Abstract
There is enormous interest in measuring amyloid fibril structures, but most structural studies measure fibril formation in vitro using aqueous buffer. Ideally, one would like to measure fibril structure and mechanism under more physiological conditions. Toward this end, we have developed a method for studying amyloid fibril structure in human serum. Our approach uses isotope labeling, antibody depletion of the most abundant proteins (albumin and IgG), and infrared spectroscopy to measure aggregation in human serum with reduced protein content. Reducing the nonamyloid protein content enables the measurements by decreasing background signals but retains the full composition of salts, sugars, metal ions, etc. that are naturally present but usually missing from in vitro studies. We demonstrate the method by measuring the two-dimensional infrared (2D IR) spectra of isotopically labeled human islet amyloid polypeptide (hIAPP or amylin). We find that the fibril structure of hIAPP formed in serum differs from that formed via aggregation in aqueous buffer at residues Gly24 and Ala25, which reside in the putative “amyloidogenic core” or FGAIL region of the sequence. The spectra are consistent with extended parallel stacks of strands consistent with β-sheet-like structure, rather than a partially disordered loop that forms in aqueous buffer. These experiments provide a new method for using infrared spectroscopy to monitor the structure of proteins under physiological conditions and reveal the formation of a significantly different polymorph structure in the most important region of hIAPP.
The formation of β-sheet rich amyloid fibrils is associated with more than 20 human diseases, including type 2 diabetes, Alzheimer’s disease, and Parkinson’s disease.1,2 There is immense interest in measuring the molecular structure of these fibrils and better understanding how amyloid fibrils form. X-ray diffraction, solid state NMR, and electron crystallography are among the most commonly used methods for measuring amyloid structure and have provided the majority of our structural insight into date.3−8 Ideally, fibril formation would be studied directly in animals or humans. However, most often fibril formation is studied in vitro by dilution into an aqueous buffer or artificial membranes, because of the difficulty to apply protein structure characterization techniques to more biologically relevant environments. The most common experiment to study the formation of amyloid proteins is in vitro aggregation using fluorescence dyes like Tht that bind to amyloid β-sheets. While those experiments provide the time scales for amyloid formation, they provide very little structural information.9,10 The structures of fibrils and the mechanism by which they form may be very different in vitro versus in vivo because amyloid fibril formation depends on salt conditions, metal ions, and can be catalyzed by proteins. Indeed, the human islet amyloid polypeptide (hIAPP or amylin) is secreted from β-cell granules into the bloodstream.11
Infrared spectroscopies, including FTIR and vibrational circular dichroism, are useful tools for studying protein structure, including those that form amyloid.13−15 Isotope labels can be used to resolve the secondary structure of specific residues or, in the case of amyloids, the formation of β-sheets or stacked columns of proteins.16−25 2D IR spectroscopy is nonlinear infrared spectroscopy that has also been utilized with amyloids.26−30,34,58,60 The sensitivity of all three methods, FTIR, VCD, and 2D IR spectroscopy, arises from vibrational couplings between the backbone amide groups. The couplings create the characteristic frequencies in FTIR, doublets in VCD, and cross peaks and nonlinear intensities in 2D IR spectroscopy.31−33,36,37
Most often, amide I is used in neat solvents. The main challenge to utilizing the amide modes in biological fluids are background signals from the overlapping absorptions of the H2O solvent bending motion at 1644 cm−1 and the amide I vibrational modes of other proteins in the sample.38 To minimize the background, the naturally present H2O is replaced with an equal volume of D2O, as is often done in infrared experiments, along with the use of antibodies to extract the two most abundant proteins present in human serum, serum albumin, and immunoglobulins. In this report, we show that this procedure enables 2D IR spectra to be measured by isotopically labeled amyloid fibrils in biologically relevant human serum. The purpose of this approach is to provide a more realistic aggregation environment, with the presence and concentrations of salt, ions, proteins, and other species more similar to those encountered in the body.
