Abstract
Epithelial‐to‐mesenchymal transition (EMT) is implicated in a wide array of malignant behaviors of cancers, including proliferation, invasion, and metastasis. Most notably, previou studies have indicated that both cancer stem‐like properties and drug resistance were associated with EMT. Furthermore, microRNAs (miRNAs) play a pivotal role in the regulation of EMT phenotype, as a result, some miRNAs impact cancer stemness and drug resistance. Therefore, understanding the relationship between EMT‐associated miRNAs and cancer stemness/drug resistance is beneficial to both basic research and clinical treatment. In this review, we preliminarily looked into the various roles that the EMT‐associated miRNAs play in the stem‐like nature of malignant cells. Then, we reviewed the interaction between EMT‐associated miRNAs and the drug‐resistant complex signaling pathways of multiple cancers including lung cancer, gastric cancer, gynecologic cancer, breast cancer, liver cancer, colorectal cancer, pancreatic cancer, esophageal cancer, and nasopharyngeal cancer. We finally discussed the relationship between EMT, cancer stemness, and drug resistance, as well as looked forward to the potential applications of miRNA therapy for malignant tumors.
Keywords: cancer, epithelial‐to‐mesenchymal transition, microRNA, cancer stem cell, cancer stemness, drug resistance
In this review, we preliminarily looked into the various roles that the EMT‐associated miRNAs play in the stem‐like nature of malignant cells. Then we reviewed the interaction between drug resistance and EMT‐associated miRNAs with elaborated signal pathways, especially the opposite roles in various cancer types. We finally arrived at a conclusion concerning the relationship between EMT, stemness and drug resistance and discussed the potential application of miRNA therapy for malignant tumors.
Abbreviations
- 5‐FU
fluorouracil
- ADGRE2
adhesion G protein‐coupled receptor E2
- BOK
Bcl2 related ovarian killer
- Cathepsin L
CTSL
- CAVIN2
caveolae‐associated protein 2
- CRC
colorectal cancer
- CSC
cancer stem cell
- CTSB
cathepsin B
- DEDD
death effector domain‐containing
- DOX
doxorubicin
- E2F2
E2F transcriptional factor 2
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EMT
epithelial‐mesenchymal‐transition
- EOC
epithelial ovarian cancer
- ERRFI1
ERBB receptor feedback inhibitor 1
- ESCRT
endosomal sorting complex that is required for transport
- EZH2
enhancer of zeste homolog 2
- FAK
focal adhesion kinase
- FOXO1
forkhead box protein O1
- FOXQ1
forkhead box Q1
- FZD7
frizzled class receptor 7
- GAS5
growth arrest‐specific 5
- GATA3
GATA binding protein 3
- GBM
glioblastoma multiforme
- GDPD5
glycerophosphodiester phosphodiesterase domain containing 5
- Gli1
GLI family zinc finger 1
- GSK‐3β
glycogen synthase kinase 3β
- HCC
hepatocellular carcinoma
- ITGB3
integrin β3
- IL‐6
interleukin 6
- IL6R
interleukin 6 receptor
- LAD
lung adenocarcinoma
- MCRS1
microspherule protein 1
- MCTS1
malignant T‐cell amplified sequence 1
- MET
mesenchymal‐epithelial transition
- miRNA
microRNA
- MMP‐2
matrix metalloproteinase‐2
- MMP‐9
matrix metalloproteinase‐9
- NFATC1
nuclear factor of activated T‐cells 1
- NRP1
neuropilin‐1
- NSCLC
non‐small‐cell lung carcinoma
- OC
ovarian cancer
- ORF
open reading frames
- PC
pancreatic cancer
- PIK3CD
phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit delta
- PEBP1
phosphatidylethanolamine binding protein 1
- PEBP4
phosphatidylethanolamine binding protein 4
- PTP1B
protein‐tyrosine phosphatase 1B
- RBICC1
RB1‐induced coiled‐coil 1
- RKIP
Raf kinase inhibitor protein
- SALL4
sal‐like protein 4
- SCLC
small cell lung cancer
- SMAD2
SMAD family member 2
- SMAD4
SMAD family member 4
- SOCS3
suppressor of cytokine signaling 3
- STAT3
signal transducer and activator of transcription‐3
- TDGF1
teratocarcinoma‐derived growth factor 1
- TKI
tyrosine kinase inhibitor
- TMZ
temozolomide
- TP53INP1
tumor protein p53 inducible nuclear protein 1
- UBE2C
ubiquitin‐conjugating enzyme E2 C
- VEGFA
Vascular endothelial growth factor A
- VitD
Vitamin D
- ZEB1
zinc finger E‐box binding homeobox 1
1. INTRODUCTION
The epithelial‐mesenchymal‐transition (EMT) represents a morphogenetic process that is associated with the invasiveness [1], metastasis [2], and chemoresistance [3] of malignant tumors. In cancer cells undergoing EMT, the expression of mesenchymal markers, including N‐cadherin [4] and vimentin [5], is up‐regulated and the expression of the epithelial markers, including E‐cadherin [4] and ZO‐1 [6], was down‐regulated. Moreover, the essential program of EMT depends on EMT‐associated transcriptional factors, such as zinc finger E‐box binding homeobox (ZEB1 and ZEB2), zinc finger proteins (Snail and Slug), and Twist‐family of basic helix‐loop‐helix (bHLH) transcription factors (Twist1, Twist2, and inhibitors of DNA binding and cell differentiation) [7, 8].
MicroRNAs (miRNAs) are a group of non‐coding single‐stranded small RNAs (18‐22 nucleotides) that suppress gene expression by binding to the 3'‐UTR of target mRNA [9, 10, 11]. Mounting evidence exhibited that miRNAs play a crucial role in the malignant behaviors of cancer cells, including EMT‐related cancer metastasis [12]. Therefore, a better understanding of the roles of EMT‐associated miRNAs can help us further explore their potential diagnostic, prognostic, and therapeutic values [13].
2. EMT‐ASSOCIATED miRNAs IN CANCER STEM CELLS (CSCs)
CSCs, a special range of cancer cells, are capable of unlimited self‐renewal and differentiation, thereby contributing to the initiation, progression, metastasis, and development of drug resistance of malignant tumors [14, 15, 16]. Over the past decade, a lingering issue regarding CSCs is how to identify and select them. Nowadays, CD44 [17] and CD133 [18] have been the two most common surface markers used to characterize CSCs. In addition, aldehyde dehydrogenase is also a critical biomarker of CSCs due to their ability to self‐renew [19]. Recently, CD73 was found to be correlated with the features of CSC since it elevated the expression of Sox9 in dual ATK‐mediated signaling pathways via regulating the expression of c‐Myc, and down‐regulated glycogen synthase kinase 3β (GSK‐3β). CD73 and Sox9 in combination could more precisely predict the prognosis of tumor, suggesting that CD73 might serve as a marker of CSCs [20]. Several major factors (i.e. Nanog, Sox2, Oct4, KLF4, c‐Myc) are essential for the maintenance of the pluripotency of CSCs, and they are regulated by miRNAs [21, 22, 23]. It was reported that EMT was intimately associated with to CSCs. EMT is a gradually adjusted process[24]. Cancer cells undergoing a partial EMT (hybrid epithelial/mesenchymal phenotype) acquire stem‐like features [25]. This epithelial/mesenchymal hybrid status is crucial for tumor initiation, in which the Wnt signaling pathway plays a key part [26]. Apart from the Wnt signaling, there are other crosstalks between EMT and CSCs, such as the Notch/Jagged [27] and hedgehog [28] signal pathway. Most EMT transcriptional factors, which are modulated by a number of miRNAs, are implicated in CSCs, indicating that miRNAs can affect EMT‐associated elements and subsequently exert an impact on the stem‐like properties of CSCs [29, 30].
2.1. EMT‐associated miRNAs that inhibit cancer stemness
miR‐99a prevents EMT progression and reduces CSC population, both in vitro and in vivo, by directly suppressing the expression of E2F transcriptional factor 2 (E2F2) and adhesion G protein‐coupled receptor E2 (ADGRE2) [29]. Zhou et al. [31] illustrated that miR‐125b overexpression could attenuate EMT phenotype and CSC generation by inhibiting SMAD family member 2 and 4 (SMAD2 and SMAD4). miR‐145 targets multiple stem cell transcription factors, and the action was found inversely correlated with EMT in colorectal cancer (CRC). Furthermore, Snail could elicit resistance to radiotherapy by repressing the expression of miR‐145 [32]. miR‐199a‐5p conferred its tumor‐suppressing function in triple‐negative breast cancer by inhibiting EMT and stemness by down‐regulating its potential target phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit delta (PIK3CD) [33]. Loss of miR‐205 was found to expand mammary stem cell populations, enhance self‐renewal, and promote EMT. Jagged1 secreted by the tumor stroma significantly suppressed miR‐205 [34]. Moreover, the loss of miR‐205 induced cell stemness by activating NOTCH2 suggests that there might be a jagged1/miR‐205/NOTCH2 signaling pathway that regulates cancer stemness [34, 35]. In prostate cancer, miR‐218 suppressed the exhibition of CSC‐like properties and EMT by binding to its potential target GLI family zinc finger 1 (Gli1) [36]. Vascular endothelial growth factor A (VEGFA) upregulated Sox2, resulting in cancer cell invasion, self‐renewal, and metastasis triggered by Slug overexpression plus miR‐452 loss [37]. miR‐504, which is involved in the Wnt‐β‐catenin pathway, was shown to suppress malignant behaviors of glioblastoma multiforme (GBM), including aggression, migration, EMT, and stemness by directly inhibiting frizzled class receptor 7 (FZD7) [38]. miRNA sponges with multiple tandem miRNA binding sites could separate miRNAs from their target mRNAs [39]. In CRC, hypoxia upregulated the expression of a newly‐identified lncRNA AK000053 in a HIF‐1α‐dependent manner, and functioned as a miR‐508 sponge. Additionally, loss of miR‐508 resulted in the overexpression of zinc finger E‐box binding homeobox 1 (ZEB1), Bmi1, and sal‐like protein 4 (SALL4), subsequently leading to EMT and cancer stemness, and poor survival of CRC patients [40]. The downregulation of miR‐1247 induced by cancer‐associated fibroblasts boosted the performance of EMT and escalated cell invasion and stemness. In addition, neuropilin‐1 (NRP1) served not only as the miR‐1247 target but also as a coreceptor of EGFR signaling [41]. Han et al. [42] illustrated that miR‐4319 repressed cell proliferation, EMT, and cancer stemness by targeting forkhead box Q1 (FOXQ1) at the post‐transcriptional level in hepatocellular carcinoma (HCC).
miR‐203 was found to be not only a stemness‐suppressing factor but also an anti‐apoptotic factor and was downregulated by the EMT‐associated transcriptional factor, ZEB1 [43]. In renal cell carcinoma, miR‐203 inhibited LncRNA HOTAIR and induced a tumor‐suppressor effect, i.e., suppressing EMT via the PTEN/PI3K/ATK pathway, which was involved in a recognized lipid kinase dubbed Acylglycerol kinase [44].and the progress was also decrease the expression of KLF4 and Nanog[44, 45]. miR‐203 inhibited the migration, endothelial cell tube formation, and stemness of prostate cancer cells, with Slug being downregulated. Moreover, miR‐203, by targeting Slug, further repressed the GSK‐3β/β‐catenin signaling pathway [46]. Besides, silencing miR‐203 enhanced the stemness of colon cancer cells, with several EMT activators up‐regulated, in which Snail could inhibit miR‐203 expression. Additionally, hyaluronan and CD44 suppressed miR‐203 expression via activating c‐Src kinase [47].
