Abstract
Angelman syndrome (AS) is a rare neurodevelopmental disorder caused by mutation or deletion of the maternally-inherited UBE3A allele. These pathogenic mutations lead to loss of maternal UBE3A expression in neurons. Antisense oligonucleotides and gene therapies are in development that activate the intact but epigenetically silenced paternal UBE3A allele. Preclinical studies indicate that treating during the prenatal period could greatly reduce the severity of symptoms or prevent AS from developing. Genetic tests can detect the chromosome 15q11-q13 deletion that is the most common cause of AS. New, highly sensitive non-invasive prenatal tests that take advantage of single cell genome sequencing technologies are expected to enter the clinic in the coming years and make early genetic diagnosis of AS more common. Efforts are needed to identify fetuses and newborns with maternal 15q11-q13 deletions and to phenotype these babies relative to neurotypical controls. Clinical and parent observations suggest AS symptoms are detectable in infants, including reports of problems with feeding and motor function. Quantitative phenotypes in the 0-1 year age range will permit a more rapid assessment of efficacy when future treatments are administered prenatally or shortly after birth. While prenatal therapies are currently not available for AS, prenatal testing combined with prenatal treatment has the potential to revolutionize how clinicians detect and treat babies before they are symptomatic. This pioneering prenatal treatment path for AS will lay the foundation for treating other syndromic neurodevelopmental disorders.
Lay summary
Prenatal treatment could benefit expectant parents whose babies test positive for the chromosome microdeletion that causes Angelman syndrome (AS). Prenatal treatment is predicted to have better outcomes than treating after symptoms develop, and may even prevent AS altogether. This approach could generally be applied to the treatment of other syndromic neurodevelopmental disorders.
1. Overview
Angelman syndrome (AS) is characterized by developmental delays, severe intellectual disabilities, lack of speech, seizures, problems with movement and balance, and microcephaly (Pelc, Cheron, & Dan, 2008; Williams et al., 2006). Many individuals with AS also meet the diagnostic criteria for autism (Peters, Beaudet, Madduri, & Bacino, 2004). With a prevalence of 1:12,000 to 1:24,000 and a need for constant care across a full lifespan, the family burden and health care costs are high, and likely exceed the now dated $2.4M lifetime estimate for an individual with autism and intellectual disability (Buescher, Cidav, Knapp, & Mandell, 2014).
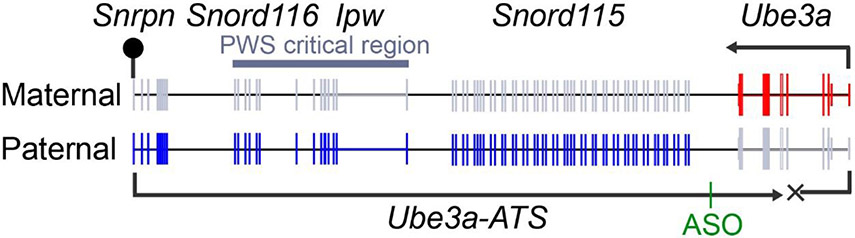
UBE3A is expressed biallelically in nearly all cells of the body, with the exception of neurons, where UBE3A is expressed only from the maternally-inherited allele (Fig. 1). In most cases, AS is caused by deletion or mutation of the maternally-inherited UBE3A allele (LaSalle, Reiter, & Chamberlain, 2015; Mabb, Judson, Zylka, & Philpot, 2011; Matsuura et al., 1997). This biology explains why loss of the maternal allele causes AS and impairs brain function. The paternal allele is silenced by UBE3A-ATS, an extremely long antisense transcript that represses the paternal UBE3A transcript in cis (Fig. 1) (Chamberlain & Brannan, 2001; Landers et al., 2004; Meng, Person, & Beaudet, 2012).
Figure 1.
In neurons, Ube3a-ATS transcriptionally blocks paternal Ube3a in cis. Grey color denotes genes that are repressed. Location of Meng et al. ASO shown. Lollipop = methylated/imprinted region.
2. Genetic tests for AS
All of the mutation types that cause AS can be detected with routine genetic tests and provide a definitive diagnosis (Committee on, 2018; Dagli, Mueller, & Williams, 1993). Deletion of the maternally inherited chromosome 15q11-q13 region is the most common cause of AS, is present in ~70% of cases, and is amenable to treatments that are in development (see below). This deletion typically occurs de novo and is identified using array comparative genome hybridization and a methylation test to determine if the deletion is on the maternal or paternal chromosome. Genetic tests can also be performed prenatally in conjunction with amniocentesis or chorionic villus sampling (CVS).
