Abstract
Background
Long non‐coding RNAs (lncRNAs) have been found to be involved in the development of many cancers. In this study, we aimed to identify the molecular mechanisms of lncRNA BAALC antisense RNA 1 (BAALC‐AS1) in regulating the malignancy of esophageal squamous cell carcinoma (ESCC).
Methods
The expression of BAALC‐AS1 in cancer patients was analyzed using a tissue microarray. The protein and RNA levels of BAALC‐AS1 were determined by Western blotting analysis and quantitative reverse transcription‐PCR (RT‐qPCR), respectively. The cell proliferation was determined by cell viability assays, bromodeoxyuridine incorporation, and flow cytometry. The relationships among BAALC‐AS1, RasGAPSH3 domain‐binding protein 2 (G3BP2), and c‐Myc were determined using RNA immunoprecipitation, RNA pull‐down assays, and luciferase assays.
Results
The expression of BAALC‐AS1 was highly up‐regulated and associated with malignant phenotypes in ESCC tissues and cell lines. In vivo and in vitro assays showed that BAALC‐AS1 promoted ESCC cell proliferation, migration, and invasion. BAALC‐AS1 directly interacted with G3BP2, and thereby inhibited the degradation of c‐Myc RNA 3'‐UTR by G3BP2, thus leading to the accumulation of c‐Myc expression. Additionally, c‐Myc acted as a transcription factor that can induce the expression of BAALC‐AS1 by directly binding to its promoter region.
Conclusions
BAALC‐AS1/G3BP2/c‐Myc feedback loop plays a critical role in the development of ESCC, which might provide a novel therapeutic target and facilitate the development of new therapeutic strategies for the treatment of ESCC.
Keywords: esophageal squamous cell carcinoma, proliferation, migration, invasion, long non‐coding RNA, BAALC‐AS1, lncFZD6, G3BP2, c‐Myc
Abbreviations
- BAALC‐AS1
BAALC antisense RNA 1
- EC
esophageal carcinoma
- ESCC
esophageal squamous cell carcinoma
- lncRNA
long non‐coding RNA
- miRNA
microRNA
- G3BP2
RasGAPSH3 domain binding protein 2
- 3′‐UTR
three prime untranslated region
- shRNA
short hairpin RNA
- GFP
green fluorescent protein
- RT‐qPCR
quantitative reverse transcription PCR
- AJCC
American Joint Committee on Cancer
- GAPDH
3‐phosphate dehydrogenase
- FISH
fluorescence in situ hybridization
- BrdU
Bromodeoxyuridine
- H&E
Hematoxylin and Eosin
- IHC
immunohistochemistry
- PBS
phosphate‐buffered saline
- FBS
fetal bovine serum
- LC‐MS/MS
liquid chromatography‐mass spectrometry/ mass spectrometry
- RIP
RNA immunoprecipitation
- CDK2
cyclin‐dependent kinase 2
- CDK4
cyclin‐dependent kinase 4
1. BACKGROUND
Esophageal carcinoma (EC) is the eighth most common cancer and the sixth leading cause of cancer‐related death worldwide, with the highest incidence in Asia [1, 2, 3]. Esophageal squamous cell carcinoma (ESCC) accounts for the majority of EC globally. It widely occurs in China [4]. Due to a lack of effective clinical approaches for earlier diagnosis and treatment, the efficacy of ESCC therapy has not significantly changed in past decades, and the 5‐year overall survival rate has been in the range of 15%–25% [5]. Thus, a better understanding of the molecular mechanism of ESCC development may help to identify new diagnostic approaches and therapeutic modalities.
The mammalian genome encodes a large number of long non‐coding RNAs (lncRNAs), which transcribe over 200 nucleotides that have no evidence of protein‐coding potential. Much of the non‐protein‐coding portion of the human genome has historically been regarded as junk DNA [6]. However, over the past decade, numerous studies have highlighted how regulatory RNAs, such as microRNAs (miRNAs) and lncRNAs, play crucial roles in the development of diseases [7, 8, 9]. Studies have confirmed that lncRNAs interact with DNA, RNA, and proteins in a cis‐ or trans‐acting manner, which can regulate gene expression at the epigenetic, transcriptional, and post‐transcriptional levels [10]. For example, lncRNA MALAT1 [7, 11, 12], ANRIL [13, 14], HOTAIR [15, 16], and H19 [17, 18] have been confirmed to be highly expressed in many cancers and act as oncogenic genes to regulate tumor progression and metastasis. An understanding of the functions of these lncRNAs provides new opportunities for the diagnosis and treatment of cancer. Nevertheless, the molecular regulatory mechanisms of lncRNAs in ESCC remain largely to be further defined.
Our group has comprehensively and systematically investigated the genomic alterations of ESCC using whole‐genome sequencing and whole‐exon sequencing [2]. We identified many somatic copy number alterations of ESCC, among which the copy number amplification of Homo sapiens BAALC antisense RNA 1 (BAALC‐AS1, NR_109954.1) attracted our attention. BAALC‐AS1 is an lncRNA with a length of 726 bp, which is also known as lncFZD6. It is located on chromosome 8q22.3 and contains three exons, which are antisense chains with the intron region of the BAALC protein‐coding gene. As a novel lncRNA, there have been few reports on its function and molecular mechanism. Only one study indicated that BAALC‐AS1 promotes self‐renewal of hepatic tumor‐initiating cells by activating the Wnt/beta‐catenin signaling pathway [19]. Hence, the functions and the underlying molecular mechanisms of BAALC‐AS 1 need to be further investigated.
lncRNAs can directly interact with protein to regulate protein activity [8, 20, 21]. We found that BAALC‐AS1 interacts with the protein RasGAPSH3 domain‐binding protein 2 (G3BP2). G3BP2 is a member of the G3BP family and plays an important role in a variety of cancers, and is closely related to lymph node metastasis and prognosis in ESCC [22]. G3BP2 has RNA‐binding sites and can regulate RNA homeostasis, such as binding to CDK7 and CDK9, stabilizing CDK7 mRNA, and degrading CDK9 mRNA [23]. G3BP2 can also degrade the 3′‐untranslated regions (3′‐UTRs) of c‐Myc mRNA in vitro, affecting the RNA stability of c‐Myc [24, 25]. However, the biological function and regulatory mechanism of lncRNAs and G3BP2 remain to be elucidated.
In this study, we identified the function and mechanism of BAALC‐AS1 in ESCC which might provide a novel therapeutic target and facilitate the development of new therapeutic strategies for the treatment of ESCC.
2. MATERIALS AND METHODS
2.1. Cell lines, cell culture and transfection
The normal esophageal epithelial cell line (Het‐1A) and the human ESCC cell lines (YES‐2, KYSE‐30, ‐150, ‐180, ‐410, ‐450, ‐510 and COLO‐680) were generously provided by Dr. Yutaka Shimada of Kyoto University (Kyoto, Japan) and cultured as previously described [26]. The cells were cultured in RPMI‐1640 (Lonza, Basel, Switzerland) or DMEM with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) at 37°C in a 5% CO2 humidified incubator.
The expressions of BAALC‐AS1, G3BP2, and c‐Myc were silenced by transfecting ESCC cells with siRNAs, and a non‐targeted control siRNA (siNC) was used as a negative control to determine and optimize the efficiency of transfection. The detailed target sequences are listed as follows: sense sequence 5'‐ CAGGAACAAAGGUUACAGA ‐3' and antisense sequence 5'‐ UCUGUAACCUUUGUUCCUG‐3' for si/BAALC‐AS1‐1; sense sequence 5'‐ AGCGAACAAUGGAGAACCU ‐3' and antisense sequence 5'‐ AGGUUCUCCAUUGUUCGCU ‐3' for si/BAALC‐AS1‐2; sense sequence 5'‐ AGUCGAAGCUAAACCAGAA ‐3' and antisense sequence 5'‐ UUCUGGUUUAGCUUCGACU ‐3' for si/G3BP‐2; sense sequence 5'‐ GAGGAGACAUGGUGAACCA ‐3' and antisense sequence 5'‐ UGGUUCACCAUGUCUCCUC ‐3' for si/c‐Myc‐1; sense sequence 5'‐ GGGUCAAGUUGGACAGUGU ‐3' and antisense sequence 5'‐ ACACUGUCCAACUUGACCC‐3' for si/c‐Myc‐2. The siRNA was transfected into ESCC cells using Lipofectamine 2000 siRNA transfection reagent (Invitrogen) following the manufacturer's protocol as previously described [27].
