Abstract
The overwhelming majority of participants in current genetic studies are of European ancestry. To elucidate disease biology in the East Asian population, we conducted a genome-wide association study (GWAS) with 212,453 Japanese individuals across 42 diseases. We detected 320 independent signals in 276 loci for 27 diseases, with 25 novel loci (P < 9.58 x 10−9). East Asian-specific missense variants were identified as candidate causal variants for three novel loci, and we successfully replicated two of them by analyzing independent Japanese cohorts; p.R220W of ATG16L2 associated with coronary artery disease and p.V326A of POT1 associated with lung cancer. We further investigated enrichment of heritability within 2,868 annotations of genome-wide transcription factor occupancy, and identified 378 significant enrichments across nine diseases (FDR < 0.05) (e.g. NKX3-1 for prostate cancer). This large-scale GWAS in a Japanese population provides insights into the etiology of complex diseases and highlights the importance of performing GWAS in non-European populations.
INTRODUCTION
Currently, large-scale genetic studies are dominated by European-descent samples, and fail to capture the level of diversity that exists globally1-5. Due to differential genetic architectures, transferability of genetic findings between populations is generally limited. Therefore, this imbalance poses a limitation in our understanding of the genetic architecture of complex diseases in non-European populations. Moreover, this imbalance could result in unequal benefits of precision medicine, as polygenic risk sores (PRS) based on large-scale genetic studies in European populations have high predictive power of clinical outcomes in European samples6-10 but poor predictive power in non-European samples1,11. Therefore, increasing the ethnic diversity of participants is an essential direction of genetic studies for the equality of genetic findings.
In addition, diversifying the ethnicity of participants is important for the discovery of novel disease etiology12. Even in large-scale European studies, causal variants might be missed if they have low frequencies or are monomorphic in European populations; such examples include p.E508K of HNF1A identified in Latino populations13 and p.R684* of TBC1D4 identified in a Greenlandic population14, both associated with type 2 diabetes (T2D). Therefore, differences in allele frequencies across populations can be an advantage for discovering genetic signals which were failed to be identified in European populations.
Here we report a GWAS of 42 common diseases in the BioBank Japan Project (BBJ)15,16, one of the largest non-European biobanks consisting of around 200,000 individuals. We provide detailed discussion of the biology of these diseases using multiple genomic annotations. We also examined inter-sex differences in genetic signals. Moreover, by incorporating previous genetic findings, we discussed the extent to which genetic signals are shared across populations while also investigating East Asian-specific genetic signals. Our study provided multiple insights into the etiology of complex traits, and highlighted the importance of conducing genetic studies in non-European populations.
RESULTS
Genome-wide association study of 42 diseases.
We conducted a genome-wide association study (GWAS) of 42 diseases in a Japanese population, comprising 179,660 patients who participated in BBJ and 32,793 population-based controls (Table 1 and Supplementary Table 1). The 42 diseases encompassed a wide-range of disease categories; 13 neoplastic diseases, five cardiovascular diseases, four allergic diseases, three infectious diseases, two autoimmune diseases, one metabolic disease, and 14 uncategorized diseases. By including patients with unrelated diagnoses into control samples, we maximized the power of our GWAS (Methods, Extended Data Figure 1, and Supplementary Table 1). We employed a generalized linear mixed model in our association analysis using SAIGE17. After imputing our genotypes with 1000 Genomes Project Phase 3 reference data (1KG Phase3)18, we tested 8,712,794 autosomal variants and 207,198 X chromosome variants for association with 42 diseases. For 35 diseases for which we have both male and female patients, we also conducted male- and female-specific GWAS.
Table 1.
Overview of the findings in this GWAS.
| Number of loci | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample size | Previous GWAS | BBJ-GWAS | ||||||
| Disease category |
Disease | Cases | Controls | All | Replicated | All | Novel | Additional signal |
| Allergic | Asthma | 8216 | 201592 | 66 | 57 | 7 | 2 | 2 |
| Allergic | Atopic dermatitis | 2385 | 209651 | 21 | 17 | 7 | 0 | 0 |
| Allergic | Drug eruption | 430 | 209651 | 0 | 0 | 0 | 0 | 0 |
| Allergic | Pollinosis | 5746 | 206707 | 28 | 24 | 0 | 0 | 0 |
| Autoimmune | Graves' disease | 2176 | 210277 | 8 | 8 | 9 | 3 | 0 |
| Autoimmune | Rheumatoid arthritis | 4199 | 208254 | 72 | 63 | 5 | 0 | 0 |
| Cardiovascular | Cerebral aneurysm | 2820 | 192383 | 8 | 7 | 4 | 2 | 0 |
| Cardiovascular | Congestive heart failure | 9413 | 203040 | 0 | 0 | 0 | 0 | 0 |
| Cardiovascular | Coronary artery disease | 29319 | 183134 | 184 | 167 | 53 | 1 | 7 |
| Cardiovascular | Ischemic stroke | 17671 | 192383 | 12 | 9 | 3 | 0 | 0 |
| Cardiovascular | Peripheral artery disease | 3593 | 208860 | 13 | 10 | 1 | 0 | 0 |
| Infectious | Chronic hepatitis B | 1394 | 211059 | 1 | 1 | 0 | 0 | 0 |
| Infectious | Chronic hepatitis C | 5794 | 206659 | 2 | 2 | 1 | 0 | 0 |
| Infectious | Pulmonary tuberculosis | 549 | 211904 | 4 | 4 | 0 | 0 | 0 |
| Metabolic | Type 2 diabetes | 40250 | 170615 | 234 | 220 | 89 | 7 | 20 |
| Neoplastic | Biliary tract cancer | 339 | 195745 | 0 | 0 | 0 | 0 | 0 |
| Neoplastic | Breast cancer | 5552 | 89731 | 121 | 102 | 7 | 0 | 0 |
| Neoplastic | Cervical cancer | 605 | 89731 | 4 | 4 | 0 | 0 | 0 |
| Neoplastic | Colorectal cancer | 7062 | 195745 | 73 | 68 | 11 | 0 | 1 |
| Neoplastic | Endometrial cancer | 999 | 89731 | 12 | 7 | 0 | 0 | 0 |
| Neoplastic | Esophageal cancer | 1300 | 195745 | 14 | 10 | 2 | 0 | 0 |
| Neoplastic | Gastric cancer | 6563 | 195745 | 10 | 9 | 4 | 1 | 1 |
| Neoplastic | Hematological malignancy | 1236 | 211217 | 45 | 32 | 0 | 0 | 0 |
| Neoplastic | Hepatocellular carcinoma | 1866 | 195745 | 2 | 0 | 1 | 1 | 0 |
| Neoplastic | Lung cancer | 4050 | 208403 | 18 | 15 | 6 | 1 | 1 |
| Neoplastic | Ovarian cancer | 720 | 89731 | 4 | 3 | 0 | 0 | 0 |
| Neoplastic | Pancreatic cancer | 442 | 195745 | 20 | 17 | 0 | 0 | 0 |
| Neoplastic | Prostate cancer | 5408 | 103939 | 107 | 97 | 20 | 0 | 9 |
| Other | Arrhythmia | 17861 | 194592 | 114 | 105 | 16 | 1 | 0 |
| Other | Cataract | 24622 | 187831 | 0 | 0 | 1 | 1 | 0 |
| Other | COPD | 3315 | 201592 | 70 | 54 | 5 | 1 | 2 |
| Other | Cirrhosis | 2184 | 210269 | 3 | 2 | 2 | 0 | 0 |
| Other | Endometriosis | 734 | 102372 | 11 | 11 | 0 | 0 | 0 |
| Other | Epilepsy | 2143 | 210310 | 4 | 1 | 0 | 0 | 0 |
| Other | Glaucoma | 5761 | 206692 | 55 | 43 | 5 | 0 | 0 |
| Other | Interstitial lung disease | 806 | 211647 | 10 | 7 | 1 | 1 | 0 |
| Other | Keloid | 812 | 211641 | 3 | 3 | 4 | 1 | 1 |
| Other | Nephrotic syndrome | 957 | 211496 | 0 | 0 | 0 | 0 | 0 |
| Other | Osteoporosis | 7788 | 204665 | 2 | 1 | 1 | 1 | 0 |
| Other | Periodontal disease | 3219 | 209234 | 2 | 0 | 0 | 0 | 0 |
| Other | Urolithiasis | 6638 | 205815 | 23 | 23 | 7 | 1 | 0 |
| Other | Uterine fibroids | 5954 | 95010 | 16 | 16 | 4 | 0 | 0 |
The sample size in this GWAS, the number of loci detected in previous GWAS, and that detected in this GWAS are provided. We considered a previous GWAS signal is replicated when the signal in the previous studies has the same effect direction in this study. We utilized a generalized linear mixed model in our GWAS, and set a genome-wide significance threshold at P < 9.58 x 10−9 for our study. We also included the variants which passed this significance threshold after meta-analyzing with the replication study. Detailed information is also provided in Supplementary Table 3-7. Additional signal, the number of independent significant signals identified by conditioning analyses. COPD, chronic obstructive pulmonary disease.
