitrov A., Boncompain G., Vielemeyer O., Perez F., and Fukata M.. 2013. Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 202:145–161. 10.1083/jcb.201302071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, R., Asano S.M., Upadhyayula S., Pisarev I., Milkie D.E., Liu T.L., Singh V., Graves A., Huynh G.H., Zhao Y., et al. 2019. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science. 363:eaau8302. 10.1126/science.aau8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrow, K., and Triller A.. 2010. Synaptic stability and plasticity in a floating world. Curr. Opin. Neurobiol. 20:631–639. 10.1016/j.conb.2010.06.010 [DOI] [PubMed] [Google Scholar]
- Greger, I.H., Ziff E.B., and Penn A.C.. 2007. Molecular determinants of AMPA receptor subunit assembly. Trends Neurosci. 30:407–416. 10.1016/j.tins.2007.06.005 [DOI] [PubMed] [Google Scholar]
- Grunditz, A., Holbro N., Tian L., Zuo Y., and Oertner T.G.. 2008. Spine neck plasticity controls postsynaptic calcium signals through electrical compartmentalization. J. Neurosci. 28:13457–13466. 10.1523/JNEUROSCI.2702-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwosch, K.C., Pape J.K., Balzarotti F., Hoess P., Ellenberg J., Ries J., and Hell S.W.. 2020. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods. 17:217–224. 10.1038/s41592-019-0688-0 [DOI] [PubMed] [Google Scholar]
- Harnett, M.T., Makara J.K., Spruston N., Kath W.L., and Magee J.C.. 2012. Synaptic amplification by dendritic spines enhances input cooperativity. Nature. 491:599–602. 10.1038/nature11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K.M., and Stevens J.K.. 1989. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J. Neurosci. 9:2982–2997. 10.1523/JNEUROSCI.09-08-02982.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, T., Noguchi J., Watanabe S., Takahashi N., Hayashi-Takagi A., Ellis-Davies G.C., Matsuzaki M., and Kasai H.. 2013. GABA promotes the competitive selection of dendritic spines by controlling local Ca2+ signaling. Nat. Neurosci. 16:1409–1416. 10.1038/nn.3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen, F., and Denk W.. 2005. Deep tissue two-photon microscopy. Nat. Methods. 2:932–940. 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- Herring, B.E., and Nicoll R.A.. 2016. Long-term potentiation: from CaMKII to AMPA receptor trafficking. Annu. Rev. Physiol. 78:351–365. 10.1146/annurev-physiol-021014-071753 [DOI] [PubMed] [Google Scholar]
- Hoffman, D.P., Shtengel G., Xu C.S., Campbell K.R., Freeman M., Wang L., Milkie D.E., Pasolli H.A., Iyer N., Bogovic J.A., et al. 2020. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science. 367:eaaz5357. 10.1126/science.aaz5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro, N., Grunditz A., and Oertner T.G.. 2009. Differential distribution of endoplasmic reticulum controls metabotropic signaling and plasticity at hippocampal synapses. Proc. Natl. Acad. Sci. USA. 106:15055–15060. 10.1073/pnas.0905110106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcman, D., and Schuss Z.. 2011. Diffusion laws in dendritic spines. J. Math. Neurosci. 1:10. 10.1186/2190-8567-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler, S., Köstinger G., Martin K.A.C., Schuhknecht G.F.P., and Stratford K.J.. 2021. Structure and function of a neocortical synapse. Nature. 10.1038/s41586-020-03134-2 [DOI] [PubMed] [Google Scholar]
- Honkura, N., Matsuzaki M., Noguchi J., Ellis-Davies G.C., and Kasai H.. 2008. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 57:719–729. 10.1016/j.neuron.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Hotulainen, P., and Hoogenraad C.C.. 2010. Actin in dendritic spines: connecting dynamics to function. J. Cell Biol. 189:619–629. 10.1083/jcb.201003008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, S., Exell J., and Welsher K.. 2020. Real-time 3D single molecule tracking. Nat. Commun. 11:3607. 10.1038/s41467-020-17444-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir, R.L., and Nicoll R.A.. 2013. AMPARs and synaptic plasticity: the last 25 years. Neuron. 80:704–717. 10.1016/j.neuron.2013.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau, Y., and Choquet D.. 2019. The next generation of approaches to investigate the link between synaptic plasticity and learning. Nat. Neurosci. 22:1536–1543. 10.1038/s41593-019-0480-6 [DOI] [PubMed] [Google Scholar]