Non-invasive prenatal tests (NIPTs) can detect fetal sex and chromosomal abnormalities as early as ten weeks post conception (Vora & Wapner, 2018). Currently available “cell-free” NIPTs entail isolation and sequencing of fetal-derived DNA that is circulating in the mother’s blood. This simplicity, low cost, and accuracy at providing useful genetic information about the fetus has promoted increasing adoption of this test by pregnant women irrespective of maternal age (Grace, Hardisty, Dotters-Katz, Vora, & Kuller, 2016; Larion et al., 2014). Cell-free NIPTs can also detect the 15q11-q13 deletion that causes AS (when the deletion is maternally inherited) or Prader-Willi syndrome (PWS; when the deletion is paternally inherited) (Liang et al., 2019; Wapner et al., 2015). However, the positive predictive value is not high enough for experts to recommend cell-free NIPT for 15q11-q13 deletion testing (Petersen et al., 2017; Vora & Wapner, 2018). Expectant mothers can “opt-in” to get 15q11-q13 deletion NIPT results, with an understanding that an invasive diagnostic procedure like CVS will be needed to confirm the diagnosis.
The increasing use of cell-free NIPTs by patients and caregivers will invariably drive rapid adoption of next generation “cell-based” NIPTs that show specificity and accuracy values that are similar to invasive procedures (Kolvraa et al., 2016; Vossaert et al., 2018). It has been known for over a decade that nucleated fetal-derived cells circulate in the mother’s blood, but only recently has it been possible to reliably identify and isolate these cells (Christensen et al., 2005). Cell-based NIPTs utilize cutting-edge single cell sequencing technologies to identify mutations in these rare circulating fetal-derived cells. It is now possible to detect deletions, duplications, DNA methylation, and single nucleotide variants using single cell DNA sequencing (Hui et al., 2018; McConnell et al., 2017). Thus, it is technically possible to examine portions of the fetal genome and epigenome at single nucleotide resolution using a non-invasive procedure. Widespread adoption of cell-based NIPTs will undoubtedly revolutionize prenatal screening and increase the demand for early treatments for AS and for other syndromic disorders.
Newborn screening for AS is also possible, but criteria largely limit newborn screening to diseases for which early diagnosis benefits the baby, such as when a treatment or an intervention exists (Pitt, 2010). There is reason for optimism with regard to newborn screening for AS and its “sister” imprinted disorders—PWS and Dup15q syndrome. A recent study found that PWS can be treated shortly after birth with growth hormone (Scheermeyer et al., 2017). It might thus be possible to justify newborn screening for paternal 15q11-q13 deletion, to identify PWS newborns who would benefit from growth hormone. This test would also identify newborns with maternal 15q11-q13 deletion or duplication. Screening of this single genetic locus could thus permit early detection of three distinct neurodevelopmental disorders.
3. Critical period for treatment
Existing genetic tests can identify individuals with a maternally-inherited deletion of 15q11-q13 at any age, so a major question for the field is when to treat, particularly given that AS therapeutics are moving towards the clinic. To address this question, Elgersma and colleagues developed a way to genetically “reinstate” Ube3a at different ages in an AS model mouse (Silva-Santos et al., 2015). AS model mice have reproducible phenotypes that recapitulate AS symptoms, including motor deficits, sleep disruption, seizures, and cognitive deficits (Ehlen et al., 2015; Huang et al., 2013; Jiang et al., 1998; Sonzogni et al., 2018). Elgersma and colleagues found that early embryonic reinstatement of Ube3a prevented AS phenotypes across multiple domains from developing, while reinstatement at birth (postnatal day 0; P0) or twenty-one days after birth (P21) rescued some phenotypes (Table 1). Extrapolating to humans, P0 in a mouse corresponds to early second trimester in humans, while P21 corresponds to ~210 days old in humans (Workman, Charvet, Clancy, Darlington, & Finlay, 2013).
Table 1.
Phenotypes that are expected to show full (FR), partial, or no rescue (NR) when paternal Ube3a is unsilenced at different developmental time periods in AS model mice. Expectations based on data in (Meng et al., 2015; Silva-Santos et al., 2015).
| Developmental time period | ||||
|---|---|---|---|---|
| Phenotype | Embryonic | P0 | P21 | Adult |
| Learning/Memory | FR | FR | FR | FR |
| Rotarod | FR | FR | Partial | NR |
| Open field | FR | FR | NR | NR |
| Marble burying | FR | NR | NR | NR |
| Nest building | FR | NR | NR | NR |
| Forced swim | FR | NR | NR | NR |
In contrast, adult Ube3a reinstatement only rescued hippocampal long-term potentiation; a learning and memory phenotype (Table 1) (Silva-Santos et al., 2015). Likewise, antisense oligonucleotides (ASOs, described below) that unsilence paternal Ube3a partially treated a fear conditioning deficit in adult AS model mice (Meng et al., 2015), a phenotype that is also related to learning and memory.