For stable transfection of BAALC‐AS1 short hairpin RNA (shRNA), ESCC cells were transfected with pLenti‐U6‐BAALC‐AS‐shRNA‐GFP‐Puro. Control cells were transfected with pLenti‐U6‐NC‐shRNA‐GFP‐Puro. For stable transfection of BAALC‐AS1 overexpressing plasmid, pCDH‐CMV‐MCS‐BAALC‐EF1‐CopGFP‐P2A‐Puro was transfected into ESCC cells as previously described [28]. Briefly, the virus solution and the final concentration of 5μg/mL polybrene were added into the fresh medium to infect the cells. All plasmids were from GenePharma (Shanghai, China). The cells transfected with pCDH‐CMV‐MCS‐EF1‐CopGFP‐P2A‐Puro were served as control. Transduction efficiency (multiplicity of infection [MOI] = 100) was determined by green fluorescent protein (GFP) expression to be over 80%, and knockdown efficiency was measured by quantitative reverse transcription‐PCR (RT‐qPCR) to be approximately 70%.
2.2. Tissue specimens
Tissue microarrays of ESCC specimens were obtained from Shanghai Outdo Biotech (Shanghai, China), with the approval of the local Institutional Review Board. Detailed clinicopathological characteristics for all specimens are presented in Supplementary Table S1. The pathological grade of ESCC was determined by pathological experts according to the American Joint Committee on Cancer (AJCC) prognosis classification standard (2009), and the disease staging of ESCC was based on the seventh edition of AJCC clinical staging system.
2.3. In situ hybridization
Digoxigenin‐labeled DNA probes complementary to BAALC‐AS1 RNA were generated using random primer labeling (Boster, Wuhan, Hubei, China). The experimental procedure was operated according to the instructions. For in situ hybridization, 5 μm thick sections were prepared and treated. After sectioned, the prehybrid solution and the hybrid solution were incubated respectively and hybridized overnight at 40°C. After full washing, blocking solution and color developing solution were added, and control the color developing under the microscope. The intensity of BAALC‐AS1 expression was graded as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. The proportion of positive tumor cells was graded as follows: 0, <5%; 1, 5%‐25%; 2, 26%‐50%; 3, 51%‐75%; and 4, >75%. The final score was derived from the multiplication of these two primary scores. Final scores of 0‐6 were defined as “low expression”, and scores of 6‐12 as “high expression”.
2.4. Cellular fractionation, RNA isolation, and RT‐qPCR analyses
To investigate the expression of target genes, total RNA was extracted and isolated from tissues and cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol.
Cytoplasmic and nuclear RNAs were isolated and purified using the PARIS™ Kit (Thermo Fisher Scientific, Scotts Valley, CA, USA) according to the manufacturer's instructions.
For the RT‐qPCR assay, the isolated RNA was reversely transcribed into cDNA using a reverse transcription kit (Takara, Dalian, Liaoning, China). The expression levels were normalized against the expression of glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). The PCR primers were as follows: forward primer sequence 5′‐AAAAGCGAACAATGGAGAACC‐3′ and reverse primer sequence 5′‐TCCACTGCACCGAATTGTATAA‐3′ for BAALC‐AS1; forward primer sequence 5′‐GAGCTGAAACCACAAGTGGAGG‐3′ and reverse primer sequence, 5′‐GGTCACTGAAGCCCAGGAGAAA‐3′ for G3BP2; forward primer sequence 5′‐CCTGGTGCTCCATGAGGAGAC‐3′ and reverse primer sequence 5′‐CAGACTCTGACCTTTTGCCAGG‐3′ for c‐Myc; forward primer sequence 5′‐CAATGACCCCTTCATTGACC‐3′ and reverse primer sequence 5′‐TGGAAGATGGTGATGGGATT‐3′ for GAPDH. Then, specific gene expression was detected by using SYBR Premix EX TaqII (Takara, Dalian, Liaoning, China). The RT‐qPCR assays were conducted in an ABI 7500 apparatus (Applied Biosystems, Foster City, CA, USA).
2.5. RNA fluorescence in situ hybridization (FISH)
Fluorescence‐conjugated lncBAALC‐AS1 probes were used for RNA‐FISH, which was performed as previously described [28]. Hybridization was performed using DNA probe sets (Ribobio, cat# lnc1101063, Guangzhou, Guangdong, China) according to the manufacturer's instructions. Images were captured and visualized by a confocal microscope (ST2; Leica, Wetzlar, Germany).
2.6. 3‐(4,5‐dimethylthiazol‐2‐yl)‐5‐(3‐carboxymethoxyphenyl)‐2‐(4‐sulfophenyl)‐2H‐tetrazolium,inner salt (MTS) assay
The Cell Titer 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) was used to evaluate the cell viability in the indicated ESCC cell lines. Briefly, 10 μL MTS solution was added to each well of the 96‐well plate and incubated for 1 h at 37°C. The absorbance was read at 490 nm for the MTS solution using an ELISA plate reader (TECAN, Infinite M200 Pro, Mannedorf, Switzerland). All experiments were performed in triplicate.
2.7. Bromodeoxyuridine (BrdU) incorporation
ESCC cells were plated into 96‐well plates at a density of 1 × 104 cells/well, and then subjected to growth arrest for 24 h before transfection. BrdU was incorporated into proliferating cells by adding 24 h prior to the end of the test reagent incubation. Following incorporation, the cells were fixed using a fixing solution at room temperature for 30 min. BrdU was added after fully washing according to the manufacturer's protocol. Finally, the plate was read using a spectrophotometer microplate reader (TECAN, Infinite M200 Pro, Mannedorf, Switzerland) at dual wavelengths of 450/550 nm. All experiments were performed in triplicate.
2.8. Western blotting analysis
Proteins were solubilized and extracted with RIPA buffer (Beyotime, Shanghai, China) and incubated for 30 min on ice. The lysates were sonicated and centrifuged at 16,099 × g for 15 min, and the insoluble fractions were discarded. Protein concentrations were determined by the BCA protein assay kit (Thermo Fisher Scientific, Scotts Valley, CA, USA). Cell samples containing 20 μg of protein were subjected to electrophoresis using an SDS‐polyacrylamide gel. After electrophoresis, the proteins were transferred to nitrocellulose membranes (Millipore, Cambridge, MA, USA). The membranes were blocked in blocking buffer (20 mmol/L Tris, pH 7.6, 150 mmol/L NaCl, and 0.1% Tween 20) containing 5% non‐fat dry milk and incubated with antibodies. Antibodies against c‐Myc (cat# 13987S), cyclin D (cat# 2978S), and CDK4 (cat# 12790S) were all from Cell Signaling Technology (Danvers, MA, USA); antibodies against CDK2 (cat# ab32147), IgG (cat# ab171870), and actin (cat# ab179467) were from Abcam (Cambridge, MA, USA); antibody against G3BP2 (cat# 16276‐1‐AP) was from Proteintech (Wuhan, Hubei, China); and antibody against cyclin E (cat# sc‐377100) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Actinomycin D was obtained from MedChem Express (Monmouth Junction, NJ, USA). The ECL Detection were from Beyotime (Shanghai, China). The chemiluminescence signals were detected with an Amersham Imager 600 (Amersham, Chalfont, UK).
2.9. Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC)
The tumor tissues from nude mice were sliced into tissue blocks, and were immersed in 4% paraformaldehyde for overnight fixation. Then fixed tissues were dehydrated, cleared, and embedded in paraffin base. The paraffin blocks were cut into sections of 5 μm thick. Some sections were stained with HE and others for IHC, which was performed as previously described [29].