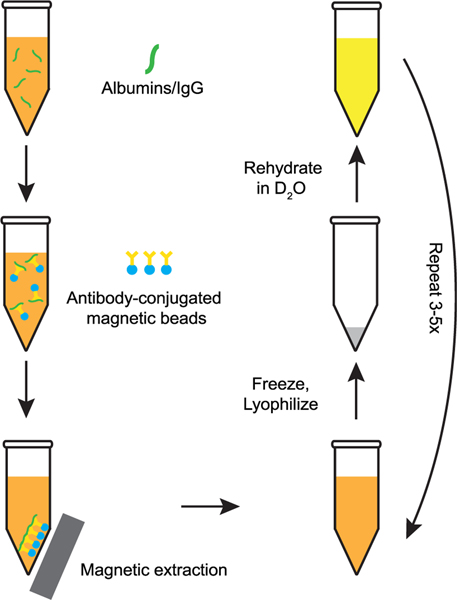
Our process for creating serum samples is as follows. Human serum naturally contains ca. 80 mg/mL of protein. Serum albumin, responsible for maintaining colloidal osmotic pressure and transport of hormones and fatty acids, accounts for ca. 50% of the protein present by mass. Serum albumins also have nonspecific chaperone activity that can affect the folding of hIAPP in the blood. Immunoglobulins, the most common antibodies in blood, account for the next most substantial mass fraction, with Immunoglobulin G (IgG) accounting for ca. 20% of the total protein present. We used a kit (PureProteomeTM Ablumin/IgG Depletion Kit) that makes use of albumin and IgG specific antibodies conjugated to magnetic beads to deplete these two most highly expressed proteins from serum. Doing so decreases the overall protein content to an estimated ca. 24 mg/mL, while maintaining the full complement of other salts, lipids, sugars, cholesterol, metal ions, etc. naturally present. After the protein depletion step, we flash freeze the serum using liquid N2 and lyophilize to remove H2O and then reconstitute the serum in an equal volume of D2O, such that the concentration of the solutes has not changed. This procedure is repeated multiple times to ensure nearly complete exchange of D2O for H2O. Our overall procedure for preparing the serum for use as a solvent for in vitro 2D IR infrared experiments is summarized in Figure 1.
Figure 1.
Schematic overview of the process used to prepare deuterated serum samples and deplete serum albumin and IgG concentrations to reduce protein background in the amide I region.
To gain residue specificity, we introduce 13C18O isotope labels into specific sites of our synthesized hIAPP peptides; this results in a ca. 60 cm−1 red shift of these residues, creating a separate spectral window with lower total background to study their secondary structure. Here, we introduce a double isotope label at residues G24A25 of hIAPP that are located in the “amyloidogenic core” of the protein. This approach is common in IR studies of amyloids to gain site-specific information on secondary structure.12,21,39−41 We compare the isotope signals observed in serum to those in aqueous buffer to assess the fibril structures formed under the two conditions. Our approach thus provides a means to gain structurally specific information on amyloids formed in biological fluids, providing more insight into amyloid formation under physiological conditions.
We use Tris buffer as a control comparison to the aggregation behavior and structure of hIAPP in serum. We have thoroughly characterized hIAPP in 20 mM Tris buffer in the past and observe reasonably consistent aggregation and structure behavior.39−41,60 The peptide samples are prepared in the same way for the buffer control and the serum experiments, a 2 μL sample at 1 mM is hydrated with either buffer or human serum.
Amyloid 2D IR Spectra in Protein-Depleted Human Serum.
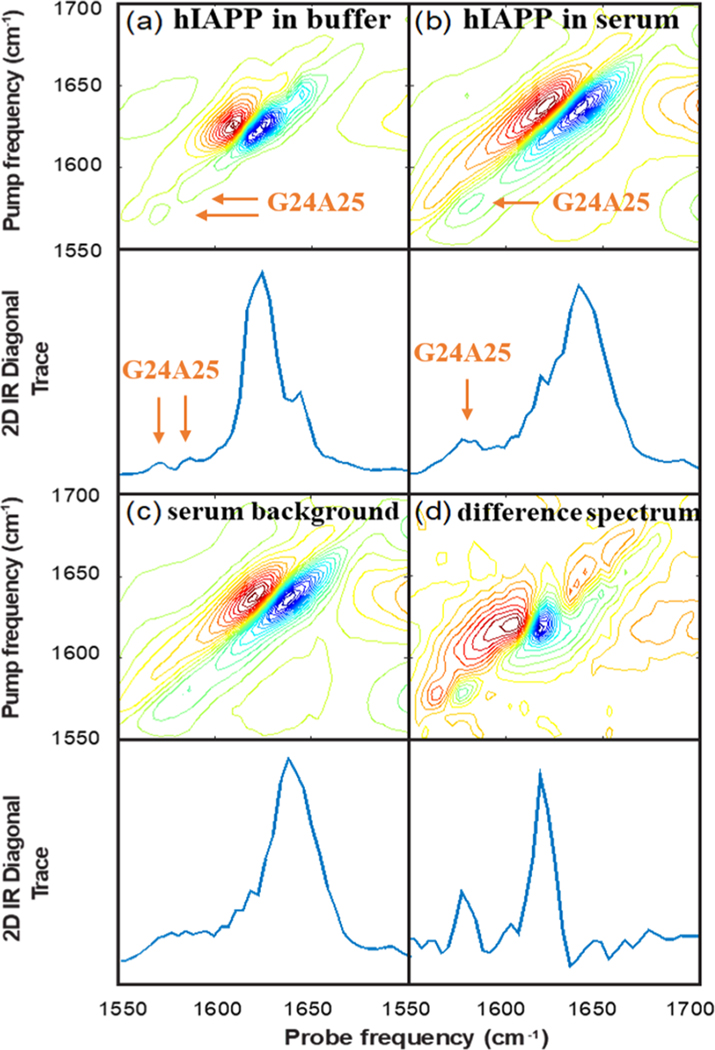
Figure 2a shows the 2D IR spectrum of G24A25 13C18O isotope labeled amylin formed by dilution into 20 mM Tris buffer and subsequently measured. The accompanying diagonal trace through the fundamental absorption is shown in the panel below. This spectrum shows an aggregated sample of amylin fibrils formed in a standard aqueous buffer and will be used for comparison to the spectra collected in protein-depleted human serum. The main β-sheet transition in the unlabeled region of the spectrum is observed at 1620 cm−1, consistent with previous 2D IR studies on human amylin.27,33,39,45 Smaller peaks are observed at ca. 1566 and 1580 cm−1 in the isotope labeled region of the spectrum. These report on the secondary structure of amylin fibrils at the G24 and A25 residues, which have previously been reported to occupy a partially disordered loop, consistent with their relatively weak intensities.48
Figure 2.