These findings were reinforced by a subsequent study from Elgersma and colleagues, in which they deleted maternal Ube3a at different ages (Sonzogni et al., 2019). They found that deletion of maternal Ube3a embryonically recapitulated all measured AS phenotypes. In contrast, deletion of maternal Ube3a postnatally or in adults had very few deleterious effects. Collectively, these data indicate that there is a critical period, early in development, when AS can most effectively be treated through restoration of Ube3a.
4. Therapeutic development
Therapeutic approaches are being pursued that restore Ube3a expression in neurons, and hence target the root cause of AS. One approach is based on unsilencing the functional, but epigenetically silenced paternal Ube3a allele in neurons. Unsilencing approaches have a key advantage over other approaches. Namely, unsilencing approaches drive expression of paternal Ube3a from the endogenous promoter, which is predicted to permit proper isoform distribution and proper UBE3A protein levels at each stage of life. Work in our lab, jointly with others, provided the first evidence that the paternal Ube3a allele can be unsilenced with a class of drugs that are used clinically to treat cancer, namely topoisomerase inhibitors (Huang et al., 2012). These drugs repress the extremely long Ube3a-ATS transcript, permitting expression of paternal Ube3a in mouse and human neurons (King et al., 2013). However, topoisomerase inhibitors are not suitable for life-long use because of known side-effects in humans and these drugs downregulate many other long gene transcripts, some of which are important for brain development and synaptic function (King et al., 2013; Mabb et al., 2014).
Subsequently, Beaudet and colleagues unsilenced paternal Ube3a by targeting Ube3a-ATS with ASOs (Fig. 1) (Meng et al., 2015). ASOs rescued one behavioral phenotype in AS model mice but required repeated invasive spinal injections every few months, which is not ideal for a pediatric onset disorder that lasts a lifetime. An artificial transcriptional repressor was also developed that binds to Ube3a-ATS and partially unsilenced paternal Ube3a in the mouse brain (Bailus et al., 2016). No behavioral studies were performed in this latter study. Like ASOs, this protein repressor had to be injected repeatedly—three times per week for four weeks, and this protein downregulated genes in the PWS critical region, which is not a desirable side-effect.
It may be possible to overcome shortcomings of topoisomerase inhibitors, ASOs, and protein-based repressors using new genome editing technologies like CRISPR/Cas9. When packaged into adeno-associated virus (AAV) gene therapy vectors, Cas9 can be delivered to animals and treat diseases for significantly longer periods of time (Nelson et al., 2016).
Another approach is based on using AAV to deliver UBE3A to individuals with AS, a so-called gene replacement therapy. AAV-based gene replacement therapies have an acceptable safety profile in humans (Hocquemiller, Giersch, Audrain, Parker, & Cartier, 2016; Hudry & Vandenberghe, 2019), and drive gene expression in the human and primate brain for a decade or more (Sehara et al., 2017; Tuszynski et al., 2015). Gene replacement therapies are used clinically to treat other single gene disorders, including spinal muscular atrophy, a pediatric-onset neurological disorder (Mendell et al., 2017). However, challenges remain, particularly with regard to selecting the appropriate UBE3A isoform (Avagliano Trezza et al., 2019), and with selecting a promoter that drives UBE3A expression at the correct levels across the lifespan, without overshooting normal expression levels. It is well-established that UBE3A levels must be tightly maintained within a narrow range to permit normal brain development, as evidenced by the fact that loss of maternal UBE3A causes AS while duplication of UBE3A increases autism and neuropsychiatric disorder risk (LaSalle et al., 2015; Noor et al., 2015). Traditional gene replacement therapies typically drive expression of a single isoform at abnormally high levels, with variability between cells influenced by vector copy number.
5. Prenatal treatment path
The benefits and risks associated with treating AS at different ages will ultimately need to be evaluated by institutional review boards and government regulators. However, the studies described above indicate that the optimal age to treat AS is prenatally. Pathogenic mutations that cause or increase risk for AS can be detected prenatally, further enabling proactive prenatal treatment. Given the necessity and sufficiency of Ube3a over a narrow prenatal window, prenatal administration of a gene therapy vector has the potential to greatly diminish the severity of AS, and if transduction efficiency is high, prevent AS from developing.
