Abstract
Backgorund
Environmental stresses including abiotic stresses and biotic stresses limit yield of plants. Stress-tolerant breeding is an efficient way to improve plant yield under stress conditions. Genome editing by CRISPR/Cas9 can be used in molecular breeding to improve agronomic traits in crops, but in most cases, with fitness costs. The plant hormone ABA regulates plant responses to abiotic stresses via signaling transduction. We previously identified AITRs as a family of novel transcription factors that play a role in regulating plant responses to ABA and abiotic stresses. We found that abiotic stress tolerance was increased in the single, double and triple aitr mutants. However, it is unclear if the increased abiotic stress tolerance in the mutants may have fitness costs.
Results
We report here the characterization of AITRs as suitable candidate genes for CRISPR/Cas9 editing to improve plant stress tolerance. By using CRISPR/Cas9 to target AITR3 and AITR4 simultaneously in the aitr256 triple and aitr1256 quadruple mutants respectively, we generated Cas9-free aitr23456 quintuple and aitr123456 sextuple mutants. We found that reduced sensitivities to ABA and enhanced tolerance to drought and salt were observed in these mutants. Most importantly, plant growth and development was not affected even in the aitr123456 sextuple mutants, in whom the entire AITR family genes have been knocked out, and the aitr123456 sextuple mutants also showed a wild type response to the pathogen infection.
Conclusions
Our results suggest that knockout of the AITR family genes in Arabidopsis enhanced abiotic stress tolerance without fitness costs. Considering that knock-out a few AITRs will lead to enhanced abiotic stress tolerance, that AITRs are widely distributed in angiosperms with multiple encoding genes, AITRs may be targeted for molecular breeding to improve abiotic stress tolerance in plants including crops.
Keywords: ABA, Abiotic stress, AITRs, Arabidopsis, Gene editing, Fitness cost
Background
The world population is expected to reach 9 billions by the year 2050, as a consequence, a 70 % increase in crop yield are needed in order to feed the population by then [1, 2]. However, yield of plants including crops is largely affected by environmental stresses, including abiotic stresses such as drought, salinity and low and/or high temperatures, and biotic stresses such as pathogens and insects [3, 4]. It is estimated that biotic stresses cause an average ~ 20 % global yield loss for most major crops, whereas abiotic stresses, ~ 50 % [5, 6]. Among the abiotic stresses, drought and salinity are occurred in many different regions, and it is predicted that more than 50 % of all arable lands on the earth will be salinized seriously by the year 2050 [3]. Accelerated climate changes and global warming will aggravate the situation, and lead to a further yield loss for plants including crops.
Breeding for crops with enhanced tolerance to abiotic stresses will increase crop yield under stress conditions, however, traditional breeding takes quite a long time, and the results are usually unpredictable [7]. Molecular breeding, on the other hand is able to largely overcome these shortages [8], whereas new developed techniques, if used properly, may help to further shorten the breeding process.
Shortly after the discovery that the endonuclease Cas9 (CRISPR -associated protein 9) is able to cleave targeted double-stranded DNA in eukaryotic cells [9, 10], CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 has been developed as a new technique and used for targeted genome editing, and has been used successfully in plants [11–13]. Currently, there are two different types of CRIPSR/Cas9 genome editing systems, i.e., the DNA cleavage system, and the base editor system. The first allows small deletions or insertions of nucleotides at the target sites [14], whereas the second enables generation of precise single-nucleotide substitution [15]. Both of them have been used to generate mutations in plants, not only in the model plant Arabidopsis, but also in some major crops such as wheat and rice, at least in some of the cases, to improve agronomic traits [16–19]. CRISPR/Cas9 genome editing enables to generate predicted mutations, and transgene-free mutants can be isolated from genome edited transgenic plants, therefore can be used to accelerate plant breeding process [17, 19–22]. Recent improvement on the CRISPR/Cas9 systems, such as expanding target space by using Cas9 variants or engineered Cas9 further expanded it capacity for using in plants breeding [23, 24]. However, appreciate candidate genes that can be targeted by CRISPR/Cas9 editing to improve plant abiotic stress tolerance in crop breeding are largely unidentified.
The plant hormone ABA (abscisic acid) is a key stress hormone, it can regulates plant responses to abiotic stresses such as drought, salinity, cold and heat via signaling transduction [25–30]. Several different types of proteins including the receptor proteins PYR1 (Pyrabactin resistance 1)/PYL (PYR1-like)/ RCAR (Regulatory component of ABA receptor), the protein kinases PP2Cs (A-group PROTEIN PHOSPHATASE 2 C) phosphatases, the SnRK2s (NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASES), and the bZIP (basic region leucine zipper) transcription factors ABF/AREB/ABI5 function as key regulators in ABA signaling [26, 30–35]. Whereas several UBQ ligases can affect protein stability of the key regulators, therefore involve in regulating ABA signaling. As examples, the E2 ligase VPS23A and the E3 ligases CUL4 and RSL1 are able to target the PYR/PYL/RCABR receptors for proteasomal degradation [36–39], the E3 ligases KEG (KEEP ON GOING), DWA1 and DWA2 are able to target ABF/AREB/ABI5 transcription factors for degradation [40–43], and the E3 ligases PUB12 and PUB13 are able to target PP2C protein kinases for degradation [44]. Because ABA plays a key role in regulating plant abiotic stress responses, expression level changes in the ABA signaling regulator genes usually affect plant abiotic stress tolerance. For example, drought tolerance was reduced in the loss-of-function mutants of the ABF/AREB/ABI5 transcription factor genes or the SnRK2s genes [45, 46], whereas enhanced drought tolerance was observed in the transgenic plants overexpressing PYL genes [47, 48]. Therefore, gene editing of these regulator genes by CRISPR/Cas9 may not able be to improve abiotic tolerance in plants.
In previous report, we identified AITRs (ABA-induced transcription repressors) as an angiosperm conserved novel family of transcription factors, and we found that AITRs function as negative regulators in regulating ABA signaling and plant response to abiotic stresses [30]. We report here the characterization of AITRs as proper genome editing targets for improving plant abiotic stress tolerance. We generated Arabidopsis aitr mutants with all the six AITR genes being knocked out, and found that the mutants showed enhanced drought and salt tolerance, but plant growth and development, and plant response to pathogen infections remaining unaffected in the mutants.
Results
Generation of aitr mutants with all the AITR genes knocked out
We have previously identified AITRs as an angiosperm conserved novel family of transcription factors, and found that tolerance to ABA and abiotic stresses such as drought and salt was enhanced in the aitr mutant plants [30], indicating that AITRs may be targeted for molecular breeding to increase plant tolerance to abiotic stresses.
In Arabidopsis, there are a total of 6 genes encoding AITRs [30], to further examine the functions of AITRs in plant tolerance to abiotic stresses, and whether loss-of-function of AITRs may have fitness costs, we decided to generate high order aitr mutants with all the AITR genes knocked out. T-DNA insertion mutants were available for AITR1, AITR2, AITR5 and AITR6, in addition to the aitr2 aitr5 aitr6 (aitr256) triple mutant [30], we thus generated aitr1 aitr2 aitr5 aitr6 (aitr1256) quadruple mutant by crossing. We then generated aitr2 aitr3 aitr4 aitr5 aitr6 (aitr23456) quintuple and aitr1 aitr2 aitr3 aitr4 aitr5 aitr6 (aitr123456) sextuple mutants by using CRISPR/Cas9 to simultaneously target AITR3 and AITR4 in the aitr256 triple and aitr1256 quadruple mutants, respectively.
