Abstract
Serotonin is a neurotransmitter that plays an important role in regulating behavior and personality in humans and other mammals. Polymorphisms in genes coding for the serotonin receptor subtype 1A (HTR1A), the serotonin transporter (SLC6A4), and the serotonin degrading enzyme monoamine oxidase A (MAOA) are associated with anxiety, impulsivity, and neurotic personality in humans. In primates, previous research has largely focused on SLC6A4 and MAOA, with few studies investigating the role of HTR1A polymorphic variation on behavior. Here, we examined variation in the coding region of HTR1A across apes, and genotyped polymorphic coding variation in a sample of 214 chimpanzees with matched measures of personality and behavior. We found evidence for positive selection at three amino acid substitution sites, one in chimpanzees-bonobos (Thr26Ser), one in humans (Phe33Val), and one in orangutans (Ala274Gly). Investigation of the HTR1A coding region in chimpanzees revealed a polymorphic site, where a C/A single nucleotide polymorphism changes a proline to a glutamine in the amino acid sequence (Pro248Gln). The substitution is located in the third intracellular loop of the receptor, a region important for serotonin signal transduction. The derived variant is the major allele in this population (frequency 0.67), and is associated with a reduction in anxiety, decreased rates of male agonistic behavior, and an increase in socio-positive behavior. These results are the first evidence that the HTR1A gene may be involved in regulating social behavior in chimpanzees and encourage further systematic investigation of polymorphic variation in other primate populations with corresponding data on behavior.
Keywords: behavioral genetics, primates, personality, display, behavioral tendencies
Introduction
Serotonin (5-Hydroxytryptamine or 5-HT) is a monoamine neurotransmitter that plays an important role in the regulation of personality and behavioral tendencies across mammals (Barchas and Usdin 1973). Serotonin levels typically have an inverse relationship with levels of anxiety, depression, and aggressiveness (Lesch and Merschdorf 2000; Azmitia 2007; Duke et al. 2013; Gottschalk and Domschke 2016). Although the serotonin pathway is present in a variety of taxa and is relatively conserved (Azmitia 2007; Curran and Sreekanth 2012), important variation exists in the genes coding for the transporter, degradation enzymes, and receptors of this neurotransmitter that have been linked to differences in species-specific and individual behavior.
In humans, repeat polymorphisms were identified in the regulatory regions of the genes encoding the serotonin transporter (SCL6A4, also known as 5HTT or SERT) and the enzyme monoamine oxidase A (MAOA) (Lesch et al. 1997; Sabol et al. 1998). The serotonin transporter is responsible for reuptake of serotonin from the synaptic cleft into the presynaptic neuron (Murphy et al. 2004), and MAOA is the key enzyme responsible for the degradation of serotonin, as well as other amine neurotransmitters, via oxidative deamination (Sabol et al. 1998). The regulatory region repeats in both genes have been shown to affect transcription of the genes in vitro (Heils et al. 1996; Denney et al. 1999; Bennett et al. 2002; Guo et al. 2008) and have been linked to several psychiatric conditions and behaviors. The SCL6A4 regulatory polymorphism (5-HTTLPR) is a microsatellite with two predominant alleles, a 14 (s or short) and 16 (l or long) repeat (Nakamura et al. 2000). There is substantial evidence that the short allele is linked to lower levels of transcription and increased anxiety, depression, and aggressiveness, as well as a cluster of personality traits like Neuroticism and Harm Avoidance (for reviews, see Murphy et al. 2004; Sen et al. 2004; Karg et al. 2011; Siegel and Crockett 2013; Gottschalk and Domschke 2016). Similar results were found for a MAOA promotor repeat (MAOA-LPR), a length polymorphism with six allelic variants, ranging from two to six repeats. Most studies have focused on the three and four repeat alleles, and found that the shorter allele is linked to lower transcriptional activity, higher anxiety, and more aggressive behavior (Sabol et al. 1998; Deckert et al. 1999; Guo et al. 2008; Raine 2008; Gottschalk and Domschke 2016; but see Fisher et al. 2017).
Although research has largely focused on the serotonin transporter and MAOA enzyme, genetic variation in serotonin receptor proteins has also been shown to be involved in governing behavior. Serotonin signals through a total of 14 receptors (Hoyer et al. 2002). The 1A receptor subtype (HTR1A) is among the most abundant of serotonin receptors in the brain, with particularly high expression in neural structures important in emotional regulation and social cognition, such as the amygdala, hippocampus, and the pyramidal cells and interneurons of the cerebral cortex (Lanfumey and Hamon 2000; Varnäs et al. 2004). This G-protein coupled receptor (GPCR) mediates inhibitory serotonin transmission and is expressed on both serotoninergic and nonserotonin-expressing neurons (Hjorth et al. 2000; Zhong et al. 2008). HTR1A knockout studies in mice reveal increases in anxiety and stress responses (Toth 2003). In humans, missense mutations and regulatory polymorphisms in HTR1A have similarly been implicated in mood and panic disorders (Akimova et al. 2009; Gottschalk and Domschke 2016), and neurotic personality (Strobel et al. 2003).
Understanding the biological basis and origin of this serotonin-related behavioral variation requires a broad comparative perspective within an evolutionary context (Rogers 2018). In nonhuman primates, research on polymorphisms in the serotonin system so far has primarily focused on the SCL6A4 and MAOA linked promotor repeats (Inoue-Murayama et al. 2000; Trefilov et al. 2000; Bennett et al. 2002; Champoux et al. 2002; Barr et al. 2004; Newman et al. 2005; Wendland et al. 2006; Claw et al. 2010; Dobson and Brent 2013; Kalbitzer et al. 2016). While 5-HTTLPR and MAOA-LPR are unique to hominoids (i.e., apes), orthologues were found in rhesus macaques (rs5-HTTLPR and rsMAOA-LPR), with similar common length variants (Lesch et al. 1997; Newman et al. 2005). The rs5-HTTLPR short allele is associated with reduced transcription (Bennett et al. 2002), higher emotional distress and anxiety in infant macaques (Champoux et al. 2002; Barr et al. 2004), and lower age of first dispersal in males, which may be an indicator of higher impulsivity and risk-taking (Trefilov et al. 2000). The rsMAOA-LPR low activity allele was linked to higher aggression in male mother-reared rhesus macaques (Newman et al. 2005). In marmosets, nucleotide polymorphisms were recently identified within the regulatory region of SLC6A4 and found to be linked to gene expression and anxiety (Santangelo et al. 2016). In great apes, studies on serotonin candidate genes lack associated behavioral phenotypic measures (Inoue-Murayama et al. 2000; Wendland et al. 2006; Garai et al. 2014). Instead, polymorphic variation is described in relation to species-specific differences in general behavioral patterns, for example, in aggressiveness (Garai et al. 2014). While length variation in these regions may be linked to known between-species differences, polymorphic regions may have been under differential selective pressures among taxa, and potentially even populations spread across different geographic regions (Kalbitzer et al. 2016). Further detailed studies are therefore needed to assess the association of within-species genetic variation with behavior.
Despite the great interest in the serotonin transporter and the MAOA enzyme, behavioral association studies investigating serotonin receptors, particularly HTR1A genetic variation, are lacking in primates (but see Shattuck et al. 2014). HTR1A is of special interest, given that previous research has documented evidence of selection and a high level of divergence among macaque species, which is suggested to explain some of the species differences in behavior, such as levels of aggression, dispersal, and exploratory behavior (Shattuck et al. 2014). Investigating the gene in great apes, humans’ closest living relatives (Prado-Martinez et al. 2013), would provide further insight into the evolutionary history of the serotonin system. For example, much like different macaque species (Shattuck et al. 2014), bonobos and chimpanzees show behavioral differences in aggressiveness, risk-taking, and social tolerance that may be explained by evolutionary selective pressures acting upon the serotonin system (Hare et al. 2012). Interspecies comparisons of HTR1A may thus shed light on the evolution of behavioral differences among these closely related species.
