Abstract
Background
The serotonin transporter gene (SLC6A4; 5-HTT; SERT) is considered a prime candidate in pharmacogenetic research in major depressive disorder (MDD). Besides genetic variation, recent advances have spotlighted the involvement of epigenetic mechanisms such as DNA methylation in predicting antidepressant treatment response in “pharmaco-epigenetic” approaches. In MDD, lower SLC6A4 promoter methylation has been suggested to predict impaired response to serotonergic antidepressants. The present study sought to replicate and extend this finding in a large, independent sample of MDD patients.
Methods
The sample comprised n = 236 Caucasian patients with MDD receiving antidepressant medication in a naturalistic treatment setting. Functional DNA methylation of 9 CpG sites located in the SLC6A4 promoter region was analyzed via direct sequencing of sodium bisulfite– treated DNA extracted from blood cells. Patients were assessed over the course of a 6-week in-patient treatment using the Hamilton Depression Scale (HAM-D).
Results
Results confirm relative SLC6A4 hypomethylation to predict impaired antidepressant response both dimensionally and categorically (HAM-D reductions < 50%) and to furthermore be indicative of nonremission (HAM-D > 7). This also held true in a homogenous subgroup of patients continuously treated with selective serotonin reuptake inhibitors or serotonin/noradrenaline reuptake inhibitors (n = 110).
Conclusions
Impaired response to serotonergic antidepressants via SLC6A4 hypomethylation may be conveyed by increased gene expression and consequently decreased serotonin availability, which may counteract the effects of serotonergic antidepressants. The present results could in the future inform clinical decision-making towards a more personalized treatment of MDD.
Keywords: epigenetics, DNA methylation, mood disorders, 5-HT, precision medicine
Significance Statement.
The high treatment resistance rates observed in major depressive disorder (MDD) stress the need for the identification of early treatment response markers informing expert decision-making towards more personalized and thus more efficacious pharmacological interventions. Biomarkers such as epigenetic profiles carry great potential in this regard. “Pharmacoepigenetic” investigations form a young but burgeoning field that has produced first promising results, which, however, still warrant replication. The present study thus sought to replicate a previous pilot finding of lower serotonin transporter gene (SLC6A4) promoter DNA methylation—assumed to result in increased gene expression and decreased serotonin availability—to predict the clinical response to antidepressant treatment. In a sample of 236 MDD patients, SLC6A4 hypomethylation was again found to predict impaired dimensional and categorical response to serotonergic antidepressants as well as nonremission after a 6-week antidepressant treatment, suggesting SLC6A4 methylation as a biomarker that in the future is hoped to translate into clinical application.
Introduction
The serotonergic system has long since been of prime interest in pharmacogenetic research in major depressive disorder (MDD). In particular, the gene coding for the serotonin transporter (SLC6A4; chr. 17q11.1–12) has garnered much attention in this regard given that it codes for the presumed site of action of antidepressants such as selective serotonin reuptake inhibitors (SSRIs) and serotonin/noradrenaline reuptake inhibitors (SNRIs). Despite the well-documented efficacy and tolerability of SSRIs and SNRIs, initial treatment nonresponse rates are reported to be as high as 60% (Fava, 2003), which has spurred research into identifying early biomarkers for the prediction of antidepressant treatment response. A functional length polymorphism in the promoter region of the SLC6A4 gene, the serotonin transporter linked polymorphic region (5-HTTLPR) has been widely investigated in this regard, although results have been mixed. Meta-analytic evidence points to impaired antidepressant treatment response in carriers of the less active 5-HTTLPR S allele in European, but not Asian populations (Serretti et al., 2007; Porcelli et al., 2012), though the effect is likely of only small magnitude (odds ratio = 1.20–1.58) (cf. McGuffin et al., 2011).
In recent years, attention has shifted to the involvement of epigenetic processes in predicting and possibly also mediating treatment response in affective disorders (Menke et al., 2012; Vialou et al., 2013; Nestler et al., 2016; Schiele et al., 2020a). Epigenetic processes such as DNA methylation crucially modulate gene function without, however, entailing changes to the DNA sequence itself (Moore et al., 2013; Schuebel et al., 2016). DNA methylation of gene promoters, enhancers, and transcription start sites is associated with gene silencing (Suzuki and Bird, 2008). With regard to SLC6A4, differential methylation of a CpG island located within the transcriptional control region was found to functionally influence SLC6A4 mRNA levels, with hypermethylation resulting in reduced mRNA expression (Philibert et al., 2007). Functional in vitro assays have furthermore shown SLC6A4 promoter hypermethylation to result in decreased reporter gene activity (Wang et al., 2012; Schiele et al., 2019). Increased SLC6A4 promoter methylation has been linked to MDD diagnosis (Philibert et al., 2008; Iga et al., 2016; Shi et al., 2017), higher depressive symptom severity (Kang et al., 2013; Zhao et al., 2013), post-stroke depression (Kim et al., 2013), and comorbid MDD in panic disorder (Schiele et al., 2019) (for review, see Palma-Gudiel and Fananas, 2017). So far, only 2 studies have adopted a pharmaco-epigenetic approach in an attempt to probe SLC6A4 methylation as a potential predictor of antidepressant treatment response in MDD. In n = 108 Korean patients with MDD, apart from a trend-level association with 1 CpG site, no overall predictive effect of SLC6A4 promoter methylation on treatment outcome after 12 weeks of antidepressant treatment in a naturalistic study design could be discerned (Kang et al., 2013). By contrast, we recently demonstrated relative SLC6A4 promoter hypomethylation to predict impaired antidepressant treatment response after 6 weeks of SSRI treatment in a study comprising n = 94 Caucasian patients with MDD (Domschke et al., 2014). The aim of the present study was to validate this pilot finding by investigating the same CpG region as in Domschke et al. (2014) as well as in Kim et al. (2013), Kang et al. (2013), and Schiele et al. (2019) (see above) in an independent but otherwise comparable sample of MDD patients, which to the best of our knowledge constitutes the largest sample investigated in this regard to date in a naturalistic treatment setting as well as in a subsample of patients continuously receiving SSRIs/SNRIs as primary treatment.