Two impendent Cas9-free homozygous aitr23456 quintuple mutant lines, i.e., aitr23456-c1 and aitr23456-c2, and two impendent Cas9-free homozygous aitr123456 sextuple mutant lines, i.e., aitr123456-c1 and aitr123456-c2, were generated. In all the four mutant lines, single nucleotide insertions were occurred at the target sites for both AITR3 and AITR4 (Fig. 1a), resulting in frame shift after the nucleotide insertion sites and premature stops of AITR3 and AITR4, respectively (Fig. 1b).
Fig. 1.
Generation of aitr23456 quintuple and aitr123456 sextuple mutants. a Alignment of the CRISPR/Cas9 target sequences of AITR3 and AITR4 in the Col wild type and the aitr23456 quintuple and aitr123456 sextuple mutants. The mutants were generated by transforming the aitr256 triple and aitr1256 quintuple T-DNA insertion mutant plants, respectively with a pHEE CRISPR/Cas9 construct that simultaneously targeting AITR3 and AITR4. Genome editing status in the selected T1 transgenic plants was examined by sequencing. Homozygous Cas9-free mutants were isolated from T2 or T3 progeny of the individual genome edited T1 plants. Numbers indicate the nucleotide position relative to the start condons of AITR3 and AITR4, respectively. Underlines indicate the PAM sites immediately after the target sites. b Amino acid sequence alignment of AITR3 and AITR4 in the Col wild type and the aitr23456 quintuple and aitr123456 sextuple mutants. The coding sequences of AITR3 and AITR4 in the aitr23456 quintuple and aitr123456 sextuple mutants were used for ORF analysis on ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/). Predicated amino acid sequences were used for alignment with wild type AITR3 and AITR4, respectively. Numbers indicate amino acid position relative to the first Met residue. Underlines indicate the fully or partial conserved LxLxL motif in AITRs
The aitr mutants are hyposensitivity to ABA
In both seed germination and cotyledon greening assays, reduced sensitivity to ABA was observed for all the single T-DNA insertion mutants of genes AITR1, AITR2, AITR5 and AITR6, and further reduced ABA sensitivity was observed in the double mutants aitr2 aitr5 (aitr25) and aitr2 aitr6 (aitr26), and triple mutant aitr256 [30], suggesting that AITRs may have redundant functions in regulating ABA responses.
Seed germination and cotyledon greening assays were used to examine ABA responses of the aitr higher order mutants generated. As shown in Fig. 2, in seed germination assays, all the mutants examined including the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants showed greatly reduced sensitivity to ABA at both the concentrations tested. However, little, if any difference on the germination rate was observed between the aitr123456 sextuple and the aitr256 triple mutants (Fig. 2).
Fig. 2.
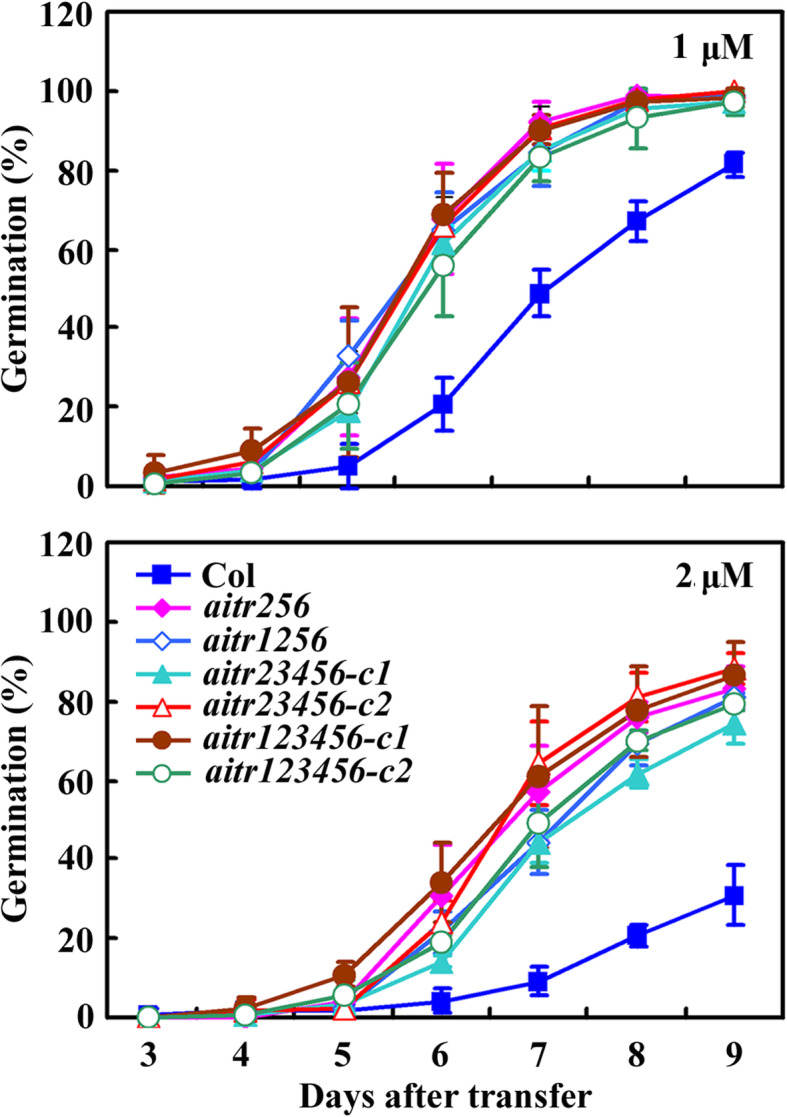
Effects of ABA on seed germination of the aitr mutants. Seeds of the Col wild type, the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were sterilized and sown on 1/2 MS plates in the presence or absence of 1 µM (up panel) or 2 µM ABA (low panel). The plates were transferred, after kept at 4 °C in darkness for 2 days, to a growth room. All the seeds on plates in the absence of ABA were germinated one day after the transfer. Seed germination on ABA-containing plates was examined daily after the transfer, germinated seeds were scored, and percentage of germination was calculated. The experiments were repeated three times, and similar results were obtained. Data represent the mean ± SD of three replicates
All the mutants examined also showed greatly reduced sensitivity to ABA in cotyledon greening assays (Fig. 3a). Almost all the seedlings of all the mutants produced green cotyledons at the presence of ABA, i.e., nearly 100 % green cotyledons for all the mutant seedlings, compared with ~ 50 % for the Col wild type seedlings (Fig. 3b).
Fig. 3.