2D IR spectra of (a) G24A25 amylin in 20 mM Tris buffer at pD 7.5, (b) G24A25 amylin in depleted human serum, (c) depleted human serum (for background subtraction), and (d) G24A25 amylin in depleted human serum with the spectrum of the depleted serum subtracted to isolate the signal from G24A25 amylin.
We turn next to the spectra of hIAPP fibrils formed by dilution into the depleted serum. Figure 2b shows the 2D IR spectrum and its diagonal trace of G24A25 amylin reconstituted in depleted human serum. Figure 2c shows the 2D IR spectrum and its diagonal trace of the depleted serum without hIAPP added. The two spectra look very similar because hIAPP only represents a small fraction of protein in the sample; the main amide I feature at ca. 1645 cm−1 reflects the distribution of secondary structures found in the array of proteins present in the serum, which are mostly denatured and α-helical. Side-chain absorptions create the broad features observed in the 1560−1600 cm−1 range.35 The absorption of hIAPP at 1620 cm−1 is observed as a small shoulder on the main band in Figure 2b, and it is difficult to discern the location of the isotope labeled bands in Figure 2b as they overlap significantly with the side chain absorptions from the serum proteins.
To remove the serum features from the spectrum in Figure 2b, we subtract the spectrum in Figure 2c from that in Figure 2b. Due to heterogeneity in the sample path length or local solute concentration, we linearly scale the spectra in Figure 2c,d such that the maxima of their fundamental absorptions match before performing the subtraction. Figure 2d shows the difference spectrum. With the serum features subtracted, there are now two clear peaks in the spectra at 1620 cm−1, corresponding to the main amyloid β-sheet transition, and at 1575 cm−1, corresponding to the isotope labeled G24 and A25 residues.
To assess whether the same fibril structure is formed in serum and in buffer, we compare the spectra in Figure 2a,d. The isotope labeled region provides the most sensitive probe of structure and reports site-specifically on the structure of the G24 and A25 residues. We clearly see a different spectrum in the isotope labeled region between Figure 2a,d. The larger intensity of the peak at 1575 cm−1 in the serum is indicative of strong and negative interstrand coupling32 and therefore a more highly ordered structure at positions G24 and A25, consistent with an ordered β-sheet secondary structure. Thus, the aggregation in human serum produces a much different fibril structure at G24 and A25 than is formed in buffer.
Significantly, this structural difference exists in the “amyloidogenic core” or FGAIL region of the amylin sequence. Nearly 3 decades ago, the FGAIL region was identified as the “amyloidogenic core” of the sequence, and it was believed to adopt a β-sheet structure in the fibril.11 This inference was based largely on comparisons of amylin sequences from different species for the location of sequence differences (often proline residues, which inhibit β-sheet formation) along with differing observed aggregation propensities.49−52 Nearly 20 years after these initial observations, solid state NMR experiments determined the FGAIL region adopts a partially disordered loop in fibrils formed in standard in vitro experiments at pH 7.22. Our results in serum indicate that the FGAIL region G24A25 residues may adopt a β-sheet in the fibril formed under biological conditions. Indeed, a new hIAPP polymorph was recently reported using cryoEM at pH 6 in which G24 and A25 form the hydrophobic core of the fibrils.53 That structure would be consistent with the large and negative coupling constants measured in the 2D IR experiments.
While in this paper our focus is on amyloid fibril structure, one other observation to note is that proteins aggregate much more rapidly in human serum than they do in buffer; in buffer we can observe the disordered, unaggregated state of amylin or other peptides prior to amyloid formation, while in serum hIAPP aggregates within the deadtime between dilution and inserting the sample into the 2D IR spectrometer, which is about 2 min. This is consistent with previous observations of more crowded environments or higher salt concentrations leading to faster rates of amyloid formation.52,54,55 A modified setup in which dilution is performed within the spectrometer would decrease the deadtime to seconds.56
Use of an Internal Standard for Background Subtraction.
In the previous section we subtracted the serum background from the foreground by normalizing the two spectra to the bleach maximum at 1645 cm−1. An alternative method is to use an internal standard. For this reason, we performed the same experiment with an internal standard of trifluoroacetic acid (TFA) added as the final step of preparation of our depleted serum. TFA has a very strong antisymmetric carboxylic acid stretch at 1680 cm−1, which is outside the spectral window of the amide I absorptions and thus easily recognizable.