Therefore, the aims of this study were to examine inter- and intraspecific coding variation and the role of selection on HTR1A in great apes, and to explore the association of polymorphic variation with individual differences in personality and behavior, particularly in chimpanzees. To this end, we genotyped a large sample of captive chimpanzees that had been the subjects of previous research quantifying variation in personality and behavioral tendencies (Freeman et al. 2013). The design of the study is exploratory, resulting in a large number of models being tested. While this may increase our chances of finding false positive results (Ranganathan et al. 2016), the benefit of using an exploratory approach is that unexpected or biologically unsound associations can be ruled out, thereby confirming the targeted effects of the candidate gene on specific aspects of personality and behavioral tendencies. Based on the well-documented serotonin effects in human and nonhuman primates, we predicted that coding variation would primarily impact variation in anxiety and aggression-related personality traits and behaviors.
Results
HTR1A Coding Variation
Interspecific Variation
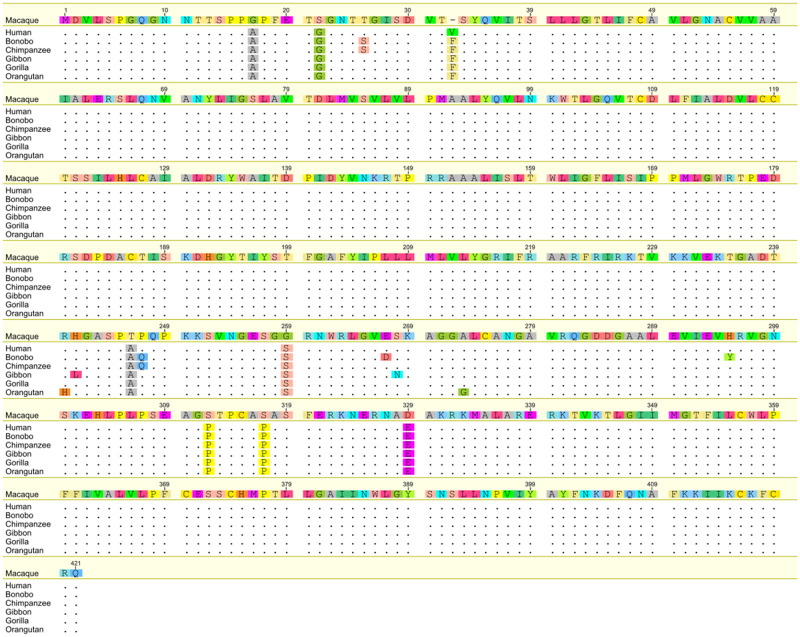
We identified nine fixed species-specific amino acid substitutions among apes (fig. 1). In humans, a phenylalanine was substituted for a valine (Phe33Val). Chimpanzees and bonobos share two substitutions, a threonine to serine (Thr26Ser) and a proline to glutamine (Pro248Gln) (fig. 2). In bonobos, two additional substitutions are present, a glutamic acid to aspartic acid (Glu268Asp) and a histidine to tyrosine (His296Tyr). In orang-utans, two substitutions were found, an arginine to histidine (Arg241His) and an alanine to glycine (Ala274Gly). Finally, gibbons had two species-specific substitutions, a histidine to leucine (His242Leu) and a serine to asparagine (Ser269Asn). In gorillas, no species-specific nonsynonymous substitutions were identified. Signals of positive diversifying selection were found for the Thr26Ser site in the Pan lineage (i.e., chimpanzees and bonobos) (ω = 275, P = 0.03), Phe33Val in humans (ω = 126, P = 0.07), and Ala274Gly in orangutans (ω = 1.4, P = 0.02). For an overview of all site-specific positive selection signals with likelihood ratio test results, see supplementary table S1, Supplementary Material online.
Fig. 1.
Alignment of the HTR1A amino-acid sequences across apes using rhesus macaque HTR1A as a reference sequence. Dots indicate similarity to consensus sequence.
Fig. 2.
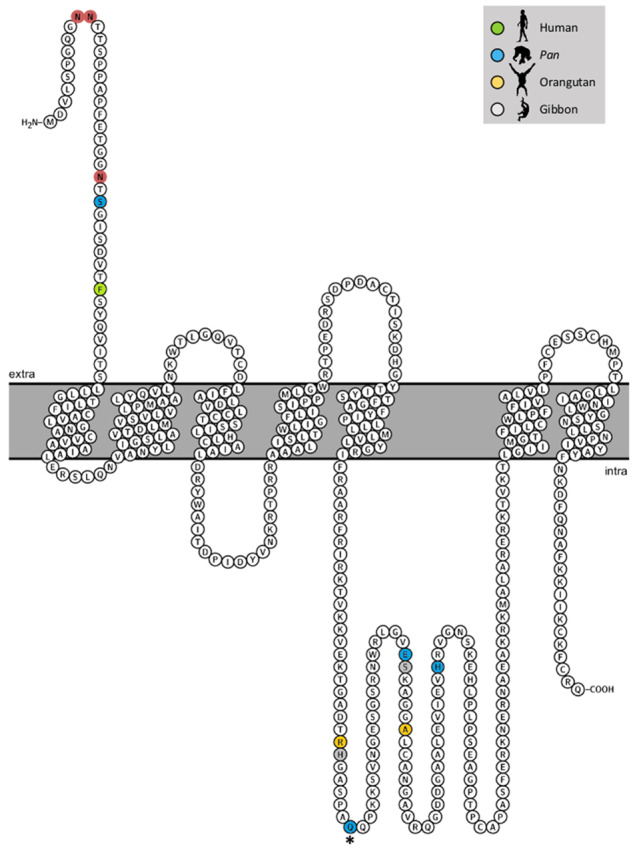
Transmembrane structure of G protein coupled 5-HT1A receptor found in chimpanzees. Mutations/polymorphisms are indicated for humans (Phe33Val), Pan (i.e., chimpanzees and/or bonobos) (Thr26Ser, Pro248Gln, Glu268Asp, His296Tyr), orangutans (Arg241His, Ala274Gly), and gibbons (His242Leu, Ser269Asn). Extracellular N-linked glycosylation motifs are indicated by borderless circles. *indicates polymorphic site in chimpanzees.
Intraspecific Variation
Further examination of the protein-coding region of HTR1A in our chimpanzee sample revealed that the Pro248Gln substitution is polymorphic (rs25209664: C/A). Solution of the 2D structure of the protein indicates that the substitution is located in the third intracellular loop of the receptor (fig. 2). Genotyping of this SNP in our total sample of 214 chimpanzees revealed that the substitution (derived A allele) is the major allele (frequency 0.67) and genotype frequencies were in Hardy–Weinberg equilibrium (χ2 = 0.20, df = 1, P = 0.656; Genotype frequency of ratings AA = 97, AC = 92, CC = 25; Genotype frequency of codings: AA = 20, AC = 24, CC = 9; Genotype frequency of experimental scratching AA = 28, AC = 25, CC = 5).