Methods
Sample
A total of 236 in-patients with MDD (138 female; mean age ± SD: 48.26 ± 15.90 years) were recruited at the Department of Psychiatry and Psychotherapy, University of Muenster, Germany, between 2004 and 2011 in a naturalistic study design. To maximize comparability, inclusion/exclusion criteria were identical to those applied by Domschke et al. (2014). Briefly, only patients treated with antidepressants for at least 6 consecutive weeks were eligible for the present analysis. Medication with monoamine oxidase inhibitors or valproate or concomitant electroconvulsive treatment led to study exclusion. Co-medication with other psychopharmacological agents was permitted and recorded; side effects were not systematically assessed. Only patients with a primary diagnosis of MDD according to DSM-IV criteria were included. Patients with bipolar disorder, cyclothymia, psychotic disorders including schizoaffective disorder, comorbid substance abuse/addiction, intellectual disability, and severe neurological, neurodegenerative, cardiological, endocrinological (apart from diabetes), and immunological disorders were excluded from analysis. Diagnoses were ascertained by experienced psychiatrists on the basis of medical records and a structured clinical interview according to DSM-IV criteria. A total of n = 11 patients had comorbid panic disorder and/or agoraphobia, n = 7 patients were additionally diagnosed with social phobia, n = 7 met criteria for somatoform disorder, n = 5 for stress-related disorders, and n = 2 for obsessive-compulsive disorder. All patients were assessed for depressive symptoms using the Hamilton Depression Scale (HAM-D-21) on a weekly basis. At admission, the Beck Depression Inventory and the Global Assessment of Functioning scale were recorded as well. Caucasian descent was ascertained by Caucasian background of both parents. Ethical approval was granted by the ethical board of the University of Muenster, Germany. Written informed consent was obtained from all patients, and the study was conducted according to the ethical principles of the Helsinki Declaration.
Medication
All patients received antidepressant treatment in a naturalistic design. In week 1, n = 69 received SNRIs, n = 32 SSRIs, n = 32 tri- or tetracyclic antidepressants (TCAs), n = 85 a noradrenergic and specific serotonergic antidepressant (NaSSA; partly in subclinical dosage for sleep promotion only), n = 8 a norepinephrine reuptake inhibitor, and n = 1 a norepinephrine-dopamine reuptake inhibitor either as stand-alone medication or in combination. In addition to antidepressants, co-medication with atypical antipsychotics (n = 77), typical antipsychotics (n = 12), anticonvulsants (n = 13), lithium (n = 15), benzodiazepines (n = 61), or zopiclone/zolpidem (n = 17) was used as partly off-label augmentation of antidepressant treatment.
In a subgroup analysis, patients receiving either an SSRI or SNRI as primary antidepressant for the duration of the observation period of 6 weeks (initiation of SSRI/SNRI treatment no later than in week 2 of in-patient treatment to account for the delay in onset of effect) were considered, resulting in a total of n = 110 patients: n = 23 were treated continuously with an SSRI, and n = 87 received continuous SNRI treatment. Co-medication commencing either at admission or during the course of treatment with TCAs (n = 14), NaSSA (n = 57; partly in subclinical dosage for sleep promotion), norepinephrine reuptake inhibitor (n = 6), atypical antipsychotics (n = 70), typical antipsychotics (n = 5), anticonvulsants (n = 11), lithium (n = 6), benzodiazepines (n = 36), or zopiclone/zolpidem (n = 13) was allowed.
SLC6A4 Methylation Analysis and Genotyping
DNA was isolated from whole blood using the FlexiGene DNA Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A 635-bp amplicon comprising part of the SLC6A4 promoter upstream of exon 1A (chr17:30 235 634-30 236 268; GRCh38.p2 Primary Assembly, UCSC Genome Browser) was chosen for DNA methylation analysis in analogy to previous studies (e.g., Kang et al., 2013; Kim et al., 2013; Domschke et al., 2014; Schiele et al., 2019). The amplicon was PCR amplified using the following set of oligonucleotide primers recognizing bisulfite modified DNA (F: 5’ TAAGGGTTTTTAAGTTGAGTTTA- TATTTTA 3′ and R: 5’ CTAATCCCRAACTAAACAAACRAACTAA 3′). Commercially available fully methylated and fully nonmethylated DNA were included in all experiments as a control. All samples were tested in duplicates to account for run variability, resulting in a mean individual methylation score for each CpG site as well as an individual SD for each duplicate (for a detailed description, see Domschke et al., 2014; Schiele et al., 2019). The resulting electropherograms were robustly readable for 9 individual CpG sites. CpG sites were numbered in analogy to previous studies on SLC6A4 methylation (Domschke et al., 2014; Schiele et al., 2019; for a detailed overview, see Palma-Gudiel and Fananas, 2017): CpG1 = chr17:30 236 071; CpG2 = chr17:30 236 083; CpG3 = chr17:30 236 088; CpG4 = chr17:30 236 090; CpG5 = chr17:30 236 101; CpG6 = chr17:30 236 120; CpG7 = chr17:30 236 125; CpG8 = chr17:30 236 141; CpG9 = chr17:30 236 156. The obtained sequences were quantitatively analyzed using the Epigenetic Sequencing Methylation analysis software (Lewin et al., 2004) as successfully applied in previous studies (e.g., Alasaari et al., 2012; Domschke et al., 2012, 2014; Tadic et al., 2014; Ziegler et al., 2016, 2018; Schartner et al., 2017; Schiele et al., 2018, 2019, 2020b, 2020c). Average SLC6A4 methylation status and methylation at single CpG sites at admission is given in Table 1.
Table 1.
SLC6A4 Methylation Levels in Patients With Major Depressive Disorder (n = 236)
| Methylation | Mean | SD |
|---|---|---|
| Average | .035 | .017 |
| CpG 1 | .052 | .044 |
| CpG 2 | .028 | .027 |
| CpG 3 | .050 | .038 |
| CpG 4 | .020 | .021 |
| CpG 5 | .016 | .026 |
| CpG 6 | .014 | .023 |
| CpG 7 | .040 | .038 |
| CpG 8 | .049 | .036 |
| CpG 9 | .047 | .041 |
Abbreviations: CpG1 = chr17:30 236 071; CpG2 = chr17:30 236 083; CpG3 = chr17:30 236 088; CpG4 = chr17:30 236 090; CpG5 = chr17:30 236 101; CpG6 = chr17:30 236 120; CpG7 = chr17:30 236 125; CpG8 = chr17:30 236 141; CpG9 = chr17:30 236 156.