To quantify the heritability and the bias in our GWAS results, we analyzed them using linkage disequilibrium score regression (LDSC) analysis19 (Supplementary Table 2). Consistent with a recent finding in the European population20, heritability estimation was improved by incorporating the baselineLD model21 which includes functional annotations, LD-dependent architectures, and minor allele frequency (MAF)-dependent architectures (Supplementary Figure 1 and Supplementary Table 2). Although we observed high genomic inflation factors (λGC) for some diseases (e.g. λGC = 1.3 for T2D; Supplementary Table 2), LDSC analysis indicated that the majority of the inflated chi-squared statistics originated from polygenic effects rather than confounding biases (e.g. intercept = 1.01 for T2D; Supplementary Table 2).
To confirm that our GWAS produced reasonable signals, we examined how much of the previously identified risk alleles were replicable in our GWAS results (Extended Data Figure 2, Table 1, and Supplementary Table 3). By analyzing all diseases together, 1,219 out of 1,396 previously reported risk alleles were replicated with the same effect direction (sign test P = 1.47x10−191). In East Asian populations of 1KG Phase3, MAF of non-replicated alleles are significantly lower than those of replicated alleles (Extended Data Figure 3). Therefore, the replication failures might be due to insufficient statistical power. The high replicability of previous GWAS signals suggested that genetic etiologies are generally shared across populations.
Considering that more than 1.5 million variants in our study are rare variants (MAF < 1%) (Supplementary Figure 2), applying the conventional genome-wide significance threshold (P < 5 x 10−8), which assumes 1 million independent tests, might increase type-I errors. Therefore, to empirically estimate the appropriate P value threshold, we conducted GWAS using 1,000 random binary phenotypes and analyzed distributions of minimum P values (Pmin) for each phenotype. The 95-th percentile of Pmin was 2.87 x 10−8, and we defined this P value as an empirical genome-wide significance threshold at a significance level of α = 0.05 (Extended Data Figure 4). In addition, we considered the multiple testing burden of analyzing sex-specific GWAS; each variant was tested for sex-combined, male-specific, and female-specific analyses. Therefore, we set the significance threshold for our GWAS at P = 2.87 x 10−8 / 3 (= 9.58 x 10−9), and considered P = 5 x 10−8 as a threshold of suggestive associations.
We defined a locus as a genomic region within ± 1 Mb from the lead variant, and we considered a locus as novel when it does not include any previously reported variants (P in previous GWAS < 5.0 x 10−8). In sex-combined analysis, we detected significant associations for 27 diseases at 260 autosomal loci (outside of the HLA region) and nine loci on the X chromosome (P < 9.58 x 10−9; Supplementary Table 4 and 5). Associations at the HLA region have been investigated in detail in a separate article22. We further performed conditional analyses in these 269 loci to explore associations independent of the lead variants. We detected 44 additional independent signals for 9 diseases (P < 9.58 x 10−9; Supplementary Table 6). The largest number of independent signals in a single locus was seven, found in the FAM84B/POU5F1B locus associated with prostate cancer. In the sex-specific analysis (male- and female-cases were analyzed separately), we detected 4 additional loci for 3 diseases which were not identified in a sex-combined analysis (P < 9.58 x 10−9; Supplementary Table 7). We tested heterogeneity between effect size estimates for males and females using Cochran’s Q test. This analysis found all of the four loci showed nominally significant differences in effect size estimates between sexes (P values of heterogeneity (Phet) < 0.003). As we will introduce below, three variants with novel suggestive associations (P < 5.0 x 10−8) passed the significance threshold after meta-analyzing with independent replication studies (P < 9.58 x 10−9). In total, we detected 320 independent significant signals in 276 loci for 27 diseases, of which 25 loci were novel (P < 9.58 x 10−9; Figure 1a, Table 1, and Table 2). At three novel significant loci, the lead variants are rare variants with large effect size (MAF < 0.01 and odds ratio (OR) > 2; Figure 1b), and two of them are missense variants.
Figure 1. Disease-associated loci detected in this GWAS.
a, Phenogram52 of 331 suggestive loci detected in this GWAS (P < 5.0 x 10−8). Pleiotropic associations were plotted at the same position (Methods). b, Allele frequencies and the odds ratios (OR) of the lead variants at 331 suggestive loci detected in this GWAS (P < 5.0 x 10−8). The odds ratio of the risk allele was used. a and b, Novel loci (◆) are annotated by the closest gene names (only genes with OR > 2 are highlighted in b). Genes with significant associations are highlighted by red (P < 9.58 x 10−9). The sample size of GWAS is provided in Table 1. We utilized a generalized linear mixed model in our GWAS. *, loci detected in sex-specific GWAS. ¶, the lead variants were linked to missense variants (see text for the criteria). c, d, and e, Trans-ethnic minor allele frequency (MAF) comparison of disease-associated variants at novel (n=41) and known loci (n=153) with suggestive significance (P < 5 x 10−8). For known loci, we restricted this analysis to loci where the closest reported variants were discovered by GWAS in European populations. Mann–Whitney U test P value is provided (two-sided test). When MAF < 0.001, MAF was adjusted to 0.001 to fit in log scale. MAFEAS, MAF in East Asian population (1KG Phase3). MAFEUR, MAF in European population (1KG Phase3). e, The center line in each box indicates the median, and the box limits indicate the upper and lower quartiles. COPD, chronic obstructive pulmonary disease.
Table 2.
25 novel loci detected in this GWAS.
| Allele frequency | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease | Variant | REF | ALT | Gene | OR | L95 | U95 | P | EAS | EUR | AFR | Distance [Mbp] |
| Loci detected in sex-combined analysis | ||||||||||||
| Arrhythmia | rs73205368 | T | C | PTCHD1 | 1.08 | 1.06 | 1.10 | 4.25E-15 | 0.281 | 0.055 | 0.047 | NA |
| Coronary artery disease | rs11235571 (rs11235604) |
G (C) |
A (T) |
ARAP1 (ATG16L2) |
0.90 (0.91) |
0.87 (0.88) |
0.93 (0.94) |
2.64E-09 (1.73E-08) |
0.083 (0.100) |
0.000 (0.000) |
0.000 (0.000) |
2.9 |
| Cataract | rs75812946 | G | A | FLRT2 | 1.35 | 1.22 | 1.50 | 3.41E-09 | 0.015 | 0.000 | 0.000 | NA |
| Cerebral aneurysm | rs12226402 | G | A | SIRT3 | 1.34 | 1.23 | 1.45 | 1.57E-12 | 0.155 | 0.033 | 0.099 | 68.9 |
| Cerebral aneurysm | rs78535549 | C | T | AEBP2, PDE3A | 0.85 | 0.81 | 0.90 | 7.97E-09 | 0.528 | 0.036 | 0.055 | 12.2 |
| COPD | rs11066008 | A | G | ACAD10 | 1.29 | 1.21 | 1.37 | 4.34E-17 | 0.275 | 0.000 | 0.001 | 3.8 |
| Gastric cancer | rs1205528 | T | C | GUCY2F, IRS4 | 0.92 | 0.89 | 0.94 | 2.80E-10 | 0.354 | 0.884 | 0.654 | NA |
| Graves’ disease | rs10673095 | T | TTC | FAM84B, POU5F1B | 0.81 | 0.76 | 0.87 | 2.11E-09 | 0.476 | 0.362 | 0.772 | 5.9 |
| Graves' disease | rs11065783 | A | G | CUX2, MYL2 | 1.34 | 1.24 | 1.44 | 7.23E-14 | 0.264 | 0.010 | 0.000 | NA |
| Graves' disease | rs1569723 | C | A | CD40, NCOA5 | 1.20 | 1.13 | 1.28 | 4.06E-09 | 0.565 | 0.743 | 0.976 | NA |
| Hepatocellular carcinoma | rs8107030 | A | G | IFNL2, IFNL3 | 1.44 | 1.28 | 1.62 | 7.96E-10 | 0.078 | 0.170 | 0.027 | NA |
| Interstitial lung disease | rs6477542 | C | T | TMEM38B, ZNF462 | 1.34 | 1.21 | 1.48 | 6.90E-09 | 0.451 | 0.207 | 0.123 | NA |
| Keloid | rs192314256 | T | C | PHLDA3 | 9.56 | 5.91 | 15.45 | 3.28E-20 | 0.010 | 0.000 | 0.000 | 20.8 |
| Osteoporosis | rs578031265 | C | T | STK39 | 10.16 | 4.74 | 21.74 | 2.38E-09 | 0.002 | 0.001 | 0.000 | 31.8 |
| Type 2 diabetes | rs7721099 | T | C | MEF2C, TMEM161B | 1.05 | 1.04 | 1.07 | 1.41E-09 | 0.512 | 0.143 | 0.255 | 1.4 |
| Type 2 diabetes | rs200525873 | GT | G | CEP120, PRDM6 | 0.91 | 0.88 | 0.94 | 4.90E-09 | 0.086 | 0.040 | 0.037 | 11.2 |
| Type 2 diabetes | rs39218 | T | C | STEAP1, ZNF804B | 1.06 | 1.04 | 1.08 | 1.28E-09 | 0.191 | 0.503 | 0.311 | 12.6 |
| Type 2 diabetes | rs5762925 | A | C | ZNRF3 | 1.05 | 1.03 | 1.07 | 3.93E-09 | 0.462 | 0.353 | 0.262 | 1.0 |
| Type 2 diabetes* | rs2277339 | T | G | PRIM1 | 1.05 | 1.04 | 1.07 | 2.67E-10 | 0.206 | 0.111 | 0.199 | 9.0 |
| Type 2 diabetes* | rs17105012 | C | A | IRF2BPL, LRRC74A | 1.04 | 1.03 | 1.06 | 8.84E-09 | 0.297 | 0.143 | 0.034 | 2.6 |
| Urolithiasis | rs12290747 | T | C | STIM1 | 0.89 | 0.85 | 0.92 | 3.24E-09 | 0.317 | 0.313 | 0.017 | 107.3 |
| Loci detected in sex-specific analysis | ||||||||||||
