Abstract
Extracellular vesicles (EVs) are now well established as important mediators of intercellular communication. EVs constitute a diverse group of secreted vesicles which function by the delivery of protein and nucleic acid cargoes from donor to recipient cells. In cancer, tumor cell-derived EVs are shown to promote disease progression by facilitating local reprogramming of the tumor microenvironment. EVs also have more distant systemic effects via transport in biofluids, and therefore have great potential as biomarkers for disease detection and monitoring. Recently, the discovery that EVs derived from glioblastoma cells can mediate immunosuppression by activation of immune checkpoint signaling and T cell dysfunction was reported. Mechanistically we showed that this occurs via direct binding of PD-L1 secreted in EVs, to its receptor PD1 expressed on the surface of activated T cells. This previously unidentified mechanism of tumor immunosuppression has been confirmed in subsequent independent studies, which have demonstrated the biologic importance of this mechanism across multiple tumor types. These studies have established a new and significant paradigm in which PD-L1 containing tumor cell-derived EVs cause immune suppression by the direct engagement of PD1 on T cells, decreasing their activation and providing a further barrier to protect tumors from T cell killing.
Keywords: cancer, extracellular vesicles, exosomes, immune checkpoints, immunosuppression, PD-L1
1. Introduction
Extracellular vesicles (EVs) encompass a diverse group of secreted membrane bound vesicles which are now well established as important mediators of intercellular communication,[1,2] ranging in size from 30–1000 nm, and include exosomes (30–150 nm). Their mechanism of action is based on the local and systemic cell-to-cell transfer of host cell-derived cargoes (including proteins, RNA, and DNA) to recipient cells. Fundamental roles in the biology of multicellular organisms have been described, but there are likely many details of EV mechanisms that are yet to be uncovered.
EVs play important roles in multiple diseases including cancer,[3,4] where they have been implicated in all cancer hallmarks, and have been shown to facilitate tumor progression,[5] angiogenesis,[6] and immune tolerance.[7,8] In addition to their fundamental mechanistic roles in biology, EVs may be useful as therapeutic gene delivery vehicles,[9,10] and also may be a rich source of blood-based disease biomarkers, for diagnosis and monitoring.[11,12] Furthermore, improved understanding of the roles of EVs in disease will facilitate the identification of novel therapeutic targets.
In cancer, the emergence of immune checkpoint blockade (ICB) as a therapeutic strategy has revolutionized the treatment of certain tumors such as advanced melanoma and lung cancer, where unprecedented responses have been observed.[13–15] Immune checkpoints refer to a number of immune inhibitory pathways which are involved in the balanced regulation of physiological T cell responses.[16–18] Expression of immune checkpoint ligands in the tumor microenvironment can engage cognate receptors on T cells, and reduce immune cell tumor killing. ICB target interactions include T cell expressed proteins such as CTLA4, TIM3, and PD1. PD-L1 expressed on the surface of tumor and antigen-presenting cells binds to PD1, which is expressed by activated T cells. In normal physiology, this machinery is used as a feedback mechanism to control T cell activation. Activated T cells release interferon gamma (IFNγ) which upregulates PD-L1 locally in surrounding cells, allowing engagement of PD1 on T cells, followed by a state of reduced activity known as T cell exhaustion, limiting non-specific damage to tissues. In tumors, activation of these pathways leads to blockade of T cell activation and protects tumor cells from T cell-mediated killing. It is now clear that tumors use various mechanisms to hijack immune checkpoints to evade immune recognition by tumor antigen-specific T cells.[16–18]
Recent studies have shown that tumor cell-derived EVs carry multiple layers of immune modulatory capacities,[7,8] and our knowledge of how these influence infiltrating lymphocytes within the tumor microenvironment is evolving rapidly. Here we highlight a relatively newly identified property of EVs which is mediated by the extracellular engagement of cell signaling pathways by direct interaction of PD-L1 on EVs with PD1 on the surface of T cells to mediate T cell dysfunction in cancer. We found that the immune checkpoint ligand PD-L1 is expressed on glioblastoma cell-derived EVs where it can block T cell activation via direct interaction with PD-1 on the T cell plasma membrane.[19] Additional studies[20–24] have reported similar observations in other tumor types and made additional insights into this phenomenon suggesting suppression of T cell activation by EV expressed PD-L1 interaction with T cell PD1 may be an important general mechanism and new paradigm in tumor immunosuppression.