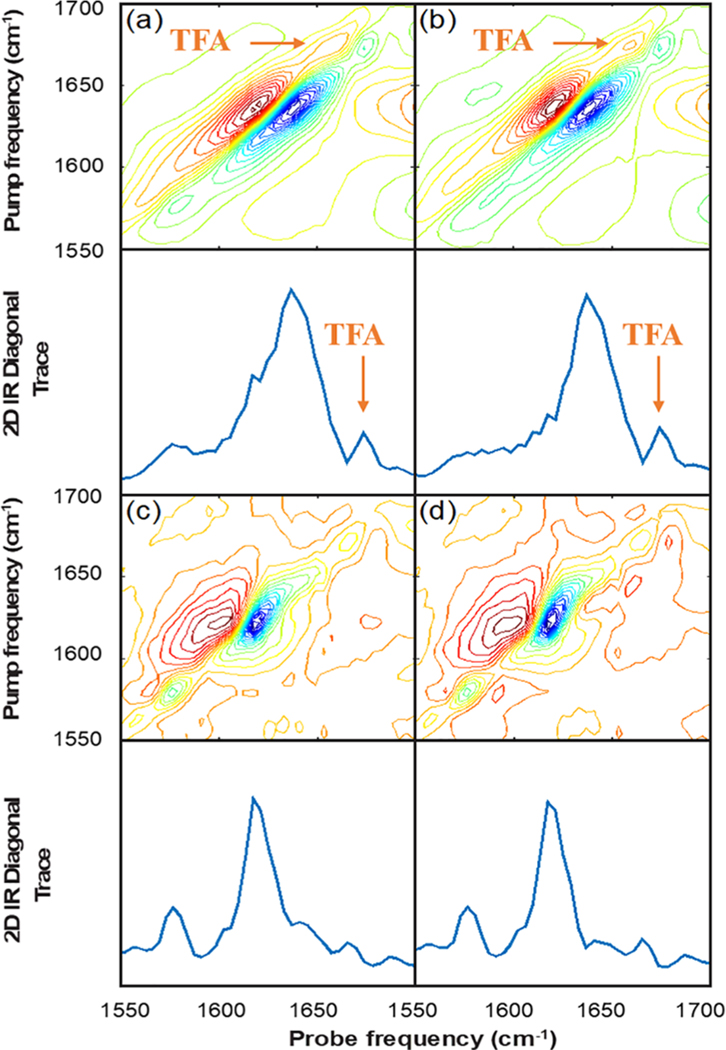
Figure 3a shows the 2D IR spectrum and diagonal trace of G24A25 amylin dissolved in depleted serum, analogous to Figure 2b, but with 0.03% trifluoroacetic acid (TFA) added as an internal standard. Figure 3b shows the spectrum of depleted human serum with 0.03% TFA added with no protein, which serves as our background spectrum. Similar to the data in Figure 2b,c, these spectra look quite similar and the amylin absorptions are only weakly visible to the eye in Figure 3a before subtraction.
Figure 3.
2D IR spectra of (a) G24A25 amylin in depleted serum with 0.03% TFA added as an internal standard, (b) depleted human serum with 0.03% TFA added as an internal standard, and (c) G24A25 amylin in depleted human serum with the internal standard TFA signal used to scale the subtraction of the depleted serum background and, for comparison, (d) the same spectral subtraction where the TFA signal was not used.
In Figure 3c we have subtracted the spectrum in Figure 3b from that in Figure 3a after applying a linear scaling factor based on the peak intensity of the TFA absorption in the diagonal trace. For comparison to the method used in the absence of an internal standard, Figure 3d shows the difference spectrum of the same data in Figure 3a,b, scaled this time to the bleach maximum analogous to the subtraction in Figure 2d. Both methods work well and produce two clearly visible peaks centered at 1575 and 1620 cm−1. These peaks have the same frequencies and relative intensities to those observed in Figure 2d, thus indicating that the presence of the small amount of TFA added has not changed the fibril structure.
The primary difference between the two subtraction methods is with regard to the intensity observed at ca. 1645 cm−1 after subtraction. Using the internal standard, which does not absorb in this region, there is a small shoulder in the 1645 cm−1 region after subtraction. Notably, this feature at 1645 cm−1 is also observed in the spectrum collected in buffer (Figure 2a), and for previously reported spectra of amylin fibrils.10 The intensity in this region of the spectrum is less clearly visible, if not absent, in Figure 3d, as well as in Figure 2d, which used the subtraction method based on scaling the spectra to the 1645 cm−1 peak. Thus, an internal standard based method can be used, but only provides a modest improvement in background subtraction, considering that the spectra in buffer and previously reported spectra indicate that amylin fibrils show intensity near 1645 cm−1 region in the 2D IR spectra.
