Abstract
High-throughput technologies have become essential in many fields of pharmaceutical and biological development and production. Such technologies were initially developed with compatibility with liquid handling-based cell culture techniques to produce large-scale 2D cell culture experiments for the compound analysis of candidate drug compounds. Over the past two decades, tools for creating 3D cell cultures, organoids, and other 3D in vitro models, such as cell supportive biomaterials and 3D bioprinting, have rapidly advanced. Concurrently, a significant body of evidence has accumulated which speaks to the many benefits that 3D model systems have over traditional 2D cell cultures. Specifically, 3D cellular models better mimic aspects such as diffusion kinetics, cell-cell interactions, cell-matrix interactions, inclusion of stroma, and other features native to in vivo tissue and as such have become an integral part of academic research. However, most high throughput assays were not developed to specifically support 3D systems. Here, we describe the need for improved compatibility and relevant advances toward deployment and adoption of high throughput 3D models to improve disease modeling, drug efficacy testing, and precision medicine applications.
I. INTRODUCTION
Both high-throughput screening (HTS) and three-dimensional (3D) bioprinting technologies have emerged in the last several decades, and these technologies have proven to be instrumental for the advancement of tissue modeling, drug development, and drug screening. HTS is well established within the pharmaceutical industry for compound and drug discovery and is relied upon as the primary method for experimentation prior to compound validation and preclinical animal model-based studies prior to clinical trials.1 However, the industry standard remains the use of two-dimensional (2D) cell culture for screening, as it has a long record of use and the cell lines employed have been well characterized. This is despite the fact that the use of 2D culture is limiting in that it is unable to capture in vivo-like cell-cell and cell-matrix interactions, and many cell types display different phenotypes and varying genomic profiles in 2D versus 3D.2–4 A recently proposed solution has been to use spheroid cultures as they are simple to implement 3D culture systems, but such cultures continue to lack cell-matrix interactions which may be vital for accurate tissue architecture and subsequent drug response and efficacy. One alternative method for employing 3D cultures is through the use of hydrogel biomaterials, which allow for chemical and biological manipulation to mimic the in vivo microenvironment of tissue. 3D cultures are complex and can be arduous to reproduce at the level required for HTS, and thus, only spheroids have been employed for some applications. Bioprinting allows for the creation of reproducible 3D cell cultures in an assay format for HTS. This has not always been the case, but in recent years, technology advances in both industry and academia have allowed for the advancement in bioprinting technologies to increase the speed and reproducibility of prints.
Researchers have been pursuing the use of bioprinting to create multiple, or parallel, 3D cultures to facilitate drug screening, toxicity, tissue and disease formation and progression, and precision medicine applications. Motivated to create more physiologically relevant structures for such studies, extrusion, inkjet, and laser-based bioprinting technologies have been developed. Commonly, using multiple cell types, unique patterns or structures, extracellular matrix (ECM) component additions, and well formulated hydrogels, there have been substantial improvements to 3D cell cultures and their utilization in tissue and disease modeling. As bioprinting allows for physiologically relevant architectures to be developed, it also has begun to support biofabrication of 3D tissue constructs and organoids in HTS formats. HTS is primarily focused on drug screening and toxicity studies, with additional interest in improving analyses of disease models and most recently precision medicine applications. With bioprinting as a tool, physiologically relevant models can be realized within these applications for the improvement of healthcare.
II. HIGH-THROUGHPUT SCREENING
HTS and automation within the pharmaceutical industry have become widely adopted capabilities required for the success of drug discovery and development, primarily established in the late 1980s.5 High-throughput assays are often directly related to metabolism, pharmacokinetics, and toxicology. Optimally, through these assays, the cellular response to candidate drug compounds can be determined and yield useful data, indicating if a compound is positively impacting the disease of focus, and, when tested on non-diseased cells, elucidate potential toxicity. After automated screening procedures are concluded and it has been indicated that a drug compound is efficacious, target validation and preclinical testing are carried out. HTS is predominantly carried out with 2D cell cultures and implemented in multi-well plates. In comparison to traditional screening, high-throughput differs in that it utilizes 96-well or greater arrays, small volumes (50–100 μl), and micrograms rather than milligrams of drug compounds and is fast and mechanically driven rather than performed manually.5 These capabilities enable the discovery of potential drug compounds from compound sets in the hundreds ranging to thousands at a high speed.
When considering HTS, it is important to understand the complex experimental design requirements and ensure that there are appropriate controls and methods for quantification. Successful designs include screens with counter screens, sequences of screens, and interpretation of results to avoid false positive or negative results.1 Specifically, considering pharmaceutical development, measures that are of importance when developing new compounds include efficacy, availability, persistence, safety, and practicality.1 Improvements in molecular biology, specifically genomics, have allowed for the creation of single cell-type disease models that can be well characterized. In turn, these disease models are used for in vitro 2D culture systems specifically in ultrahigh-throughput screening when screening for new drug compounds.1
Qualifications for useful HTS are well outlined and practiced within industry. To meet such demands, 2D cell culture has been primarily used as individual cell lines are well characterized and easy to use. Bioreactors or large culture containers and media are readily available, and the cell phenotype and genotype are reproducible on a scale suitable for HTS.4 For screening, cells are grown in multi-well (96-well or greater) tissue culture plastic well plates until confluent or near-confluent, after which screening studies are initiated. Results are often yielded through quantitative assay read outs, such as supernatant absorbance, fluorescence, or luminescence, and cell staining.1 However, it has been documented that 2D cell culture is not always adequate for the representation of complex diseases and tissue behaviors and that advancement to 3D models may be necessary.4,6,7 Traditional 2D cultures can be limiting when trying to replicate tissue-level physiology.8 Alternatively, 3D culture specifically allows for more nuanced control of cell-cell and cell-matrix interactions, mechanical properties such as stiffness and fluid flow, ECM composition, addition of biochemical factors, and modulation of tissue density—altogether allowing tailoring of the microenvironment to fit the tissue or organ of interest.4,7,9 The use of 3D culture over traditional 2D tissue culture has become broadly accepted in recent years as differences in the genotype, phenotype, and cellular behavior are apparent between the culture types.10,11 Each of these differences can contribute to cellular and tissue-level changes which affect the drug response, disease progression, and overall function.10 Thus, for applications directly related to compound screening and drug development, it is increasingly important that 3D culture be considered, allowing the potential to incorporate the characteristics of in vivo tissue to yield more representative models. The use of D culture demands that the functionality of tissue over single cells must be considered and, when performed in compound and drug development, may result in more human subject-mimicking data.12
Three-dimensional cell cultures and organoids can contain single or multiple cell types that are normally found within the target tissue.9 The ratio of cell types can be optimized to induce the tissue function which can be measured using organ-specific biomarkers or other assays.2,11,13 Tissues can be formed through simple cell-cell aggregation in hanging drop or round-bottom non-adherent culture plates, which yield spheroids, which can be deployed in studies as is or placed into hydrogels. Alternatively, 3D constructs may be created using hydrogels in which the cells are embedded within synthetic polymers or native ECM-derived components.14,15 3D cultures, if not formed through aggregation, are often created using biomaterials that suspend cells in 3D within polymer or protein-networked matrices. Biomaterial-based approaches have an advantage over spheroid cultures as they allow for greater control of the tissue microenvironment with regard to environmental and physical parameters, such as stiffness, addition of ECM components, and spatial organization of cell types.16 The biomaterials used for tissue and tumor organoids are selected based on particular properties for a given tissue type. Biomaterials can be tailored to present different porosities, elastic moduli, cell adherent motifs, and viscosities, each of which can drive more specific tissue design, resulting in more accurate cell and tissue functions.17,18 Common biomaterials for use with 3D cultures include collagen, hyaluronic acid, gelatin, and chitosan, among others, which can be deployed by a variety of biofabrication techniques for creating 3D structures.19,20 These can be used as hydrogels in which cells are encapsulated or as scaffolds in which cells are directly seeded into. Hybrid approaches, such as embedding aggregated tissue spheroids within hydrogels to form larger multi-colonies, and highly functional tissue construct models also exist.21
III. 3D BIOPRINTING TECHNOLOGY
In the past thirty years, biofabrication and 3D printing have advanced from the first patented device, a stereolithography apparatus, to low-cost bioprinting devices now widely available in research laboratories today.22 Tissue engineering itself is also a new area of study with its own origin dating back to approximately the same time period, with both technologies emerging in unison.22 Bioprinting can generally be defined as the printing of biological or bio-friendly material-based 3D structures.22,23 This encompasses materials printed containing cells, as scaffolds for tissue to reside or grow within, or materials to be placed within the body. At the inception of 3D bioprinting, materials of interest were those that would be able to act as scaffolds for cell culture and evolve to recreation of portions or entire organs.23,24 Printing of functional tissues and organs continues to be of interest as they could serve as alternatives to donor organs for transplant, thereby addressing the donor organ shortage and allowing patients to receive immune-matched organs with assumed lower risk of rejection. Although this is one major interest within the field of bioprinting—perhaps the “holy grail”—bioprinting has evolved to encompass the creation of 3D structures and patterns for miniaturization, biomimicry, and complexity of cell experimentation.22
In addition to simplified tissue fabrication, 3D bioprinting allows for an extensive study of cell behavior and the creation of new methods for experimentation. As the in vivo cell microenvironment is a complex 3D structure with numerous elements including multiple cell types, ECM, and pathways for oxygen and nutrient exchange, there is a high demand for 3D culture systems. Bioprinting is one of the most promising technologies for producing such 3D structures en masse. With the advancement of tissue engineering, scientists have become increasingly interested in cell-to-cell interactions, individual cell behavior based on chemical gradients, and substrate surface topography effects, all of which can be better understood and more replicative of the in vivo environment when in 3D culture. With these interests, it has been widely shown that 3D environments can better emulate the in vivo cell microenvironment in comparison to traditional 2D tissue culture environments.9 These 3D environments can be better modulated through the use of biofabrication technologies such as bioprinting which allow for construct stiffness, topography, composition, and architecture to be controlled. Such characteristics are minimally or not controlled in 2D tissue culture.26 In addition to the benefits yielded from 3D versus 2D culture, bioprinting also has the potential to support rapid prototyping and high throughput capabilities.27–29 By automating fabrication of 3D tissue constructs commonly deposited manually using pipets, experimentation and assays can be carried out at a faster rate with more consistency and increased sample sizes.