2. Glioblastoma EVs Block T Cell Activation via PD-1/PD-L1 Interactions (Ricklefs et al.)[19]
One of the defining characteristics of glioblastoma, a universally fatal brain cancer,[25] is its profound local and systemic immunosuppression.[26,27] Glioblastoma patients show severe T cell dysfunction, both intratumorally and systemically.[28] Glioblastoma cells release anti-inflammatory cytokines, such as IL10 and TGFβ, and express immune checkpoint ligands including PD-L1,[29,30] which engage immune checkpoint-activating regulatory receptors on T cells. Recent studies have shown that in glioblastoma patients T cells become sequestered in the bone marrow, further quenching their anti-tumor potential.[31] There are increasing efforts to target glioblastoma clinically using immune therapies including ICB, which despite its unprecedented activity in some tumors,[13–15] has not yet shown broad efficacy in glioblastoma.[32–34]
Prior studies had shown that EVs derived from glioblastoma cells may have immunosuppressive functions, although mechanisms were not clear.[35,36] In our study,[19] we examined EVs derived from multiple distinct glioblastoma cell lines, and found that they all suppressed the activation of both CD4+ and CD8+ T cells. This was shown in various experimental settings, using flow cytometry to evaluate T cell activation markers CD69 and CD25, as well as T cell proliferation (data is summarized in Figure 1). First, human glioblastoma-patient stem-like cell (GSC) derived EVs were co-incubated with peripheral blood mononuclear cells (PBMCs) from healthy human donors stimulated with anti-CD3 to mimic antigen recognition through the T-cell receptor (TCR), showing a significant blockade of T cell activation. EVs derived from human neural stem cells had no effect in these assays, suggesting cancer cell EV specificity. Also, glioblastoma-derived EVs did not alter activation marker expression in unstimulated T cells or change the overall CD3+ population. Similar inhibition was observed using both CD8+ and CD4+ T cells purified from donor PBMCs. To ensure that the observed EV-mediated suppression of T cell activation could occur in the context of TCR recognition of MHC-bound antigen, we used mouse CD8+ T cells that react to the potent LCMV gp33 model antigen, isolated from transgenic P14 mice.[37] To present the gp33 antigen, mouse dendritic cells (DCs) were pulsed with gp33 peptide and then co-incubated with CD8+ T cells from p14 mice in the presence or absence of mouse glioblastoma EVs. Similar to the experiment in human cells, EVs were able to suppress T cell activation. These findings show that glioblastoma cell-derived EVs can specifically suppress TCR-mediated T cell activation.
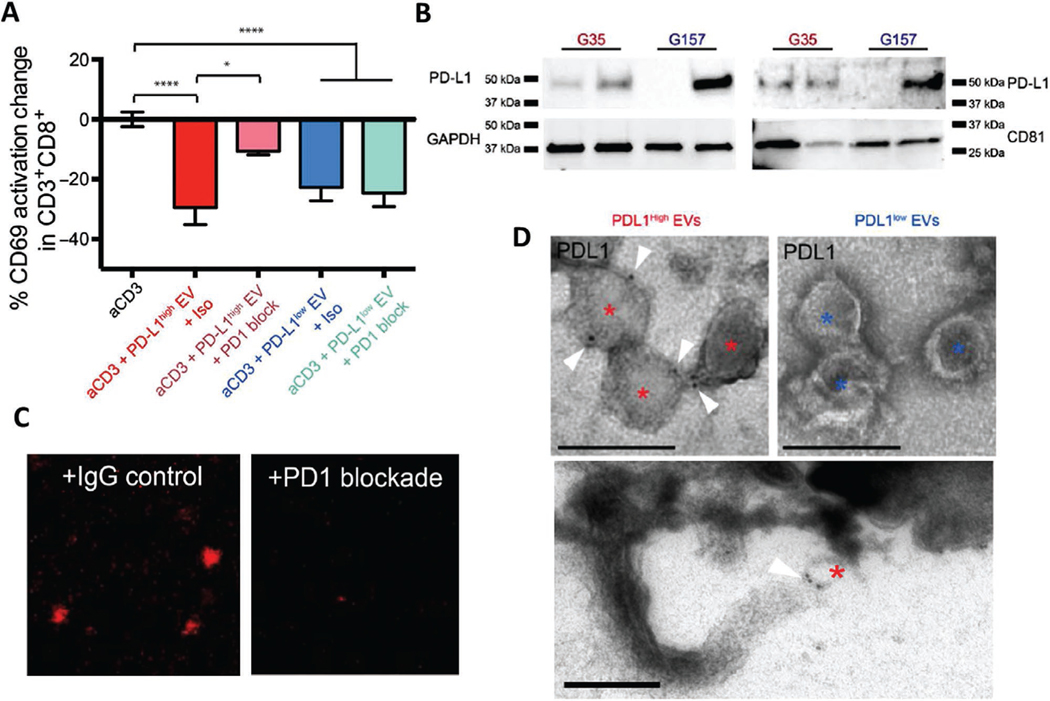
Figure 1.
