Abstract
Science funders are increasingly requiring evidence of the broader impacts of even basic research. Initiatives such as NIH’s CTSA program are designed to shift the research focus toward more translational research. However, tracking the effectiveness of such programs depends on developing indicators that can track the degree to which basic research is influencing clinical research. We propose a new bibliometric indicator, the TS score, that is relatively simple to calculate, can be implemented at scale, is easy to replicate, and has good reliability and validity properties. This indicator is broadly applicable in settings where the goal is to estimate the degree to which basic research is used in more applied downstream research, relative to use in basic research. The TS score should be of use for a variety of policy analysis and research evaluation purposes.
Keywords: translational research, indicators, citation analysis, research evaluation
Introduction
Every year, the U.S. National Institutes of Health spends about $40B to promote the NIH mission of “turning discovery into health”. Much of this money supports upstream basic research, with the goal of building up the scientific basis for the development of new drugs and medical treatments as well as other means of promoting health. However, there have been longstanding concerns about the challenge of translating this basic research into improved health outcomes (Zerhouni 2005; R. S. Williams et al. 2015). While there have been significant advances in basic biomedical knowledge, concerns have been raised that this knowledge – primarily generated in academic research labs – is not being integrated effectively into the clinical research system, potentially slowing the discovery and commercialization of new drugs and treatments.
Recognizing this challenge, countries across the globe have instituted programs that aim to facilitate the movement of promising basic research from the laboratory bench to the bedside (Butler 2008; Blümel 2017). In 2005, the US NIH announced the establishment of the Clinical & Translational Science Award (CTSA) program (Zerhouni 2005). As NIH director Zerhouni said when announcing the program, the goal is to “ensure that extraordinary scientific advances of the past decade will be rapidly captured, translated, and disseminated for the benefit of all Americans.” In 2010, the UK increased the budget of the Medical Research Council (MRC), with the aim of increasing support for translational research (Medical Research Councils UK 2017). This was a part of the MRC’s Translational Research Strategy that was announced in the same year (Lander and Atkinson-Grosjean 2011; Blümel 2017). Similarly, the £27 million Scottish Universities Life Sciences Alliance Assay Development Fund (SULSA), established by the Scottish Funding Council, has the aim of unlocking the translational potential of biology to address industry needs (McElroy et al. 2017). In 2013, the EU created the European Infrastructure for Translational Medicine (EATRIS), a network of European biomedical translation hubs incorporating over eighty academic research centers (EATRIS ERIC n.d.; Blümel 2017). Similar movements exist in Asian countries as well. In 2002 Japan established the Translational Research Informatics Center, later renamed the Translational Research Center for Medical Innovation, to facilitate the translation of findings from basic medical research to clinical practice (Translational Research Informatics Center 2003; Fukushima and Kimura n.d.). In Korea, the concept of translational research was introduced in the 2000s and the Ministry of Health and Welfare started supporting translational research in 2005 (S. Kim 2013). China’s National Center for Translational Medicine and several other institutions provide facilities and funding opportunities specific to translational research (S. Williams 2016).
The NIH CTSA program, as one example, supports a network of medical research institutions – called CTSA Hubs – that provide a variety of services in support of translational research, with significant flexibility across centers in the mix of services provide. These services include providing: shared research infrastructure, collaboration tools (such as web-based systems for data sharing or finding experts), education and training in translational research (such as developing a master’s degree in translational research, or providing training programs for clinical researchers), administrative support (such as streamlining IRB approvals or contract negotiations), as well supporting pilot programs to help researchers collect preliminary data, with different CTSA hubs putting greater or lesser emphasis on one or another of these services. Notably, the CTSA program allows significant flexibility in the design and implementation of their centers. This design flexibility and the resulting variation among CTSA hubs provides a valuable opportunity to expand our understanding of how specific aspects of these centers contribute to the goal of advancing translational research. Similarly, the introduction of such programs across the globe provides opportunities for cross-national studies comparing programs and outcomes. However, such evaluations are hampered by difficulties in developing useful measures for estimating impacts. While the ultimate goal is to have translational research programs (as a whole) improve overall health, policy researchers also need more proximate outcomes to see the impact of funding initiatives on the types of science being conducted (Myers forthcoming). Since the process of translating fundamental science into improved treatments and health outcomes often spans decades (R. S. Williams et al. 2015), it is important to also gather shorter-term indicators tracking the movement of basic research portfolios. For example, we may want to assess if basic research advances produced by a single lab or a larger group of researchers associated with a CTSA hub are used primarily by other basic researchers or by clinical researchers and observe how this balance between basic and clinical use changes over time or across programs with different components. Hence, although the goal of the CTSA and similar programs is converting fundamental biomedical research funding into improved health outcomes, scientific publications are often used as a more proximal indicator of the impact of the funding program (Smith et al. 2017; Luke et al. 2015; Hutchins et al. 2019).
The goal of this paper is to describe a newly developed measure of the degree to which a basic science publication is “translational”, defined as the extent to which it is used by downstream clinical research, relative to its use by basic research. The downstream clinical use of basic research is a critical first step in the translational process, providing a foundation for subsequent clinical research, with the ultimate goal being successful clinical research leading to new drugs and treatments that improve community health. The measure we propose has several advantages (cf. Trochim et al. 2011): it is designed to capture the short-term use of basic research in downstream clinical research, relative to its use in other basic research; its operationalization and interpretation are straightforward; it can be used stand alone or to complement other metrics of translational impact; and it can be generalized to other settings (e.g., to capture the use of basic research in physics or chemistry by engineering sciences) to estimate broader impacts of scientific findings in a variety of fields.
In the rest of the paper we discuss the concept of translational research and prior work on tracking translational research, describe the proposed measure and illustrate the estimation process by calculating the measure for a sample of papers funded by a CTSA hub and then provide several indicators for testing the reliability and validity of the measure.
The concept of translational research
Policymakers have argued that the remarkable advances in basic biomedical research have not led to significant increasese in new medicines and treatments, and many pointed to a “valley of death” as the core reason for this phenomenon. As a result, the concept of translational research attracted the interest of the biomedical research community, as a means to bridge the gap between basic science research and clinical science research (President’s Council of Advisors on Science and Technology 2012). Having started to gain popularity in the 1990s (Lander and Atkinson-Grosjean 2011), translational research has been promoted over the past couple of decades as a potential tool to speed up the process of moving basic science research innovations into clinical interventions and acceptance (Fishburn 2013; Stevenson et al. 2013).1
Translational research also helps connect the scientific research entreprise with societal needs. Translational research responds, in part, to demands that the scientific community justify the substantial contribution of public funds to research and respond to changing public concerns (Blümel 2017). Translational research can be seen as a response to this desire for societal impact from basic science and it aims to produce knowledge that benefits patients and ultimately society as a whole (Blümel 2017). The goal of translational research is to create research outcomes that are closely related to patient needs and translate discoveries in the laboratory into new therapies and improved community health (Fishburn 2013; Surkis et al. 2016). The concept and definition of translational research are provided by institutes such as NIH, National Academy of Medicine (formerly the Institute of Medicine (IOM)), and the Translational Research Working Group of the National Cancer Institute (NCI). They all share similar themes reflecting the goals of reducing the time for and increasing the efficiency of the transfer of fundamental scientific research into clinical research by bridging the gap between the bench and the bedside (Rubio et al. 2010; Han et al. 2018).
Scholars and practitioners sometimes characterize translational research as consisting of multiple stages. One approach divides translational research into two stages (Drolet and Lorenzi 2011; Han et al. 2018; Rubio et al. 2010; S. Kim 2013; National Library of Medicine 2017). The first stage (T1) involves transferring results from early-stage basic research into clinical research and the second stage (T2) captures the use of research outcomes from clinical studies into actual practice and diffusion into local communities (Rubio et al. 2010; Woolf 2008). In this paper, we will follow this approach, and to signify this two-part staging, we will designate the stages as 2ST1 (two stage time 1) and 2ST2 (two stage time 2). A second approach views translational research as a four-stage process: diagnosis or treatment (T1), evidence-based research (T2), clinical practice (T3) and verification for actual practice (T4) (Lander and Atkinson-Grosjean 2011; Weber 2013). A third approach, originally from reports from NCATS and Institute of Medicine and used by Surkis et al. (2016), breaks the process into five steps: basic biomedical research (T0), translation to humans (T1), translation to patients (T2), translation to practice (T3) and translation to communities (T4). Trochim et al. (2011) summarize these various stage models, highlighting that they share a common framework of tracing the process from basic science research to health practices, and also that these processes are not linear, but contain a variety of feedback loops. Here we are focusing on the stages related to research activities and the movement of information from basic to clinical research (e.g., 2ST1 in the two-stage approach). While the second stage (2ST2) is clearly critical for the ultimate improvement of community health, the bulk of the research funding in the area of translational research focuses on 2ST1 (Woolf 2008). And, consistent with Trochim et al.’s process marker model, we are developing an indicator that can provide a marker for the movement of knowledge from basic research to clinical research (Trochim et al. 2011). One advantage of Trochim et al’s process marker model is that it is operationally tractable, emphasizing the development of clearly operationalized markers capturing movement along a particular part of the process, as well as focusing on not just movement but also volume of flows.