Indeed, studies with rare “AS mosaics” suggest some behavioral recovery can be achieved even if UBE3A is restored in a small percentage of all neurons. AS individuals that are mosaic show variable loss of methylation at the maternal imprinting center, which leads to inactivation of maternal UBE3A in a variable number of cells. AS mosaics show symptoms that are classified as “exceptionally mild” when as few as 10% of all blood cells contain normal levels of methylation (Brockmann, Bohm, & Burger, 2002; Carson, Bird, Childers, Wheeler, & Duis, 2019; Le Fevre et al., 2017; Nazlican et al., 2004). Mild phenotypes include near normal speech, near normal motor performance, and lack of seizures. Assuming mosaicism in blood cells is reflective of mosaicism in the brain, restoring functional UBE3A in as few as 10% of all brain neurons could largely prevent severe AS symptoms from developing. Changes of this magnitude would greatly improve the quality of life for individuals with AS and their caregivers.
Prenatal testing can be performed around the end of the first trimester, and results are returned early in the second trimester. Thus, not inclusive of preimplantation genetic testing, the earliest practical age to treat is during the second trimester. This is the period when cortical neurogenesis ends (Meyer, Schaaps, Moreau, & Goffinet, 2000). A minimally invasive ultrasound guided injection procedure was recently developed to deliver an AAV gene therapy vector to the brain of second trimester rhesus macaques—a non-human primate whose gestation period is similar to that of humans (Massaro et al., 2018).
Prenatal treatment with a gene therapy vector has other advantages. Mammals, including humans, develop an immune responses to AAV and its cargo when injected into adults (Vandamme, Adjali, & Mingozzi, 2017). This immune response is known to reduce treatment efficacy over time. However, prenatal or early postnatal injection of AAV results in immune tolerance—meaning the virus and cargo are seen as self, allowing for persistent expression with no immune response (Hinderer et al., 2015; Hordeaux et al., 2019; Tai et al., 2015). Treatment during this immune tolerant period is ideal, especially if non-mammalian proteins like CAS9 are used as part of the gene therapy (Nelson et al., 2019). As additional advantages, the brain is smaller in a fetus, so significantly less gene therapy vector will need to be manufactured and administered, potentially reducing costs. And, AAV is effective at transducing >50% of all neurons when injected intracerebroventricularly into newborn mice (Chakrabarty et al., 2013). As noted above, this age in mice is equivalent to the second trimester in humans. Whether AAV transduces neurons as efficiently when injected prenatally into non-human primates is currently unclear.
There are parallels between prenatal treatment of AS and prenatal treatment of spina bifida—a neurodevelopmental disorder that, like AS, results in severe, life-long neurological deficits if not treated (Fletcher & Brei, 2010). Prenatal neurosurgeries to treat spina bifida have been performed for over two decades (Tulipan & Bruner, 1998), the risks are tolerated, and patient outcomes are significantly better than when surgeries are performed after birth (Adzick et al., 2011). This procedure involves creating an incision in the mother’s abdomen and uterus to access the fetus. The bioethics associated with performing invasive maternal-fetal surgeries to treat nonlethal fetal conditions have been extensively reviewed (Chervenak & McCullough, 2007; Lyerly, Gates, Cefalo, & Sugarman, 2001). The bioethics associated with treating AS prenatally, using a less invasive intracerebroventricular injection procedure (Massaro et al., 2018), is expected to be comparable.
6. Limitations to early treatment
One of the main limitations to prenatal treatment of AS is that the regulatory path and safety profile for the fetus and mother has not been established. But as biomedical history has taught us, the prenatal period is not off-limits for new treatments that benefit the baby, including fetal blood transfusions and fetal neurosurgeries. Regulators base decisions on safety and efficacy data. Encouragingly, recent draft guidance from the United States Food and Drug Administration seeks to broaden the definition of the neonatal period to include preterm newborns born during the second trimester (Administration, 2019). As a logical next step, clinicians may be able to evaluate the safety and efficacy of drug and biological therapeutics in second- or third trimester fetuses, provided risks to the mother are considered and assessed. The reproducible and disease-modeling phenotypes in AS model mice, coupled with the ability to deliver gene therapies to fetal non-human primates (Conlon et al., 2016; Massaro et al., 2018), should allow for the necessary safety data to be collected.