Canonical tumor suppressor miRNAs, such as miR‐34a, miR‐200 family, and let‐7, interact in various signaling pathways involving inhibition of CSCs and EMT. In ovarian cancer, let‐7a, miR‐200c, and miR‐186 could significantly reverse resistin‐induced EMT and stemness [48]. Dong et al. [49] reported that miR‐34a and miR‐137 directly targeted Snail, thus suppressing EMT and sphere‐forming capability of ovarian cancer cells, and leading to more favorable survival outcome of the patients. In high grade serous ovarian cancer cells with Snail knockdown, let‐7 expression was up‐regulated, and Nanog and Lin28 were down‐regulated, suggesting that Snail/Let‐7 axis might be an intersection between stemness and EMT [50]. miR‐204, miR‐200c, and miR‐34a inhibited cancer stemness and EMT, leading to self‐renewal and metastasis of breast cancer [14]. EMT and CSC properties were involved in the lung cancer risk of PM2.5, and chronic PM2.5 could significantly downregulate the levels of three stemness‐associated microRNAs, Let‐7a, miR‐16 and miR‐34a [51]. Weng et al. [52] identified an oncogene called malignant T‐cell amplified sequence 1 (MCTS1), which mediated cancer stemness and EMT in triple‐negative breast cancer by up‐regulating interleukin 6 (IL‐6) expression, elevating interleukin 6 receptor (IL6R) level and increasing the population of tumor‐promoting M2 macrophages. Nonetheless, miR‐34a could reverse the carcinogenic effect of MCTS1 by inhibiting IL‐6R expression and triggering M1 polarization. ZNF281 not only interacted with Nanog, OCT4, Sox2, and c‐Myc, but also induced cancer stemness markers LGR5 and CD33 in CRC. The expression of ZNF281 was up‐regulated by Snail but down‐regulated by tumor suppressor miR‐34a [53]. And miR‐200c and miR‐141could regulate the expression of Bmi1 and ZEB1 in HCC with bile duct tumor thrombus [54]. The inflammation‐induced transcriptional factor, that is a nuclear factor of activated T‐cells 1 (NFATC1) functioned as a paramount regulator of cell plasticity in pancreatic cancer[55]. Particularly, NFATC1 drove EMT to reprogram and bestowed pancreatic cancer cells with the phenotype that CSCs possessed via Sox2‐dependent transcription of EMT and stemness factors, which was antagonized by antithetical p53‐miR200c signaling [55].
Together, it is demonstrated that EMT and stemness share the similar signal pathways, which are mediated by miRNAs. EMT‐associated miRNAs could inhibit the stemness features in various cancer types due to suppressing the specific gene and the downstream signal pathways.
2.2. EMT‐associated miRNAs that promote cancer stemness
miR‐10b promoted CSC features, such as stemness and self‐renewal. It is regulated by TWIST and TGF‐β, and they are both associated with CSCs [56, 57, 58]. Moreover, miR‐10b indirectly affected stem markers OCT4 and SNAIL expression in breast cancer through the PTEN/PI3K/AKT pathway [58]. In breast cancer, the up‐regulation of TGF‐β‐induced miR‐10b‐5p contributed to tumor‐related myoepithelial cells acquiring invasiveness phenotype and CSCs occurring through targeting RB1‐induced coiled‐coil 1 (RBICC1) [57]. Thus, miR‐10b is a bona fide regulator of the clonal potential and migration capability of CSCs. Hypoxic microenvironment‐induced miR‐210 up‐regulation, in breast cancer stem cells, inhibited E‐cadherin by binding to its open reading frames (ORF) and inducing the over‐expression of its transcription repressor Snail [59]. miR‐577 is involved in the metastasis of TGF‐β‐induced gastric cancer by targeting caveolae‐associated protein 2 (CAVIN2). In addition, TGF‐β activated miR‐577 via the NF‐κB signaling pathway [60]. In normal human colonic epithelial cells, an elevated expression of miR‐1207‐5p could reinforce stemness of the cancer cells as demonstrated by significantly enhanced morphological phenotype of EMT and increased levels of mesenchymal and CSC markers [61]. miR‐5188 targeted forkhead box protein O1 (FOXO1) and reduced the nuclear translocation of β‐catenin directly, it could promote the activation of Wnt signaling to downstream EMT, cancer stemness, and c‐Jun both in HCC and breast cancer. In addition, c‐Jun activated miR‐5188 expression at the transcriptional level, forming a positive feedback loop, which could be induced by hepatitis X protein in HCC [62, 63].
Though the majority of EMT‐associated miRNAs inhibit the stemness, there still exsit some EMT‐associated miRNAs promiting the cancer stemness, which is mentioned above. For the most cases, these miRNAs facilitate the stemness properties by blocking related tumor‐suppressive signals.
3. EMT‐ASSOCIATED miRNAs INVOLVED IN THE DEVELOPMENT OF DRUG RESISTANCE OF CANCER
Long‐term drug therapy tends to result in drug resistance of cancer, a tough challenge facing clinicians. The development of drug resistance is multifactorial and EMT is a key factor [64]. Moreover, several miRNAs co‐regulate EMT and drug resistance. Therefore, understanding the function of relevant miRNAs and the pathways involved will help us gain insight into and eventually sort out the problem.
3.1. Lung cancer
In docetaxel‐resistant lung adenocarcinoma (LAD), over‐expression of miR‐26a could suppress cellular proliferation, increase apoptosis rate and switch EMT to mesenchymal‐epithelial transition (MET), both in vitro and in vivo, by downregulating the enhancer of zeste homolog 2 (EZH2) that was reported to induce EMT via binding to the PTEN promoter to a certain extent [65, 66]. Down‐regulated miR‐130a was associated with multidrug resistance in various cancers [67, 68, 69]. Moreover, miR‐130a reportedly targeted MET and enhanced TRAIL‐sensitivity in non‐small‐cell lung carcinoma (NSCLC) cells [70]. miR‐146b targets protein‐tyrosine phosphatase 1B (PTP1B), and miR‐218 directly targets Slug/ZEB2 signaling pathway, as well as let‐7c suppresses ABCC2‐transporter and Bcl‐xl were all shown to be capable of reversing EMT, thereby lowering the resistance of lung cancer cells to cisplatin [69, 71, 72]. Overexpression of teratocarcinoma‐derived growth factor 1 (TDGF1), an epidermal growth factor (EGF)‐related gene, generated a phenotype of erlotinib resistance, both in epidermal growth factor receptor (EGFR)‐mutated and EGFR‐tyrosine kinase inhibitor (TKI)‐sensitive NSCLC cells, which was confirmed by in‐vitro studies, in murine xenograft models and clinical patients. Mechanistically, SRC and ZEB1 activated by TDGF1 stimulate EMT by down‐regulating miR‐205. As a consequence, up‐regulated miR‐205 might repress SRC and ZEB1 activation in a TDGF1‐dependent fashion, restoring the sensitivity to erlotinib. Furthermore, targeting both EGFR and SRC might overcome inherent EGFR‐inhibitor resistance in EGFR‐mutated NSCLC patients positive for TDGF1 [73]. When PRKCA was directly targeted to repress FAK/Ras/c‐Myc signaling pathway, miR‐296‐3p stimulated its own expression, forming a feedback loop that blocked cisplatin chemoresistance and EMT signaling [74]. Additionally, miR‐296‐3p was inactivated by DDX5/HDGF/β‐catenin signaling, leading to a more aggressive metastasis and stronger chemoresistance in lung adenocarcinoma (LAD) [74]. It was also reported that silencing c‐Myc regulated by miR‐451‐induced MET in docetaxel‐resistant LAD cells through decreasing the expression level of matrix metalloproteinase‐2 (MMP‐2), matrix metalloproteinase‐9 (MMP‐9), Snail, p‐ERK as well as p‐GSK‐3β and increasing E‐cadherin expression. Furthermore, patients with high miR‐451 expression had significantly)P <0.05(more favorable prognosis compared with those with low miR‐451 expression. These findings suggested that miR‐451/c‐Myc/ERK/GSK‐3β axis played a crucial role in suppressing EMT phenotype in docetaxel‐resistant LAD [75]. Yue et al. [76] reported that miR‐483‐3p reversed EMT to MET and inhibited the invasion, migration, and metastasis of lung cancer cells resistant to gefitinib. In molecular terms, miR‐483‐3p directly targeted integrin β3 (ITGB3), and thereby inhibited downstream focal adhesion kinase (FAK)/ERK signaling pathways. Moreover, the miR‐483‐3p deficiency in gefitinib‐resistant lung cancer cells might be ascribed to the hypermethylation of its own promoter. It was reported that miR‐495 served as an oncogenic miRNA or a tumor‐suppressor in a variety of cancers [77, 78, 79]. Intriguingly, different theories are proposed about its function in NSCLC and small cell lung cancer (SCLC). In NSCLC, miR‐495 decreased vimentin but increased E‐cadherin at both transcriptional and translational levels [80]. Additionally, miR‐495 reversed cisplatin resistance by suppressing drug resistance genes ERCC1 and ABCG2 in cisplatin‐resistant NSCLC cells [80].Of note, ubiquitin‐conjugating enzyme E2 C (UBE2C), which promotes EMT, was found to mediate miR‐495 in the reversal of cisplatin resistance [80].Furthermore, the combination of siUBE2C and cisplatin caused the in vitro down‐regulation of vimentin and up‐regulation of E‐cadherin in mRNA and protein levels. miR‐495 was also found able to inhibit tumor growth in vivo. These results indicated that the miR‐495‐UBE2C‐ERCC1/ABCG2 axis could restore the sensitivity to cisplatin by down‐regulating anti‐drug genes and inhibiting EMT in cisplatin‐resistant NSCLC [80]. However, miR‐495 promoted the EMT‐related chemoresistance of SCLC via ETK/BMX. This study provided a promising strategy of restoring the sensitivity of SCLC to multiple drugs, including doxorubicin, cisplatin, and VP‐16 in SCLC: i.e., by expressing miR‐495 or depleting ETK/BMX [79].
Canonically, the miR‐200 family, consisting of miR‐141, miR‐200a, miR‐200b, miR‐200c, and miR‐429, serves as tumor suppressors in assorted cancer types, including lung cancer. It is well‐known that miR‐200 participates in the TGF‐β‐induced EMT. Typically, the inhibitory effect of miRNA depends on the number of binding sites in the 3'UTR of the target mRNA. Burk et al. [81] found that TGF‐β2 was a direct target of miR‐200. However, Gregory et al. [82] reported that miR‐200 worked on all the three TGF‐β isoforms, indicating that miR‐200 influences the expression of TGF‐β in both direct and indirect manners due to the lack of binding sites in TGF‐β1 and TGF‐β3. Besides, the prolonged exposure to TGF‐β significantly inhibited the level of miR‐200 because there were more methylated cytosine phosphate guanine)CpG) in the miR‐200 promoter. By up‐regulating miR‐200b and miR‐141, and downregulating ZEB1 in NSCLC cells, nintedanib was capable of reversing TGF‐β1‐induced EMT and resistance to gefitinib. Thus, the combined use of gefitinib and nintedanib promises to be a new alternative for the treatment of NSCLC cells, since it takes care of both the resistance to gefitinib and EMT phenotype [83]. In TGF‐β‐mediated EMT, the miR‐200 family depletion led to an up‐regulated expression of ERBB receptor feedback inhibitor 1 (ERRFI1), a negative regulator of EGFR. The ERRFI1‐mediated decrease of EGFR took place simultaneously with a TGF‐β‐induced EMT‐related kinase switch of cancer cells to an EGFR‐independent state with AKT activated.
In primary tumor xenografts of patient‐derived lung and pancreatic cancers that carried wild‐type EGFR, the tumor MIG6 (mRNA)/miR200 ratio was negatively associated with the responsiveness to erlotinib in vivo. This indicated that a low ratio of ERRFI1 to miR‐200 might serve as a potential predictor of the tumor responsiveness to EGFR‐TKIs [84]. In addition to the TGF‐β signaling, miR‐200c might be related to the obstruction of paclitaxel resistance in lung cancer cells via cathepsin L (CTSL)‐mediated EMT. Moreover, miRNA‐200c and CTSL were mutually attached in a feedback loop [85]. Interestingly, Krentz Gober et al. [86] indicated that the signal containing miR‐140/141/200c was probably regulated by the cell cycle instead of TGF‐β. Particularly, this study revealed that the inhibition of TGF‐β did not suppress EMT in lung cancer cells but induced an EMT‐intermediate state, which overturns the traditional notion about TGF‐β‐mediated EMT. Proliferation/growth signals by constitutively‐activated EGFR might depend on TGF‐β and, in this context; there might be an interaction between TGF‐β and EGFR signaling pathways that obstruct EMT progression instead of stimulating it. This assumption needs to be further verified with more researches, preferably involving cellular or tumoral field.
Researchers have failed to reach a consensus about the function of miR‐155 either. On the one hand, conspicuous discrepancies were observed in the expression levels of miR‐155 and miR‐200c, which were dramatically decreased in gefitinib‐resistant NSCLC cells. Apart from this finding, the expression of SMAD2 and ZEB1 were identified as the target of miR‐155 and miR‐200c, respectively, were substantially up‐regulated. As expected, the E‐cadherin expression was down‐regulated upon restrictive histone modification, whereas vimentin was up‐regulated after active histone modification. Besides, this deficiency of miR‐155 and miR‐200c might be correlated with the epigenetic modifications‐induced EMT and might promote the loss of sensitivity to gefitinib irrespective of the secondary EGFR mutation, which some gefitinib‐resistant cells possess [87]. On the other hand, miR‐155 was found to induce EMT by targeting RHOA at the post‐transcriptional level [88]. Moreover, microspherule protein 1 (MCRS1) promotes TGF‐β1‐induced EMT and triggers resistance to cisplatin and cetuximab by up‐regulating ABCB1 (a multidrug‐resistance gene) at the transcriptional level. Nevertheless, MCRS1 was directly mediated by miR‐129, indicating that miR‐129 was a tumor suppressor that impacted cellular behaviors by regulating the expression of MCRS1 in NSCLC cells. To sum up, the miR‐129/MCRS1/miR‐155 signal axis offers a new perspective for us to understand the molecular mechanism of EMT and drug resistance development, two events that are indicative of the invasion and metastasis of tumor [89]. More researches are warranted to understand the diversified role of miR‐155.