| Asthma | rs13227841 | T | C | WBSCR28 | 0.86 | 0.81 | 0.90 | 2.04E-09 | 0.650 | 0.677 | 0.334 | 32.4 |
| Asthma | rs9836823 | A | G | LRRC3B, NEK10 | 0.86 | 0.82 | 0.91 | 5.19E-09 | 0.337 | 0.362 | 0.116 | 6.2 |
| Lung cancer* | rs75932146 | A | G | POT1 | 2.42 | 1.87 | 3.13 | 1.69E-11 | 0.003 | 0.000 | 0.000 | NA |
| Type 2 diabetes | rs202209118 | T | TCC | SETBP1 | 1.16 | 1.10 | 1.22 | 7.78E-09 | 0.023 | 0.019 | 0.002 | 6.1 |
Summary data of the lead variants in the novel loci in this GWAS. Detailed information of these variants is provided in Supplementary Table 4, 5, and 7. For variants detected in sex-specific GWAS, statistics of sex with significant associations are provided. For a lead variant of coronary artery disease (rs11235571), we also provided data of a missense variant (rs11235604) in LD with the lead variant in parenthesis (r2 = 0.68 in East Asian populations of 1KG Phase3; Table 3). The sample size is provided in Table 1. We utilized a generalized linear mixed model in our GWAS, and set a genome-wide significance threshold at P < 9.58 x 10−9. Disease names are marked by asterisk (*) when the variants passed the significance threshold after meta-analyzing with replication studies (Supplementary Table 10 and 13), and statistics of meta-analysis are provided for such variants. The distance between the lead variant in this study and the closest reported variant in the previous GWAS is also provided. When there are no reported associations on the same chromosome, distance information is set to NA. Allele frequencies of 1KG Phase3 are provided. REF, reference allele; ALT, alternative allele; OR, odds ratio relative to the alternative allele; L95, lower 95% confidence interval; U95, upper 95% confidence interval; EAS, East Asian populations; EUR, European populations; and AFR, African populations. COPD, chronic obstructive pulmonary disease.
To understand the characteristics of novel and known disease-associated variants in our study, we examined their allele frequencies in East Asian and European populations of 1KG Phase3. Intra-population MAF comparison showed that novel variants have significantly lower allele frequencies than known variants in European populations but not in East Asian populations (Extended Data Figure 5). Trans-ethnic MAF comparison showed that both novel and known variants have higher MAF in East Asian populations than in European populations (Figure 1c and d). However, trans-ethnic MAF differences are more pronounced in novel variants (Figure 1e). These observations suggested that the high allele frequencies of disease-associated variants in our cohorts increased the statistical power to detect their significance, especially for novel variants. This highlights the importance of performing GWAS in non-European populations.
We sought to refine the previously identified association signals in European GWAS. We counted the number of variants in LD with the lead variants in our GWAS and those of previous European GWAS (r2 > 0.8 in respective populations in 1KG Phase3) (Supplementary Table 4). The average number of variants in LD with the lead variants is 25.9 in European GWAS and 29.3 in our GWAS. On the other hand, the average number of variants in LD with both lead variants is 12.9. Therefore, our study successfully limited the number of potential causative variants.
Since a disproportionate number of patients with T2D and coronary artery disease (CAD) were included in the controls of GWAS for other diseases, our study design might create spurious associations mirroring the effects of risk alleles of T2D and CAD. However, this possibility was ruled out by the following observations; (i) excluding all patients from control samples did not affect effect size estimates (Extended Data Figure 1); (ii) risk loci detected in our GWAS for other diseases were not enriched within T2D or CAD known loci (Supplementary Figure 3); and (iii) effect directions of the known protective alleles of T2D or CAD were not significantly biased to positive values in our GWAS for other diseases (Supplementary Figure 4). Thus, we confirmed that our study results were not biased by having many patient samples in control groups.
Biological interpretation of disease-associated variants.
We next investigated the potential impact of the disease-associated variants on protein functions (Supplementary Table 8). We linked the GWAS association and the missense variant when the lead variant and the missense variant are in LD (r2 > 0.6 in East Asians of 1KG Phase3) and the missense variant is included in 95% credible set (Methods). Using these criteria, seven novel significant signals (P < 9.58 x 10−9) are linked to missense variants. Although four missense variants are not the lead variant, conditioning on these missense variants cancelled the signal of the lead variant (Figure 2a and Supplementary Figure 5). Importantly, three missense variants are monomorphic in Europeans and Africans (1KG Phase3); p.R220W of ATG16L2 (rs11235604) associated with CAD; p.V326A of POT1 (rs75932146) associated with lung cancer; and p.E62G of PHLDA3 (rs192314256) associated with keloid (Figure 2, Extended Data Figure 6, and Table 3). Considering the relevance of these findings, we additionally included two independent cohorts in a Japanese population (2,855 CAD cases and 15,211 controls; and 2,440 lung cancer cases and 467 controls). This replication study successfully confirmed the associations at ATG16L2 and POT1 loci, and fixed-effect meta-analysis improved statistical significance; the suggestive association at POT1 locus passed significance threshold (P < 9.58 x 10−9) (Supplementary Table 9 and 10). Here, we discuss each of the three East Asian-specific missense variants in detail. First, ATG16L2 is an autophagy-related gene highly expressed in immune cells, and previous studies reported that p.R220W of ATG16L2 is also associated with immune related traits; serum level of non-albumin protein in a Japanese population23 and Crohn's disease in a Chinese population24. Previous GWAS for CAD in European populations did not detect significant associations at ATG16L2 locus25 (Figure 2a), suggesting that p.R220W of ATG16L2, absent in Europeans, may be the causal variant. Therefore, dysregulated autophagy in immune cells might have an important role in CAD. Second, POT1 is a member of the telombin family and this protein binds to telomeres, regulating telomere length. Missense variants of POT1 have been described as being responsible for several familial cancers26-28. In addition, our study showed that p.V326A of POT1 is also positively associated with the risk of five other neoplastic diseases (P < 0.05; Extended Data Figure 7). These findings suggest this variant might increase the risk of neoplastic diseases in general. p.V326A of POT1 is more strongly associated with lung cancer in females than males; OR for female is 2.29 and OR for male is 1.26 (Phet = 7.7x10−4) (Figure 2b and Supplementary Table 7). We sought to figure out whether the sex-dependent effect can be explained by other factors, and conducted an association test stratified by histological and smoking status (Supplementary Table 10). However, we could not reach a definitive conclusion due to limited statistical power, and hence further large-scale studies will be required to answer this question. Together with a known association at the TERT locus (Supplementary Table 4), we provide additional evidence that telomere dysregulation is pathogenic for lung cancer. Third, p.E62G of PHLDA3 is predicted to have a deleterious effect to its protein function (SIFT score29=0; CADD score30=33), and we detected a large effect size for keloid (odds ratio = 9.56; 95% CI 5.91-15.45). We confirmed that genotyping of rs192314256 (p.E62G of PHLDA3) was not biased by batches of genotyping experiments or geographic areas (Supplementary Figure 6). PHLDA3 is known to be a suppressor of AKT31, and upregulated AKT signaling pathway is related to increased collagen production from dermal fibroblasts32. Therefore, damaged PHLDA3 may activate the AKT pathway, promoting the development of keloid. Together, our study successfully identified novel potential causal genes which would be hard to be discovered by GWAS in European populations due to restrictive European allele frequencies.
Figure 2. Novel associations which can be explained by East Asian-specific missense variants.
Regional association plots are provided. a, coronary artery disease (29,319 cases vs 183,134 controls). b, lung cancer (2,710 male cases vs 106,637 male controls; 1,340 female cases vs 101,766 female controls). For coronary artery disease (a), P values from conditional analysis and those in European GWAS25 were plotted separately.