Bioprinting technologies have advanced significantly in recent years and now comprise a range of modalities, such as laser, inkjet, stereolithography, and extrusion printing.22 Laser bioprinting, similar to a common laser printer, is the deposition of the cell material guided by a laser beam in contact with the substrate creating dots of the printed material that once printed can form a continuous line or design.30,31 One such benefit of this form is that it can have precision up to 30 μm but is limited by low speeds in comparison to other printer modalities.32 Inkjet bioprinting, similar to inkjet printing, is actuated thermally, piezoelectrically, or mechanically using solenoid vales pulsed when printing is desired.33 Pressure is applied to the material reservoir which causes ejection from the print head. It is then deposited drop wise by the actuator mechanism to print the desired design and can have multiple reservoirs. Extrusion printing, also considered robotic dispensing, prints an unbroken stream of materials when a force is applied to a cartridge of ink.34,35 The force is applied most commonly via pneumatic pressure, mechanical screw pistons, or screws with pneumatic pressure.22 Many different print heads can be used at one time via this modality each with different solutions or mixtures. This method of bioprinting carries the highest printing speed of the three methods and allows for a high cell density.36 Extrusion printing has also been pursued as a low-cost technology. Considering all modes of bioprinting, limitations with regard to high-throughput screening include the speed of printing and resolution. As many high-throughput applications require the use of 384-well plates or greater, speed in which precise designs, more complicated than those simply made of a single bioink or cell suspension, can be created is limited. Bioprinting is currently better suited for use in lesser throughput, larger scale models in 96-well or fewer plates when complex designs are created. To address these challenges, bioprinters and bioinks are being rapidly improved to meet the growing needs of the biomedical industry outside of the academic laboratory setting. These limitations do not exist when considering simple designs with just one bioink containing cells. Both large and small companies have realized the opportunities available and are creating extrusion and inkjet printers for both research and commercial use. Bioprinters for laboratory research are available commercially at low-cost and are accessible around the world, no longer requiring researchers to build their own devices for experimentation.36 However, many labs do continue to create their own printers with the intention of continued improvement to hardware parameters, features, and resolution. In addition to the standard bioprinting modalities described above, novel bioprinting systems have been developed to challenge the status quo and introduce new approaches within the field. Micromirror, microfluidic, direct-write loading, magnetic, and light-assisted printing, as well as the use of sacrificial or hybrid support materials, are all examples of new methods for bioprinting.37–43
The use of bioprinting has been primarily limited to single models or experiments with a focus placed on the structure or design rather than throughput. The utility of 3D bioprinting is two-fold. It first allows for the actual deposition of 3D structures, but it can also be leveraged for automation of bioink-based 3D cell cultures or tissue construct biofabrication. Although 3D bioprinters have the capabilities to produce complex structures, they can also be leveraged to automate and upscale simple structures to yield large datasets using 3D culture systems rather than traditional 2D cultures typically employed in high throughput screening. Such throughput can be created via multiple means and includes utilizing multi-well plates for printing within each well, fluidic devices with multiple or parallel chambers for individualized experiments, and printing of unique designs with boundaries within one plate or device. It should be noted that there remain a number of hurdles to overcome to fully realize high throughput screening with 3D cell models, both on the biofabrication and assay implementation ends of the process.
Challenges currently faced by the integration of 3D cell models and bioprinting into high throughput screening are multi-fold. In creating an appropriate model, the appropriate microenvironment for cells must be in place, as well as the measurement of relevant outputs via assays and quantitative measures. These are challenges that must be overcome when moving from 2D to 3D cultures as many of the techniques previously used do not appropriately translate to additional dimensions. Appropriate modeling is addressed below as much research effort has been made towards both improving tissue and disease models and creating them for throughput applications.
IV. BENCH TOP CONSIDERATIONS
The utilization of 3D cell culture has become well established within organ and disease modeling to create more physiologically relevant tissue in comparison to 2D cultures.44,45 Historically, 2D culture has been used to better understand cellular mechanisms, organ development and function, and disease modeling, amongst many other topics. However, a more holistic approach to modeling has been taken by considering the cell microenvironment and the ECM components that tissue is composed of. Importantly, 3D models have been shown to be express more representative biomarkers related to drug and stress responses in comparison to 2D.2,46,47 For example, in tumor specific studies, cells have presented epithelial to mesenchymal transition markers which indicated more mesenchymal, invasive behavior when grown in 3D models in comparison to 2D.2 These behaviors are more closely aligned to what would be expected in patient tumor cell populations. With each tissue system being unique, considerations for 3D models include cell types, growth factors, ECM proteins, stiffness, and porosity of 3D materials to list a few. Such conditions allow cells to behave in a more in vivo like manner by facilitating remodeling, cell adhesion, and migration.8,48,49 Collectively, it is important to consider not only the dimensions of the cell culture but also the actual components comprising the cell culture playing a role in replicating each tissue type or disease. In pursuing the use of bioprinting, understanding of the cell microenvironment and the components required to recapitulate the disease state must be greater than current models and methods and be of greater throughput.49 The cell microenvironment is vital in maintaining the appropriate cell genotype and phenotype. The importance of the microenvironment has been well discussed in the previous literature, and it is required for creating tissue and disease models that are representative of in vivo conditions.50–52 The drug discovery and development process can measurably benefit from the integration of 3D bioprinting, from validation through clinical trials, but the qualification of a good model must be benchmarked (Fig. 1).
FIG. 1. Interface between pharmaceutical and diagnostic development and 3D bioprinting.