2.10. Cell cycle analyses
The proportions of cells in the G0/G1, S, and G2/M phases were detected by flow cytometry. The detection kit was from Beyotime (Shanghai, China). Briefly, the cells were harvested by trypsinization and then fixed with 70% ethanol at 4°C. The fixed cells were centrifuged at 300 × g for 5 min and resuspended in 500 μL staining buffer before detection. A total of 10 μL of RNase A was added and mixed, then 25 μL of propidium iodide was added, and the suspension was incubated at 37°C in a water bath for 30 min. Finally, the cells were filtered once through 400 mesh sieves and detected by accuri C6 flow cytometry (Corning, New York, NY, USA).
2.11. Wound healing assay
Six‐well plates, containing fully confluent ESCC cells, were scratched with a fine sterile pipette tip to generate a cell‐free gap with a width of 1 mm, and the damaged cells were removed using phosphate‐buffered saline (PBS). The cells were then treated with vehicle or the chemical of interest in 5% FBS‐Dulbecco's Modified Eagle's Medium. The cells in all study groups were photographed in the same area of the culture plate, immediately after and 24 h after the wound was made.
2.12. Invasion assay
Cell invasion was measured using a Boyden chamber (Corning, New York, NY, USA) with a polycarbonate filter with 8 μm inserts coated with Matrigel (Corning, New York, NY, USA). After 24 h of incubation, the non‐migrating cells in the upper chamber were removed. The cells on the underside of the membrane were incubated in 4% formaldehyde solution for 10 min, followed by incubation in 0.4% crystal violet in 10% ethanol for 5 min. The number of cells was measured by counting the number of stained nuclei per high power field using a microscope (Olympus, Tokyo, Japan). Each sample was counted randomly at nine separate fields in the center of the membrane, and the cell invasion activity was reported as the number of cells migrating per field of view. The experiments were performed at least three times in quadruplicate.
2.13. Colony formation assay
The transduced and control cells were placed in 6‐well plates at a density of 1000 cells/well and maintained in medium containing 10% FBS. The culture medium was replaced every 4 days during the growth of colonies. After 14 days, the cell colonies were washed with PBS, fixed with 4% paraformaldehyde for 10 min, and stained with crystal violet for 8 min. Triplicate wells were measured for each treatment group.
2.14. Tumor formation assay in nude mice
Male BALB/c nude mice (6‐week‐old) were purchased from Vital River Laboratories (Beijing, China) and maintained in pathogen‐free conditions. The mice were injected subcutaneously into the right flanks with 1 × 106 cells/mL (0.1 mL) of ESCC cells (KYSE‐450 or KYSE‐410), which were stably transfected with sh/NC, sh/BAALC‐AS1, BAALC‐AS1 overexpression plasmid, or the empty vector. Tumor growth was examined every 7 days, and tumor volumes were calculated using the equation, V = 0.5 × length × width2. At 28 days after injection, the mice were euthanized by CO2, and the subcutaneous growth of each tumor was examined by H&E staining and immunohistochemical analysis. All animal care and experiments were conducted in accordance with national and institutional policies for animal health and well‐being. The protocol was approved by the Institutional Animal Care and Use Committee.
2.15. mRNA stability assay
Cells were incubated for the indicated time following the addition of 5 μg/mL actinomycin D (MedChem Express; Monmouth Junction, NJ, USA) as previously described [30]. Total RNA was subsequently extracted, and RT‐qPCR was conducted to quantify the levels of c‐Myc mRNA.
2.16. RNA pull‐down assay
The RNA pull‐down assay was performed using the Magnetic RNA‐Protein Pull‐Down kit (Pierce, Waltham, MA, USA) according to the manufacturer's instructions. First, the full‐length BAALC‐AS1 was synthesized using the RiboMAX™ Large Scale RNA Production System (Promega). After biotin labeling, the BAALC‐AS1 was bound to the beads for protein binding. The cell protein lysate was added to RNA‐bound beads for immunoprecipitation, and the beads were washed three times and boiled in SDS buffer, followed by detection of the retrieved protein using Western blotting analyses.
2.17. Liquid chromatography‐mass spectrometry/mass spectrometry (LC‐MS/MS) analysis
The peptide of each sample was desalted on C18 Cartridges (Empore™ SPE Cartridges, Sigma‐Aldrich, St. Louis, MO, USA), then concentrated by vacuum centrifugation (Eppendorf Concentrator 5301) and reconstituted in 10 μL of 0.1% (v/v) Formic acid (HPLC Grade, Sigma‐Aldrich, St. Louis, MO, USA). MS experiments were performed on a Q Exactive HF mass spectrometer that was coupled to Easy nLC (Thermo Scientific, Scotts Valley, CA, USA). The MS data were analyzed using MaxQuant software version 1.6.0.16. MS data were searched against the UniProtKB Human database (162989 total entries; https://www.uniprot.org/uniprot/?query = taxonomy:9606; downloaded 04/14/2018). This part of work was supported by Shanghai Bioprofile Technology Company Ltd.
2.18. RNA immunoprecipitation (RIP) assay
RIP was performed using the Magna RIP RNA‐Binding Protein Immunoprecipitation kit (Merck Millipore, Oakville, Ontario, Canada) following the manufacturer's instructions as previously described [31]. G3BP2 and IgG antibodies were used. Final analysis was performed using RT‐qPCR and shown as fold enrichment of BAALC‐1.
2.19. Luciferase reporter assay
ESCC cells in a 96‐well plate were transfected with RNA duplex for 24 h and then transfected with pGL3‐BAALC‐AS1‐WT or psi‐check2‐c‐Myc‐WT. The cells were collected 48 h after transfection and analyzed by using the Dual‐Luciferase reporter assay (Promega). Luciferase activity was measured using a Synergy H1 microplate fluorescence reader (BioTek, Winooski, VT, USA). Firefly activity was normalized to Renilla luciferase activity.
2.20. Statistical analysis
All data were obtained from at least three independent experiments and are expressed as the mean ± standard error of mean (SEM). Statistical analysis was performed with one‐way analysis of variance followed by Dunnett's test, the two‐way analysis of variance, or the Student's t‐test. Overall survival curves were plotted using the Kaplan‐Meier method and compared using the log‐rank test. The chi‐square test was used to compare indicated proteins between cancer and adjacent normal tissues, or various clinical parameters. A value of P < 0.05 was considered statistically significant.
3. RESULTS
3.1. BAALC‐AS1 was up‐regulated in ESCC and associated with a poor prognosis
We recently conducted whole‐genome sequencing and whole‐exome sequencing in ESCC cases [2] and observed a large number of genes with copy number amplification [32], including 20 lncRNAs located in the gene copy number amplification regions. Among those 20 lncRNAs, BAALC‐AS1, which was amplified in about 25% of ESCCs, was confirmed to be highly expressed in ESCC using Cancer Cell Line Encyclopedia (CCLE; https://portals.broadinstitute.org/ccle/page?gene = RP11‐318M2.2) and Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer‐pku.cn/detail.php?gene = BAALC‐AS1) approaches (Supplementary Figure S1) and verified by RT‐qPCR. We then detected BAALC‐AS1 expressions in 112 ESCC tissues and 68 adjacent normal esophageal tissues by in situ hybridization. BAALC‐AS1 staining was barely detectable in most normal esophageal tissues, while ESCC tumor tissues exhibited moderate to intense diffuse BAALC‐AS1 staining (Figure 1A). The expression of BAALC‐AS1 was significantly higher in ESCC tissues than in adjacent normal esophageal tissues (Figure 1B). We next analyzed the relationship between BAALC‐AS1 expression level and the clinicopathological features of 112 ESCC patients. Notably, the expression level of BAALC‐AS1 was significantly associated with prognoses in ESCC patients (P = 0.037; Figure 1C). Additionally, BAALC‐AS1 expression was correlated with overall survival and pathological stage (Figure 1D). We also detected BAALC‐AS1 expression in cell lines, as shown in Figure 1E. Its expression in ESCC cell lines (YES‐2, KYSE‐30, ‐150, ‐450, ‐510, and COLO‐680) was significantly higher than that in the normal esophageal epithelial cell line (Het‐1A), but the increases in ESCC cell lines KYSE‐180 and 410 cells were not significant. We next performed cellular fractionation to analyze the subcellular localization of BAALC‐AS1 in KYSE‐510 cells (Figure 1F) and KYSE‐450 cells (Supplementary Figure S2A). BAALC‐AS1 was almost evenly distributed in the cytoplasm and nucleus. The results were further confirmed by RNA‐FISH, showing that BAALC‐AS1 expression was distributed in both the cytoplasm and nucleus of KYSE‐510 cells (Figure 1G) and KYSE‐450 cells (Supplementary Figure S2B).