- Jayant, K., Hirtz J.J., Plante I.J., Tsai D.M., De Boer W.D., Semonche A., Peterka D.S., Owen J.S., Sahin O., Shepard K.L., and Yuste R.. 2017. Targeted intracellular voltage recordings from dendritic spines using quantum-dot-coated nanopipettes. Nat. Nanotechnol. 12:335–342. 10.1038/nnano.2016.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, N. 2017. Adaptive optical fluorescence microscopy. Nat. Methods. 14:374–380. 10.1038/nmeth.4218 [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Xu B., Melnykov A., Genin G.M., and Elson E.L.. 2020. Fluorescence correlation spectroscopy and photon counting histograms in finite, bounded domains. Biophys. J. 119:265–273. 10.1016/j.bpj.2020.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis, E.D., Kang J.S., Shin T.W., Emenari A., Asano S., Lin L., Costa E.K., Marblestone A.H., Kasthuri N., and Boyden E.S.. 2019. Expansion microscopy of lipid membranes. BioRxiv. 10.1101/829903. (Preprint posted November 4, 2019). [DOI]
- Kasai, H., Fukuda M., Watanabe S., Hayashi-Takagi A., and Noguchi J.. 2010. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 33:121–129. 10.1016/j.tins.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Kashiwagi, Y., Higashi T., Obashi K., Sato Y., Komiyama N.H., Grant S.G.N., and Okabe S.. 2019. Computational geometry analysis of dendritic spines by structured illumination microscopy. Nat. Commun. 10:1285. 10.1038/s41467-019-09337-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrukha, E.A., Mikhaylova M., van Brakel H.X., van Bergen En Henegouwen P.M., Akhmanova A., Hoogenraad C.C., and Kapitein L.C.. 2017. Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Nat. Commun. 8:14772. 10.1038/ncomms14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus, C.J., Raghunathan K., DiBenedetto E., and Kenworthy A.K.. 2016. Analysis of diffusion in curved surfaces and its application to tubular membranes. Mol. Biol. Cell. 27:3937–3946. 10.1091/mbc.E16-06-0445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokolaki, M.L., Fauquier A., and Renner M.. 2020. Molecular crowding and diffusion-capture in synapses. iScience. 23:101382. 10.1016/j.isci.2020.101382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommaddi, R.P., Das D., Karunakaran S., Nanguneri S., Bapat D., Ray A., Shaw E., Bennett D.A., Nair D., and Ravindranath V.. 2018. Aβ mediates F-actin disassembly in dendritic spines leading to cognitive deficits in Alzheimer’s disease. J. Neurosci. 38:1085–1099. 10.1523/JNEUROSCI.2127-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova, F., and Svitkina T.. 2010. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell. 21:165–176. 10.1091/mbc.e09-07-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik, Y.D., Watson D.J., Cao G., Kuwajima M., and Harris K.M.. 2019. Structural plasticity of dendritic secretory compartments during LTP-induced synaptogenesis. eLife. 8:e46356. 10.7554/eLife.46356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters, R., Kapitein L.C., Hoogenraad C.C., and Storm C.. 2013. Shape-induced asymmetric diffusion in dendritic spines allows efficient synaptic AMPA receptor trapping. Biophys. J. 105:2743–2750. 10.1016/j.bpj.2013.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi, A., Fujiwara T.K., Chadda R., Xie M., Tsunoyama T.A., Kalay Z., Kasai R.S., and Suzuki K.G.. 2012. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 28:215–250. 10.1146/annurev-cellbio-100809-151736 [DOI] [PubMed] [Google Scholar]