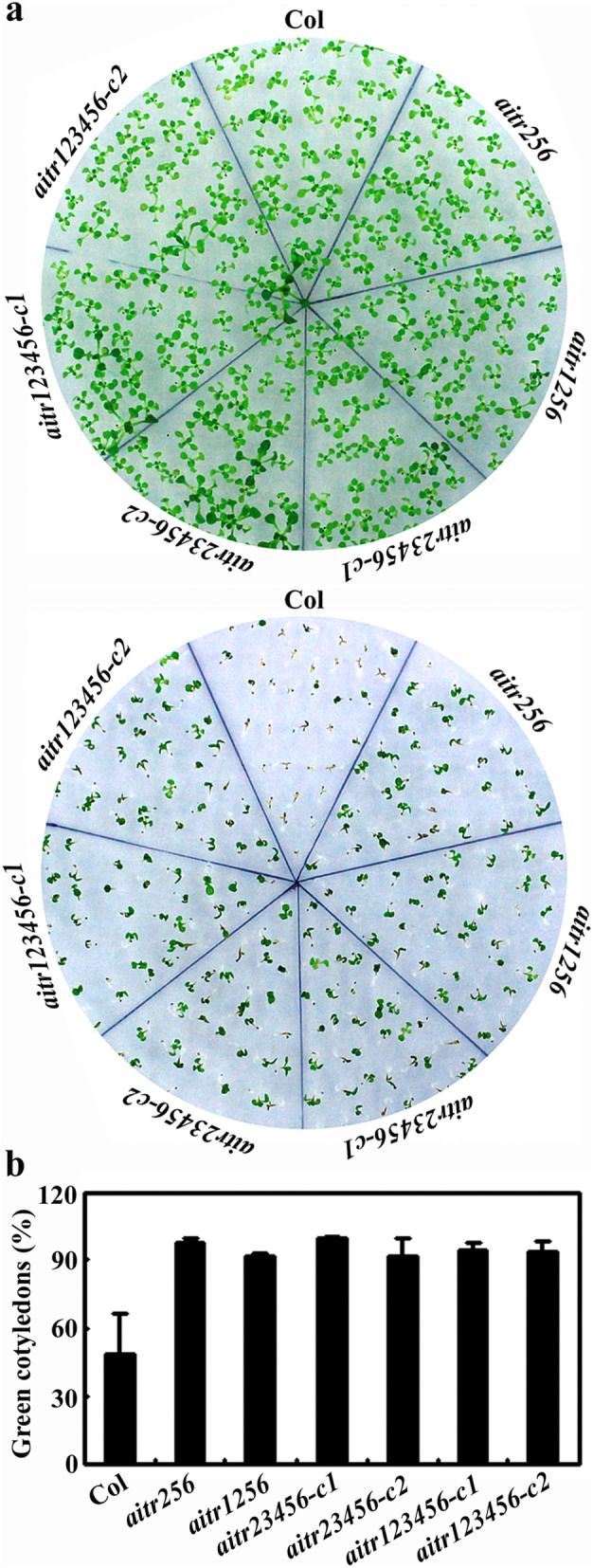
Effects of ABA on cotyledon greening of the aitr mutants. a Cotyledon greening of the Col wild type and the aitr mutants in response to ABA treatment. Seeds of the Col wild type, the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were sterilized and sown on 1/2 MS plates in the presence or absence of 2.5 µM ABA. The plates were transferred, after kept at 4 °C in darkness for 2 days, to a growth room. Pictures were taken 17 days after the transfer. b Quantitative assays of cotyledon greening of the Col wild type and the aitr mutants in response to ABA. Green seedlings were scored 17 days after the transfer, and percentage of green seedlings was calculated. The experiments were repeated three times, and similar results were obtained. Data represent the mean ± SD of three replicates
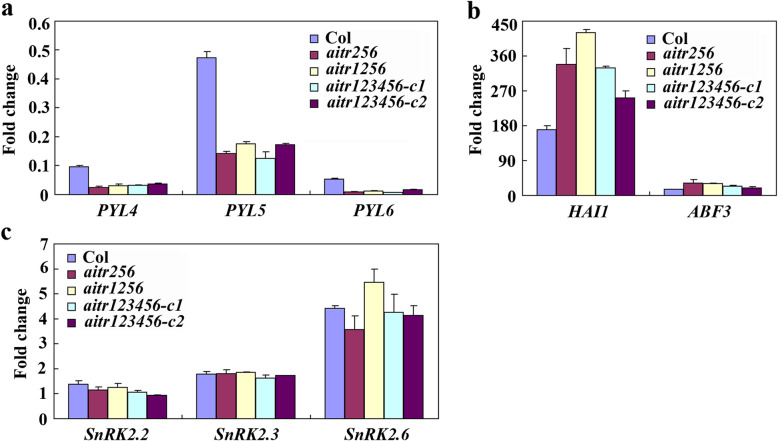
We then compared ABA response of the core ABA signaling regulator genes in the Col wild type, and the aitr256, aitr1256 and aitr123456 mutants. We found that the expression levels of the PYL receptor genes PYL4, PYL5 and PYL6 in response to ABA were further decreased in the aitr mutants when compare with the Col wild type seedlings, but no difference was observed between the aitr256, aitr1256 and aitr123456 mutants (Fig. 4a). In contrast, the expression levels of PP2C phosphatase gene HAI1 and ABF/AREB/ABI5 transcription factor gene ABF3 in response to ABA were further increased (Fig. 4b), whereas that of the SnRK2 kinase genes SnRK2.2, SnRK2.3 and SnRK2.6 in response to ABA remained largely unchanged (Fig. 4c).
Fig. 4.
ABA response of some ABA signaling component genes in the Col wild type and the aitr256, aitr1256 and aitr123456 mutants. a Fold changs of the PYR/PYL receptor gene PYL4, PYL5 and PYL6. b Fold changs of PP2C phosphatase gene HAI1 and ABF transcription factor gene ABF3. c Fold changs of SnRK2 kinase gene SnRK2.2, SnRK2.3 and SnRK2.6. Twelve-day-old seedlings of the Col wild type and the aitr256, aitr1256 and aitr123456 mutants were treated with 50 µM ABA, total RNA was isolated and quantitative RT-PCR was used to examine the expression of key regulator genes in ABA signaling. The expression of ACT2 was used as an inner control, and the fold changes were calculated by comparing the expression levels of the corresponding genes in ABA treated and un-treated samples. The experiments were repeated three times, and similar results were obtained. Data represent the mean ± SD of three replicates
The aitr mutant plants show enhanced tolerance to drought and salt stresses
Expression level changes of the regulator genes in ABA signaling usually affect plant abiotic stress tolerance [45–48]. That is also the case with AITRs, the aitr single, double and triple mutants examined showed enhanced drought and salt tolerance [30]. We examined abiotic stress tolerance in the high order aitr mutants generated. Soil grown mature plants were used for drought and salt treatments. As shown in Fig. 5, after re-watering, most of the mutant plants including the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were recovered, whereas all the Col wild type plants were not (Fig. 5).
Fig. 5.
