Abstract
RBM45 is an RNA-binding protein involved in neural development, whose aggregation is associated with neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD). However, the mechanisms of RNA-binding and aggregation of RBM45 remain unelucidated. Here, we report the crystal structure of the N-terminal tandem RRM domains of human RBM45 in complex with single-stranded DNA (ssDNA). Our structural and biochemical results revealed that both the RRM1 and RRM2 of RBM45 recognized the GAC sequence of RNA/ssDNA. Two aromatic residues and an arginine residue in each RRM were critical for RNA-binding, and the interdomain linker was also involved in RNA-binding. Two RRMs formed a pair of antiparallel RNA-binding sites, indicating that the N-terminal tandem RRM domains of RBM45 bound separate GAC motifs in one RNA strand or GAC motifs in different RNA strands. Our findings will be helpful in the identification of physiologic targets of RBM45 and provide evidence for understanding the physiologic and pathologic functions of RBM45.
INTRODUCTION
RNA-binding proteins (RBPs) interact with RNAs sequence-specifically and/or structure-specifically and regulate RNA metabolism, including splicing, maturation, localization, translation and degradation (1–4). Aberrant expression and dysfunction of RBPs correlate with several serious human diseases, such as cancers, infertility and neurodegenerative diseases (3–6). The cytoplasmic inclusions, or liquid-liquid phase separation (LLPS), formed by the aggregation of RBPs, such as TAR DNA-binding protein 43 (TDP-43) (7) and fused-in sarcoma (FUS) (8,9), is a common feature of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD) (10–14). LLPS is driven by both multivalent protein−RNA interactions and multivalent protein−protein interactions (15,16). The RNA-binding motif protein 45 (RBM45), also known as developmentally regulated RNA-binding protein 1 (Drb1), is an RBP that plays an important role in neural development (17,18). RBM45 is predominantly localized in the nucleus and can shuttle between the nucleus and cytoplasm (19,20). RBM45 interacts with >100 proteins in vivo, with most of them being RBPs (21), and it is hypothesized to regulate RNA splicing and processing in the nucleus (22). RBM45 aggregates and co-localizes with the well-understood ALS-linked RBP TDP-43 in the cytoplasmic inclusions in neurons and glia of ALS and FTLD patients (20,23). Investigators have recently found that RBM45 also formed nuclear inclusions in neurons and glia in ALS, FTLD and Alzheimer's disease (AD) (22). These results suggested that the aggregation of RBM45 was associated with neurodegenerative diseases (24).
RBM45 contains three RNA recognition motif (RRM) domains: the RRM1 and RRM2 at the N-terminus, and the RRM3 near the C-terminus. RRM is a common RNA-binding module characterized by a typical βαββαβ topology and two conserved aromatic residues (25,26). All the three RRMs in RBM45 contain the characteristic aromatic residues, suggesting that they may all manifest RNA-binding capability. RBM45 contains a nuclear localization sequence (NLS) at the C-terminus, and mutations in NLS lead to the cytoplasmic aggregation of RBM45 (19,20). The linker region between RRM2 and RRM3, which was predicted to be a pseudo-RRM domain, was reported to promote the homo-oligomerization of RBM45 and was therefore termed the homo-oligomer assembly (HOA) domain (20). The aggregation of RBM45 also involves interactions between RBM45 and other ALS-linked proteins. The interaction between RBM45 and TDP-43 is not well defined. Although RBM45 can bind to TDP-43 without RNA in vitro (19), the interaction becomes weaker when RNA is absent (20), suggesting that the interaction is most likely RNA-dependent. In contrast, RBM45 binds FUS, another well-understood ALS-linked RBP, in an RNA/DNA-independent manner (20,27). RBM45 was also reported to compete with histone deacetylase 1 (HDAC1) for binding to FUS in the DNA-damage response (DDR), suggesting that RBM45 may regulate the FUS-related DDR signaling (27).
Although the relationship between RBM45 and neurodegenerative diseases has been well established, the mechanisms underlying RBM45’s roles in these diseases remain unclear. The RNA-binding property of RBM45 is critical in understanding its physiologic and pathologic functions. It was reported that the full-length RBM45 bound the GACGAC (28) and ACGC (29) sequences of RNA. Recent work also showed that the full-length RBM45 bound GGGACGGU RNA with a dissociation constant (KD) of 18.2 ± 9.3 nM and that the RRM1-truncated RBM45 bound an 18-mer GGGACGGU-containing RNA with higher affinity (21.8 ± 1.5 nM) than the full-length protein (36.9 ± 6.3 nM) (30). Although these results suggested that RBM45 might bind ACG-containing RNA, the RNA-binding preference of each RRM module remains unclear. In the present work, we demonstrated that both the RRM1 and RRM2 of RBM45 recognized the GAC sequence of RNA or ssDNA, and we determined the crystal structure of the tandem RRM1 and RRM2 of human RBM45 in complex with ssDNA. Our structural and biochemical results uncovered the mechanism of the recognition of RNA sequences by the N-terminal tandem RRMs of RBM45. Moreover, the three-dimensional arrangement of the two N-terminal RRMs provided additional information for understanding the physiologic and pathologic functions of RBM45.