As it generally takes many years to go from scientific discoveries to products in the market (Morris et al. 2011; Contopoulos-Ioannidis et al. 2008), more proximate measures of the translation process are needed. Patents are one potential measure that can serve as an indicator of successful translation, but the lag time for patents to issue along with uncertainty about which patents will be used commercially complicate use of this measure. On the other hand, it takes much less time for publications describing novel research to appear, with more than 80% of publications being published within six years from the year of the funding directly related to that publication (Ihli 2016). This makes publication-related measures attractive for the near-term evaluation of grants that support translational research. Although publications have less commercial value than patents and drugs, they can be used to understand the dissemination of research and their forward citation data can be used to understand their usage across academic disciplines (Llewellyn et al. 2019) and by firms (F Narin et al. 1997; Chen and Hicks 2004). Because of these desirable characteristics, several attempts have been made to develop an index measuring translational features of publications using bibliometric information (e.g., Weber 2013; Han et al. 2018; Surkis et al. 2016; Fontelo and Liu 2011). However, thus far, no measure has acquired broad consensus within the research community. Hence, we propose a novel index that measures translational features of publications, and which, we believe, has the potential to be used widely.
Tracking translational research
NIH has supported translational research through the CTSA award for over ten years (Weber, 2013). As these efforts are ongoing and represent a substantial investment, it is important to assess their effectiveness. Several studies have assessed various aspects of the CTSA program (e.g., Liu et al. 2013; Knapke et al. 2015; Schneider et al. 2017; Llewellyn et al. 2018; Y. H. Kim 2019). Some studies focus on research productivity and the scholarly impact of publications. Schneider et al. (2017), for example, used bibliometric tools to measure the impact of articles that CTSA hubs published. They found the six CTSA hubs they analyzed experienced an increase in publications and forward citations. Another example is Llewellyn et al. (2018), which looked at articles citing CTSA hub awards. They found publications supported by CTSA hubs are cited more than papers not from CTSA hubs that are matched on characteristics such as publication year, disciplines, etc. and this relationship strengthens when a publication is from a multi-institutional CTSA hub. Liu et al. (2013) find that the CTSA award had a positive impact on the number of patients enrolled in clinical trials. CTSA awards are also associated with the probability of receiving subsequent NIH grants (Knapke et al. 2015).
These and other studies examine important aspects of the CTSA program, but do not directly assess whether it pushed the research community to focus more on translational research. Fewer studies have addressed this question directly. One study is by Weber (2013), who used Medical Subject Headings (MeSH) to divide publications into three groups [publications on animals (group A), publications on cells (group C), and publications on humans (group H)], and tracked the location of groups of publication in the space spanned by these three poles over time. If a group of publications (e.g. those related to the topic “Cloning, Organism”) moved toward the H pole over time, this set of publications was classified as becoming more translational. Weber (2013) also introduced the concept of “generations” of translation lag. An article on animals or cells is classified as a first-generation article if it was cited directly by a group-H paper, as a second-generation article if it was cited by another animal or cell paper that was itself cited by a group-H paper and so on. For the citation data, Weber used PubMed Central (PMC). However, as he notes, PMC only includes a subset of the papers in PubMed and hence undercounts citations. To correct for this, Weber calculates a ‘corrected citation count’ by weighting the citations in PMC by the representativeness of PMC (as a share of PubMed) for each class of papers (Human, Animal, Cellular). Weber notes that while other databases, which may have broader coverage, are available, such as Web of Science, Scopus or Google, PMC has the advantage that it is freely available, and also linked directly to PubMed and to the MeSH terms. He also shows that the distributions across MeSH categories is similar for Web of Science and the corrected PMC data, suggesting that WoS and PMC would generate similar results. Han et al. (2018) provide another example using MeSH keywords for the classification of translational work. They classified a publication as “primary translational research” if the publication type was in one of the clinical science fields (Han et al., 2018, p. 5).2 The authors also introduced a category named “secondary translational research” and classified a paper into this group if a paper was not associated with clinical research but got citations from papers that deal with clinical issues (Han et al. 2018). This category of secondary translational research is similar to our TS score, discussed below. Using this approach, they showed that 13.4% of CTSA supported articles published in the field of behavioral and social science could be classified as (primary or secondary) translational research.
In some studies, methods not related to MeSH keywords were used for the classification of translational publications. For example, Fontelo and Liu (2011) introduced a web application filter that can be used to retrieve articles that have potential clinical applications. They created this filter by manually reviewing words and phrases that frequently appear in articles published in clinical and translational science journals. A study by Grant et al. (2000) looked at the citation links between publications and clinical guidelines. They assumed that publications cited by guidelines for clinical practice have an impact on the field of health. Based on the type of journal in which an article was published, they classified cited publications as 1) clinical observation, 2) basic, 3) clinical mix, or 4) clinical investigation. Their result showed that publications in the “basic” category are not often cited (only 8% were cited) by clinical guidelines. Similarly, Williams et al. (2015) traced the backward citation networks associated with two recent important advances in drug development: ipilimumab in oncology and ivacaftor for cystic fibrosis (R. S. Williams et al. 2015). They show that such networks are broad, spanning a large number of authors and institutions and many decades, suggesting that translation is a complicated process that requires aggregating multiple pathways of information flows. An alternative approach was developed by Surkis et al. (2016), who developed a checklist that can be used to manually categorize publications into a specific stage in the five-stage process of translational research (T0 to T4). Then, they used this manually labelled data as training data for a machine learning algorithm to categorize publications within these five stages. Due to the low frequency of T1, T2, T3 and T4 articles relative to T0 articles (in their schema), they combined T2 with T3 articles and T4 with T5 articles during the classification. The authors reported good performance and reliability in all groups of articles, with the machine learning models closely matching manual coding of articles from each of the three stages.
Recently, Hutchins et al. (2019) used machine learning to predict the future use of a research publications in clinical trials or guidelines. Their machine learning model incorporates 22 features such as MeSH terms in the focal paper (building from Weber, 2013), categorized into Human, Animal and Molecular/Cellular using fractional counts, plus additional MeSH terms representing Chemical/Drug, Disease, and Therapeutic/Diagnostic Approaches, as well as forward citations per year from Web of Science and data on the MeSH terms of the citing papers (max, mean, and standard deviations for each MeSH category). They developed a labelled training dataset (using forward citations in the 5–20 year post publication window, as the measures tend to stabilize by this point) to predict “cited by clinical articles or not”. They tested a variety of machine learning algorithms (logistic regression, Support Vector Machines, Random Forest, Neural Networks, Maxent, LibLinear) and found that the Random Forest method has the best predicting capabilities. They showed that their model predicted use in clinical research at least as well as expert judgements of whether the paper “could ultimately have a substantial positive impact on human health outcomes”, which suggests that such bibliometric measures are promising as early indicators of the translationalness of the research. They also found that even limiting the training data to only 2 years of post-publication information produced reasonably accurate predictions of a binary clinical research citation outcome. Finally, they found that among those articles that are cited in clinical research, the clinical papers citing higher predicted Approximate Potential to Translate (APT) score publications were more likely to have positive clinical outcomes and were more likely to move to the next stage in clinical development, suggesting that such bibliometric predictors of citation by clinical research may be a valid indicator for research with high translation potential. Hutchins et al. also note the need for more research that estimates how early citation patterns predict future use of research papers by clinical research. Fortunately, Hutchins et al. scaled their measure to the whole of PubMed and made the APT and related indicators available on their iCite web page (https://icite.od.nih.gov/analysis), which can become a valuable resource for scholars in this area. We will use this dataset to help validate our measure below.
Together these prior studies illustrate a growing interest in measuring and understanding the conduct of translational research. They show the research assessment community exploring a variety of different approach from labor-intensive small-scale studies to automated machine learning strategies and they highlight the challenges associated with developing measures that can be feasibly calculated and widely used among the research evaluation and science policy communities. Although a variety of approaches have been developed, there is no single consensus indicator to measure the translational feature of research articles (Blümel 2017; Surkis et al. 2016). Thus, when a researcher wants to conduct research on the change of translational features of research, they can either select an existing measure or develop a novel measure that fits the context of the planned research well (Trochim et al. 2011). To help offer a viable alternative, we propose a new measure called the TS score that we argue has a variety of strengths for capturing the translational nature of basic research.
A new measure: the TS score
To measure how translational a publication is, we propose a new index that is related to the share of forward citations from clinical science that a non-clinical article receives (cf. Han et al. 2018). Like Han et al.’s secondary translational research measure, we are using citations by clinical research papers as evidence of use of the basic research. However, rather than a binary (cited by a clinical journal or not), or a count variable (number of times cited in clinical journals), we use the share of citations by clinical journals. Using the share allows us to capture the relative emphasis of the knowledge in the paper in influencing clinical versus basic research. As we will show below, this method also has the advantage of generating a stable indicator that reflects the fundamental characteristics of the paper in terms of its relative value to clinical research, which has advantages for evaluation or prediction (Hutchins et al. 2019).