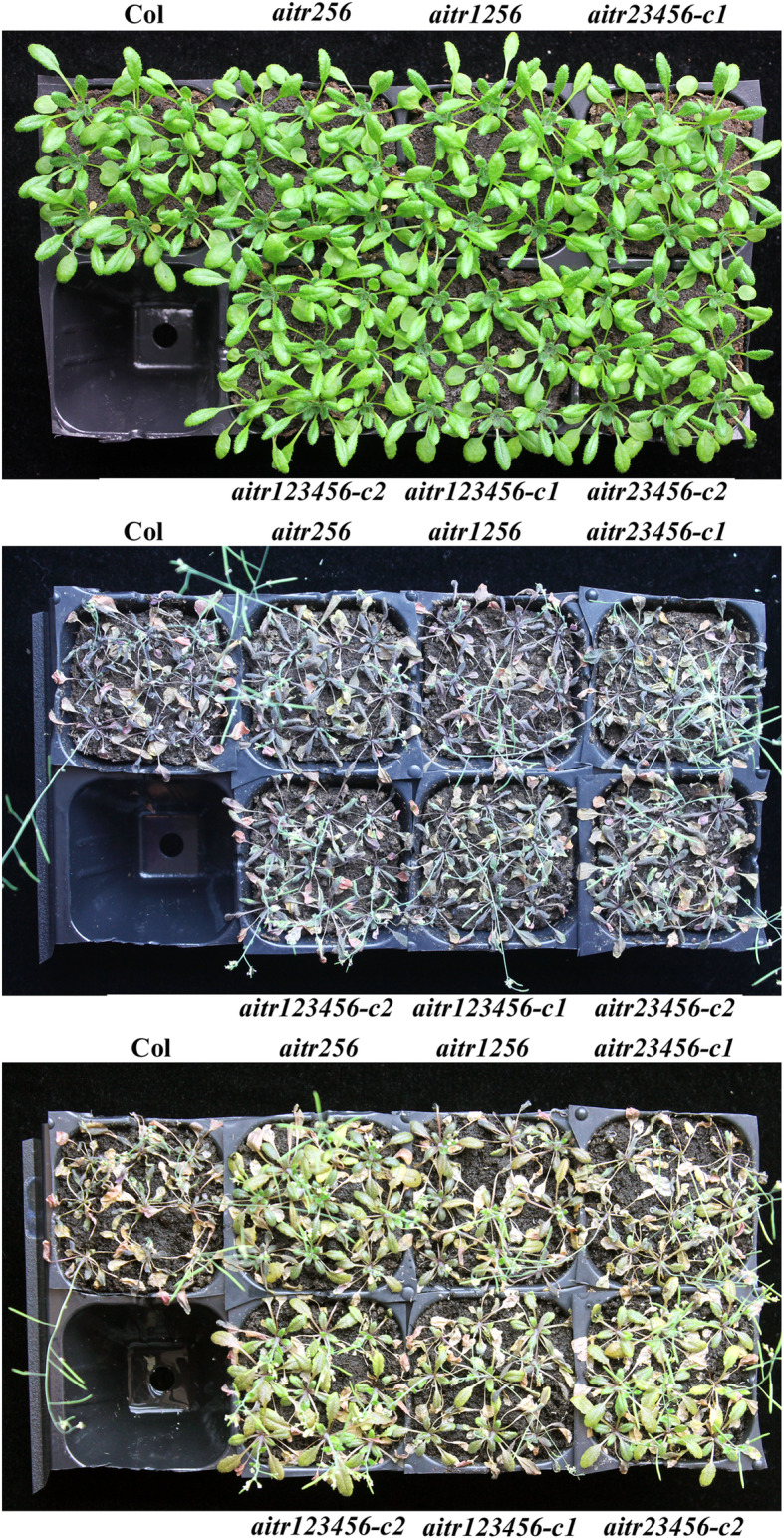
The aitr mutants are tolerant to drought treatment. Seeds of the Col wild type, the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth room with sufficient watering for 30 days. The plants were subjected to drought treatment by withholding watering for 12 days. Watering was then resumed. Pictures were taken before (up panel), and after drought treatment (middle panel), and 2 day after watering was resumed (low panel). The experiments were repeated three times, and similar results were obtained.
All the mutants are also showed enhanced tolerance to salt treatment. As shown in Fig. 6, salt treatment severely affected plant growth and development in the Col wild type plants, which failed to produce seeds. On the other hand, growth and development of the aitr mutants including the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants was less affected, as most of the mutant plants were still be able to produce seeds (Fig. 6).
Fig. 6.
The aitr mutants are tolerant to salt treatment. Seeds of the Col wild type, the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth room with sufficient watering for 30 days. The plants were then subjected to salt treatment by watering every other day with 150 mM NaCl for 10 times. Pictures were taken before treatment (up panel), and 1 day after the last salt watering (low panel). The experiments were repeated three times, and similar results were obtained
Growth and development are not affected in the aitr mutants
Enhanced stress tolerance usually accompanied by fitness costs on growth and development [49, 50]. The results that enhanced tolerance to abiotic stresses including drought and salt were observed in the aitr mutants indicate that AITRs may be targeted for plant breeding to enhance plant tolerance to abiotic stresses. However, fitness costs should be evaluated before AITRs can be targeted for plant breeding. We therefore examined plant growth and development of the aitr mutants under normal growth conditions.
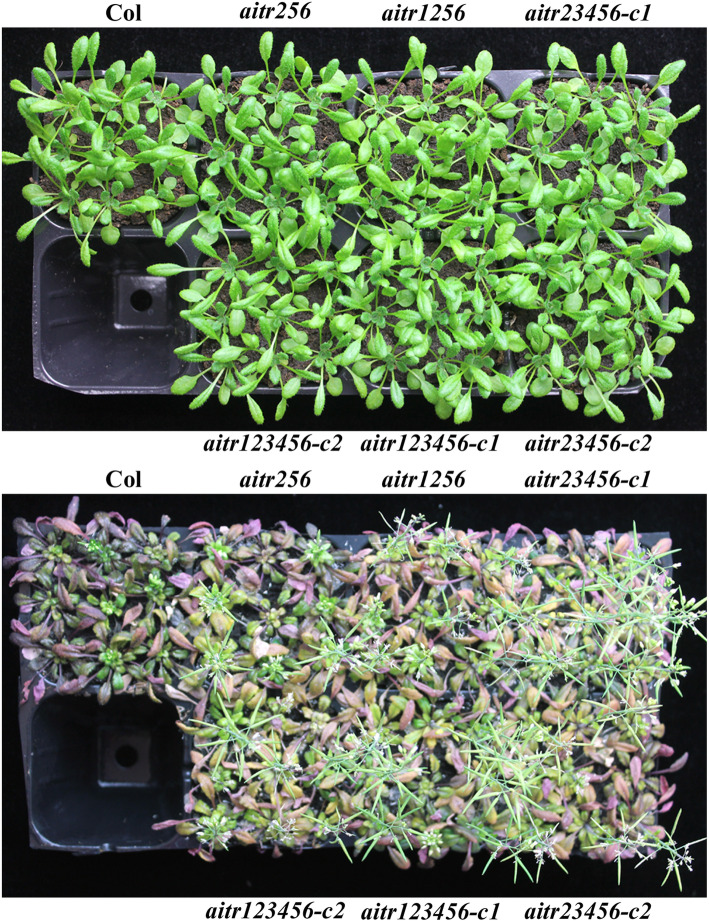
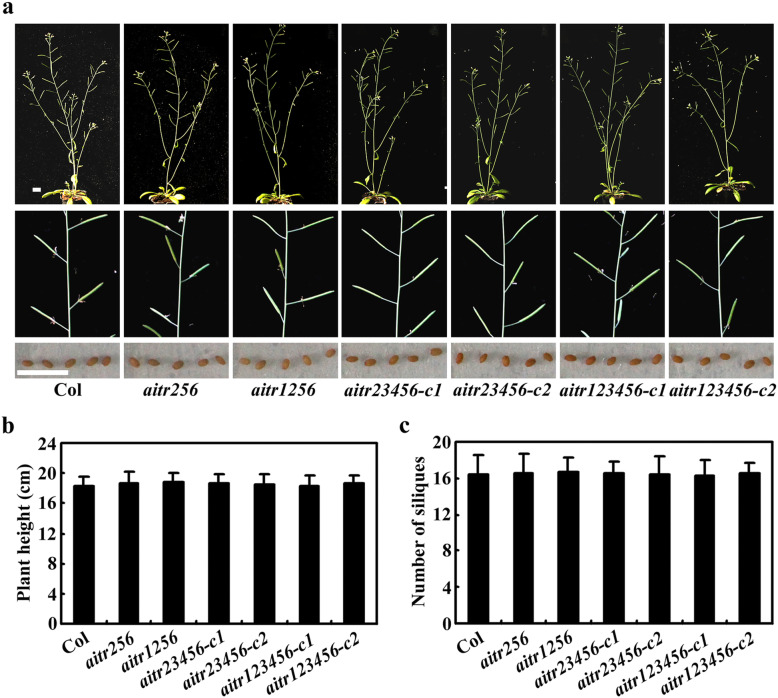
For direct comparison, the Col wild type and the aitr mutants including the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were geminated directly and grown in soil pots side by side in a growth room. No obviously different was found between the mutants and the Col wild type plants at all the growth stages during their whole life cycle, including vegetative growth, flowering and seed producing stages (Fig. 7a). Quantitive analysis results show that all the aitr mutants reached a same height as that of the Col wild type plants (Fig. 7b), and the numbers of the siliques produced by the plants were also similar (Fig. 7c).