FIGURE 1.
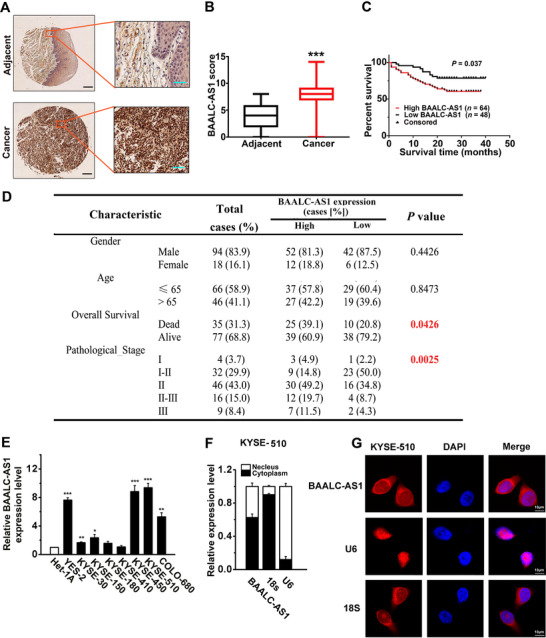
The expression of BAALC‐AS1 in ESCC. A. In situ hybridization of BALC‐AS1 in ESCC tissue samples and their corresponding adjacent normal tissues. Scale bar: left, 100 μm; right, 200 μm. Adjacent, adjacent normal tissues; Cancer, ESCC tissues. B. Comparison of BAALC‐AS1 scores of ESCC and adjacent normal tissues. C. ESCC patients with high levels of BAALC‐AS1 expression showed reduced survival compared with those with low levels of BAALC‐AS1 expression (P = 0.037; log‐rank test). D. Differences in the overall survival and pathological stage between patients with high BAALC‐AS1 expression level and low BAALC‐AS1 expression level were highly significant. E. Expression of BAALC‐AS1 quantified by RT‐qPCR in ESCC cell lines (YES‐2, KYSE‐30, ‐150, ‐180, ‐410, ‐450, ‐510, and COLO‐680) and normal esophageal epithelial cell line (Het‐1A). F. Analysis of BAALC‐AS1 distribution by cellular fractionation followed RT‐qPCR in KYSE‐510 cells. U6 and 18s mRNAs were served as controls for nuclear and cytoplasmic RNAs, respectively. G. RNA‐ FISH was performed to detect BAALC‐AS1 expression in KYSE‐510 cells. The nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI). Scale bar, 10 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All of the values are expressed as the mean ± SEM of 3 experiments
3.2. BAALC‐AS1 regulates cell proliferation by accelerating the cell cycle
To further investigate the function of increased BAALC‐AS1 levels in ESCC cells, we examined the effects of BAALC‐AS1 knockdown and overexpression on ESCC cell lines. Two independent siRNAs were identified that could potently knock down BAALC‐AS1 expression in BAAALC‐AS1 high expression cell lines (KYSE‐450 and KYSE‐510) (Figure 2A). When BAALC‐AS1 was silenced, cell viability (Figure 2B) and proliferation (Figure 2C) in both KYSE‐450 and KYSE‐510 cells were substantially decreased. To investigate the effects of BAALC‐AS1 on cell cycle, the number of cells in different cell cycle phases was detected by flow cytometry. Figure 2D shows that the percentage of cells in S and G2/M phases was decreased in both KYSE‐450 and KYSE‐510 cells in which BAALC‐AS1 was knocked down. After siRNA transfection for BAALC‐AS1 in ESCC cells, the expression levels of Cyclin D, Cyclin E, Cyclin‐dependent kinase 2 (CDK2), and CDK4 were also decreased (Figure 2E). In contrast, BAALC‐AS1 overexpression increased cell viability and proliferation in both KYSE‐180 and KYSE‐410 cells(Figure 3A‐C). The percentage of cells in S and G2/M phases was increased in both KYSE‐180 and KYSE‐410 cells in which BAALC‐AS1 was overexpressed (Figure 3D). In addition, the expressions of Cyclin D, Cyclin E, CDK2, and CDK4 were also increased (Figure 3E).
FIGURE 2.
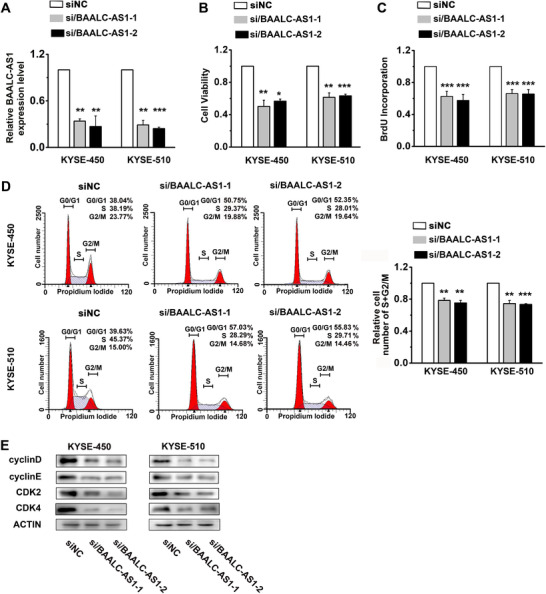
BAALC‐AS1 knockdown reduces cell proliferation and regulates the cell cycle in KYSE‐450 and KYSE‐510 cells. A. The expression levels of BAALC‐AS1 in si/BAALC‐AS1‐transfected KYSE‐450 and KYS‐510 cells were assessed by RT‐qPCR. B. MTS assay was used to determine the cell viability in si/BAALC‐AS1‐transfected KYSE‐450 and KYS‐510 cells. C. BrdU incorporation showed the synthesis of DNA in si/BAALC‐AS1‐transfected KYSE‐450 and KYS‐510 cells. D. Flow cytometry detected the number of si/BAALC‐AS1‐transfected KYSE‐450 and KYS‐510 cells in each phase of the cell cycle (left panel). The percentages of si/BAALC‐AS1‐transfected KYSE‐450 and KYS‐510 cells are decreased in S and G2/M phases (right panel). E. Western blotting analyses of the protein expression levels of Cyclin D, Cyclin E, CDK2, and CDK4 in si/BAALC‐AS1‐transfected KYSE‐450 and KYS‐510 cells. NC, negative control; siRNA/BAALC‐AS1, small interfering RNA for BAALC‐S1. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All values are expressed as mean ± SEM of 3 experiments.
FIGURE 3.