In this manuscript we monitored aggregation of isotope labeled amylin in both 20 mM Tris buffer and human serum. Most significantly, we observe formation of a distinct fibril structure when aggregation takes place in serum, relative to aqueous buffer. This fibril structure is characterized by increased coupling strength in the G24/A25 residues, consistent with well-ordered parallel stacking of strands. The 2D IR spectra appear to be consistent with a recently published cryoEM polymorph in which these residues form the amyloidogenic core of the fibrils,53 which is a region of particular interest for hIAPP. The fact that different structures are observed in serum than in buffer highlights the importance of studying aggregation under as-close-to-possible biologically relevant conditions, which this new infrared method enables. The method is not limited to amyloids but might be used to study other proteins or processes, such as the impact of crowding on biological structures.57,59 It might also be extended to VCD or other infrared spectroscopies to reduce background signals.
Protein Samples and Synthesis.
Amylin with 13C18O isotope labels at the G24 and A25 positions was synthesized using a previously reported protocol. Briefly, a CEM Liberty Blue microwave peptide synthesizer was used to carry out the synthesis on a 0.1 mmol scale using 9-fluorenylmethoxycarbonyl (Fmoc) chemistry. 5-(4′-fmoc-aminomethyl-3′, 5-dimethoxyphenol) valeric acid (PAL−PEG-PS) resin was used to form the naturally occurring amidated C-terminus. Pseudoproline dipeptide derivatives were used at positions 9−10 and 19−20 to facilitate the synthesis.42,43 The 13C18O isotope labeled glycine and alanine used at positions 24 and 25 were prepared using 13C labeled amino acids purchased from Cambridge Isotope Laboratories, and a previously reported oxygen exchange reaction was used to incorporate the 18O label.44 Peptides were cleaved using standard trifluoroacetic acid (TFA) methods. The crude peptide was dissolved in acetic acid and lyophilized, after which they were dissolved in DMSO to promote formation of the disulfide bond between residues 2 and 7 that is found in naturally occurring amylin. Purification was achieved using reversed-phase high performance liquid chromatography on an XSelect CSH C18 OBD Prep Column. A two-buffer gradient was used: buffer A consisted of 100% H2O and 0.045% HCl (v/v) and buffer B included 80% acetonitrile, 20% H2O, and 0.045% HCl. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry confirmed the correct molecular mass.
Sample Preparation.
Lyophilized proteins were dissolved in deuterated hexafluoroisopropanol (HFIP-d) to deuterate exchangeable sites. Protein concentrations were determined using a NanoDrop 2000 and 280 nm extinction coefficient of 1615 M−1 cm−1 for amylin. Aliquots were prepared for reconstitution into buffer or serum.
The human serum used for reconstitution of protein samples was purchased from Sigma-Aldrich and is from AB male plasma. Aliquots (100 μL) of serum were flash frozen using liquid N2 and lyophilized to remove H2O, and the resulting solid was reconstituted in 100 μL of D2O to promote deuteration of exchangeable sites and to reduce background from H2O in the amide I region of the infrared spectrum. This procedure was repeated five times to minimize H2O content while maintaining the same overall solute concentration present in the original serum.
Serum albumins and IgG proteins were depleted from the serum using a commercially available kit from Millipore (PureProteomeTM Ablumin/IgG Depletion Kit). This procedure makes use of albumin and IgG specific antibodies conjugated to magnetic beads, which selectively extracts these proteins from the serum. This procedure was used as described in the text to reduce protein background from the serum, improving our ability to subtract the serum background to obtain the 2D IR spectrum of the protein of interest.
To initiate aggregation, a hIAPP aliquot is diluted with either the buffer or the depleted serum to a final concentration of 1 mM. Freshly dissolved samples were sealed between a pair of 2 × 25 mm CaF2 windows separated by a 56 μm Teflon spacer for 2D IR measurements.
2D IR Spectroscopy.
The details of the 2D IR spectrometer have been described previously.45−47 Briefly, a 3.2 W regenerative amplifier (Spectra Physics, Solstice) produces a 1 kHz train of 100 fs pulses centered at 800 nm. These pump an optical parametric amplifier (Light Conversion, TOPAS), producing signal and idler pulses centered at 1417 and 1845 nm, which then undergo difference frequency generation to produce pulses in the mid-IR centered at ca. 1600 cm−1. The mid-IR pulses are split roughly 95:5 into pump and probe beams respectively using a CaF2 wedge. The pump beam passes through a horizontal reflective germanium acousto-optic modulator pulse shaper,46 which uses acoustic waves to generate a collinear pair of compressed pump pulses with variable delays that are scanned to generate the pump axis of the 2D IR spectrum. The pump and probe beams are overlapped at the sample and their relative timing is adjusted to set t2 = 0 using a motorized stage (Newport). The probe is dispersed onto a 64-element mercury−cadmium−telluride (MCT) detector array (Infrared Associates), providing ca. 3 cm−1 spectral resolution along the probe axis.