Fig. 7.
Growth and development of the aitr mutants. a Plant morphology of the Col wild type and the aitr mutants. Up panel, 7-week-old plants, middle panel, main inflorescence stem sliliques, low panel, seeds. b Height of the Col wild type plants and the aitr mutants. c Number of siliques on the main inflorescence stems of the Col wild type and the aitr mutants. Seeds of the Col wild type, the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were generated directly and grown in soil pots in a growth room. Pictures for 7-week-old whole plants and the main inflorescences were taken. Height for 12-week-old plants was measured, and numbers of siliques on the main inflorescence stems were counted. The experiments were repeated three times, and similar results were obtained. Data represent the mean ± SD of 12 plants
The aitr mutants show a wild type response to pathogen infection
Accumulated experiment evidence suggest that ABA can also plays a role in regulating biotic stress tolerance [51–53], and trade-offs between biotic and abiotic stress responses were observed [54, 55]. Having shown that AITRs are able to regulate ABA response and enhanced tolerance to abiotic stresses including drought and salt were observed in the aitr mutants, we examined if plant response to pathogen infection may be affected in the aitr mutants.
The Col wild type plants and the aitr mutants were challenged with the virulent bacterial pathogen Pseudomonas syringae pv tomato (Pto) DC3000, Pto DC3000 hrcC− and Pto DC3000 AvrRpt2, respectively, and growth of the bacterial was examined. As shown in Fig. 8, all the aitr mutants showed a response similar to that in the Col wild type plants.
Fig. 8.
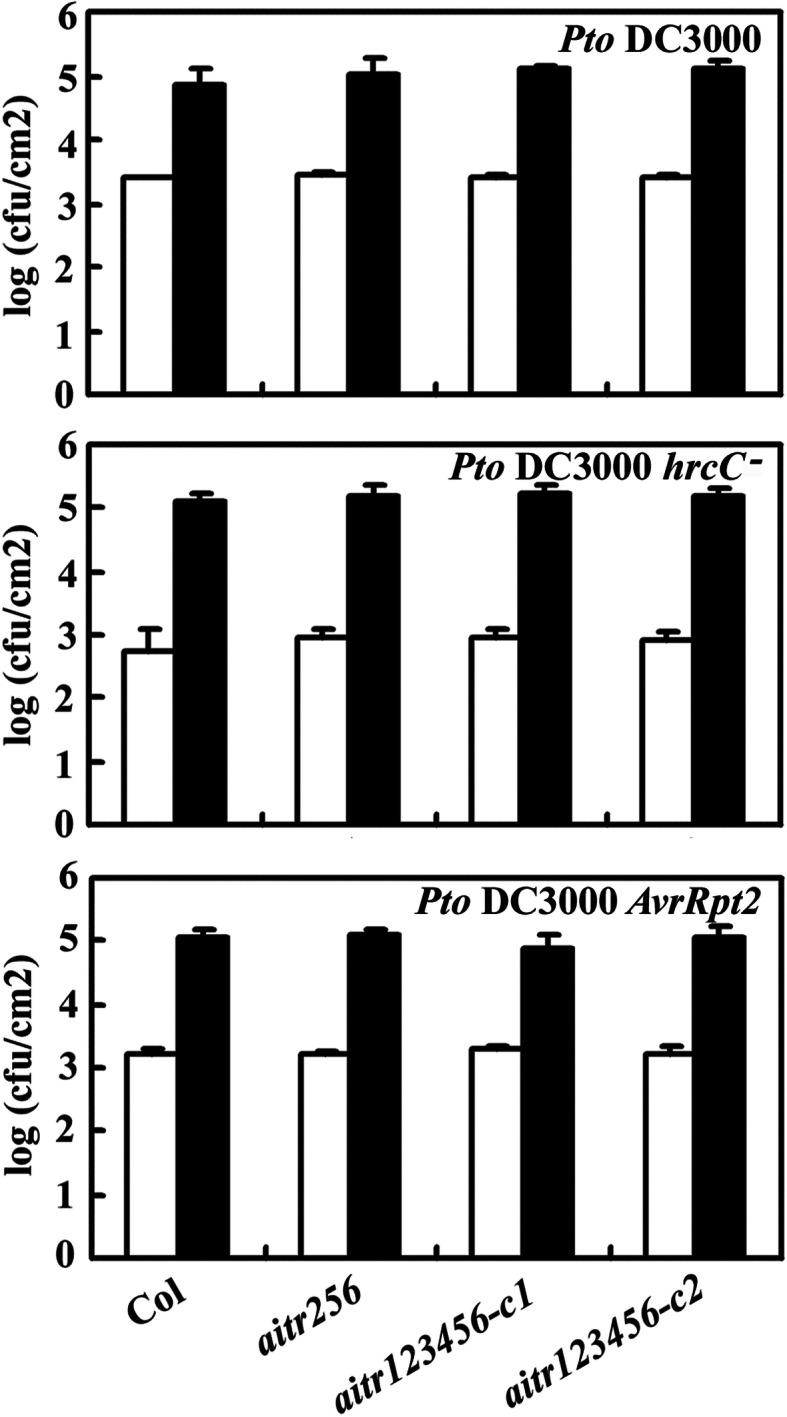
Response of the aitr mutants to pathogen infection. Seeds of the Col wild type and the aitr256 triple and aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth chamber at short-day condition. Plants about four-week-old were infiltrated with Pto DC3000, Pto DC3000 hrcC− and Pto DC3000 AvrRpt2 respectively. Samples were collected at day 0 and day 3, and bacterial growth was examined. The experiments were repeated twice, and similar results were obtained. Data represent the mean ± SD of 4 samples
Discussion
Genome editing by CRISPR/Cas9 is able to generate transgene-free mutants in different plant species, and has been successfully used to improve important agronomic traits in several different crops [17, 20–22, 56]. However, identification of suitable candidate genes is a challenge for using CRISPR/Cas9 genome editing techniques in plant breeding. We provide evidence in this study that AITR genes maybe targeted for genome editing to improve plant tolerance to abiotic stresses without compromising their agronomic performance.
Environmental stresses, especially abiotic stresses such drought and salt are a global problem causing yield lost in plants including the most important crops [5, 6]. As a key stress hormone, ABA regulates plant tolerance to abiotic stresses [25–30]. As a result, changes the expression levels of the core regulator genes in ABA signaling usually affected plant responses to abiotic stresses. As examples, manipulation of the expression of the PYL receptor genes, the SnRK2 protein kinase genes, and/or the ABF/AREB transcription factor genes affected drought tolerance in Arabidopsis [45, 46, 48]. However, enhanced abiotic tolerance are commonly observed in plants overexpressing these core regulator genes, whereas loss-of-function mutation usually led to decreased abiotic stress tolerance [45, 46, 48]. Therefore it is unlikely that these core regulator genes can be targeted by CRISPR/Cas9 for genome editing to enhance plant tolerance to abiotic stresses.