Several miRNAs also play oncogenic parts in lung cancer. The miR‐134/487b/655 cluster located on chromosome 14q32 was also found to cause the TGF‐β1‐induced EMT and influence the gefitinib resistance by directly repressing MAGI2 and its repression subsequently led to the depletion of PTEN in lung cancer. EMT was related to the loss of drug sensitivity and acquisition of resistance to EGFR‐TKIs, whereas preservation of an epithelial phenotype ensured a favorable response to EGFR‐TKIs even in LAD patients harboring wild‐type EGFR genes [90, 91, 92]. These researches indicated that EMT was responsible for the resistance to EGFR‐TKIs, independent of EGFR status. The miR‐134/miR‐487b/miR‐655 cluster promises to be a therapeutic strategy for patients with advanced LAD in the case of EMT phenotype [93]. Upregulated miR‐15b and miR‐27a contributed to EMT and the resistance to cisplatin both in vivo and in vitro by targeting phosphatidylethanolamine binding protein 4 (PEBP4) and phosphatidylethanolamine binding protein 1 (PEBP1/RKIP), respectively [94, 95]. By modulating EMT, miR‐21 reinforced the invasiveness and migrating ability of cisplatin‐ and paclitaxel‐resistant LAD cells by targeting HBP1 [96]. A noticeable shift was observed from the epithelial to the mesenchymal phenotype after the miR‐127 level was elevated in lung cancer cells, and this shift was related to the increased resistance to the EGFR inhibitor and the tumor‐propagating potential [97]. In cancer cells, up‐regulated miR‐127 led to an evident change from the epithelial to the mesenchymal phenotype, and this change was related to their stem‐like features, enhanced resistance to the EGFR receptor inhibitor, and tumor‐spreading potential. On the other hand, suppressing miR‐127 could substantially reverse this malignant transition, impaired the stem‐like traits and the in vivo tumorigenicity of malignant cells.
3.2. Gastric cancer
The expression of miR‐200c and ZEB2 was down‐ and up‐regulated, respectively, in gastric cancer cells, with an evident decline of sensitivity to trastuzumab after treatment with TGF‐β. Besides, miR‐200c was able to restore the sensitivity to trastuzumab and repress the migration and invasion of cancer cells by inhibiting ZEB1 and ZEB2 [98]. miR‐204 was inhibited in fluorouracil (5‐FU)‐resistant GC cells with the epithelial markers decreased and the mesenchymal markers increased simultaneously. In addition, restoration of TGFBR2, a target of miR‐204, could recover resistance to 5‐FU in GC cells with miR‐204 upregulated [99]. Like miR‐204, miR‐574‐3p could antagonize cisplatin resistance in gastric cancer by targeting ZEB1 at both transcriptional and translational levels [100]. Nonetheless, the resistance to cisplatin or 5‐FU in GC cells could be dramatically reduced by suppressing miR‐17, which impaired EMT in GC cells via death effector domain‐containing (DEDD) [101].
Therefore, different miRNAs are associated with antineoplastic drug in gastric cancer. More miRNAs are needed to explore to reduce the drug resistance in gastric cancer.
3.3. Gynecologic cancer
Owing to the aberrant methylation engendered by DNMT1 over‐expression, miR‐30a‐5p, and miR‐30c‐5p levels dropped significantly in cisplatin‐resistant ovarian cancer (OC) cells. On the contrary, miR‐30a/c‐5p inhibited Snail and DNMT1 directly. Hence, a feedback loop between DNMT1 and miR‐30a/c‐5p could be a potential signature for addressing EMT and cisplatin resistance in OC, thereby providing a therapeutic strategy for epigenetically improving the responsiveness to anti‐cancer agents [102].
Elevated miR‐363 restored the sensitivity to cisplatin of cisplatin‐resistant epithelial ovarian cancer (EOC) cells, both in vitro and in vivo. Moreover, studies showed that Snail, identified as a functional target of miR‐363, was greatly elevated, not only in epithelial ovarian cancer(EOC) cell lines resistant to cisplatin but also in EOC patients. Moreover, the over‐expression of Snail dramatically inhibited the repressing effect of miR‐363 on cisplatin resistance of EOC cells, indicating that miR‐363 modulates cisplatin resistance through Snail‐induced EMT [96]. Zhang et al. [103] found that miR‐1294 dysregulation affected OC cisplatin resistance by regulating IGF1R. IGF1R knockdown could suppress the proliferation, migration, invasion, and EMT of SKOVP/DDP cells. Further, elevated miR‐1294 expression inhibited the development of resistance to cisplatin in OC. The miR‐200 family also played a major role in the inhibition of EMT and sensitivity to carboplatin and paclitaxel of OC [104]. Besides, miR‐200b and miR‐200c were inhibited in taxane‐resistant OC cells, and the inhibition was correlated with EMT progression as evidenced by the elevated expression level of MMP2, MMP9, and vimentin [105]. It was found that, in cervical cancer, miR‐25‐3p reversed EMT to MET with enhanced sensitivity to cisplatin in cisplatin‐resistant cells by targeting Sema4C [106].
Knockdown of iASPP, a newly‐identified key EMT inducer, sensitized cervical cancer cells to cisplatin and repressed cell proliferation in vivo. Moreover, iASPP promoted the expression of miR‐20a targeting FBXL5 and BTG3 in a p53‐dependent manner. miR‐20a expression was increased and FBXL5 and BTG3 expression decreased in cervical cancer samples and the results were found to be related to a poor prognosis of the patients [64].
3.4. Breast cancer
Enhanced miR‐129‐5p expression significantly increased E‐cadherin and suppressed vimentin and N‐cadherin expression in MCF‐7/doxorubicin (DOX)‐treated cells. EMT has been seen as an important mechanism responsible for the increased multidrug resistance in breast cancer [107, 108]. As expected, miR‐129‐5p substantially reduced IC50 of DOX, vincristine, and paclitaxel in the MCF‐7/DOX‐treated cells. However, the level of miR‐129‐5p in MCF‐7 cells was lowered by EZH2 and SOX4, which act, respectively, as an epigenetic modification‐silencing gene and a master control gene of EMT, respectively [109]. ZEB1 and ZNF217, identified as a transcriptional activator of TGF‐β, were inhibited by miR‐200c that could restore trastuzumab sensitivity and repress invasion and migration of breast cancer cells. Given that ZEB1 reportedly inhibited miR‐200c, presumably, ZNF217 might participate in a feedback suppression of miR‐200c through TGF‐β/ZEB1 signaling [81].
Introducing miR‐200c, inhibiting TGF‐β signaling pathways, or silencing either ZEB1 or ZNF217 repressed the invasive capability and enhanced the sensitivity of breast cancer cells to trastuzumab. Therefore, the complicated interaction between miR‐200c/ZNF217/TGF‐β/ZEB1 and miR‐200c/ZEB1 suppressed the metastasis and trastuzumab resistance of cancer cells, suggesting that EMT might be involved in the molecular induction of the malignant behaviors of breast cancers [110]. Recently, an lncRNA, termed Linck, was found to exaggerate the expression of ZEB1, and both were negatively related to the miR‐200 family [111]. Due to the inhibitory capability of EMT, miR‐708‐3p was deemed as a tumor‐suppressor miRNA in breast cancer. In addition, Lee et al. [112] suggested that reintroduction of miR‐708‐3p might be a promising therapeutic option for overcoming the chemoresistance of breast cancer cells and, at the same time, suppressing breast cancer metastasis.
miR‐106b, miR‐93, and miR‐25 collectively form the miR‐106b‐25 cluster, and all target a transcriptional stimulator of E‐cadherin, i.e., EP300. They were up‐regulated in doxorubicin‐resistant cells, with miR‐25 playing the leading role in this phenotype. With this cluster, upregulation of a single miRNA would result in target cells obtaining the EMT phenotype, along with the proliferative ability upon treatment with doxorubicin [113].
EMT might be closely related to the malignant behaviors and increase multidrug resistance of breast cancers,while miRNAs could regulate the formation and development of EMT. miRNAs might be a promising therapeutic option for overcoming the chemoresistance of breast cancer cells.
3.5. Liver cancer
In HCC, miR‐125b could overcome the resistance to oxaliplatin through a mechanism involving the reduction of EVA1A‐mediated autophagy, with a simultaneous loss of EMT phenotype [114]. Depletion of Smad4, a target of miR‐130‐3p, reversed EMT to MET in gemcitabine‐resistant HCC cells. Furthermore, miR‐130a‐3p could restore the sensitivity to gemcitabine and inhibit cell growth in gemcitabine‐resistant cells [115]. Vitamin D (VitD) has been deemed as a new regulator of the mTOR pathway [116]. Donatella et al. [117] demonstrated that, in a molecular network, VitD reduced oncogene expression and modulated EMT by up‐regulating the expression of miR‐375, and subsequently resulted in a reversal of the sensitivity to everolimus in everolimus‐resistant HCC cell lines. Particularly, c‐Myc was recently identified as a novel target of miR‐375. The aforementioned results might provide a new approach to restore the sensitivity to mTOR inhibitor sensitivity in the treatment of HCC. The tumor‐suppressing function of miR‐612 was validated by inhibiting EMT and resistance to cisplatin and 5‐FU through the PI3K/AKT2 signaling pathway. Recently, it could reportedly promote HCC metastasis via influencing the morphological formation of invadopodia and EMT. Studies showed that this phenomenon involved the HADHA‐dependent lipid reprogramming [118, 119].
Nevertheless, miR‐27a and miR‐32‐5p were highly expressed in liver cancer patients, particularly in cisplatin‐resistant patients, predicting a poor prognosis. Further studies found that miR‐27a regulated EMT partially by targeting the Raf kinase inhibitor protein (RKIP) and miR‐32‐5p triggered the activation of the PI3K/AKT pathway by inhibiting PTEN and generated exosome‐mediated multidrug resistance by prompting EMT and angiogenesis [114, 120]. It has been known that Fbw7, which is regarded as a miR‐233 target, suppressed EMT, and subsequently increased chemosensitivity of hepatocellular carcinoma cells [121, 122, 123]. Notch‐1 has been confirmed to be one of the targets of FBW7 [124] and to induce EMT in human cancers [125]. Interestingly, genistein could promote the antitumor effect of miR‐223 inhibitor by regulating EMT and Notch‐1 pathway [124]. Genistein in combination with miR‐223 inhibitor can be a potential therapeutic strategy for the treatment of pancreatic cancer.
The production of EMT phenotype and the reversion from EMT to MET are related to tumor resistance. So in liver cancer, miRNAs could regulate EMT and improve the sensitivity of tumor drugs in liver cancer.
3.6. CRC
miR‐134 was reported to increase the sensitivity of CRC to oxaliplatin. However, it was found that astragaloside IV could inhibit the EMT of CRC by promoting the expression of miR‐134, which obviously down‐regulated the CREB1 signaling pathway, and further restored the sensitivity to chemotherapeutic agents [126]. miR‐139‐5p and miR‐195‐5p significantly suppressed the metastasis potential and chemo‐resistance of CRC through EMT by targeting BCL2 and glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5), respectively [127, 128]. miR‐195‐5p bound to its direct target Notch2 to repress IL‐4 secretion modulated by GATA3, ultimately leading to the inhibition of M2‐like tumor‐associated macrophage polarization [129]. Low miR‐145 expression was related to poor responsiveness of rectal cancer patients to neoadjuvant chemoradiation on the basis of 5‐FU chemotherapy. Mechanistically, Slug repressed the activity of the miR‐145 promoter in CRC cells. In addition, the ectopic expression of Slug lowered the sensitivity to 5‐FU, and inversely, the reappearance of miR‐145 dramatically increased 5‐FU sensitivity in vitro [130].