Additionally, we currently lack a quantitative understanding of symptoms that differ between maternal 15q11-q13 deletion positive and neurotypical babies in the 0-1 year range, which limits rapid assessment of efficacy following prenatal treatment. Feeding problems and muscle hypotonia are evident in AS infants (Dagli et al., 1993; Pelc et al., 2008), but these symptoms were identified retrospectively and hence need to be quantitatively evaluated in a prospective manner. In the absence of neonatal natural history data, commonly studied AS symptoms that are apparent by 1-2 years of age, like motor deficits, language impairment and cognitive delays, could serve as primary endpoints in clinical trials. Extending the follow-up window would permit the use of standard developmental assessment tools. A small number of maternal 15q11-q13 deletion positive fetuses are being detected through traditional invasive prenatal genetic testing, and could be enrolled in natural history studies focused on the 0-1 year old range. The number of fetuses that test positive for this deletion is expected to increase as next generation cell-based NIPTs come online, which will increase the demand for prenatal treatment options.
7. Applicability to other syndromic neurodevelopmental disorders
Significant progress has been made at identifying new syndromic forms of autism. As examples, de novo loss-of-function mutations in CHD8 and DYRK1 cause syndromic forms of autism (Bernier et al., 2014; van Bon et al., 2016). A variant of CRISPR/Cas9 that does not cut the genome was recently packaged into an AAV gene therapy vector and used to rescue phenotypes caused by Sim1 haploinsufficiency in mice (Matharu et al., 2019). Assuming the penetrance of syndromic autism gene mutations is high and the critical period for treatment is early, it may be possible to develop gene therapies to treat these and other syndromic forms of autism caused by gene haploinsufficiency. Ultimately, efforts to treat AS prenatally could pioneer a future where sensitive non-invasive prenatal genetic tests are coupled with gene therapies, to offer disease-modifying treatments for a diversity of syndromic neurodevelopmental disorders.
Acknowledgments
The author has no conflicts of interest to declare. I thank Drs. Neeta Vora and Anne Lyerly for helpful discussions on prenatal screening technologies and the bioethics of maternal-fetal surgeries, respectively. M.J.Z. is supported by grants from The Angelman Syndrome Foundation, the National Institute of Environmental Health Sciences (R35ES028366-01A1), and The Simons Foundation.
References
- Administration, F. a. D. (2019). General Clinical Pharmacology Considerations for Neonatal Studies for Drugs and Biological Products Guidance for Industry.
- Adzick NS, Thom EA, Spong CY, Brock JW 3rd, Burrows PK, Johnson MP, … Investigators, M. (2011). A randomized trial of prenatal versus postnatal repair of myelomeningocele. N Engl J Med, 364(11), 993–1004. doi: 10.1056/NEJMoa1014379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avagliano Trezza R, Sonzogni M, Bossuyt SNV, Zampeta FI, Punt AM, van den Berg M, … Elgersma Y (2019). Loss of nuclear UBE3A causes electrophysiological and behavioral deficits in mice and is associated with Angelman syndrome. Nat Neurosci. doi: 10.1038/s41593-019-0425-0 [DOI] [PubMed] [Google Scholar]
- Bailus BJ, Pyles B, McAlister MM, O'Geen H, Lockwood SH, Adams AN, … Segal DJ (2016). Protein Delivery of an Artificial Transcription Factor Restores Widespread Ube3a Expression in an Angelman Syndrome Mouse Brain. Mol Ther, 24(3), 548–555. doi: 10.1038/mt.2015.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, … Eichler EE (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell, 158(2), 263–276. doi: 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann K, Bohm R, & Burger J (2002). Exceptionally mild Angelman syndrome phenotype associated with an incomplete imprinting defect. J Med Genet, 39(9), e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher AV, Cidav Z, Knapp M, & Mandell DS (2014). Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA Pediatr, 168(8), 721–728. doi: 10.1001/jamapediatrics.2014.210 [DOI] [PubMed] [Google Scholar]