To address a potential influence of 5-HTT gene variation on DNA methylation status, all samples were genotyped according to published protocols (see Schiele et al., 2016, 2020d) for 5-HTTLPR and the functionally related single nucleotide polymorphism rs25531 functionally modifying 5-HTTLPR (Wendland et al., 2006). Genotype information was unavailable for n = 14 patients due to genotyping failures. Genotypes were grouped into a “low expression” group containing SS, SLG, SLA, LGLG, or LALG genotypes (n = 166) and a “high expression” group comprising LALA genotype carriers (n = 56) as done in previous studies (e.g., Baune et al., 2008a; Klauke et al., 2011; Odgerel et al., 2013; Schiele et al., 2016). Hardy-Weinberg criteria as determined by the online program DeFinetti (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) were fulfilled for the 5-HTTLPR/rs25531 triallelic model (Hu et al., 2006) (LALA, n = 56; LALG/SLA, n = 123; LGLG/SLG/SS, n = 43; P = .108).
Statistical Analysis
Dimensional treatment response was defined as the intra-individual relative change (%) of HAM-D-21 scores after week 6 relative to HAM-D at week 1 according to the American College of Neuropsychopharmacology (ACNP) task force guidelines on response and remission in MDD (Rush et al., 2006). Initial changes in HAM-D scores occurring during week 1 were not included given that HAM-D changes during this period were likely unrelated to the presently evaluated antidepressant medication. Therefore, in the present naturalistic study design, HAM-D score at week 1 was considered the pre-treatment HAM-D-21 baseline score as done in previous pharmacogenetics studies (cf. Baune et al., 2008b; Domschke et al., 2008, 2010, 2014; Baffa et al., 2010). DNA methylation was included as continuous variable in all analyses. For confounder analysis, dimensional data were analyzed using Pearson correlations, and methylation differences between categorical variables were tested by means of independent t tests. The influence of SLC6A4 methylation on dimensional treatment response (Δ% HAM-D) was investigated via linear regression analysis corrected for HAM-D score at admission. Categorical treatment response was defined as a HAM-D reduction of 50% or more from week 1 to week 6 (cf. Fava et al., 2008). Remission was defined as a HAM-D total score of 7 points or less after 6 weeks of antidepressant treatment (cf. Fava et al., 2008). For analysis of categorical response prediction and prediction of remitter status, logistic regression analyses were applied with HAM-D at admission entered as a covariate. The significance level was set at P < .05. For secondary analyses (9 single CpG sites), Bonferroni correction for multiple testing was applied, which corrected the significance level to P = .006.
Results
Confounder Analysis
Average SLC6A4 methylation status was not related to age (r = .107, P = .101), sex (t234 = 1.402, P = .162), Global Assessment of Functioning at admission (r = −.054, P = .410), Beck Depression Inventory at admission (r = .054, P = .465), HAM-D-21 at admission (r = .068, P = .298), age of onset (r = .127, P = .087), illness duration (r = −.076, P = .313), number of hospitalizations (F14 = .667, P = .804), number of suicide attempts (F3 = .240, P = .868), or 5-HTTLPR/rs25531 genotype (t220 = .216, P = .829). Additionally, medication at the beginning of treatment with SNRIs (n = 69; t234 = −.035, P = .972), SSRIs (n = 48; t234 = −.701, P = .484), TCAs (n = 32; t234 = 1.105, P = .270), NaSSA (n = 85; t234 = −.220, P = .826), anticonvulsants (n = 13; t234 = −1.252, P = .212), benzodiazepines (n = 61; t234 = −.140, P = .889), zopiclone/zolpidem (n = 17; t234 = −.451, P = .653), lithium (n = 15; t234 = .873, P = .383), atypical antipsychotics (n = 77; t234 = −1.659, P = .098), or typical antipsychotics (n = 12; t234 = −.880, P = .380) did not affect SLC6A4 average methylation status.
SLC6A4 Methylation and Dimensional Treatment Response
Full Sample
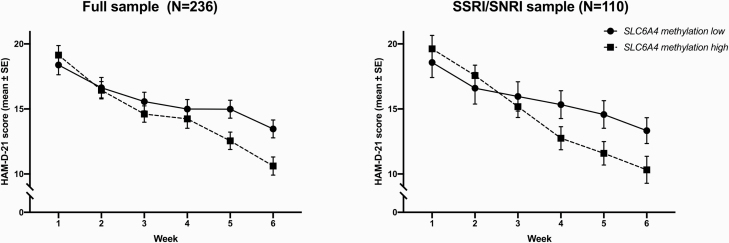
Average SLC6A4 methylation significantly predicted relative changes in HAM-D scores from week 1 to week 6 (β = −.152, P = .015; overall model fit R2 = .116) (see Figure 1A). That is, a decrease in methylation values went along with fewer differences (i.e., fewer reductions) or even increases in HAM-D scores and vice versa. Secondary analysis on a single-CpG level revealed—although not withstanding correction for multiple testing—nominally significant associations with HAM-D change for CpGs 8 and 9 (for details, see Table 2).
Figure 1.
Hamilton Depression Scale (HAM-D; 21 items) scores over the course of 6-week antidepressant treatment by SLC6A4 methylation status. For graphical representation only, average methylation was dichotomized into a low and high methylation group by means of median split (Domschke et al., 2014). Statistical analyses were performed using dimensional methylation data.
Table 2.