By targeting adenomatous polyposis coli (APC), miR‐125b, which is up‐regulated by the CXCL12/CXCR4 axis, promotes the progression of EMT, thereby further activating the Wnt/β‐catenin signaling pathway. Importantly, there existed a reciprocal positive feedback loop between miR‐125b and CXCR4. Further in‐vitro and in‐vivo experiments on CRC verified a possible role of miR‐125b, i.e., promoting EMT and autophagy, in the development of the resistance to 5‐FU [131]. Intriguingly, although miR‐514b‐3p and miR‐514b‐5p are both derived from the same RNA hairpin, they each have different influence on the invasion and metastasis of CRC. miR‐514b‐3p inhibited migration and drug resistance of CRC cells by decreasing the expression of mesenchymal markers and increasing the expression of epithelial markers. On the contrary, miR‐514b‐3p played a pro‐metastatic role by speeding up the process of EMT. However, the underlying mechanism remains unclear [40]. Apart from chemotherapeutic application, miRNAs can also regulate the effect of radiotherapy on CRC. For instance, miR‐124 could enhance the sensitivity of CRC cells to radiation via inhibiting the expression of a recently‐identified EMT regulator and stemness inducer, PRRX1 [132].
3.7. Pancreatic cancer
Hiramoto et al. [133] and Funamizu et al. [134] illustrated that miR‐200b, miR‐509‐5p or miR‐1243 overexpression could each increase the sensitivity to gemcitabine by suppressing EMT‐associated gene expression, thereby upregulating the E‐cadherin expression in pancreatic cancer. Moreover, over‐expression of miR‐125a‐3p or miR‐3656 played similar role by targeting Fyn and RHOF, respectively [128, 135]. However, TWIST1 overexpression attenuated the enhanced chemotherapeutic effects of miR‐3656 [135]. On the locus of miR‐203, the suppressive histone mark H3K27me3 was reduced by the loss of ZEB1. Mocetinostat, belonging to class I HDAC inhibitor, could affect drug resistance by down‐regulating of ZEB1 expression and up‐regulating miR‐203. Remarkably, mocetinostat did not exert its effect of anti‐resistance to gemcitabine on cancer cells, where ZEB1 had been low and miR‐203 expression was high, suggesting that the effect of mocetinostat would diminish if miR‐203 was already present and ZEB1 had not appeared. However, whether the effect of mocetinostat depends on miR‐203 had not been proved since MTT activity was increased in gemcitabine‐treated cells in either the presence or the absence of mocetinostat [43]. Epigenetic drugs for restoring chemo‐sensitivity of cancers trapped in EMT phenotype, Short‐time treatment of tumor cells with clinically‐used nanomolar doses, without causing immediate cytotoxicity, could result in an antitumor “memory” reaction [136, 137].
On the contrary, miR‐301 both regulated EMT and induced gemcitabine resistance by down‐regulating E‐cadherin expression [134]. An apoptosis‐facilitating gene, dubbed Bcl2 related ovarian killer (BOK), was a target of miR‐296‐5p in PC cells. miR‐296‐5p mimic transfectants also had an aberrant expression of mesenchymal markers. In addition, these transfectants displayed an obviously low apoptosis ratio in reaction to gemcitabine and 5‐FU, with the absence of BOK expression. These results suggested that miR‐296‐5p/BOK signaling axis did play a crucial part in the invasion, EMT, and drug resistance development in pancreatic ductal adenocarcinoma cells (PDAC) cells [138]. Growth arrest‐specific 5 (GAS5), a lncRNA, was identified as a tumor suppressor due to its ability to inhibit the malignant behavior of various cancers [139]. Upregulated GAS5 repressed the stem cell‐like features, EMT, and gemcitabine resistance of PC cells through directly binding the 3’UTR of miR‐221 and subsequently enhancing the expression of its target, suppressor of cytokine signaling 3 (SOCS3) [140].
3.8. Esophageal cancer
In esophageal squamous cell carcinoma, miR‐125a‐5p up‐regulated the E‐cadherin and down‐regulated the N‐cadherin and vimentin expression, with an enhanced cytotoxic effect of cisplatin, whose tumor‐suppressive effects on patients were further confirmed by longer survival time and earlier tumor stage. Remarkably, signal transducer and activator of transcription‐3 (STAT3) were targeted by miR‐125a‐5p. However, IL‐6, which was extensively reported to activate the STAT3 signaling pathway, could block the tumor‐repressing effect of miR‐125a‐5p [141, 142]. miR‐221 promoted the resistance of esophageal adenocarcinoma to 5‐FU, in part, by regulating Wnt/β‐catenin‐EMT pathways in a DKK2‐dependent way [143]. There are few researches of EMT‐associated miRNAs in esophageal cancer resistance and they are need to further explore.
3.9. Nasopharyngeal cancer
After transfection with miR‐139‐5p mimics, the expression of mesenchymal markers, such as MMP‐9 and Vimentin, was decreased while the expression of epithelial markers, such as ZEB1, β‐cadherin, and E‐cadherin, was upregulated in cisplatin‐resistant NPC cells. These results exhibited that miR‐139‐5p might act as a tumor suppressor in the restoration of the sensitivity of NPC cells to cisplatin by regulating EMT [144]. miR‐296‐3p, which was negatively regulated by nicotine, inhibited PI3K/AKT/c‐Myc or Ras/BRAF/ERK/MEK/c‐Myc pathways to prompt its own expression in an MK2‐dependent manner. Thus, the upregulation of miR‐296‐3p due to the feedback loop ultimately suppressed NPC cell metastasis and drug resistance partially via EMT. Besides, NPC patients with higher miR‐296‐3p expression had longer overall survival than those with lower miR‐296‐3p expression [145]. miR‐205‐5p facilitated the migration and invasion of cisplatin‐resistant NPC cells by inhibiting PTEN expression, resulting in a decrease expression in E‐cadherin and an increase expression in vimentin, N‐cadherin, MMP‐2, and MMP‐9. These findings suggested that PTEN, deemed as a candidate target of miR‐205‐5p, exerted its tumor‐repressive EMT‐regulating function through the PI3K/AKT signaling pathway in NPC cells that acquire cisplatin‐resistant phenotype [146]. In contrast, miR‐374a inactivated pPI3K/pAKT/c‐JUN network by directly targeting CCND1, inhibiting the downstream EMT‐related genes and cell cycle progression. Intriguingly, this feedback loop was modulated by tumor suppressor PDCD4, which was further confirmed in clinical specimens [147]. However, it is possible that miR‐374a plays a complicated part in context‐dependent carcinogenesis since it has been reported to serve as an oncogene in breast cancer progression. Nonetheless, its roles remain poorly understood in lung cancer pathogenesis [148, 149, 150]. The function of miR‐374a needs to be further explored in various cancer types.
3.10. Other cancers
miR‐26b could reverse temozolomide resistance‐mediated EMT in glioma by targeting Wee1 [151]. miR‐140 plays a pivotal role in tumor‐suppression for it protracted the survival of patients who had some tumors, including glioblastoma. Up‐regulated miR‐140‐5p and down‐regulated cathepsin B (CTSB) were strongly associated with enhanced temozolomide (TMZ) sensitivity in GBM. Knocking down CTSB inhibited mesenchymal transition. These results suggested that not only miR‐140 targeted the CTSB signaling pathway, this signaling was also crucial in the inhibition of the innate resistance to TMZ [152]. Several studies suggested that miR‐125a‐3p was involved in the modulation of EMT and chemoresistance in prostate cancer cells [40, 147, 153]. In oral squamous cell carcinoma, it could promote EMT by upregulate PAK1 and occurred the resistance to cisplatin by upregulating YAP and ERCC1 protein Besides, miR‐485‐5p lowered the protein expression of PAK1 in OSCC cells. Contrary to the function of PAK1, miR‐485‑5p could reverse EMT and greatly obstructed the invasion and migration and sensitized cisplatin‐resistant cells [154]. TGFβ‐miR‐499a‐SHKBP1 axis orchestrated the EMT‐related kinase switch that induced the resistance of CD166+ osteosarcoma cancer cells to EGFR inhibitors, implying that the suppression of EMT‐related kinase switch induced by TGFβ may reverse the chemoresistance to EGFR inhibitors [155]. Various miRNAs in drug resistance of different cancers are not clear entirely and they need further study in the future.
4. EMT‐ASSOCIATED miRNAs MEDIATE DRUG RESISTANCE BY THE DELIVERY OF EXOSOMES
Exosomes are a subgroup of extracellular vesicles and their diameter ranges from 40 nm to 160 nm [156]. Multiple studies have reported exosomes could mediate cellular communication under physiological and pathological conditions via transferring miRNAs [157]. Recently, exosomal miRNAs were found to play a pivotal role in EMT‐mediated drug resistance. miRNAs can be loaded into exosomes by means of the endosomal sorting complex required for transport (ESCRT) [158]. Apart from the ESCRT‐dependent manner, RNA‐binding proteins could also recognize the specific motif in the 3′ portion of miRNA to facilitate its loading into exosomes [159]. Furthermore, ceramide might take part in the sorting of the bioactive molecules into exosomes and promoting domain‐induced budding. The ceramide‐rich domains curved spontaneously to form the invaginations, resulting in the generation of exosomes [160].
Exosomes containing miR‐155 from paclitaxel‐resistant gastric cancer cells were adequately absorbed by paclitaxel‐sensitive GC cells, resulting in an exhibition of EMT and chemoresistance phenotypes. Mechanistically, miR‐155 exerts its oncogenic effect by targeting tumor protein p53 inducible nuclear protein 1 (TP53INP1) and GATA binding protein 3 (GATA3) [161]. Besides, Santos JC et al. [162, 163, 164] demonstrated that exosome‐mediated miR‐155 was positively related to breast cancer cells with EMT‐associated chemoresistance by mediating the depletion of C/EBP‐β activity and targeting FOXO‐3a‐3′‐UTR directly. However, the miR‐128‐3p could increase intracellular oxaliplatin accumulation, by suppressing the EMT pathway. Importantly, lower expression of miR‐128‐3p in patients with advanced CRC was associated with weaker responsiveness to oxaliplatin with poor prognosis. Moreover, after transfected into human normal colorectal epithelial cells, miR‐128‐3p was effectively parceled into secreted exosomes, which could be directly transferred to oxaliplatin‐resistant cells, leading to an improvement in oxaliplatin response. The possible mechanism might be that miR‐128‐3p suppressed oxaliplatin‐induced EMT via inhibiting Bmi1 expression and decreased effluent oxaliplatin inside the cell through suppressing the expression of MRP5, a drug transporter [165].
According to the studies concerning EMT‐associated exosomal miRNAs in drug resistance, some critical problems remain to be solved. For instance, how the exosomes target specific recipient cells is still unclear. Importantly, miRNAs can be loaded into exosomes for therapeutic use. miR‐374a‐5p and miR‐214 inhibitors were incorporated into exosomes to reverse drug resistance by rescuing Neurod1 and PTEN in GC, respectively [166, 167]. However, the therapeutic effect and the potential side‐effects should be further evaluated in large‐sized clinical trials. More efforts have to be made to translate these research results into clinically effective therapies [168, 169].
FIGURE 1.
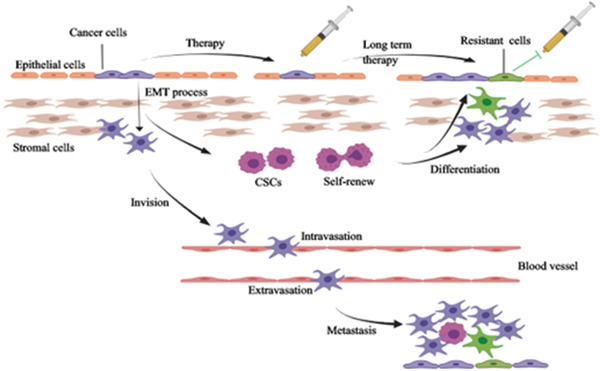
The relationship between EMT, stemness, and drug resistance. On one hand, the cancer cells harboring EMT phenotype may transform into drug‐resistant cells after long‐term therapy. On the other hand, the EMT may induce the generation of CSCs, which may go through a process of differentiation. Then the differentiated cells have the potential heterogeneity of being resistant to drug. Furthermore, EMT can generate the distant metastasis of cancer cells through the approaches of intravasation and extravasation
5. CONCLUSION AND FUTURE PERSPECTIVE
Given the nature of EMT, it is feasible to target the EMT to overcome the resistance. The therapeutic strategies can fall principally into three categories: (1) inhibiting the EMT initiation, (2) eliminating the cancer cells undergoing EMT, and (3) reversing the EMT to its opposite state, i.e., MET, since miRNAs play an important role in EMT‐induced drug resistance. Targeting miRNAs might be a promising approach in the treatment of cancer . In recent years, a great many studies intensively examined miRNAs and they have helped us better understand the role of miRNAs in the development, progression, and metastasis of tumors. Nonetheless, we still have a long way to go to fully elucidate the role of miRNAs in the development of drug resistance of tumor cells. In view of a large number researches of antitumor drugs, a database should be set up to cover the myriad information about drug targets and miRNAs, even the whole non‐coding RNAs to provide support to researchers.In this review, we highlighted the pivotal part of miRNAs in EMT, cancer stemness, and drug resistance. The relationship among them is illustrated in Figure legends. Those EMT‐associated miRNAs have showed complex functions in the regulation of cancer stemness and drug resistance phenotype: They bind to their targets and further impact the downstream pathways. (Table 1. The target of EMT‐associated miRNAs in different cancer types) Even the same miRNA plays an opposite role in different cancer types.