This measure aligns with the method of using disciplines of journals of the citing article to characterize features of the cited article (Qin et al. 1997; Grant et al. 2000). To construct the TS score, we categorized journals into four groups: clinical science (Clinical), non-clinical science (Non-Clinical), multidisciplinary (Multi), and non-science and engineering (Non-S&E). This categorization is done using Web of Science Categories (WOSC) and the NSF classification of fields of study (National Science Foundation 2018). In the Web of Science, each journal is assigned up to five WOSCs based on the topics covered by the journal. Each WOSC can be matched with the NSF classification of fields of study using name similarity. After the name matching, we classified the Web of Science category of journals into clinical science and non-clinical science (see Appendix A for the concordance). Specifically, when a field was listed in the science and engineering section (e.g., biomedical engineering) we classified it as non-clinical science, and when the field was listed in the health section (e.g., pediatrics), we classified it as clinical science. This is similar to Weber’s classification of articles into Human versus Cellular or Animal (see Weber 2013; Hutchins et al. 2019), although we use a binary rather than triangular classification. As a journal can have up to five WOSCs, a journal could have a mix of non-clinical science WOSCs and clinical science WOSCs. For the purpose of the TS score, we classified a journal as a clinical journal if the journal had only clinical WOSCs. The fields that were not listed under science and engineering or health were classified into either Multidisciplinary science (e.g., Science, Nature) or Other Science (e.g., social science), and these are also included in the denominator (see Equation (1)).
Following, for example, Hutchins, et al. (2019), we use Web of Science to gather citation data, in part because of the broad coverage and because of the journal field codes. Other citation data could also be used, such as Scopus, Google Scholar, or PubMed Central. Among Web of Science, Scopus and Google Scholar, prior work suggests that the correlations in citations are extremely high. Martín-Martín et al. (2018) collected the article level citation counts for a sample of over 2000 highly cited papers spanning 252 Google Scholar subject categories. Their data shows that the correlations among ln(1+x) citation counts across WoS, Scopus and Google Scholar range from .88 to .98 (based on a data table in the supplemental materials, calculations by authors). Field by field analyses reported in Martin-Martin, et al. (2018) show that these correlations are especially high (.98 or above) in fields such as Basic Life Sciences, Biomedical Sciences and Immunology/Microbiology, in other words, in the basic biomedical research fields used in our sample. PubMed Central could also be used as a source for citations, although it would require an adjustment to account for the lower coverage (Weber 2013). Weber notes that PMC has much more limited coverage, although it has the advantage of being freely downloadable. It is also linked to the NLM journal coding, which could be used to categorize citations for calculating the TS score. Hence, given the very high correlations in citation counts across different databases, we suspect that the TS score would be robust to the use of other sources of citation data (cf. Weber 2013). The main requirement is some field-coding of the journals (which WoS, Scopus, GS and PMC all provide).
To calculate the TS score, we begin with a corpus of publications (for example, all the publications from a given CTSA hub). Once we have the journal classification system, two steps are required to calculate the new measure. Figure 1 illustrates these steps.
Figure 1.
Classifying publications into the ones with and without translational feature based on journal discipline of forward citing publications.
Step one requires collecting from this corpus all the non-clinical publications, categorized as such by following these journal groupings. This process allows us to select from our corpus the set of papers published in non-clinical (basic) science journals (for example, Publication A in Figure 1). As the second step, we then collect and categorize all the forward citations to these non-clinical science journal publication. Each of those forward citations can be classified into coming from clinical (e.g., Publication B) or non-clinical journals (e.g., Publication C). Using this set of categorized citations, the Translational Science score (TS score) is calculated using the simple equation shown below.
| (1) |
Hence, for each publication, we calculate an individual TS score that captures the translationalness of the science in the focal paper.
Using the portion of cross-stage citations builds on the approach that Luke et al. (2015) used. In their study on the cross-disciplinary collaboration of an individual CTSA receiving institution, Luke et al. (2015) introduced the cross-disciplinary density ratio, a measure calculated by dividing the density of cross-disciplinary collaboration by the density of within-disciplinary ties. However, their focus was not on articles but on the composition of research teams. Our method is also related to Hutchins et al. (2019), who used the field coding of citing articles to help categorize focal papers as higher or lower on translationalness.
The intuition behind the TS score is that if an article published in a basic science journal is reporting on translational science, it is likely that it receives a larger portion of forward citations from clinical science journals. In other words, this research spans the 2ST1 stage (from basic to clinical research). One could use this measure at the paper, individual, research organization, or whole field level, to see how, for example, changes in funding might affect the degree of translational research in some population (cf. Myers forthcoming; Kim 2019). For example, Y. H. Kim (2019) uses this measure to estimate the impact of CTSA awards on the degree of translation among Carnegie R1 (doctoral--very high research activity) universities in the US.
Verification tests on the measure
When a new measure is developed, we need to assess how credible the measure is. In particular, we want to test its reliability and validity. We begin with a discussion of reliability, and then discuss several validity tests for the TS score.
Reliability tests
One way of estimating the reliability of a measure is to test its stability over time (Golafshani 2003). As the estimate is calculated at the paper level, it should not be vulnerable to various exogenous shocks that might change the direction of the researcher’s or university’s publication focus over time. In other words, while a particular researcher or institution might become more or less translational over time (for example, due to her institution receiving a CTSA award), an individual paper contains an inherent amount of translationalness, and that is what is being captured by the TS score. Hence, we checked whether the TS score remains stable once a minimal number of forward citations have been included in the estimate (to reduce the volatility due to sampling error) (Hutchins et al. 2019). We observe the accumulation of citations over time (each year) and examine the stability of the TS score. An alternative would be to track on a citation by citation basis as opposed to a yearly basis. However, this puts an additional burden on the data collection, and so we adopted the simpler approach described here. To estimate the stability (and the speed of convergence), we took the following steps to determine when the TS score stabilizes.
Check the TS score of each publication by year and observe how many years are needed for the TS score to stabilize, which we call the stabilizing year.
Calculate the change in the TS scores of each publication-group by year (i.e., N years) for the years after the stabilizing year (calculated in step 1), which we name TS scores after N-year.
Extract groups of papers with a certain number of forward citations in a reference starting year (i.e., the year in which it had acquired, for example, 1 through 10 citations) and calculate the TS scores of each publications-group. This group of TS scores will be named as TS scores at starting year.
Check the correlation of TS scores at starting year (acquired in step 3) and TS scores after N year (acquired in step 2). Using a correlation cutoff of 0.7 for inter-temporal stability, if the correlation is larger than 0.7 for all TS scores at starting year to TS scores after N-year pairs, we can consider the TS score stable from the starting year. While the cutoff of 0.7 is arbitrary, it is analogous to a commonly used threshold for reliability using Cronbach’s alpha (Lance et al. 2006). We also discuss the reliability using cutoffs of 0.8 and 0.9.
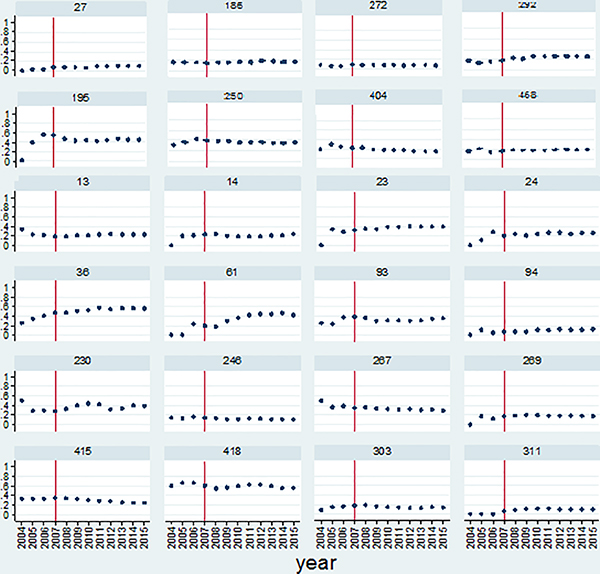
We illustrate the stability of the TS scores using a sample of 286 non-clinical publications that were published in 2003 and acknowledged NIH grants with a PI from Emory University. The cumulative counts of total forward citations and forward citations from the clinical science journals that occurred between 2004 and 2015 were used to calculate the TS scores. Figure 2 shows the variations in TS scores by year for 24 selected publications, to illustrate some of the observed patterns. These 24 publications are examples of those that received forward citations for almost all years (11 or more years) during the period of interest.3 Several patterns of change in the TS score over time are visible in Figure 2. Some publications have stable TS scores from the beginning until the end (e.g., ID 272, ID 415), some papers fluctuate in the early years (e.g., ID 24, ID 269), some publications start with very low TS scores but exhibit increasing TS scores as time passes (e.g., ID 36, ID 195) and some publications start with high TS scores in the early years and show decreases over time (e.g., ID 13, ID 267). In fact, one strength (but also possible weakness) of the TS score is that it can increase or decrease over time, which gives the possibility of using the indicator to estimate either the short-term or long-term impact of a paper on future clinical research (similar to Weber 2013). In contrast, binary measures (e.g., Han et al. 2018) cannot go back down once the paper has been cited by a clinical paper. However, despite this variation, we observe that, in almost all cases, the TS scores for these 2003 cohort publications are fairly stable after 2007, which is marked as the vertical line in each graph. These results suggest that, at least for this sample, the TS score of a publication tends to stabilize within four years after a paper is published. This means that we should wait at least four years to classify a basic science publication into one with high or low translational features. This can provide a guideline for researchers who want to use this indicator in their research. Further research is needed to assess how robust this 4-year threshold is across different populations of basic research papers. Furthermore, it might be interesting to estimate if this threshold varies systematically by the characteristics of the publications or researchers. For example, is it the case that stability is reached sooner (or later) for interdisciplinary papers, or those that span organizations, or those by larger teams, or papers published in high impact factor journals? Hence, there are many opportunities to develop this TS score in order to further calibrate the measure.