- Kwon, T., Sakamoto M., Peterka D.S., and Yuste R.. 2017. Attenuation of synaptic potentials in dendritic spines. Cell Rep. 20:1100–1110. 10.1016/j.celrep.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzanò, L., Scipioni L., Di Bona M., Bianchini P., Bizzarri R., Cardarelli F., Diaspro A., and Vicidomini G.. 2017. Measurement of nanoscale three-dimensional diffusion in the interior of living cells by STED-FCS. Nat. Commun. 8:65. 10.1038/s41467-017-00117-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier, C. 2018. The axon initial segment: an updated viewpoint. J. Neurosci. 38:2135–2145. 10.1523/JNEUROSCI.1922-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Okamoto K., Hayashi Y., and Sheng M.. 2004. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 119:873–887. 10.1016/j.cell.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Li, T.P., Song Y., MacGillavry H.D., Blanpied T.A., and Raghavachari S.. 2016. Protein crowding within the postsynaptic density can impede the escape of membrane proteins. J. Neurosci. 36:4276–4295. 10.1523/JNEUROSCI.3154-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Zhang Q., Chou S.W., Newman Z., Turcotte R., Natan R., Dai Q., Isacoff E.Y., and Ji N.. 2020. Fast widefield imaging of neuronal structure and function with optical sectioning in vivo. Sci. Adv. 6:eaaz3870. 10.1126/sciadv.aaz3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Snapp E.L., and Phair R.D.. 2018. The development and enhancement of FRAP as a key tool for investigating protein dynamics. Biophys. J. 115:1146–1155. 10.1016/j.bpj.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H.E., MacGillavry H.D., Frost N.A., and Blanpied T.A.. 2014. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J. Neurosci. 34:7600–7610. 10.1523/JNEUROSCI.4364-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckner, M., Burgold S., Filser S., Scheungrab M., Niyaz Y., Hummel E., Wanner G., and Herms J.. 2018. Label-free 3D-CLEM using endogenous tissue landmarks. iScience. 6:92–101. 10.1016/j.isci.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry, H.D., and Hoogenraad C.C.. 2015. The internal architecture of dendritic spines revealed by super-resolution imaging: What did we learn so far? Exp. Cell Res. 335:180–186. 10.1016/j.yexcr.2015.02.024 [DOI] [PubMed] [Google Scholar]
- MacGillavry, H.D., Song Y., Raghavachari S., and Blanpied T.A.. 2013. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 78:615–622. 10.1016/j.neuron.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maco, B., Holtmaat A., Cantoni M., Kreshuk A., Straehle C.N., Hamprecht F.A., and Knott G.W.. 2013. Correlative in vivo 2 photon and focused ion beam scanning electron microscopy of cortical neurons. PLoS One. 8:e57405. 10.1371/journal.pone.0057405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, J.C., and Grienberger C.. 2020. Synaptic plasticity forms and functions. Annu. Rev. Neurosci. 43:95–117. 10.1146/annurev-neuro-090919-022842 [DOI] [PubMed] [Google Scholar]
- Manley, S., Gillette J.M., and Lippincott-Schwartz J.. 2010. Single-particle tracking photoactivated localization microscopy for mapping single-molecule dynamics. Methods Enzymol. 475:109–120. 10.1016/S0076-6879(10)75005-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., Ellis-Davies G.C., Nemoto T., Miyashita Y., Iino M., and Kasai H.. 2001. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4:1086–1092. 10.1038/nn736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., Honkura N., Ellis-Davies G.C., and Kasai H.. 2004. Structural basis of long-term potentiation in single dendritic spines. Nature. 429:761–766. 10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, D., Bonhoeffer T., and Scheuss V.. 2014. Balance and stability of synaptic structures during synaptic plasticity. Neuron. 82:430–443. 10.1016/j.neuron.2014.02.031 [DOI] [PubMed] [Google Scholar]