ACKNOWLEDGMENTS
Justin Lomont is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
Funding
Support for this research was provided by NIH through R01DK79895 and R21AG061602.
Footnotes
The authors declare the following competing financial interest(s): Martin Zanni is co-owner of PhaseTech Spectroscopy, Inc., which sells mid-IR and visible pulse shapers and 2D spectrometers like those used in this publication.
Contributor Information
Caitlyn R. Fields, Department of Chemistry, University of Wisconsin—Madison, Madison, Wisconsin 53706, United States.
Sidney S. Dicke, Department of Chemistry, University of Wisconsin—Madison, Madison, Wisconsin 53706, United States
Megan K. Petti, Department of Chemistry, University of Wisconsin—Madison, Madison, Wisconsin 53706, United States.
Martin T. Zanni, Department of Chemistry, University of Wisconsin—Madison, Madison, Wisconsin 53706, United States.
Justin P. Lomont, Department of Chemistry, University of Wisconsin—Madison, Madison, Wisconsin 53706, United States.
REFERENCES
- (1).Chiti F; Dobson CM Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [DOI] [PubMed] [Google Scholar]
- (2).Bucciantini M; Giannoni E; Chiti F; Baroni F; Formigli L; Zurdo J; Taddei N; Ramponi G; Dobson CM; Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [DOI] [PubMed] [Google Scholar]
- (3).Sunde M; Serpell LC; Bartlam M; Fraser PE; Pepys MB; Blake CCF Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [DOI] [PubMed] [Google Scholar]
- (4).Jaikaran ET; Higham CE; Serpell LC; Zurdo J; Gross M; Clark A; Fraser PE Identification of a novel human islet amyloid polypeptide beta-sheet domain and factors influencing fibrillogenesis. J. Mol. Biol. 2001, 308, 515–525. [DOI] [PubMed] [Google Scholar]
- (5).Petkova AT; Ishii Y; Balbach JJ; Antzutkin ON; Leapman RD; Delaglio F; Tycko R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 16742−16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Luca S; Yau W-M; Leapman R; Tycko R. Peptide confirmation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry 2007, 46, 13505−13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Andronesi OC; von Bergen M; Biernat J; Seidel K; Griesinger C; Mandelkow E; Baldus M. Characterization of Alzheimer’s-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 5922–5928. [DOI] [PubMed] [Google Scholar]
- (8).Smith PES; Brender JR; Ramamoorthy A. Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: The case of islet amyloid polypeptide. J. Am. Chem. Soc. 2009, 131, 4470–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Biancalana M; Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta, Proteins Proteomics 2010, 1804, 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Groenning M. Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils − current status. J. Chem. Biol. 2010, 3, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lukinius A; Wilander A; Westermark GT; Engström U; Westermark P. Co-localization of islet amyloid polypeptide and insulin in the B cell secretory granules of the human and pancreatic islets. Diabetologia 1989, 32, 240–244. [DOI] [PubMed] [Google Scholar]
- (12).Moran SD; Zanni MT How to Get Insights into Amyloid Structure and Formation from Infrared Spectroscopy. J. Phys. Chem. Lett. 2014, 5, 1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dzwolak W; Smirnovas V; Jansen R; Winter R. Insulin forms amyloid in a strain-dependent manner: An FT-IR spectroscopic study. Protein Sci. 2004, 13, 1927–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kurouski D; Lu X; Popova L; Wan W; Shanmugasundaram M; Stubbs G; Dukor RK; Lednev IK; Nafie LA Is supramolecular filament chirality the underlying cause of major morphology differences in amyloid fibrils? J. Am. Chem. Soc. 2014, 136 (6), 2302–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jansen R; Dzwolak W; Winter R. Amyloidogenic self-assembly of insulin aggregates probed by high resolution force microscopy. Biophys. J. 2005, 88 (2), 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Decatur SM; Antonic J. Isotope-edited infrared spectroscopy of helical peptides. J. Am. Chem. Soc. 1999, 121, 11914−11915. [Google Scholar]
- (17).Tadesse L; Nazarbaghi R; Walters L. Isotopically enhanced infrared spectroscopy: a novel method for examining secondary structure at specific sites in conformationally heterogeneous peptides. J. Am. Chem. Soc. 1991, 113, 7036–7037. [Google Scholar]
- (18).Haris PI; Robillard GT; Van Dijk AA; Chapman D. Potential of carbon-13 and nitrogen-15 labeling for studying protein-protein interactions using Fourier-transform infrared spectroscopy. Biochemistry 1992, 31, 6279–6284. [DOI] [PubMed] [Google Scholar]
