Abstract
N6-Methyladenosine (m6A) modification is one of the most widely distributed RNA modifications in eukaryotes. It participates in various RNA functions and plays vital roles in tissue development, stem cell formation and differentiation, heat shock response control, and circadian clock controlling, particularly during tumor development. The reversible regulation of m6A modification is affected by the so-called ‘reader’, ‘writer’ and ‘eraser’. As a required component and the largest methyltransferase, vir-like m6A methyltransferase associated (VIRMA) can promote the progression of cancer and is associated with poor survival in multiple types of cancer. The present review investigated the role of VIRMA in various types of cancer. In an m6A-dependent or -independent manner, VIRMA can play an oncogenic role by regulating cancer cell proliferation, migration and invasion, metastasis, apoptosis resistance and tumor growth in different pathways by targeting stem factors, CCAT1/2, ID2, GATA3, CDK1, c-Jun, etc. VIRMA can also predict better prognosis in kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP) and papillary thyroid carcinoma by TCGA analysis. The obvious oncogenic roles of VIRMA observed in different types of cancer and the mechanisms of VIRMA promoting cancers provided the basis for potential therapeutic targeting for cancer treatments.
Keywords: VIRMA, KIAA1429, M6A, Cancer, Therapy
Background
Chemical modification of nucleic acids is an important part of biological processes, including RNA transcription, protein translation and signaling pathway. N6-Methyladenosine (m6A) modification is one of the most widely distributed RNA modifications in eukaryotes [1, 2]. m6A modification refers to methylation occurring at the sixth position of nitrogen atoms of adenosine at the post-transcriptional level, with S-adenosylmethionine serving as the methyl donor for m6A formation [3–5]. m6A modification exists in mammalian mRNAs [6, 7], long-noncoding RNAs [8], and microRNAs [9, 10], and participates in various RNA functions such as mRNA stability [6, 11], splicing [12], transport [13], translation [7, 14], primary microRNA processing [9] and RNA–protein interactions [15].
The reversible regulation of m6A modification is performed by the so-called ‘reader’, ‘writer’ and ‘eraser’ [16, 17] (Fig. 1). The ‘writers’ are also called methyltransferases, including methyltransferase like 3 (METTL3) [18], METTL14 [18], Wilms’ tumor 1-associating protein (WTAP) [19], vir-like m6A methyltransferase associated (VIRMA; also known as KIAA1429) [18, 20], METTL16 [21], RNA-binding motif protein 15 (RBM15) [22, 23], zinc finger CCCH-type containing protein 13 (Zc3h13) [22] and Hakai [24]. Studies have shown that m6A modification plays a vital role in tissue development, stem cell formation and differentiation [25, 26], heat shock response control [27] and circadian clock control [28], particularly during tumor development.
Fig. 1.
Roles of the N6-methyladenosine ‘writer’, ‘eraser’ and ‘reader’ complexes in regulating mRNA. METTL3 methyltransferase like 3, METTL14 methyltransferase like 14, WTAP Wilms tumor 1-associated protein, VIRMA vir-like m6A methyltransferase associated, METTL16 methyltransferase like 16, ZC3H13 zinc finger CCCH-type containing 13, RBM15 RNA binding motif protein 15, ALKBH5 alkB homolog 5, FTO the fat mass and obesity-associated protein, IGF2BP IGF2 mRNA binding proteins, eIF3 eukaryotic translation initiation factor 3, HNRNP heterogeneous nuclear ribonucleoprotein, YTH YT521-B homology domain-containing protein
The largest component of ‘writers’, VIRMA, has been verified to promote cancer progression or is associated with poor survival in multiple cancer types, including liver cancer [29], gastric cancer [30] and breast cancer [20], head and neck squamous cell carcinoma (HNSC) [31], and testicular germ cell tumors (TGCT) [32] and etc. VIRMA can participate in cancer progression in an m6A-dependent manner or m6A-independent manner. The obvious oncogenic roles of VIRMA observed in these cancer types suggested that VIRMA may serve as a potential therapeutic target in cancer treatment.
Main text
VIRMA
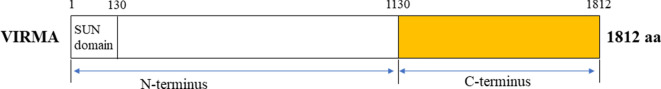
It was first reported that VIRMA is required for the entire process of mRNA methylation in human cells in 2015 [18]. As a VIRMA homologue in Drosophila melanogaster, virilizer plays an essential role in sex-lethal splicing, male and female viability, and the capability of eggs production in embryonic development [33]. In human, VIRMA is located in the nuclear speckles, the same location as that of WTAP [34, 35]. As the largest known component of the methyltransferase complex (202 kDa), VIRMA contains an N-terminal (1130 aa) as N-VIRMA and a C-terminal (1131–1812 aa) as C-VIRMA, and begins from a SUN domain (130 aa) (Fig. 2) [22, 36, 37]. A fourfold decrease in m6A peak scores has been reported following VIRMA knockdown, more obvious than that observed in human cells following METTL3 and METTL14 knockdown [18]. It was reported that in human HeLa cells, VIRMA could recruit the m6A methyltransferase components METTL3/METTL14/WTAP to guide region-selective methylations [38], suggesting that VIRMA plays a significant role in m6A modification. The m6A modification by VIRMA is enriched in the 3′-untranslated region (3′-UTR) and near the stop codon of RNA substrates [20, 38]. Virilizer contains an RNA-binding protein (RBP) domain, similar with that in RNA and DNA helicases, and ribonucleoproteins, and is involved in the RNA processes of metabolism, transport and translation [33]. Studies have demonstrated that the abnormal expression of RBPs could lead to the upregulation of certain oncogenes or the downregulation of certain tumor suppressor genes [39]. VIRMA has been reported to mainly act as the oncogene in several types of cancer, as listed in Table 1. Wang et al. [40] found that hsa_circ_0084922 (also named circ_KIAA1429), coming from VIRMA, was upregulated in hepatocellular carcinoma (HCC), which may promote cancer progression.
Fig. 2.
Schematic of domain architecture of VIRMA. aa amino acids; N-terminus (1–1130 aa, N-VIRMA); C-terminus (1131–1812 aa, C-VIRMA)
Table 1.