TABLE 1.
The target of EMT‐associated miRNAs in different cancer types
| miRNAs | Cancer types | Direct/indirect targets | Reference |
|---|---|---|---|
| Tumor suppresser miRNAs | |||
| miR‐25‐3p | CC | Sema4C | [106] |
| miR‐26a | LC | EZH2 | [65] |
| miR‐30a/c | OC | DNMT1 | [102] |
| miR‐34a | OC, LC, BC, CRC | Snail, MCTS1, ZNF281 | [49, 51‐53] |
| miR‐99a | LC | E2F2, ADGRE2 | [29] |
| miR‐125b | HCC | Smad2, Samd4, EVA1A | [31, 114] |
| miR‐125a‐3p | PC, EC, PRC | Fyn, STAT3 | [141, 170, 153] |
| miR‐128‐3p | CRC | Bmi1, MRP5 | [165] |
| miR‐129‐5p | BC, LC | MCRS1 | [89, 109] |
| miR‐130a | LC, HCC | Smad4 | [70, 115] |
| miR‐134 | CRC | CREB1 | [126] |
| miR‐137 | OC | Snail | [49] |
| miR‐139‐5p | CRC, NPC | BCL2 | [127, 144] |
| miR‐140 | LC, GBM | CTSB | [86, 152] |
| miR‐145 | CRC | Snail | [32] |
| miR‐146b | LC | PTP1B | [71] |
| miR‐155 | LC, | Smad2 | [87] |
| miR‐195‐5p | CRC | GDPD5, Notch2 | [128, 129] |
| miR‐199a‐5p | BC | PIK3CD | [33] |
| miR‐200 family | OC, HCC, LC, GC, BC, PC | NFATC1, CTSL, LIN28B, ERRFI1 ZEB1, ZEB2, ZNF17 | [48, 54, 55, 85, 171, 172, 81, 98, 133] |
| miR‐203 | RCC, PRC, CRC, PC | PTEN, Slug, ZEB1 | [43, 44, 46, 47] |
| miR‐204 | BC, GC | TGFBR2 | [14, 99] |
| miR‐205 | BC, LC, NPC | Notch2, PTEN | [35, 73, 146] |
| miR‐218 | PRC | Gli1, Slug | [36, 71] |
| miR‐221 | EC | DKK2 | [143] |
| miR‐296‐3p | LC, NPC | PRKCA, MK2 | [74, 145] |
| miR‐363 | OC | Snail | [173] |
| miR‐374a | NPC | CCND1 | [147] |
| miR‐375 | HCC | c‐Myc | [117] |
| miR‐451 | LC | c‐Myc | [75] |
| miR‐452 | BC | Slug | [37] |
| miR‐483‐3p | LC | ITGB3 | [76] |
| miR‐485‐5p | OSCC | PAK1 | [154] |
| miR‐495 | LC | UBE2C | [80] |
| miR‐499a | OS | SHKBP1 | [155] |
| miR‐504 | GBM | FZD7 | [38] |
| miR‐508 | CRC | ZEB1, Bmi1, SALL4 | [98] |
| miR‐574‐3p | GC | ZEB1 | [96] |
| miR‐612 | HCC | AKT2 | [118] |
| miR‐1247 | PRC | NRP1 | [41] |
| miR‐1294 | OC | IGF1R | [103] |
| miR‐3656 | PC | RHOF | [135] |
| miR‐4319 | HCC | FOXQ1 | [42] |
| Let‐7 | OC, LC | ABCC2, Bcl‐xl | [48, 51, 174] |
| Oncogenic miRNAs | |||
| miR‐10b | BC | PTEN, RBICC1 | [57, 58] |
| miR‐15b | LC | PEBP4 | [94] |
| miR‐17 | GC | DEDD | [101] |
| miR‐20a | CC | FBXL5, BTG3 | [64] |
| miR‐21 | LC | HBP1 | [96] |
| miR‐27a | LC, HCC | RKIP | [95, 175] |
| miR‐32‐5p | HCC | PTEN | [120] |
| miR‐106b cluster | BC | EP300 | [113] |
| miR‐124 | CRC | PRRX1 | [132] |
| miR‐134 cluster | LC | MAGI2 | [93] |
| miR‐155 | LC, GC, BC | RHOA, GATA3, P53INP1, FOXO‐3a | [88, 161, 164] |
| miR‐196a | HCC | NA | [176] |
| miR‐210 | BC | E‐cadherin, Snail | [59] |
| miR‐221 | PC | SOCS3 | [140] |
| miR‐233 | HCC | Fbw7 | [121] |
| miR‐296‐5p | PC | BOK | [138] |
| miR‐495 | LC | ETK | [79] |
| miR‐577 | GC | CAVIN2 | [60] |
| miR‐5188 | HCC, BC | FOXO1 | [62, 63] |
Abbreviations: BC, breast cancer; CC, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer, GC, gastric cancer; GBM, glioblastoma; HCC, Hepatic carcinoma; LC, lung cancer; PC, pancreatic cancer; PRC, prostate cancer; NPC, nasopharyngeal cancer; OC, ovarian cancer; OS, osteosarcoma; OSCC, oral squamous cell carcinoma; RCC, renal cell carcinoma.
Therefore, miRNAs can not only be used as potential diagnostic or prognostic markers but also are of therapeutic value. Targeting miRNAs to antagonize certain malignant properties of cancer may have more extensive clinical implications. The miRNA‐based therapies are still confronted with some challenges, such as the off‐target effect and lack of an optimal delivering system. The in vivo delivery of miRNAs remains a challenge due to their speedy excretion, incorrect intracellular release, poor biostability, endosomal escape, poor cellular ingestion, and immunogenicity. Thus, miRNA‐based therapeutics will not be clinically available for cancer treatment until these problems are fully resolved.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This study was supported by grants from the National Natural Science Foundation of China (81673760 and 81874397).
AUTHORS' CONTRIBUTIONS
S.Y. provided direction and guidance throughout the preparation of this manuscript. G.P. wrote and edited the manuscript. Y.L., L.S., and F.Z. reviewed and revised the manuscript. G.P. and F.Z. collected data. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
Not applicable.
Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT‐associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021;41:199–217. 10.1002/cac2.12138
REFERENCES
- 1. Serrano‐Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial‐mesenchymal transition through epigenetic and post‐translational modifications. Mol Cancer. 2016;15:18. 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liao TT, Yang MH. Revisiting epithelial‐mesenchymal transition in cancer metastasis: the connection between epithelial plasticity and stemness. Mol Oncol. 2017;11(7):792‐804. 10.1002/1878-0261.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial‐to‐mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472‐6. 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loh CY, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK, et al. The E‐Cadherin and N‐Cadherin Switch in Epithelial‐to‐Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells. 2019;8(10). 10.3390/cells8101118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033‐46. 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, et al. Hypoxia‐induced down‐regulation of microRNA‐34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS One. 2012;7(2):e30771. 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diepenbruck M, Christofori G. Epithelial‐mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7‐13. 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 8. Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol. 2018;13:395‐412. 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281‐97. 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10. Lim LP, Lau NC, Garrett‐Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769‐73. 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835‐40. 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jafri MA, Al‐Qahtani MH, Shay JW. Role of miRNAs in human cancer metastasis: Implications for therapeutic intervention. Semin Cancer Biol. 2017;44:117‐31. 10.1016/j.semcancer.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 13. Gong L, Yan Q, Zhang Y, Fang X, Liu B, Guan X. Cancer cell reprogramming: a promising therapy converting malignancy to benignity. Cancer Commun (Lond). 2019;39(1):48. 10.1186/s40880-019-0393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rahimi M, Sharifi‐Zarchi A, Firouzi J, Azimi M, Zarghami N, Alizadeh E, et al. An integrated analysis to predict micro‐RNAs targeting both stemness and metastasis in breast cancer stem cells. J Cell Mol Med. 2019;23(4):2442‐56. 10.1111/jcmm.14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou G, Latchoumanin O, Bagdesar M, Hebbard L, Duan W, Liddle C, et al. Aptamer‐Based Therapeutic Approaches to Target Cancer Stem Cells. Theranostics. 2017;7(16):3948‐61. 10.7150/thno.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Robertis M, Poeta ML, Signori E, Fazio VM. Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin Cancer Biol. 2018;53:232‐47. 10.1016/j.semcancer.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 17. Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11(1):64. 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manic G, Sistigu A, Corradi F, Musella M, De Maria R, Vitale I. Replication stress response in cancer stem cells as a target for chemotherapy. Semin Cancer Biol. 2018;53:31‐41. 10.1016/j.semcancer.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 19. Papadaki MA, Kallergi G, Zafeiriou Z, Manouras L, Theodoropoulos PA, Mavroudis D, et al. Co‐expression of putative stemness and epithelial‐to‐mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma XL, Hu B, Tang WG, Xie SH, Ren N, Guo L, et al. CD73 sustained cancer‐stem‐cell traits by promoting SOX9 expression and stability in hepatocellular carcinoma. J Hematol Oncol. 2020;13(1):11. 10.1186/s13045-020-0845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252‐63. 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 22. Ashley N. Regulation of intestinal cancer stem cells. Cancer Lett. 2013;338(1):120‐6. 10.1016/j.canlet.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 23. Guo Y, Bao Y, Yang W. Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis. Int J Mol Sci. 2017;18(4). 10.3390/ijms18040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463‐8. 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 25. Jolly MK, Jia D, Boareto M, Mani SA, Pienta KJ, Ben‐Jacob E, et al. Coupling the modules of EMT and stemness: A tunable ‘stemness window’ model. Oncotarget. 2015;6(28):25161‐74.doi: 10.18632/oncotarget.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kröger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A. 2019;116(15):7353‐62. 10.1073/pnas.1812876116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bocci F, Jolly MK, George JT, Levine H, Onuchic JN. A mechanism‐based computational model to capture the interconnections among epithelial‐mesenchymal transition, cancer stem cells and Notch‐Jagged signaling. Oncotarget. 2018;9(52):29906‐20.doi: 10.18632/oncotarget.25692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381‐95. 10.1002/jcp.27740. [DOI] [PubMed] [Google Scholar]
- 29. Feliciano A, Garcia‐Mayea Y, Jubierre L, miR C, Hummel M, Castellvi J, et al. miR‐99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8(10):e3141. 10.1038/cddis.2017.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fang YX, Chang YL, Gao WQ. MicroRNAs targeting prostate cancer stem cells. Exp Biol Med (Maywood). 2015;240(8):1071‐8. 10.1177/1535370215584935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou JN, Zeng Q, Wang HY, Zhang B, Li ST, Nan X, et al. MicroRNA‐125b attenuates epithelial‐mesenchymal transitions and targets stem‐like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology. 2015;62(3):801‐15. 10.1002/hep.27887. [DOI] [PubMed] [Google Scholar]
- 32. Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, et al. miR‐145 Antagonizes SNAI1‐Mediated Stemness and Radiation Resistance in Colorectal Cancer. Mol Ther. 2018;26(3):744‐54. 10.1016/j.ymthe.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Shin VY, Siu MT, Ho JC, Cheuk I, Kwong A. miR‐199a‐5p confers tumor‐suppressive role in triple‐negative breast cancer. BMC Cancer. 2016;16(1):887. 10.1186/s12885-016-2916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged‐1. Cancer Cell. 2013;23(2):171‐85. 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, et al. MicroRNA‐205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest. 2014;124(7):3093‐106. 10.1172/JCI73351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guan B, Mu L, Zhang L, Wang K, Tian J, Xu S, et al. MicroRNA‐218 inhibits the migration, epithelial‐mesenchymal transition and cancer stem cell properties of prostate cancer cells. Oncol Lett. 2018;16(2):1821‐6. 10.3892/ol.2018.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim M, Jang K, Miller P, Picon‐Ruiz M, Yeasky TM, El‐Ashry D, et al. VEGFA links self‐renewal and metastasis by inducing Sox2 to repress miR‐452, driving Slug. Oncogene. 2017;36(36):5199‐211. 10.1038/onc.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Q, Guan Y, Li Z, Wang Y, Liu Y, Cui R, et al. miR‐504 suppresses mesenchymal phenotype of glioblastoma by directly targeting the FZD7‐mediated Wnt‐beta‐catenin pathway. J Exp Clin Cancer Res. 2019;38(1):358. 10.1186/s13046-019-1370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J, Le TD, Liu L, Li J. Identifying miRNA sponge modules using biclustering and regulatory scores. BMC Bioinformatics. 2017;18(Suppl 3):44. 10.1186/s12859-017-1467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ren LL, Yan TT, Shen CQ, Tang JY, Kong X, Wang YC, et al. The distinct role of strand‐specific miR‐514b‐3p and miR‐514b‐5p in colorectal cancer metastasis. Cell Death Dis. 2018;9(6):687. 10.1038/s41419-018-0732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taddei ML, Cavallini L, Ramazzotti M, Comito G, Pietrovito L, Morandi A, et al. Stromal‐induced downregulation of miR‐1247 promotes prostate cancer malignancy. J Cell Physiol. 2019;234(6):8274‐85. 10.1002/jcp.27679. [DOI] [PubMed] [Google Scholar]