Relationship between HTR1A Genotype and Personality
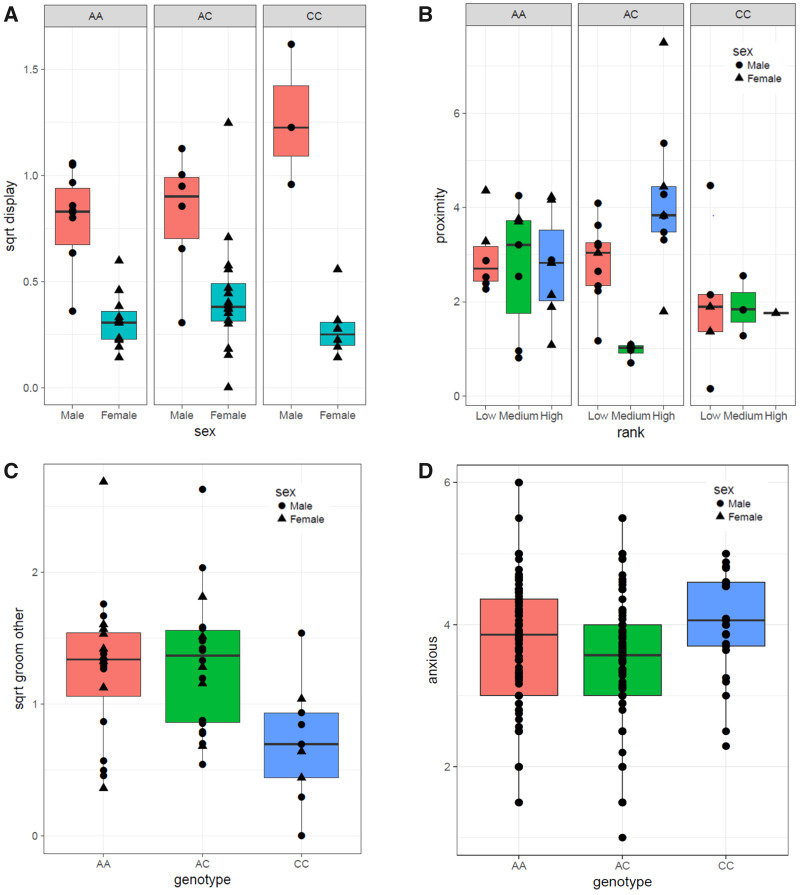
For coded personality and behavior, our model selection approach yielded models that include genotype effects for three behavioral variables: display, proximity, and groom other (see supplementary table S2, Supplementary Material online, for model information and factor estimates). For display behavior, a significant genotype by sex interaction effect (F(2, 43) = 3.920, P = 0.021, fig. 3A) was found. Males homozygous for two C alleles show higher levels of display behavior compared with males with one or two A alleles, while there is no significant difference in females. For proximity, a genotype by rank effect was found (F(4, 41) = 3.279, P = 0.020, fig. 3B). Proximity scores showed no difference between individuals from different ranks in individuals with two ancestral or derived alleles, while in chimpanzees with an AC genotype, individuals of medium rank had lower proximity scores than individuals with high or low rank. For groom other, a significant difference was found between genotypes (F(4, 44) = 4.931, P = 0.012, fig. 3C). Individuals homozygous for two C alleles show significantly lower levels of social grooming than individuals with one (B = −0.599, P = 0.009) or two A alleles (B = −0.573, P = 0.011). For an overview of genotype means for all behavioral variables and coded personality dimensions, see supplementary table S3, Supplementary Material online.
Fig. 3.
Graphical representation of (A) HTR1A genotype by sex interaction effect on display, (B) HTR1A genotype by rank interaction effect on proximity, (C) HTR1A genotype effect on groom other, and (D) HTR1A genotype effect on anxious. Personality data taken from Freeman et al. (2013).
For rated personality trait adjectives, a genotype effect was found solely on the trait adjective anxious (F(2, 209) = 4.286, P = 0.015), with individuals homozygous for two C alleles scoring higher on anxious than individuals with one A allele (B = 0.591, P = 0.006) (fig. 3D). For model information and factor estimates, see supplementary table S2, Supplementary Material online. No significant associations were found between genotype and any of the six personality dimensions or additional 40 trait adjectives. For an overview of genotype means on additional adjectives and rated personality dimensions, see supplementary table S4, Supplementary Material online.
Relationship between HTR1A Genotype and Experimental Measure for Anxiety
HTR1A genotype was significantly associated with levels of self-scratching (F(2, 53) = 9.754, P < 0.001), with CC individuals engaging in significantly more scratching compared with AC (B = 12.380, P = 0.001) and AA (B = 11.182, P = 0.003) individuals (fig. 4).
Fig. 4.
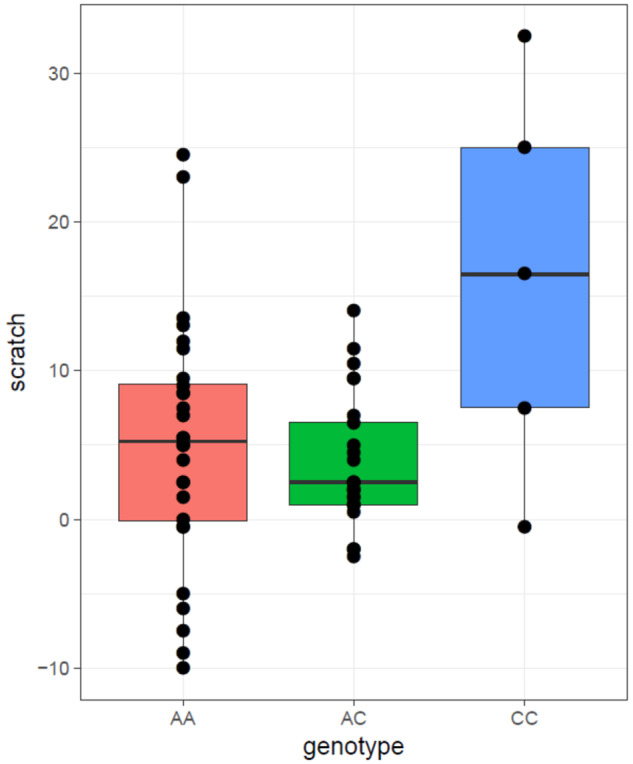
Graphical representation of HTR1A genotype effects on anxious mean differences in self-scratching between the HTR1A genotype groups. Behavioral data taken from Hopkins et al. (2006).
Discussion
Interspecies comparison of HTR1A in apes revealed a relatively conserved coding sequence. Nine fixed species-specific amino acid substitutions were identified (figs. 1 and 2). Analysis of site-based protein evolution across mammals revealed signals of directional selection for Phe33Val, Thr26Ser, and Ala274Gly, indicating that these are potential candidates for further studies of functional changes. In our sample of chimpanzees, Pro248Gln appeared to be polymorphic, and associated with anxiety, display, and potentially grooming and proximity.
The polymorphism leading to the Pro248Gln substitution is located in the third intracellular loop of the receptor. G-protein coupled receptors typically have seven transmembrane alpha helices that create three extracellular and three intracellular loops (fig. 2). The third intracellular loop plays an important role in serotonin signaling as it couples to heterotrimeric G proteins and intracellular second messengers like calmodulin to function in signal transduction (Turner et al. 2004; Masson et al. 2012). A multitude of signaling pathways have been described in which HTR1A participates, including adenylyl cyclase inhibition, phospholipase C stimulation, and K+ channel regulation (for review, see Masson et al. 2012). Although it is unclear what particular pathway might be affected by Pro248Gln, due to its location in the receptor and its association with expected behavioral phenotypes in this study, we predict that it could have an impact on intracellular signaling. Transfection experiments where wild-type and mutant receptors are investigated for differential coupling to multiple signaling pathways can shed light on the exact functional impact of the polymorphism in future work (Lembo et al. 1997).
Investigation of the relationship between the Pro248Gln polymorphism and variation in chimpanzee personality revealed a reduction in anxious, a rated personality adjective. Chimpanzees with two ancestral alleles were perceived as more anxious by human observers compared with individuals that are heterozygous. The result is in line with known regulatory effects of serotonin in other primate species, including humans (Barr et al. 2004; Lanzenberger et al. 2007; Akimova et al. 2009; Fakra et al. 2009; Gottschalk and Domschke 2016; Santangelo et al. 2016). We did not find an association with the overarching personality factor Dominance, which includes the adjective anxious. Human studies typically find an association between serotonin and Neuroticism, a personality dimension that includes the personality adjectives anxiety, angry, depression, self-consciousness, impulsiveness, and vulnerability (Hu et al. 2000; Sen et al. 2004; Ward et al. 2017). This dimension is, however, different from any of the chimpanzee dimensions included in this study. The chimpanzee Dominance dimension contains adjectives related to the shy-bold axis and assertiveness, for example, bold, shy, and dominant, which are not expected to be regulated by serotonin.