For lung cancer (b), P values from female- and male-specific GWAS were plotted separately. We utilized a generalized linear mixed model in our GWAS.
Table3.
Population-specific missense variants linked to disease-associated variants.
| BBJ-GWAS | Replication analysis | Meta-analysis | Allele frequency | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease | Variant | Gene | Amino acid change |
REF | ALT | Case | Ctrl | OR | L95 | U95 | P | Case | Ctrl | OR | L95 | U95 | P | OR | L95 | U95 | P | Phet | EAS | EUR | AFR |
| Loci detected in sex-combined analysis | |||||||||||||||||||||||||
| Coronary artery disease | rs11235604 | ATG16L2 | p.R220W | C | T | 29319 | 183134 | 0.91 | 0.88 | 0.94 | 1.73E-08 | 2855 | 15211 | 0.83 | 0.74 | 0.94 | 3.33E-03 | 0.91 | 0.88 | 0.93 | 5.69E-10 | 0.16 | 0.100 | 0.000 | 0.000 |
| Keloid | rs192314256 | PHLDA3 | p.E62G | T | C | 812 | 211641 | 9.56 | 5.91 | 15.45 | 3.28E-20 | - | - | - | - | - | - | - | - | - | - | - | 0.010 | 0.000 | 0.000 |
| Loci detected in sex-specific analysis | |||||||||||||||||||||||||
| Lung cancer | rs75932146 | POT1 | p.V326A | A | G | 1340 | 101766 | 2.29 | 1.71 | 3.05 | 2.21E-08 | 2440 | 467 | 2.99 | 1.71 | 5.24 | 1.26E-04 | 2.42 | 1.87 | 3.13 | 1.69E-11 | 0.40 | 0.003 | 0.000 | 0.000 |
We conducted a meta-analysis with a fixed effect model using independent Japanese cohorts (Supplementary Table 9 and 10), and tested heterogeneity using Cochran’s Q test (Phet). We utilized a generalized linear mixed model in BBJ-GWAS and the replication study of CAD. We utilized a generalized linear model for the replication analysis of lung cancer. We set a genome-wide significance threshold at P < 9.58 x 10−9. In addition to the statistics, the sample size and allele frequencies of 1KG Phase3 are provided. Detailed information about missense variants is provided in Supplementary Table 8. REF, reference allele; ALT, alternative allele; Case, the number of case samples; Ctrl, the number of control samples; OR, odds ratio relative to the alternative allele; L95, lower 95% confidence interval; U95, upper 95% confidence interval; EAS, East Asian populations; EUR, European populations; and AFR, African populations.
We also investigated the potential impacts of the disease-associated variants on the mRNA levels using the GTEx database of expression quantitative trait loci (eQTL)33. Since the eQTL data are generated in European populations, we could not apply formal colocalization tests34,35 which assume the same LD-structures between GWAS and eQTL studies. Therefore, we linked the GWAS association and the eQTL variant when the GWAS lead variant and the eQTL variant are in LD (r2 > 0.6 both in East Asian and European populations of 1KG Phase3) and the eQTL variant is included in 95% credible set. We found that seven novel significant signals (P < 9.58 x 10−9) and five novel suggestive signals (P < 5 x 10−8) can be explained by at least one eQTL variant (Supplementary Table 11). Among them, the eQTL signals for ATP2B1 which were linked to a novel, suggestive variant of cerebral aneurysm (rs11105352; P = 1.22 x 10−8) is highly specific to arterial tissues (Figure 3). Since the loss of ATP2B1 in vascular smooth muscle cells induced blood pressure elevation in mice36, decreased expression of ATP2B1 in arteries might induce hypertension, which leads to increased risk of cerebral aneurysm.
Figure 3. A novel suggestive association of cerebral aneurysm can be explained by artery-specific expression quantitative trait loci (eQTL) signals for ATP2B1.
a. Regional association plots of cerebral aneurysm GWAS (2,820 cases vs 192,383) at ATP2B1 locus (top) and those of eQTL signals for ATP2B1 in the tibial artery (bottom) are provided. The lead variant of GWAS (rs11105352; ◆ dot) and the lead variant of eQTL (rs2681492; ■ dot) are indicated by different shapes. Variants in LD with rs11105352 are highlighted by red (r2 > 0.6 both in East Asian and European populations of 1KG Phase3). We utilized a generalized linear mixed model in our GWAS. b, Tissue-specificity of eQTL signals for ATP2B1 at rs2681492 (the lead variant of eQTL in the tibial artery (■ dot in a)). P values in eQTL analysis and M values (the posterior probability that an eQTL effect exist in each tissue tested in the cross-tissue meta-analysis) in all tissues in GTEx project33 are provided. Each dot indicates each tissue. All statistics of eQTL analysis were derived from release v7 of GTEx project33.
Replication with European GWAS results.
Replication analysis in the same population is a critical part of genetic studies. Although we included two independent replication studies for CAD and lung cancer in a Japanese population, we were not able to prepare replication cohorts in a Japanese population for other diseases. Therefore, we conducted replication studies using previous European GWAS results. We utilized publicly available GWAS summary statistics of European populations for 10 diseases (asthma, atrial fibrillation, breast cancer, CAD, congestive heart failure, glaucoma, ischemic stroke, prostate cancer, rheumatoid arthritis, and T2D; see Methods for selection of diseases), and tested for consistency in direction of effect. For these 10 diseases, our GWAS detected suggestive associations at 218 known and 19 novel loci (P < 5 x 10−8); among them, statistics of European GWAS were available at 149 known and 15 novel loci. We first conducted replication analysis at the known loci. We restricted this analysis to 112 known loci with significant associations also in European GWAS (P < 5 x 10−8) to exclude loci where the European GWAS had insufficient power. Effect directions are consistent between BBJ- and European-GWAS at 109 out of 112 loci; but opposite at 3 loci (Extended Data Figure 8 and Supplementary Table 12). These three replication failures are probably due to differences in LD structure between populations (Extended Data Figure 8). We then conducted replication analysis at the novel loci. Among 15 novel variants, 12 were replicated with the same effect direction (Supplementary Table 13). Meta-analysis using fixed-effect model increased the level of significance in six of them; and two suggestive novel variants passed significance threshold (P < 9.58 x 10−9) (rs2277339 and rs17105012 associated with T2D; Table 2 and Supplementary Table 13). Among the three variants that failed replication, rs13227841 is a missense variant originally identified as a potential causal variant at this locus (p.W78R of WBSCR28; Supplementary Table 8), which suggests that variants in LD with rs13227841, not rs13227841 itself, may be responsible for the observed associations. The other replication failures might be due to different LD-structures or the absence of the causal variants in European populations. Further efforts to conduct a replication analysis in a Japanese population will be required to confirm the associations which we failed to replicate in these European studies.
Genetic correlation between male- and female-specific GWAS.
To understand differences in the genetic risks between males and females, we assessed genetic correlations using LDSC37 between the results of sex-specific GWAS for the 20 diseases (see Methods for selection of diseases). Although most correlations are close to one, the correlation of asthma was significantly smaller than one (genetic correlation = 0.63 (S.E. = 0.12) and P = 2.2 x 10−3 < 0.05/20; Extended Data Figure 9). This finding suggested that genetic risks of asthma might be different between males and females. To explore the biological mechanism underlying this finding, we estimated the enrichment of the heritability of male or female asthma in the 220 cell-type specific regulatory regions using stratified LD-score regression (S-LDSC)38. We found significant enrichments for either male or female asthma in three annotations; Th0, Th1, and colonic mucosa (P < 0.05/220; Extended Data Figure 9). Among them, the colonic mucosa annotation showed significant heterogeneity in the enrichment of heritability (Phet = 0.006 < 0.05/3). Recent studies suggested that host-microbiome interactions at intestinal mucosa (gut-lung axis) have important roles in the development of asthma39,40, and our study suggested that the gut-lung axis might have sex-dependent roles in asthma. Considering their marginal significance, a replication study will be required to confirm these findings.
Transcription factors underlying the etiology of diseases.