- Mikhaylova, M., Bär J., van Bommel B., Schätzle P., YuanXiang P., Raman R., Hradsky J., Konietzny A., Loktionov E.Y., Reddy P.P., et al. 2018. Caldendrin directly couples postsynaptic calcium signals to actin remodeling in dendritic spines. Neuron. 97:1110–1125.E14. 10.1016/j.neuron.2018.01.046 [DOI] [PubMed] [Google Scholar]
- Morise, J., Suzuki K.G.N., Kitagawa A., Wakazono Y., Takamiya K., Tsunoyama T.A., Nemoto Y.L., Takematsu H., Kusumi A., and Oka S.. 2019. AMPA receptors in the synapse turnover by monomer diffusion. Nat. Commun. 10:5245. 10.1038/s41467-019-13229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, J.B., Zaegel V., Coultrap S.J., Miller A.P., Bayer K.U., and Reichow S.L.. 2017. The CaMKII holoenzyme structure in activation-competent conformations. Nat. Commun. 8:15742. 10.1038/ncomms15742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägerl, U.V., Willig K.I., Hein B., Hell S.W., and Bonhoeffer T.. 2008. Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl. Acad. Sci. USA. 105:18982–18987. 10.1073/pnas.0810028105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, D., Hosy E., Petersen J.D., Constals A., Giannone G., Choquet D., and Sibarita J.B.. 2013. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33:13204–13224. 10.1523/JNEUROSCI.2381-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, C., Ritchie K., Oba Y., Nakamura M., Hotta Y., Iino R., Kasai R.S., Yamaguchi K., Fujiwara T., and Kusumi A.. 2003. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5:626–632. 10.1038/ncb1009 [DOI] [PubMed] [Google Scholar]
- Nakahata, Y., and Yasuda R.. 2018. Plasticity of spine structure: local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10:29. 10.3389/fnsyn.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, J., and Yasuda R.. 2015. Biochemical computation for spine structural plasticity. Neuron. 87:63–75. 10.1016/j.neuron.2015.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, J., Nagaoka A., Hayama T., Ucar H., Yagishita S., Takahashi N., and Kasai H.. 2019. Bidirectional in vivo structural dendritic spine plasticity revealed by two-photon glutamate uncaging in the mouse neocortex. Sci. Rep. 9:13922. 10.1038/s41598-019-50445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, I.L., Kraikivski P., and Slepchenko B.M.. 2009. Diffusion in cytoplasm: effects of excluded volume due to internal membranes and cytoskeletal structures. Biophys. J. 97:758–767. 10.1016/j.bpj.2009.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusser, Z., Lujan R., Laube G., Roberts J.D.B., Molnar E., and Somogyi P.. 1998. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 21:545–559. 10.1016/S0896-6273(00)80565-6 [DOI] [PubMed] [Google Scholar]
- Obashi, K., Matsuda A., Inoue Y., and Okabe S.. 2019. Precise temporal regulation of molecular diffusion within dendritic spines by actin polymers during structural plasticity. Cell Rep. 27:1503–1515.e8. 10.1016/j.celrep.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Oh, W.C., Hill T.C., and Zito K.. 2013. Synapse-specific and size-dependent mechanisms of spine structural plasticity accompanying synaptic weakening. Proc. Natl. Acad. Sci. USA. 110:E305–E312. 10.1073/pnas.1214705110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, W.C., Parajuli L.K., and Zito K.. 2015. Heterosynaptic structural plasticity on local dendritic segments of hippocampal CA1 neurons. Cell Rep. 10:162–169. 10.1016/j.celrep.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe, S. 2007. Molecular anatomy of the postsynaptic density. Mol. Cell. Neurosci. 34:503–518. 10.1016/j.mcn.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Okabe, S. 2020a. Recent advances in computational methods for measurement of dendritic spines imaged by light microscopy. Microscopy (Oxf.). 69:196–213. 10.1093/jmicro/dfaa016 [DOI] [PubMed] [Google Scholar]
- Okabe, S. 2020b. Regulation of actin dynamics in dendritic spines: Nanostructure, molecular mobility, and signaling mechanisms. Mol. Cell. Neurosci. 109:103564. 10.1016/j.mcn.2020.103564 [DOI] [PubMed] [Google Scholar]