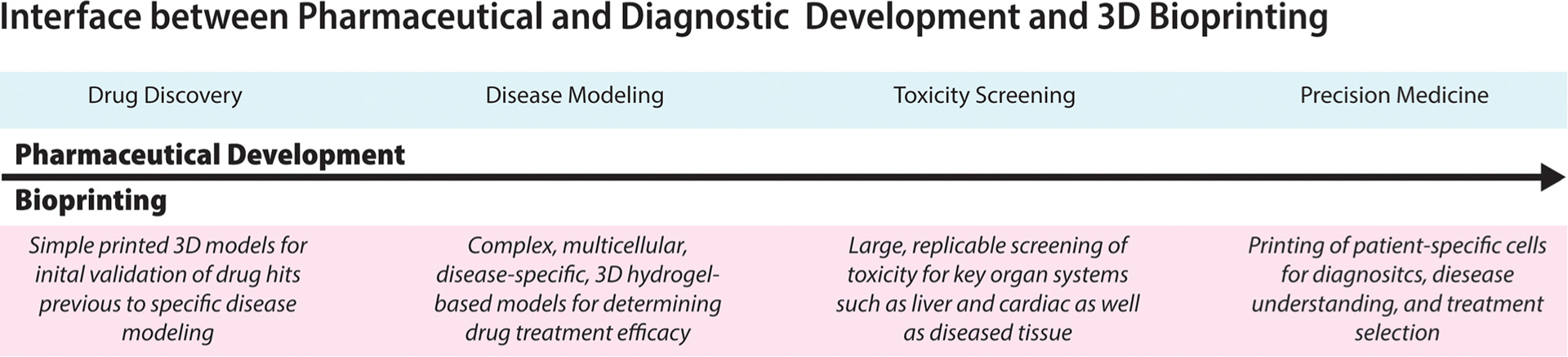
Overview of the overall drug development process from initial drug screening of drug candidates to approval and use in precision medicine applications. Over the course of pharmaceutical development (blue), corresponding bioprinted models are outlined (red). In development, initial drug screening can take place using simple 3D models in comparison to 2D to increase the number of accurate “successes” found in HTS which deem the drug useful. When drugs are found to be efficacious through initial HTS, more complex disease models can be created in large replicate to better understand the drug mechanism of action and potential dose response. Using organ specific models, such as liver and cardiac, the toxicity of the drug can be tested on complex 3D models to more completely capture the organ response. Once drugs have been screened and tested on relevant disease models and toxicity is profiled, their use in personalized or precision medicine applications can be determined to select the appropriate treatment from approved drugs.
A. Bioinks
When specifically considering bioprinting, environmental factors specific to bioprinting should be considered. Bioinks are broadly described as printable polymer- or protein-based hydrogels and more specifically defined as materials used in 3D bioprinting which allow for spatial control and patterning of cells and biocompatible materials. These materials include hydrogels, which are polymer-based materials that are commonly crosslinked from aqueous solutions to non-soluble macromolecular networks.53,54 Currently, common bioinks are composed of polyethylene glycol (PEG), collagen, alginate, hyaluronic acid, gelatin, fibrin, or polycaprolactone (PCL).53,55 Each bioink has unique properties that can either benefit or hinder printability or tissue viability and function, and these considerations should be adequately balanced to produce optimal tissue constructs. With the exception of collagen, each biomaterial described above requires the addition of a crosslinker, enzyme, or chemical modification to induce crosslinking. Furthermore, crosslinking speed is dependent on the chemical reaction taking place and can vary significantly.56,57 Both crosslinking components and time required for crosslinking can adversely affect cells and are often balanced with the benefits of bioink biocompatibility.58,59 Additionally, not all materials provide physiologically relevant cell adhesion points and can force reliance on cell-cell interactions (e.g., alginate and PEG).60,61 Although this is an improvement over standard 2D culture, it is still limiting and may not be fully representative of the cellular microenvironment of the target in vivo tissue, in which cells dynamically interact with a variety of ECM components and their cell adhesion motifs. To address this, scientific advancements have been made towards improving such hydrogels which include integrating chemically modified peptides with cell-oriented adhesion sites such as arginylglycylaspartic acid (RGD).62,63 Tissue-specific studies have also been conducted by considering the addition of multiple, different peptides in comparison to using only RGDs to represent various components within the ECM.64 These advancements are measurable as materials that allow for more traditional, ideal cell-matrix interactions can often times be challenging to print—an example of this is collagen.61 Although it is an ideal material for supporting cell-matrix interactions, it is a poor bioink because it has time-, pH-, and temperature-sensitive crosslinking.57 These factors must be collectively considered when selecting a bioink for bioprinting. Additionally, time and temperature can play a major role in bioink selection not only for collagen inks but also for each of those mentioned previously, as they are each sensitive to changes in the environment with regard to gelation kinetics.61 When bioprinting, temperature can commonly be controlled within the printing environment. This temperature, however, also has to be balanced with the demands of cells to maintain viability. Commonly sensitive to extreme heat or cold for extended times, the bioprinting environment must cautiously meet the needs of both the bioink and the cells being used. Considerations must be carefully made when selecting a bioink for use in replicative, cell-based studies but when done correctly can lead to useful bioprinted tissue constructs.
B. Cell microenvironment for tissue recapitulation
Creating a model that accurately recapitulates the tissue of interest is important in progressing drug development beyond the point at which it currently lies. Physiological accuracy of in vitro models can be measured through both the phenotype and genotype of the cells in the system in comparison to those in vivo.65,66 To better achieve in vivo-mimicking cultures, many components must be in place to create the ideal microenvironment often beyond 2D and include ECM components, mechanical and physical parameters, cell types and combinations, and organization of these.52,67 Independently, each will create unique microenvironments and must be carefully orchestrated to produce accurate tissue and disease models.
The ECM is broadly composed of proteins and polysaccharides secreted by cells to create niche microenvironments that structurally and biochemically support cells to create tissue.68 ECM components commonly include collagen, laminin, fibronectin, hyaluronan, proteoglycans, glycosaminoglycans, elastin, tenascin C, and others.69 ECM components such as these vary between tissue types and disease state. For example, native liver is composed of approximately 0.5% collagen type-1, but in a fibrotic state, it is 5–8 times greater. When creating 2D or simple 3D models such as spheroids, these components are not easily incorporated, and when added, it is often done so through surface coatings or as additions as soluble factors in media, in which case their availability to cells may be limited by diffusion. In utilizing ECM-based bioinks, many of these components can be added directly into the hydrogel through mixing, physical immobilization, or chemically added through various bonds (covalent, ionic, or hydrogen bonding, for example). ECM components have notably been found to affect cell proliferation, apoptosis, differentiation, and metabolic behavior.68,69 Each of these is key cellular output metrics for quantitative assays, and thus, the incorporation of ECM components may directly impact results, making them more representative of the in vivo response. These components have found themselves to be so important that the use of decellularized, solubilized tissue ECM is being used to create new bioink and hydrogel formulations.35,70,71
Mechanical and physical cues independent of chemical properties have been well studied initially in 2D cell culture and in 3D cell culture as hydrogel development has advanced. Properties include stiffness, microstructure (material spacing and pore size), and material size related to the contact area.72–74 Matrix mechanical properties have been well defined and have been directly related to many areas of study such as cell migration, differentiation, focal adhesion, and population expansion amongst others.74 Each of these outputs is directly related to cell type, but one notable example is that of mesenchymal stem cells. Matrix stiffness has been shown to directly impact the softer materials being neuro- or adipogenic and stiffer materials being osteogenic.75,76 Attention to these properties is essential for creating representative matrices and must be considered when selecting a bioink for bioprinting models for tissue and disease screening.
The cell types being used within the cultures must also be carefully considered for creating physiologically accurate models. Although disease models must be indicative of the specific cell type in which the disease inflicts, the surrounding cells and microenvironment produced will have substantial effects on the disease tissue behavior.44 The cell type is predominantly dependent on the type of tissue being modeled, and many cell types may be required to best replicate the tissue. In vitro, it is often times impossible to incorporate each of the cell types necessary to make an in vivo tissue. To make the most relevant model while still maintaining a reasonable number of cell types, the quantitative outcomes being measured should be considered and directly related to which cells will produce the results. From this point, other cell types that may impact that results should also be considered. This is in addition to the ECM components that have been added. Bioprinting also allows for unique structures and purposeful organization of tissue components to take place. Using multiple print heads, complex structures with zones of varying components can be created to produce disease models that are more representative of the in vivo condition.
V. APPLICATIONS AND INDUSTRIAL INTEGRATION
An important goal of in vitro models is to better determine the impact of drug treatment on diseased and healthy cells to determine drug efficacy and potential off-target effects. As previously highlighted, in many scenarios, 3D models have been well characterized as advantageous over traditional 2D culture assay. Through the use of 3D models, potential compounds can be screened in a more in vivo-representative setting, therefore increasing the chance of success reducing the overall cost of drug development. Unfortunately, many drugs have passed through both in vitro and in vivo testing to reach the market, only to have off target effects or toxicities that had not previously been characterized. The use of 3D cell models not only offers the ability to test the targeted human tissue but also allows for the off target and toxicity effects in other organs such as the heart, lungs, and liver to be studied. This advantage over simple cell culture models and animal models is being able to prevent patient toxicity and drug recall. Models for this type of testing have been well developed and are often referred to as body-on-a-chip or organ-on-a-chip technologies due to their small size but complex cellular construction, comprising multiple tissue types in a single system.25,77 These models, while extremely useful, can become limited during scale up processes due to the low throughput nature of their current manufacturing in the lab.78 However, with the advances being made in the precision, accuracy, and scale of 3D bioprinters, they may become well suited for use throughout the drug development pipeline and may offer solutions for many current challenges.