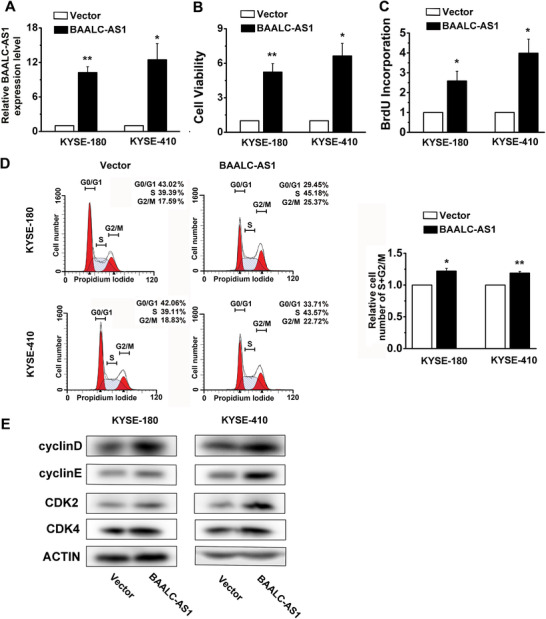
BAALC‐AS1 overexpression promotes cell proliferation in KYSE‐180 and KYSE‐410 cells. A. The expression levels of BAALC‐AS1 in BAALC‐AS1‐overexpressing KYSE‐180 and KYS‐410 cells were assessed by RT‐qPCR. B. MTS assay was used to determine the cell viability in BAALC‐AS1‐overexpressing KYSE‐180 and KYS‐410 cells. C. BrdU incorporation shows the synthesis of DNA in BAALC‐AS1‐overexpressing KYSE‐180 and KYS‐410 cells. D. Flow cytometry detected the number of BAALC‐AS1‐overexpressing KYSE‐180 and KYS‐410 cells in each phase of the cell cycle (left panel). The percentages of BAALC‐AS1‐overexpressing KYSE‐180 and KYS‐410 cells are increased in S and G2/M phases (right panel). E. Western blotting analyses of the protein expression levels of Cyclin D, Cyclin E, CDK2 and CDK4 in BAALC‐AS1‐overexpressing KYSE‐180 and KYS‐410 cells. Vector, cells were infected with empty lentiviruses; BAALC‐AS1, cells were infected with lentiviruses expressing BAALC‐AS1. *, P < 0.05; **, P < 0.01. All values are expressed as mean ± SEM of 3 experiments.
3.3. BAALC‐AS1 promotes cell migration, invasion, and colony formation in vitro
We conducted wound healing assays and observed that BAALC‐AS1 knockdown decreased cell migration in both KYSE‐450 and KYSE‐510 cells (Figure 4A). In contrast, BAALC‐AS1 overexpression increased cell migration in both KYSE‐180 and KYSE‐410 cells (Supplementary Figure S3A). Similarly, BAALC‐AS1 knockdown decreased cell invasion in both KYSE‐450 and KYSE‐510 cells (Figure 4B), while BAALC‐AS1 overexpression increased invasion in both KYSE‐180 and KYSE‐410 cells (Supplementary Figure S3B). We further established a stable BAALC‐AS1 down‐regulated ESCC cell line by using BAALC‐AS1‐shRNA. The colony formation ability was decreased following depletion of BAALC‐AS1 in both KYSE‐450 and KYSE‐510 cells (Figure 4C), while BAALC‐AS1 overexpression promoted colony formation in both KYSE‐180 and KYSE‐410 cells (Supplementary Figure S3C).
FIGURE 4.
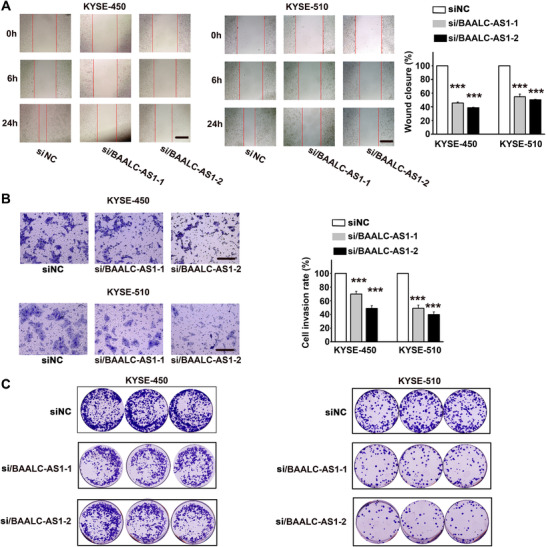
BAALC‐AS1 knockdown inhibits cell migration, invasion, and colony formation in KYSE‐450 and KYSE‐510 cells. A. Representative images of si/BAALC‐AS1‐ and control siRNA (siNC)‐transfected KYSE‐450 and KYSE‐510 cells taken at the time of wounding (0 h) and after 6 h and 24 h incubation (left panel). The percentage of wound closure at 24 h is shown in right panel. Scale bar, 200 μm. B. Representative images of the invasion assay in si/BAALC‐AS1 and siNC‐transfected KYSE‐450 and KYSE‐510 cells (left panel). The cell invasion rate at 24 h is shown in right panel. Scale bar, 200 μm. C. Images of colony formation assay in si/BAALC‐AS1‐ and siNC‐transfected KYSE‐450 and KYSE‐510 cells. NC, negative control; siRNA/BAALC‐AS1, small interfering RNA for BAALC‐AS1; shRNA/BAALC‐AS1, small hairpin RNA for BAALC‐AS1. ***, P < 0.001. All values are expressed as the mean ± SEM.
3.4. BAALC‐AS1 promotes tumor growth in vivo
Next, ESCC cells (KYSE‐450 or KYSE‐410) transfected with the control empty vectors (Vector), BAALC‐AS1‐overexpressing plasmids (BAALC‐AS1), control shRNAs (NC), or BAALC‐AS1 shRNAs (sh/BAALC‐AS1) were subcutaneously injected into nude mice. As shown in Figure 5A‐C, the tumors derived from the BAALC‐AS1 group had significantly higher tumor weights and larger tumor volumes compared with the Vector group. In contrast, tumors derived from sh/BAALC‐AS1‐transfected cells had significantly lower tumor weights and smaller tumor volumes compared with the mice in NC group. Moreover, we concerned about the health and body weight of the mice, but found that there was no obvious toxicity after BAALC‐AS1 knockdown or overexpression (Figure 5D). In addition, we detected the expression of Ki67 in tumor tissues from nude mice by IHC. After BAALC‐AS1 was knocked down, the expression of Ki67 was significantly decreased and the degree of differentiation was relatively high, and the opposite results were observed after the overexpression of BAALC‐AS1 (Figure 5E).
FIGURE 5.
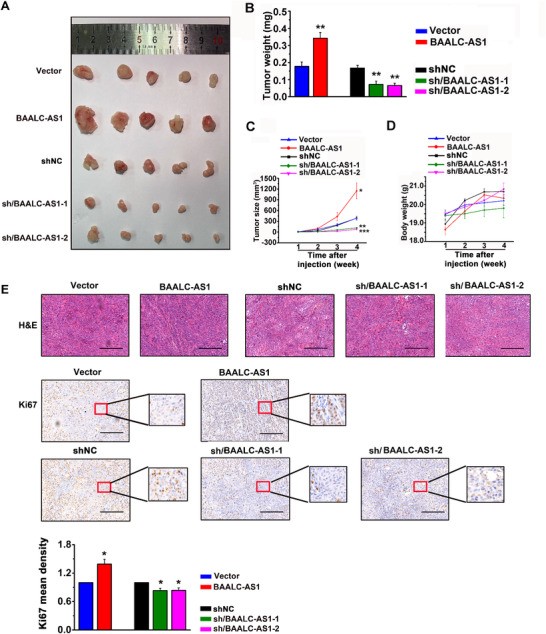
BAALC‐AS1 promotes tumor growth of ESCC in vivo. A. Images of the xenograft tumors formed in nude mice injected with BAALC‐AS1‐overexpressing (BAALC‐AS1) or ‐silencing (sh/BAALC‐AS1‐1 and ‐2) KYSE‐450 or KYSE‐410 cells. The nude mice injected with control cells (Vector and siNC) were used as control. B. Tumor weight in nude mice injected with the abovementioned cells. C. Growth curve of xenograft tumors in nude mice injected with the abovementioned cells. Five mice per group were used in the tumor formation assay. D. The body weight of the nude mice in tumor formation assay. E. Representative images of H&E staining and Ki67 IHC staining for tumor tissues harvested from nude mice in tumor formation assay. Scale bar, 50 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All values are expressed as the mean ± SEM
3.5. BAALC‐AS1 regulates c‐Myc expression via interacting with G3BP2
To determine the molecular mechanism of how BAALC‐AS1 promoted the malignant progression of ESCC, RNA pull‐down assays were performed to identify BAALC‐AS1‐binding proteins (Figure 6A and Supplementary Figure S4). G3BP2 was selected as the potential BAALC‐AS1‐binding protein. The directly binding of BAALC‐AS1 and G3BP2 was further validated by Western blotting in the retrieved RNA pull‐down samples of KYSE‐450 cells (Figure 6B). RIP results also showed that BAALC‐AS1 directly interacted with G3BP2 protein in KYSE‐450 cells (Figure 6C). Moreover, the mRNA and protein expression of G3BP2 changed little upon BAALC‐AS1 knockdown in KYSE‐450 cells (Figure 6D‐E). Considering that G3BP2 is a RNase that shares characteristic structural features with RNA‐binding proteins and has been proven to be able to regulate the steady‐state levels of c‐Myc mRNA through c‐Myc 3′‐UTR [24], we detected c‐Myc RNA (Figure 6F) and protein levels (Supplementary Figure S5A) after interference with G3BP2. We found that c‐Myc expression was up‐regulated, while BAALC‐AS1 was down‐regulated after interference with G3BP2. We also detected down‐regulated c‐Myc mRNA and protein levels after BAALC‐AS1 knockdown in KYSE‐450 cells (Figure 6G‐H). In addition, BALC‐AS1 knockdown shortened the half‐life of c‐Myc mRNA (Figure 6I).