Expression, clinical significance, and biological functions of VIRMA in various cancers
| Cancer type | Expression | Clinical significance | Biological functions | Methodology | In vivo or in vitro | References |
|---|---|---|---|---|---|---|
| Breast cancer | Upregulated (46 pairs) | Oncogene | Proliferation, metastasis, tumorigenesis | mRNA high-throughput sequencing; RIP-seq; m6A dot blot assay; mRNA FISH; mRNA stability analysis; | In vivo and in vitro | [20] |
| Upregulated (1109 BRC tissues and 113 normal breast tissues from TCGA; TMA containing 20 BRC specimens and 20 normal breast tissue specimens) | – | – | TMA cohorts; Immunohistochemical staining; Immunofluorescence assay | – | [42] | |
| Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] | |
| Liver cancer | Upregulated (70 pairs) | Oncogene | Cell proliferation, tumor growth and metastasis, cell apoptosis resistance | RNA-seq; RIP-seq; MeRIP-seq | In vivo and in vitro | [29] |
| Upregulated (371 liver cancer tissues and 50 normal liver tissues from TCGA) | Oncogene | Cell proliferation, cell migration and invasion | MeRIP-PCR | In vitro | [43] | |
| Upregulated (374 HCC and 50 normal tissues from TCGA; 243 HCC and 202 normal tissues from ICGC) | Oncogene | – | – | – | [45] | |
| Gastric cancer | Upregulated (20 pairs) | Oncogene | Proliferation | Dual‐luciferase reporter gene assay; RIP; RNA-seq | In vivo and in vitro | [30] |
| Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] | |
| Kidney cancer | Upregulated | Oncogene | – | – | – | [44] |
| – | – | – | – | [41] | ||
| Kidney renal papillary cell carcinoma | Downregulated (289 kidney renal papillary cell carcinoma and 32 normal kidney samples from the TCGA database) | – | – | – | – | [49] |
| Bladder cancer | – | Oncogene | – | – | – | [44] |
| No difference (408 bladder cancer patients from TCGA) | Oncogene | – | – | – | [48] | |
| Prostate cancer | Upregulated (TCGA + GTEx, 492 prostate adenocarcinoma and 152 adjacent normal specimens) | Oncogene | Proliferation, metastasis | RNA methylation quantification; immunofluorescence Analysis; RT2 lncRNA PCR Array Human Cancer PathwayFinder; N6-methyladenosine immunoprecipitation | In vitro | [58] |
| Upregulated | – | – | – | – | [44] | |
| Testicular cancer | Upregulated | Oncogene | – | qRT-PCR; Immunohistochemistry | In vitro | [32] |
| Upregulated | Oncogene | – | – | – | [44] | |
| Head and neck squamous cell carcinoma | TCGA | – | – | – | – | [31] |
| Osteosarcoma | Upregulated | Oncogene | Proliferation, invasion, and migration | Tissue microarray (TMA) construction and immunohistochemistry (IHC); dual luciferase reporter gene assay | In vivo and in vitro | [50] |
| Lung squamous cell carcinoma | Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| Colon adenocarcinoma | Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| Rectum adenocarcinoma | Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| Lung adenocarcinoma | Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| Cholangiocarcinoma | Upregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| Uterine corpus endometrial carcinoma | Downregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| Thyroid carcinoma | Downregulated (~ 7400 samples representing 11 cancer types) | – | – | – | – | [41] |
| papillary thyroid carcinoma | Downregulated (499 tumor tissues and 58 normal thyroid carcinoma) | Tumor suppressor gene | – | – | – | [52] |
| Ovarian cancer | Downregulated (TCGA, 370 ovarian cancer tissues and 88 normal tissues) | – | – | – | – | [51] |
VIRMA in cancers
m6A regulators, including ‘writers’, ‘erasers’, ‘readers’, play key roles in cancer. VIRMA was found to be correlated with the most positive oncogenic pathways among the ‘writers’ [41], indicating that VIRMA may has multiple different functions and plays important roles in cancer pathways.
Expression of VIRMA in cancers
The difference of VIRMA expression among different tissues was shown in Li’s study [41]. The analysis revealed that VIRMA expression was higher in HNSC, lung squamous cell carcinoma (LUSC), liver hepatocellular carcinoma (LIHC), colon adenocarcinoma (COAD), rectum adenocarcinoma (READ), lung adenocarcinoma (LUAD), stomach adenocarcinoma (STAD), cholangiocarcinoma (CHOL), breast invasive carcinoma (BRCA), kidney chromophobe (KICH) and kidney renal clear cell carcinoma (KIRC). VIRMA was more lowly expressed in uterine corpus endometrial carcinoma (UCEC), thyroid carcinoma (THCA), prostate adenocarcinoma (PRAD) and kidney renal papillary cell carcinoma (KIRP).
Prognostic potential of VIRMA in cancer
To investigate the association between VIRMA expression and cancer prognosis, the impact of VIRMA expression on survival rate was determined by Kaplan–Meier survival curves. VIRMA was found to be highly expressed and to predict poor overall survival (OS) in breast cancer [20, 41, 42], liver cancer [29, 43] and kidney cancer [44] (Table 2). However, the high VIRMA expression predicted the better OS in KIRC [41] (Table 2). In addition, VIRMA was highly expressed and predicted poor disease-free survival (DFS) in liver cancer [29] and kidney cancer (Table 2) [44].
Table 2.