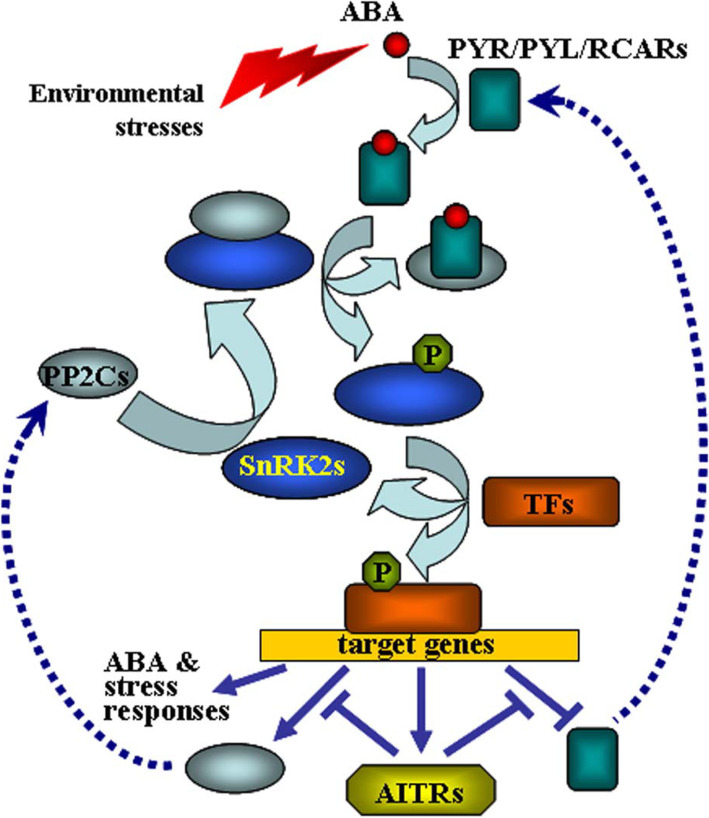
We have previously found that AITRs, a family of novel transcription factors function as feedback regulators in ABA signaling and plant response to abiotic stresses (Fig. 9), and loss-of-function of AITRs is able to enhance abiotic tolerance in Arabidopsis [30]. By using CRISPR/Cas9 to edit AITR3 and AITR4 in T-DNA insertation mutants aitr256 and aitr1256, respectively, we generated aitr23456 quintuple and aitr123456 sextuple mutants (Fig. 1). We found that, in both seed germination and green cotyledon assays, all the aitr higher order mutants showed hyposensitivity to ABA (Figs. 2 and 3), and enhanced tolerance to drought and salt treatments (Figs. 5 and 6). However, when compared with the aitr256 triple mutant, little if any enhanced ABA and abiotic stress tolerance were observed in the high order mutants including the aitr123456 sextuple mutants, in which all the 6 AITR genes have been knocked out. Consistent with these observation, ABA response of the PYL receptor genes PYL4, PYL5 and PYL6, the PP2C phosphatase gene HAI1 and ABF/AREB/ABI5 transcription factor gene ABF3, and the SnRK2 kinase genes SnRK2.2, SnRK2.3 and SnRK2.6 in response to ABA remained largely similar in the aitr123456 sextuple mutants and the aitr256 triple and the aitr1256 quadruple mutants (Fig. 4). This is likely because the basal expression levels of AITRs were very low [30], loss-of-function of AITR2, AITR5 and AITR6 may have already reached the expression level threshold where highest degree of redundancy functions of AITRs can be achieved. Nevertheless, these results suggest that AITRs could be good candidate genes for CRISPR/Cas9 genome editing to improve plant abiotic stress tolerance trait.
Fig. 9.
A model of action of AITRs in ABA and abiotic stress response. In the absent of ABA, PP2C phosphatases bind and deactivate SnRK2 kinases. Environmental stimuli and developmental cues promote ABA biosynthesis. ABA binds to PYR/PYL/RCAR receptors and suppresses the activity of PP2Cs. SnRK2s thus become activated, and they, in turn, phosphorylate down-stream transcription factors (TFs), resulting in the activation or repression of their target genes, including PP2Cs, PYR/PYL/RCARs and AITRs. Activation of PP2Cs and repression of PYR/PYL/RCARs represent negative feedback regulatory loops. AITRs regulate the expression of PP2Cs and PYR/PYL/RCARs, therefore function as feed back regulators in ABA signaling and abiotic stress responses
In order to survive under stress conditions, plants usually make trade-off between growth and development and stress tolerance, as a result, enhanced tolerance usually accompanied by a fitness penalty [49, 50, 57]. Therefore it is a challenge to improve plant stress tolerance without fitness costs. So far a few approaches have been used to overcome this problem. As examples, by using a pathogen-inducible promoter to drive the expression of IPA1(Ideal Plant Architecture 1), a transcription factor gene involved in the regulation of yield and immunity response in rice [58], enhanced disease resistance and increased yield were obtained in the transgenic rice plants [57]. By using a uORF (upstream open reading frame)-containing immune-inducible promoter to drive the expression of key immune regulator genes, improved disease resistance with no fitness costs was observed in Arabidopsis and rice [50]. However, these strategies are still relied on the generation of transgenic plants.
Recently, It has been reported MYC2-TARGETED BHLH1 (MTB1), MTB2 and MTB3, three JA (jasmonate)-inducible bHLH transcription factors functioned as negative regulators of JA mediated plant biotic stress response [59]. The mtb mutants generated by CRISPR/Cas9 genome editing showed enhance resistance to insect attack, but overall growth and development of the mutants was not affected [59]. To our knowledge, MTBs may be the first gene family identified so far that can be targeted by CRISPR/Cas9 to improve biotic stress tolerance in plants without fitness costs. Our results showed that AITRs functioned as negative regulators of ABA mediated plant abiotic stress response (Figs. 5 and 6). Knockout of all the 6 AITR genes did not affect plant growth and development (Fig. 7), and plant response to pathogen infection was also not affected in the aitr123456 sextuple mutants (Fig. 8). These results suggest that AITRs may represent the first gene family identified so far that can be targeted by CRISPR/Cas9 to improve abiotic stress tolerance in plants without fitness costs.
Considering that AITRs are only presented in angiosperms including all the crop species, and AITRs examined in other plants such as soybean and tomato shared similar features with the Arabidopsis AITRs [30], it is likely that AITRs can be targeted for genome editing to improve abitic stress tolerance in crops without worrying about fitness costs. Considering that both ABA and JA signaling pathways are conserved in plants, it may be interesting to examine if simultaneously knock out of AITRs and MTBs may enhance biotic and abiotic stresses in crops. Considering that current crops varieties grown in different areas may have different desired agronomic traits such as high yield and good quality, but showed low tolerance to abiotic stresses, enome editing of AITR genes in these crops may enhance their abiotic stress tolerance without affecting other traits, therefore integrate the desired agronomic traits in crops. Most crops have multiple AITR genes [30], it may not easy to knockout all the AITR genes in a crop. However, our observation showed that there is no significant difference between the aitr256 triple mutants and aitr123456 quadruple mutants in response to abiotic stresses, suggest that knocking out a few AITR genes knockout may sufficient to improve abiotic stress tolerance in crops, which makes the editing of AITRs to improve abiotic stress tolerance in crops more practicable.
Conclusions
In summary, we found that knock-out of a few AITR genes in Arabidopsis are sufficient to enhance abiotic stress tolerance, and knock-out the entitle family of AITR genes do not have fitness costs. Because AITRs family is conserved in crops with multiple encoding genes, AITRs may be good candidate genes for molecular breeding to improve abiotic stress tolerance in plants. This discovery is likely to usher a new wave of manipulation of plant abiotic stress tolerance in agricultural settings via CRISPR/Cas9 genome editing based molecular breeding.
Methods
Plant materials and growth conditions
The Col ecotype Arabidopsis stored in our laboratory was used as wild type, and the mutants were all in the Col ecotype background. The aitr1 single and the aitr2 aitr5 aitr6 (aitr256) triple mutants stored in our laboratory were as described previously [30]. The aitr1 aitr2 aitr5 aitr6 (aitr1256) quadruple mutant was generated by crossing the aitr1 single mutant and aitr2 aitr5 aitr6 triple mutant, and genotyping the F2 progeny.