A. Drug optimization and disease modeling
In the drug development pipeline, prior to the use of preclinical animal models, validation of a drug compound on more complex models is done to better understand drug efficacy and mechanism. With initial HTS drug discovery utilizing 2D cell culture to allow for rapid screening, validation is the step in which drug efficacy is characterized and a safety profile is created from which animal studies can be pursued. At this step, drug formulations can be altered and cellular and system level biological mechanisms are studied to more comprehensively understand how the drug works. Also known as lead optimization, many drug compounds are not pursued past this point in the drug development pipeline. Such work is carried out using more complex disease models, in either 2D or 3D, to study the treatment beyond what was performed in HTS. This portion of drug development has been characterized as slow, low-throughput and often yields few usable drugs after both time and money have been spent.79
Therefore, better disease models are crucial for better understanding behavior, development, treatment, prevention, and cure of disease. Currently, in industry-based drug development, models are often leveraged from academic research and innovation teams to best replicate each of the diseases being studied.79 When increased in throughput via bioprinting, they could be used before animal testing to optimize dosage and better understand toxicity and once drugs have been best formulated in conjunction with animal testing to confirm the results. Such models range from genetically altered cell lines expressing disease-related genotypes or phenotypes to genetically modified animals which can best approximate the disease. These models have been able to yield impressive results but remain limited in that they are unable to recapitulate symptoms of the patient disease due to lack of tissue level effects in single cell type models and the inability of animals to produce human responses. For improved cell culture specific disease modeling, the use of 3D culture is being leveraged. As highly tunable systems, researchers utilize 3D culture systems to create disease models that can be referred to as organoid disease models or disease-on-a-chip models when combined with microfluidic devices. These models can incorporate many external features such as fluid or air flow and a combination of tissue types to model the body wide disease. Due to their complexity, advanced technology is required to create these culture systems that are both reproducible and complex enough in design to replicate the disease. Bioprinting has been able to meet these demands by creating 3D cell-hydrogel constructs in a reproducible manner yielding complex, high-throughput disease models. These models allow for modulation of extracellular components and stressors, drug treatments, and disease progression studies each valuable for better understanding and ultimately treating the disease. Nano3D Biosciences, a privately owned company, has created a system for high-throughput bioprinting spheroids for disease modeling. Utilizing magnetic 3D bioprinting, their commercially available kits are able to produce spheroids in up to 384-well plates. Publications based on their 3D bioprinting system technology include disease models related to cancer and toxicity screening amongst many others.27,80,81
Disease modeling utilizing bioprinting for HTS has been widely studied to ensure that the extra measures required to produce 3D models over 2D are more representative of patient disease. One such study has been conducted by Hou et al. regarding the development of primary pancreatic organoids for drug screening.27 Using the previously described Nano3D Biosciences magnetic 3D bioprinting system and non-adherent culture plastic, researchers were able to create organoids in 384-well and 1536-well plates. Organoids were formed using patient-derived pancreatic and colorectal carcinoma cell lines with cancer associated fibroblasts and characterized using anticancer agents in a high-throughput manor. Three-dimensional assays were done in parallel with 2D, and the results indicated that 3D ECM based resistance to cytotoxicity was present as researchers expected. The resistance varied between cell lines, and authors created a “resistance factor” to account for the difference in the IC50 of 2D and 3D models. Phenotypically, it was found that 3D models more closely represented the in vivo tumor environment. Combined, researchers were able to show the 3D models to be preferential over 2D for chemotherapy screening.
B. Drug toxicity
Considering both the systemic (toxic) response and disease response previous to patient implementation is critical. When developing new drug treatments, toxic effects must be well characterized and monitored during animal studies. The opportunity to more effectively model these effects previous to animal studies may allow for drug to be modified or withdrawn previous to in vivo modeling if toxic effects are measured (Fig. 2). In conjunction with animal models, toxic effects can be compared to make aware any discrepancies between the two. Such short-comings may indicate areas of focus when moving into patient clinical trials. On considering toxicity, the liver is specifically important because of its role in metabolizing drugs. As the primary organ responsible for metabolism, it breaks down drug compounds, resulting in more or less active drug variants.82 In the process of metabolizing the drug, the liver is exposed to the drug compound and its by-products after metabolism which may be toxins.82 Liver toxicity can result from over-exposure to toxins, leading to toxic hepatitis, inflammation of the liver, and eventual cirrhosis if exposed over a long period of time.83 These side effects further lead to what is known as drug-induced liver injury which results in liver failure, permanent cirrhosis, and hepatocellular carcinoma, with each being life-threatening condition.83 For these reasons, liver metabolism of drug (known as pharmacokinetics) and liver toxicity are heavily investigated when developing new compounds for drug treatment. Currently, 2D culture or 3D spheroids are utilized within drug discovery and screening for toxicity after which animal models are used although widely recognized for being misrepresentative of human metabolism due to a lack of overlapping cytochrome p450 enzymes across species.84,85 The complexity of the liver combined with the substantial role it plays in drug metabolism has created a demand for physiologically improved liver models capable of being scaled for use within HTS. In bioprinting specifically, many novel approaches have been taken to improve liver models for HTS.85,86
FIG. 2. Drug toxicity screening—Current vs integrated with 3D bioprinted models.
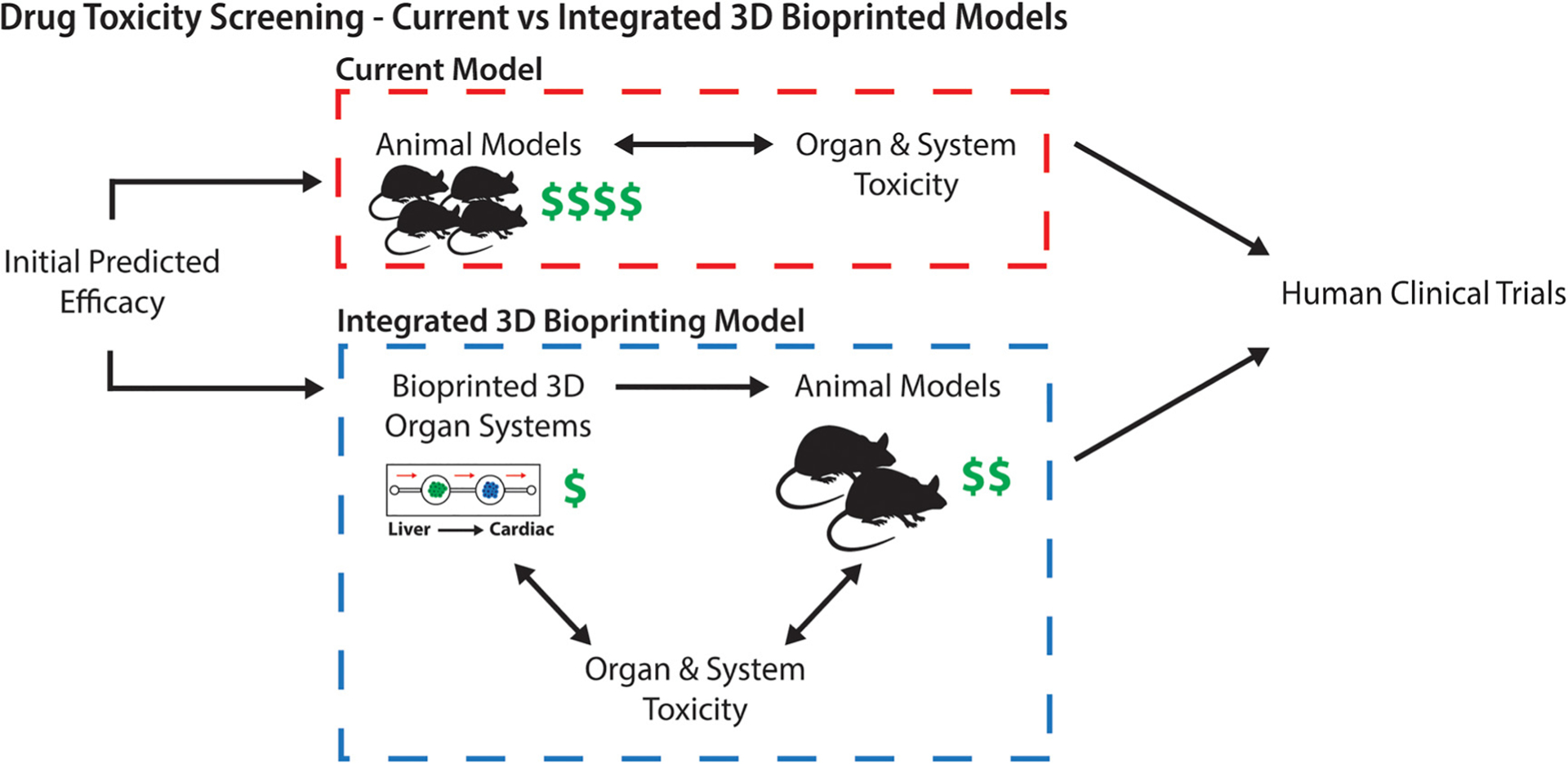
Drug toxicity screening is currently carried out through the use of animals which are not always representative of the human response, can require large quantities of animals, and can be costly. Through an integrated system utilizing representative human liver, cardiac, and other target tissue organoids, toxicity can be better predicted and requires fewer animals to determine drug toxicity for human clinical trials.