PD-L1 carried on extracellular vesicles inhibits T cell activation via direct binding to PD-1. A) PD1 blockade prevents the inhibition of PD-L1high GSC EVs on PBMCs. Percent change in CD69 expression CD8+ T cells is shown as a measure of T cell activation. PD1 blocking antibody or isotype control was added at day 0 and CD69 levels were determined 48 h later (n = 7 PBMC donors, means ± SD). B) IFNγ-mediated increase of PD-L1 expression levels in PD-L1high and PD-L1low GSCs as shown by Western blots. C) Representative confocal microscopy images showing that fluorescently labeled EVs isolated from PD-L1high GSCs can bind to wells coated with recombinant PD1 (left panel). EV binding is displaced by co-incubation with an anti-PD1 antibody (right panel), indicating specificity of binding. D) Immuno-electron microscope images of PD-L1 immunolabeled PD-L1high and PD-L1low glioblastoma EVs are shown. Asterisks mark each EV, and the white triangle indicates gold-immunolabeled PD-L1, scale bar = 200 nm. Lower panel shows a PD-L1-labelled EV apparently ligated to a process at the lymphocyte surface. Reproduced under the terms and conditions of the Creative Commons Attribution, Non Commercial Licence 4.0.[19]
To further understand these observations we asked whether EV-mediated suppression of T cell activation involved PD-L1. T cell/EV co-incubation studies in the presence of a PD1/PD-L1 interaction blocking antibody showed that anti-PD1 was able to block the effects of EVs on TCR activation to a large extent but only in a subset of GSCs. We then went on to perform several further experiments which provided a deeper and more robust understanding of these observations. First, Western blotting showed variability in the expression of PD-L1 between different GSC lines, and between their respective EVs. PD-L1 was readily detectable in GSCs with a more mesenchymal transcriptional subtype[5,38] and their EVs (PD-L1high), but was expressed at low or undetectable levels in GSCs with a more proneural transcriptional subtype and their EVs (PD-L1low). As expected anti-PD1 treatment had no effect on T cell activation with PD-L1low EVs. Interestingly, PD-L1 expression could be strongly upregulated by treatment with IFNγ in PD-L1low GSCs and their EVs, whereas PD-L1high EVs had a constitutively high expression. EVs from IFNγ-treated PD-L1low GSCs also inhibited T cell activation, and blockade of PD1/PD-L1 interaction significantly reversed the inhibition of T cell activation. Analysis of RNA sequencing databases using patient glioblastoma specimens showed that the mesenchymal transcriptional glioblastoma subtype[38] was associated with higher PD-L1 levels, compared with other subtypes. Additionally expression of other IFNγ-responsive genes[39] such as interferon regulatory factor 1 (IRF1), HLA-A, and ICAM1, was correlated with PD-L1 in agreement with a model where glioblastoma evades immune recognition by expression of PD-L1 and engagement of PD1.
Immuno-electron microscopy (EM) showed that PD-L1 was present on the surface of PD-L1high EVs but was barely detectable on the surface of PD-L1low EVs. The overall EV size distribution did not reveal significant differences between PD-L1high and PD-L1low vesicles. During our EM studies we identified binding of PD-L1high EVs bound to the surface of lymphocytes (Figure 1). EVs fluorescently labeled with palmitoylated tdTomato (palmtdT) and palmitoylated green-fluorescent protein (GFP) (palmGFP)[40] could be visualized to localize to the surface of PBMCs and CD3+ sorted cells. To investigate whether there was in vivo co-localization of labeled EVs to infiltrating lymphocytes in mouse glioblastoma, we intracranially injected palmGFP-labeled murine CT2A glioma cells that constantly produce palmGFP-EVs in mice.[40] This showed visual evidence of co-localization of these EVs to CD3+ cells in vivo in mouse glioblastoma. We then transduced GSCs with a vector encoding a PD-L1/red fluorescent protein (RFP) fusion protein to visualize PD-L1 expression. Purified PD-L1/RFP EVs from GSCs could be observed bound to the outer surface of PBMCs, which was reduced by addition of anti-PD1. Further support for direct PD1 /PD-L1-EV interaction was provided by biochemical analysis showed that PD-L1high EVs bound to recombinant PD-1 in a dose-dependent manner that could be inhibited in the presence of a PD-1 antibody (Figure 1). Taken together, these studies established for the first time that PD-L1 on the surface of tumor-derived EVs can engage immune checkpoint pathways to block T cell activation, suggesting an important new mechanism of immune suppression in cancer. Additional studies have since confirmed and expanded on the importance of exosomal PD-L1 in other cancer types as described in detail below.
3. Exosomal PD-L1 Promotes T Cell Dysfunction in Metastatic Melanoma (Chen et al.)[20]
Advanced melanoma has become the poster-child of immunotherapy, with remarkable responses seen in response to ICB and other immunostimulatory therapies including oncolytic viruses.[13,14,41,42] These high response rates, thought to be at least partly due to the high mutational loads in this type of tumor which lead to an increased probability of overcoming tumor-mediated immunosuppression and raising a potent immune response in the presence of ICB.[43]
However, despite these breakthroughs, resistance to immunotherapy remains a problem in the majority of patients, and understanding in greater detail underlying mechanisms remains an area of intense interest. In a study of metastatic melanoma, Chen et al.[20] used reverse-phase protein assays (RPPA) to analyze the protein composition of melanomaderived EVs, focusing on the exosomal fraction. They found that metastatic melanoma cells release exosomes containing PD-L1 at significantly higher levels than primary melanoma cells, and PD-L1 is also expressed in EVs from murine B16-F10 melanoma cells. Similar to our study, the authors used immuno-EM to show that exosomal PD-L1 is expressed at the vesicle surface, and also that the extracellular domain was exposed on the external face of the exosome in a similar orientation to cell surface PD-L1. Exosomal PD-L1 was able to bind to recombinant PD1 in a concentration-dependent manner, and the interaction could be disrupted by anti-PD-L1 blocking antibodies. Also similar to our study, IFNγ upregulated PD-L1 in melanoma cells and their EVs, leading to increased PD-1 binding. Blockade of exosome generation by genetic knockdown of RAB27A or the ESCRT subunit HRS confirmed that PD-L1 secretion was dependent on the exosome releasing machinery. PD-L1 was also detected on circulating exosomes in vivo in melanoma xenograft models, and its levels correlated with tumor volume. The authors also showed that metastatic melanoma patient EVs purified from plasma had significantly higher PD-L1 levels than in healthy controls, with the greatest differences seen in exosomes rather than microvesicle fractions.