- Perez-Alvarez, A., Yin S., Schulze C., Hammer J.A., Wagner W., and Oertner T.G.. 2020. Endoplasmic reticulum visits highly active spines and prevents runaway potentiation of synapses. Nat. Commun. 11:5083. 10.1038/s41467-020-18889-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, T., Poll S., Bancelin S., Angibaud J., Inavalli V.K., Keppler K., Mittag M., Fuhrmann M., and Nägerl U.V.. 2018. Chronic 2P-STED imaging reveals high turnover of dendritic spines in the hippocampus in vivo. eLife. 7:e34700. 10.7554/eLife.34700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic, M.A., Carnevale N., Rozsa B., and Zecevic D.. 2015. Electrical behaviour of dendritic spines as revealed by voltage imaging. Nat. Commun. 6:8436. 10.1038/ncomms9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, S.A., Raghavachari S., and Lew D.J.. 2015. Dendritic spine geometry can localize GTPase signaling in neurons. Mol. Biol. Cell. 26:4171–4181. 10.1091/mbc.E15-06-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetniak, S., Fernández-Busnadiego R., Müller M., Rizzoli S.O., and Tetzlaff C.. 2020a. Quantitative synaptic biology: a perspective on techniques, numbers and expectations. Int. J. Mol. Sci. 21:7298. 10.3390/ijms21197298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetniak, S., Ußling J.E., Perego E., Rammner B., Schikorski T., Fornasiero E.F., Truckenbrodt S., Köster S., and Rizzoli S.O.. 2020b. A comparative analysis of the mobility of 45 proteins in the synaptic bouton. EMBO J. 39:e104596. 10.15252/embj.2020104596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribrault, C., Sekimoto K., and Triller A.. 2011. From the stochasticity of molecular processes to the variability of synaptic transmission. Nat. Rev. Neurosci. 12:375–387. 10.1038/nrn3025 [DOI] [PubMed] [Google Scholar]
- Sala, C., and Segal M.. 2014. Dendritic spines: the locus of structural and functional plasticity. Physiol. Rev. 94:141–188. 10.1152/physrev.00012.2013 [DOI] [PubMed] [Google Scholar]
- Saneyoshi, T., Matsuno H., Suzuki A., Murakoshi H., Hedrick N.G., Agnello E., O’Connell R., Stratton M.M., Yasuda R., and Hayashi Y.. 2019. Reciprocal activation within a kinase-effector complex underlying persistence of structural LTP. Neuron. 102:1199–1210.e6. 10.1016/j.neuron.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, D., Kang J., Wassie A.T., Schroeder M.E., Peng Z., Tarr T.B., Tang A.-H., Niederst E., Young J.Z., Tsai L.-H., et al. 2020. Expansion revealing: decrowding proteins to unmask invisible brain nanostructures. BioRxiv. 10.1101/2020.08.29.273540. (Preprint posted September 26, 2020). [DOI]
- Schätzle, P., Esteves da Silva M., Tas R.P., Katrukha E.A., Hu H.Y., Wierenga C.J., Kapitein L.C., and Hoogenraad C.C.. 2018. Activity-dependent actin remodeling at the base of dendritic spines promotes microtubule entry. Curr. Biol. 28:2081–2093.e6. 10.1016/j.cub.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Sezgin, E., Levental I., Mayor S., and Eggeling C.. 2017. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18:361–374. 10.1038/nrm.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Tauzin L.J., Baiyasi R., Wang W., Moringo N., Shuang B., and Landes C.F.. 2017. Single particle tracking: from theory to biophysical applications. Chem. Rev. 117:7331–7376. 10.1021/acs.chemrev.6b00815 [DOI] [PubMed] [Google Scholar]
- Sheng, M., and Hoogenraad C.C.. 2007. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76:823–847. 10.1146/annurev.biochem.76.060805.160029 [DOI] [PubMed] [Google Scholar]
- Sidenstein, S.C., D’Este E., Böhm M.J., Danzl J.G., Belov V.N., and Hell S.W.. 2016. Multicolour multilevel STED nanoscopy of actin/spectrin organization at synapses. Sci. Rep. 6:26725. 10.1038/srep26725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal, Y.M., Zhou R., and Zhuang X.. 2018. Visualizing and discovering cellular structures with super-resolution microscopy. Science. 361:880–887. 10.1126/science.aau1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, C.M., Hepburn I., Chen W., and De Schutter E.. 2014. The role of dendritic spine morphology in the compartmentalization and delivery of surface receptors. J. Comput. Neurosci. 36:483–497. 10.1007/s10827-013-0482-4 [DOI] [PubMed] [Google Scholar]