Figure 2.
Change in TS score by year for 24 non-clinical publications.
Table 1 shows the results of step 2 through step 4 of the reliability test, broken out by the number of citations the paper received in the reference starting year. By starting year, we mean the year by which the paper had accumulated N citations (with N varying from 1 to 10).4 If we look at the case of publications with only one forward citation in its starting year, 63 cases in our sample, the correlations of the TS scores at the starting point and four years and five years after the starting point are 0.730 and 0.721, respectively. However, the correlation between the starting point’s TS scores and six years after the starting point is 0.634, which is below the 0.7 threshold. Furthermore, the correlation value stays below 0.7 if we increase the post year to 7 or 8. Hence, having only one forward citation in the initial period does not provide a stable TS score, which is not surprising. This is also the case for the publications with two citations. The correlations are smaller than the 0.7 threshold for all starting point and post starting point pairs. For citations with three citations, the correlation values are all larger than 0.7, but for four citations, the correlation values are smaller than 0.7 in most pairs. By the time we get to papers with 5 initial citations, the reliability is consistently above the 0.7 threshold. This suggests that one could also use the reference starting year citations, with a floor of at least 5 citations, to get a fairly reliable initial indicator of future translationalness. In other words, although it, in general, takes four years for the indicator to stabilize (see Figure 2), once the paper has accumulated at least 5 citations (possibly in the first year), the TS score is fairly stable, which may allow use of the TS score even before four years of data have accumulated. For the TS score, even 3 or 4 initial citations seems to be sufficient for a moderately strong estimate of the future translationalness. Similarly, Hutchins et al. (2019) tested their model using data in a +5 to +20 year window, and found that collecting data from a +7 year window from the publication date allowed for the most reliable estimator of their Approximate Potential to Translate (APT) score. But, importantly, they also found that using only a +2 year window of citation data fed into the model (trained on the 5–20 year citation window) gave quite good predictive performance.
Table 1.
Correlations between the TS score of the starting point and the TS scores after four or more years, by number of citations in the starting year.
| Forward citation counts at starting year | Correlation between TS scores at the start and corresponding years |
Sample size | ||||
|---|---|---|---|---|---|---|
| After 4 years | After 5 years | After 6 years | After 7 years | After 8 years | ||
| 2 | 0.661 | 0.504 | 0.533 | 0.512 | 0.462 | 62 |
| 3 | 0.794 | 0.799 | 0.770 | 0.732 | 0.727 | 71 |
| 4 | 0.690 | 0.707 | 0.637 | 0.645 | 0.622 | 81 |
| 5 | 0.786 | 0.745 | 0.789 | 0.793 | 0.808 | 69 |
| 6 | 0.836 | 0.792 | 0.785 | 0.770 | 0.743 | 59 |
| 7 | 0.827 | 0.807 | 0.800 | 0.810 | 0.812 | 74 |
| 8 | 0.904 | 0.891 | 0.879 | 0.874 | 0.849 | 50 |
| 9 | 0.878 | 0.865 | 0.853 | 0.833 | 0.831 | 64 |
| 10 | 0.915 | 0.923 | 0.918 | 0.931 | 0.937 | 55 |
Hence, similar to other citation-based indicators of translationalness (e.g., Hutchins et al. 2019), the TS score seems quite reliable, even with only very modest initial information (5 initial citations). If a higher threshold of reliability is desired, 7 initial cites produces an estimate with correlations of 0.8 or above from +4 to +8 years, and 10 initial cites produces correlations above 0.9 (Lance et al. 2006). And, with a longer time series (4 years or more), the results seem quite stable. This suggests that this relatively simple score can be a reliable estimate of the relative use of basic research findings by clinical research publications.
Validity tests
In addition to establishing the reliability of a measure, it is also important to assess its validity (i.e. whether an indicator is measuring what it is intended to measure (Golafshani 2003)). In the case of the TS score, we used multiple approaches to evaluate the extent to which it captures the degree to which a basic research publication is being translated into later stage clinical research and/or practice. These include calculating an alternative department affiliation based translational score, and calculating the TS scores of publications associated with other indicators of translation (e.g. articles that resulted in a patent; were cited by patents; or led to the development of a new drug). We also compare the TS score to Hutchins et al.’s (2019) APT and to the percent of human MeSH terms in the focal publication (building from Weber 2013).
Publication classified based on the affiliation of the first author
The proposed TS score is based on the classification of the disciplines of journals (into clinical and non-clinical science journals). If an alternative forward-citation-based translation score has similar values (a high correlation with the original TS score), our confidence in the validity of the measure will increase. A variety of prior research has examined citations outside the category an article was published in, mainly for assessing the multidisciplinarity of articles (e.g., Morillo et al. 2001; Ortega and Antell 2006; Porter and Chubin 1985). One of the approaches adopted in these studies is using the first author’s affiliation to categorize papers into different disciplines (Ortega and Antell 2006). This approach relies on the long-standing norm in the life sciences that first authors of publications are likely to have the largest contribution to the paper (Carpenter et al. 2014, p. 1162).5 Drawing on the same set of articles, we calculated a new translational science score, which we label “dTS” [department-based Translational Science] score using the discipline classification of the first author’s affiliation (clinical versus non-clinical basic science). We then use this dTS to help assess the validity of our original TS score measure. For creating the dTS measure, we re-classified forward citing publications using the first author’s affiliation (rather than the journal’s subject code), such that those with lead authors in clinical science departments are classified as clinical science publications and classify forward citing articles as non-clinical publications otherwise. Figure 3 depicts the process of classifying publications based on the first author’s affiliation.
Figure 3.
The dTS score, classifying publications into those with and without translational feature based on citing papers’ first author’s affiliations.
This validity test involves the following steps:
Select a sample of publications for the analysis. We use all journal articles that were published in a non-clinical science journal in 2003 that acknowledge any NIH grant that has a PI from Emory University. This yields a sample of 114 articles (with between 10 and 42 citations).
Gather the bibliometric information of the articles that cited the publications in this sample. This yields a total of 2,089 forward citing articles.
Classify disciplines of forward citing publications based on the affiliation of the first author.6 There were approximately 2,000 unique affiliations, each of which was classified manually.
For each article, calculate the share of forward citing publications that have the first author’s affiliation classified as a clinical department (based on equation 1 above).
Several difficulties complicate the process of manually classifying author affiliations and, in general, we used classification rules that erred on the conservative side (i.e., not classifying as clinical unless there was clear information suggesting such a classification).7 This approach would tend toward lower values for the dTS scores. One could use other methods that were more liberal in classifying papers as translational on this dTS score. However, given that the address formatting details of different authors/institutions/journals citing the paper are likely unrelated to the degree of translationalness of the focal paper, any errors are unlikely to systematically affect the analysis. This difference between the efforts involved in the coding process in the dTS score versus the TS score highlights one advantage of the original TS score. For the TS score, the journal classifications already exist in the bibliometric databases, and these can be readily converted to clinical versus non-clinical fields, while department descriptions are not standardized and require time-consuming hand coding, or sophisticated computerized text processing methods to scale up the measure.
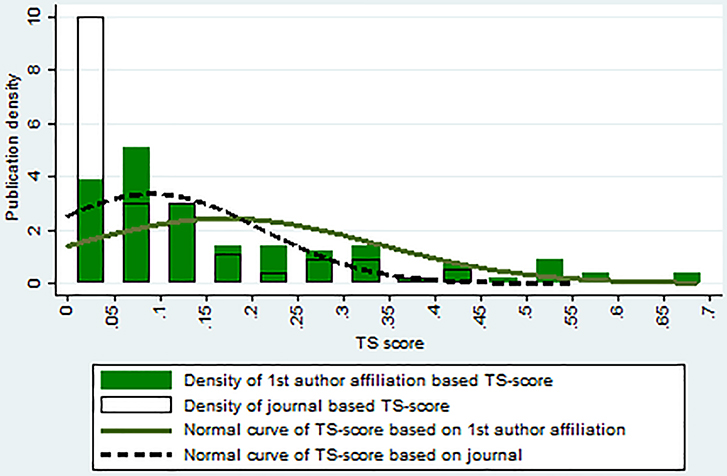
Table 2 compares summary statistics for two different approaches, one using the journal classification of forward citing articles (TS score) and the other using the affiliations of first authors in forward citing articles (dTS score). We can see that, even with the conservative classification rule, the value of the dTS score based on first author’s affiliation (first row) is larger than the TS score calculated based on journal classification (second row). A total of 81 out of 144 articles (71.1%) in the sample had larger translational science scores when the first author’s affiliation-based approach was used. Figure 4 shows the distribution of translational science scores calculated using the two different approaches. We can see that the dTS scores, calculated using the citing paper’s first author’s affiliation, have a larger mean value and are more dispersed. However, and most importantly for our purpose, the correlation between the two scores is 0.75, suggesting that these two indicators are closely related. The similarity between the scores calculated using these two different approaches provides some evidence of the validity of our TS score based on journal classification.