Arabidopsis seedlings used for ABA or abiotic treatments and RNA isolation were germinated on ½ MS plates. Arabidopsis plants used for genome editing, phenotypic observation and pathogen infection were germinated directly and grown in soil pots. All the plants were grown in a growth room with growth conditions described previously [30, 60], except that Arabidopsis plants used for pathogen infection were grown in a growth chamber at short-day conditions. Since the plants were grown by ourselves, we have all the right to collect the plants materials.
Construct
CRISPR/Cas9 genome editing construct for AITR3 and AITR4 editing was generated using the pHEE401E vector. Appropriate target sites on the genome sequences of the single exon of AITR3 and AITR4 were identify on CRISPRscan (www.crisprscan.org), and then evaluated on Cas-OFFinder (www.rgenome.net/cas-offinder/). The target sequences used for AITR3 and AITR4 editing were 5’-GGGTAAACCGGGCCTACCGG(AGG)-3’, and 5’- TGGTTAACGAGGCTTACCGG(AGG)-3’, respectively. The CRISPR/Cas9 construct was generated by following a procedure described previously [61]. The primers used to insert the target sequences into the pHEE401E vector were, AITR3-DT1-BsF, 5’-ATATATGGTCTCGATTGGGTAAACCGGGCCTACCGGGTT-3’, AITR3-DTI-F0, 5’-TGGGTAAACCGGGCCTACCGGGTTTTAGAGCTAGAAATAGC-3’, AITR4-DT2-R0, 5-AACCCGGTAAGCCTCGTTAACCCAATCTCTTAGTCGACTCTAC-3’, and AITR4-DT2-BsR, 5’-ATTATTGGTCTCGAAACCCGGTAAGCCTCGTTAACCC-3’. The U626-IDF and U629-IDR primers used for clone PCR and sequencing of the sgRNA expression cassette in the CRISPR/Cas9 construct were described previously [62].
Plant transformation, transgenic plant identification and Cas9-free mutant isolation
About 5-week-old aitr256 triple and aitr1256 quadruple mutant plants, when several mature flowers were produced on the main inflorescence stems, were used for transformation. Plants were transformed with the CRISPR/Cas9 construct generated by using the floral dip method [63]. To select transgenic plants, collected T1 seeds from the transformed plants were plated on 1/2 MS plates conaining 30 µg/ml hygromycin and 50 µg/ml carbenicillin. To examine gene editing status in the transgenic plants, genome sequences of AITR3 and AITR4 in T1 plants were amplified and sequenced. To select Cas9-free homozygous mutants, T2 progeny of confirmed gene edited T1 plants were germinated directly in soil pots, and gene editing status and absence of Cas9 were examined. Two confirmed independent homozygous Cas9-free mutants were used for the experiments.
DNA isolation and PCR
To examine genome editing status of AITR3 and AITR4, leaves of the T1 transgenic plants or T2 progeny of individual gene edited T1 plants were collected, and used for DNA isolation. Isolated DNA was used for PCR amplification of the genome sequence of AITR3 and AITR4, respectively. PCR products was isolated and sent for sequencing, and sequencing results was aligned with the wild type sequences of AITR3 and AITR4, respectively. To isolate Cas9-free mutants, leaves of the T2 progeny of individual gene edited T1 plants were collected, DNA was isolated and used to amply Cas9 by PCR. The primers used for amplification of AITR3 were, AITR3-MF, 5’-AATGGAGATAAAGCTGGTGAGT-3’ and AITR3-R, 5’- TCACATGCCAAGCCTTAGAG-3’. For AITR4 were, AITR4-MF, 5’- TGGAGTCCGTTAACAGTGG-3’ and AITR4-R, 5’-TCAAAGCCAAAGAGT-3’. The Cas9-F and Cas9-R primers used for amplification of Cas9 were described previously [62].
RNA isolation and quantitative RT-PCR (qRT-PCR)
Twelve-day-old seedlings of the Col wild type, the aitr256, aitr1256 and aitr123456 mutants were used for ABA treatment as described previously [30]. Total RNA was isolated by using an EasyPure Plant RNA Kit (Transgene), and 1 µg RNA was subjected to cDNA synthesis by using an EazyScript First-Strand DNA Synthesis Super Mix Kit (TransGen Biotech).
The reponse of ABA signaling component genes in response to ABA treatment were examined by using qRT-PCR, and calculated as described previously [30]. All the primers used were as repotted previously [30, 64–67].
ABA sensitivity analysis
Seed germination and cotyledon greening assays of ABA sensitivity were performed as described by previously [68]. Briefly, seeds of the Col wild type, the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were sterilized and plated on 1/2 MS plates with or without indicated concentrations of ABA. After kept in darkness at 4 °C for 2 days, the plates were transferred to a growth room.
For seed germination assays, seed germinated was examined and counted under a dissection microscopy daily after the transfer, and germination rate was calculated. For cotyledon greening assays, pictures of the seedlings were taken 17 days after the transfer, green seedlings were counted, and percentage of green seedlings was calculated.
Drought tolerance analysis
Drought tolerance was assayed as previously described [30] with modifications. In brief, the seeds of the Col wild type and the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth room for 30 days with normal watering. Pictures were taken and the plants were subjected to drought treated for 12 days by withholding watering. Pictures were taken and watering was resumed after drought treatment. Pictures were taken again 2 day after watering was resumed.
Salt tolerance analysis
Salt tolerance of the aitr mutants was assayed as previously described [30] with modifications. In brief, seeds of the Col wild type and the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth room for 30 days with sufficient watering. Pictures were taken and the plants were then watered every other day with 150 mM NaCl for 10 times. Pictures were taken after salt treatment.
Pathogen infection assays
For pathogen infection assays, seeds of the Col wild type and the aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth chamber at short-day condition. Plants ~ 4-week-old were infiltrated and bacterial growth was assayed as described previously [69]. The pathogens used for infiltration and their corresponding concentrations were Pseudomonas syringae pv tomato (Pto) DC3000, Pto DC3000 hrcC− and Pto DC3000 AvrRpt2 at a dose of OD600 = 0.0002, 0.001 and 0.001, respectively.
Morphological assays
For plant growth and development assays, seeds of the Col wild type and the aitr256 triple, aitr1256 quadruple, aitr23456 quintuple and aitr123456 sextuple mutants were germinated directly and grown in soil pots in a growth room. Morphology of the plants at different growth stages was examined. Pictures for the plants and inflorescences at indicated growth stages were taken by using a digital camera, and pictures for seeds were taken under a dissection microscopy equipped with a digital camera.
Acknowledgements
We thank all the lab members for their help discussion.
Abbreviations
- ABA
Abscisic acid
- AITRs
ABA-induced transcription repressors
- bZIP
Basic region leucine zipper
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats/CRISPR -associated protein 9
- IPA1
Ideal Plant Architecture 1
- JA
Jasmonate
- KEG
KEEP ON GOING
- MTB1
MYC2-TARGETED BHLH1
- PP2Cs
PROTEIN PHOSPHATASE 2C
- PYL
PYR1-like
- PYR1
Pyrabactin resistance 1
- RCAR
Regulatory component of ABA receptor
- SnRKs
NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASES
- uORF
Upstream open reading frame
Authors’ contributions
SW conceived the study. SC, HT and SW designed the experiments. SC, NZ, GZ, SH, SA and HT performed the experiments. SC, HT and SW analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32071938), the National Key R&D Program of China (2016YFD0101900), and a startup funding from Linyi University ( LYDX2019BS039). The funders have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hainan Tian, Email: tianhainan2012@126.com.