In addition, no significant associations were found with behavioral variables that potentially reflect anxiety, like exhibiting a fear-grimace and self-grooming. Fear-grimace was only observed in five individuals, limiting its reliability, but even so, it did follow the direction of the anxiety effect we found, with individuals with an ancestral allele scoring higher than individuals with derived alleles (supplementary fig. S1A, Supplementary Material online). Self-grooming did not follow the pattern of the anxiety effect (supplementary fig. S1B, Supplementary Material online), but although some studies include it as an indicator off anxiety in chimpanzees (Fraser et al. 2008; Koski 2011), others have found no support for this (Aureli and de Waal 1997; Baker and Aureli 1997). However, self-scratching and yawning, the behaviors that are suggested to be the most reliable indicators of anxiety in chimpanzees (Baker and Aureli 1997; Yamanashi and Matsuzawa 2010), were not part of the personality study from which we drew data (Freeman et al. 2013). Therefore, in addition, we chose to investigate the effects of HTR1A genotype on another existing data set of recorded self-scratching behavior of chimpanzees during an experimental set-up that aimed to induce negative arousal (Hopkins et al. 2006). In line with our findings, CC apes engaged in significantly more anxiety-related scratching compared with AC and AA individuals. These results confirm the association between the Pro248Gln substitution and anxiety in chimpanzees. The higher abundance of the AA genotype in the population may be indicative of selective pressures favoring lower levels of stress and/or better stress coping behavior in these chimpanzees. This species has a wide distribution, ranging from East to West Africa (Humle et al. 2016), and genetic modifications that allow for lower levels of anxiety may have facilitated chimpanzee dispersal throughout time. It is also worth noting that biased sampling is a potential issue when studying the behavioral genetics of captive animals. That is, more anxious individuals might be less likely to end up in captivity (Carter et al. 2012). Such a bias would not impact our findings regarding phenotypic associations, but the CC genotype might be underrepresented in the captive population. Genotype frequencies from wild populations are needed to clarify this issue.
An effect of the Pro248Gln polymorphism was also found on male display behavior. Males with one or two derived alleles showed a significant decrease in display behavior compared with males with two ancestral alleles. Display behavior was a coded behavioral variable defined as “a behavioral sequence that incorporates elements such as drumming, repeated swaying, exaggerated, often bipedal locomotion, charging, and pilo-erection,” and is the most frequent form of male aggression in this study and the wild (Muller 2002). Genotype was not significantly associated with observed behaviors contact aggression, noncontact aggression, or with the rated adjective aggressiveness. However, as with fear-grimace, aggression rates were low in the population and therefore more prone to large error distributions. Nevertheless, inspection of the plots for these behaviors and aggressiveness revealed a similar direction of HTR1A genotype effects (supplementary fig. S2, Supplementary Material online), supporting the potential association between serotonin receptor genotype and agonistic behavior.
In chimpanzees, male display rates have been documented to show individual differences and are positively correlated with rank (Muller 2002; Foster et al. 2009). Therefore, these results suggest that HTR1A genotype could play a role in shaping individual variation in agonistic dominance styles of high-ranking chimpanzees (Foster et al. 2009). Because the number of males with two rare ancestral alleles was low in our study, our genotype by rank interaction effect was not significant. We can, however, not fully rule out that rank effects are driving the display effect in our study. If true, the male biased effect of genotype is likely due to the interaction of serotonin with testosterone (Birger et al. 2003). In humans, androgens like testosterone have been shown to downregulate serotonin receptor mRNA expression and serotonin turnover (Ambar and Chiavegatto 2009). Furthermore, high levels of competition-induced testosterone combined with low levels of serotonin have been shown to augment levels of impulsive aggression (Birger et al. 2003). In chimpanzees, testosterone is known to increase with rank (Muller and Wrangham 2004; Anestis 2006). Increased testosterone levels in high-raking males, in combination with the genotype effects on serotonin receptor signaling, possibly leads to augmented levels of agonistic behavior. Male aggression is characteristic of chimpanzee society, with males sometimes showing extreme coercion of females, and intergroup interactions that are typically aggressive (Wilson and Wrangham 2003; Wilson et al. 2014). On rare occasions, high-ranking males in the wild have been documented to show extreme aggression, even leading to within-group lethal attacks on lower ranking males (Watts 2004; Kaburu et al. 2013). Further studies investigating HTR1A genotype effects may offer insight into proximate mechanisms associated with such behaviors.
Finally, a significant effect of genotype was found on social grooming behavior. Individuals with two ancestral C alleles showed lower levels of social grooming than individuals with a derived allele. While previous research has been more oriented toward investigating the role of serotonin in regulating socio-negative behaviors like aggression and anxiety, recent studies have also highlighted its role in prosociality, empathy, and social bonding (Tost and Meyer-Lindenberg 2010; Tibi-Elhanany and Shamay-Tsoory 2011; Kiser et al. 2012; Siegel and Crockett 2013; Stoltenberg et al. 2013; Larke et al. 2016). In humans, individuals with the long 5-HTTLPR allele showed lower levels of social avoidance and higher levels of helping others, indicating that reduced social anxiety in long allele carriers may enhance prosocial behavior (Stoltenberg et al. 2013). Our results are in line with these findings, with higher levels of social behavior found in individuals with the derived alleles. For proximity, a genotype by rank effect was found. However, the data reveal no clear pattern for this interaction effect. In line with the grooming effect, individuals with two ancestral C alleles do show lower levels of proximity behavior, independent of their rank. While these associations with socio-positive behavior are interesting, they have not previously been discussed in primates and thus require further investigation.
Notably, Pan showed more lineage-specific variation than any other ape lineage, with four nonsynonymous substitutions found in bonobos. Despite their close phylogenetic relationship, bonobos and chimpanzees are known to differ in social behaviors. Bonobos show lower levels of severe aggression and risk taking behavior, and are more sensitive to social cues than are chimpanzees (Kano 1992; Doran et al. 2002; Herrmann et al. 2010; Furuichi 2011; Haun et al. 2011; Rosati and Hare 2012; Hopkins et al. 2017), all of which are potentially influenced by serotonin. While the function and polymorphic nature of these amino acid substitutions remains to be investigated, they could be informative for our understanding of the evolutionary mechanisms behind these behavioral differences. Given the association of the Pro248Gln polymorphism with agonistic behavior in chimpanzees, we genotyped an additional six bonobos to investigate polymorphic variation at this site and found that all individuals were homozygous for the derived Pan specific allele. While further testing in larger samples of bonobos is warranted, this result is interesting given the reported behavioral differences in the two species. Hare et al. (2012) have hypothesized that selection pressures against aggression have occurred in bonobos and that serotonin system-regulating genes offer potential candidates (Hare et al. 2012). If the HTR1A allele associated with reduced agonistic behavior has been fixed in bonobos, our results may support a model of selection against aggression. It has already been shown that bonobos show greater serotonergic innervation selectively in the amygdala, a brain region important for the regulation of emotional response (Stimpson et al. 2016). Further systematic investigation of variation in serotonin-related genes (HTR1A, SLC6A4, MAOA) and their effects on brain expression patterns and serotonin signaling would be informative to answer questions regarding evolutionary patterns underlying the behavioral differences in these two closely related species.
To conclude, these findings provide the first evidence for HTR1A genotype associations with anxiety, agonistic, and potentially socio-positive behavioral tendencies in chimpanzees. While our results warrant replication and confirmation of the exact physiological impact of the Pro248Gln polymorphism, they offer an expanded framework for understanding genetic variation underlying behavioral variation in chimpanzee populations. In addition, further systematic assessment of HTR1A polymorphic variation with behavior in the other primate species could provide insights into the evolutionary pathways underlying serotonergic signaling.
Materials and Methods
Subjects
Matched DNA/blood samples and personality rating scores were available for 214 adult and subadult chimpanzees (127 females and 87 males, age range: 8–53 years). All chimpanzees were members of the colony of apes housed at the Yerkes National Primate Research Center (YNPRC) (N = 77, 55 females and 22 males, age range: 9–53 years) or the National Center for Chimpanzee Care at the Michale E. Keeling Center for Comparative Medicine and Research, The University of Texas MD Anderson Cancer Center, Bastrop, TX (KCCMR) (N = 137, 72 females and 65 males, age range: 8–51 years). All aspects of this research adhered to the American Psychological Association’s guidelines for the ethical treatment of animals in research.