Table 2.
Descriptive statistics for and correlation between translationalness scores calculated with different methods.
| Approach of classifying discipline of forward citing article | Mean | Std Dev | Min | Max | Correlation with TS score based on journal classification |
|---|---|---|---|---|---|
| TS score: based on journal classification | 0.092 | 0.119 | 0 | 0.553 | ---- |
| dTS score: based on first author affiliation | 0.172 | 0.164 | 0 | 0.684 | 0.748 |
Note: The number of observations is 114 for both groups
Figure 4.
Distribution of translational science scores calculated using different methods.
TS scores of publications that result in patents
In addition to testing if the TS score is correlated with other ways of measuring the translationalness of specific publications, we can also calculate TS scores for papers that other evidence suggests are translational in nature. To the extent that these papers have high TS scores (or higher TS scores than other papers without this external evidence of translation), this would support the validity of the TS score. We focus initially on publications cited by one or more patents, because the citation of a journal article by a patent suggests that the publication is relevant to potentially commercializable inventions (F Narin et al. 1997; Roach and Cohen 2012; Meyer 2000). Indeed, unlike journal to journal citations, which serve a wide variety of purposes, patent to journal citations play a specific legal role, highlighting the importance of existing knowledge to a new invention. For this reason, a patent’s citation of a basic science publication may well represent direct knowledge-transfer from basic research to technological outcome (Meyer 2000; Huang et al. 2015; Sung et al. 2015). While patent examiners do add citations to patents, Cotropia et al. (2013) found that 94% of the non-patent literature (NPL) citations are added by the applicant. Hence, this analysis can serve as an additional measure of the validity of the TS score as an indicator of the eventual contribution to 2ST2 translation.
US patents list citations to NPL on their front pages and the citation link between publications and patents can be identified using the NPL list (Meyer 2000). Drawing on this evidence of patenting and translation of research, we can test the validity of the proposed TS score using publication-patent citation links, using both a prospective approach and a retrospective approach.
Prospective approach of publication – patent citation analysis
As a first step to test the validity of the TS score using a prospective approach to publication-patent citation analysis, we selected a group of basic science publications. As in the reliability test above, we used a set of 286 non-clinical articles published in 2003 that received support from NIH and had a PI affiliated with Emory University. We then searched the DOI of these 286 articles in LENS.ORG (http://www.lens.org).8 The Lens database returned information on 217 of the articles, a retrieval rate of 76%. Not all publication information is retrieved, as the Lens database does not cover all scholarly databases, but only covers three datasets (PubMed, Crossref and Microsoft Academics).9 These papers were then checked to see if any of these were cited by a patent or published patent application (for this discussion, we will say cited by a “patent” for simplicity).
Among the total sample of 217 publications, 144 publications (66%) did not receive any patent citation, while 73 publications (34%) had at least one citation from a patent. Following the reliability results above, for this analysis, we calculated the 2007 TS score, in other words 4 years after publication. The findings are nearly identical if we use a TS score calculated on 2019 data (16 years after publication). The average (2007) TS score of the publications with at least one patent citation (0.157) is higher than those without any patent citations (0.110). A two-tailed t-test also shows that the difference in mean TS scores is statistically significant (t=2.12, p=0.035). This result provides additional support for the validity of the TS score as a measure of the translational features of publications.
To further calibrate the TS score, we compare these results with the results from using Hutchins et al.’s (2019) APT, which is a prediction of the likelihood of being cited by clinical articles (taking binned values between 5% and 95%). We also tested a measure based on the share of human MeSH terms in the focal paper (building from Weber’s triangle of biomedicine).10 In the same way as for the TS score, we calculated the difference in APT and percent human MeSH terms for papers that were cited and not cited by patents for this same sample of 2003 publication year papers. The results show that the APT is also higher for papers that are eventually cited by patents (.256) than for those not cited by patents (.166), (t=2.88, p=.004). The correlation between the 2007 TS (4 year window) score and the APT is .40 (p<.001), suggesting that these indicators are related, but also that they capture somewhat distinct aspects of the paper’s translational characteristics. For the percent human MeSH terms, there is no difference between the patent cited and not patent cited publications (.098 versus .109, respectively, t=0.36, p=.72), suggesting that this measure does not capture this aspect of translation (being used by a patented invention). Table 3 shows probit regression models predicting citation by a patent for the three measures.11 The table shows that TS and APT each independently predicts patent citation, while percent human MeSH does not predict citation by a patent. We also tested (not shown) percent Animal and percent Molecular/Cellular and these also did not predict patent citation (either individually or in combination with Human). If both TS and APT are included, only the APT measure is statistically significant, likely due to the high collinearity between the two measures. However, we cannot reject the null hypothesis that the effects are equal between these two measures (chi-square=.09, p=.77). All of these results are very similar if we use a binary version of the TS score (Han et al. 2018), based on the 2007 (+4 years after publication) TS score (TS>0 v. TS=0) (results available from contact author).
Table 3:
Probit regression results for being cited by a patent, by TS, APT and Percent Human MeSH scores
| (1) | (2) | (3) | (4) | |
|---|---|---|---|---|
| PatentCite | PatentCite | PatentCite | PatentCite | |
| TS | 1.156** (0.555) | 0.645 (0.612) | ||
| APT | 1.082*** (0.391) | 0.903** (0.426) | ||
| Pct Human MeSH | −0.174 (0.465) | |||
| _cons | −0.574*** (0.115) | −0.643*** (0.120) | −0.404*** (0.100) | −0.691*** (0.129) |
| Obs. | 217 | 217 | 217 | 217 |
| Pseudo R2 | 0.016 | 0.028 | 0.001 | 0.032 |
Standard errors are in parenthesis
p<0.01
p<0.05
p<0.1
Hence, we find that the TS score is a significant predictor of the publication being cited in a patent document. We also find that it is significantly correlated with the recently developed Hutchins APT (based on a machine learning algorithm using a combination of forward citations, MeSH keywords in the focal paper and MeSH keywords in the citing papers). We also find that while each predicts being cited in a patent document, when we use both in the same model, only the APT is still statistically significant, although a Chi-square test shows that we cannot reject the null hypothesis that the effects for each measure are equal. These results show that the TS score can predict one dimension of translation, the research being used in a patent (F Narin et al. 1997). In addition, these analyses provide an additional validation for the Hutchins APT, showing that it not only predicts being cited in clinical literature, but that it also predicts being cited by a patent document. The percent human MeSH terms, however, does not predict being cited by a patent (and is also uncorrelated with TS score, r=.03). These results suggest that forward citation based measures (TS, APT) are reasonable indicators of future use of the publication by patented inventions. The percent human MeSH terms in the focal publication, however, is not a good predictor of this indicator of translationalness, although as Weber (2013) shows, it is useful for tracking trends in translationalness over generations of citations.
Retrospective approach of publication – patent citation analysis
For an additional validity test of the TS scores, we used a retrospective approach to examine publication-patent linkages. Here, we started with a group of patents and gathered the publications cited in those patents, and then checked to see if they had above average TS scores. To do this analysis, we gathered the following information:
Using the patent ID - NIH project ID link CSV file downloaded from NIH RePORTER, we collect the patents issued during 2001–2015 that acknowledge any NIH project with an Emory University affiliated PI (List 1).
Using PatentsView (2018)’s “otherreferences” file, we extracted bibliometric information (e.g., author, title, journal names) for “Non-patent citations” from List 1 patents (List 2).
Using this bibliometric information, we manually search Google Scholar and Web of Science to get their DOIs (List 3).
Find the List 3 publications that are from basic science journals, our population of interest (List 4).
Using Web of Science, collect the DOIs of publications citing the basic science papers in List 4 (List 5).
Calculate TS scores of focal publications in List 4 using the Web of Science Category classifications of List 5 citing publications.
The result from Step 1 produced a sample of 177 patents (List 1). This group of patents had a total of 186 non-patent citations (List 2), of which 88 were scholarly publications (List 3). Among 88 scholarly publications, 68 of them were published in basic science journals based on our classification rule using the Web of Science category and NSF Classification of fields of study (List 4) (see above). These 68 basic science publications received 29,503 forward citations (List 5), which were divided into clinical publications and non-clinical publications based on the Web of Science category and NIH classification of fields of study. Table 4 shows the summary of all basic science publications (sample size 68) and a subset of publications that received more than five forward citations, where we have more confidence in the reliability of the estimated TS score (sample size of 66). One problem with this test is that while we know the average TS score for this sample of papers cited by patents, we need some way of calibrating whether this is higher than a typical basic science paper. As a comparison, we calculate the average TS score for all basic science articles published during 2001 – 2015 with support from NIH grants that have PIs affiliated with one of the 115 Carnegie R1 universities. We can think of this as an estimate of the baseline translationalness of the overall population of NIH funded basic research. The mean values on the TS score for the patent-cited basic research publications (0.135 for all and 0.140 for publications with more than 5 cites) were larger than the average value for the whole group of publications, which was 0.117. However, we cannot reject the null hypothesis in a one-tailed test (p=.18 for all publications and p=.12 for those with more than 5 citations). Hence, we find additional evidence consistent with the claim that TS scores capture the translational character of publications, using a retrospective test, although we cannot rule out that the difference is due to chance.