Prediction of Treatment Response and Remission by SLC6A4 Methylation Status
| Full sample (n = 236) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimensional response | Categorical response | Remission | |||||||||||||||
| Methylation | β | t | P | b | S.E. | Wald | df | P | OR | 95% CI | b | S.E. | Wald | df | P | OR | 95% CI |
| Average | −0.152 | −2.452 | .015 | 0.241 | 0.086 | 7.947 | 1 | .005 | 1.273 | 1.049–1.140 | 0.213 | 0.083 | 6.593 | 1 | .010 | 1.237 | 1.052–1.455 |
| CpG 1 | −0.107 | −1.721 | .087 | 0.057 | 0.032 | 3.118 | 1 | .077 | 1.059 | 0.994–1.128 | 0.092 | 0.032 | 8.253 | 1 | .004 | 1.096 | 1.030–1.167 |
| CpG 2 | −0.082 | −1.319 | .188 | 0.146 | 0.053 | 7.480 | 1 | .006 | 1.157 | 1.042–1.284 | 0.110 | 0.050 | 4.731 | 1 | .030 | 1.116 | 1.011–1.232 |
| CpG 3 | −0.117 | −1.882 | .061 | 0.052 | 0.037 | 1.934 | 1 | .164 | 1.053 | 0.979–1.133 | 0.056 | 0.036 | 2.383 | 1 | .123 | 1.058 | 0.985–1.135 |
| CpG 4 | 0.037 | 0.587 | .558 | -0.029 | 0.071 | 0.167 | 1 | .683 | 0.971 | 0.845–1.116 | −0.001 | 0.068 | <0.001 | 1 | .988 | 0.999 | 0.874–1.142 |
| CpG 5 | −0.011 | −0.183 | .855 | <0.001 | 0.055 | <0.001 | 1 | .996 | 0.100 | 0.898–1.114 | 0.029 | 0.053 | 0.301 | 1 | .583 | 1.030 | 0.927–1.143 |
| CpG 6 | 0.002 | 0.033 | .974 | 0.092 | 0.060 | 2.340 | 1 | .126 | 1.097 | 0.974–1.235 | −0.040 | 0.065 | 0.369 | 1 | .544 | 0.961 | 0.846–1.092 |
| CpG 7 | −0.070 | −1.117 | .265 | 0.011 | 0.037 | 0.095 | 1 | .758 | 1.011 | 0.941–1.087 | 0.001 | 0.037 | 3.58 x 10E-5 | 1 | .985 | 1.001 | 0.931–1.075 |
| CpG 8 | −0.127 | −2.047 | .042 | 0.128 | 0.041 | 9.732 | 1 | .002 | 1.137 | 1.049–1.232 | 0.078 | 0.039 | 4.098 | 1 | .043 | 1.082 | 1.002–1.167 |
| CpG 9 | −0.136 | −2.186 | .030 | 0.068 | 0.034 | 3.947 | 1 | .047 | 1.070 | 1.001–1.145 | 0.068 | 0.034 | 4.113 | 1 | .043 | 1.071 | 1.002–1.144 |
| SSRI/SNRI sample (n = 110) | |||||||||||||||||
| Methylation | Dimensional Response | Categorical Response | Remission | ||||||||||||||
| β | t | P | b | S.E. | Wald | df | P | OR | 95% CI | b | S.E. | Wald | df | P | OR | 95% CI | |
| Average | −0.211 | −2.350 | .021 | 0.399 | 0.133 | 8.979 | 1 | .003 | 1.490 | 1.148–1.934 | 0.243 | 0.121 | 4.058 | 1 | .044 | 1.275 | 1.007–1.614 |
| CpG 1 | −0.115 | −1.266 | .208 | 0.092 | 0.042 | 4.737 | 1 | .030 | 1.096 | 1.009–1.191 | 0.104 | 0.042 | 6.092 | 1 | .014 | 1.109 | 1.022–1.205 |
| CpG 2 | −0.147 | −1.623 | .108 | 0.143 | 0.076 | 3.503 | 1 | .061 | 1.153 | 0.993–1.339 | 0.076 | 0.073 | 1.101 | 1 | .294 | 1.079 | 0.936–1.244 |
| CpG 3 | −0.147 | −1.626 | .107 | 0.076 | 0.050 | 2.349 | 1 | .125 | 1.079 | 0.979–1.189 | 0.089 | 0.049 | 3.336 | 1 | .068 | 1.093 | 0.994-1-203 |
| CpG 4 | 0.070 | 0.763 | .447 | -0.026 | 0.109 | 0.057 | 1 | .811 | 0.974 | 0.786–1.207 | −0.022 | 0.108 | 0.042 | 1 | .837 | 0.978 | 0.792–1.208 |
| CpG 5 | −0.087 | −0.950 | .344 | 0.101 | 0.097 | 1.094 | 1 | .296 | 1.106 | 0.915–1.337 | 0.070 | 0.093 | 0.565 | 1 | .452 | 1.073 | 0.893–1.288 |
| CpG 6 | 0.031 | 0.340 | .734 | 0.073 | 0.078 | 0.890 | 1 | .346 | 1.076 | 0.924–1.254 | −0.213 | 0.134 | 2.517 | 1 | .113 | 0.808 | 0.321–1.051 |
| CpG 7 | −0.241 | −2.720 | .008 | 0.137 | 0.064 | 4.532 | 1 | .033 | 1.147 | 1.011–1.302 | 0.054 | 0.060 | 0.803 | 1 | .370 | 1.055 | 0.938–1.187 |
| CpG 8 | −0.212 | −2.369 | .020 | 0.260 | 0.074 | 12.436 | 1 | <.001 | 1.297 | 1.123–1.500 | 0.096 | 0.061 | 2.440 | 1 | .118 | 1.101 | 0.976–1.242 |
| CpG 9 | −0.065 | −0.709 | .480 | 0.039 | 0.048 | 0.649 | 1 | .421 | 1.039 | 0.946–1.142 | 0.045 | 0.047 | 0.911 | 1 | .340 | 1.046 | 0.953–1.148 |
Abbreviations: b, unstandardized regression coefficient; CI, confidence interval; df, degrees of freedom; OR, odds ratio; SNRI, serotonin-norepinephrine reuptake inhibitors; SSRI, selective serotonin reuptake inhibitors; β, standardized regression coefficient.
Dimensional response was defined as intraindividual changes in HAM-D-21 scores (in %) from week 1 to week 6 of antidepressant treatment; categorical response was defined as a reduction of 50% or greater from week 1 to week 6 in HAM-D scores; remission was defined as HAM-D-scores ≤7 after treatment week 6 (see Methods). Average methylation levels significant at P < .05 are bolded. For single CpGs, P-values significant after Bonferroni correction (Bonferroni-corrected P = .006) are bolded.
SSRI/SNRI Sample
In patients primarily and continuously treated with SSRIs or SNRIs, SLC6A4 average methylation also emerged as a significant predictor of HAM-D reduction after 6 weeks of treatment in the same direction as in the full sample (β = −.211, P = .021; overall model fit R2 = .139) (see Figure 1B). On a single CpG level, trendwise associations between CpGs 7 and 8 and HAM-D changes emerged (for details, see Table 2).