To investigate the mechanistic underpinnings of these observations the authors then showed a physical interaction between tumor exosomes and CD8+ T cells purified from human peripheral blood. This interaction increased with CD8+ T cell activation, and with the use of exosomes from IFNγ treated cells, consistent with PD-L1/PD1 interaction. To test this further, the authors then chose a melanoma cell line (MEL264) which does not express PD-L1 and showed that overexpression of PD-L1 led to a PD-L1-dependent reduction in CD8+ T cell activation, as measured by proliferation, cytokine production, and cytotoxicity, and this was inhibited for the most part by an anti-PD-L1 blocking antibody. Similar observations were made using exosomes from PD-L1 expressing human melanoma cells, and from murine B16-F10 melanoma. PD-L1 was also seen in exosomes from breast and lung cancer cells, suggesting that this maybe a widespread mechanism.
This study then went on to examine the effects of PD-L1 exosomes in vivo. Intravenous injection of exosomes from wild type B16 melanoma cells into PD-L1 knockdown B16 tumor-bearing mice promoted tumor growth and reduced CD8+ T cell infiltrates. Reduced T cell proliferation was observed in both spleens and lymph nodes in treated animals. These effects were blocked by preincubation of exosomes with anti-PD-L1 antibodies. These data support a role of PD-L1 on tumor-derived exosomes in systemic tumor driven immunosuppression in vivo. Studies in human melanoma patients showed that the level of circulating exosomal PD-L1 positively correlated with serum IFNγ levels and overall tumor burden, and dynamically changed during the course of anti-PD1 therapy. Pre-treatment levels of circulating exosomal PD-L1 were higher in non-responders. Total circulating PD-L1, or microvesicle associated PD-L1 were not as significantly correlated with response as exosomal PD-L1. These data suggest that circulating exosomal PD-L1 may therefore be a relevant target of ICB and could be a predictor of clinical outcome for anti-PD1 therapies.
4. Targeting of Exosomal PD-L1 Can Improve Immunotherapeutic Approaches (Poggio et al.)[21]
Poggio et al. came upon the role of exosomal PD-L1 in tumor immunosuppression by investigating the regulation of PD-L1 expression in cancer cells. They made the initial observation that PD-L1 mRNA and protein levels were discordant across a panel of cancer cell lines including prostate and melanoma cells. In PC3 prostate cancer cells, although protein levels were similar as measured by Western blot, PD-L1 mRNA levels were 15-fold higher compared with SK-MEL-28 melanoma cells. Translation rates and protein degradation rates were similar between cell lines. Further investigation revealed that prostate cancer cells had a higher level of PD-L1 protein in EVs than SK-MEL-28 cells. This differential secretion of PD-L1 was due to prostate cancer cells packaging greater amounts of PD-L1 into their EVs, and accounted for the differences observed between them. Similar to the two reports described above,[19,20] IFNγ upregulated the amount of PD-L1 in tumor cells and in their EVs. The authors then fractionated the vesicles further and similar to Chen et al. they saw a preferential PD-L1 enrichment in the exosomal fraction compared with other EVs. Genetic disruption of exosome secretion via CRISPR/Cas9-mediated deletion of RAB27A or NSMASE2 genes in PC3 prostate cancer cells blocked exosome formation and PD-L1 secretion, supporting the notion that exosome release provides a major pathway for extracellular PD-L1 secretion. Further biochemical analysis showed that exosomal PD-L1 originates from the surface of PC3 cells, rather than being directly packaged from the ER or early Golgi. Similar to the other two studies, the authors tested whether exosomal PD-L1 was functional, this time measuring suppression of IL2 secretion in Jurkat T cells. After CRISPR/Cas9 deletion of PDL1 in PC3 cells, exosomes were unable to suppress IL2 secretion, thus, similar to the previous two studies, data supports the concept that exosomal PD-L1 could suppress T cell activation in vitro.