Table 4.
Summary of TS scores of publications cited by Emory University NIH funded patents, 2001–2015.
| Group | Obs | Mean | Std Dev | Min | Max |
|---|---|---|---|---|---|
| All publications | 68 | 0.135 | 0.163 | 0 | 0.624 |
| Publications with more than 5 forward citations | 66 | 0.140 | 0.163 | 0 | 0.624 |
This test also gives information on the broader population of biomedical research. We see that among all Carnegie R1 universities, NIH funded basic research has a mean TS score of 0.117. In other words, at the base rate, about 10% of forward cites to NIH funded basic research conducted at major research universities are due to the research being cited in clinical research journals. This analysis also demonstrates that the TS score can be scaled up (covering not just one university in one year, but can be readily calculated for all research universities over a 15 year period). This also highlights the ready interpretability of the TS score, since it is simply the share of total citations that are from clinical research journals.
Several examples of the TS scores of publications that led to a drug
One strong indicator of whether a publication is translational is checking if the publication contributed to the development of a drug. Several scholars have focused on this characteristic of publications and tried to find articles that result in drugs (R. S. Williams et al. 2015; Sampat and Pincus 2015; Li et al. 2017). For instance, Sampat and Pincus (2015) used a machine learning technique to match publication, patents and drugs. They found about half of new molecular entities (NME) approved by the FDA between 2000 and 2009 are associated with publications from NIH-funded Academic Medical Center projects. Another study by Li et al. (2017) linked publications with patents associated with approved drugs. They found that approximately 5% of NIH grants result in publications that are cited by patents related to drugs in the market.
If the TS scores of basic science publications from the above-mentioned studies turn out to be high, it can be an additional sign that the measure is valid. Full lists of the publication – drug pairings were not included in the previous studies. However, the supplementary material of Sampat and Pincus (2015) included some examples of publications that led to newly developed drugs. In particular, they provided a list of 15 publication-patent-drug link examples. With this information, we calculated the TS scores of the four publications on the list that were published in non-clinical science journals based on our classification criteria. Table 5 shows the TS scores for these publications. We can see that, in general, the TS scores of these publications are high, with the largest value of 0.552. The average TS score of these four publications, which were translated into drugs, is 0.394. This value is much higher than the mean value of the total population of NIH funded basic research papers, which is 0.117. In fact, even with n=4, the difference is statistically significant in a one-tailed test (p=.03). Though the number of publications analyzed is very small, this finding provides additional support for the validity of our proposed TS score.
Table 5.
TS score of non-clinical science publications that resulted in drugs
| Brand | Drug | Cited Article | TS score |
|---|---|---|---|
| Tasigna | Nilotinib Hydrochloride Monohydrate | (Bhat et al. 1997) | 0.552 |
| Entereg | Alvimopan | (Bagnol et al. 1997) | 0.528 |
| Inomax | Nitric Oxide | (Garg and Hassid 1990) | 0.345 |
| Velcade | Bortezomib | (Rock et al. 1994) | 0.149 |
Note: Table recreated from a table in supplementary appendix of Sampat and Pincus (2015)
Conclusion
In recent years, encouraging the efficient movement of biomedical research “from bench to bedside” has been a priority of science funding agencies and policymakers. As a result, a variety of policy initiatives have been launched and a substantial investment has been made to increase the translational character of basic science. These efforts have occurred in an environment where there has also been increased interest in developing indicators that can track the broader impacts of basic research. While moving basic science to clinical practice is often a long and complex process (R. S. Williams et al. 2015), there is also a strong interest in focusing on the early steps in the process, when basic research is integrated into ongoing clinical research (2ST1 in the Weber (2013) classification). We focus on this meaning of translational science, recognizing that this is only an early stage in the process of improving overall clinical practice and community outcomes. To get a clearer lens on this process, we need a proximate measure that is simple to calculate; captures the idea of translational at the 2ST1 stage; and that is reliable and valid.
We propose a novel measure, the TS score, to assess the translationalness of a non-clinical publication and provide evidence that this measure meets these desired criteria. The measure is calculated using the share of clinical forward citations among all forward citations a non-clinical article receives. This measure is based on the assumption that if a non-clinical article is cited by an article in a clinical science journal, a knowledge flow from basic science to clinical science is taking place. The measure borrows concepts from and builds upon previous studies using journal discipline of forward citing articles (Qin et al. 1997), distinguishing basic science research and clinical science (Grant et al. 2000; F. Narin et al. 1976), and using cross-disciplinary density ratios (Luke et al. 2015; Porter and Chubin 1985).
The TS score has a variety of attractive features. First, the measure (percent of citations that are from clinical journals) is relatively simple and easy to understand, which may make it especially useful for policy discussions involving those who are not specialists in bibliometrics. Potential users only need to know the concept of forward citation and the classification of journals (clinical v. non-clinical basic science) based on fields of study. Second, the measure is relatively easy to implement (as it does not require any hand coding of documents nor calibrating and evaluating among machine learning algorithms). In this way it is similar to Weber’s triangle of biomedicine indicator (which is based on categorization of MeSH keywords associated with the focal document). Also, the calculation process for the TS score could be automated easily given the classification of journals into clinical and non-clinical groups. And, the classification rules can be easily described (e.g., appendix A in this article) allowing others to exactly replicate the scores, or to modify the measure for their own purposes. The TS score may also have low susceptibility to gaming, as it is a function of future citations (not any characteristic of the focal paper/author). As Hutchins et al. argue regarding their APT, another indicator that depends heavily on the information found in forward citations, since such indicators depend on the aggregated behavior of the scientific community, it is very difficult for the focal authors to significantly manipulate the final score (Hutchins et al. 2019). If gaming was a significant concern (for example if the TS score became an evaluation metric for individual researchers or research centers), the TS score could be further refined by excluding self-citations.
One limitation of the measure is that it depends on forward citations, a limitation shared by other forward citation-based measures such as Hutchins’ APT or Han’s secondary translational research measure. This results in some time lag in order to estimate the measure reliably (we estimate about 4 years from publication for the TS score). Furthermore, calculating the measure requires a citation database that allows the coding of journals by field. We used Web of Science, as this is commonly used in bibliometric analyses. However, Google Scholar or Scopus would likely give similar results, as the correlations among citation counts for each of these databases is extremely high (Martín-Martín et al. 2018). PubMed Central could also be used although the coverage is significantly less than these other citation databases (Weber 2013).
Trochim et al. (2011) argue for a process marker model for evaluating the rate and volume of translational research. They argue that such markers have the following advantages: they are pragmatic, conceptually clear, replicable, independent of debates of the scope of ‘translational’ research, and applicable both prospectively and retrospectively. Such markers also provide the basis for generating hypotheses on what characteristics change the values on the marker, as well as allowing for studying translational research while focusing on some rather than other particular markers. The TS score benefits from all of these strengths. Hence, the TS score we propose here follows the call for developing markers that can be used to examine the drivers of the rates and volumes of translation outcomes (Trochim et al. 2011).
Preliminary assessments presented suggest the measure is both reliable and valid. The reliability tests showed that if a publication receives only five or more forward citations, the TS score of the publication tends to reaches a stable value. This suggests that the score is capturing an inherent feature of the publication, the relative degree to which it is useful for clinical research (relative to basic research). Results from the validity tests that compared the TS score with a dTS score calculated using the citing publication’s first author’s departmental affiliation showed a correlation of approximately .75 between these two measures, with the TS score being much easier to calculate. Comparing the TS score to Hutchins’ APT shows a correlation of .40, with each of these predicting the probability of the publication being cited in a patent document, and, again, the TS score is substantially simpler. Similarly, the TS scores of publications cited in patents or leading to new drugs are higher than the general population of NIH-funded basic research publications (although the differences are not always statistically significant). The sample sizes in these validity tests are modest and in some of the tests are linked to a specific university. However, even with modest sample sizes, we can still see significant effects, which gives us some confidence in the measure. Still, further tests are needed to see how well the TS score performs under a variety of tests with other samples.
All in all, the proposed TS score has many advantages and this measure has the potential to be used widely for the evaluation of the translation of biomedical research. There are a variety of available measures for estimating the translationalness of basic research (e.g., Weber’s triangle of biomedicine, Hutchins’ APT, and Surkis’ checklist approach) and each has different strengths. The choice may depend partially on available data and on the purpose of the indicator. In addition, multiple indicators can be used to test the robustness of research findings.