Nguyen et al. have utilized bioprinting to create 3D primary liver tissues for the study of the organ-level response to drug induced toxicity.85 Composed of primary human hepatocytes, hepatic stellate cells, and human umbilical vein endothelial cells (HUVECs), the 3D models were formed using a commercially available extrusion bioprinter printing directly into 24-well transwell culture inserts. The model had two regions, the center in which the parenchymal cells were included and a border in which the non-parenchymal cells were printed. Cells were supported over the course of 28 days during which time metabolism and toxicity studies were conducted. The results from toxicity studies were compared to the current industry standard, 2D hepatocyte cultures. The 3D bioprinted structures yielded tissue and cell-type specific responses above simple cytotoxicity results from 2D culture and allowed for histological assessment over time. The studies conducted have been able to demonstrate the importance of a 3D culture environment and the utilization of many tissue specific cell types rather than the use of only the primary cells to be affected.
Ma et al. have also leveraged 3D bioprinting to deterministically pattern human induced pluripotent stem cell (hiPSC)-derived hepatocytes to create a biomimetic model.87 Trying to mimic the complex microarchitecture of the liver, a lobule design was printed using a Gelma-based hydrogel and custom digital light-processing based 3D bioprinter. This unique bioprinting system combines digital masks containing specified designs with light sensitive biomaterials to produce light-polymerized structures as small as a micrometer in width.88 A triculture consisting of hiPSC hepatic cells, HUVECs, and adipose-derived stem cells was used for printing and compared to hiPSC hepatic cells in 2D culture. It was found that the bioprinted liver constructs had greater morphological organization, higher liver-specific gene expression, and increased metabolic product secretion in comparison to 2D, proving advantageous over industry standard models.
In addition to liver models, cardiac models are also important for considering cardiotoxicity which can be measured through the change in the function (i.e., beating rate) and potentially cell death. Examples of such toxicity have been measured in 3D cell culture models and show an appropriate response to drugs both toxic and non-toxic.21 Importantly, toxicity manifests itself in different ways in different tissues. This understanding makes it ever more important that drug testing is conducted on healthy tissue models that accurately represent the dynamic tissue system to determine potential toxicities.
Organovo, a publicly held company working within the pharmaceutical industry, has produced multiple tissue-engineered, bioprinted high-throughput models for drug screening including a liver model for toxicity testing.89 The product, exVive3D™ Human Liver Tissue, was specifically launched for use by pharmaceutical companies in their investigation of toxicity of new drug compounds in development and has been used as such.90 Using primary hepatocytes, stellate cells, and endothelial cells, a hexagonal shape was printed and after 60 h of incubation, microcapillaries are formed. Organovo has conducted toxicity screenings on drugs known to be either toxic or non-toxic to the liver and has been able to yield an appropriate liver response from their systems.89,91 They have also created models for cancer disease modeling and screening. Using multiple cell types and relevant ECM components, the models are bioprinted and allowed to form microcapillaries before being tested with chemotherapeutics from which results can be drawn which are given in Ref. 91.
C. Precision medicine
Advancement in technology, as well as increased public interest in precision medicine, has amplified the need for patient specific models. Precision medicine as a current clinical application is the tailoring of therapy based on a patient’s genetic information.92 Closely tied to other symptoms and disease pathologies present, the genetic information acts as another tool for which clinicians are able to use in determining patient treatment. Specifically, in terms of cancer, this means targeting specific mutations that may be directly related to treatments.92 However, as is the case with cancer, not all mutations are related to treatment but can be informative of disease behavior as they are primary drivers of pathology (Fig. 3). The use of 3D cell culture models in this case allows for cells from the patient to cultured and is expanded to study the disease ex vivo while still maintaining in vivo genotype.93,94 Using the patient’s own cells, 3D bioprinting can be leveraged to create a large quantity of patient specific disease models for use in parallel with clinical trials or independently through precision medicine initiatives. Broadly considered a two-fold initiative, precision medicine aids patients immediately in the short term and allows for an amount of data to be collected for long term studies of disease progression and drug treatment response.95 Thus, precision medicine is defined as individualized diagnosis and treatment utilizing strategies for targeting patient or disease specific genetic, proteomic, and phenotypic characteristics.7,96 Vital for patient-oriented diagnosis and treatment success, 3D cell culture models have been leveraged to grow and expand patient cell population in culture to observe their behavior, progression, and response to drug treatments.97 These models require a minimal number of cells to recapitulate the in vivo microenvironment and can be used for many applications to determine primary cell and patient results.21 Utilizing methods and approaches described previously, patient cells can be used in place of commercially available cell lines to create personalized models more representative of the patient specific disease. These studies are advantageous over disease models using cell lines as each patient has unique mutations that allow insights into the genetic variation, cell type mixtures, and patient-specific variations.7 This reduces generalization of the disease and gives researchers and patients a more individualized understanding of their specific disease state. Unlike today, these models would allow for clinicians to determine if the patient is an appropriate candidate for the study and further if they are at increased risk during the clinical trial for adverse outcomes. Additionally, such models may yield substantial financial benefits as they may reduce poor patient outcomes and allow for improved monitoring of disease progression and response.
FIG. 3. Precision medicine for diagnostics and treatment prediction.