- 42. Han S, Shi Y, Sun L, Liu Z, Song T, Liu Q. miR‐4319 induced an inhibition of epithelial‐mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1. Int J Biol Sci. 2019;15(13):2936‐47. 10.7150/ijbs.38000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meidhof S, Brabletz S, Lehmann W, Preca BT, Mock K, Ruh M, et al. ZEB1‐associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7(6):831‐47.doi: 10.15252/emmm.201404396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dasgupta P, Kulkarni P, Majid S, Shahryari V, Hashimoto Y, Bhat NS, et al. MicroRNA‐203 Inhibits Long Noncoding RNA HOTAIR and Regulates Tumorigenesis through Epithelial‐to‐mesenchymal Transition Pathway in Renal Cell Carcinoma. Mol Cancer Ther. 2018;17(5):1061‐9. 10.1158/1535-7163.MCT-17-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu Q, Zhong AL, Hu H, Zhao JJ, Weng DS, Tang Y, et al. Acylglycerol kinase promotes tumour growth and metastasis via activating the PI3K/AKT/GSK3beta signaling pathway in renal cell carcinoma. J Hematol Oncol. 2020;13(1):2. 10.1186/s13045-019-0840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tian X, Tao F, Zhang B, Dong JT, Zhang Z. The miR‐203/SNAI2 axis regulates prostate tumor growth, migration, angiogenesis and stemness potentially by modulating GSK‐3beta/beta‐CATENIN signal pathway. IUBMB Life. 2018;70(3):224‐36. 10.1002/iub.1720. [DOI] [PubMed] [Google Scholar]
- 47. Ju SY, Chiou SH, Su Y. Maintenance of the stemness in CD44(+) HCT‐15 and HCT‐116 human colon cancer cells requires miR‐203 suppression. Stem Cell Res. 2014;12(1):86‐100. 10.1016/j.scr.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 48. Qiu L, Zhang GF, Yu L, Wang HY, Jia XJ, Wang TJ. Novel oncogenic and chemoresistance‐inducing functions of resistin in ovarian cancer cells require miRNAs‐mediated induction of epithelial‐to‐mesenchymal transition. Sci Rep. 2018;8(1):12522. 10.1038/s41598-018-30978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dong P, Xiong Y, Watari H, Hanley SJ, Konno Y, Ihira K, et al. miR‐137 and miR‐34a directly target Snail and inhibit EMT, invasion and sphere‐forming ability of ovarian cancer cells. J Exp Clin Cancer Res. 2016;35(1):132. 10.1186/s13046-016-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hojo N, Huisken AL, Wang H, Chirshev E, Kim NS, Nguyen SM, et al. Snail knockdown reverses stemness and inhibits tumour growth in ovarian cancer. Sci Rep. 2018;8(1):8704. 10.1038/s41598-018-27021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei H, Liang F, Cheng W, Zhou R, Wu X, Feng Y, et al. The mechanisms for lung cancer risk of PM2.5 : Induction of epithelial‐mesenchymal transition and cancer stem cell properties in human non‐small cell lung cancer cells. Environ Toxicol. 2017;32(11):2341‐51. 10.1002/tox.22437. [DOI] [PubMed] [Google Scholar]
- 52. Weng YS, Tseng HY, Chen YA, Shen PC, Al Haq AT, Chen LM, et al. MCT‐1/miR‐34a/IL‐6/IL‐6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple‐negative breast cancer. Mol Cancer. 2019;18(1):42. 10.1186/s12943-019-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hahn S, Hermeking H. ZNF281/ZBP‐99: a new player in epithelial‐mesenchymal transition, stemness, and cancer. J Mol Med (Berl). 2014;92(6):571‐81. 10.1007/s00109-014-1160-3. [DOI] [PubMed] [Google Scholar]
- 54. Yeh TS, Wang F, Chen TC, Yeh CN, Yu MC, Jan YY, et al. Expression profile of microRNA‐200 family in hepatocellular carcinoma with bile duct tumor thrombus. Ann Surg. 2014;259(2):346‐54. 10.1097/SLA.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 55. Singh SK, Chen NM, Hessmann E, Siveke J, Lahmann M, Singh G, et al. Antithetical NFATc1‐Sox2 and p53‐miR200 signaling networks govern pancreatic cancer cell plasticity. EMBO J. 2015;34(4):517‐30.doi: 10.15252/embj.201489574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li X, Xu F, Chang C, Byon J, Papayannopoulou T, Deeg HJ, et al. Transcriptional regulation of miR‐10a/b by TWIST‐1 in myelodysplastic syndromes. Haematologica. 2013;98(3):414‐9. 10.3324/haematol.2012.071753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lo PK, Zhang Y, Yao Y, Wolfson B, Yu J, Han SY, et al. Tumor‐associated myoepithelial cells promote the invasive progression of ductal carcinoma in situ through activation of TGFbeta signaling. J Biol Chem. 2017;292(27):11466‐84. 10.1074/jbc.M117.775080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bahena‐Ocampo I, Espinosa M, Ceballos‐Cancino G, Lizarraga F, Campos‐Arroyo D, Schwarz A, et al. miR‐10b expression in breast cancer stem cells supports self‐renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016;17(5):648‐58.doi: 10.15252/embr.201540678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tang T, Yang Z, Zhu Q, Wu Y, Sun K, Alahdal M, et al. Up‐regulation of miR‐210 induced by a hypoxic microenvironment promotes breast cancer stem cells metastasis, proliferation, and self‐renewal by targeting E‐cadherin. FASEB J. 2018:fj201801013R. 10.1096/fj.201801013R. [DOI] [PubMed] [Google Scholar]
- 60. Luo Y, Wu J, Wu Q, Li X, Wu J, Zhang J, et al. miR‐577 Regulates TGF‐beta Induced Cancer Progression through a SDPR‐Modulated Positive‐Feedback Loop with ERK‐NF‐kappaB in Gastric Cancer. Mol Ther. 2019;27(6):1166‐82. 10.1016/j.ymthe.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Farhana L, Antaki F, Anees MR, Nangia‐Makker P, Judd S, Hadden T, et al. Role of cancer stem cells in racial disparity in colorectal cancer. Cancer Med. 2016;5(6):1268‐78. 10.1002/cam4.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin X, Zuo S, Luo R, Li Y, Yu G, Zou Y, et al. HBX‐induced miR‐5188 impairs FOXO1 to stimulate beta‐catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics. 2019;9(25):7583‐98. 10.7150/thno.37717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zou Y, Lin X, Bu J, Lin Z, Chen Y, Qiu Y, et al. Timeless‐Stimulated miR‐5188‐FOXO1/beta‐Catenin‐c‐Jun Feedback Loop Promotes Stemness via Ubiquitination of beta‐Catenin in Breast Cancer. Mol Ther. 2020;28(1):313‐27. 10.1016/j.ymthe.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiong Y, Sun F, Dong P, Watari H, Yue J, Yu MF, et al. iASPP induces EMT and cisplatin resistance in human cervical cancer through miR‐20a‐FBXL5/BTG3 signaling. J Exp Clin Cancer Res. 2017;36(1):48. 10.1186/s13046-017-0520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen J, Xu Y, Tao L, Pan Y, Zhang K, Wang R, et al. miRNA‐26a Contributes to the Acquisition of Malignant Behaviors of Doctaxel‐Resistant Lung Adenocarcinoma Cells through Targeting EZH2. Cell Physiol Biochem. 2017;41(2):583‐97. 10.1159/000457879. [DOI] [PubMed] [Google Scholar]
- 66. Gan L, Xu M, Hua R, Tan C, Zhang J, Gong Y, et al. The polycomb group protein EZH2 induces epithelial‐mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J Hematol Oncol. 2018;11(1):9. 10.1186/s13045-017-0547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug‐resistant ovarian cancer cells. Gynecol Oncol. 2008;111(3):478‐86. 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 68. Dai Y, Xie CH, Neis JP, Fan CY, Vural E, Spring PM. MicroRNA expression profiles of head and neck squamous cell carcinoma with docetaxel‐induced multidrug resistance. Head Neck. 2011;33(6):786‐91. 10.1002/hed.21540. [DOI] [PubMed] [Google Scholar]
- 69. Zhang X, Huang L, Zhao Y, Tan W. Downregulation of miR‐130a contributes to cisplatin resistance in ovarian cancer cells by targeting X‐linked inhibitor of apoptosis (XIAP) directly. Acta Biochim Biophys Sin (Shanghai). 2013;45(12):995‐1001. 10.1093/abbs/gmt113. [DOI] [PubMed] [Google Scholar]
- 70. Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, et al. miR‐130a targets MET and induces TRAIL‐sensitivity in NSCLC by downregulating miR‐221 and 222. Oncogene. 2012;31(5):634‐42. 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Han Q, Cheng P, Yang H, Liang H, Lin F. miR‐146b Reverses epithelial‐mesenchymal transition via targeting PTP1B in cisplatin‐resistance human lung adenocarcinoma cells. J Cell Biochem. 2019. 10.1002/jcb.29554. [DOI] [PubMed] [Google Scholar]
- 72. Shi ZM, Wang L, Shen H, Jiang CF, Ge X, Li DM, et al. Downregulation of miR‐218 contributes to epithelial‐mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene. 2017;36(18):2577‐88. 10.1038/onc.2016.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Park KS, Raffeld M, Moon YW, Xi L, Bianco C, Pham T, et al. CRIPTO1 expression in EGFR‐mutant NSCLC elicits intrinsic EGFR‐inhibitor resistance. J Clin Invest. 2014;124(7):3003‐15. 10.1172/JCI73048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C, et al. miRomics and Proteomics Reveal a miR‐296‐3p/PRKCA/FAK/Ras/c‐Myc Feedback Loop Modulated by HDGF/DDX5/beta‐catenin Complex in Lung Adenocarcinoma. Clin Cancer Res. 2017;23(20):6336‐50. 10.1158/1078-0432.CCR-16-2813. [DOI] [PubMed] [Google Scholar]
- 75. Chen D, Huang J, Zhang K, Pan B, Chen J, De W, et al. MicroRNA‐451 induces epithelial‐mesenchymal transition in docetaxel‐resistant lung adenocarcinoma cells by targeting proto‐oncogene c‐Myc. Eur J Cancer. 2014;50(17):3050‐67. 10.1016/j.ejca.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 76. Yue J, Lv D, Wang C, Li L, Zhao Q, Chen H, et al. Epigenetic silencing of miR‐483‐3p promotes acquired gefitinib resistance and EMT in EGFR‐mutant NSCLC by targeting integrin beta3. Oncogene. 2018;37(31):4300‐12. 10.1038/s41388-018-0276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cao M, Nie W, Li J, Zhang Y, Yan X, Guan X, et al. MicroRNA‐495 induces breast cancer cell migration by targeting JAM‐A. Protein Cell. 2014;5(11):862‐72. 10.1007/s13238-014-0088-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jiang X, Huang H, Li Z, He C, Li Y, Chen P, et al. miR‐495 is a tumor‐suppressor microRNA down‐regulated in MLL‐rearranged leukemia. Proc Natl Acad Sci U S A. 2012;109(47):19397‐402. 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wei T, Zhu W, Fang S, Zeng X, Huang J, Yang J, et al. miR‐495 promotes the chemoresistance of SCLC through the epithelial‐mesenchymal transition via Etk/BMX. Am J Cancer Res. 2017;7(3):628‐46. [PMC free article] [PubMed] [Google Scholar]