Survival influence of VIRMA in various cancers
| Cancer type | Sample size | Survival influence | References |
|---|---|---|---|
| Breast cancer | low-VIRMA (n = 528); high-VIRMA (n = 528) | Worse OS | [20] |
| low-VIRMA (n = 598); high-VIRMA (n = 472) | Worse OS | [42] | |
| low-VIRMA (n = 611); high-VIRMA (n = 457) | Worse RFS | [42] | |
| TCGA | Worse OS | [41] | |
| Liver cancer | 70 HCC patients | Worse OS and DFS | [29] |
| low-VIRMA (n = 182); high-VIRMA (n = 182) | Worse OS | [43] | |
| 374 HCC and 50 normal tissues from TCGA; 243 HCC and 202 normal tissues from ICGC | Worse OS | [45] | |
| Kidney cancer | 897 samples from 895 patients | Worse OS and DFS | [44] |
| Kidney renal clear cell carcinoma | TCGA | Better OS | [41] |
| Kidney renal papillary cell carcinoma | 289 kidney renal papillary cell carcinoma and 32 normal kidney samples from the TCGA database | Better OS and DFS | [49] |
| Prostate cancer | TCGA + GTEx, 492 prostate adenocarcinoma and 152 adjacent normal specimens | Worse DFS | [58] |
| Gliomas | 309 glioma patients from CGGA and 595 glioma patients from TCGA | Worse OS | [75] |
| Ovarian cancer | 370 ovarian cancer tissues and 88 normal tissues from TCGA | Worse OS | [51] |
| Papillary thyroid carcinoma | 499 tumor tissues and 58 normal thyroid carcinoma | Better OS | [52] |
Alteration frequency of VIRMA in 33 cancer types
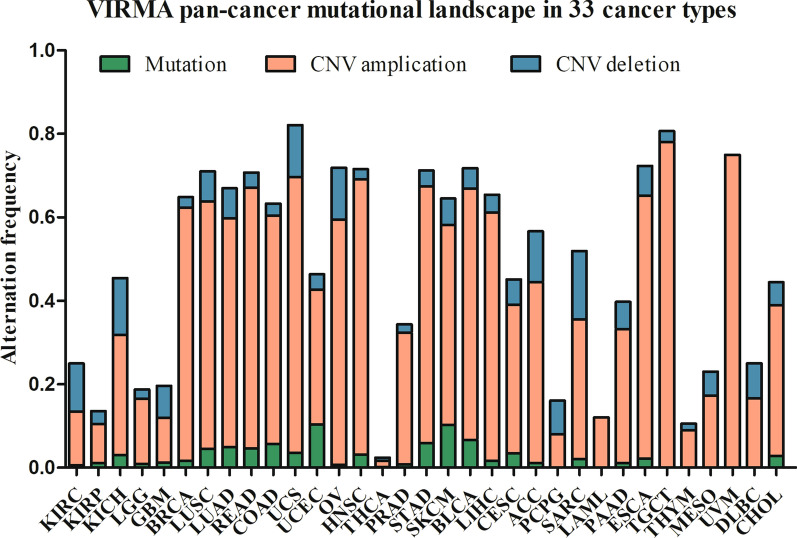
To explore the alteration frequency of VIRMA in 33 cancers, we extracted the result data from the study by Li et al. [42]. The original data in Li’s study was from TCGA MAF file (“MC3”) and Broad GDAC Firehose (https://gdac.broadinstitute.org/) [41]. The overall copy number variation (CNV) amplification frequency of VIRMA in cancer was 1.40–78%, with a high frequency in most tumors, such as uveal melanoma (UVM), TGCT, and low frequency in THCA, PCPG (Fig. 3) [41]. The overall CNV deletion frequency of VIRMA in cancers was 0–16.34%, with high frequency in sarcoma (SARC), kidney Chromophobe (KICH), and low frequency in UVM, THCA (Fig. 3) [41]. The overall VIRMA mutation frequency in cancers was 0–10.35%, with a high frequency in SKCM and UCEC, and low frequency in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), UVM, mesothelioma (MESO), THTM and TGCT (Fig. 3).
Fig. 3.
VIRMA pan-cancer mutational landscape in 33 cancer types. CNV copy number variation, KIRC kidney renal clear cell carcinoma, KIRP kidney renal papillary cell carcinoma, KICH kidney chromophobe, LGG brain lower grade Glioma, GBM glioblastoma multiforme, BRCA breast invasive carcinoma, LUSC lung squamous cell carcinoma, LUAD lung adenocarcinoma, READ rectum adenocarcinoma, COAD colon adenocarcinoma, UCS uterine carcinosarcoma, UCEC uterine corpus endometrial carcinoma, OV ovarian serous cystadenocarcinoma, HNSC head and neck squamous carcinoma, THCA thyroid carcinoma, PRAD prostate adenocarcinoma, STAD stomach adenocarcinoma, SKCM skin cutaneous melanoma, BLCA bladder urothelial carcinoma, LIHC liver hepatocellular carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, ACC adrenocortical carcinoma, PCPG pheochromocytoma and paraganglioma, SARC sarcoma, LAML acute myeloid leukemia, PAAD pancreatic adenocarcinoma, ESCA esophageal carcinoma, TGCT testicular germ cell tumors, THYM thymoma, MESO mesothelioma, UVM uveal melanoma, DLBC lymphoid neoplasm diffuse large B-cell lymphoma, CHOL cholangiocarcinoma
VIRMA in breast cancer
The functions of VIRMA in breast cancer tumorigenesis and the related mechanisms have been previously reported [20]. VIRMA was found to be more highly expressed in breast cancer tissues than in non-tumor breast tissues in clinical samples [20, 42]. The high expression of VIRMA also predicted poor OS in patients with breast cancer [20, 42]. VIRMA could promote breast cancer proliferation and metastasis in vivo and in vitro, which indicated that VIRMA could promote breast cancer progression and was associated with pathogenesis. In addition, 5′-fluorouracil (5-FU) could decrease the expression of VIRMA and its downstream target in breast cancer cells [20].
IRMA and circ_KIAA1429 in liver cancer
A higher expression of VIRMA was observed in liver cancer tissues, as compared with adjacent normal tissues, both in the TCGA database and clinical samples, which predicted poor OS and DFS in patients with liver cancer [29, 43, 45]. Knockdown of VIRMA could inhibit cancer cell proliferation and metastasis in vitro [43]. Lan et al. [29] further demonstrated that VIRMA markedly facilitated the cell cycle progression, cell proliferation, invasion, and migration, as well as apoptosis resistance in vitro. It also promoted tumor growth, as well as pulmonary and intrahepatic metastasis in vivo, which confirmed and identified VIRMA as a formidable driver of liver tumor growth and metastasis. To further understand whether the differential expression of VIRMA is caused by genetic changes related to the corresponding genes, Qu et al. [45] then analyzed CNV and single-nucleotide polymorphism (SNP) data from the TCGA database and found that the CNV in HCC tissues was significantly different from that in normal tissues. More specifically, VIRMA mainly exhibited increased copy numbers in HCC tissues [45]. It was also observed that SNP mutations was very low in HCC tissues, which indicated that upregulation of VIRMA is not entirely caused by CNV or SNP mutations in the corresponding genes [45].
circ_KIAA1429 (chr8:95547066–95550574, 305 bp spliced sequence, http://www.circbase.org/), which comes from VIRMA, was obviously upregulated in HCC cancer cells and tissues. Wang et al. [40] demonstrated that the upregulation of circ_KIAA1429 could promote migration, invasion and EMT in HCC cancer cells in vitro and in vivo, whereas the knockdown of circ_KIAA1429 yielded the opposite results.