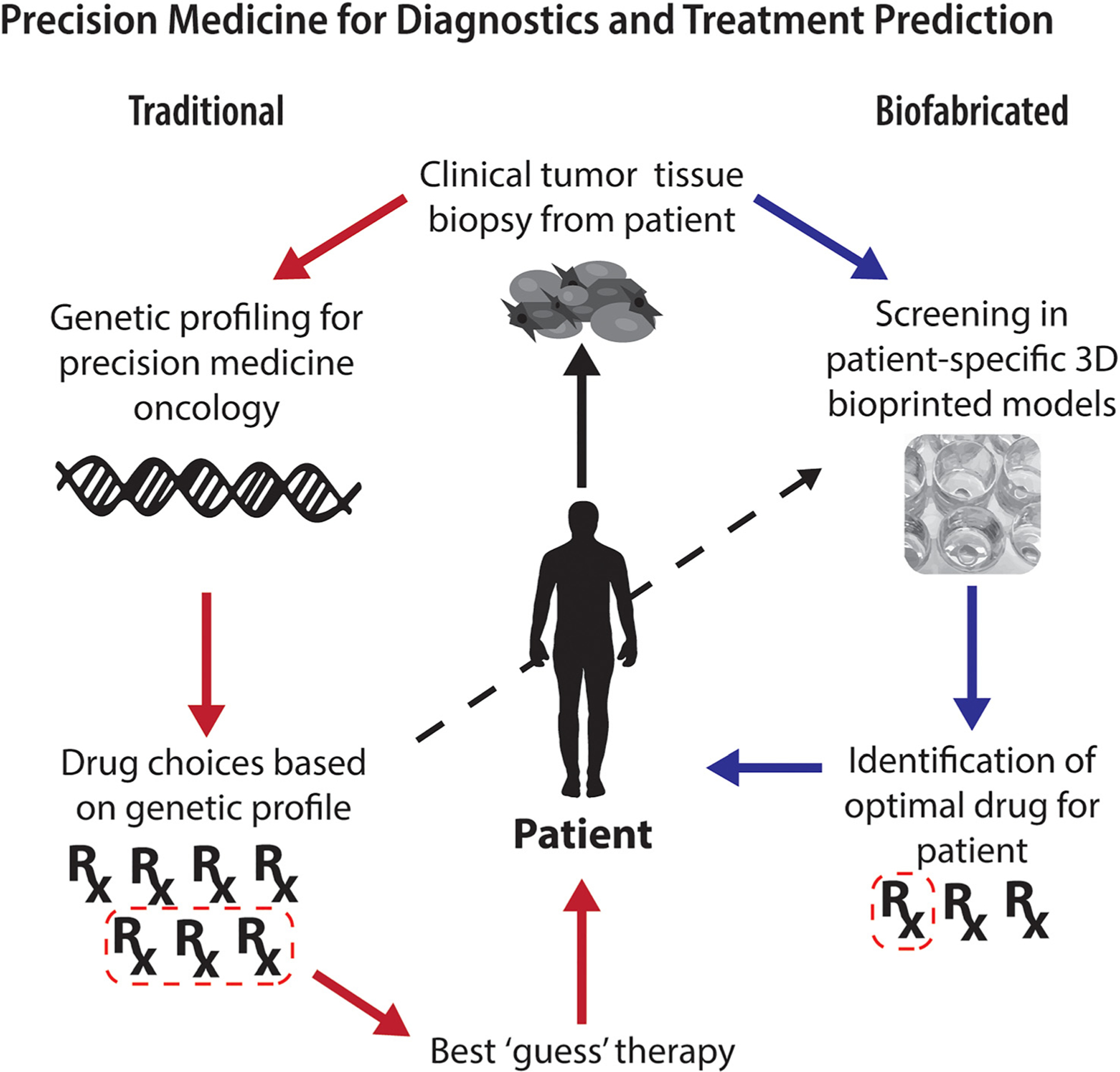
Utilizing 3D culture models via bioprinting will allow for a large number of patient specific samples to be tested for drug efficacy to determine the best treatments based on the response and not genetics alone. Although still in its infancy, traditional methods for precision medicine include collecting DNA from patient samples, carrying out genetic characterization, reducing drug selection through mutations, and finally selecting a drug based on clinician best guess. Through the use of biofabricated 3D cell cultures and bioprinting, drug screening can be carried out on patient-specific models to determine best treatments.
Current research initiatives, while still in their infancy for precision medicine, are step-wise as patient cells are isolated and integrated into the model system, and subsequent experiments are carried out. For in vitro 3D cell culture, tissue is isolated directly from the patient and then processed to further have single cell separation and ECM recovery. Cell isolation is carried out using diseased tissue resections or biopsy from which many cell types and ECM components can be recovered. Models that are unable to acquire tissue from patients may use hiPSC isolation methods by collecting easy to isolate cells from patients, de-differentiating the cells into hiPSCs, and then differentiating the cells into the desired diseased cell types for experimentation. This method of culturing patient cells can have its own challenges due to the nature of hiPSCs which often have variability within the differentiation process, and the results can be unpredictable to unrepresentative of the desired disease state.98 When considering high throughput processes, however, it is important to remember the scale in which cell production must occur. For high throughput 3D bioprinted precision medicine applications, patient cell expansion must be done to allow for many reproducible 3D culture systems to be made at one time for experimentation. Using patient diseased tissue or biopsy, cells can be isolated and expanded in 2D culture until there is great enough volume to carry out 3D experimentation.99,100 Expansion of patient cells from tissue can be challenging as it has been found that not all cells will adhere to tissue culture plastic which will limit cell yield and potentially the cell type populations that are preserved.94 This has created the opportunity to take advantage of hiPSCs for culture expansion prior to differentiation to yield millions of cells to be differentiated into the desired diseased cell type.101,102 Both of these methods are limited due to the amount of time required for expansion and the potential to have non-representative populations but remain as best options. In industry as in research, precision medicine is a new but growing field in which clinicians, patients, and researchers are invested. For both predictive disease modeling and drug screening capabilities, there is new found engagement in leveraging technologies to make precision medicine possible for a large audience.
VI. CONCLUSION
In the near future, advances in both 3D bioprinting and HTS technologies will continue to allow for more complex, representative models to be utilized for drug screening, disease modeling, and precision medicine. 3D bioprinting is currently limited by speed and print size, but with mechanical and chemistry-based advances, these areas continue to improve and will become more suitable for high-throughput applications. Additionally, HTS is currently limited by traditional well plates; this too is an area in which 3D bioprinting will play an important role. Microfabrication techniques currently used to make on-chip, multi-well, or channel-containing devices can be leveraged to make large-scale multi-well devices. Bioprinted cultures could then be placed within each of the wells and, with modified HTS devices, could produce drug and compound efficacy results previously discussed. It is also important to consider that in advancing HTS with 3D bioprinted cultures, the quantifiable outcomes will also need to be improved. Current outcomes are quantified using 2D models and rely on monolayers or cell suspensions in media to yield data. These outputs will need to be adapted for 3D cell culture and consider the additional components within the culture and the limitations and advantages they offer. Although these new processes may be radical and require updates to previously well-defined protocols, they are proving necessary for the creation of physiological relevant tissue models in drug development and screening that better recapitulate human physiology and human drug responses.
Footnotes
Note: This paper is part of the Special Topic on 3D Bioprinting: Physical and Chemical Processes.