SLC6A4 Methylation and Categorical Treatment Response
Full Sample
Average SLC6A4 methylation significantly predicted categorical treatment response (HAM-D reduction ≥50% in week 6 relative to week 1) (P = .005, OR = 1.273; for full statistics, see Table 2). That is, each unit increase in methylation values increased the odds of being classified as a responder, and vice versa. Follow-up analyses regarding methylation at single CpG sites revealed significant associations between CpG 2, CpG 8, and—on a trend-level (after Bonferroni correction)—CpG 9 with treatment responder status (for statistics, see Table 2).
SSRI/SNRI Sample
In the subgroup of patients receiving continuous SSRI/SNRI treatment, average SLC6A4 methylation predicted categorical treatment response in the same direction (P = .003, OR = 1.490). Secondary analyses regarding single CpGs revealed a significant effect for CpG 8 and, while not withstanding surviving Bonferroni correction for multiple testing, suggestive effects for CpGs 1 and 7 (for statistics, see Table 2).
SLC6A4 Methylation and Remission Status
Full Sample
Average SLC6A4 methylation significantly predicted remission status (HAM-D ≤ 7) at week 6 (P = .010, OR = 1.237). That is, each unit increase in methylation values increased the odds of being categorized as a remitter and vice versa. Secondary analyses revealed significant effects in particular for CpG 1 and, on a nominally significant level, for CpGs 2, 8, and 9 (for statistics, see Table 2).
SSRI/SNRI Sample
In the group of patients receiving continuous SSRI/SNRI treatment, remitter status also was predicted by average SLC6A4 methylation (P = .044, OR = 1.275). On a single CpG level, although not surviving Bonferroni-correction for multiple testing, methylation at CpG 1 was most strongly related to remission status.
Discussion
In the present study, the predictive value of SLC6A4 promoter methylation on naturalistic antidepressant response was investigated in n = 236 patients with MDD in what is the largest sample investigated in this regard to date. Average SLC6A4 emerged as a significant predictor of dimensional antidepressant treatment response (relative reductions in HAM-D scores), categorical response (HAM-D reduction ≥50%), and remission status (HAM-D ≤ 7) after 6 weeks of antidepressant treatment, with relative hypomethylation being associated with nonresponse and nonremission, respectively. The predictive effect of SLC6A4 promoter methylation on dimensional and categorical treatment response as well as on remission status also held true in the more homogenous subgroup of patients continuously treated with SSRIs or SNRIs (n = 110), which supports the conclusion of SLC6A4 methylation to predict treatment outcome in relation to treatment with serotonergic antidepressants. The present results confirm a previous finding of SLC6A4 hypomethylation of the same region as investigated presently to predict impaired dimensional treatment response (Domschke et al., 2014) and extend it by providing additional evidence for SLC6A4 promoter methylation to also predict categorical responder as well as remission status in MDD.
Functionally, DNA promoter methylation is generally assumed to result in gene silencing (Suzuki and Bird, 2008), and methylation of the presently investigated region has been shown to go along with reduced 5-HTT mRNA levels (Philibert et al., 2007) as well as with decreased reporter gene activity in functional in vitro assays (Wang et al., 2012; Schiele et al., 2019). Accordingly, the observation of SLC6A4 promoter hypomethylation to confer impaired antidepressant response may be due to increased gene transcription and consequently decreased 5-HT availability in the synaptic cleft, which, on a mechanistic level, may counteract the serotonergic effects of serotonergic antidepressants and thus impair treatment response. However, the suggested mechanistic effect of lower methylation resulting in higher SLC6A4 expression to confer impaired treatment response is in contrast to previous studies reporting association of impaired antidepressant treatment response in carriers of the less active 5-HTTLPR S allele (see Serretti et al., 2007; Porcelli et al., 2012) as well as lower in vivo SERT binding with depression itself (Yeh et al., 2015) and nonremission (e.g., Miller et al., 2008). Both clinical (e.g., history of suicide attempts [cf. Yeh et al., 2015]) and biological factors (e.g., alterations on several other levels of the serotonergic system such as the 5-HT1A receptor [cf. Parsey et al., 2006], or different polygenic backgrounds [cf. Biernacka et al., 2015; García-González et al., 2017]) might constitute potential confounders, which could explain diverging findings. Also, in the present study, 9 CpG sites were selected for analysis as those have been subject to investigation in previous studies on 5-HTT DNA methylation with respect to stress-related measures and depression (Devlin et al., 2010; Alasaari et al., 2012; Kang et al., 2013; Schiele et al., 2019) as well as on the relation of 5-HTT methylation status with antidepressant treatment response (Kang et al., 2013; Domschke et al., 2014). However, other CpG sites further upstream or downstream towards exon IA, within exon IA, or even in intron I have not been considered in the present analysis but might crucially impact functionality (for a detailed schematic representation of CpG sites assessed by 49 studies on 5-HTT methylation: see Figure 4 in Palma-Gudiel and Fananas, 2017). Furthermore, trait and state characteristics of the serotonergic system as potentially differentially conferred by genetic and epigenetic factors might partly account for discrepancies across studies. Finally, given the present blood-based analysis, region specificity of SERT distribution in the brain as demonstrated in human positron emission tomography (PET) studies in the context of antidepressant treatment response (cf. Lanzenberger et al., 2012) cannot be and thus was not taken into account in the present study, but might be of high relevance in explaining the contradictory findings on the role of SERT activity in conferring antidepressant treatment response in the literature.