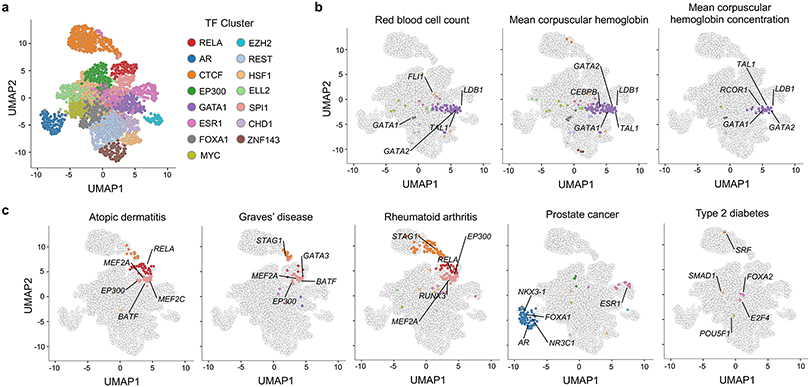
To acquire more insights to disease biology, we estimated the heritability enrichments in the binding sites of a variety of transcription factors (TFs) using S-LDSC. We included TF binding sites defined by 2,868 publicly available chromatin immunoprecipitation sequencing (ChIP-seq) datasets for 410 unique TFs (Supplementary Table 14). To make mutually comparable data, we began our analysis from the raw sequencing data, and defined TF binding sites using a uniform protocol (Methods). Using LD-scores of all TF binding sites, we grouped them into 15 clusters (cluster name was defined by the most dominant TF), and performed uniform manifold approximation and projection (UMAP)41 to project all TF binding sites into a two-dimensional space (Methods; Figure 4a and Supplementary Figure 7). To scale the performance of this analysis, we first analyzed previously reported GWAS for red blood cell-related traits23 where the critical role of GATA1 was supported by multiple pieces of evidence42-46, and we successfully recapitulated this biology (Figure 4b). We then applied this analysis to our 24 GWAS results (see Methods for selection of diseases), and detected 378 significant enrichments for nine diseases (FDR < 0.05) (Figure 4c, Extended Data Figure 10, and Supplementary Table 15). Biologically plausible TFs were highlighted by this analysis; RELA, a subunit of NF-κB, for atopic dermatitis, rheumatoid arthritis (RA), and Graves’ disease; sex hormone receptors (AR and ESR1) for prostate cancer; and FOXA2, which regulates insulin secretion in pancreatic beta-cells47, for T2D (Figure 4c). This analysis also suggested that NKX3-1, a prostate-specific homeobox gene, has an important role in the biology of prostate cancer (Figure 4c). In addition to this polygenic analysis, the importance of NKX3-1 was also suggested by the regional analysis integrating eQTL databases; the risk allele of prostate cancer at the NKX3-1 locus (rs4872174-C) was suggested to decrease the expression of NKX3-1 (Supplementary Table 11). Consistently, loss of NKX3-1 expression in human prostate cancers was reported to be correlated with tumor progression48. Together, our results confirmed and expanded our current understanding of complex traits in the context of TF activity.
Figure 4. Transcription factors (TF) whose binding sites were enriched for heritability of diseases.
a, All of the 2,868 sets of TF binding sites grouped into 15 clusters were plotted in the UMAP space. b and c, The results of S-LDSC were plotted on the UMAP space. The significant results (FDR < 0.05) are highlighted by cluster-specific colors. The names of the top five most significant TFs are also shown on the plot. b, The results of red blood cell-related traits. c, The results of diseases in this GWAS which had more than five significant TF binding site tracks (the results of the other diseases are provided in Extended Data Figure 10).
DISCUSSION
Our study demonstrated the advantages of conducting genetic studies in non-European populations. Typically, LD acts as a major hurdle limiting the identification of causal variants in GWAS. However, jointly analyzing GWAS results from populations with different LD structures can narrow down causal variants12. Indeed, when we consider variants in LD with a lead variant as candidate causal variants (r2 > 0.8), our study successfully reduced the number of candidate causal variants at 68 loci which were originally discovered in previous European GWAS (Supplementary Table 4). In addition, some novel variants in our study have been missed in larger GWAS in European populations due to restrictive European allele frequencies. Therefore, diversifying the ethnicity of participants is important not only for the equality of genetic findings but also for the discovery of novel disease etiology.
Although previous studies already reported important roles of TFs in the etiology of complex traits49-51, our TF enrichment analysis has two distinguishing features from previous studies. One feature is the comprehensiveness; we included 2,868 TF annotations, more than those used in most previous studies. The second feature is the method of the enrichment test; we utilized S-LDSC, whereas most previous studies utilized naïve enrichment tests using genome-wide significant variants. S-LDSC evaluates enrichment of GWAS signals irrespective of significance, and it is robust to the biases coming from the overlapping annotations. Therefore, by incorporating a comprehensive catalog of TF annotations with a sophisticated method to test heritability enrichment, we provided evidence of TF importance in complex diseases from a polygenic angle.
The critical limitation of this study is insufficient replication analyses to validate novel signals. Among 25 novel loci (P < 9.58 x 10−9), we were able to prepare East-Asian replication datasets for only two of them; p.R220W of ATG16L2 associated with CAD and p.V326A of POT1 associated with lung cancer. To supplement this insufficiency, we utilized European GWAS results when data was available; we tested replicability of eight novel signals (P < 9.58 x 10−9) and observed evidence of heterogeneity in effect size estimates for three of them (Phet < 0.05; Supplementary Table 13). This may be the case for several reasons; the locus might possess different LD structures between populations and the variant might tag the causal variant only in East Asian populations (as illustrated in Extended Data Figure 8); effect sizes might be truly different between populations; or they might be false positives. Therefore, until further replication studies in East-Asian populations are conducted, we need to be cautious about the validity of these putatively novel variants since we were not able to provide evidence of replicability.
In summary, we conducted a large-scale GWAS of 42 diseases in a non-European population and provided rich public resources for genetic studies. Our study provided multiple insights into the etiology of complex traits by integrating annotations of missense variants, eQTL variants, and transcription factor binding site tracks. Currently, genetic studies are overwhelmed by European-descent samples, making the clinical translation of genetic findings far more beneficial to European individuals than other populations1. Our study contributed to broaden the population diversity in genetic studies and should potentially mitigate the problems originating from this imbalance.
ONLINE METHODS
Subjects
All case samples in this GWAS were collected in the BioBank Japan Project (BBJ; https://biobankjp.org/english/index.html)15,16, which is a biobank that collaboratively collects DNA and serum samples from 12 medical institutions in Japan and recruited approximately 200,000 patients with the diagnosis of at least one of 47 diseases. Among them, cases with dyslipidemia were not analyzed in this study because it was already reported as a quantitative trait in our previous study23. Amyotrophic lateral sclerosis and febrile seizure were also not analyzed due to limited sample size. Cases with myocardial infarction, stable angina, and unstable angina were re-classified into a single disease category (coronary artery disease). Thus, we analyzed 42 disease in this study. For control samples, we used samples from the population-based prospective cohorts; the Tohoku University Tohoku Medical Megabank Organization (ToMMo), Iwate Medical University Iwate Tohoku Medical Megabank Organization (IMM)53, the Japan Public Health Center–based Prospective Study and the Japan Multi-institutional Collaborative Cohort Study. In addition, we also included samples in BBJ without related diagnoses into control group (Extended Data Figure 1 and Supplementary Table 1). The sample sizes and the demographic data are provided in Supplementary Table 1. All participating studies obtained informed consent from all participants by following the protocols approved by their institutional ethical committees. We obtained approval from ethics committees of RIKEN Center for Integrative Medical Sciences, and the Institute of Medical Sciences, The University of Tokyo. We have complied with all relevant ethical regulations.
Genotyping
We genotyped samples with the Illumina HumanOmniExpressExome BeadChip or a combination of the Illumina HumanOmniExpress and HumanExome BeadChips. For quality control (QC) of samples, we excluded those with (i) sample call rate < 0.98 and (ii) outliers from East Asian clusters identified by principal component analysis using the genotyped samples and the three major reference populations (Africans, Europeans, and East Asians) in the International HapMap Project54. For QC of genotypes, we excluded variants meeting any of the following criteria: (i) call rate < 99%, (ii) P value for Hardy Weinberg equilibrium (HWE) < 1.0 × 10−6, and (iii) number of heterozygotes less than five. Using 939 samples whose genotypes were also analyzed by whole genome sequencing (WGS), we added additional QC based on the concordance rate between genotyping array and WGS. Variants with a concordance rate < 99.5% or a non-reference discordance rate ≥ 0.5% were excluded. We note that the allele frequency of rs671 (the East Asian-specific functional missense variant at ALDH2) substantially varies among the domestic regions within Japan due to strong selection pressure55 and that genotypes of rs671 did not follow HWE. We thus did not apply the HWE QC for rs671. We had confirmed the 100% concordance of rs671 genotypes between the SNP microarray data used in this study and our internal WGS data (n = 2,798; see details in the discussion in ref56).
Imputation
We utilized all samples in the 1000 Genomes Project Phase 3 (version 5; www.1000genomes.org/)18 as a reference for imputation. We first pre-phased the genotypes with SHAPEIT2 (v2.778; https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html) and then imputed dosages with minimac3 (v2.0.1; https://genome.sph.umich.edu/wiki/Minimac). After imputation, we excluded variants with imputation quality of Rsq < 0.7. For the X chromosome, we performed prephasing and imputation separately for males and females, and we excluded variants with imputation quality of Rsq < 0.7 in either of them.