FIGURE 6.
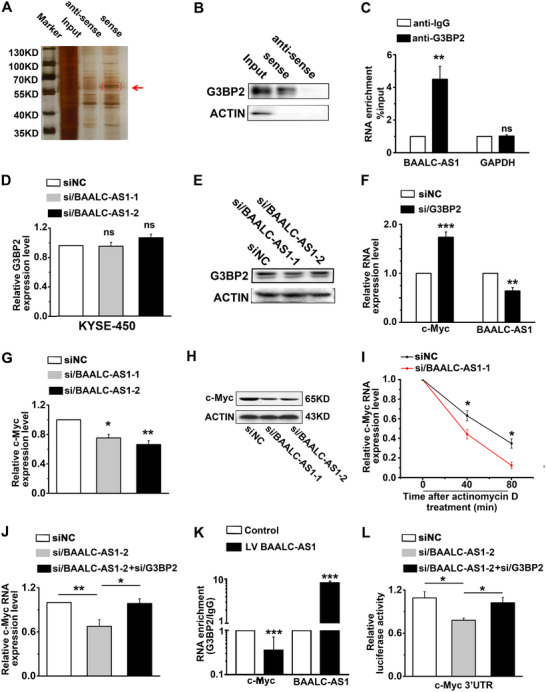
BAALC‐AS1 directly interacts with G3BP2 to regulate the expression of c‐Myc. A. Identification of BAALC‐AS1‐binding proteins in KYSE‐450 cells using RNA pull‐down assay. The band specific to sense BAALC‐AS1 (pointed by the red arrow) was detected using LC‐MS/MS. B. Western blotting in the retrieved RNA pull‐down samples of KYSE‐450cells confirmed the direct interaction of BAALC‐AS1 and G3BP2. C. RIP assays were performed using G3BP2 and IgG antibody in KYSE‐450 cells. RT‐qPCR was performed to detect the RNA enhancement after BAALC‐AS1 knockdown. IgG antibody was used as negative control. D. Relative expression of G3BP2 quantified by RT‐qPCR after BAALC‐AS1 knockdown in KYSE‐450 cells. E. Western blotting analyses of the protein expression levels of G3BP2 after BAALC‐AS1 knockdown in KYSE‐450 cells. F. Expression of c‐Myc and BAALC‐AS1 quantified by RT‐qPCR after G3BP2 knockdown in KYSE‐450 cells. G. Expression of c‐Myc quantified by RT‐qPCR after BAALC‐AS1 knockdown in KYSE‐450 cells. H. Western blotting analyses of the protein expression levels of c‐Myc after BAALC‐AS1 knockdown in KYSE‐450 cells. I. BAALC‐AS1 knockdown reduced the stability of c‐Myc mRNA. si/BAALC‐AS1‐transfected KYSE‐450 cells were treated with 5 μg/mL actinomycin D for the indicated times. Relative c‐Myc mRNA expression levels were detected by RT‐qPCR. siNC‐transfected KYSE‐450 cells were used as control. J. Expression of c‐Myc quantified by RT‐qPCR after BAALC‐AS1 and G3BP2 knockdown in KYSE‐450 cells. K. RIP experiments were performed using G3BP2 antibody and IgG antibody in KYSE‐450 cells after BAALC‐AS1 overexpression. RT‐qPCR was performed to detect pulled‐down c‐Myc and BAALC‐AS1. IgG antibody was used as negative control. L. Silencing G3BP2 rescued the siBAALC‐AS1‐reduced luciferase activity of c‐Myc 3‐'UTR. NC, negative control; siRNA/BAALC‐AS1, small interfering RNA for BAALC‐AS1; siRNA/G3BP2, small interfering RNA for G3BP2; Vector, cells were infected with empty lentiviruses; BAALC‐AS1, cells were infected with lentiviruses expressing BAALC‐AS1. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All values are expressed as the mean ± SEM of 3 experiments.
We therefore hypothesized that BAALC‐AS1 regulated c‐Myc expression through G3BP2. We then determined whether BAALC‐AS1 enhanced c‐Myc expression by binding to G3BP2 and found that G3BP2 knockdown significantly abrogated the si/BAALC‐AS1‐induced c‐Myc down‐regulation (Figure 6J and Supplementary Figure S5B). We next investigated whether BAALC‐AS1 bound competitively to G3BP2 and sequestered it from c‐Myc mRNA. RIP assays showed that c‐Myc was significantly enriched in the anti‐G3BP2‐precipitated complexes, whereas BAALC‐AS1 overexpression significantly decreased the enrichment of c‐Myc in G3BP2 precipitates (Figure 6K). Furthermore, silencing G3BP2 rescued the si/BAALC‐AS1‐reduced luciferase activity via the c‐Myc 3'‐UTR‐containing reporter (Figure 6L). These results suggested that BAALC‐AS1 directly interacted with G3BP2 and disrupted the association between G3BP2 and c‐Myc 3′‐UTR.
3.6. BAALC‐AS1 promotes cell proliferation by up‐regulating c‐Myc expression
BAALC‐AS1 overexpression increased the mRNA and protein levels of c‐Myc (Figure 7A‐B). Silencing c‐Myc abrogated the stimulatory effect of BAALC‐AS1 overexpression on cell growth (Figure 7C) and proliferation (Figure 7D). Flow cytometry showed that the increased percentage of cells in S and G2/M phases caused by BAALC‐AS1 overexpression was successfully rescued by si/c‐Myc (Figure 7E). Similar results were found in the expression of Cyclin D, Cyclin E, CDK‐2, and CDK4 (Figure 7F). Together, these results suggested that BAALC‐AS1 promoted cell proliferation and G1/S transition by enhancing c‐Myc expression.
FIGURE 7.