HTR1A Variant Identification and Protein Modeling
Interspecific Variation
To identify coding variation in the HTR1A gene in apes, we used the Ensembl genome browser to extract coding sequences for humans (Homo sapiens, GRCh38: CM000667.2), chimpanzees (Pan troglodytes, Pan_tro_3.0: CM000319.3), bonobos (Pan paniscus, panpan1.1: CM003388.1), western lowland gorillas (Gorilla gorilla gorilla, gorGor4: FR853088.3), Sumatran orangutans (Pongo abelii, PPYG2: ENSPPYG00000015500), and white-cheeked gibbons (Nomascus leucogenys, Nleu_3.0: CM001664.1). To investigate further potential functional implications of variants, we also tested for positive and purifying selection on individual codon sites across a large alignment of 27 mammals, including 25 primate species (supplementary table S4, Supplementary Material online) using the mixed effects model of evolution (MEME) in HyPhy (Murrell et al. 2012). Positive selection can be inferred whenever the estimated ratio (ω) of site-specific nonsynonymous (β) to synonymous (α) substitution rates significantly exceeds one (Murrell et al. 2012).
Intraspecific Variation
For chimpanzees, the reference genome Pan_tro_3.0 (Kuderna et al. 2017) is largely based on a single western chimpanzee (Pan troglodytes verus) male, but includes additional lower-coverage genomic sequence data from seven other individuals (The Chimpanzee Sequencing and Analysis Consortium 2005). Using these combined data for eight individuals, we identified nonsynonymous single nucleotide polymorphisms as candidates for assessing within-species variation potentially associated with aspects of personality and behavior. Given that the genome data are based on western chimpanzees, it may not capture the potential genetic variation in the other three subspecies of chimpanzees. However, the chimpanzees living in captivity in United States descended from a founder population that was >95% from the western subspecies (Ely et al. 2005), so the genome data are expected to be representative for the individuals included in this study. We then modeled the chimpanzee receptor structure to identify the location of noncoding polymorphisms using protter, a web-based tool that supports interactive protein data analysis by visualizing annotated sequence features in the context of protein topology (Omasits et al. 2014).
Personality Assessment
We used the personality measures reported by Freeman et al. (2013), which were collected using two methodologies. The first method, referred to as the rating method, used a chimpanzee-specific personality questionnaire that was developed by the authors and extensively tested for validity. Rating data were collected for all 214 chimpanzees from both YNPRC and KCCMR included in the study. The questionnaire consists of 41 personality trait adjectives that were rated on a Likert scale ranging from 1 (least descriptive of chimpanzee) to 7 (most descriptive of chimpanzee). Trait adjectives were rated by observers who were familiar with the individuals to collect overall impressions of the chimpanzee’s behaviors. For example, the adjective “anxious” is defined as an individual that is hesitant, indecisive, tentative, and jittery. Interrater reliabilities were strong for all but one adjective (predictable), which was excluded from subsequent analysis. Principal component analysis on the means of the 40 rating adjectives, combined with expert evaluations of the factor solution, resulted in a personality structure comprising six dimensions: Reactivity/Undependability, Dominance, Extraversion, Openness, Agreeableness, and Methodical (table 1). For detailed factor adjective scores, see supplementary table S5, Supplementary Material online.
Table 1.
Adjectives Loading onto Varimax-Rotated Chimpanzee Personality Traits.
| Methods | Trait | Adjectives/Behaviors |
|---|---|---|
| Rating | Reactivity/undependability | + Irritable + Temp./moody + Deceptive + Impulsive + Defiant + Mischievous + Jealous + Manipulative + Stingy + Bullying + Aggressive + Eccentric + Socially inept + Excitable + Autistic – Calm |
| Dominance | + Bold + Relaxed + Dominant – Fearful – Timid – Cautious – Dependent – Anxious | |
| Extraversion | + Active + Playful + Sexual + Affiliative – Solitary – Depressed | |
| Openness | + Human oriented + Inq./curious + Inventive + Intelligent + Aff./ Friendly + Persistent | |
| Agreeableness | + Protective + Considerate | |
| Methodical | + Self-caring + Methodical | |
| Coding | Dominance | + Solicit + Sexual + Contact Aggression + Noncontact Aggression + Displace |
| Affiliation | + Postconflict Affiliation + Affiliation + Contact | |
| Proximity | + Groom Other + Proximity – Play – Begging | |
| Solitary | + Display + Fear Grimace + Groom self – Submissive |
Note.—The data are taken from Freeman et al. (2013).
The second method, referred to as the coding method, is based on using coded behaviors that are observed in the social group. Coding data were collected from 2012 to 2014 for a subset of 54 individuals only from the KCCMR group (34 females, 20 males, age range: 8–53 years). Within the KCCMR group, subjects were housed in six smaller social groups with relatively stable group compositions across the entire period of observation (supplementary table S7, Supplementary Material online). Each group was observed for an average of 41 h (range: 10–61 h). Behaviors were coded using an extensive ethogram. Behavioral variables and their definitions are shown in supplementary table S6, Supplementary Material online. The behaviors were collected 2 years prior to the collection of the personality ratings as part of a separate study (Silk et al. 2013; unpublished data). Three observers collected the behavioral data and did not rate the personalities of the chimpanzees. The behavioral ethogram used for the observations included both scan sampling (for common state behaviors) and ad libitum (for rare behaviors and/or point behaviors) data. Each observation session lasted 60 min with scan samples taken every three min. Scan data included the behaviors groom, proximity, contact, play, and begging. Ad libitum observations included the groupings of behavior: aggressive (display, non‐contact aggression, contact aggression, solicit, displace, and intervene) and sexual, postcontact affiliation, submissive (begging, fear grimace, flee, and submissive), and food sharing. Principal component analysis on the means of the 17 coding variables, combined with expert evaluations of the factor solution, resulted in a personality structure comprising four dimensions: Dominance, Affiliation, Proximity, and Solitary (table 1). For detailed factor behavioral item scores, see supplementary table S8, Supplementary Material online. In addition, dominance rank was available for all 54 chimpanzees that were observed. Rank was assigned by a researcher based on >20 years of past experience with the chimpanzees at the facility. All chimpanzees were classified as either high, medium, or low ranking. For a subset of 25 individuals, dominance data were also collected using behavioral observations, including displacement, displays, aggression, and grooming received (Lewis 2002), which were used to validate the ranking data. The correlation between the rated ranking data and the data based on behavioral observations was r = 0.66 (Freeman 2010). All our analyses of SNP associations were based on these previously collected behavioral data and calculations of personality dimensions; we did not perform any additional behavioral studies for the current analyses.
Experimental Measure Scratching
In addition to personality data and behavioral tendencies, we also tested for genotype effects on previously collected experimental measures of self-scratching, an anxiety-related behavior (Baker and Aureli 1997), for a subset of 58 individuals. For a detailed description of the methods used to collect these data, see Hopkins et al. (2006). In brief, the frequency of self-scratching was recorded during a baseline and experimental condition where chimpanzees were shown videos of unfamiliar chimpanzees sharing, fighting over and consuming a watermelon. Difference scores were computed by subtracting the number of responses in the baseline condition from the frequency recorded when shown videos of unfamiliar chimpanzees.
DNA Extraction and Genotyping
For the 214 chimpanzees with matched personality data, genomic DNA was extracted from 200-µl blood samples using the QIAampDNA Mini Kit automated on a QiaCube (Qiagen) and following the manufacturer’s instructions. The DNA extract was recovered in 200 µl of elution buffer, and kept frozen at −20 °C. Negative extraction controls showed no evidence of DNA contamination. DNA concentrations were quantified using a Nanodrop 2000 (Thermo-Fisher Scientific) spectrophotometer. Subsequent genotyping of the SNP of interest was done using High Resolution Melt Analysis (Smith et al. 2010). Primer pairs flanking the SNP (forward: 5′-CATGCTGGTTCTCTATGGGC-3′ and reversed 5′-ACTCTCCATTCACACTCTTCTTG-3′) were designed to target a short segment (100 bp) containing the single polymorphic site. All qPCR reactions and melting curves were done on a Rotor-Gene Q (Qiagen) platform. The 25-µl PCR reaction mix contained 12.5 µl HRM MasterMix (Qiagen: HotStarTaq plus DNA polymerase, EvaGreen dye, Q-Solution, deoxynucleotides, and MgCl2), 1.75 µl primer mix (10 mM concentration forward and reverse primers) and ∼20 ng of genomic DNA. PCR started with an initial activation at 95 °C (5 min), followed by 40 cycles of denaturing at 95 °C (10 s), annealing at 54 °C (30 s) 72 °C (40 s) and a final extension period of 10 min at 72 °C. In each reaction, no-template controls were included. Data on melting patterns of qPCR products were generated immediately following amplification by increasing temperatures from 65 to 95 °C, rising by 0.1 °C/2 s. Fluorescence data were plotted as a function of temperature during DNA denaturation (melting) and visualized and compared using the Rotor-Gene Q HRM software package (Qiagen). The resulting melting temperatures and curve shapes were assigned to different genotypes and up to four independent qPCR and high resolution melt analyses were repeated per sample (mean = 2.76 replicates per sample). Additionally, for each melting curve with matching genotype, we Sanger sequenced ten percent of the samples to validate and confirm the genotype (Applied Biosystems Genetic Analyzer, DNA Analysis Facility at Yale University). Multiple alignments of the resulting DNA sequences were performed using Geneious (Version 6.0.6).