- 80. Guo J, Jin D, Wu Y, Yang L, Du J, Gong K, et al. The miR 495‐UBE2C‐ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin‐resistant non‐small cell lung cancer cells. EBioMedicine. 2018;35:204‐21. 10.1016/j.ebiom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81. Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR‐200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582‐9. 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An autocrine TGF‐beta/ZEB/miR‐200 signaling network regulates establishment and maintenance of epithelial‐mesenchymal transition. Mol Biol Cell. 2011;22(10):1686‐98. 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nishijima N, Seike M, Soeno C, Chiba M, Miyanaga A, Noro R, et al. miR‐200/ZEB axis regulates sensitivity to nintedanib in non‐small cell lung cancer cells. Int J Oncol. 2016;48(3):937‐44. 10.3892/ijo.2016.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Izumchenko E, Chang X, Michailidi C, Kagohara L, Ravi R, Paz K, et al. The TGFbeta‐miR200‐MIG6 pathway orchestrates the EMT‐associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014;74(14):3995‐4005. 10.1158/0008-5472.CAN-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhao YF, Han ML, Xiong YJ, Wang L, Fei Y, Shen X, et al. A miRNA‐200c/cathepsin L feedback loop determines paclitaxel resistance in human lung cancer A549 cells in vitro through regulating epithelial‐mesenchymal transition. Acta Pharmacol Sin. 2018;39(6):1034‐47. 10.1038/aps.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Krentz Gober M, Collard JP, Thompson K, Black EP. A microRNA signature of response to erlotinib is descriptive of TGFbeta behaviour in NSCLC. Sci Rep. 2017;7(1):4202. 10.1038/s41598-017-04097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Narita M, Shimura E, Nagasawa A, Aiuchi T, Suda Y, Hamada Y, et al. Chronic treatment of non‐small‐cell lung cancer cells with gefitinib leads to an epigenetic loss of epithelial properties associated with reductions in microRNA‐155 and ‐200c. PLoS One. 2017;12(2):e0172115. 10.1371/journal.pone.0172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA‐155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28(22):6773‐84. 10.1128/mcb.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu MX, Zhou KC, Cao Y. MCRS1 overexpression, which is specifically inhibited by miR‐129*, promotes the epithelial‐mesenchymal transition and metastasis in non‐small cell lung cancer. Mol Cancer. 2014;13:245. 10.1186/1476-4598-13-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11(24 Pt 1):8686‐98. 10.1158/1078-0432.Ccr-05-1492. [DOI] [PubMed] [Google Scholar]
- 91. Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non‐small‐cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65(20):9455‐62. 10.1158/0008-5472.Can-05-1058. [DOI] [PubMed] [Google Scholar]
- 92. Rho JK, Choi YJ, Lee JK, Ryoo BY, Na, II , Yang SH, et al. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non‐small cell lung cancer cell line. Lung Cancer. 2009;63(2):219‐26. 10.1016/j.lungcan.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 93. Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, et al. miR‐134/487b/655 cluster regulates TGF‐beta‐induced epithelial‐mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13(2):444‐53. 10.1158/1535-7163.Mct-13-0448. [DOI] [PubMed] [Google Scholar]
- 94. Zhao Z, Zhang L, Yao Q, Tao Z. miR‐15b regulates cisplatin resistance and metastasis by targeting PEBP4 in human lung adenocarcinoma cells. Cancer Gene Ther. 2015;22(3):108‐14. 10.1038/cgt.2014.73. [DOI] [PubMed] [Google Scholar]
- 95. Martinho O, Pinto F, Granja S, miRanda‐Goncalves V, Moreira MA, Ribeiro LF, et al. RKIP inhibition in cervical cancer is associated with higher tumor aggressive behavior and resistance to cisplatin therapy. PLoS One. 2013;8(3):e59104. 10.1371/journal.pone.0059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Su C, Cheng X, Li Y, Han Y, Song X, Yu D, et al. miR‐21 improves invasion and migration of drug‐resistant lung adenocarcinoma cancer cell and transformation of EMT through targeting HBP1. Cancer Med. 2018;7(6):2485‐503. 10.1002/cam4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shi L, Wang Y, Lu Z, Zhang H, Zhuang N, Wang B, et al. miR‐127 promotes EMT and stem‐like traits in lung cancer through a feed‐forward regulatory loop. Oncogene. 2017;36(12):1631‐43. 10.1038/onc.2016.332. [DOI] [PubMed] [Google Scholar]
- 98. Zhou X, Men X, Zhao R, Han J, Fan Z, Wang Y, et al. miR‐200c inhibits TGF‐beta‐induced‐EMT to restore trastuzumab sensitivity by targeting ZEB1 and ZEB2 in gastric cancer. Cancer Gene Ther. 2018;25(3‐4):68‐76. 10.1038/s41417-017-0005-y. [DOI] [PubMed] [Google Scholar]
- 99. Li LQ, Pan D, Chen Q, Zhang SW, Xie DY, Zheng XL, et al. Sensitization of Gastric Cancer Cells to 5‐FU by MicroRNA‐204 Through Targeting the TGFBR2‐Mediated Epithelial to Mesenchymal Transition. Cell Physiol Biochem. 2018;47(4):1533‐45. 10.1159/000490871. [DOI] [PubMed] [Google Scholar]
- 100. Wang M, Zhang R, Zhang S, Xu R, Yang Q. MicroRNA‐574‐3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene. 2019;700:110‐9. 10.1016/j.gene.2019.03.043. [DOI] [PubMed] [Google Scholar]
- 101. Wu DM, Hong XW, Wang LL, Cui XF, Lu J, Chen GQ, et al. MicroRNA‐17 inhibition overcomes chemoresistance and suppresses epithelial‐mesenchymal transition through a DEDD‐dependent mechanism in gastric cancer. Int J Biochem Cell Biol. 2018;102:59‐70. 10.1016/j.biocel.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 102. Han X, Zhen S, Ye Z, Lu J, Wang L, Li P, et al. A Feedback Loop Between miR‐30a/c‐5p and DNMT1 Mediates Cisplatin Resistance in Ovarian Cancer Cells. Cell Physiol Biochem. 2017;41(3):973‐86. 10.1159/000460618. [DOI] [PubMed] [Google Scholar]
- 103. Zhang Y, Huang S, Guo Y, Li L. miR‐1294 confers cisplatin resistance in ovarian Cancer cells by targeting IGF1R. Biomed Pharmacother. 2018;106:1357‐63. 10.1016/j.biopha.2018.07.059. [DOI] [PubMed] [Google Scholar]
- 104. Brozovic A, Duran GE, Wang YC, Francisco EB, Sikic BI. The miR‐200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR‐3 and MES‐OV cells. Mol Oncol. 2015;9(8):1678‐93. 10.1016/j.molonc.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Duran GE, Wang YC, Moisan F, Francisco EB, Sikic BI. Decreased levels of baseline and drug‐induced tubulin polymerisation are hallmarks of resistance to taxanes in ovarian cancer cells and are associated with epithelial‐to‐mesenchymal transition. Br J Cancer. 2017;116(10):1318‐28. 10.1038/bjc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Song J, Li Y. miR‐25‐3p reverses epithelial‐mesenchymal transition via targeting Sema4C in cisplatin‐resistance cervical cancer cells. Cancer Sci. 2017;108(1):23‐31. 10.1111/cas.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mallini P, Lennard T, Kirby J, Meeson A. Epithelial‐to‐mesenchymal transition: what is the impact on breast cancer stem cells and drug resistance. Cancer Treat Rev. 2014;40(3):341‐8. 10.1016/j.ctrv.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 108. Saxena M, Stephens MA, Pathak H, Rangarajan A. Transcription factors that mediate epithelial‐mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011;2:e179. 10.1038/cddis.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Luan QX, Zhang BG, Li XJ, Guo MY. miR‐129‐5p is downregulated in breast cancer cells partly due to promoter H3K27m3 modification and regulates epithelial‐mesenchymal transition and multi‐drug resistance. Eur Rev Med Pharmacol Sci. 2016;20(20):4257‐65. [PubMed] [Google Scholar]
- 110. Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ, Huang X, et al. miR‐200c suppresses TGF‐beta signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J Cancer. 2014;135(6):1356‐68. 10.1002/ijc.28782. [DOI] [PubMed] [Google Scholar]
- 111. Li J, Hao Y, Mao W, Xue X, Xu P, Liu L, et al. LincK contributes to breast tumorigenesis by promoting proliferation and epithelial‐to‐mesenchymal transition. J Hematol Oncol. 2019;12(1):19. 10.1186/s13045-019-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lee JW, Guan W, Han S, Hong DK, Kim LS, Kim H. MicroRNA‐708‐3p mediates metastasis and chemoresistance through inhibition of epithelial‐to‐mesenchymal transition in breast cancer. Cancer Sci. 2018;109(5):1404‐13. 10.1111/cas.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhou Y, Hu Y, Yang M, Jat P, Li K, Lombardo Y, et al. The miR‐106b∼25 cluster promotes bypass of doxorubicin‐induced senescence and increase in motility and invasion by targeting the E‐cadherin transcriptional activator EP300. Cell Death Differ. 2014;21(3):462‐74. 10.1038/cdd.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ren WW, Li DD, Chen X, Li XL, He YP, Guo LH, et al. MicroRNA‐125b reverses oxaliplatin resistance in hepatocellular carcinoma by negatively regulating EVA1A mediated autophagy. Cell Death Dis. 2018;9(5):547. 10.1038/s41419-018-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu Y, Li Y, Wang R, Qin S, Liu J, Su F, et al. miR‐130a‐3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res. 2016;35:19. 10.1186/s13046-016-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lisse TS, Hewison M. Vitamin D: a new player in the world of mTOR signaling. Cell Cycle. 2011;10(12):1888‐9. 10.4161/cc.10.12.15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Provvisiero DP, Negri M, de Angelis C, Di Gennaro G, Patalano R, Simeoli C, et al. Vitamin D reverts resistance to the mTOR inhibitor everolimus in hepatocellular carcinoma through the activation of a miR‐375/oncogenes circuit. Sci Rep. 2019;9(1):11695. 10.1038/s41598-019-48081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tang J, Tao ZH, Wen D, Wan JL, Liu DL, Zhang S, et al. miR‐612 suppresses the stemness of liver cancer via Wnt/beta‐catenin signaling. Biochem Biophys Res Commun. 2014;447(1):210‐5. 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- 119. Liu Y, Lu LL, Wen D, Liu DL, Dong LL, Gao DM, et al. miR‐612 regulates invadopodia of hepatocellular carcinoma by HADHA‐mediated lipid reprogramming. J Hematol Oncol. 2020;13(1):12. 10.1186/s13045-019-0841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, et al. Exosomal microRNA‐32‐5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37(1):52. 10.1186/s13046-018-0677-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121. Xu Y, Sengupta T, Kukreja L, Minella AC. MicroRNA‐223 regulates cyclin E activity by modulating expression of F‐box and WD‐40 domain protein 7. J Biol Chem. 2010;285(45):34439‐46. 10.1074/jbc.M110.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li J, Guo Y, Liang X, Sun M, Wang G, De W, et al. MicroRNA‐223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol. 2012;138(5):763‐74. 10.1007/s00432-012-1154-x. [DOI] [PubMed] [Google Scholar]
- 123. Yu J, Zhang W, Gao F, Liu YX, Chen ZY, Cheng LY, et al. FBW7 increases chemosensitivity in hepatocellular carcinoma cells through suppression of epithelial‐mesenchymal transition. Hepatobiliary Pancreat Dis Int. 2014;13(2):184‐91. 10.1016/s1499-3872(14)60029-1. [DOI] [PubMed] [Google Scholar]
- 124. Ma J, Zeng F, Ma C, Pang H, Fang B, Lian C, et al. Synergistic reversal effect of epithelial‐to‐mesenchymal transition by miR‐223 inhibitor and genistein in gemcitabine‐resistant pancreatic cancer cells. Am J Cancer Res. 2016;6(6):1384‐95. [PMC free article] [PubMed] [Google Scholar]
- 125. Gao XJ, Liu JW, Zhang QG, Zhang JJ, Xu HT, Liu HJ. Nobiletin inhibited hypoxia‐induced epithelial‐mesenchymal transition of lung cancer cells by inactivating of Notch‐1 signaling and switching on miR‐200b. Pharmazie. 2015;70(4):256‐62. [PubMed] [Google Scholar]
- 126. Ye Q, Su L, Chen D, Zheng W, Liu Y. Astragaloside IV Induced miR‐134 Expression Reduces EMT and Increases Chemotherapeutic Sensitivity by Suppressing CREB1 Signaling in Colorectal Cancer Cell Line SW‐480. Cell Physiol Biochem. 2017;43(4):1617‐26. 10.1159/000482025. [DOI] [PubMed] [Google Scholar]