Genome-wide association analysis
We conducted GWAS by employing a generalized linear mixed model (GLMM) using SAIGE (v0.29.4.2; https://github.com/weizhouUMICH/SAIGE)17. This strategy enabled us to maintain related samples in our GWAS, and the sample sizes were increased by 6% on average compared to removing related samples. Briefly, there are two steps in SAIGE. In step 1, we fit a null logistic mixed model using genotype data, and we added covariates in this step (see below). In step 2, we performed the single-variant association tests using imputed variant dosages. We applied the leave-one-chromosome-out (LOCO) approach. For the X chromosome, we conducted GWAS separately for males and females, and merged their results by inverse-variance fixed-effect meta-analysis. We used only female control samples for GWAS of female-specific diseases; breast cancer, cervical cancer, endometrial cancer, ovarian cancer, endometriosis, and uterine fibroids. Similarly, we used only male control samples for GWAS of prostate cancer. We incorporated age and top 5 principal component (PC) as covariates. We also used sex as covariate for GWAS of diseases which include both of male and female samples. We also conducted male-specific and female-specific GWAS using the same pipeline as described above, and estimated heterogeneity in the effect size estimates using Cochran’s Q test. In each GWAS, we excluded variants with minor allele count (MAC) < 10 based on the recommendation from thee developers of SAIGE. We created regional association plots by LocusZoom (v1.2; http://locuszoom.sph.umich.edu/locuszoom/)57. We performed stepwise conditional analysis within ± 1 Mb from the lead variant; we repeated the association test by additionally incorporating the dosages of the identified variants as covariates in SAIGE step 1 until we do not detect any significant associations.
For each disease, we defined a significantly associated locus as a genomic region within ± 1 Mb from the lead variant. When a locus did not include any variants which were previously reported to be significantly associated with the same disease (P < 5.0 × 10−8), we defined it as a novel locus. Since we tested each variant for disease association three times (sex-combined, female-specific, and male-specific analysis), we considered multiple-testing burden on the empirical significance threshold (P = 2.87 x 10−8, see next paragraph), and we set the genome-wide significance threshold for our study at P = 2.87 x 10−8 / 3 (= 9.58 x 10−9).
Estimation of empirical significance threshold by permutation test
Using the identical statistical method and imputed genotype data as used in the main analysis, we conducted GWAS using 1,000 simulated phenotypes. We utilized down-sampled individuals (n=10,000) because permutation test using all samples (~200,000) was not computationally tractable. We simulated binary phenotypes with 1,920 cases and 8,080 controls; the same case-control ratio as in T2D GWAS in our study. For each of the 1,000 simulated phenotypes, the minimum P values (Pmin) were recorded, and the distributions of 1,000 Pmin were analyzed. This analysis showed that the 95-th percentile of Pmin is 2.87 x 10−8 (Extended Data Figure 4). We defined this value as an empirical genome-wide significance threshold at a significance level of α=0.05. 95% confidence interval was estimated by 1,000 bootstraps using the R package boot (v1.3-20).
To test the potential effect of down-sampling on the Pmin distributions, we compared the Pmin distributions using all samples (n=198,137) with those using 10,000 samples. To increase computational efficiency, we restricted this analysis to imputed genotype data in chromosome 22. For this analysis, we utilized Plink2 (https://www.cog-genomics.org/plink/2.0/)58 because SAIGE requires whole genotype data to estimate relatedness even when we restrict the analysis to chromosome 22. This analysis confirmed that down-sampling does not have substantial impact on the Pmin distributions (Extended Data Figure 4).
Estimation of heritability
We estimated heritability and confounding bias in our GWAS results with LDSC (v1.0.0; https://github.com/bulik/ldsc/)19 using the baselineLD model (v2.1; https://data.broadinstitute.org/alkesgroup/LDSCORE/)21 which includes 86 annotations, including 10 MAF- and 6 LD-related annotations that correct for bias in heritability estimates20, and were calculated using 481 East Asian samples in 1KG Phase3. For the analysis using LDSC, we excluded variants in the HLA region (chr6:26 Mb-34 Mb). We also calculated heritability Z-score to assess the reliability of heritability estimation.
Absolute quantification of heritability estimation using GWAS results using GLMM can be biased because effective sample size could be different from the true sample size (relative quantification is not biased, and hence GWAS results using GLMM can be applied for genetic correlation analysis and S-LDSC safely). Therefore, to confirm the robustness of heritability estimation in our analysis, we also performed GWAS using generalized linear regression model (GLM). As simple GLM does not account for the bias caused by genetic relationships, we further excluded related samples (Pi-hat by > 0.187), and we analyzed genotype data with Plink2 using the same covariates as described above. Heritability estimates based on GWAS using two different methods (SAIGE vs PLINK) were comparable (Supplementary Table 2).
Replication of the previously reported variants by this GWAS
We included data in the GWAS Catalog (https://www.ebi.ac.uk/gwas/) that satisfy the following criteria; (i) P in previous GWAS < 5 x 10−8, (ii) risk allele information is reported, (iii) outside of MHC region (Chr6: 23Mb-37Mb), and (iv) variants were analyzed in this study. When multiple variants were reported within 1Mb window, we included one variant for each disease. We considered a previous GWAS signal as replicated when the signal in the previous GWAS has the same effect direction in our GWAS.
Replication of the findings in this GWAS by independent cohorts in a Japanese population
We included an independent Japanese cohort of CAD and controls who enrolled in the Osaka Acute Coronary Insufficiency Study (OACIS)59 and the National Center for Geriatrics and Gerontology (NCGG) Biobank60. OACIS is a study that examined patients with myocardial infarction at 25 collaborating hospitals in Osaka, Japan, from April 1998 to April 2006. The NCGG Biobank is one of the facilities belonging to the National Center Biobank Network (NCBN; https://ncbiobank.org/en/home.php). It has been running since 2012. The participants were recruited from NCGG hospital, which is located in Obu city, and the other nearby medical institutes. We also included 1,392 control DNAs from the Health Science Research Resources Bank (HSRRB), Osaka, Japan. Samples in NCGG were genotyped by Infinium Asian Screening Array-24 v1.0 (Illumina), and samples in OACIS were genotyped using the same platform as in BBJ samples. We extracted bi-allelic, shared variants genotyped in these studies. We excluded variants with 1) hardy Weinberg disequilibrium (P < 1 x 10−6), 2) low call rate (< 99%). We excluded samples using the following criteria: samples with low call rate (< 99%), PCA outliers, heterozygosity outliers, and sex discordant samples. After QC, 2,855 CAD cases, 15,211 controls, and 111,041 SNPs remained. After pre-phasing with Eagle (v2.3), we performed imputation by minimac4 (v1.0.0) using 1KG phase3 reference panel. Association test was conducted using SAIGE (v0.36.3) including age, sex, top 5 PCs as covariates. We tested the influence of bias using LDSC; intercept was 1.008 (S.E. = 0.014), and lambda GC was 1.053, suggesting there is no substantial bias in the association results.
We also included a Japanese cohort with 2,440 female lung cancer cases and 467 female controls enrolled in the study of the National Cancer Center Hospital (NCCH). All cases are adenocarcinoma. Genotyping of rs75932146 was conducted by invader assay. Association test was conducted by logistic regression. Meta-analysis was conducted using fixed effect model via inverse-variance weighting; heterogeneity of effect size estimates was tested by using Cochran’s Q test.
Replication of the findings in this GWAS by the previous European GWAS
We searched for European GWAS whose summary statistics are publicly available and whose disease affection status were based on physician diagnosis (excluding GWAS based on self-reported phenotypes). The latter criterion was added because all cases in BBJ were diagnosed by a physician, and we wanted to prepare European GWAS of comparable phenotypes. We were able to prepare European GWAS summary statistics for 10 diseases. Summary statistics for eight diseases were downloaded from GWAS Catalog (https://www.ebi.ac.uk/gwas/) and their names and their PMIDs were as follows; atrial fibrillation (30061737), breast cancer (29059683), coronary artery disease (29212778), glaucoma (29891935), ischemic stroke (29531354), prostate cancer (29892016), rheumatoid arthritis (24390342), and type 2 diabetes (30054458). Summary statistics of two diseases were downloaded from UK Biobank GWAS summary statistics at Neale Lab (http://www.nealelab.is/uk-biobank) and their names and their phenotype code were as follows; asthma (22127), and congestive heart failure (I50). Meta-analysis was conducted using fixed effect model via inverse-variance weighting, and tested heterogeneity in effect size estimates using Cochran’s Q test.
Pleiotropy
We utilized the following variants detected in GWAS for each disease; (i) lead variants in the significantly associated loci, (ii) independent signals detected by conditional analysis, and (iii) lead variants detected in sex-specific GWAS. We defined pleiotropic association when these variants were in LD (r2 > 0.6). We calculated r2 using East Asian samples in the 1KG Phase318 by PLINK58.
Functional annotation of associated variants
We calculated r2 using East Asian samples (r2EAS) and European samples (r2EUR) in the 1KG Phase318 by PLINK58. We also identified 95% credible sets using R package corrcoverage (v1.2.1). We linked the GWAS association and the missense variant when the lead variant and the missense variant are in LD (r2EAS > 0.6) and the missense variant is included in 95% credible set. For the annotation of nonsynonymous variants, we used ANNOVAR (http://annovar.openbioinformatics.org/en/latest/)61. GRCh37 (hg19) coordinates were used in this study.