VIRMA in gastric cancer
A higher expression of VIRMA was observed in gastric cancer tissues, as compared with their adjusted normal tissues, which was associated with tumor grade. The knockdown of VIRMA could inhibit gastric cancer cell proliferation by arresting the cell cycle in vivo and in vitro. Miao et al. [30] demonstrated that VIRMA promotes gastric cancer, which may serve as a therapeutic target in the future.
VIRMA in urological cancers
VIRMA was upregulated in four major urogenital neoplasms: including kidney cancer, bladder cancer, prostate cancer and testicular cancer [44]. TGCT account for > 95% of all types of testicular neoplasms, and consist of two major families: the germ-cell neoplasia in situ (GCNIS)-related tumors [the most frequent; include seminomas (SEs) and non-seminomatous tumors (NSTs), which have an obvious different clinical behavior and impact], and the GCNIS-unrelated ones [46].
In prostate cancer, the high expression of VIRMA was correlated with cancer progression. In TGCTs, VIRMA appeared to be a promising biomarker for distinguishing between SEs and NSTs, and had an impact disease stage [32, 44]. In bladder cancer, VIRMA seems to be a useful marker, as it is amongst the most commonly deregulated and significantly upregulated in high grade tumors [44]. VIRMA was significantly upregulated in non-papillary tumors (the most aggressive, more prone to progress and metastasize) among different types of bladder cancers [47]. Chen. et al. [48] latter confirmed that VIRMA was highly expressed in high grade bladder cancer and further found that the expression of VIRMA did not differ between BC and normal tissues. According to the current World Health Organization (WHO) 2016 classification, kidney cancer (also named renal cell carcinoma, RCC) are mainly classified into three subtypes: KICH, KIRC and KIRP [46]. In kidney cancer, the VIRMA expression could be a biomarker to discriminate these RCC subtypes, and was associated with OS and DFS [44]. Sun et al. further focused on KIRP study and found that VIRMA was downregulated in, compared with in normal kidney tissue samples [49]. High expression of VIRMA was correlated with high grade of KIRP and predicted poor OS and DFS, which indicated that VIRMA is a potential prognostic biomarker that can accurately predict survival outcomes of KIRP patients [49]. However, the data of the above studies were from TCGA database, that needs the validation of clinical results and further mechanism research, especially to explain the different results between kidney cancer and KIRP need further clinical confirm and further mechanism research.
Of note, VIRMA and m6A methylation reader YTHDF3 were upregulated and showed a strong positive correlation in prostate cancer and TGCTs, as they jointly facilitated poor prognosis [32, 44].
HNSC
VIRMA was more highly expressed in HNSC tissues, as compared with their adjacent normal tissues, as shown by TCGA analysis [31]. The result was from TCGA database, which need clinical confirm and mechanism research.
VIRMA in osteosarcoma
VIRMA was upregulated and associated with unfavorable outcomes in patients with osteosarcoma. Han et al. [50] revealed that knockdown of VIRMA could suppress osteosarcoma cancer cell migration, invasion, and proliferation in vitro, as well as suppress tumor growth in vivo. It was also found that VIRMA was the direct target of miR-143, and could partly reverse the anti-proliferative effect of miR-143 [50]. miR-143 and VIRMA may become potential predictive biomarkers in the treatment of osteosarcoma.
VIRMA in ovarian cancer
Ovarian cancer is one of the deadliest gynecological malignancies. VIRMA protein was found to be downregulated in ovarian cancer tissues than normal tissues and positively correlated with worse OS [51]. The expression of VIRMA in different types of ovarian cancers in the Oncomine database is different and the highest in ovarian serous adenocarcinoma [51]. The expression of VIRMA in grade 3 is significantly higher than that in grade 2 [51]. The samples in grade 1 and grade 4 were insufficient, which need further collected and analyzed. However, the protein expression and the mRNA expression of VIRMA in the normal control group and tumor group was not consistent, which suggests that the mRNA may serve as a reservation [51]. Only under certain circumstances, such as hypoxia or immune stimulation, can the protein be translated. A similar phenomenon could also be observed in the production of some cytokines [51]. The mRNA may also regulate the expression of other proteins by generating microRNAs. The studies on ovarian cancer were from TCGA database, which need further experiments and analysis in detail.
VIRMA in papillary thyroid carcinoma
VIRMA was downregulated and predicted better OS in papillary thyroid carcinoma [41, 52]. Univariate and multivariate analyses demonstrated that the risk score of VIRMA is an independent prognostic factor in papillary thyroid carcinoma, indicating that VIRMA might act as a tumor suppressor [52]. Studies on VIRMA in papillary thyroid carcinoma were from TCGA database, which need further exploration of clinical confirm and the underlying molecular mechanisms.
Regulatory mechanisms of VIRMA in cancer
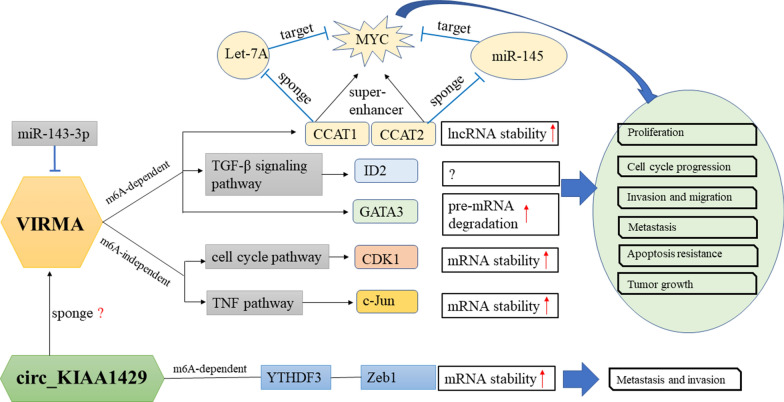
As shown in Fig. 4, several m6A modification regulators participate in cancer development in an m6A-dependent manner by targeting related mRNAs. As regards the regulatory role of VIRMA in cancer, it was found that, in addition to the m6A-dependent pathway, VIRMA could also regulate downstream m6A-independent pathways. Circ_KIAA1429 could also regulate downstream m6A-dependent pathways. The expression of VIRMA may also be regulated by miR-143-3p in cancer cells.