- (19).Huang C-Y; Getahun Z; Wang T; DeGrado WF; Gai F. Time-resolved infrared study of the helix-coil transition using 13C-labeled helical peptides. J. Am. Chem. Soc. 2001, 123, 12111−12112. [DOI] [PubMed] [Google Scholar]
- (20).Brewer SH; Song B; Raleigh DP; Dyer RB Residue specific resolution of protein folding dynamics using isotope-edited infrared temperature jump spectroscopy. Biochemistry 2007, 46, 3279–3285. [DOI] [PubMed] [Google Scholar]
- (21).Goldblatt G; Matos JO; Gornto J; Tatulian SA Isotope-edited FTIR reveals distinct aggregation structural behaviors of unmodified and pyroglutamylated amyloid β peptides. Phys. Chem. Chem. Phys. 2015, 17, 32149−32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hauser K; Engelhard M; Friedman N; Sheves M; Siebert F. Interpretation of amide I differences bands observed during protein reactions using site-directed isotopically labeled bacteriorhodopsin as a model system. J. Phys. Chem. A 2002, 106, 3553–3559. [Google Scholar]
- (23).Scheerer D; Chi H; McElheny D; Keirderling TA; Hauser K. Enhanced sensitivity to local dynamics in peptides by use of temperature-jump IR spectroscopy and isotope labeling. Chem. - Eur. J. 2020, 26, 3524–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Manor J; Arkin IT Gaining insight into membrane protein structure using isotope-edited FTIR. Biochim. Biophys. Acta, Biomembr. 2013, 1828, 2256–2264. [DOI] [PubMed] [Google Scholar]
- (25).Chu H-A; Hillier W; Debus RJ Evidence of the C-terminus of the D1 polypeptide of photosystem II is ligated to the manganese ion that undergoes oxidation during the S1 to S2 transition: An isotope-edited FTIR study. Biochemistry 2004, 43, 3152–3166. [DOI] [PubMed] [Google Scholar]
- (26).Shim SH; Strasfeld DB; Ling YL; Zanni MT Automated 2D IR spectroscopy using a mid-IR pulse shaper and application of this technology to the human islet amyloid polypeptide. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 14197−14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shim SH; Gupta R; Ling YL; Strasfeld DB; Raleigh DP; Zanni MT Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6614–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kim YS; Liu L; Axelsen PH; Hochstrasser RM Two-dimensional infrared spectra of isotopically diluted amyloid fibrils from Aβ40. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kim YS; Liu L; Hochstrasser RM 2D IR provides evidence for mobile water molecules in beta-amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 17751−17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pazos IM; Ma J; Mukherjee D; Gai F. Ultrafast hydrogen-bonding dynamics in amyloid fibrils. J. Phys. Chem. B 2018, 122, 11023−11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Krummel AT; Zanni MT Interpreting DNA vibrational circular dichroism spectra using a coupling model from two-dimensional infrared spectroscopy. J. Phys. Chem. B 2006, 110, 24720−24727. [DOI] [PubMed] [Google Scholar]
- (32).Hamm P; Zanni M. Concepts and Methods of 2D Infrared Spectroscopy; Cambridge University Press, 2011. [Google Scholar]
- (33).Dunkelberger EB; Grechko M; Zanni MT Transition Dipoles from 1D and 2D Infrared Spectroscopy Help Reveal the Secondary Structures of Proteins: Application to Amyloids. J. Phys. Chem. B 2015, 119, 14065−14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Cheatum CM; Tokmakoff A; Knoester J. Signatures of β-sheet secondary structures in linear and two-dimensional infrared spectroscopy. J. Chem. Phys. 2004, 120, 8201–8215. [DOI] [PubMed] [Google Scholar]
- (35).Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta, Bioenerg. 2007, 1767, 1073–1101. [DOI] [PubMed] [Google Scholar]
- (36).Grechko M; Zanni MT Quantification of transition dipole strengths using 1D and 2D spectroscopy for the identification of molecular structures via exciton delocalization: application to α-helices. J. Chem. Phys. 2012, 137, 184202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lomont JP; Ostrander JS; Ho J-J; Petti MK; Zanni MT Not All β-Sheets Are the Same: Amyloid Infrared Spectra, Transition Dipole Strengths, and Couplings Investigated by 2D IR Spectroscopy. J. Phys. Chem. B 2017, 121 (38), 8935–8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Max JJ; Chapados C. Isotope effects in liquid water by infrared spectroscopy. III. H2O and D2O spectra from 6000 to 0 cm−1. J. Chem. Phys. 2009, 131, 184505. [DOI] [PubMed] [Google Scholar]