One promising implication of the TS score is that an analogous indicator can be calculated for other fields (since it does not use field-specific classifiers such as MeSH terms), after categorizing the relevant journals into basic and more applied. Hence, the TS score can be used to estimate the broader impacts for a wide variety of areas of science. For example, this may be used for evaluating broader impacts for NSF-funded projects.
We find that, overall, the TS score for NIH-funded basic research papers produced at Carnegie R1 universities is about 0.10, suggesting a base rate of about 10% of forward citations from clinical research. Also, as the TS score is calculated at the article level, it can be readily aggregated to calculate individual, organizational or even field-level indicators of translationalness. This opens the opportunity for using the variations in this indicator to show, for example, if translationalness is changing over time or, another example, if junior people or senior people are more likely to engage in translational science. Using this approach, Y. H. Kim (2019) shows that publications from CTSA hubs have higher translational characteristics. We hope that others find a variety of uses for the indicator in order to develop our understandings of the process of translational research in biomedical sciences, as well as in other fields of science.
Supplementary Material
Acknowledgements
The authors would like to thank Nicole Llewellyn for her help in providing detailed information on the CTSA program, as well as help in data collection and data coding. The authors also thank Alan Porter and Weihua An for helpful suggestions. The research was supported by NSF award numbers SMA-1646689; DRL-1150114 and EEC-1648035, as well as the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR000454. The content is solely the responsibility of the authors and does not represent the official views of the U.S. National Institutes of Health, the U.S. National Science Foundation or the Ministry of Science and ICT, Republic of Korea. An earlier version of this work appeared in Kim (2019).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
One specific example of translational research is a study by Bhat et al. (1997) that found binding of fusion genes (BCR-ABL) and certain type of protein (c-CBL) only occurs when phosphate (PO43−) is added to acid (amino tyrosine) on a protein. Their finding was applied in treating genetic abnormality in chromosome 22 of leukemia cancer cell and led to the invention of Tasiga® (Sampat and Pincus, 2015). Another example of translational research is a study by Garg and Hassid (1990). They found that proliferation of cell lines developed from disaggregated mouse embryos (BALB/c 3T3) are more active when muscle smoother is not present (CGMP-independent mechanism). Their finding was applied in solving respiratory failure problem and this led to the invention of INOmax® (Sampat and Pincus, 2015).
Examples include clinical study, clinical trial, phase 1 clinical trial, phase 2 clinical trial, phase 3 clinical trial, phase 4 clinical trial, controlled clinical trial, practice guideline, observational study and randomized controlled trial.
More examples are available on request.
In other words, if the paper was published in 2003, and had 3 cites in the first year (2004), it would appear in the 3 cite row, and the correlations are for the scores for the years 4+ in the future from 2004. If in the following year, it had reached 8 citations, then it would reappear in the 8 cite row, with a starting year of 2005, and the columns representing 4+ years from 2005. This means that the same paper can appear in multiple rows (but not generally every row), and also that a paper can drop out of the table if it acquires a total of 11+ cites. Hence, the Ns can fluctuate up and down across the rows.
To the extent that this is not true, then our measure will include greater measurement error, biasing the correlations toward zero and hence giving us a more conservative test of the validity of our TS score.
For simplicity, we ignore additional “co-first authors”. We do not expect there are a large share of such cases, and, furthermore, if the co-first authors are in the same department, or even in the same class of departments (clinical v. non-clinical), the results would be unchanged if we included them.
There were some issues to consider when classifying publications into clinical papers and non-clinical papers, which led to doing the classification manually. First, the department information did not exist in some listings (e.g., Univ St Andrews, St Andrews KY 16 9ST, Fife, Scotland; RAND Corp, Santa Monica, CA 90401 USA). In these cases, these were coded as missing. Second, the order that university name, department or school name and city are listed were different between articles. Though the department information was listed second in most cases, it was listed first in some cases, later in other cases. Third, the details of the affiliation were different across publications. For instance, some publications provided very detailed information (e.g., Sch Med & Dent, Dept Biostat & Computat Bio) whereas some publications only provided information at the school level (e.g., Sch Life Sci & Technol). To be conservative on classifying a publication into clinical papers, we classified the publications into a clinical article only if there was clear and sufficient information on the discipline. For instance, we did not classify a publication as clinical if the most detailed information of the first author’s affiliation is “School of Medicine”. This is because, in many universities, the School of Medicine is composed of departments conducting basic science work (e.g., Department of Microbiology) as well as clinical work. Therefore, we classified a publication as a clinical paper only if the sub-division of the school is listed and it is closely related to fields of clinical research (e.g., Department of Pediatrics).
LENS.ORG is an open public website managed by Cambia, an independent non-profit organization, that provides linkages between scholarly works, patents and biological sequences (LENS.ORG, n.d.). Its patent database covers patent datasets from the USPTO, European Patent Office, WIPO and IP Australia and its scholarly dataset includes PubMed, Crossref and Microsoft Academics (LENS.ORG, n.d.). With the collaboration with NIH Pubmed and Crossref teams, the Lens links publications’ Digital Object Identifier (DOI) with NPL in their patent database. Hence, using the DOI of publications, we could check if the publications were cited by patents or not.
Using Pubmed ID instead of DOI for the search may give a higher retrieval rate. However, it would then be more difficult to track the forward citation links of publications with only Pubmed ID but without the DOI.
Both of these measures are available from the iCite website (https://icite.od.nih.gov/analysis). It is not clear what data window is used for calculating the APT scores published on the website. The data were downloaded June 18, 2020.
The inferences from a linear probability model are the same (results available from contact author).
Contributor Information
Yeon Hak KIM, Ministry of Science and ICT, Sejong, Republic of Korea.
Aaron D. LEVINE, School of Public Policy, Georgia Institute of Technology, Atlanta, GA USA
Eric J. NEHL, Rollins School of Public Health, Emory University, Atlanta, GA USA
John P. WALSH, School of Public Policy, Georgia Institute of Technology, Atlanta, GA USA)
References
- Bagnol D, Mansour A, Akil H, & Watson SJ (1997). Cellular localization and distribution of the cloned mu and kappa opioid receptors in rat gastrointestinal tract. Neuroscience, 81(2), 579–591. [DOI] [PubMed] [Google Scholar]
- Bhat A, Kolibaba K, Oda T, Ohno-Jones S, Heaney C, & Druker BJ (1997). Interactions of CBL with BCR-ABL and CRKL in BCR-ABL-transformed myeloid cells. Journal of Biological Chemistry, 272(26), 16170–16175. [DOI] [PubMed] [Google Scholar]
- Blümel C (2017). Translational research in the science policy debate: a comparative analysis of documents. Science and Public Policy, 45(1), 24–35. [Google Scholar]
- Butler D (2008). Translational research: Crossing the valley of death. Nature, 453, 840–842, doi: 10.1038/453840a. [DOI] [PubMed] [Google Scholar]
- Carpenter CR, Cone DC, & Sarli CC (2014). Using publication metrics to highlight academic productivity and research impact. Academic Emergency Medicine, 21(10), 1160–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, & Hicks D (2004). Tracing knowledge diffusion. Scientometrics, 59(2), 199–211, doi: 10.1023/B:SCIE.0000018528.59913.48. [DOI] [Google Scholar]
- Contopoulos-Ioannidis DG, Alexiou GA, Gouvias TC, & Ioannidis JP (2008). Life cycle of translational research for medical interventions. Science, 321(5894), 1298–1299. [DOI] [PubMed] [Google Scholar]
- Cotropia CA, Lemley MA, & Sampat B (2013). Do applicant patent citations matter? Research Policy, 42(4), 844–854, doi: 10.1016/j.respol.2013.01.003. [DOI] [Google Scholar]
- Drolet BC, & Lorenzi NM (2011). Translational research: understanding the continuum from bench to bedside. Translational Research, 157(1), 1–5. [DOI] [PubMed] [Google Scholar]
- EATRIS ERIC (n.d.). About EATRIS European Infrastructure for translational medicine. http://eatris.eu/about/.
- Fishburn CS (2013). Translational research: the changing landscape of drug discovery. Drug discovery today, 18(9), 487–494. [DOI] [PubMed] [Google Scholar]
- Fontelo P, & Liu F (2011). Finding translational science publications in MEDLINE/PubMed with translational science filters. Clinical and translational Science, 4(6), 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima M, & Kimura Y (n.d.). Translational Research Informatics Center (TRI) takes the initiative to reduce disability through Japanese academic research. http://s3-service-broker-live-19ea8b98-4d41-4cb4-be4c-d68f4963b7dd.s3.amazonaws.com/uploads/ckeditor/attachments/8326/TRi.pdf.