Fig. 4.
Mechanisms of VIRMA and circ_KIAA1429 involved in human cancer progression. VIRMA expression could be regulated by miR-143-3p. VIRMA played an important role in tumor cell proliferation, invasion, migration, cell cycle, metastasis, apoptosis resistance, tumorigenesis in an m6A-dependent or m6A-independent manner. The TGF-β, cell cycle, TNF and other signaling pathways were involved in the underlying mechanism. Circ_KIAA1429 can promote HCC through the mechanism of m6A-YTHDF3-Zeb1 pathway. VIRMA, vir-like m6A methyltransferase associated
VIRMA regulates downstream targets in an m6A-dependent manner
In the VIRMA-depleted cell lines, m6A modification around the 3′-UTR and near the stop codon significantly disappeared, indicating that VIRMA can exert its effects by mediating mRNA m6A methylation in 3′UTR and near stop codon region [38]. Yue et al. [38] revealed VIRMA functioned by recruiting the methyltransferase core components and interacting with polyadenylation cleavage factors CPSF5 and CPSF6, which suggested interactions between the m6A methylation and polyadenylation during mRNA processing and mRNA metabolism.
ID2 target
VIRMA promoted cell migration and invasion in liver cancer by increasing the m6A modification of ID2 mRNA, which then led to the decrease in ID2 expression [43]. Decreasing ID2 could regulate the secretion of vascular endothelial growth factor to promote liver cancer metastasis [53]. The ID gene family, including ID2, was found to be upregulated in various kinds of cancers [54, 55], and ID2 in particular was associated with the development of several diseases. VIRMA regulated the migration and invasion of liver cancer by regulating the m6A modification of ID2.
GATA3 target
GATA3 was identified as the direct downstream target of VIRMA-mediated m6A modification in liver cancer through the combination of immunoprecipitation sequencing (RIP-seq), and high-throughput methylated RNA immunoprecipitation sequencing (MeRIP-seq) [29]. VIRMA methylate in the 3′-UTR of GATA3 pre-mRNA to separate the RNA-binding protein HuR and promote GATA3 pre-mRNA degradation. GATA3-AS, a long-noncoding RNA (lncRNA), transcribed from the antisense strand of the GATA3 gene, acts as a cis-acting element in the interaction between VIRMA and GATA3 pre-mRNA. Therefore, VIRMA promote the growth, metastasis and malignant phenotypes of liver cancer cells by regulating the GATA expression, which is mediated by the m6A modification of GATA3 pre-mRNA [29].
lncRNA target
lncRNAs are important factors in prostate cancer. m6A is considered one of the most prevalent modifications in lncRNAs [56, 57]. VIRMA knockdown could decrease the m6A levels and stability of CCAT1 and CCAT2 lncRNA in prostate cancer. Another study demonstrated that knockdown of VIRMA could decrease the stability of CCAT1 and CCAT2 lncRNA in an m6A-dependent manner to regulate MYC transcription, promoting progression of prostate cancer [58]. A direct correlation between CCAT1/2 and MYC transcript levels was found in prostate cancer [59]. Therefore, Daniela et al. hypothesized that stabilization of lncRNAs CCAT1/2 by m6A modification amplified the effect of MYC expression levels in cancer cells by 2 separate mechanisms: (i) directly, both lncRNAs acted as super-enhancers for positive regulation of MYC mRNA [60]; (ii) indirectly, CCAT1/2 actedas microRNA sponges for MYC-targeting microRNAs let-7A and miR-145, respectively [61–64].
Circ_KIAA1429 and Zeb1
CircRNAs have attracted considerable attention in multiple cancer studies in recent years. Overexpression of circ_KIAA1429 could promote HCC cancer cell migration and invasion, whereas the knockdown of circ_KIAA1429 could inhibit these effects. Zeb1 was identified as the downstream target of circ_KIAA1429. It was also found that m6A reader YTHDF3 could stabilize Zeb1 mRNA and prolong its half-life. Overall, circ_KIAA1429 could promote HCC progression through the m6A-YTHDF3-Zeb1 pathway to stabilize Zeb1 expression, which may represent a new target in cancer treatment [40]. As mentioned above, VIRMA is also upregulated and promote cancer progression in HCC [29, 43]. The present study has not clarified whether circ_KIAA1429 affects the expression, biological function, and tumorigenesis effect of VIRMA. Previous studies showed circRNAs might act as a sponge to increase the expression of its host gene [65–67], which would provide possible pathways that how circ_KIAA1429 influence VIRMA expression.
VIRMA regulates downstream target in an m6A-independent manner
CDK1 target
The cell cycle pathway played a significant role in the oncogenic activities of VIRMA, and ranked in the top two in breast cancer cell RIP-seq [20]. Among cell cycle-related proteins, CDK1 acts as an oncogene in cancers and is most associated with VIRMA in different breast cells. Reverse experiments, RIP-seq, and RT-qPCR confirmed that CDK1 is the main target of VIRMA in breast cancer. METTL3 knockdown could decrease CDK1 mRNAs with m6A modification, while VIRMA knockdown did not change the level of m6A modification in CDK1 mRNAs, indicating that m6A modification did not disturb the interaction between VIRMA and CDK1 in cancer cells [20]. VIRMA facilitated breast cancer by regulating the CDK1 mRNA expression in an m6A-independent manner [20].
c-Jun target
It was revealed that transcripts in gastric cancer cells were most enriched in the TNF signaling pathway, as compared with several other cancer‐related pathways, as shown by the KEGG pathway analysis in MGC803 cells [30]. After combining the associated genes by RIP-seq and mRNA-seq, c-Jun was identified as potential downstream target directly regulated by VIRMA in gastric cancer [20, 38]. c‐Jun, a transcriptional activator and member of the AP‐1 family, has been shown to participate in cell proliferation and apoptosis, tumorigenesis, and tissue morphogenesis [68–70]. As the regulation of CDK1 mRNA in breast cancer, no significant difference was observed in luciferase activity between the reporter carrying mutant m6A site of 3′-UTR in c-Jun and that carrying the non-mutant m6A site. It was demonstrated that VIRMA promoted gastric cell progression mainly by directly binding to the 3′-UTR of c‐Jun mRNA to regulate c‐Jun expression in an RNA-binding activity rather than the m6A-dependent manner [30]. However, more experiments should be completed to verify the m6A-independent manner, such as whether METTL3 affect the m6A modification level of c-Jun mRNA in gastric cancer cells.