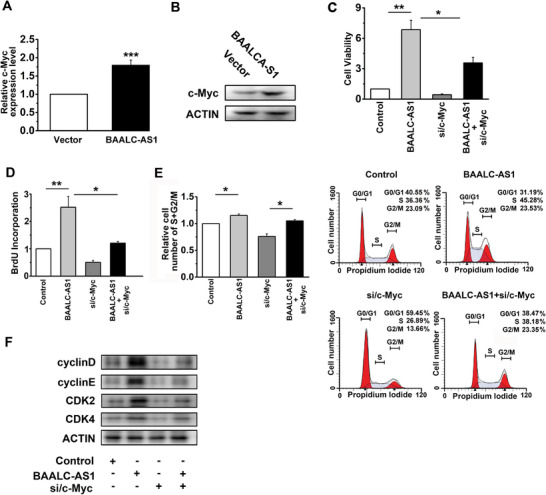
BAALC‐AS1 promotes cell proliferation by up‐regulating c‐Myc. A. Expression of c‐Myc quantified by RT‐qPCR after BAALC‐AS1 overexpression in KYSE‐450 cells. B. Western blotting analyses of the protein expression levels of c‐Myc after BAALC‐AS1 overexpression in KYSE‐450 cells. C. MTS assay was used to determine the viability of KYSE‐450 cells transfected with BAALC‐AS1 overexpression plasmids and c‐Myc siRNAs. D. BrdU incorporation showed the synthesis of DNA in KYSE‐450 cells transfected with BAALC‐AS1 overexpression plasmids and c‐Myc siRNAs. E. Flow cytometry were performed to determine the percentage of cells in S and G2/M phases in KYSE‐450 cells transfected with BAALC‐AS1 overexpression plasmids and c‐Myc siRNAs. F. Western blotting analyses of the protein expression levels of Cyclin D, Cyclin E, CDK2, and CDK4 in KYSE‐450 cells transfected with BAALC‐AS1 overexpression plasmids and c‐Myc siRNAs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All values are expressed as the mean ± SEM.
3.7. c‐Myc regulates BAALC‐AS1 expression by directly binding to its promoter region
Considering that c‐Myc is a transcription factor [33], we examined the expression of BAALC‐AS1 by RT‐qPCR after silencing c‐Myc expression. We found that BAALC‐AS1 expression was decreased following c‐Myc knockdown (Figure 8A). Importantly, BAALC‐AS1 promoter activity was decreased by transfection with c‐Myc siRNAs (Figure 8B). Overall, our results showed that c‐Myc regulates BAALC‐AS1 expression by directly binding to its promoter region in ESCC cells.
FIGURE 8.
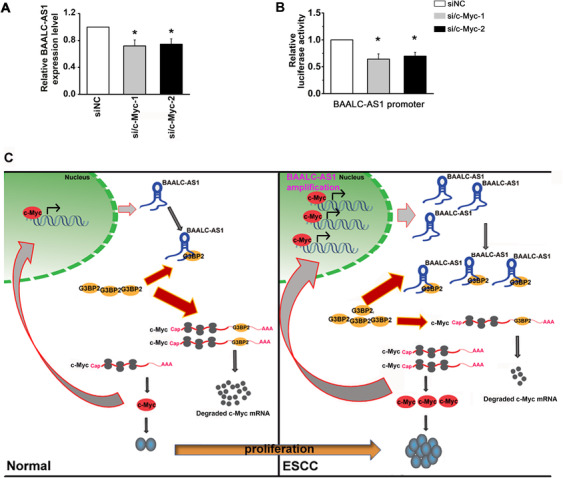
c‐Myc acts directly with the BAALC‐AS1 promoter. A. Expression of BAALC‐AS1 quantified by RT‐qPCR after c‐Myc knockdown in KYSE‐450 cells. B. Silencing c‐Myc decreased the luciferase activity controlled by BAALC‐AS1 promoter. C. Summary diagram describing how BAALC‐AS1 regulates proliferation in ESCC cells. NC, negative control; si/c‐Myc, small interfering RNA for c‐Myc; *, P < 0.05. All values are expressed as the mean ± SEM of 3 experiments
In summary, we demonstrated that BAALC‐AS1 interacted with G3BP2 and inhibited the degradation of c‐Myc RNA 3'‐UTR by G3BP2, leading to the accumulation of c‐Myc expression. The c‐Myc also acted as a transcription factor on the promoter region of BAALC‐AS1, which induced the expression of BAALC‐AS1, and formed a positive feedback loop of BAALC‐AS1/G3BP2/c‐Myc (Figure 8C).
4. DISCUSSION
At present, we have found a large number of lncRNAs, but their functions remain largely unknown [7, 10, 34, 35, 36, 37, 38, 39, 40, 41, 42]. In the present study, we identified a novel lncRNA BAALC‐ASS1 that played a vital role in ESCC progression. BAALC‐AS1 regulated cell proliferation by accelerating the cell cycle and promoting cell migration and invasion in ESCC.
BAALC‐AS1 is an antisense transcript, and previous studies have reported that natural antisense transcripts have important roles in many physiological or pathological processes involving the regulation of sense gene expression [9, 43, 44]. However, because BAALC‐AS1 is located at the antisense chain of the BAALC protein‐coding gene intron region, it has little effect on the regulation of BAALC. The mechanisms of lncRNAs in regulating gene expression might be partially dependent on its interaction with proteins [8, 45]. We therefore used an RNA pull‐down assay to identify the protein(s) interacting with BAALC‐AS1 and confirmed the BAALC‐AS1 interaction with G3BP2. In general, the interaction between lncRNAs and proteins affects protein activity [9, 46], while we found that neither the RNA nor the protein level of G3BP2 was changed when BAALC‐AS1 was knocked down. Considering that both BAALC‐AS1 and G3BP2 are expressed in the cytoplasm and that G3BP has been reported to cleave the 3′‐UTR of the c‐Myc mRNA, thereby affecting c‐Myc stability, we examined the expression of c‐Myc when BAALC‐AS1 was knocked down. We were surprised that the RNA and protein levels of c‐Myc were down‐regulated when BAALC‐AS1 was knocked down. RIP and luciferase activity confirmed that BAALC‐AS1 up‐regulation abolished the inhibitory effects of G3BP2 on c‐Myc expression. We found that BAALC‐AS1 was down‐regulated while c‐Myc was up‐regulated after G3BP2 was knocked down. We therefore concluded that G3BP bound to various mRNA species and was either degraded or stabilized, as dictated by the bound sequences and associated proteins. Similar results occurred in its regulation with CDK7 and CDK9 [23]. The regulatory network of G3BP2 appears to be complex and may be highly dependent on the cellular context.
Further investigations are warranted to delineate the mechanistic consequences of G3BP2 in tumorigenesis. A previous study has reported that G3BP exhibited endonuclease activity on a target in a phosphorylation‐dependent manner [25], so we speculated that BAALC‐AS1 may affect the phosphorylation state of G3BP2 and thus its regulation of c‐Myc. We will then explore the site and specific mechanism of interaction between G3BP2 and BAALC‐AS1 in the future.
We confirmed that high levels of BAALC‐AS1 were associated with cell proliferation, which regulated cell cycle distribution by up‐regulating c‐Myc at the mRNA and protein levels. We also showed that c‐Myc bound to the promoter of BAALC‐AS1 to activate its transcription. A study has shown that c‐Myc regulated the promoter activity of lncRNA, such as those of lncRNA NEAT1 [33]. These results along with our results might reflect a reciprocal regulatory mechanism between transcriptional factors and lncRNAs, which consequentially amplified their mutual oncogenic functions in somatic malignancies.
Our findings identified the role and mechanism of BAALC‐AS1 in the regulation of cell proliferation in ESCC, which should facilitate the development of new therapeutic strategies for the treatment of this disease.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Tissue microarrays (TMAs) of ESCC specimens were obtained from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China) with the approval of the Institutional Review Board. Written informed consent was obtained from all patients prior to the study. The use of the animals for research purposes was approved by the Peking University Cancer Hospital Institutional Research Ethics Committee (Approval No. EAEC 2020‐07).
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the National Natural Science Foundation of China (81830086, 81988101, 81702748, and 81902835), China Postdoctoral Science Foundation (2020M670067), Beijing Municipal Commission of Health and Family Planning Project (PXM2018_026279_000005), and Guangdong Basic and Applied Basic Research Foundation (2019B030302012).
AUTHORS' CONTRIBUTIONS
Q.M.Z. and H.Y.Z. conceived the idea. Y.W. and W.M.Z. conducted the analyses. H.Y.Z., Q.N.W. and J.W.F. provided the data. All authors contributed to the writing and revisions. All authors read and approved the final manuscript.
Supporting information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
ACKNOWLEDGEMENTS
Not applicable.