Statistical Analysis
Statistical analysis was performed using the statistical software program R (www.r-project.org, version 3.3.2; last accessed March 22, 2019). We tested for population adherence to Hardy–Weinberg equilibrium using the “HWAlltests” function in the HardyWeinberg package in R (https://cran.r-project.org/web/packages/HardyWeinberg/index.html; last accessed March 22, 2019). To test for HTR1A genotype effects on personality, we ran general linear models using the lm function in the lme4 package in R (https://cran.r-project.org/web/packages/lme4/index.html; last accessed March 22, 2019) . Sex, genotype, rank, and all two-way interactions were included as fixed effects and age and relatedness coefficient were entered as covariates. Relatedness coefficients were used to correct for the degree of relatedness of each individual to all other individuals in the colony. Relatedness coefficients were extracted from pedigree data using the kinship2 package in R (https://cran.r-project.org/package=kinship2; last accessed March 22, 2019). Given that we correct all our models for relatedness, all means reported are estimated marginal means corrected for the effect of relatedness. Model selection was done using the Akaike Information Criterion (AIC). Diagnostic plots (residuals vs. leverage and QQ plots), Shapiro–Wilk tests, and variance inflation factors were used to confirm the assumptions of linear models. The Levene’s test was used to assess the homogeneity of variances assumption of linear models. When any of the assumptions were not met, we used square root or log transformations of our variables. Given that the chimpanzee personality dimensions are different from human personality dimensions and therefore the expected associations between genotype and personality may vary between studies, we use an exploratory approach that includes testing all ten personality dimensions (PCA components) and all 40 personality adjectives and 14 behavioral variables. Four behavioral variables were not individually tested for genotype effect, as the behaviors were rare and only observed in a small proportion of individuals (fear grimace N = 5, solicit N = 9, intervene N = 6, and flee N = 2). Group was not added as a random effect, as not all genotype-rank combinations were present in all subgroups (see supplementary table S8, Supplementary Material online). Our study is therefore limited in that we cannot fully rule out a potential confound between group and genotype-rank effects.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank colleagues for help with providing DNA samples, specifically: the Yerkes National Primate Research Center, The Michale E. Keeling Center for Comparative Medicine and Research, and John J. Ely. We thank Andrew Barr for his statistical advice, and Arash Panjwani for help with data compilation. We also thank all members of the Primate Genomics Lab and the Laboratory for Evolutionary Neuroscience at the George Washington University for helpful feedback on the project. This work was supported by the National Science Foundation (SMA 1542848 to N.S., C.C.S., W.D.H., S.J.S., and B.J.B., and SES 1425216 to S.F.B.), and the James S. McDonnell Foundation (220020293 to C.C.S.). The collection of the behavioral data was funded by a grant from the MacArthur Foundation Preferences Network to Joan B. Silk, Joseph Henrich, and Daniel Povinelli. The authors declare that there are no conflicts of interest to report.
References
- Akimova E, Lanzenberger R, Kasper S. 2009. The serotonin-1A receptor in anxiety disorders. Biol Psychiatry. 66(7):627–635. [DOI] [PubMed] [Google Scholar]
- Ambar G, Chiavegatto S. 2009. Anabolic-androgenic steroid treatment induces behavioral disinhibition and downregulation of serotonin receptor messenger RNA in the prefrontal cortex and amygdala of male mice. Genes Brain Behav. 8(2):161–173. [DOI] [PubMed] [Google Scholar]
- Anestis SF. 2006. Testosterone in juvenile and adolescent male chimpanzees (Pan troglodytes): effects of dominance rank, aggression, and behavioral style. Am J Phys Anthropol. 130(4):536–545. [DOI] [PubMed] [Google Scholar]
- Aureli F, de Waal F. 1997. Inhibition of social behavior in chimpanzees under high-density conditions. Am J Primatol. 41(3):213–228. [DOI] [PubMed] [Google Scholar]
- Azmitia EC. 2007. Serotonin and brain: evolution, neuroplasticity, and homeostasis. Int Rev Neurobiol. 77:31–56. [DOI] [PubMed] [Google Scholar]
- Baker KC, Aureli F. 1997. Behavioural indicators of anxiety: an empirical test in chimpanzees. Behaviour 134(13–14):1031–1050. [Google Scholar]
- Barchas J, Usdin E. 1973. Serotonin and behavior. New York: Elsevier Science. [Google Scholar]
- Barr CS, Newman TK, Schwandt M, Shannon C, Dvoskin RL, Lindell SG, Taubman J, Thompson B, Champoux M, Lesch KP. 2004. Sexual dichotomy of an interaction between early adversity and the serotonin transporter gene promoter variant in rhesus macaques. Proc Natl Acad Sci U S A. 101(33):12358–12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, Shoaf SE, Champoux M, SUOMI SJ, Linnoila MV, Higley JD. 2002. Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatry. 7(1):118–122. [DOI] [PubMed] [Google Scholar]
- Birger M, Swartz M, Cohen D, Alesh Y, Grishpan C, Kotelr M. 2003. Aggression: the testosterone-serotonin link. Isr Med Assoc J. 5(9):653–658. [PubMed] [Google Scholar]
- Carter AJ, Heinsohn R, Goldizen AW, Biro PA. 2012. Boldness, trappability and sampling bias in wild lizards. Anim Behav. 83(4):1051–1058. [Google Scholar]
- Champoux M, Bennett a, Shannon C, Higley JD, Lesch KP, Suomi SJ. 2002. Serotonin transporter gene polymorphism, differential early rearing, and behavior in rhesus monkey neonates. Mol Psychiatry. 7(10):1058–1063. [DOI] [PubMed] [Google Scholar]
- Claw KG, Tito RY, Stone AC, Verrelli BC. 2010. Haplotype structure and divergence at human and chimpanzee serotonin transporter and receptor genes: implications for behavioral disorder association analyses. Mol Biol Evol. 27(7):1518–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran KP, Sreekanth CH. 2012. Serotonin circuits and anxiety: what can invertebrates teach us? Invertabret Neurosci. 12(2):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Catalano M, Syagailo YV, Bosi M, Okladnova O, Di Bella D, Nothen MM, Maffei P, Franke P, Fritze J, et al. 1999. Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet. 8(4):621–624. [DOI] [PubMed] [Google Scholar]
- Denney RM, Koch H, Craig IW. 1999. Association between monoamine oxidase A activity in human male skin fibroblasts and genotype of the MAOA promoter-associated variable number tandem repeat. Hum Genet. 105(6):542–551. [DOI] [PubMed] [Google Scholar]
- Dobson SD, Brent LJN. 2013. On the evolution of the serotonin transporter linked polymorphic region (5-HTTLPR) in primates. Front Hum Neurosci. 7:588.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran D, Jungers W, Sugiyama Y, Fleagle J, Heesy C. 2002. Multivariate and phylogenetic approaches to understanding chimpanzee and bonobo behavioural diversity. In: Boesch C, Hohmann G, Marchant LF, editors. Behavioral diversity in chimpanzees and bonobos. Cambridge: Cambridge University Press. p. 14–34. [Google Scholar]
- Duke AA, Bègue L, Bell R, Eisenlohr-Moul T. 2013. Revisiting the serotonin–aggression relation in humans: a meta-analysis. Psychol Bull. 139(5):1148–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely JJ, Dye B, Frels WI, Fritz J, Gagneux P, Khun HH, Switzer WM, Lee DR. 2005. Subspecies composition and founder contribution of the captive U.S. chimpanzee (Pan troglodytes) population. Am J Primatol. 67(2):223–241. [DOI] [PubMed] [Google Scholar]
- Fakra E, Hyde LW, Gorka A, Fisher PM, Muñoz KE, Kimak M, Halder I, Ferrell RE, Manuck SB, Hariri AR. 2009. Effects of HTR1A C(−1019)G on amygdala reactivity and trait anxiety. Arch Gen Psychiatry. 66(1):33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher PM, Ozenne B, Svarer C, Adamsen D, Lehel S, Baaré WFC, Jensen PS, Knudsen GM. 2017. BDNF val66met association with serotonin transporter binding in healthy humans. Transl Psychiatry. 7(2):e1029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Gilby IC, Murray CM, Johnson A, Wroblewski EE, Pusey AE. 2009. Alpha male chimpanzee grooming patterns: implications for dominance “style.” Am J Primatol. 71(2):136–144. [DOI] [PubMed] [Google Scholar]
- Fraser ON, Stahl D, Aureli F. 2008. Stress reduction through consolation in chimpanzees. Proc Natl Acad Sci U S A. 105(25):8557–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman HD, Brosnan SF, Hopper LM, Lambeth SP, Schapiro SJ, Gosling SD. 2013. Developing a comprehensive and comparative questionnaire for measuring personality in chimpanzees using a simultaneous top-down/bottom-up design. Am J Primatol. 75(10):1042–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman HD. 2010. Scale development and construct validation of a chimpanzee rating scale.