We also annotated GWAS variants with eQTL detected in the European population (release v7 of the GTEx project)33 in the following conditions; (i) the lead variants of the eQTL study are in LD (r2EAS > 0.6 and r2EUR > 0.6) with GWAS variants, (ii) the missense variant is included in 95% credible set, and (iii) Q values of the lead variants in the eQTL study are less than 0.05.
Genetic correlations between sex-specific GWAS
We estimated genetic correlations between our GWAS results by LDSC (v1.0.0)19 using East Asian LD scores which we presented in our previous study23. We excluded variants in the HLA region (chr6:26 Mb-34 Mb). We analyzed 20 diseases based on two criteria; (i) heritability was reliably estimated (heritability Z-score > 2; Supplementary Table 2); and (ii) both of male and female patients were included.
Transcription factor binding sites
We obtained 3,158 raw human ChIP-seq data files in SRA format from the GEO database. We converted them to FASTQ format using the fastq-dump function of SRA Toolkit (https://www.ncbi.nlm.nih.gov/sra/). We performed QC of sequence reads using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We mapped these reads to the genome assembly GRCh37 using Bowtie2 (v2.2.5; http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml) with default parameters. We called peaks using MACS (v2.1; https://github.com/taoliu/MACS) with default parameters (q < 0.01) and defined them as TF binding sites. We excluded TF binding site tracks which do not have at least one binding region on every chromosome, and 2,868 genome-wide TF binding site tracks remained (Supplementary Table 14).
Stratified LD score regression
We conducted stratified LD score regression (S-LDSC)38 to partition heritability. For S-LDSC analysis of sex-specific GWAS of asthma, we used 220 cell-type specific annotations used in previous articles23,38. For other S-LDSC analysis, we used TF binding site tracks which were described in the previous paragraph. For all sites of TF binding, we empirically extended sites by 500 bp at the both ends for this analysis. We computed annotation-specific LD scores using the 1000 Genomes Project Phase 3 (version 5) East Asian reference haplotypes18. We estimated heritability enrichment of binding sites of each TF, while controlling for the merged binding sites of all TFs and the 53 categories of the full baseline model available at the authors’ website (https://data.broadinstitute.org/alkesgroup/LDSCORE/). We did not use the baselineLD model (v2.1)21 in this analysis to increase the power of detecting significant enrichment. We excluded variants in the HLA region (chr6:26 Mb-34 Mb). We analyzed 24 diseases whose heritability was reliably estimated (heritability Z-score > 2; Supplementary Table 2). We calculated the P value of the regression coefficient. For each trait, we calculate FDR using the Benjamini-Hochberg method. We set a significance threshold at FDR < 0.05 for this analysis.
Visualization of TF binding sites
There is a complex correlation structure among 2,868 TF binding site tracks used for S-LDSC analysis. In S-LDSC, we regress GWAS chi-squared statistics on LD-scores of each TF binding site (TF LD-score), and hence we focused on correlations between TF LD-scores, not correlations between TF binding sites. We first performed PCA using all TF LD-scores. To classify them into mutually correlated TF groups, we performed k-means clustering (k=15) using the top 15 PCs. We named each cluster by the most dominant TF in each cluster (Figure 4). The list of each TF binding site and its assigned cluster name was provided in Supplementary Table 14. We then performed uniform manifold approximation and projection (UMAP)41 using the top 15 PCs to project all TF binding sites into a two-dimensional space. UMAP was conducted using the R package umap (v.0.2.0.0). Our workflow was illustrated in Supplementary Figure 7.
Extended Data
Extended Data Fig. 1. Study design of this GWAS.
a, Study designs in this GWAS. Study design 1 (top) was used in the main analysis. An example of study design 1 is provided; in GWAS of disease 3, we included all other patients (except those have related diseases) into control group. The definition of related diseases is provided in Supplementary Table 1. Study design 2 (bottom) was used to discuss the appropriateness of study design selection. b, Effect size estimates and S.E. at the 309 autosomal disease-associated variants detected in sex-combined analysis (P < 5 x 10−8). We compared the effect size estimates in study design 1 with those in study design 2. Heterogeneity between two studies was tested using Cochran’s Q test. The identity line is shown in blue. The red dot (rs373205748 associated with arrhythmia) indicates a variant with significant heterogeneity in effect size estimates between two study designs (P = 0.00012 < 0.05/309).
Extended Data Fig. 2. Replication analysis of previous GWAS findings using this GWAS results.
We compared effect sizes reported in the previous GWAS with those in this GWAS. Effect size and S.E. are shown. The identity line is shown in blue. The sample size of GWAS is provided in Table 1. We utilized a generalized linear mixed model in our GWAS.
Extended Data Fig. 3. Low allele frequency might contribute to replication failure.
We first compared effect sizes reported in the previous GWAS with those in our GWAS (Supplementary Table 3 and Extended Data Figure 2); 1,219 out of 1,396 previously reported risk alleles were replicated with the same effect direction (177 alleles were not replicated). We compared MAF of replicated variants (n=1,219) and MAF of not replicated variants (n=177). Mann-Whitney U test P value is provided (two-sided test).
Extended Data Fig. 4. Permutation test to estimate appropriate P value threshold to control type I errors.
Using 1,000 simulated binary phenotypes with down-sampled samples (n=10,000), we conducted GWAS utilizing the same strategy as used in the main analysis. a, The distribution of minimum P values in each phenotype (Pmin). The 95-th percentile of Pmin was 2.87 x 10-8. The 95% confidence interval was estimated by 1,000 bootstraps. b, The distributions of Pmin using all samples (n=198,137) and those using 10,000 samples. To increase computational efficiency, we restricted this analysis to imputed genotype data in chromosome 22. For this analysis in b, we utilized Plink2.
Extended Data Fig. 5. Allele frequency comparison between novel and known disease-associated variants.
MAF comparison at disease-associated variants at novel (n=41) and known loci (n=153) with suggestive significance (P < 5 x 10−8) (a, East Asian populations; b, European populations in 1KG phase3). For known loci, we restricted this analysis to loci where the closest reported variants were discovered by GWAS in European populations. Mann-Whitney U test P value is provided (two-sided test).
Extended Data Fig. 6. A novel association which can be explained by an East Asian-specific missense variant.
A regional association plot for keloid (812 cases vs 211,641 controls) at the PHLDA3 region is provided. We utilized a generalized linear mixed model in our GWAS.
Extended Data Fig. 7. The association of p.V326A of POT1 for all diseases in this GWAS.
Effect size and S.E. are provided for neoplastic diseases (a) and non-neoplastic diseases (b). The sample size of GWAS is provided in Table 1. We utilized a generalized linear mixed model in our GWAS.
Extended Data Fig. 8. Comparison of allelic directions between this GWAS and previous European GWAS at known loci.
a, Schematic explanations how we compared statistics between BBJ-GWAS and GWAS conducted in European populations (EUR-GWAS). We utilized two inclusion criteria of known loci: (i) EUR-GWAS has significant associations (P < 5 x 10−8) within 1Mb from the BBJ-lead variants and (ii) the BBJ-lead variant is in LD with the lead variant in the European-GWAS (r2 > 0.4 in European samples in 1KG phase3). The first criterion was added to exclude loci where EUR-GWAS has insufficient power (112 known loci remained after applying the first criterion). The second criterion was added because EUR-GWAS statistics at the BBJ-lead variant is not representing those at the EUR-lead variant when they are not in LD. b, effect sizes of BBJ- and EUR-GWAS at the BBJ-lead variants. All variants which passed the first criterion were used (n=112). Variants which passed the second criterion are shown in red (n=65). Since two variants have extremely large effect size, we provided two plots in different scales. The three variants with the opposite effect directions are marked by large dots, and their details are also provided. c, Regional association of T2D around rs12031188. Variants in LD (r2 > 0.4) with BBJ-lead variant (rs12031188) but not with EUR-lead variant are shown in red; Variants in LD (r2 > 0.4) with both lead variants are shown in blue. East Asians and Europeans in 1KG phase3 were used for LD calculation of the BBJ- and the EUR-lead variant, respectively.
Extended Data Fig. 9. Genetic correlations between male- and female-specific GWAS.
a. Genetic correlations between male- and female-specific GWAS. Estimates of genetic correlation and standard errors are provided. *: genetic correlation was significantly different from one (two-sided t test P = 2.2 x 10−3 < 0.05/20). b. The results of S-LDSC analysis based on sex-specific GWAS of asthma using 220 cell-type specific annotations. Significant annotations in either male or female asthma were shown (P < 0.05/220). Heterogeneity was tested by Cochran’s Q test, and its P values (Phet) were also provided. Black dashed line indicates P value = 0.05/220; grey dashed line indicates P value = 0.05.