When interpreting the current results, it should be noted that in the full sample antidepressant treatment was administered in a naturalistic setting, which entails that, although all patients received serotonergic antidepressants as the primary medication, different drug classes were used. Additionally, in both the full sample and the SSRI/SNRI subsample, dosages varied between patients and were not held constant over the course of treatment, drug switches (initiation/discontinuation) were permitted, and co-medication with other pharmacological agents (e.g., mood stabilizers, atypical antipsychotics) was allowed, which may have confounded the present results. Of note, patients receiving a MAO inhibitor, which might act as an epigenetic drug (Binda et al., 2010), or valproic acid, which in its function as a histone deacetylase inhibitor indirectly also affects DNA methylation (Veronezi et al., 2017) (for review, see Boks et al., 2012), were not included in this study. Furthermore, given that the naturalistic study design did not allow for a placebo control arm, medication effects on the course of MDD cannot presently be dissected. Also, no healthy control group was investigated in parallel, precluding conclusions about SLC6A4 methylation as a trait or state marker of MDD. Given that no follow-up data were available, conclusions cannot be drawn regarding the long-term stability of treatment response and remission effects and the predictive quality of SLC6A4 methylation thereon. Also, information on smoking status was unavailable, which may have confounded the present results given that smoking has been found to affect global DNA methylation levels (Gao et al., 2015). However, a number of studies investigating SLC6A4 promoter methylation have observed methylation levels to be unaffected by smoking status (e.g., Domschke et al., 2014; Booij et al., 2015; Schiele et al., 2019). Therefore, while an effect of smoking cannot be fully excluded, it appears to be highly unlikely that the presents findings are a result of nicotine consumption. Additionally, although SLC6A4 methylation status was presently observed to be unaffected by 5-HTTLPR genotype, an interactive effect of genotype and methylation status on treatment response cannot be fully excluded. While inclusion of genotype or the interaction term of genotype × methylation did not increase, or even decreased the predictive value of the model, detecting cross-level interactions would require a much larger number of cases and therefore should be addressed in future studies in larger, sufficiently powered samples. Finally, a general limitation pertains to the fact that DNA methylation levels were derived from whole blood, which precludes conclusions about brain methylation status. However, accumulating evidence points to peripheral methylation patterns to constitute valid proxies for central processes (see, e.g., Ursini et al., 2011; Davies et al., 2012; Provencal et al., 2012). With regard to SLC6A4, employing PET, methylation levels measured in peripheral cells were shown to be inversely related to 5-HT synthesis (Wang et al., 2012) and 5-HTT availability (Drabe et al., 2017) in the human brain, supporting the notion of peripheral SLC6A4 methylation to possibly mirror central processes.
In conclusion, the present study applying a pharmaco-epigenetic approach to investigate the predictive quality of SLC6A4 promoter methylation on treatment response in MDD replicates and extends a previous pilot finding of SLC6A4 promoter hypomethylation to drive impaired response to antidepressant treatment and to furthermore be related to nonremission in a large independent but phenotypically comparable cohort of patients. The detrimental effect of SLC6A4 hypomethylation on treatment outcome might be conveyed by increased gene expression and, in turn, decreased 5-HT availability, which may counteract the effects of serotonergic antidepressants. The present results are hoped to inform future clinical decision-making and to enable early treatment modification towards a more personalized treatment of MDD.
Acknowledgments
We gratefully acknowledge the technical assistance by Dr M. Gottschalk, U. Götzinger-Berger, B. Günter, H. Lutz, P. Veratti, and U. Wering.
This work was supported by a grant by the Dr Robert Pfleger Stiftung (to K.D.).
Statement of Interest
K.D. is a member of the Janssen Pharmaceuticals, Inc. Steering Committee Neurosciences. B.T.B. is a member of Advisory Boards: Lundbeck, Janssen-Cilag; Consultant: National Health and Medical Research Council, Australia; Grant/Research Support: AstraZeneca, Fay Fuller Foundation, James & Diana Ramsay Foundation, National Health and Medical Research Council, Australia; German Research Council (DFG), European Union, Sanofi, Lundbeck; Honoraria: AstraZeneca, Bristol-Myers Squibb, Lundbeck, Pfizer, Servier Laboratories, Wyeth Pharmaceuticals, Takeda, Janssen, LivaNova PLC. V.A. has received compensations for his contributions as member of advisory boards and for presentations for the following companies: Astra-Zeneca, Eli Lilly, Janssen-Cilag, Lundbeck, Otsuka, Sanofi, Servier, and Trommsdorff. These co-operations have no relevance to the work that is covered in the manuscript. P.Z. has received speaker fees or honoraria for advisory board participation from Janssen Pharmaceuticals, Schwabe, Servier, and Neuraxpharm. All other authors have no conflicts of interest to declare.
References
- Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimäki M, Vahtera J, Kronholm E, Härmä M, Puttonen S, Paunio T (2012) Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. Plos One 7:e45813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffa A, Hohoff C, Baune BT, Müller-Tidow C, Tidow N, Freitag C, Zwanzger P, Deckert J, Arolt V, Domschke K (2010) Norepinephrine and serotonin transporter genes: impact on treatment response in depression. Neuropsychobiology 62:121–131. [DOI] [PubMed] [Google Scholar]
- Baune BT, Hohoff C, Mortensen LS, Deckert J, Arolt V, Domschke K (2008a) Serotonin transporter polymorphism (5-HTTLPR) association with melancholic depression: a female specific effect? Depress Anxiety 25:920–925. [DOI] [PubMed] [Google Scholar]
- Baune BT, Hohoff C, Berger K, Neumann A, Mortensen S, Roehrs T, Deckert J, Arolt V, Domschke K (2008b) Association of the COMT val158met variant with antidepressant treatment response in major depression. Neuropsychopharmacology 33:924–932. [DOI] [PubMed] [Google Scholar]
- Biernacka JM, et al. (2015) The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl Psychiatry 5:e553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A (2010) Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc 132:6827–6833. [DOI] [PubMed] [Google Scholar]