- Smith, K.R., Kopeikina K.J., Fawcett-Patel J.M., Leaderbrand K., Gao R., Schürmann B., Myczek K., Radulovic J., Swanson G.T., and Penzes P.. 2014. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron. 84:399–415. 10.1016/j.neuron.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, A.H., Wang D., Chen G., Li Y., Luo J., Duan S., and Poo M.M.. 2009. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 136:1148–1160. 10.1016/j.cell.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Sun, Y., Smirnov M., Kamasawa N., and Yasuda R.. 2019. Rapid ultrastructural changes of PSD and extrasynaptic axon-spine interface membrane during LTP induced in single dendritic spine. BioRxiv. 10.1101/840629. (Preprint posted November 13, 2019). [DOI]
- Sun, D.E., Fan X., Shi Y., Zhang H., Huang Z., Cheng B., Tang Q., Li W., Zhu Y., Bai J., et al. 2021. Click-ExM enables expansion microscopy for all biomolecules. Nat. Methods. 18:107–113. 10.1038/s41592-020-01005-2 [DOI] [PubMed] [Google Scholar]
- Svoboda, K., Tank D.W., and Denk W.. 1996. Direct measurement of coupling between dendritic spines and shafts. Science. 272:716–719. 10.1126/science.272.5262.716 [DOI] [PubMed] [Google Scholar]
- Takasaki, K., and Sabatini B.L.. 2014. Super-resolution 2-photon microscopy reveals that the morphology of each dendritic spine correlates with diffusive but not synaptic properties. Front. Neuroanat. 8:29. 10.3389/fnana.2014.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada, H., Blanc J., Korogod N., Petersen C.C., and Knott G.W.. 2020. Ultrastructural comparison of dendritic spine morphology preserved with cryo and chemical fixation. eLife. 9:e56384. 10.7554/eLife.56384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, A.H., Chen H., Li T.P., Metzbower S.R., MacGillavry H.D., and Blanpied T.A.. 2016. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature. 536:210–214. 10.1038/nature19058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, C.-L., Liu Y.-T., Sun R., Zhang B., Qi L., Shivakoti S., Tian C.-L., Zhang P., Lau P.-M., Zhou Z.H., et al. 2018. Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. J Neurosci. 38:1493–1510. 10.1523/JNEUROSCI.1548-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraska, J.W. 2015. Cell biology of the future: Nanometer-scale cellular cartography. J. Cell Biol. 211:211–214. 10.1083/jcb.201508021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraska, J.W. 2019. A primer on resolving the nanoscale structure of the plasma membrane with light and electron microscopy. J. Gen. Physiol. 151:974–985. 10.1085/jgp.201812227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazerart, S., Mitchell D.E., Miranda-Rottmann S., and Araya R.. 2020. A spike-timing-dependent plasticity rule for dendritic spines. Nat. Commun. 11:4276. 10.1038/s41467-020-17861-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg, P.W., Chen F., Piatkevich K.D., Zhao Y., Yu C.C., English B.P., Gao L., Martorell A., Suk H.J., Yoshida F., et al. 2016. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34:987–992. 10.1038/nbt.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønnesen, J., and Nägerl U.V.. 2016. Dendritic spines as tunable regulators of synaptic signals. Front. Psychiatry. 7:101. 10.3389/fpsyt.2016.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønnesen, J., Katona G., Rózsa B., and Nägerl U.V.. 2014. Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17:678–685. 10.1038/nn.3682 [DOI] [PubMed] [Google Scholar]