Extended Data Fig. 10. S-LDSC results of four diseases in our GWAS.
The results of S-LDSC were plotted on the UMAP space. The significant results (FDR<0.05) were highlighted by cluster-specific colors (the same colors as used in Figure 4). The names of the top five most significant TFs were also shown on the plot. The results of diseases with less than five significant TF binding site tracks were shown.
Supplementary Material
ACKNOAWLEDGEMENTS
We acknowledge the staff of BBJ for their outstanding assistance. We express our heartfelt gratitude to Tohoku University Tohoku Medical Megabank Organization (ToMMo), Iwate Medical University Iwate Tohoku Medical Megabank Organization (IMM), the Japan Public Health Center–based Prospective (JPHC) Study, the Japan Multi-Institutional Collaborative Cohort (J-MICC) Study, and National Center for Geriatrics and Gerontology (NCGG) Biobank for their invaluable contributions to collecting control samples. We also express our gratitude to the Osaka Acute Coronary Insufficiency Study (OACIS) for the contribution to the replication study of coronary artery disease, and the National Cancer Center Hospital (NCCH) for the contribution to the replication study of lung cancer. We also express our gratitude to E.K. and H.S. for kindly sharing their results of ChIP-seq data analysis. We extend our appreciation to Y.Yukawa, Y.Yokoyama, and other members of the Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences for their great support. This research was supported by the Tailor-Made Medical Treatment Program (the BioBank Japan Project) of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), the Japan Agency for Medical Research and Development (AMED) under Grant Numbers JP17km0305002 (M.Kubo) and JP17km0305001 (M.M, S.Nagayama, Y.D, Y.Miki, T.Katagiri, O.O, W.O, H.I, T.Yoshida, I.I, T.Takahashi, J.I. and K.M), JST KAKENHI Grants (18H02932, S.I), and Research Program on Hepatitis from AMED (JP19fk0310109 and JP19fk0210020, K.C). ToMMo has been supported in part by MEXT-JST and AMED; most recent grant numbers are JP19km0105001 and JP19km0105002 (M.Y). IMM has been supported in part by MEXT-JST and AMED; most recent Grant Numbers are JP19km0105003 and JP19km0105004. The JPHC Study has been supported by the National Cancer Center Research and Development Fund since 2011 (the latest grant number: 29-A-4, S.T), and was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan from 1989 to 2010. The J-MICC Study has been supported by Grants-in-Aid for Scientific Research for Priority Areas of Cancer (17015018, N.H) and Innovative Areas (221S0001, H.T) and by JSPS KAKENHI Grants (CoBiA, 16H06277) from MEXT (H.T and K.W). The NCGG study was partly supported by AMED under Grant Number JP18kk0205009 (S.Niida) and JP20dk0207045 (K.O). OACIS has been supported by AMED (JP19ek0210081, Yasuhiko Sakata). Lung cancer study at NCCH was supported by The National Cancer Center Research and Development Fund (NCC Biobank), AMED (JP16ck0106096, T.K), and The Ministry of Health, Labour and Welfare (MHLW) program (H29-Gantaisaku-Ippann-025, T.K). The study at Fujita Health University was supported by AMED under Grant Numbers JP20dm0107097 (M.Ikeda and N.I), JP20km0405201 (N.I) and JP20km0405208 (M.Ikeda).
Data availability
GWAS summary statistics of the 42 diseases are publicly available at our website (JENGER; http://jenger.riken.jp/en/) and the National Bioscience Database Center (NBDC; https://humandbs.biosciencedbc.jp/en/) Human Database (Research ID: hum0014) without any access restrictions. GWAS genotype data for case samples were deposited at the NBDC Human Database (Research ID: hum0014).
REFERENCES
- 1.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popejoy AB & Fullerton SM Genomics is failing on diversity. Nature 538, 161–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morales J et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol. 19, 21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diversity matters. Nature Reviews Genetics 20, 495 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Sirugo G, Williams SM & Tishkoff SA The Missing Diversity in Human Genetic Studies. Cell 177, 26–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maas P et al. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA Oncol. 2, 1295–1302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher FR et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet 50, 928–936 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kullo IJ et al. Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk EstimatesCLINICAL PERSPECTIVE. Circulation 133, 1181–1188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan P et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 135, 2091–2101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilhjálmsson BJ et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am. J. Hum. Genet 97, 576–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wojcik GL et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estrada K et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a latino population the SIGMA Type 2 Diabetes Consortium. JAMA - J. Am. Med. Assoc 311, 2305–2314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moltke I et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512, 190–193 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Nagai A et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol 27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirata M et al. Cross-sectional analysis of BioBank Japan clinical data: A large cohort of 200,000 patients with 47 common diseases. J. Epidemiol 27, S9–S21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou W et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet 50, 1335–1341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazal S, Marquez-Luna C, Finucane HK & Price AL Reconciling S-LDSC and LDAK functional enrichment estimates. Nat. Genet 51, 1202–1204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazal S et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet 49, 1421–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata J et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat. Genet 51, 470–480 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Kanai M et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet 50, 390–400 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Ma T, Wu S, Yan W, Xie R & Zhou C A functional variant of ATG16L2 is associated with Crohn’s disease in the Chinese population. Color. Dis 18, O420–O426 (2016). [DOI] [PubMed] [Google Scholar]
- 25.van der Harst P & Verweij N The Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res CIRCRESAHA.117.312086 (2017). doi: 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvete O et al. The wide spectrum of POT1 gene variants correlates with multiple cancer types. Eur. J. Hum. Genet 25, 1278–1281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bainbridge MN et al. Germline Mutations in Shelterin Complex Genes Are Associated With Familial Glioma. J Natl Cancer Inst 107, 384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robles-Espinoza CD et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet 46, 478–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng PC & Henikoff S Predicting deleterious amino acid substitutions. Genome Res. 11, 863–74 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rentzsch P, Witten D, Cooper GM, Shendure J & Kircher M CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawase T et al. PH Domain-Only Protein PHLDA3 Is a p53-Regulated Repressor of Akt. Cell 136, 535–550 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Bujor AM et al. Akt Blockade Downregulates Collagen and Upregulates MMP1 in Human Dermal Fibroblasts. J. Invest. Dermatol 128, 1906–1914 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Aguet F et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–7 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Giambartolomei C et al. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi Y et al. Mice Lacking Hypertension Candidate Gene ATP2B1 in Vascular Smooth Muscle Cells Show Significant Blood Pressure Elevation. Hypertension 59, 854–860 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frati F et al. The Role of the Microbiome in Asthma: The Gut–Lung Axis. Int. J. Mol. Sci 20, 123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokholm J et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun 9, 141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInnes L, Healy J & Melville J UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. (2018). [Google Scholar]
- 42.Matsuda M, Sakamoto N & Fukumaki Y Delta-thalassemia caused by disruption of the site for an erythroid-specific transcription factor, GATA-1, in the delta-globin gene promoter. Blood 80, 1347–51 (1992). [PubMed] [Google Scholar]
- 43.De Gobbi M et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science 312, 1215–7 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Pevny L et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349, 257–260 (1991). [DOI] [PubMed] [Google Scholar]
- 45.Elhanati Y, Marcou Q, Mora T & Walczak AM RepgenHMM: A dynamic programming tool to infer the rules of immune receptor generation from sequence data. Bioinformatics 32, 1943–1951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welch JJ et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Lantz KA et al. Foxa2 regulates multiple pathways of insulin secretion. J. Clin. Invest 114, 512–520 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowen C et al. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 60, 6111–5 (2000). [PubMed] [Google Scholar]
- 49.Deplancke B, Alpern D & Gardeux V The Genetics of Transcription Factor DNA Binding Variation. Cell 166, 538–554 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Maurano MT et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science (80-.). 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaulton KJ et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet 47, 1415–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfe D, Dudek S, Ritchie MD & Pendergrass SA Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 6, 18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES (for method)
- 53.Kuriyama S et al. The Tohoku Medical Megabank Project: Design and Mission. J. Epidemiol 26, 493–511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altshuler DM et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okada Y et al. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat. Commun 9, 1631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matoba N et al. GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat. Hum. Behav (2020). doi: 10.1038/s41562-019-0805-1 [DOI] [PubMed] [Google Scholar]
- 57.Pruim RJ et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizuno H et al. Impact of atherosclerosis-related gene polymorphisms on mortality and recurrent events after myocardial infarction. Atherosclerosis 185, 400–5 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Asanomi Y et al. A rare functional variant of SHARPIN attenuates the inflammatory response and associates with increased risk of late-onset Alzheimer’s disease. Mol. Med 25, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS summary statistics of the 42 diseases are publicly available at our website (JENGER; http://jenger.riken.jp/en/) and the National Bioscience Database Center (NBDC; https://humandbs.biosciencedbc.jp/en/) Human Database (Research ID: hum0014) without any access restrictions. GWAS genotype data for case samples were deposited at the NBDC Human Database (Research ID: hum0014).