- Boks MP, de Jong NM, Kas MJ, Vinkers CH, Fernandes C, Kahn RS, Mill J, Ophoff RA (2012) Current status and future prospects for epigenetic psychopharmacology. Epigenetics 7:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij L, Szyf M, Carballedo A, Frey EM, Morris D, Dymov S, Vaisheva F, Ly V, Fahey C, Meaney J, Gill M, Frodl T (2015) DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. Plos One 10:e0119061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J (2012) Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol 13:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AM, Brain U, Austin J, Oberlander TF (2010) Prenatal exposure to maternal depressed mood and the MTHFR C677T variant affect SLC6A4 methylation in infants at birth. Plos One 5:e12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domschke K, Hohoff C, Mortensen LS, Roehrs T, Deckert J, Arolt V, Baune BT (2008) Monoamine oxidase A variant influences antidepressant treatment response in female patients with major depression. Prog Neuropsychopharmacol Biol Psychiatry 32:224–228. [DOI] [PubMed] [Google Scholar]
- Domschke K, Dannlowski U, Hohoff C, Ohrmann P, Bauer J, Kugel H, Zwanzger P, Heindel W, Deckert J, Arolt V, Suslow T, Baune BT (2010) Neuropeptide Y (NPY) gene: impact on emotional processing and treatment response in anxious depression. Eur Neuropsychopharmacol 20:301–309. [DOI] [PubMed] [Google Scholar]
- Domschke K, Tidow N, Kuithan H, Schwarte K, Klauke B, Ambrée O, Reif A, Schmidt H, Arolt V, Kersting A, Zwanzger P, Deckert J (2012) Monoamine oxidase A gene DNA hypomethylation - a risk factor for panic disorder? Int J Neuropsychopharmacol 15:1217–1228. [DOI] [PubMed] [Google Scholar]
- Domschke K, Tidow N, Schwarte K, Deckert J, Lesch KP, Arolt V, Zwanzger P, Baune BT (2014) Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int J Neuropsychopharmacol 17:1167–1176. [DOI] [PubMed] [Google Scholar]
- Drabe M, Rullmann M, Luthardt J, Boettcher Y, Regenthal R, Ploetz T, Becker GA, Patt M, Schinke C, Bergh FT, Zientek F, Hilbert A, Bresch A, Fenske W, Hankir MK, Sabri O, Hesse S(2017) Serotonin transporter gene promoter methylation status correlates with in vivo prefrontal 5-HTT availability and reward function in human obesity. Transl Psychiatry 7:e1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava M (2003) Diagnosis and definition of treatment-resistant depression. Biol Psychiatry 53:649–659. [DOI] [PubMed] [Google Scholar]
- Fava M, Rush AJ, Alpert JE, Balasubramani GK, Wisniewski SR, Carmin CN, Biggs MM, Zisook S, Leuchter A, Howland R, Warden D, Trivedi MH (2008) Difference in treatment outcome in outpatients with anxious versus nonanxious depression: a STAR*D report. Am J Psychiatry 165:342–351. [DOI] [PubMed] [Google Scholar]
- Gao X, Jia M, Zhang Y, Breitling LP, Brenner H (2015) DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-González J, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomic Consortium (2017) Pharmacogenetics of antidepressant response: a polygenic approach. Prog Neuropsychopharmacol Biol Psychiatry 75:128–134. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, Goldman D (2006) Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet 78:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iga J, Watanabe SY, Numata S, Umehara H, Nishi A, Kinoshita M, Inoshita M, Shimodera S, Fujita H, Ohmori T (2016) Association study of polymorphism in the serotonin transporter gene promoter, methylation profiles, and expression in patients with major depressive disorder. Hum Psychopharmacol 31:193–199. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, Shin IS, Shin MG, Yoon JS (2013) Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol Biol Psychiatry 44:23–28. [DOI] [PubMed] [Google Scholar]
- Kim JM, Stewart R, Kang HJ, Kim SW, Shin IS, Kim HR, Shin MG, Kim JT, Park MS, Cho KH, Yoon JS (2013) A longitudinal study of SLC6A4 DNA promoter methylation and poststroke depression. J Psychiatr Res 47:1222–1227. [DOI] [PubMed] [Google Scholar]
- Klauke B, Deckert J, Reif A, Pauli P, Zwanzger P, Baumann C, Arolt V, Glöckner-Rist A, Domschke K (2011) Serotonin transporter gene and childhood trauma–a G × E effect on anxiety sensitivity. Depress Anxiety 28:1048–1057. [DOI] [PubMed] [Google Scholar]
- Lanzenberger R, Kranz GS, Haeusler D, Akimova E, Savli M, Hahn A, Mitterhauser M, Spindelegger C, Philippe C, Fink M, Wadsak W, Karanikas G, Kasper S (2012) Prediction of SSRI treatment response in major depression based on serotonin transporter interplay between median raphe nucleus and projection areas. Neuroimage 63:874–881. [DOI] [PubMed] [Google Scholar]
- Lewin J, Schmitt AO, Adorján P, Hildmann T, Piepenbrock C (2004) Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics 20:3005–3012. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Alsabban S, Uher R (2011) The truth about genetic variation in the serotonin transporter gene and response to stress and medication. Br J Psychiatry 198:424–427. [DOI] [PubMed] [Google Scholar]
- Menke A, Klengel T, Binder EB (2012) Epigenetics, depression and antidepressant treatment. Curr Pharm Des 18:5879–5889. [DOI] [PubMed] [Google Scholar]
- Miller JM, Oquendo MA, Ogden RT, Mann JJ, Parsey RV (2008) Serotonin transporter binding as a possible predictor of one-year remission in major depressive disorder. J Psychiatr Res 42:1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Peña CJ, Kundakovic M, Mitchell A, Akbarian S (2016) Epigenetic basis of mental illness. Neuroscientist 22:447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odgerel Z, Talati A, Hamilton SP, Levinson DF, Weissman MM (2013) Genotyping serotonin transporter polymorphisms 5-HTTLPR and rs25531 in European- and African-American subjects from the National Institute of Mental Health’s Collaborative Center for Genomic Studies. Transl Psychiatry 3:e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Gudiel H, Fañanás L (2017) An integrative review of methylation at the serotonin transporter gene and its dialogue with environmental risk factors, psychopathology and 5-HTTLPR. Neurosci Biobehav Rev 72:190–209. [DOI] [PubMed] [Google Scholar]
- Parsey RV, Olvet DM, Oquendo MA, Huang YY, Ogden RT, Mann JJ (2006) Higher 5-HT1A receptor binding potential during a major depressive episode predicts poor treatment response: preliminary data from a naturalistic study. Neuropsychopharmacology 31:1745–1749. [DOI] [PubMed] [Google Scholar]
- Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H (2007) Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet 144B:101–105. [DOI] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A (2008) The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet 147B:543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli S, Fabbri C, Serretti A (2012) Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol 22:239–258. [DOI] [PubMed] [Google Scholar]
- Provençal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, Bennett AJ, Pierre PJ, Friedman DP, Côté SM, Hallett M, Tremblay RE, Suomi SJ, Szyf M (2012) The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci 32:15626–15642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush AJ, Kraemer HC, Sackeim HA, Fava M, Trivedi MH, Frank E, Ninan PT, Thase ME, Gelenberg AJ, Kupfer DJ, Regier DA, Rosenbaum JF, Ray O, Schatzberg AF; ACNP Task Force (2006) Report by the ACNP Task Force on response and remission in major depressive disorder. Neuropsychopharmacology 31:1841–1853. [DOI] [PubMed] [Google Scholar]
- Schartner C, Ziegler C, Schiele MA, Kollert L, Weber H, Zwanzger P, Arolt V, Pauli P, Deckert J, Reif A, Domschke K (2017) CRHR1 promoter hypomethylation: an epigenetic readout of panic disorder? Eur Neuropsychopharmacol 27:360–371. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Ziegler C, Holitschke K, Schartner C, Schmidt B, Weber H, Reif A, Romanos M, Pauli P, Zwanzger P, Deckert J, Domschke K (2016) Influence of 5-HTT variation, childhood trauma and self-efficacy on anxiety traits: a gene-environment-coping interaction study. J Neural Transm (Vienna) 123:895–904. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Ziegler C, Kollert L, Katzorke A, Schartner C, Busch Y, Gromer D, Reif A, Pauli P, Deckert J, Herrmann MJ, Domschke K (2018) Plasticity of functional MAOA gene methylation in acrophobia. Int J Neuropsychopharmacol 21:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiele MA, Kollert L, Lesch KP, Arolt V, Zwanzger P, Deckert J, Ziegler C, Domschke K (2019) Hypermethylation of the serotonin transporter gene promoter in panic disorder-Epigenetic imprint of comorbid depression? Eur Neuropsychopharmacol 29:1161–1167. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Gottschalk MG, Domschke K (2020a) The applied implications of epigenetics in anxiety, affective and stress-related disorders - a review and synthesis on psychosocial stress, psychotherapy and prevention. Clin Psychol Rev 77:101830. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Thiel C, Deckert J, Zaudig M, Berberich G, Domschke K (2020b) Monoamine Oxidase A hypomethylation in obsessive-compulsive disorder: reversibility by successful psychotherapy? Int J Neuropsychopharmacol 23:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiele MA, Thiel C, Kollert L, Fuerst L, Putschin L, Kehle R, Mahr M, Hauke W, Reinhold E, Gottschalk MG, Heinrichs M, Zaudig M, Berberich G, Domschke K (2020c) Oxytocin receptor gene DNA methylation – a biomarker of treatment response in obsessive-compulsive disorder? Psychother Psychosom In press. doi:10.1159/000509910. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Herzog K, Kollert L, Bohnlein J, Repple J, Rosenkranz K, Leehr EJ, Ziegler C, Lueken U, Dannlowski U, Pauli P, Arolt V, Zwanzger P, Deckert J, Erfurth A, Domschke K (2020d) Affective temperaments (TEMPS-A) in panic disorder and healthy probands: genetic modulation by 5-HTT variation. World J Biol Psychiatry In press. doi:10.1080/15622975.2019.1705999. [DOI] [PubMed] [Google Scholar]
- Schuebel K, Gitik M, Domschke K, Goldman D (2016) Making sense of epigenetics. Int J Neuropsychopharmacol 19:pyw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Kato M, De Ronchi D, Kinoshita T (2007) Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry 12:247–257. [DOI] [PubMed] [Google Scholar]
- Shi M, Sun H, Xu Y, Wang Z, Cui H, Wang C, Liu W, An G, Hu J (2017) Methylation status of the serotonin transporter promoter CpG island is associated with major depressive disorder in Chinese Han population: a case-control study. J Nerv Ment Dis 205:641–646. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9:465–476. [DOI] [PubMed] [Google Scholar]
- Tadić A, Müller-Engling L, Schlicht KF, Kotsiari A, Dreimüller N, Kleimann A, Bleich S, Lieb K, Frieling H (2014) Methylation of the promoter of brain-derived neurotrophic factor exon IV and antidepressant response in major depression. Mol Psychiatry 19:281–283. [DOI] [PubMed] [Google Scholar]
- Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, Sinibaldi L, Gelao B, Romano R, Rampino A, Taurisano P, Mancini M, Di Giorgio A, Popolizio T, Baccarelli A, De Blasi A, Blasi G, Bertolino A (2011) Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J Neurosci 31:6692–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronezi GM, Felisbino MB, Gatti MS, Mello ML, Vidal BC (2017) DNA methylation changes in valproic acid-treated hela cells as assessed by image analysis, immunofluorescence and vibrational microspectroscopy. Plos One 12:e0170740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialou V, Feng J, Robison AJ, Nestler EJ (2013) Epigenetic mechanisms of depression and antidepressant action. Annu Rev Pharmacol Toxicol 53:59–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Szyf M, Benkelfat C, Provençal N, Turecki G, Caramaschi D, Côté SM, Vitaro F, Tremblay RE, Booij L (2012) Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. Plos One 7:e39501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL (2006) Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry 11:224–226. [DOI] [PubMed] [Google Scholar]
- Yeh YW, Ho PS, Chen CY, Kuo SC, Liang CS, Yen CH, Huang CC, Shiue CY, Huang WS, Ma KH, Lu RB, Huang SY (2015) Suicidal ideation modulates the reduction in serotonin transporter availability in male military conscripts with major depression: a 4-[18F]-ADAM PET study. World J Biol Psychiatry 16:502–512. [DOI] [PubMed] [Google Scholar]
- Zhao J, Goldberg J, Bremner JD, Vaccarino V (2013) Association between promoter methylation of serotonin transporter gene and depressive symptoms: a monozygotic twin study. Psychosom Med 75:523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, Richter J, Mahr M, Gajewska A, Schiele MA, Gehrmann A, Schmidt B, Lesch KP, Lang T, Helbig-Lang S, Pauli P, Kircher T, Reif A, Rief W, Vossbeck-Elsebusch AN, Arolt V, Wittchen HU, Hamm AO, Deckert J, Domschke K (2016) MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Transl Psychiatry 6:e773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, et al. (2018) Monoamine oxidase a gene methylation and its role in posttraumatic stress disorder: first evidence from The South Eastern Europe (SEE)-PTSD Study. Int J Neuropsychopharmacol 21:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]