- Furuichi T. 2011. Female contributions to the peaceful nature of bonobo society. Evol Anthropol. 20(4):131–142. [DOI] [PubMed] [Google Scholar]
- Garai C, Furuichi T, Kawamoto Y, Ryu H, Inoue-Murayama M. 2014. Androgen receptor and monoamine oxidase polymorphism in wild bonobos. Meta Gene 2:831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk MG, Domschke K. 2016. Novel developments in genetic and epigenetic mechanisms of anxiety. Curr Opin Psychiatry. 29(1):32–38. [DOI] [PubMed] [Google Scholar]
- Guo G, Ou X-M, Roettger M, Shih JC. 2008. The VNTR 2 repeat in MAOA and delinquent behavior in adolescence and young adulthood: associations and MAOA promoter activity. Eur J Hum Genet. 16(5):626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare B, Wobber V, Wrangham R. 2012. The self-domestication hypothesis: evolution of bonobo psychology is due to selection against aggression. Anim Behav. 83(3):573–585. [Google Scholar]
- Haun DBM, Nawroth C, Call J. 2011. Great apes’ risk-taking strategies in a decision making task. PLoS One 6(12):e28801.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heils A, Petri S, Stöber G, Riederer P, Bengel D, Lesch K, Teufel A. 1996. Allelic variation of human serotonin transporter gene expression. J Neurochem. 66(6):2621–2624. [DOI] [PubMed] [Google Scholar]
- Herrmann E, Hare B, Call J, Tomasello M. 2010. Differences in the cognitive skills of bonobos and chimpanzees. PLoS One 5:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth S, Bengtsson HJ, Kullberg A, Carlzon D, Peilot H, Auerbach S. 2000. Serotonin autoreceptor function and antidepresent drug actions. J Psychopharmacol. 14(2):177–185. [DOI] [PubMed] [Google Scholar]
- Hopkins WD, Russell JL, Freeman H, Reynolds EAM, Griffis C, Leavens DA. 2006. Lateralized scratching in chimpanzees (Pan troglodytes): Evidence of a functional asymmetry during arousal. Emotion 6(4):553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins WD, Stimpson CD, Sherwood CC. 2017. Social cognition and brain organization in chimpanzees (Pan troglodytes) and bonobos (Pan paniscus). In: Hare B, Yamamoto S, editors. Bonobos: unique in mind, brain and behavior. Oxford: Oxford University Press. p. 199–213. [Google Scholar]
- Hoyer D, Hannon JP, Martin GR. 2002. Molecular, pharmacological and functional diversity of 5-HT receptors. Pharmacol Biochem Behav. 71(4):533–554. [DOI] [PubMed] [Google Scholar]
- Hu S, Brody CL, Fisher C, Gunzerath L, Nelson ML, Sabol SZ, Sirota LA, Marcus SE, Greenberg BD, Murphy DL, et al. 2000. Interaction between the serotonin transporter gene and neuroticism in cigarette smoking behavior. Mol Psychiatry. 5(2):181–188. [DOI] [PubMed] [Google Scholar]
- Humle T, Maisels F, Oates JF, Plumptre A, Williamson EA. 2016. Pan troglodytes (errata version published in 2018). The IUCN Red List of Threatened Species 2016: e.T15933A129038584. 10.2305/IUCN.UK.2016-2.RLTS.T15933A17964454.en [DOI]
- Inoue-Murayama M, Niimi Y, Takenaka O, Okada K, Matsuzaki I, Ito S, Murayama Y. 2000. Allelic variation of the serotonin transporter gene polymorphic region in apes. Primates 41(3):267–273. [DOI] [PubMed] [Google Scholar]
- Kaburu SSK, Inoue S, Newton-Fisher NE. 2013. Death of the alpha: within-community lethal violence among chimpanzees of the Mahale mountains national park. Am J Primatol. 75(8):789–797. [DOI] [PubMed] [Google Scholar]
- Kalbitzer U, Roos C, Kopp GH, Butynski TM, Knauf S, Zinner D, Fischer J. 2016. Insights into the genetic foundation of aggression in Papio and the evolution of two length-polymorphisms in the promoter regions of serotonin-related genes (5-HTTLPR and MAOALPR) in Papionini. BMC Evol Biol. 16:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y. 1992. The last ape. Stanford ( CA: ): Stanford University Press. [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. 2011. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 68(5):444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser D, Steemer S B, Branchi I, Homberg JR. 2012. The reciprocal interaction between serotonin and social behaviour. Neurosci Biobehav Rev. 36(2):786–798. [DOI] [PubMed] [Google Scholar]
- Koski SE. 2011. Social personality traits in chimpanzees: temporal stability and structure of behaviourally assessed personality traits in three captive populations. Behav Ecol Sociobiol. 65(11):2161–2174. [Google Scholar]
- Kuderna LFK, Tomlinson C, Hillier LW, Tran A, Fiddes IT, Armstrong J, Laayouni H, Gordon D, Huddleston J, Garcia Perez R, et al. 2017. A 3-way hybrid approach to generate a new high-quality chimpanzee reference genome (Pan_tro_3.0). Gigascience 6(11):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfumey L, Hamon M. 2000. Central 5-HT1A receptors: regional distribution and functional characteristics. Nucleic Med Biol. 27(5):429–435. [DOI] [PubMed] [Google Scholar]
- Lanzenberger RR, Mitterhauser M, Spindelegger C, Wadsak W, Klein N, Mien LK, Holik A, Attarbaschi T, Mossaheb N, Sacher J, et al. 2007. Reduced serotonin-1A receptor binding in social anxiety disorder. Biol Psychiatry. 61(9):1081–1089. [DOI] [PubMed] [Google Scholar]
- Larke RH, Maninger N, Ragen BJ, Mendoza SP, Bales KL. 2016. Serotonin 1A agonism decreases affiliative behavior in pair-bonded Titi monkeys. Horm Behav. 86:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo PM, Ghahremani MH, Morris SJ, Albert PR. 1997. A conserved threonine residue in the second intracellular loop of the 5-hydroxytryptamine 1A receptor directs signaling specificity. Mol Pharmacol. 52(1):164–171. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Merschdorf U. 2000. Impulsivity, aggression, and serotonin: a molecular psychobiological perspective. Behav Sci Law. 18(5):581–604. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Meyer J, Glatz K, Flügge G, Hinney A, Hebebrand J, Klauck SM, Poustka A, Poustka F, Bengel D, et al. 1997. The 5-HT transporter gene-linked polymorphic region (5-HTTLPR) in evolutionary perspective: alternative biallelic variation in rhesus monkeys. J Neural Transm. 104(11–12):1259–1266. [DOI] [PubMed] [Google Scholar]
- Lewis RJ. 2002. Beyond dominance: the importance of leverage. Q Rev Biol. 77(2):149–164. [DOI] [PubMed] [Google Scholar]
- Masson J, Emerit MB, Hamon M, Darmon M. 2012. Serotonergic signaling: multiple effectors and pleiotropic effects. Wiley Interdiscip Rev Membr Transp Signal. 1(6):685–713. [Google Scholar]
- Muller MN, Wrangham RW. 2004. Dominance, aggression and testosterone in wild chimpanzees: a test of the “challenge hypothesis.” Anim Behav. 67(1):113–123. [Google Scholar]