Zhang H, Wang Y, Zhang W, et al. BAALC‐AS1/G3BP2/c‐Myc feedback loop promotes cell proliferation in esophageal squamous cell carcinoma. Cancer Commun. 2021;41:240–257. 10.1002/cac2.12127
REFERENCES
- 1. Xu Y, Yu X, Chen Q, Mao W. Neoadjuvant versus adjuvant treatment: which one is better for resectable esophageal squamous cell carcinoma? World journal of surgical oncology. 2012;10:173. 10.1186/1477-7819-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91‐5. 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68(6):394‐424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4. Lin C, Wang Y, Wang Y, Zhang S, Yu L, Guo C et al. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene. 2017;36(38):5392‐406. 10.1038/onc.2017.133. [DOI] [PubMed] [Google Scholar]
- 5. Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400‐12. 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 6. Matsui M, Corey DR. Non‐coding RNAs as drug targets. Nature reviews Drug discovery. 2017;16(3):167‐79. 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. The New England journal of medicine. 2016;374(23):2287‐9. 10.1056/NEJMcibr1603785. [DOI] [PubMed] [Google Scholar]
- 8. Engreitz JM, Ollikainen N, Guttman M. Long non‐coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nature reviews Molecular cell biology. 2016;17(12):756‐70. 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 9. Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561(7721):132‐6. 10.1038/s41586-018-0453-z. [DOI] [PubMed] [Google Scholar]
- 10. Gil N, Ulitsky I. Regulation of gene expression by cis‐acting long non‐coding RNAs. Nature reviews Genetics. 2020;21(2):102‐17. 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 11. Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nature genetics. 2018;50(12):1705‐15. 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang R, Sun Y, Li L, Niu Y, Lin W, Lin C, et al. Preclinical Study using Malat1 Small Interfering RNA or Androgen Receptor Splicing Variant 7 Degradation Enhancer ASC‐J9((R)) to Suppress Enzalutamide‐resistant Prostate Cancer Progression. European urology. 2017;72(5):835‐44. 10.1016/j.eururo.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pasmant E, Sabbagh A, Masliah‐Planchon J, Ortonne N, Laurendeau I, Melin L, et al. Role of noncoding RNA ANRIL in genesis of plexiform neurofibromas in neurofibromatosis type 1. Journal of the National Cancer Institute. 2011;103(22):1713‐22. 10.1093/jnci/djr416. [DOI] [PubMed] [Google Scholar]
- 14. Yap KL, Li S, Munoz‐Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular cell. 2010;38(5):662‐74. 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocrine reviews. 2015;36(1):25‐64. 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071‐6. 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christofori G, Naik P, Hanahan D. Deregulation of both imprinted and expressed alleles of the insulin‐like growth factor 2 gene during beta‐cell tumorigenesis. Nature genetics. 1995;10(2):196‐201. 10.1038/ng0695-196. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, Tycko B. Monoallelic expression of the human H19 gene. Nature genetics. 1992;1(1):40‐4. 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]
- 19. Chen Z, Gao Y, Yao L, Liu Y, Huang L, Yan Z, et al. LncFZD6 initiates Wnt/beta‐catenin and liver TIC self‐renewal through BRG1‐mediated FZD6 transcriptional activation. Oncogene. 2018;37(23):3098‐112. 10.1038/s41388-018-0203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uszczynska‐Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non‐coding RNA transcriptome. Nature reviews Genetics. 2018;19(9):535‐48. 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nair L, Chung H, Basu U. Regulation of long non‐coding RNAs and genome dynamics by the RNA surveillance machinery. Nature reviews Molecular cell biology. 2020;21(3):123‐36. 10.1038/s41580-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang HZ, Liu JG, Wei YP, Wu C, Cao YK, Wang M. Expression of G3BP and RhoC in esophageal squamous carcinoma and their effect on prognosis. World journal of gastroenterology. 2007;13(30):4126‐30. 10.3748/wjg.v13.i30.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lypowy J, Chen IY, Abdellatif M. An alliance between Ras GTPase‐activating protein, filamin C, and Ras GTPase‐activating protein SH3 domain‐binding protein regulates myocyte growth. The Journal of biological chemistry. 2005;280(27):25717‐28. 10.1074/jbc.M414266200. [DOI] [PubMed] [Google Scholar]
- 24. Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, et al. A novel phosphorylation‐dependent RNase activity of GAP‐SH3 binding protein: a potential link between signal transduction and RNA stability. Molecular and cellular biology. 1998;18(7):3956‐65. 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, et al. RasGAP‐associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation‐dependent localization. Molecular and cellular biology. 2001;21(22):7747‐60. 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu M, Wang Y, Yang D, Gong Y, Rao F, Liu R, et al. A PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma. EBioMedicine. 2019;41:244‐55. 10.1016/j.ebiom.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H, Liu Y, Yan L, Du W, Zhang X, Zhang M, et al. Bone morphogenetic protein‐7 inhibits endothelial‐mesenchymal transition in pulmonary artery endothelial cell under hypoxia. Journal of cellular physiology. 2018;233(5):4077‐90. 10.1002/jcp.26195. [DOI] [PubMed] [Google Scholar]
- 28. Zhang H, Liu Y, Yan L, Zhang M, Yu X, Du W, et al. Increased levels of the long noncoding RNA, HOXA‐AS3, promote proliferation of A549 cells. Cell death & disease. 2018;9(6):707. 10.1038/s41419-018-0725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang LY, Wu JL, Qiu HB, Dong SS, Zhu YH, Lee VH, et al. PSCA acts as a tumor suppressor by facilitating the nuclear translocation of RB1CC1 in esophageal squamous cell carcinoma. Carcinogenesis. 2016;37(3):320‐32. 10.1093/carcin/bgw010. [DOI] [PubMed] [Google Scholar]
- 30. Niknezhad Z, Hassani L, Norouzi D. Investigating actinomycin D binding to G‐quadruplex, i‐motif and double‐stranded DNA in 27‐nt segment of c‐MYC gene promoter. Materials science & engineering C, Materials for biological applications. 2016;58:1188‐93. 10.1016/j.msec.2015.09.072. [DOI] [PubMed] [Google Scholar]
- 31. Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox‐3 and PSF interact to activate neural cell‐specific alternative splicing. Nucleic acids research. 2011;39(8):3064‐78. 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang W, Hong R, Li L, Wang Y, Du P, Ou Y, et al. The chromosome 11q13.3 amplification associated lymph node metastasis is driven by miR‐548k through modulating tumor microenvironment. Molecular cancer. 2018;17(1):125. 10.1186/s12943-018-0871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng C, Liu S, Lu S, Yu X, Lai J, Wu Y, et al. The c‐Myc‐regulated lncRNA NEAT1 and paraspeckles modulate imatinib‐induced apoptosis in CML cells. Molecular cancer. 2018;17(1):130. 10.1186/s12943-018-0884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei LH, Guo JU. Coding functions of “noncoding” RNAs. Science. 2020;367(6482):1074‐5. 10.1126/science.aba6117. [DOI] [PubMed] [Google Scholar]
- 35. Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nature reviews Genetics. 2019;20(10):599‐614. 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- 36. Schmitt AM, Chang HY. Gene regulation: Long RNAs wire up cancer growth. Nature. 2013;500(7464):536‐7. 10.1038/nature12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, et al. Genome‐scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343‐6. 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rheinbay E, Parasuraman P, Grimsby J, Tiao G, Engreitz JM, Kim J, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547(7661):55‐60. 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344‐52. 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wahlestedt C. Targeting long non‐coding RNA to therapeutically upregulate gene expression. Nature reviews Drug discovery. 2013;12(6):433‐46. 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 41. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172(3):393‐407. 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell‐cycle promoters. Nature genetics. 2011;43(7):621‐9. 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514(7520):102‐6. 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin alpha Promoter Choice. Cell. 2019;177(3):639‐53 e15. 10.1016/j.cell.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. lncRNA‐dependent mechanisms of androgen‐receptor‐regulated gene activation programs. Nature. 2013;500(7464):598‐602. 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang W, Wan H, Feng G, Qu J, Wang J, Jing Y, et al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature. 2018;560(7720):661‐5. 10.1038/s41586-018-0437-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