Shucai Wang, Email: wangshucai@yahoo.com.
References
- 1.Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS One. 2013;8:e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanlliyodan B, Ye H, Song L, Murphy M, Shannon JG, Nguyen HT. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J Exp Bot. 2017;68:1835–49. doi: 10.1093/jxb/erw433. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 4.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Bio. 2006;9:436–42. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Boyer JS. Plant productivity and environment. Science. 1982;218:443–8. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh D, Xu J. Abiotic stress responses in plant roots: a proteomics perspective. Front Plant Sci. 2014;5:6. doi: 10.3389/fpls.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manavalan LP, Guttikonda SK, Tran LS, Nguyen HT. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50:1260–76. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Lu Y, Xie C, Gao S, Wan J, Prasanna BM. Whole-genome strategies for marker-assisted plant breeding. Mol Breeding. 2012;29:833–54. doi: 10.1007/s11032-012-9699-6. [DOI] [Google Scholar]
- 9.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–91. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–3. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 13.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–8. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 14.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–9. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19:770–88. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441–3. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant. 2018;11:1210–3. doi: 10.1016/j.molp.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, et al. De novo domestication of wild tomato using genome editing. Nat Biotechnol. 2018;36:1211–6. doi: 10.1038/nbt.4272. [DOI] [PubMed] [Google Scholar]
- 19.Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–97. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;1274–1284. [DOI] [PubMed]
- 21.Gao X, Chen J, Dai X, Zhang D, Zhao Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by RISPR/Cas9-mediated genome editing. Plant Physiol. 2016;171:1794–800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu HP, Liu SM, Xu SL, Chen WY, Zhou X, Tan YY, et al. CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol J. 2017;15:1371–3. doi: 10.1111/pbi.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–62. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–5. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–39. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–9. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, et al. A transcription factor hierarchy defines an environmental stress response network Science. 2016;354:aag1550. doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian H, Chen S, Yang W, Wang T, Zheng K, Wang Y, et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 2017;40:2958–71. doi: 10.1111/pce.13058. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez PL, Leube MP, Grill E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2 C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol. 1998;38:879–83. doi: 10.1023/A:1006012218704. [DOI] [PubMed] [Google Scholar]
- 32.Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2 C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–94. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, Yang X, Weston DJ, Chen JG. Abscisic acid receptors: past, present and future. J Integr Plant Biol. 2011;53:469–79. doi: 10.1111/j.1744-7909.2011.01044.x. [DOI] [PubMed] [Google Scholar]
- 35.Dong T, Park Y, Hwang I. Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015;58:29–48. doi: 10.1042/bse0580029. [DOI] [PubMed] [Google Scholar]
- 36.Bueso E, Rodriguez L, Lorenzo-Orts L, Gonzalez-Guzman M, Sayas E, Muñoz-Bertomeu J, et al. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014;80:1057–71. doi: 10.1111/tpj.12708. [DOI] [PubMed] [Google Scholar]
- 37.Seo KI, Lee JH, Nezames CD, Zhong S, Song E, Byun MO, et al. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell. 2014;26:695–711. doi: 10.1105/tpc.113.119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu F, Lou L, Tian M, Li Q, Ding Y, Cao X, et al. ESCRT-I Component VPS23A Affects ABA Signaling by Recognizing ABA Receptors for Endosomal Degradation. Mol Plant. 2016;9:1570–82. doi: 10.1016/j.molp.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Belda-Palazon B, Rodriguez L, Fernandez MA, Castillo MC, Anderson EA, Gao C, et al. FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell. 2016;28:2291–311. doi: 10.1105/tpc.16.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–28. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YT, Liu HX, Stone S, Callis J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013;75:965–76. doi: 10.1111/tpj.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Stone SL. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell. 2010;22:2630–41. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H, Stone SL. Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J Biol Chem. 2013;288:20267–79. doi: 10.1074/jbc.M113.465369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong L, Cheng J, Zhu Y, DingY, Meng J, Chen Z, et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun. 2015;6:8630. doi: 10.1038/ncomms9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–32. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signaling in response to osmotic stress. Plant Cell Environ. 2015;38:35–49. doi: 10.1111/pce.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015;520:545–8. doi: 10.1038/nature14123. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA. 2016;113:1949–54. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol. 2005;23:283–90. doi: 10.1016/j.tibtech.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545:91–494. doi: 10.1038/nature22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alzaem M, He MH, Moffett P, Lin NS. Abscisic acid induces resistance against Bamboo Mosaic Virus through Argonaute 2 and 3. Plant Physiol. 2017;174:339–55. doi: 10.1104/pp.16.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alzaem M, Lin NS. Antiviral roles of abscisic acid in plants. Front Plant Sci. 2017;8:1760. doi: 10.3389/fpls.2017.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xun H, Yang X, He H, Wang M, Guo P, Wang Y, et al. Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol Biol. 2019;99:95–111. doi: 10.1007/s11103-018-0804-z. [DOI] [PubMed] [Google Scholar]
- 54.Berens ML, Wolinska KW, Spaepen S, Ziegler J, Nobori T, Nair A, et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc Natl Acad Sci USA. 2019;116:2364–73. doi: 10.1073/pnas.1817233116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bostock RM, Pye MF, Roubtsova TV. Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response. Annu Rev Phytopathol. 2014;52:517–49. doi: 10.1146/annurev-phyto-081211-172902. [DOI] [PubMed] [Google Scholar]
- 56.Li X, Xie Y, Zhu Q, Liu YG. Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol Plant. 2017;10:1368–70. doi: 10.1016/j.molp.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 57.Liu M, Shi Z, Zhang X, Wang M, Zhang L, Zheng K, et al. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat Plants. 2019;5:389–400. doi: 10.1038/s41477-019-0383-2. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, et al. A single transcription factor promotes both yield and immunity in rice. Science. 2018;361:1026–8. doi: 10.1126/science.aat7675. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, et al. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell. 2019;31:106–27. doi: 10.1105/tpc.18.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai X, Zhou L, Zhang W, Cai L, Guo H, Tian H, et al. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2016;39:897–907. doi: 10.1111/pce.12695. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Xing H, Dong L, Zhang H, Han C, Wang X, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S, Zhang N, Zhang Q, Zhou G, Tian H, Hussain S, et al. Genome Editing to Integrate Seed Size and Abiotic Stress Tolerance Traits in Arabidopsis Reveals a Role for DPA4 and SOD7 in the Regulation of Inflorescence Architecture. Int J Mol Sci. 2019;20:2695. doi: 10.3390/ijms20112695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 64.Xu ZY, Kim SY, Kim DH, Dong T, Park Y, Jin JB, et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell. 2013;25:4708–24. doi: 10.1105/tpc.113.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S, Hu Q, Luo S, Li Q, Yang X, Wang X, et al. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front Plant Sci. 2015;6:388. doi: 10.3389/fpls.2015.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian H, Guo H, Dai X, Cheng Y, Zheng K, Wang X, et al. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci Rep. 2015;5:17587. doi: 10.1038/srep17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y, Feng CZ, Ye Q, Wu WH, Chen YF. Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 2016;12:e1005833. doi: 10.1371/journal.pgen.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo J, Wang J, Xi L, Huang WD, Liang J, Chen JG. RACK1 is a negative regulator of ABA responses in Arabidopsis. J Exp Bot. 2009;60:3819–33. doi: 10.1093/jxb/erp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong Q, Sun T, Qu N, Ma J, Li M, Cheng YT, et al. Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 2016;171:1344–54. doi: 10.1104/pp.15.01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.