- Carson RP, Bird L, Childers AK, Wheeler F, & Duis J (2019). Preserved expressive language as a phenotypic determinant of Mosaic Angelman Syndrome. Mol Genet Genomic Med, e837. doi: 10.1002/mgg3.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Rosario A, Cruz P, Siemienski Z, Ceballos-Diaz C, Crosby K, … Levites Y (2013). Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One, 8(6), e67680. doi: 10.1371/journal.pone.0067680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, & Brannan CI (2001). The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics, 73(3), 316–322. doi: 10.1006/geno.2001.6543 [DOI] [PubMed] [Google Scholar]
- Chervenak FA, & McCullough LB (2007). Ethics of maternal-fetal surgery. Semin Fetal Neonatal Med, 12(6), 426–431. doi: 10.1016/j.siny.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Christensen B, Philip J, Kolvraa S, Lykke-Hansen L, Hromadnikova I, Gohel D, … Djursing H (2005). Fetal cells in maternal blood: a comparison of methods for cell isolation and identification. Fetal Diagn Ther, 20(2), 106–112. doi: 10.1159/000082432 [DOI] [PubMed] [Google Scholar]
- Committee on G (2018). ACOG Technology Assessment in Obstetrics and Gynecology No. 14: Modern Genetics in Obstetrics and Gynecology. Obstet Gynecol, 132(3), e143–e168. doi: 10.1097/AOG.0000000000002831 [DOI] [PubMed] [Google Scholar]
- Conlon TJ, Mah CS, Pacak CA, Rucker Henninger MB, Erger KE, Jorgensen ML, … Byrne BJ (2016). Transfer of Therapeutic Genes into Fetal Rhesus Monkeys Using Recombinant Adeno-Associated Type I Viral Vectors. Hum Gene Ther Clin Dev, 27(4), 152–159. doi: 10.1089/humc.2016.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli AI, Mueller J, & Williams CA (1993). Angelman Syndrome. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, & Amemiya A (Eds.), GeneReviews((R)). Seattle (WA). [PubMed] [Google Scholar]
- Ehlen JC, Jones KA, Pinckney L, Gray CL, Burette S, Weinberg RJ, … DeBruyne JP (2015). Maternal Ube3a Loss Disrupts Sleep Homeostasis But Leaves Circadian Rhythmicity Largely Intact. J Neurosci, 35(40), 13587–13598. doi: 10.1523/JNEUROSCI.2194-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JM, & Brei TJ (2010). Introduction: Spina bifida--a multidisciplinary perspective. Dev Disabil Res Rev, 16(1), 1–5. doi: 10.1002/ddrr.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace MR, Hardisty E, Dotters-Katz SK, Vora NL, & Kuller JA (2016). Cell-Free DNA Screening: Complexities and Challenges of Clinical Implementation. Obstet Gynecol Surv, 71(8), 477–487. doi: 10.1097/OGX.0000000000000342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer C, Bell P, Louboutin JP, Zhu Y, Yu H, Lin G, … Wilson JM (2015). Neonatal Systemic AAV Induces Tolerance to CNS Gene Therapy in MPS I Dogs and Nonhuman Primates. Mol Ther, 23(8), 1298–1307. doi: 10.1038/mt.2015.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquemiller M, Giersch L, Audrain M, Parker S, & Cartier N (2016). Adeno-Associated Virus-Based Gene Therapy for CNS Diseases. Hum Gene Ther, 27(7), 478–496. doi: 10.1089/hum.2016.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J, Hinderer C, Buza EL, Louboutin JP, Jahan T, Bell P, … Wilson JM (2019). Safe and Sustained Expression of Human Iduronidase After Intrathecal Administration of Adeno-Associated Virus Serotype 9 in Infant Rhesus Monkeys. Hum Gene Ther. doi: 10.1089/hum.2019.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, … Philpot BD (2012). Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature, 481(7380), 185–189. doi: 10.1038/nature10726nature10726 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Burns AJ, Nonneman RJ, Baker LK, Riddick NV, Nikolova VD, … Moy SS (2013). Behavioral deficits in an Angelman syndrome model: effects of genetic background and age. Behav Brain Res, 243, 79–90. doi: 10.1016/j.bbr.2012.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry E, & Vandenberghe LH (2019). Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron, 102(1), 263. doi: 10.1016/j.neuron.2019.03.020 [DOI] [PubMed] [Google Scholar]
- Hui T, Cao Q, Wegrzyn-Woltosz J, O'Neill K, Hammond CA, Knapp D, … Hirst M (2018). High-Resolution Single-Cell DNA Methylation Measurements Reveal Epigenetically Distinct Hematopoietic Stem Cell Subpopulations. Stem Cell Reports, 11(2), 578–592. doi: 10.1016/j.stemcr.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, … Beaudet AL (1998). Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron, 21(4), 799–811. [DOI] [PubMed] [Google Scholar]
- King IF, Yandava CN, Mabb AM, Hsiao JS, Huang HS, Pearson BL, … Zylka MJ (2013). Topoisomerases facilitate transcription of long genes linked to autism. Nature, 501(7465), 58–62. doi: 10.1038/nature12504nature12504 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolvraa S, Singh R, Normand EA, Qdaisat S, van den Veyver IB, Jackson L, … Beaudet AL (2016). Genome-wide copy number analysis on DNA from fetal cells isolated from the blood of pregnant women. Prenat Diagn, 36(12), 1127–1134. doi: 10.1002/pd.4948 [DOI] [PubMed] [Google Scholar]
- Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, … Lalande M (2004). Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res, 32(11), 3480–3492. doi: 10.1093/nar/gkh670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larion S, Warsof SL, Romary L, Mlynarczyk M, Peleg D, & Abuhamad AZ (2014). Uptake of noninvasive prenatal testing at a large academic referral center. Am J Obstet Gynecol, 211(6), 651 e651–657. doi: 10.1016/j.ajog.2014.06.038 [DOI] [PubMed] [Google Scholar]
- LaSalle JM, Reiter LT, & Chamberlain SJ (2015). Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics, 7(7), 1213–1228. doi: 10.2217/epi.15.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fevre A, Beygo J, Silveira C, Kamien B, Clayton-Smith J, Colley A, … Dudding-Byth T (2017). Atypical Angelman syndrome due to a mosaic imprinting defect: Case reports and review of the literature. Am J Med Genet A, 173(3), 753–757. doi: 10.1002/ajmg.a.38072 [DOI] [PubMed] [Google Scholar]
- Liang D, Cram DS, Tan H, Linpeng S, Liu Y, Sun H, … Wu L (2019). Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet Med. doi: 10.1038/s41436-019-0467-4 [DOI] [PubMed] [Google Scholar]
- Lyerly AD, Gates EA, Cefalo RC, & Sugarman J (2001). Toward the ethical evaluation and use of maternal-fetal surgery. Obstet Gynecol, 98(4), 689–697. [DOI] [PubMed] [Google Scholar]
- Mabb AM, Judson MC, Zylka MJ, & Philpot BD (2011). Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci, 34(6), 293–303. doi:S0166-2236(11)00056-7 [pii] 10.1016/j.tins.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Kullmann PH, Twomey MA, Miriyala J, Philpot BD, & Zylka MJ (2014). Topoisomerase 1 inhibition reversibly impairs synaptic function. Proc Natl Acad Sci U S A, 111(48), 17290–17295. doi: 10.1073/pnas.1413204111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro G, Mattar CNZ, Wong AMS, Sirka E, Buckley SMK, Herbert BR, … Rahim AA (2018). Fetal gene therapy for neurodegenerative disease of infants. Nat Med, 24(9), 1317–1323. doi: 10.1038/s41591-018-0106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A, … Ahituv N (2019). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science, 363(6424). doi: 10.1126/science.aau0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, … Beaudet AL (1997). De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet, 15(1), 74–77. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, … Brain Somatic Mosaicism, N. (2017). Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science, 356(6336). doi: 10.1126/science.aal1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, … Kaspar BK (2017). Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N Engl J Med, 377(18), 1713–1722. doi: 10.1056/NEJMoa1706198 [DOI] [PubMed] [Google Scholar]
- Meng L, Person RE, & Beaudet AL (2012). Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet, 21(13), 3001–3012. doi: 10.1093/hmg/dds130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, & Rigo F (2015). Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature, 518(7539), 409–412. doi: 10.1038/nature13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Schaaps JP, Moreau L, & Goffinet AM (2000). Embryonic and early fetal development of the human neocortex. J Neurosci, 20(5), 1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazlican H, Zeschnigk M, Claussen U, Michel S, Boehringer S, Gillessen-Kaesbach G, … Horsthemke B (2004). Somatic mosaicism in patients with Angelman syndrome and an imprinting defect. Hum Mol Genet, 13(21), 2547–2555. doi: 10.1093/hmg/ddh296 [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, … Gersbach CA (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science, 351(6271), 403–407. doi: 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, … Gersbach CA (2019). Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med, 25(3), 427–432. doi: 10.1038/s41591-019-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Dupuis L, Mittal K, Lionel AC, Marshall CR, Scherer SW, … Stavropoulos DJ (2015). 15q11.2 Duplication Encompassing Only the UBE3A Gene Is Associated with Developmental Delay and Neuropsychiatric Phenotypes. Hum Mutat, 36(7), 689–693. doi: 10.1002/humu.22800 [DOI] [PubMed] [Google Scholar]