REFERENCES
- 1.White RE, “High-throughput screening in drug metabolism and pharmacokinetic support of drug discovery,” Annu. Rev. Pharmacol. Toxicol 40, 133–157 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Skardal A, Devarasetty M, Rodman C, Atala A, and Soker S, “Liver-tumor hybrid organoids for modeling tumor growth and drug response in vitro,” Ann. Biomed. Eng 43(10), 2361–2373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, and Takayama S, “High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array,” Analyst 136(3), 473–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breslin S and O’Driscoll L, “Three-dimensional cell culture: The missing link in drug discovery,” Drug Discovery Today 18(5–6), 240–249 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Pereira DA and Williams JA, “Origin and evolution of high throughput screening,” Br. J. Pharmacol 152(1), 53–61 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auer RC, White RR, Kemeny NE, Schwartz LH, Shia J, Blumgart LH, Dematteo RP, Fong Y, Jarnagin WR, and D’Angelica MI, “Predictors of a true complete response among disappearing liver metastases from colorectal cancer after chemotherapy,” Cancer 116(6), 1502–1509 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Devarasetty M, Mazzocchi AR, and Skardal A, “Applications of bioengineered 3D tissue and tumor organoids in drug development and precision medicine: Current and future,” BioDrugs 32(1), 53–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pampaloni F, Reynaud EG, and Stelzer EH, “The third dimension bridges the gap between cell culture and live tissue,” Nat. Rev. Mol. Cell Biol 8(10), 839–845 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Skardal A, Shupe T, and Atala A, “Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling,” Drug Discovery Today 21(9), 1399–1411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schafer KL, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, Krieg A, and Stoecklein NH, “Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines,” PLoS One 8(3), e59689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messner S, Agarkova I, Moritz W, and Kelm JM, “Multi-cell type human liver microtissues for hepatotoxicity testing,” Arch. Toxicol 87(1), 209–213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Astashkina A and Grainger DW, “Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments,” Adv. Drug Delivery Rev 69–70, 1–18 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Devarasetty M, Wang E, Soker S, and Skardal A, “Mesenchymal stem cells support growth and organization of host-liver colorectal-tumor organoids and possibly resistance to chemotherapy,” Biofabrication 9(2), 021002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, and Rizzi SC, “Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells,” Biomaterials 31(32), 8494–8506 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Fukuda J, Khademhosseini A, Yeo Y, Yang X, Yeh J, Eng G, Blumling J, Wang CF, Kohane DS, and Langer R, “Micromolding of photocros-slinkable chitosan hydrogel for spheroid microarray and co-cultures,” Biomaterials 27(30), 5259–5267 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Yamada M, Utoh R, Ohashi K, Tatsumi K, Yamato M, Okano T, and Seki M, “Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions,” Biomaterials 33(33), 8304–8315 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Skardal A, Smith L, Bharadwaj S, Atala A, Soker S, and Zhang Y, “Tissue specific synthetic ECM hydrogels for 3-D in vitro maintenance of hepatocyte function,” Biomaterials 33(18), 4565–4575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck JN, Singh A, Rothenberg AR, Elisseeff JH, and Ewald AJ, “The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination,” Biomaterials 34(37), 9486–9495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Place ES, Evans ND, and Stevens MM, “Complexity in biomaterials for tissue engineering,” Nat. Mater 8(6), 457–470 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Tibbitt MW and Anseth KS, “Hydrogels as extracellular matrix mimics for 3D cell culture,” Biotechnol. Bioeng 103(4), 655–663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skardal A, Murphy SV, Devarasetty M, Mead I, Kang HW, Seol YJ, Shrike Zhang Y, Shin SR, Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Jin Lee S, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, and Atala A, “Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform,” Sci. Rep 7(1), 8837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy SV and Atala A, “3D bioprinting of tissues and organs,” Nat. Biotechnol 32(8), 773–785 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Mironov V, Reis N, and Derby B, “Review: Bioprinting: A beginning,” Tissue Eng. 12(4), 631–634 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, and Atala A, “A 3D bioprinting system to produce human-scale tissue constructs with structural integrity,” Nat. Biotechnol 34(3), 312–319 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Perestrelo AR, Aguas AC, Rainer A, and Forte G, “Microfluidic organ/body-on-a-chip devices at the convergence of biology and micro-engineering,” Sensors(Basel) 15(12), 31142–31170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stowers RS, Allen SC, and Suggs LJ, “Dynamic phototuning of 3D hydrogel stiffness,” Proc. Natl. Acad. Sci. U. S. A 112(7), 1953–1958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou S, Tiriac H, Sridharan BP, Scampavia L, Madoux F, Seldin J, Souza GR, Watson D, Tuveson D, and Spicer TP, “Advanced development of primary pancreatic organoid tumor models for high-throughput phenotypic drug screening,” SLAS Discovery 23(6), 574–584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen CJ, Saksena R, Kolesky DB, Vericella JJ, Kranz SJ, Muldowney GP, Christensen KT, and Lewis JA, “High-throughput printing via microvascular multinozzle arrays,” Adv. Mater 25(1), 96–102 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Xu F, Celli J, Rizvi I, Moon S, Hasan T, and Demirci U, “A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform,” Biotechnol. J 6(2), 204–212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch L, Gruene M, Unger C, and Chichkov B, “Laser assisted cell printing,” Curr. Pharm. Biotechnol 14(1), 91–97 (2013). [PubMed] [Google Scholar]
- 31.Guillemot F, Souquet A, Catros S, Guillotin B, Lopez J, Faucon M, Pippenger B, Bareille R, Remy M, Bellance S, Chabassier P, Fricain JC, and Amedee J, “High-throughput laser printing of cells and biomaterials for tissue engineering,” Acta Biomater. 6(7), 2494–2500 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Guillemot F, Souquet A, Catros S, and Guillotin B, “Laser-assisted cell printing: Principle, physical parameters versus cell fate and perspectives in tissue engineering,” Nanomedicine (London) 5(3), 507–515 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Kim YK, Park JA, Yoon WH, Kim J, and Jung S, “Drop-on-demand inkjet-based cell printing with 30-lm nozzle diameter for cell-level accuracy,” Biomicrofluidics 10(6), 064110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Zhang YS, Heinrich MA, De Ferrari F, Jang HL, Bakht SM, Alvarez MM, Yang J, Li YC, Trujillo-de Santiago G, Miri AK, Zhu K, Khoshakhlagh P, Prakash G, Cheng H, Guan X, Zhong Z, Ju J, Zhu GH, Jin X, Shin SR, Dokmeci MR, and Khademhosseini A, “Rapid continuous multimaterial extrusion bioprinting,” Adv. Mater 29(3), 1604630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skardal A, Devarasetty M, Kang HW, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo J, Soker S, and Atala A, “A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs,” Acta Biomater. 25, 24–34 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Ozbolat IT and Hospodiuk M, “Current advances and future perspectives in extrusion-based bioprinting,” Biomaterials 76, 321–343 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR, and Khademhosseini A, “Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels,” Biofabrication 6(2), 024105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, and Khademhosseini A, “Microfluidic bioprinting of heterogeneous 3D tissue constructs using low-viscosity bioink,” Adv. Mater 28(4), 677–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanjani Y, Pan CC, Elomaa L, and Yang Y, “A novel bioprinting method and system for forming hybrid tissue engineering constructs,” Biofabrication 7(4), 045008 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Hong S, Song SJ, Lee JY, Jang H, Choi J, Sun K, and Park Y, “Cellular behavior in micropatterned hydrogels by bioprinting system depended on the cell types and cellular interaction,” J. Biosci. Bioeng 116(2), 224–230 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, Ramadan MH, Hudson AR, and Feinberg AW, “Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels,” Sci. Adv 1(9), e1500758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adine C, Ng KK, Rungarunlert S, Souza GR, and Ferreira JN, “Engineering innervated secretory epithelial organoids by magnetic three-dimensional bioprinting for stimulating epithelial growth in salivary glands,” Biomaterials 180, 52–66 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Ma X, Liu J, Zhu W, Tang M, Lawrence N, Yu C, Gou M, and Chen S, “3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling,” Adv. Drug Delivery Rev 132, 235–251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatsura T, and Minami H, “Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer,” Oncol. Rep 33(4), 1837–1843 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Ramaiahgari SC, den Braver MW, Herpers B, Terpstra V, Commandeur JN, van de Water B, and Price LS, “A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies,” Arch. Toxicol 88(5), 1083–1095 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Skardal A, Devarasetty M, Forsythe S, Atala A, and Soker S, “A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening,” Biotechnol. Bioeng 113(9), 2020–2032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, and Kim DY, “A three-dimensional human neural cell culture model of Alzheimer’s disease,” Nature 515(7526), 274–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Justice BA, Badr NA, and Felder RA, “3D cell culture opens new dimensions in cell-based assays,” Drug Discovery Today 14(1–2), 102–107 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Cox TR and Erler JT, “Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer,” Dis. Models Mech 4(2), 165–178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metallo CM, Mohr JC, Detzel CJ, de Pablo JJ, Van Wie BJ, and Palecek SP, “Engineering the stem cell microenvironment,” Biotechnol. Prog 23(1), 18–23 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Clark AG and