In vivo, the authors examined the anti-PD-L1 resistant TRAMP-C2 prostate cancer mouse model. TRAMP-C2 tumors deleted for RAB27A, NSMASE2, or PDL1 showed a high level of growth suppression compared with wild type controls in immunocompetent mice. Also, spleen sizes were not reduced in mice bearing mutant tumors, changes in the draining lymph nodes were not observed and no anti-tumor memory was established. When exosomal biogenesis-deficient or PD-L1 knockdown tumors were grown in immune-deficient NSG mice, no such differences were observed, supporting an immune-mediated effect. Similar, but less pronounced results were seen in the MC38 colon cancer model, which is partially resistant to anti-PD-L1. The authors then used engineered MC38 cells to dissect the role of exosomal PD-L1 in vivo. They found that PD-L1 knockout cells had a similar survival to RAB27A knockout cells after anti-PD-L1 treatment. This led them to conclude that exosomal PD-L1 must be somehow resistant to anti-PD-L1. When wild-type or mutant TRAMP-C2 cells were injected on opposite flanks of the same animal, the growth of the wild type cells was always suppressed, with increased tumor immune infiltrates. This shows that PD-L1 expressing exosomes secreted by wild type cells could not rescue the growth of mutant tumors; rather the mutant tumors appeared able to induce an anti-tumor immune response blocking growth of wild type cells. Finally, exosomes from the wild type tumors were able to suppress the anti-tumor immune response, and promoted tumor growth whereas PD-L1 deficient exosomes had no effect. Thus exogenous exosomal PD-L1 suppresses the anti-tumor immune response thus promoting tumor growth, and may require targeting modalities in addition to anti-PD-L1.
5. Exosomal PD-L1 in Breast Cancer (Yang et al.)[22]
In a brief letter to the editor, Yang et al. described a series of observations supporting a role of exosomal PD-L1 in immune evasion in breast cancer.[22] Exosomes from human MDA-MB-231 and mouse 4T1 breast cancer cells were isolated and shown to contain PD-L1, and to mediate PD-L1-dependent blockade of T cell activation in vitro. Similar to the above studies, injection of PD-L1 expressing EVs into animal models led to increased tumor growth compared with PD-L1 knockdown controls suggesting a systemic immunosuppressive effect. Accordingly, PD-L1 was also expressed at higher levels in more advanced human breast cancers. Treatment of 4T1 breast cancer cells with the exosome secretion inhibitor GW4869 in vivo inhibited exosome secretion and tumor growth, and significantly enhanced anti-PD-L1 therapeutic efficacy. Tet-on inducible RAB27A knockdown 4T1 cells showed a similar effect. EV PD-L1 was also delivered to APCs, suggesting that in addition to suppressing T cells directly, exosomal PD-L1 can be delivered to multiple cell types in the tumor microenvironment. The data suggests that combining inhibitors of exosomal secretion and anti-PD-L1 could be a useful therapeutic approach.
6. Promotion of Tumor Growth via Exosomal PD-L1 in Non-Small Cell Lung Cancer (Kim et al.)[23]
Further support for the role of exosomal PD-L1 in the suppression of T cell-mediated anti-tumor immunity was provided by a study focused on lung cancer. Similar to the studies above, exosomes derived from lung cancer cells contained PD-L1 and inhibited IFNγ secretion by Jurkat T cells which could be reversed by PD-L1 knockout or by specific PD-L1 blocking antibodies. In vivo, expression of PD-L1 on tumor cells, or injection of PD-L1 containing EVs promoted tumor growth. Also, PD-L1 was detected in circulating exosomes from non-small cell lung cancer patients, and its abundance was correlated with PD-L1 positivity in tumor tissues. These findings provide further support for the concept of exosomal PD-L1 as a mediator of tumor immune escape.
7. Exosomal PD-L1 Functions in Head and Neck Cancer (Theodoraki et al.)[24]
Immunosuppression is important in head and neck squamous cell cancer (HNSCC), where tumor PD-L1 is associated with patient survival, and exosomes have T cell suppressive functions.[44] HNSCC has relatively low response rates to ICB. In this study, published at a similar time to our paper,[19] Theodoraki et al. isolated exosomes from patients’ serum using anti-CD63 beads and measured PD-L1 levels. They found that exosomal PD-L1 correlated with tumor grade, and that PD1-dependent CD8+ T cell suppression could be mediated by PD-L1high exosomes but not PD-L1low EVs, as measured by T cell surface CD69 levels. The authors also examined the link between tumor characteristics with soluble “free” serum PD-L1 and exosomal PD1 but did not establish any correlations with clinicopathological parameters. Importantly, this study showed that PD-L1high EVs derived directly from patients could mediate PD1-dependent immunosuppressive effects on CD8+ T cells, supporting the relevance of exosomal PD-L1 in in human cancer immune evasion.