Others
The Notch signaling pathway plays an important role in cell development and differentiation. In recent years, accumulating evidence have suggested that abnormal activation of the Notch signaling pathway participate in tumor progression. The high expression of the Notch signaling pathway has been observed in several types of cancer, an expression that results in Notch signal enhancement, which promotes cancer cell survival [71]. In osteosarcoma, upregulation of VIRMA was found to be associated with the activation of the Notch signaling pathway. It was revealed that VIRMA knockdown could suppress the expression of stem factors Notch1, Oct4, Nanog and CD44 in osteosarcoma cells, which suggested that VIRMA promoted osteosarcoma progression by activating Notch signaling pathway [50]. However, that study did not elucidate whether VIRMA regulates these downstream targets in an m6A-dependent manner.
A study by Han et al. revealed that VIRMA and miR-143-3p were negatively correlated in osteosarcoma samples [50]. The overexpression of miR-143-3p could inhibit VIRMA expression in osteosarcoma cells. The overexpression of VIRMA could reverse the anti-proliferation caused by miR-143-3p. It was proven that miR-143-3p could suppress VIRMA expression by directly targeting its 3ʹ-UTR region in osteosarcoma cells [50].
Prospect
While the roles of m6A modifications in cancers have been extensively reviewed elsewhere, the crucial functions of VIRMA in various types of cancer, as well as the potential targeting of VIRMA as cancer treatment, have not yet been highlighted. Our review is to summarize and analyze the present research on VIRMA, provide more ideas for the future research.
More studies are needed to demonstrate the biological function of VIRMA and its impact on tumor progression and survival, as well as the mechanisms involved, in more cancer types and phenotypes. High expression of VIRMA has been verified to predict poor survival in multiple cancers, such as breast cancer [20, 41, 42], liver cancer [29, 43], kidney cancer [44] and prostate cancer [58]. The expression of VIRMA is up-regulated in KIRC, but it indicates a poor prognosis. The study did not explain the reason for this conflict, which need further exploration [41]. VIRMA not only regulates the expression of downstream mRNA, but also regulates the stability of pre-mRNAs and lncRNAs. Inhibiting these downstream targets could reverse the oncogenic effect of VIRMA in cancer progression. Generally, VIRMA is an oncogene in most types of cancer, and downregulating it could inhibit cancer progression, indicating that VIRMA can become a diagnostic/prognostic biomarker in certain cancer types. Based on the emerging evidence of the roles and the molecular mechanisms in cancers, m6A regulators have attracted growing investigation as therapeutic targets [72]. Given the critical roles of the m6A regulatory proteins in cancers, m6A modification ‘eraser’ FTO appears to be a good drug target in cancer therapy. Several FTO small-molecule inhibitors, including meclofenamic acid [73] and MO-I-500 [74] have shown effective activity to inhibit the survival and growth of GBM and breast cancer cells by inhibiting the catalytic activity of FTO. Thus, inhibiting VIRMA and these downstream targets are considered potent cancer therapies. Studies have not yet elucidated the mechanism underlying the downstream regulation of VIRMA and whether it occurs through the m6A pathway in several tumor models, which is crucial for the development of treatments. The underlying mechanisms of VIRMA in cancer should be further addressed. Novel therapeutic strategies for m6A RNA methylation should be further explored in the treatment of cancer.
Conclusions
m6A modification has been shown to act by influencing RNA transcript, splicing, processing, translation and decay, and to participate in the development of various cancer types [6, 7, 9, 11–15]. VIRMA is a required component of the methyltransferase complex in m6A modification. It mediates methylation in the 3′-UTR and near the stop codon region of mRNAs [38]. The data included in this review suggested that VIRMA is mainly upregulated in various cancer types, and is associated with poor survival [20, 29–32, 42–44]. VIRMA has been shown to be upregulated and r associated with a more favorable survival in KIRC [41]. Further studies on the biological functions and mechanisms of VIRMA are required to identify its role in KIRC. Hou et al. showed that VIRMA was downregulated and predicted better prognosis in papillary thyroid carcinoma and acted as a tumor suppressor gene [52]. But the result was only from TCGA database and lacked the clinical confirm. VIRMA was also downregulated in KIRP and predicted better prognosis, which also need clinical confirm. By the m6A-dependent manner or the m6A-independent manner, VIRMA has been shown to promote cancer cell proliferation, apoptosis resistance, invasion, migration, and tumor progression through different pathways, including the TGF-β signaling pathway by targeting ID2, cell cycle pathway by targeting CDK1, CCAT1/2 pathway by upregulating its lncRNA stability, GATA3 pathway by upregulating pre-GATA3 mRNA degradation, and TNF pathway by targeting c-Jun [20, 29–32, 42–44]. Since circ_KIAA1429 can also promote tumor progression in hepatocellular carcinoma. VIRMA and circ_KIAA1429 both have great potential for clinical application by serving as a new treatment target. Therefore, more studies on VIRMA and m6A modification are needed to clarify the functions of VIRMA in the biological progress of various types of human cancer.
Acknowledgements
Not applicable.