- 127. Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai S, et al. miR‐139‐5p Inhibits the Epithelial‐Mesenchymal Transition and Enhances the Chemotherapeutic Sensitivity of Colorectal Cancer Cells by Downregulating BCL2. Sci Rep. 2016;6:27157. 10.1038/srep27157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Feng C, Zhang L, Sun Y, Li X, Zhan L, Lou Y, et al. GDPD5, a target of miR‐195‐5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed Pharmacother. 2018;101:945‐52. 10.1016/j.biopha.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 129. Lin X, Wang S, Sun M, Zhang C, Wei C, Yang C, et al. miR‐195‐5p/NOTCH2‐mediated EMT modulates IL‐4 secretion in colorectal cancer to affect M2‐like TAM polarization. Journal of Hematology & Oncology. 2019;12(1). 10.1186/s13045-019-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130. Findlay VJ, Wang C, Nogueira LM, Hurst K, Quirk D, Ethier SP, et al. SNAI2 modulates colorectal cancer 5‐fluorouracil sensitivity through miR145 repression. Mol Cancer Ther. 2014;13(11):2713‐26. 10.1158/1535-7163.MCT-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Yu X, Shi W, Zhang Y, Wang X, Sun S, Song Z, et al. CXCL12/CXCR4 axis induced miR‐125b promotes invasion and confers 5‐fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci Rep. 2017;7:42226. 10.1038/srep42226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, et al. miR‐124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9(4):e93917. 10.1371/journal.pone.0093917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hiramoto H, Muramatsu T, Ichikawa D, Tanimoto K, Yasukawa S, Otsuji E, et al. miR‐509‐5p and miR‐1243 increase the sensitivity to gemcitabine by inhibiting epithelial‐mesenchymal transition in pancreatic cancer. Sci Rep. 2017;7(1):4002. 10.1038/s41598-017-04191-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Funamizu N, Lacy CR, Kamada M, Yanaga K, Manome Y. MicroRNA‐200b and ‐301 are associated with gemcitabine response as biomarkers in pancreatic carcinoma cells. Int J Oncol. 2019;54(3):991‐1000. 10.3892/ijo.2019.4676. [DOI] [PubMed] [Google Scholar]
- 135. Yang RM, Zhan M, Xu SW, Long MM, Yang LH, Chen W, et al. miR‐3656 expression enhances the chemosensitivity of pancreatic cancer to gemcitabine through modulation of the RHOF/EMT axis. Cell Death Dis. 2017;8(10):e3129. 10.1038/cddis.2017.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Baylin SB, Jones PA. A decade of exploring the cancer epigenome ‐ biological and translational implications. Nat Rev Cancer. 2011;11(10):726‐34. 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Transient low doses of DNA‐demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21(3):430‐46. 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Okazaki J, Tanahashi T, Sato Y, Miyoshi J, Nakagawa T, Kimura T, et al. MicroRNA‐296‐5p Promotes Cell Invasion and Drug Resistance by Targeting Bcl2‐Related Ovarian Killer, Leading to a Poor Prognosis in Pancreatic Cancer. Digestion. 2019:1‐13. 10.1159/000503225. [DOI] [PubMed] [Google Scholar]
- 139. Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long Noncoding RNA GAS5 Suppresses Cell Growth and Epithelial‐Mesenchymal Transition in Osteosarcoma by Regulating the miR‐221/ARHI Pathway. J Cell Biochem. 2017;118(12):4772‐81. 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 140. Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, et al. lncRNA GAS5 Reverses EMT and Tumor Stem Cell‐Mediated Gemcitabine Resistance and Metastasis by Targeting miR‐221/SOCS3 in Pancreatic Cancer. Mol Ther Nucleic Acids. 2018;13:472‐82. 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zhao Y, Ma K, Yang S, Zhang X, Wang F, Zhang X, et al. MicroRNA‐125a‐5p enhances the sensitivity of esophageal squamous cell carcinoma cells to cisplatin by suppressing the activation of the STAT3 signaling pathway. Int J Oncol. 2018;53(2):644‐58. 10.3892/ijo.2018.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kishimoto T. IL‐6: from its discovery to clinical applications. Int Immunol. 2010;22(5):347‐52. 10.1093/intimm/dxq030. [DOI] [PubMed] [Google Scholar]
- 143. Wang Y, Zhao Y, Herbst A, Kalinski T, Qin J, Wang X, et al. miR‐221 Mediates Chemoresistance of Esophageal Adenocarcinoma by Direct Targeting of DKK2 Expression. Ann Surg. 2016;264(5):804‐14. 10.1097/sla.0000000000001928. [DOI] [PubMed] [Google Scholar]
- 144. Shao Q, Zhang P, Ma Y, Lu Z, Meng J, Li H, et al. MicroRNA‐139‐5p affects cisplatin sensitivity in human nasopharyngeal carcinoma cells by regulating the epithelial‐to‐mesenchymal transition. Gene. 2018;652:48‐58. 10.1016/j.gene.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 145. Deng X, Liu Z, Liu X, Fu Q, Deng T, Lu J, et al. miR‐296‐3p Negatively Regulated by Nicotine Stimulates Cytoplasmic Translocation of c‐Myc via MK2 to Suppress Chemotherapy Resistance. Mol Ther. 2018;26(4):1066‐81. 10.1016/j.ymthe.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhang P, Lu X, Shi Z, Li X, Zhang Y, Zhao S, et al. miR‐205‐5p regulates epithelial‐mesenchymal transition by targeting PTEN via PI3K/AKT signaling pathway in cisplatin‐resistant nasopharyngeal carcinoma cells. Gene. 2019;710:103‐13. 10.1016/j.gene.2019.05.058. [DOI] [PubMed] [Google Scholar]
- 147. Zhen Y, Fang W, Zhao M, Luo R, Liu Y, Fu Q, et al. miR‐374a‐CCND1‐pPI3K/AKT‐c‐JUN feedback loop modulated by PDCD4 suppresses cell growth, metastasis, and sensitizes nasopharyngeal carcinoma to cisplatin. Oncogene. 2017;36(2):275‐85. 10.1038/onc.2016.201. [DOI] [PubMed] [Google Scholar]
- 148. Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA‐374a activates Wnt/beta‐catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123(2):566‐79. 10.1172/jci65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Vosa U, Vooder T, Kolde R, Fischer K, Valk K, Tonisson N, et al. Identification of miR‐374a as a prognostic marker for survival in patients with early‐stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50(10):812‐22. 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 150. Li JY, Zhang Y, Zhang WH, Jia S, Kang Y, Tian R. Effects of differential distribution of microvessel density, possibly regulated by miR‐374a, on breast cancer prognosis. Asian Pac J Cancer Prev. 2013;14(3):1715‐20. 10.7314/apjcp.2013.14.3.1715. [DOI] [PubMed] [Google Scholar]
- 151. Wang L, Su J, Zhao Z, Hou Y, Yin X, Zheng N, et al. miR‐26b reverses temozolomide resistance via targeting Wee1 in glioma cells. Cell Cycle. 2017;16(20):1954‐64. 10.1080/15384101.2017.1367071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ho KH, Cheng CH, Chou CM, Chen PH, Liu AJ, Lin CW, et al. miR‐140 targeting CTSB signaling suppresses the mesenchymal transition and enhances temozolomide cytotoxicity in glioblastoma multiforme. Pharmacol Res. 2019;147:104390. 10.1016/j.phrs.2019.104390. [DOI] [PubMed] [Google Scholar]
- 153. Ninio‐Many L, Grossman H, Shomron N, Chuderland D, Shalgi R. microRNA‐125a‐3p reduces cell proliferation and migration by targeting Fyn. J Cell Sci. 2013;126(Pt 13):2867‐76. 10.1242/jcs.123414. [DOI] [PubMed] [Google Scholar]
- 154. Lin XJ, He CL, Sun T, Duan XJ, Sun Y, Xiong SJ. hsa‐miR‐485‐5p reverses epithelial to mesenchymal transition and promotes cisplatin‐induced cell death by targeting PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med. 2017;40(1):83‐9. 10.3892/ijmm.2017.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Wang T, Wang D, Zhang L, Yang P, Wang J, Liu Q, et al. The TGFbeta‐miR‐499a‐SHKBP1 pathway induces resistance to EGFR inhibitors in osteosarcoma cancer stem cell‐like cells. J Exp Clin Cancer Res. 2019;38(1):226. 10.1186/s13046-019-1195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li S, Yi M, Dong B, Tan X, Luo S, Wu K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer. 2020. 10.1002/ijc.33386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478). 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo‐loading and synthetic exosome‐mimics as potential therapeutic tools. Acta Pharmacol Sin. 2018;39(4):542‐51. 10.1038/aps.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Villarroya‐Beltri C, Gutiérrez‐Vázquez C, Sánchez‐Cabo F, Pérez‐Hernández D, Vázquez J, Martin‐Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Xiao C, Song F, Zheng YL, Lv J, Wang QF, Xu N. Exosomes in Head and Neck Squamous Cell Carcinoma. Front Oncol. 2019;9:894. 10.3389/fonc.2019.00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Wang M, Qiu R, Yu S, Xu X, Li G, Gu R, et al. Paclitaxelresistant gastric cancer MGC803 cells promote epithelialtomesenchymal transition and chemoresistance in paclitaxelsensitive cells via exosomal delivery of miR1555p. Int J Oncol. 2019;54(1):326‐38. 10.3892/ijo.2018.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM. Exosome‐mediated breast cancer chemoresistance via miR‐155 transfer. Sci Rep. 2018;8(1):829. 10.1038/s41598-018-19339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Johansson J, Berg T, Kurzejamska E, Pang MF, Tabor V, Jansson M, et al. miR‐155‐mediated loss of C/EBPbeta shifts the TGF‐beta response from growth inhibition to epithelial‐mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. 2013;32(50):5614‐24. 10.1038/onc.2013.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA‐155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2016;291(43):22855. 10.1074/jbc.A110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome‐transmitted miR‐128‐3p increase chemosensitivity of oxaliplatin‐resistant colorectal cancer. Mol Cancer. 2019;18(1):43. 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 166. Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J, et al. miR‐374a‐5p: A New Target for Diagnosis and Drug Resistance Therapy in Gastric Cancer. Mol Ther Nucleic Acids. 2019;18:320‐31. 10.1016/j.omtn.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T, et al. Exosomes Serve as Nanoparticles to Deliver Anti‐miR‐214 to Reverse Chemoresistance to Cisplatin in Gastric Cancer. Mol Ther. 2018;26(3):774‐83. 10.1016/j.ymthe.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Li L, Wu P, Wang Z, Meng X, Zha C, Li Z, et al. NoncoRNA: a database of experimentally supported non‐coding RNAs and drug targets in cancer. J Hematol Oncol. 2020;13(1):15. 10.1186/s13045-020-00849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Chaudhary AK, Mondal G, Kumar V, Kattel K, Mahato RI. Chemosensitization and inhibition of pancreatic cancer stem cell proliferation by overexpression of microRNA‐205. Cancer Lett. 2017;402:1‐8. 10.1016/j.canlet.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Liu G, Ji L, Ke M, Ou Z, Tang N, Li Y. miR‐125a‐3p is responsible for chemosensitivity in PDAC by inhibiting epithelial‐mesenchymal transition via Fyn. Biomed Pharmacother. 2018;106:523‐31. 10.1016/j.biopha.2018.06.114. [DOI] [PubMed] [Google Scholar]
- 171. Sato H, Shien K, Tomida S, Okayasu K, Suzawa K, Hashida S, et al. Targeting the miR‐200c/LIN28B axis in acquired EGFR‐TKI resistance non‐small cell lung cancer cells harboring EMT features. Sci Rep. 2017;7:40847. 10.1038/srep40847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Izumchenko E, Chang X, Michailidi C, Kagohara L, Ravi R, Paz K, et al. The TGFβ‐miR200‐MIG6 pathway orchestrates the EMT‐associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014;74(14):3995‐4005. 10.1158/0008-5472.Can-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Cao L, Wan Q, Li F, Tang C‐e. miR‐363 inhibits cisplatin chemoresistance of epithelial ovarian cancer by regulating snail‐induced epithelial‐mesenchymal transition. BMB Reports. 2018;51(9):456‐61. 10.5483/BMBRep.2018.51.9.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhan M, Qu Q, Wang G, Zhou H. Let‐7c sensitizes acquired cisplatin‐resistant A549 cells by targeting ABCC2 and Bcl‐XL. Pharmazie. 2013;68(12):955‐61. [PubMed] [Google Scholar]
- 175. Li W, Yu ZX, Ma BF. The increase of miR‐27a affects the role of cisplatin on proliferation and migration capacities of liver cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(17):5490‐8. 10.26355/eurrev_201809_15809. [DOI] [PubMed] [Google Scholar]
- 176. Wang SY, Chen CL, Hu YC, Chi Y, Huang YH, Su CW, et al. High Expression of MicroRNA‐196a is Associated with Progression of Hepatocellular Carcinoma in Younger Patients. Cancers (Basel). 2019;11(10). 10.3390/cancers11101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