- Muller MN. 2002. . Agonistic relations among Kanyawara chimpanzees. In:Boesch C, Hohmann G, Marchant L, editors. Behavioural diversity in chimpanzees and bonobos. Cambridge: Cambridge University Press. p. 112–124. [Google Scholar]
- Murphy DL, Lerner A, Rudnick G, Lesch K. 2004. Serotonin transporter: gene, genetic disorders, and pharmacogenetics. Mol Interv. 4(2):109–123. [DOI] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8(7):e1002764.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Ueno S, Sano A, Tanabe H. 2000. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol Psychiatry. 5(1):32–38. [DOI] [PubMed] [Google Scholar]
- Newman TK, Syagailo YV, Barr CS, Wendland JR, Champoux M, Graessle M, Suomi SJ, Higley JD, Lesch KP. 2005. Monoamine oxidase a gene promoter variation and rearing experience influences aggressive behavior in rhesus monkeys. Biol Psychiatry. 57(2):167–172. [DOI] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B. 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30(6):884–886. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature 499(7459):471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A. 2008. From genes to brain to antisocial behavior. Curr Dir Psychol Sci. 17(5):323–328. [Google Scholar]
- Ranganathan P, Pramesh CS, Buyse M. 2016. Common pitfalls in statistical analysis: the perils of multiple testing. Perspect Clin Res. 7(2):106–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. 2018. The behavioral genetics of nonhuman primates: status and prospects. Am J Phys Anthropol. 165:23–36. [DOI] [PubMed] [Google Scholar]
- Rosati AG, Hare B. 2012. Decision making across social contexts: competition increases preferences for risk in chimpanzees and bonobos. Anim Behav. 84(4):869–879. [Google Scholar]
- Sabol SZ, Hu S, Hamer D. 1998. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 103(3):273–279. [DOI] [PubMed] [Google Scholar]
- Santangelo AM, Ito M, Shiba Y, Clarke HF, Schut EHS, Cockcroft G, Ferguson-Smith AC, Roberts AC. 2016. Novel primate model of serotonin transporter genetic polymorphisms associated with gene expression, anxiety and sensitivity to antidepressants. Neuropsychopharmacology 41(9):2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Burmeister M, Ghosh D. 2004. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet. 127B(1):85–89. [DOI] [PubMed] [Google Scholar]
- Shattuck MR, Satkoski-Trask J, Deinard A, Tito RY, Smith DG, Melnick DJ, Malhi RS. 2014. Patterns of genetic variation and the role of selection in HTR1A and HTR1B in macaques (Macaca). BMC Genet. 15:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JZ, Crockett MJ. 2013. How serotonin shapes moral judgment and behavior. Ann N Y Acad Sci. 1299:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk JB, Brosnan SF, Henrich J, Lambeth SP, Shapiro S. 2013. Chimpanzees share food for many reasons: the role of kinship, reciprocity, social bonds and harassment on food transfers. Anim Behav. 85(5):941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BL, Lu C-P, Alvarado Bremer JR. 2010. High-resolution melting analysis (HRMA): a highly sensitive inexpensive genotyping alternative for population studies. Mol Ecol Resour. 10(1):193–196. [DOI] [PubMed] [Google Scholar]
- Stimpson CD, Barger N, Taglialatela JP, Gendron-Fitzpatrick A, Hof PR, Hopkins WD, Sherwood CC. 2016. Differential serotonergic innervation of the amygdala in bonobos and chimpanzees. Soc Cogn Affect Neurosci. 11(3):413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltenberg SF, Christ CC, Carlo G. 2013. Afraid to help: social anxiety partially mediates the association between 5-HTTLPR triallelic genotype and prosocial behavior. Soc Neurosci. 8(5):400–406. [DOI] [PubMed] [Google Scholar]
- Strobel A, Gutknecht L, Rothe C, Reif A, Messner R, Zeng Y, Brocke B, Lesch K-P. 2003. Allelic variation in 5-HT 1A receptor expression is associated with anxiety- and depression-related personality traits. J Neural Transm. 110(12):1445–1453. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Tibi-Elhanany Y, Shamay-Tsoory SG. 2011. Social cognition in social anxiety: first evidence for increased empathic abilities. Isr J Psychiatry Relat Sci. 48(2):98–106. [PubMed] [Google Scholar]
- Tost H, Meyer-Lindenberg A. 2010. I fear for you: a role for serotonin in moral behavior. Proc Natl Acad Sci U S A. 107(40):17071–17072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M. 2003. 5-HT1A receptor knockout mouse as a genetic model of anxiety. Eur J Pharmacol. 463(1–3):177–184. [DOI] [PubMed] [Google Scholar]
- Trefilov A, Berard J, Krawczak M, Schmidtke J. 2000. Natal dispersal in rhesus macaques is related to serotonin transporter gene promoter variation. Behav Genet. 30(4):295–301. [DOI] [PubMed] [Google Scholar]
- Turner JH, Gelasco AK, Raymond JR. 2004. Calmodulin interacts with the third intracellular loop of the serotonin 5-hydroxytryptamine 1A receptor at two distinct sites: putative role in receptor phosphorylation by protein kinase C. J Biol Chem. 279(17):17027–17037. [DOI] [PubMed] [Google Scholar]
- Varnäs K, Halldin C, Hall H. 2004. Autoradiographic distribution of serotonin transporters and receptor subtypes in human brain. Hum Brain Mapp. 22(3):246–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R, Sreenivas S, Read J, Saunders KEA, Rogers RD. 2017. The role of serotonin in personality inference: tryptophan depletion impairs the identification of neuroticism in the face. Psychopharmacology 234(14):2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DP. 2004. Intracommunity coalitionary killing of an adult male chimpanzee at Ngogo, Kibale National Park, Uganda. Int J Primatol. 25(3):507–521. [Google Scholar]
- Wendland JR, Hampe M, Newman TK, Syagailo Y, Meyer J, Schempp W, Timme A, Suomi SJ, Lesch KP. 2006. Structural variation of the monoamine oxidase a gene promoter repeat polymorphism in nonhuman primates. Genes Brain Behav. 5(1):40–45. [DOI] [PubMed] [Google Scholar]
- Wilson ML, Boesch C, Fruth B, Furuichi T, Gilby IC, Hashimoto C, Hobaiter CL, Hohmann G, Itoh N, Koops K, et al. 2014. Lethal aggression in Pan is better explained by adaptive strategies than human impacts. Nature 513(7518):414–417. [DOI] [PubMed] [Google Scholar]
- Wilson ML, Wrangham R. 2003. Intergroup relations in chimpanzees. Ann Rev Anthropol. 32(1):363–392. [Google Scholar]
- Yamanashi Y, Matsuzawa T. 2010. Emotional consequences when chimpanzees (Pan troglodytes) face challenges: individual differences in self-directed behaviours during cognitive tasks. Anim Welfare. 19:25–30. [Google Scholar]
- Zhong P, Yuen EY, Yan Z. 2008. Modulation of neuronal excitability by serotonin-NMDA interactions in prefrontal cortex. Mol Cell Neurosci. 38(2):290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.