Abbreviations
- m6A
N6-Methyladenosine
- METTL3
Methyltransferase like 3
- METTL14
Methyltransferase like 14
- WTAP
Wilms tumor 1-associated protein
- VIRMA
Vir like m6A methyltransferase associated
- METTL16
Methyltransferase like 16
- ZC3H13
Zinc finger CCCH-type containing 13
- RBM15
RNA binding motif protein 15
- ALKBH5
AlkB homolog 5
- FTO
The fat mass and obesity-associated protein
- IGF2BP
IGF2 mRNA binding proteins
- eIF3
Eukaryotic translation initiation factor 3
- HNRNP
Heterogeneous nuclear ribonucleoprotein
- YTH
YT521-B homology domain-containing protein
- 3′-UTR
3′-Untranslated region
- CNV
Copy number variation
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- KICH
Kidney Chromophobe
- LGG
Brain lower grade glioma
- GBM
Glioblastoma multiforme
- BRCA
Breast invasive carcinoma
- LUSC
Lung squamous cell carcinoma
- LUAD
Lung adenocarcinoma
- READ
Rectum adenocarcinoma
- COAD
Colon adenocarcinoma
- UCS
Uterine carcinosarcoma
- UCEC
Uterine corpus endometrial carcinoma
- OV
Ovarian serous cystadenocarcinoma
- HNSC
Head and neck squamous carcinoma
- THCA
Thyroid carcinoma
- PRAD
Prostate adenocarcinoma
- STAD
Stomach adenocarcinoma
- SKCM
Skin cutaneous melanoma
- BLCA
Bladder urothelial carcinoma
- LIHC
Liver hepatocellular carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- ACC
Adrenocortical carcinoma
- PCPG
Pheochromocytoma and paraganglioma
- SARC
Sarcoma
- LAML
Acute myeloid leukemia
- PAAD
Pancreatic adenocarcinoma
- ESCA
Esophageal carcinoma
- TGCT
Testicular germ cell tumors
- THYM
Thymoma
- MESO
Mesothelioma
- UVM
Uveal melanoma
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- CHOL
Cholangiocarcinoma
- OS
Overall survival
- DSS
Disease-specific survival
- DFI
Disease-free interval
Authors’ contributions
CL and JFW designed the study, WZ and JZW collected and analyzed the data, wrote, and edited the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets analysed during the current study are available in the sanger box (http://sangerbox.com/) and GTEx database (http://commonfund.nih.gov/GTEx/). The mutation data during this study are included in Li’s published article (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6744659/).
Declarations
Ethics approval and consent to participate
Data we used in this study were from publicly available datasets under ethics approval and consent.
Consent for publication
All authors are aware of and agree to the content of the paper and their being listed as a co-author of the paper.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wei Zhu and Jing-Zi Wang contributed equally to this work
Contributor Information
Ji-Fu Wei, Email: weijifu@hotmail.com.
Chen Lu, Email: gzfylc@163.com.
References
- 1.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Narayan P, Rottman FM. Methylation of mRNA. Adv Enzymol Relat Areas Mol Biol. 1992;65:255–285. doi: 10.1002/9780470123119.ch7. [DOI] [PubMed] [Google Scholar]
- 4.Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2(10):1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haugland RA, Cline MG. Post-transcriptional modifications of oat coleoptile ribonucleic acids. 5'-Terminal capping and methylation of internal nucleosides in poly(A)-rich RNA. Eur J Biochem. 1980;104(1):271–277. doi: 10.1111/j.1432-1033.1980.tb04425.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-Methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18(1):87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu W, Si Y, Xu J, Lin Y, Wang JZ, Cao M, Sun S, Ding Q, Zhu L. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020;24(6):3521–3533. doi: 10.1111/jcmm.15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature. 2015;518(7540):560. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 2014;5:5630. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W, Wang JZ, Xu Z, Cao M, Hu Q, Pan C, Guo M, Wei JF, Yang H. Detection of N6methyladenosine modification residues (Review) Int J Mol Med. 2019;43(6):2267–2278. doi: 10.3892/ijmm.2019.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8(1):284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38(33):6123–6141. doi: 10.1038/s41388-019-0861-z. [DOI] [PubMed] [Google Scholar]
- 21.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835.e814. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villasenor R, Hess D, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5–6):415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215(1):157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Li Y, Wang T, Zhong X. Modification of N6-methyladenosine RNA methylation on heat shock protein expression. PLoS ONE. 2018;13(6):e0198604. doi: 10.1371/journal.pone.0198604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fustin JM, Kojima R, Itoh K, Chang HY, Ye S, Zhuang B, Oji A, Gibo S, Narasimamurthy R, Virshup D. Two Ck1delta transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc Natl Acad Sci USA. 2018;115(23):5980–5985. doi: 10.1073/pnas.1721371115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, Yan X, Liao H, Chen X, Xie K, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18(1):186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao R, Dai CC, Mei L, Xu J, Sun SW, Xing YL, Wu LS. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J Cell Physiol. 2020;235(10):7420–7432. doi: 10.1002/jcp.29645. [DOI] [PubMed] [Google Scholar]
- 31.Zhao X, Cui L. Development and validation of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am J Cancer Res. 2019;9(10):2156–2169. [PMC free article] [PubMed] [Google Scholar]
- 32.Lobo J, Costa AL, Cantante M, Guimarães R, Lopes P, Antunes L, Braga I, Oliveira J, Pelizzola M, Henrique R, et al. m(6)A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance. J Transl Med. 2019;17(1):79. doi: 10.1186/s12967-019-1837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niessen M, Schneiter R, Nothiger R. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene sex-lethal in Drosophila melanogaster. Genetics. 2001;157(2):679. doi: 10.1093/genetics/157.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum Mol Genet. 2000;9(15):2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 35.Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288(46):33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, Tang HW, Li J, Perrimon N, Yan D. Xio is a component of the Drosophila sex determination pathway and RNA N(6)-methyladenosine methyltransferase complex. Proc Natl Acad Sci USA. 2018;115(14):3674–3679. doi: 10.1073/pnas.1720945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69(6):1028–1038.e1026. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yue Y, Liu J, Cui X, Cao J, Luo G. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong S. RNA binding protein as an emerging therapeutic target for cancer prevention and treatment. J Cancer Prev. 2017;22(4):203–210. doi: 10.15430/JCP.2017.22.4.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Yang Y, Yang J, Yang J, Han S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 2020;257:118082. doi: 10.1016/j.lfs.2020.118082. [DOI] [PubMed] [Google Scholar]