- (39).Buchanan LE; Dunkelberger EB; Tran HQ; Cheng PN; Chiu CC; Cao P; Raleigh DP; Pablo J. J. de; Nowick JS; Zanni MT Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 19285−19290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Serrano AL; Lomont JP; Tu LH; Raleigh DP; Zanni MT A Free Energy Barrier Caused by the Refolding of an Oligomeric Intermediate Controls the Lag Time of Amyloid Formation by hIAPP. J. Am. Chem. Soc. 2017, 139, 16748−16758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Maj M; Lomont JP; Rich KL; Alperstein AM; Zanni MT Site-specific detection of protein secondary structure using 2D IR dihedral indexing: a proposed assembly mechanism of oligomeric hIAPP. Chem. Sci. 2018, 9, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Marek P; Woys A; Sutton K; Zanni M; Raleigh D. Efficient Microwave-Assisted Synthesis of Human Islet Amyloid Polypeptide Designed to Facilitate the Specific Incorporation of Labeled Amino Acids. Org. Lett. 2010, 12, 4848–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Abedini A; Raleigh DP Incorporation of Pseudoproline Derivatives Allows the Facile Synthesis of Human IAPP, a Highly Amyloidogenic and Aggregation-Prone Polypeptide. Org. Lett. 2005, 7, 693–696. [DOI] [PubMed] [Google Scholar]
- (44).Marecek J; Song B; Brewer S; Belyea J; Dyer RB; Raleigh DP A Simple and Economical Method for the Production of 13C,18O-Labeled Fmoc-Amino Acids with High Levels of Enrichment: Applications to Isotope-Edited IR Studies of Proteins. Org. Lett. 2007, 9, 4935–4937. [DOI] [PubMed] [Google Scholar]
- (45).Strasfeld DB; Ling YL; Shim SH; Zanni MT Tracking Fiber Formation in Human Islet Amyloid Polypeptide with Automated 2D-IR Spectroscopy. J. Am. Chem. Soc. 2008, 130, 6698–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Middleton CT; Woys AM; Mukherjee SS; Zanni MT Residue-specific structural kinetics of proteins through the union of isotope labeling, mid-IR pulse shaping, and coherent 2D IR spectroscopy. Methods 2010, 52, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ghosh A; Serrano AL; Oudenhoven TA; Ostrander JS; Eklund EC; Blair AF; Zanni MT Experimental implementations of 2D IR spectroscopy through a horizontal pulse shaper design and a focal plane array detector. Opt. Lett. 2016, 41, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Luca S; Yau WM; Leapman R; Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry 2007, 46, 13505−13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Westermark P; Wernstedt C; Wilander E; Hayden DW; O’Brien TD; Johnson KH Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Westermark P; Engströ U; Johnson KH; Westermark GT; Betsholtz C. Islet amyloid polypeptide: pinpointing amino acid residues linked to amyloid fibril formation. Proc. Natl. Acad. Sci. U. S. A. 1990, 87, 5036–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Westermark P; Wernstedt C; O’Brien TD; Hayden DW; Johnson KH Islet amyloid in type 2 human diabetes mellitus and adult diabetic cats contains a novel putative polypeptide hormone. Am. J. Pathol. 1987, 127, 414–417. [PMC free article] [PubMed] [Google Scholar]
- (52).Marek PJ; Patsalo V; Green DF; Raleigh DP Ionic Strength Effects on Amyloid Formation by Amylin Are a Complicated Interplay between Debye Screening, Ion Selectivity, and Hofmeister Effects. Biochemistry 2012, 51, 8478–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Röder C; Kupreichyk T; Gremer L; Schafer LU; Pothula KR; Ravelli RBG; Willbold D; Hoyer W; Schröder GF Amyloid fibril structure of islet amyloid polypeptide by cryo-electron microscopy reveals similarities with amyloid beta bioRxiv 2020.02.11.944546; 2020. DOI: 10.1101/2020.02.11.944546. [DOI] [PubMed] [Google Scholar]
- (54).White DA; Buell AK; Knowles TPJ; Welland ME; Dobson CM Protein Aggregation in Crowded Environments. J. Am. Chem. Soc. 2010, 132, 5170–5175. [DOI] [PubMed] [Google Scholar]
- (55).Ma Q; Fan J-B; Zhou Z; Zhou B-R; Meng SR; Hu J-Y; Chen J; Liang Y. The contrasting effect of macromolecular crowding on amyloid fibril formation. PLoS One 2012, 7, No. e36288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Tracy KM; Barich MV; Carver C; Krummel AT High Throughput 2D IR Spectroscopy Achieved by Interfacing Microfluidic Technology with a High Repetition Rate 2D IR Spectrometer. J. Phys. Chem. Lett. 2016, 7, 4865–4870. [DOI] [PubMed] [Google Scholar]
- (57).Zhai Y; Winter R. Effect of molecular crowding on the temperature-pressure stability diagram of ribonuclease A. ChemPhysChem 2013, 14, 386–393. [DOI] [PubMed] [Google Scholar]
- (58).Middleton CT; Buchanan LE; Dunkelberger EB; Zanni MT Utilizing Lifetimes to Suppress Random Coil Features in 2D IR Spectra of Peptides. J. Phys. Chem. Lett. 2011, 2, 2357–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Oh K; Baiz CR Crowding stabilizes DMSO-water hydrogen-bonding interactions. J. Phys. Chem. B 2018, 122, 5984–5990. [DOI] [PubMed] [Google Scholar]
- (60).Buchanan LE; Maj M; Dunkelberger EB; Cheng P; Nowick JS; Zanni MT Structural Polymorphs Suggest Competing Pathways for the Formation of Amyloid Fibrils That Diverge from a Common Intermediate Species. Biochemistry 2018, 57 (46), 6470–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]