Vignjevic DM, “Modes of cancer cell invasion and the role of the microenvironment,” Curr. Opin. Cell Biol 36, 13–22 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Huh D, Hamilton GA, and Ingber DE, “From 3D cell culture to organs-on-chips,” Trends Cell Biol. 21(12), 745–754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmed EM, “Hydrogel: Preparation, characterization, and applications: A review,” J. Adv. Res 6(2), 105–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holzl K, Lin S, Tytgat L, Van Vlierberghe S, Gu L, and Ovsianikov A, “Bioink properties before, during and after 3D bioprinting,” Biofabrication 8(3), 032002 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Hospodiuk M, Dey M, Sosnoski D, and Ozbolat IT, “The bioink: A comprehensive review on bioprintable materials,” Biotechnol. Adv 35(2), 217–239 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Murphy SV, Skardal A, and Atala A, “Evaluation of hydrogels for bioprinting applications,” J. Biomed. Mater. Res., Part A 101(1), 272–284 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Skardal A and Atala A, “Biomaterials for integration with 3-D bioprinting,” Ann. Biomed. Eng 43(3), 730–746 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Delgado LM, Bayon Y, Pandit A, and Zeugolis DI, “To cross-link or not to cross-link? Cross-linking associated foreign body response of collagen-based devices,” Tissue Eng., Part B 21(3), 298–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouyang L, Highley CB, Sun W, and Burdick JA, “A generalizable strategy for the 3D bioprinting of hydrogels from nonviscous photo-crosslinkable inks,” Adv. Mater 29(8), 1604983 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, Rizzi SC, Weber FE, and Lutolf MP, “Elucidating the role of matrix stiffness in 3D cell migration and remodeling,” Biophys. J 100(2), 284–293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chimene D, Lennox KK, Kaunas RR, and Gaharwar AK, “Advanced bioinks for 3D printing: A materials science perspective,” Ann. Biomed. Eng 44(6), 2090–2102 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Cruz-Acuna R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, Garcia-Hernandez V, Miller AJ, Spence JR, Nusrat A, and Garcia AJ, “Synthetic hydrogels for human intestinal organoid generation and colonic wound repair,” Nat. Cell Biol 19(11), 1326–1335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kudva AK, Luyten FP, and Patterson J, “RGD-functionalized polyethylene glycol hydrogels support proliferation and in vitro chondrogenesis of human periosteum-derived cells,” J. Biomed. Mater. Res., Part A 106(1), 33–42 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Worthington P, Pochan DJ, and Langhans SA, “Peptide hydrogels - Versatile matrices for 3D cell culture in cancer medicine,” Front. Oncol 5, 92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, and Ingber DE, “Reconstituting organ-level lung functions on a chip,” Science 328(5986), 1662–1668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huh D, Torisawa YS, Hamilton GA, Kim HJ, and Ingber DE, “Microengineered physiological biomimicry: Organs-on-chips,” Lab Chip 12(12), 2156–2164 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Yum K, Hong SG, Healy KE, and Lee LP, “Physiologically relevant organs on chips,” Biotechnol. J 9(1), 16–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hynes RO, “The extracellular matrix: Not just pretty fibrils,” Science 326(5957), 1216–1219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonnans C, Chou J, and Werb Z, “Remodelling the extracellular matrix in development and disease,” Nat. Rev. Mol. Cell Biol 15(12), 786–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pati F, Jang J, Ha DH, Won Kim S, Rhie JW, Shim JH, Kim DH, and Cho DW, “Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink,” Nat. Commun 5, 3935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skardal A, Devarasetty M, Kang HW, Seol YJ, Forsythe SD, Bishop C, Shupe T, Soker S, and Atala A, “Bioprinting cellularized constructs using a tissue-specific hydrogel bioink,” J. Visualized Exp 110, e53606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, and Spatz JP, “Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands,” Biophys. J 92(8), 2964–2974 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zinger O, Zhao G, Schwartz Z, Simpson J, Wieland M, Landolt D, and Boyan B, “Differential regulation of osteoblasts by substrate microstructural features,” Biomaterials 26(14), 1837–1847 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Byfield FJ, Reen RK, Shentu TP, Levitan I, and Gooch KJ, “Endothelial actin and cell stiffness is modulated by substrate stiffness in 2D and 3D,” J. Biomech 42(8), 1114–1119 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Engler AJ, Sen S, Sweeney HL, and Discher DE, “Matrix elasticity directs stem cell lineage specification,” Cell 126(4), 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, and Engler AJ, “Interplay of matrix stiffness and protein tethering in stem cell differentiation,” Nat. Mater 13(10), 979–987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Esch MB, King TL, and Shuler ML, “The role of body-on-a-chip devices in drug and toxicity studies,” Annu. Rev. Biomed. Eng 13, 55–72 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Figeys D and Pinto D, “Lab-on-a-chip: A revolution in biological and medical sciences,” Anal. Chem 72(9), 330A–335A (2000). [DOI] [PubMed] [Google Scholar]
- 79.Khanna I, “Drug discovery in pharmaceutical industry: Productivity challenges and trends,” Drug Discovery Today 17(19–20), 1088–1102 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Timm DM, Chen J, Sing D, Gage JA, Haisler WL, Neeley SK, Raphael RM, Dehghani M, Rosenblatt KP, Killian TC, Tseng H, and Souza GR, “A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis,” Sci. Rep 3, 3000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan Y, Robertson G, Pedersen L, Lim E, Hernandez-Herrera A, Rowat AC, Patil SL, Chan CK, Wen Y, Zhang X, Basu-Roy U, Mansukhani A, Chu A, Sipahimalani P, Bowlby R, Brooks D, Thiessen N, Coarfa C, Ma Y, Moore RA, Schein JE, Mungall AJ, Liu J, Pecot CV, Sood AK, Jones SJ, Marra MA, and Gunaratne PH, “miR-509–3p is clinically significant and strongly attenuates cellular migration and multi-cellular spheroids in ovarian cancer,” Oncotarget 7(18), 25930–25948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Almazroo OA, Miah MK, and Venkataramanan R, “Drug metabolism in the liver,” Clin. Liver Dis 21(1), 1–20 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Fisher RA and Strom SC, “Human hepatocyte transplantation: Worldwide results,” Transplantation 82(4), 441–449 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Bale SS, Vernetti L, Senutovitch N, Jindal R, Hegde M, Gough A, McCarty WJ, Bakan A, Bhushan A, Shun TY, Golberg I, DeBiasio R, Usta BO, Taylor DL, and Yarmush ML, “In vitro platforms for evaluating liver toxicity,” Exp. Biol. Med. (Maywood) 239(9), 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nguyen DG, Funk J, Robbins JB, Crogan-Grundy C, Presnell SC, Singer T, and Roth AB, “Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro,” PLoS One 11(7), e0158674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Faulkner-Jones A, Fyfe C, Cornelissen DJ, Gardner J, King J, Courtney A, and Shu W, “Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D,” Biofabrication 7(4), 044102 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CS, Zanella F, Feng GS, Sheikh F, Chien S, and Chen S, “Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting,” Proc. Natl. Acad. Sci. U. S. A 113(8), 2206–2211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hribar KC, Soman P, Warner J, Chung P, and Chen S, “Light-assisted direct-write of 3D functional biomaterials,” Lab Chip 14(2), 268–275 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Charbe N, McCarron PA, and Tambuwala MM, “Three-dimensional bio-printing: A new frontier in oncology research,” World J. Clin. Oncol 8(1), 21–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelson B, “3-Dimensional bioprinting makes its mark: New tissue and organ printing methods are yielding critical new tools for the laboratory and clinic,” Cancer Cytopathol. 123(4), 203–204 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Peng W, Datta P, Ayan B, Ozbolat V, Sosnoski D, and Ozbolat IT, “3D bioprinting for drug discovery and development in pharmaceutics,” Acta Biomater. 57, 26–46 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Friedman AA, Letai A, Fisher DE, and Flaherty KT, “Precision medicine for cancer with next-generation functional diagnostics,” Nat. Rev. Cancer 15(12), 747–756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CG, van der Velden DL, Peeper DS, Cuppen EP, Vries RG, Clevers H, and Voest EE, “Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases,” Proc. Natl. Acad. Sci. U. S. A 112(43), 13308–13311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mazzocchi AR, Rajan SAP, Votanopoulos KI, Hall AR, and Skardal A, “In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening,” Sci. Rep 8(1), 2886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Collins FS and Varmus H, “A new initiative on precision medicine,” N. Engl. J. Med 372(9), 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jameson JL and Longo DL, “Precision medicine–personalized, problematic, and promising,” N. Engl. J. Med 372(23), 2229–2234 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Sasai Y, “Next-generation regenerative medicine: Organogenesis from stem cells in 3D culture,” Cell Stem Cell 12(5), 520–530 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Wu SM and Hochedlinger K, “Harnessing the potential of induced pluripotent stem cells for regenerative medicine,” Nat. Cell Biol 13(5), 497–505 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, Giacomello A, Abraham MR, and Marban E, “Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens,” Circulation 115(7), 896–908 (2007). [DOI] [PubMed] [Google Scholar]
- 100.Sonnaert M, Papantoniou I, Bloemen V, Kerckhofs G, Luyten FP, and Schrooten J, “Human periosteal-derived cell expansion in a perfusion bioreactor system: Proliferation, differentiation and extracellular matrix formation,” J. Tissue Eng. Regener. Med 11(2), 519–530 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Meng G, Liu S, Poon A, and Rancourt DE, “Optimizing human induced pluripotent stem cell expansion in stirred-suspension culture,” Stem Cells Dev. 26(24), 1804–1817 (2017). [DOI] [PubMed] [Google Scholar]
- 102.Zweigerdt R, Olmer R, Singh H, Haverich A, and Martin U, “Scalable expansion of human pluripotent stem cells in suspension culture,” Nat. Protoc 6(5), 689–700 (2011). [DOI] [PubMed] [Google Scholar]