- Garg UC, & Hassid A (1990). Nitric oxide-generating vasodilators inhibit mitogenesis and proliferation of BALBc 3T3 fibroblasts by a cyclic GMP-independent mechanism. Biochemical and Biophysical Research Communications, 171(1), 474–479. [DOI] [PubMed] [Google Scholar]
- Golafshani N (2003). Understanding reliability and validity in qualitative research. The Qualitative Report, 8(4), 597–606. [Google Scholar]
- Grant J, Cottrell R, Cluzeau F, & Fawcett G (2000). Evaluating “payback” on biomedical research from papers cited in clinical guidelines: applied bibliometric study. BMJ, 320(7242), 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Williams SR, & Zuckerman BL (2018). A snapshot of translational research funded by the National Institutes of Health (NIH): A case study using behavioral and social science research awards and Clinical and Translational Science Awards funded publications. PLoS ONE, 13(5), e0196545, doi: 10.1371/journal.pone.0196545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MH, Yang HW, & Chen DZ (2015). Increasing science and technology linkage in fuel cells: A cross citation analysis of papers and patents. Journal of Informetrics, 9(2), 237–249. [Google Scholar]
- Hutchins B, Davis M, Meseroll R, & Santangelo G (2019). Predicting translational progress in biomedical research. PLoS Biology, 17(10), e3000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihli M. I. R. f. (2016). Acknowledgement Lag and Impact: Domain Differences in Published Research Supported by the National Science Foundation (master’s thesis). University of Tennessee-Knoxville, Knoxville, TN. [Google Scholar]
- Kim S (2013). The concept of translational research and the requirements for the success (in Korean). STEPI Insight, 15(115), 1–25. [Google Scholar]
- Kim YH (2019). Bridging the Valley of Death in Biomedicine with Translational Research: Assessing the Impact of National Institutes of Health’s Clinical and Translational Science Award. Georgia Institute of Technology, Atlanta, GA. [Google Scholar]
- Knapke JM, Haynes EN, Kuhnell P, & Tsevat J (2015). NIH grant awards as a metric of clinical and translational research training effectiveness. Clinical and translational Science, 8(1), 52–56, doi: 10.1111/cts.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance CE, Butts MM, & Michels LC (2006). The Sources of Four Commonly Reported Cutoff Criteria: What Did They Really Say? Organizational Research Methods, 9(2), 202–220, doi: 10.1177/1094428105284919. [DOI] [Google Scholar]
- Lander B, & Atkinson-Grosjean J (2011). Translational science and the hidden research system in universities and academic hospitals: A case study. Social Science & Medicine, 72(4), 537–544. [DOI] [PubMed] [Google Scholar]
- Li D, Azoulay P, & Sampat BN (2017). The applied value of public investments in biomedical research. Science, 356(63333), 78–81. [DOI] [PubMed] [Google Scholar]
- Liu G, Chen G, Sinoway LI, & Berg A (2013). Assessing the impact of the NIH CTSA program on institutionally sponsored clinical trials. Clinical and translational Science, 6(3), 196–200, doi: 10.1111/cts.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn N, Carter DR, DiazGranados D, Pelfrey C, Rollins L, & Nehl EJ (2019). Scope, Influence, and Interdisciplinary Collaboration: The Publication Portfolio of the NIH Clinical and Translational Science Awards (CTSA) Program From 2006 Through 2017. Evaluation & the Health Professions, 0163278719839435, doi: 10.1177/0163278719839435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn N, Carter DR, Rollins L, & Nehl EJ (2018). Charting the Publication and Citation Impact of the NIH Clinical and Translational Science Awards (CTSA) Program From 2006 Through 2016. Academic medicine : journal of the Association of American Medical Colleges, 93(8), 1162–1170, doi: 10.1097/ACM.0000000000002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke DA, Carothers BJ, Dhand A, Bell RA, Moreland-Russell S, Sarli CC, et al. (2015). Breaking down silos: Mapping growth of cross-disciplinary collaboration in a translational science initiative. [Article]. Clinical and translational Science, 8(2), 143–149, doi: 10.1111/cts.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Martín A, Orduna-Malea E, Thelwall M, & Delgado López-Cózar E (2018). Google Scholar and Scopus: A systematic comparison of citations in 252 subject categories. Journal of Informetrics, 12(4), 1160–1177, doi: 10.1016/j.joi.2018.09.002. [DOI] [Google Scholar]
- McElroy SP, Jones PS, & Barrault DV (2017). The SULSA Assay Development Fund: accelerating translation of new biology from academia to pharma. Drug discovery today, 22(2), 199–203. [DOI] [PubMed] [Google Scholar]
- Medical Research Councils UK (2017). Translational research. https://www.mrc.ac.uk/funding/science-areas/translation/.
- Meyer M (2000). Does science push technology? Patents citing scientific literature. Research Policy, 29(3), 409–434. [Google Scholar]
- Morillo F, Bordons M, & Gómez I (2001). An approach to interdisciplinarity through bibliometric indicators. Scientometrics, 51(1), 203–222. [Google Scholar]
- Morris ZS, Wooding S, & Grant J (2011). The answer is 17 years, what is the question: understanding time lags in translational research. Journal of the Royal Society of Medicine, 104(12), 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K (forthcoming). The Elasticity of Science. American Economic Journal: Applied Economics. [Google Scholar]
- Narin F, Hamilton K, & Olivastro D (1997). The increasing linkage between U.S. technology and public science. Research Policy, 26(3), 317–330. [Google Scholar]
- Narin F, Pinski G, & Gee HH (1976). Structure of the biomedical literature. Journal of the American Society for Information Science, 27(1), 25–45. [Google Scholar]
- National Library of Medicine (2017). Translation Medical Research MeSH Descriptor Data 2017. https://meshb.nlm.nih.gov/record/ui?name=Translational%20Medical%20Research.
- National Science Foundation (2018). Classification of field of study. https://www.nsf.gov/statistics/nsf13327/pdf/tabb1.pdf.
- Ortega L, & Antell K (2006). Tracking cross-disciplinary information use by author affiliation: Demonstration of a method. College & Research Libraries, 67(5), 446–462. [Google Scholar]
- Porter A, & Chubin D (1985). An indicator of cross-disciplinary research. Scientometrics, 8(3–4), 161–176. [Google Scholar]
- President’s Council of Advisors on Science and Technology (2012). Executive Summary Report to the President on Propelling Innovation in Drug Discovery, Development and Evaluation. https://www.brookings.edu/wp-content/uploads/2012/11/PCAST-Executive-Summary.pdf.
- Qin J, Lancaster FW, & Allen B (1997). Types and levels of collaboration in interdisciplinary research in the sciences. Journal of the American Society for Information Science, 48(10), 893–916. [Google Scholar]
- Roach M, & Cohen WM (2012). Lens or Prism? Patent Citations as a Measure of Knowledge Flows from Public Research. Management Science, 59(2), 504–525, doi: 10.1287/mnsc.1120.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell, 78(5), 761–771, doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Rubio DM, Schoenbaum EE, Lee LS, Schteingart DE, Marantz PR, Anderson KE, et al. (2010). Defining translational research: implications for training. Academic medicine : journal of the Association of American Medical Colleges, 85(3), 470–475, doi: 10.1097/ACM.0b013e3181ccd618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampat BN, & Pincus HA (2015). Citations in life science patents to publicly funded research at academic medical centers. Clinical and translational Science, 8(6), 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Kane CM, Rainwater J, Guerrero L, Tong G, Desai SR, et al. (2017). Feasibility of common bibliometrics in evaluating translational science. Journal of Clinical and Translational Science, 1(1), 45–52, doi: 10.1017/cts.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Baveja R, Grieb T, & Mashour GA (2017). Toward a science of translational science. Journal of Clinical and Translational Science, 1(4), 253–255, doi: 10.1017/cts.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson DK, Shaw GM, Wise PH, Norton ME, Druzin ML, Valantine HA, et al. (2013). Transdisciplinary translational science and the case of preterm birth. Journal of Perinatology, 33(4), 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HY, Wang CC, Huang MH, & Chen DZ (2015). Measuring science-based science linkage and non-science-based linkage of patents through non-patent references. Journal of Informetrics, 9(3), 488–498. [Google Scholar]
- Surkis A, Hogle JA, DiazGranados D, Hunt JD, Mazmanian PE, Connors E, et al. (2016). Classifying publications from the clinical and translational science award program along the translational research spectrum: a machine learning approach. Journal of translational medicine, 14(1), 235–235, doi: 10.1186/s12967-016-0992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Translational Research Informatics Center (2003). https://www.tri-kobe.org/english/index.html.
- Trochim W, Kane C, Graham MJ, & Pincus HA (2011). Evaluating translational research: a process marker model. Clinical and translational Science, 4(3), 153–162, doi: 10.1111/j.1752-8062.2011.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GM (2013). Identifying translational science within the triangle of biomedicine. Journal of translational medicine, 11(1), 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Lotia S, Holloway AK, & Pico AR (2015). From Scientific Discovery to Cures: Bright Stars within a Galaxy. Cell, 163(1), 21–23, doi: 10.1016/j.cell.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Williams S (2016). Building a translational medicine powerhouse in China. Science, 352(6293), 1596. [Google Scholar]
- Woolf SH (2008). The Meaning of Translational Research and Why It Matters. JAMA, 299(2), 211–213, doi: 10.1001/jama.2007.26. [DOI] [PubMed] [Google Scholar]
- Zerhouni EA (2005). Translational and Clinical Science — Time for a New Vision. New England Journal of Medicine, 353(15), 1621–1623, doi: 10.1056/NEJMsb053723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.