- Trimble, W.S., and Grinstein S.. 2015. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol. 208:259–271. 10.1083/jcb.201410071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bommel, B., Konietzny A., Kobler O., Bär J., and Mikhaylova M.. 2019. F-actin patches associated with glutamatergic synapses control positioning of dendritic lysosomes. EMBO J. 38:e101183. 10.15252/embj.2018101183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela, J.A., Dupuis J.P., Etchepare L., Espana A., Cognet L., and Groc L.. 2016. Targeting neurotransmitter receptors with nanoparticles in vivo allows single-molecule tracking in acute brain slices. Nat. Commun. 7:10947. 10.1038/ncomms10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan, R., Rowan M.P., Lee C.T., Johnson G.R., Rangamani P., and Holst M.. 2020. Applications and challenges of machine learning to enable realistic cellular simulations. Front. Phys. 7:247. 10.3389/fphy.2019.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicidomini, G., Bianchini P., and Diaspro A.. 2018. STED super-resolved microscopy. Nat. Methods. 15:173–182. 10.1038/nmeth.4593 [DOI] [PubMed] [Google Scholar]
- Vlachos, A. 2012. Synaptopodin and the spine apparatus organelle-regulators of different forms of synaptic plasticity? Ann. Anat. 194:317–320. 10.1016/j.aanat.2011.10.014 [DOI] [PubMed] [Google Scholar]
- Wang, L., Dumoulin A., Renner M., Triller A., and Specht C.G.. 2016. The role of synaptopodin in membrane protein diffusion in the dendritic spine neck. PLoS One. 11:e0148310. 10.1371/journal.pone.0148310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassie, A.T., Zhao Y., and Boyden E.S.. 2019. Expansion microscopy: principles and uses in biological research. Nat. Methods. 16:33–41. 10.1038/s41592-018-0219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler, B., Forscher P., and Mellman I.. 1999. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 397:698–701. 10.1038/17806 [DOI] [PubMed] [Google Scholar]
- Wu, Y., and Shroff H.. 2018. Faster, sharper, and deeper: structured illumination microscopy for biological imaging. Nat. Methods. 15:1011–1019. 10.1038/s41592-018-0211-z [DOI] [PubMed] [Google Scholar]
- Wu, Y., Whiteus C., Xu C.S., Hayworth K.J., Weinberg R.J., Hess H.F., and De Camilli P.. 2017. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl. Acad. Sci. USA. 114:E4859–E4867. 10.1073/pnas.1701078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, L., Chen K., Yan R., Li W., and Xu K.. 2020. Single-molecule displacement mapping unveils nanoscale heterogeneities in intracellular diffusivity. Nat. Methods. 17:524–530. 10.1038/s41592-020-0793-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K., Zhong G., and Zhuang X.. 2013. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 339:452–456. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, R. 2012. Studying signal transduction in single dendritic spines. Cold Spring Harb. Perspect. Biol. 4:a005611. 10.1101/cshperspect.a005611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, R. 2017. Biophysics of biochemical signaling in dendritic spines: implications in synaptic plasticity. Biophys. J. 113:2152–2159. 10.1016/j.bpj.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste, R. 2013. Electrical compartmentalization in dendritic spines. Annu. Rev. Neurosci. 36:429–449. 10.1146/annurev-neuro-062111-150455 [DOI] [PubMed] [Google Scholar]
- Zaccard, C.R., Shapiro L., Martin-de-Saavedra M.D., Pratt C., Myczek K., Song A., Forrest M.P., and Penzes P.. 2020. Rapid 3D enhanced resolution microscopy reveals diversity in dendritic spinule dynamics, regulation, and function. Neuron. 107:522–537.e6. 10.1016/j.neuron.2020.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, G., He J., Zhou R., Lorenzo D., Babcock H.P., Bennett V., and Zhuang X.. 2014. Developmental mechanism of the periodic membrane skeleton in axons. eLife. 3:e04581. 10.7554/eLife.04581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Homma K.J., and Poo M.M.. 2004. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 44:749–757. 10.1016/j.neuron.2004.11.011 [DOI] [PubMed] [Google Scholar]