- 41.Li YK, Xiao J, Bai J, Tian Y, Xu J. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer. 2019;18(1):1–16. doi: 10.1186/s12943-019-1085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R. N6-Methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10(22):5447–5459. doi: 10.7150/jca.35053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng X, Li M, Rao X, Zhang W, Li X, Wang L, Huang G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Photochem Photobiol. 2019;12:3421–3428. doi: 10.2147/OTT.S180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lobo J. The emerging role of epitranscriptomics in cancer: focus on urological tumors. Genes. 2018;9(11):552. doi: 10.3390/genes9110552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu N, Qin S, Zhang X, Bo X, Liu Z, Tan C, Wen G, Jiang H. Multiple m(6)A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer. 2020;20(1):165. doi: 10.1186/s12885-020-6638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-Part A: renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 47.Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, Lotan Y. Bladder cancer. Nat Rev Dis Prim. 2017;3:17022. doi: 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Nie ZY, Wen XH, Gao YH, Cao H, Zhang SF. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep. 2019;39(12):BSR20192892. doi: 10.1042/BSR20192892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Z, Jing C, Xiao C, Li T, Wang Y. Prognostic risk signature based on the expression of three m6A RNA methylation regulatory genes in kidney renal papillary cell carcinoma. Aging. 2020;12(21):22078–22094. doi: 10.18632/aging.104053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Q, Yang J, Yang H, Li C, Li J, Cao Y. KIAA1429 promotes osteosarcoma progression by promoting stem cell properties and is regulated by miR-143-3p. Cell Cycle. 2020;19(10):1172–1185. doi: 10.1080/15384101.2020.1749465. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Fan L, Lin Y, Lei H, Shu G, He L, Yan Z, Rihan H, Yin G. A newly defined risk signature, consisting of three m(6)A RNA methylation regulators, predicts the prognosis of ovarian cancer. Aging. 2020;12(18):18453–18475. doi: 10.18632/aging.103811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hou J, Shan H, Zhang Y, Fan Y, Wu B. m(6)A RNA methylation regulators have prognostic value in papillary thyroid carcinoma. Am J Otolaryngol. 2020;41(4):102547. doi: 10.1016/j.amjoto.2020.102547. [DOI] [PubMed] [Google Scholar]
- 53.Tsunedomi R, Iizuka N, Tamesa T, Sakamoto K, Hamaguchi T, Somura H, Yamada M, Oka M. Decreased ID2 promotes metastatic potentials of hepatocellular carcinoma by altering secretion of vascular endothelial growth factor. Clin Cancer Res. 2008;14(4):1025–1031. doi: 10.1158/1078-0432.CCR-07-1116. [DOI] [PubMed] [Google Scholar]
- 54.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14(2):77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 55.Havrda MC, Paolella BR, Ran C, Jering KS, Wray CM, Sullivan JM, Nailor A, Hitoshi Y, Israel MA. Id2 mediates oligodendrocyte precursor cell maturation arrest and is tumorigenic in a PDGF-rich microenvironment. Can Res. 2014;74(6):1822–1832. doi: 10.1158/0008-5472.CAN-13-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shafik A, Schumann U, Evers M, Sibbritt T, Preiss T. The emerging epitranscriptomics of long noncoding RNAs. Biochem Biophys Acta. 2016;1859(1):59–70. doi: 10.1016/j.bbagrm.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 57.Jacob R, Zander S, Gutschner T. The dark side of the epitranscriptome: chemical modifications in long non-coding RNAs. Int J Mol Sci. 2017;18(11):2387. doi: 10.3390/ijms18112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barros-Silva D, Lobo J, Guimarães-Teixeira C, Carneiro I, Oliveira J, Martens-Uzunova ES, Henrique R, Jerónimo C. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers. 2020;12(4):771. doi: 10.3390/cancers12040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barros-Silva D, Costa-Pinheiro P, Duarte H. MicroRNA-27a-5p regulation by promoter methylation and MYC signaling in prostate carcinogenesis. Cell Death Dis. 2018;9(2):167. doi: 10.1038/s41419-017-0241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamilton MJ, Young MD, Sauer S, Martinez E. The interplay of long non-coding RNAs and MYC in cancer. AIMS Biophys. 2015;2(4):794–809. doi: 10.3934/biophy.2015.4.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhuang K, Wu Q, Jiang S, Yuan H, Huang S, Li H. CCAT1 promotes laryngeal squamous cell carcinoma cell proliferation and invasion. Am J Transl Res. 2016;8(10):4338–4345. [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Y, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16(1):155. doi: 10.1186/s12943-017-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Can Res. 2007;67(20):9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, Hu J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29(1):151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G, Zhang L, Chen ZS, Xie X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019;18(1):23. doi: 10.1186/s12943-019-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong Y, Yang L, Wei W, Lyu N, Zou Y, Gao G, Ou X, Xie X, Tang H. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics. 2019;11(10):1163–1176. doi: 10.2217/epi-2019-0093. [DOI] [PubMed] [Google Scholar]
- 68.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochem Biophys Acta. 1991;1072(2–3):129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 69.Abate C, Curran T. Encounters with Fos and Jun on the road to AP-1. Semin Cancer Biol. 1990;1(1):19–26. [PubMed] [Google Scholar]
- 70.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20(19):2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 71.Rizzo P, Miao H, D'Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Can Res. 2008;68(13):5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boriack-Sjodin PA, Ribich S, Copeland RA. RNA-modifying proteins as anticancer drug targets. Nat Rev Drug Discov. 2018;17(6):435–453. doi: 10.1038/nrd.2018.71. [DOI] [PubMed] [Google Scholar]
- 73.Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43(1):373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng G, Cox T, Tribbey L, Wang GZ, Iacoban P, Booher ME, Gabriel GJ, Zhou L, Bae N, Rowles J, et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem Neurosci. 2014;5(8):658–665. doi: 10.1021/cn500042t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chai RC, Wu F, Wang QX, Zhang S, Zhang KN, Liu YQ, Zhao Z, Jiang T, Wang YZ, Kang CS. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging. 2019;11(4):1204–1225. doi: 10.18632/aging.101829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed during the current study are available in the sanger box (http://sangerbox.com/) and GTEx database (http://commonfund.nih.gov/GTEx/). The mutation data during this study are included in Li’s published article (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6744659/).