- Pelc K, Cheron G, & Dan B (2008). Behavior and neuropsychiatric manifestations in Angelman syndrome. Neuropsychiatr Dis Treat, 4(3), 577–584.18830393 [Google Scholar]
- Peters SU, Beaudet AL, Madduri N, & Bacino CA (2004). Autism in Angelman syndrome: implications for autism research. Clin Genet, 66(6), 530–536. doi: 10.1111/j.1399-0004.2004.00362.x [DOI] [PubMed] [Google Scholar]
- Petersen AK, Cheung SW, Smith JL, Bi W, Ward PA, Peacock S, … Breman AM (2017). Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. Am J Obstet Gynecol, 217(6), 691 e691–691 e696. doi: 10.1016/j.ajog.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Pitt JJ (2010). Newborn screening. Clin Biochem Rev, 31(2), 57–68. [PMC free article] [PubMed] [Google Scholar]
- Scheermeyer E, Harris M, Hughes I, Crock PA, Ambler G, Verge CF, … collaboration, O. (2017). Low dose growth hormone treatment in infants and toddlers with Prader-Willi syndrome is comparable to higher dosage regimens. Growth Horm IGF Res, 34, 1–7. doi: 10.1016/j.ghir.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Sehara Y, Fujimoto KI, Ikeguchi K, Katakai Y, Ono F, Takino N, … Muramatsu SI (2017). Persistent Expression of Dopamine-Synthesizing Enzymes 15 Years After Gene Transfer in a Primate Model of Parkinson's Disease. Hum Gene Ther Clin Dev, 28(2), 74–79. doi: 10.1089/humc.2017.010 [DOI] [PubMed] [Google Scholar]
- Silva-Santos S, van Woerden GM, Bruinsma CF, Mientjes E, Jolfaei MA, Distel B, … Elgersma Y (2015). Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J Clin Invest, 125(5), 2069–2076. doi: 10.1172/JCI80554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonzogni M, Hakonen J, Bernabe Kleijn M, Silva-Santos S, Judson MC, Philpot BD, … Elgersma Y (2019). Delayed loss of UBE3A reduces the expression of Angelman syndrome-associated phenotypes. Mol Autism, 10, 23. doi: 10.1186/s13229-019-0277-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonzogni M, Wallaard I, Santos SS, Kingma J, du Mee D, van Woerden GM, & Elgersma Y (2018). A behavioral test battery for mouse models of Angelman syndrome: a powerful tool for testing drugs and novel Ube3a mutants. Mol Autism, 9, 47. doi: 10.1186/s13229-018-0231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai DS, Hu C, Lee CC, Martinez M, Cantero G, Kim EH, … Lipshutz GS (2015). Development of operational immunologic tolerance with neonatal gene transfer in nonhuman primates: preliminary studies. Gene Ther, 22(11), 923–930. doi: 10.1038/gt.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulipan N, & Bruner JP (1998). Myelomeningocele repair in utero: a report of three cases. Pediatr Neurosurg, 28(4), 177–180. doi: 10.1159/000028645 [DOI] [PubMed] [Google Scholar]
- Tuszynski MH, Yang JH, Barba D, U HS, Bakay RA, Pay MM, … Nagahara AH (2015). Nerve Growth Factor Gene Therapy: Activation of Neuronal Responses in Alzheimer Disease. JAMA Neurol, 72(10), 1139–1147. doi: 10.1001/jamaneurol.2015.1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bon BW, Coe BP, Bernier R, Green C, Gerdts J, Witherspoon K, … Eichler EE (2016). Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol Psychiatry, 21(1), 126–132. doi: 10.1038/mp.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme C, Adjali O, & Mingozzi F (2017). Unraveling the Complex Story of Immune Responses to AAV Vectors Trial After Trial. Hum Gene Ther, 28(11), 1061–1074. doi: 10.1089/hum.2017.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora NL, & Wapner RJ (2018). Introducing new and emerging genetic tests into prenatal care. Semin Perinatol, 42(5), 283–286. doi: 10.1053/j.semperi.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossaert L, Wang Q, Salman R, Zhuo X, Qu C, Henke D, … Beaudet A (2018). Reliable detection of subchromosomal deletions and duplications using cell-based noninvasive prenatal testing. Prenat Diagn, 38(13), 1069–1078. doi: 10.1002/pd.5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, … Benn P (2015). Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol, 212(3), 332 e331–339. doi: 10.1016/j.ajog.2014.11.041 [DOI] [PubMed] [Google Scholar]
- Williams CA, Beaudet AL, Clayton-Smith J, Knoll JH, Kyllerman M, Laan LA, … Wagstaff J (2006). Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A, 140(5), 413–418. [DOI] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, & Finlay BL (2013). Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci, 33(17), 7368–7383. doi: 10.1523/JNEUROSCI.5746-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]