8. Discussion
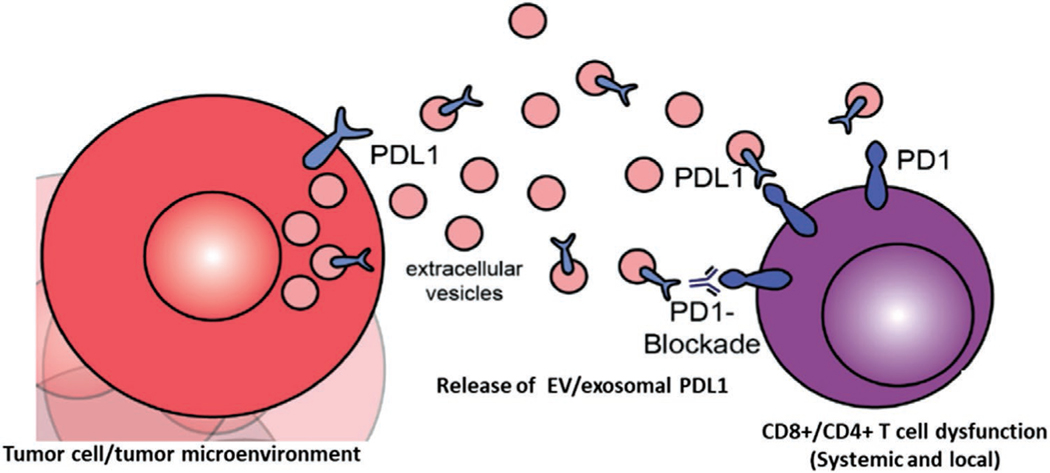
The studies outlined in this review establish that exosomes released by cancer cells can suppress T cell activation by direct binding of exosomal PD-L1 to PD1 on the surface of T cells. A simple scheme for this interaction is shown in Figure 2. This is clearly demonstrable in vitro in multiple tumor types. Evidence from a range of animal cancer models using PD-L1 and exosome deficient cell lines provides strong support of the relevance observations to human cancer. This is further supported by studies on human patients that show circulating exosomal PD-L1 may reflect tumor volumes and responses to immunotherapy, as well as their direct effects on CD8+ T cells.[24] The major conclusions from these studies so far can be summarized as follows:
Figure 2.
Diagrammatic representation of T cell suppressive effects of EV/exosomal PD-L1. Tumor cells and potentially other cells in the tumor microenvironment release extracellular vesicles including exosomes. These may carry PD-L1 depending on its expression status in the originating cell. PD-L1 is presented on the surface of EVs, where is can act systemically and locally to impair T cell activation via binding of PD1, and subsequent blockade of T cell receptor signaling and tumor cell killing.
PD-L1 expressing tumor cells release EVs containing PD-L1. Data suggests that PD-L1 is present mainly in the exosomal fraction.
IFNγ-mediated upregulation of PD-L1 in tumor cells leads to proportionally greater PD-L1 content in the associated EVs.
PD-L1 is present on the surface of these vesicles with its extracellular domain exposed allowing suppression of T cell activation in vitro via direct binding to PD-1. This interaction can be targeted by anti-PD1/PD-L1 interaction blocking antibodies.
Exosomal PD-L1 is immunosuppressive in vivo, leading to faster tumor growth.
Targeting tumor exosomes, an/or tumor exosomal PD-L1 may be a useful therapeutic strategy
High levels of PD-L1 on exosomes may be an indicator of non-response to ICB in patients.
These data provide compelling evidence of an immunosuppressive role for PD-L1 in EVs in cancer. A role as a biomarker is suggested but would need assessment in larger patient cohorts, and in multiple tumor types. An increase of exosomal PD-L1 during treatment could indicate the establishment of immunity and provides a rationale for developing circulating exosomal PD-L1 as a predictor for outcomes of anti-PD1 therapy. This is less clear in the case of glioblastoma where our published data showed that we were not able to detect a significant correlation between PD-L1 in circulating EVs in glioblastoma patients as it relates to tumor volume or patient versus healthy serum.[19] However, we did not look solely in the exosome fraction, which does appear to be a stronger indicator of response.[20,21] We were able to detect a potential relationship between PD-L1 DNA content in EVs and tumor volume. This is under further investigation.
Mechanistically, it seems that exosomal PD-L1 may be acting locally, where it could directly suppress activation of CD4+ and CD8+ T cells in the tumor microenvironment, effectively shielding tumor cells from destruction, and also systemically, by shutting down T cells at various sites. The papers above provide evidence for both of these mechanisms. One interesting and novel feature of these studies is the observation that binding can occur between EVs and lymphocytes at the lymphocyte surface. Thus rather than internalization and cargo delivery exosomal PD-L1 may be able to engage ligands at the cell surface to illicit their functions.
The biochemical structure of this exosome/lymphocyte synapse requires further study to elucidate fully whether other ligand/receptor interactions are involved, which cell signaling pathways are engaged and whether cargo delivery is also of importance. Data from the study by Poggio et al.[21] suggested that it may be necessary to target exosomal PD-L1 independently of cellular PD-L1/PD1 interactions. The use of drugs or genetic means to prevent exosome secretion support this concept,[23] although mechanistically it is not well understood why this is the case.
Importantly, our data show that there are additional mechanisms of T cell suppression by EVs as well as PD1 activation. As we showed, PD-L1low EVs were able to suppress T cell activation independently of PD1, and anti-PD1 was not able to fully reverse T cell inhibition even in PD-L1high EVs suggesting additional mechanisms are at play. This was supported by our observation of upregulated IL10 and IDO1 in T cells treated with PD-L1low EVs. Thus there may be multiple layers of immunosuppression mediated by EVs, depending on their target cell,[45] and also on the subpopulation of EVs involved (i.e., exosomes versus microvesicles). In vivo, multiple cells express PD-L1 in the tumor microenvironment,[45] their contribution to this phenomenon has not yet been examined. Finally, the role of the second PD1 ligand, PD-L2,[46] which is also expressed in tumors and tumor cell-derived EVs (our unpublished data) has not yet been investigated in this context and would be expected to play a significant role.
In summary, these data show that PD-L1 on tumor-derived exosomes may have important immunosuppressive functions in cancer. This has implications therapeutically, in patient stratification and treatment monitoring. The only caveat being that many of these studies are performed with relatively high EV concentrations in experimental settings, thus at this stage we should consider these studies a proof-of-principle, opening the door for further more detailed studies.
Acknowledgements
This work was supported by NCI P01CA069246 (PI: Breakefield/Chiocca). Michal Nowicki is supported by National Cancer Institute R50CA243706.
Biography
Sean E. Lawler is a Principal Investigator at the Harvey Cushing Neurooncology Laboratories in the Department of Neurosurgery at Brigham and Women’s Hospital, and faculty member at Harvard Medical School. He is a molecular and cellular biologist and his main research interest is translational approaches for the treatment of brain tumors, bridging the research laboratory and the clinic, to ultimately improve outcomes for patients faced with this devastating disease.

Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Contributor Information
Sean E. Lawler, Harvey Cushing Neurooncology Laboratories Department of Neurosurgery Brigham and Women’s Hospital Boston, MA 02115, USA
Dr. Michal O. Nowicki, Harvey Cushing Neurooncology Laboratories Department of Neurosurgery Brigham and Women’s Hospital Boston, MA 02115, USA
Dr. Franz L. Ricklefs, Department of Neurosurgery University Medical Center Hamburg-Eppendorf Hamburg, 20246, Germany
Dr. E. Antonio Chiocca, Harvey Cushing Neurooncology Laboratories Department of Neurosurgery Brigham and Women’s Hospital Boston, MA 02115, USA
References
- [1].van Niel G, D’Angelo G, Raposo G, Nat. Rev. Mol. Cell Biol 2018, 19, 213. [DOI] [PubMed] [Google Scholar]
- [2].Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas E, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers EM, Laitinen S, Lässer C, Lener T, et al. , J. Extracell. Vesicles 2015, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jaiswal R, Sedger LM, Front. Oncol 2019, 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ, Nat. Rev. Clin. Oncol 2018, 15, 617. [DOI] [PubMed] [Google Scholar]
- [5].Ricklefs F, Mineo M, Rooj AK, Nakano I, Charest A, Weissleder R, Breakefield XO, Chiocca EA, Godlewski J, Bronisz A, Cancer Res. 2016, 76, 2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ludwig N, Whiteside TL, Expert Opin. Ther. Targets 2018, 22, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Whiteside TL, Adv. Exp. Med. Biol 2017, 1036, 81. [DOI] [PubMed] [Google Scholar]
- [8].Han L, Lam EW, Sun Y, Mol. Cancer 2019, 18, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maas SLN, Breakefield XO, Weaver AM, Trends Cell Biol. 2017, 27, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rufino-Ramos D, Albuquerque PR, Carmona V, Perfeito R, Nobre RJ, Pereira de Almeida L, Controlled Release J 2017, 262, 247. [DOI] [PubMed] [Google Scholar]
- [11].Shao H, Im H, Castro CM, Breakefield XO, Weissleder R, Lee H, Chem. Rev 2018, 118, 1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shah R, Patel T, Freedman JE, N. Engl. J. Med 2018, 379, 958. [DOI] [PubMed] [Google Scholar]
- [13].Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, Dronca R, Gangadhar TC, Patnaik A, Zarour H, Joshua AM, Gergich K, Elassaiss-Schaap J, Algazi A, Mateus C, Boasberg P, Tumeh PC, Chmielowski B, Ebbinghaus SW, Li XN, Kang SP, Ribas A, N. Engl. J. Med 2013, 369, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, Shaheen M, Ernstoff MS, Minor D, Salama AK, Taylor M, Ott PA, Rollin LM, Horak C, Gagnier P, Wolchok JD, Hodi FS, N. Engl. J. Med 2015, 372, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Garon EB, Rizvi NA, Hui R, Leigh IN, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, Lee JS, Hellmann MD, Hamid O, Goldman JW, Soria JC, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford LK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K K, Gandhi L, KEYNOTE-001 Investigators, N. Engl. J. Med 2015, 372, 2018.25891174 [Google Scholar]
- [16].Pardoll DM, Nat. Rev. Cancer 2012, 12, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mahoney KM, Rennert PD, Freeman GJ, Nat. Rev. Drug Discovery 2015, 14, 561. [DOI] [PubMed] [Google Scholar]
- [18].Topalian SL, Taube JM, Anders RA, Pardoll DM, Nat. Rev. Cancer 2016, 16, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ricklefs FL, Alayo Q, Krenzlin H, Mahmoud AB, Speranza MC, Nakashima H, Hayes JL, Lee K, Balaj L, Passaro C, Rooj AK, Krasemann S, Carter BS, Chen CC, Steed T, Treiber J, Rodig S, Yang K, Nakano I, Lee H, Weissleder R, Breakefield XO, Godlewski J, Westphal M, Lamszus K, Freeman GJ, Bronisz A, Lawler SE, Chiocca EA, Sci. Adv 2018, 4, eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, et al. , Nature 2018, 560, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R, Cell 2019, 177, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, Cha JH, Hou J, Hsu JL, Sun L, Hung MC, Cell Res. 2018, 28, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim DH, Kim H, Choi YJ, Kim SY, Lee JE, Sung KJ, Sung YH, Pack CG, Jung MK, Han B, Kim K, Kim WS, Nam SJ, Choi CM, Yun M, Lee JC, Rho JK, Exp. Mol. Med 2019, 51, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL, Clin. Cancer Res 2018, 24, 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alexander BM, Cloughesy TF, J. Clin. Oncol 2017, 35, 2402. [DOI] [PubMed] [Google Scholar]
- [26].Filbin MG, Suvà ML, Annu. Rev. Pathol.: Mech. Dis 2016, 11, 497. [DOI] [PubMed] [Google Scholar]
- [27].Tomaszewski W, Sanchez-Perez L, Gajewski TF, Sampson JH, Clin. Cancer Res 2019, 25, 4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Woroniecka KI, Rhodin KE, Chongsathidkiet P, Keith KA, Fecci PE, Clin. Cancer Res 2018, 24, 3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Heiland DH, Haaker G, Delev D, Mercas B, Masalha W, Heynckes S, Gäbelein A, Pfeifer D, Carro MS, Weyerbrock A, Prinz M, Schnell O, Oncotarget 2017, 8, 42214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nduom EK, Wei J, Yaghi NK, Huang N, Kong LY, Gabrusiewicz K, Ling X, Zhou S, Ivan C, Chen JQ, Burks JK, Fuller GN, Calin GA, Conrad CA, Creasy C, Ritthipichai K, Radvanyi L, Heimberger AB, Neuro-Oncology 2016, 18, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, Woroniecka K, Elsamadicy AA, Dechant CA, Kemeny HR, Sanchez-Perez L, Cheema TA, Souders NC, Herndon JE, Coumans JV, Everitt JI, Nahed BV, Sampson JH, Gunn MD, Martuza RL, Dranoff G, Curry WT, Fecci PE, Nat. Med 2018, 24, 1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lukas RV, Rodon J, Becker K, Wong ET, Shih K, Touat M, Fassò M, Osborne S, Molinero L, O’Hear C, Grossman W, Baehring J, J. Neuro-Oncology 2018, 140, 317. [DOI] [PubMed] [Google Scholar]
- [33].Omuro A, Vlahovic G, Lim M, Sahebjam S, Baehring J, Cloughesy T, Voloschin A, Ramkissoon SH, Ligon KL, Latek R, Zwirtes R, Strauss L, Paliwal P, Harbison CT, Reardon DA, Sampson JH, Neuro-Oncology 2018, 20, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, Wang AC, Ellingson BM, Rytlewski JA, Sanders CM, Kawaguchi ES, Du L, Li G, Yong WH, Gaffey SC, Cohen AL, Mellinghoff IK, Lee EQ, Reardon DA, O’Brien BJ, Butowski NA, Nghiemphu PL, Clarke JL, Arrillaga I, Romany C, Colman H, Kaley TJ, de Groot JF, Liau LM, Wen PY, Prins RM, Nat. Med 2019, 25, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Domenis R, Cesselli D, Toffoletto B, Bourkoula E, Caponnetto F, Manini I, Beltrami AP, Ius T, Skrap M, Di Loreto C, Gri G, PLoS One 2017, 12, e0169932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hellwinkel JE, Redzic JS, Harland TA, Gunaydin D, Anchordoquy TJ, Graner MW, Neuro-Oncology 2016, 18, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM, Nature 1989, 342, 559. [DOI] [PubMed] [Google Scholar]
- [38].Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, CarlottiJr CG, Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, et al. , Cell 2016, 164, 550.26824661 [Google Scholar]
- [39].Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ, Nucleic Acids Res. 2013, 41, D1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, Breakefield XO, Nat. Commun 2015, 6, 7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Andtbacka RH, Ross M, Puzanov I, Milhem M, Collichio F, Delman KA, Amatruda T, Zager JS, Cranmer L, Hsueh E, Chen L, Shilkrut M, Kaufman HL, Annals of Surgical Oncology 2016, 23, 4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RH, Michielin O, Olszanski AJ, Malvehy J, Cebon J, Fernandez E, Kirkwood JM, Gajewski TF, Chen L, Gorski KS, Anderson AA, Diede SJ, Lassman ME, Gansert J, Hodi FS, Long GV, Cell 2017, 170, 1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA, N. Engl. J. Med 2014, 371, 2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lin W, Chen M, Hong L, Zhao H, Chen Q, Front. Oncol 2018, 8, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee-Chang C, Rashidi A, Miska J, Zhang P, Pituch KC, Hou D, Xiao T, Fischietti M, Kang SJ, Appin CL, Horbinski C, Platanias LC, Lopez-Rosas A, Han Y, Balyasnikova IV, Lesniak MS, Cancer Immunol. Res 2019, 7, 1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, Lunceford J, Cheng J, Chow LQ, Seiwert TY, Handa M, Tomassini JE, McClanahan T Clin. Cancer Res 2017, 23, 3158. [DOI] [PubMed] [Google Scholar]