MATERIALS AND METHODS
Cloning, protein expression and purification
The DNA encoding the RRM1 and RRM2 domains of human RBM45 (RBM45RRM1–2, residues 23–202) was obtained from full-length RBM45 gene synthesized from Sangon Biosystems (China) and cloned into pET-28a (+) and pET-22b (+) with an N-terminal or C-terminal His-tag, respectively, by using restriction enzymes NdeI and XhoI. Mutant plasmids were produced from the wild-type pET-28a (+) construction via site-directed mutant PCR. The recombinant plasmids were verified by DNA sequencing. All recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells in LB medium. After the growth of bacterial cells up to an OD600 of 0.6–0.8, protein expression was induced by 0.3 mM isopropyl-β-d-thiogalactoside (IPTG) and continued at 16°C for 20 h. For protein purification, the cells were suspended in a buffer containing 20 mM Tris–HCl, pH 7.5 and 300 mM NaCl and lysed by sonication. RBM45RRM1–2 or its mutants were initially purified by Ni-NTA (GE) affinity column and eluted in a buffer containing 20 mM Tris–HCl pH 7.5, 300 mM NaCl and 200 mM imidazole. The proteins were further purified by a pre-equilibrated HiLoad 16/600 Superdex 75 PG column (GE Healthcare) in a buffer containing 20 mM Tris–HCl pH 7.5, 300 mM NaCl and 1 mM DTT. The peak fractions containing highly purified RBM45RRM1–2 or its mutants were collected for later use in crystallization and biochemical assays.
RNA and DNA oligonucleotides
The RNA oligonucleotides used for gel filtration binding assays and SEC-MALS assays, the 5′-FAM RNA and 5′-FAM DNA oligonucleotides used for fluorescent polarization assays, and the DNA oligonucleotides used for crystallization were purchased from General Biosystems (China).
Crystallization, data collection and structure determination
The crystals were grown using the sitting-drop vapor diffusing method at 16°C. For the crystallization of apo RBM45RRM1–2, the C-terminal His-tagged RBM45RRM1–2 protein was concentrated to 3 mg/ml in a buffer containing 20 mM Tris–HCl pH 7.5 and 95 mM NaCl. The crystals were obtained in the condition containing 0.2 M ammonium phosphate dibasic and 20% (w/v) polyethylene glycol (PEG) 3350. For co-crystallization with single-stranded DNA (ssDNA), the N-terminal His-tagged protein was concentrated to 7 mg/ml in a buffer containing 20 mM Tris–HCl pH 7.5 and 300 mM NaCl. The RBM45RRM1–2–ssDNA complex was prepared by mixing the protein with 11-nt 5′-CGACGGGACGC-3′ ssDNA at a molar ratio of 1:1.2. The complex crystals were obtained in a reservoir solution consisting of 0.2 M sodium formate and 20% (w/v) PEG 3350. The crystals were soaked in reservoir solution with 10% (v/v) glycerol and flash-cooled in liquid nitrogen before data collection.
The X-ray diffraction data of the RBM45RRM1–2 crystal were collected at beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF) at a wavelength of 0.9792 Å using a Dectris Eiger 16 M detector (31). The data were processed with the HKL2000 package (32). The structure was solved by the molecular replacement method with PHASER (33) in the CCP4 suite (34), using the crystal structure of human RBM38 (PDB code: 6JVX) (35) as the search model. The model was rebuilt with PHENIX (36) and COOT (37), and refined with PHENIX. The RBM45RRM1–2−ssDNA complex data were collected at beamline BL18U of SSRF at a wavelength of 0.9793 Å using a Dectris Pilatus 6 M detector. The data were processed with the HKL3000 package (32). The structure was solved by the molecular replacement method with PHASER, using the protein alone structure as the search model. The electron density of DNA was clear after several rounds of refinement with REFMAC5 (38) and COOT, allowing an unambiguous DNA model building with COOT. The final model was refined with PHENIX. The statistics of data collection and structure refinement are shown in Table 1.
Table 1.
Data collection and structure refinement statistics
| RBM45RRM1–2 | RBM45RRM1–2−ssDNA | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9792 | 0.9793 |
| Space group | P43 | P43 |
| Cell dimensions | ||
| a, b, c (Å) | 62.48, 62.48, 52.02 | 86.55, 86.55, 27.34 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.00−2.50 (2.59−2.50)* | 50.00−1.80 (1.86−1.80) |
| R merge | 0.160 (0.950) | 0.091(1.391) |
| I/σI | 16.4 (3.0) | 21.3 (1.7) |
| CC1/2 | 0.982 (0.810) | 0.999 (0.764) |
| Completeness (%) | 99.7 (99.3) | 100.0 (100.0) |
| Redundancy | 10.9 (9.2) | 7.3 (7.4) |
| Total/Unique reflections | 76 581/7056 | 141 140/19410 |
| Refinement | ||
| Resolution (Å) | 44.18−2.50 | 21.64−1.80 |
| No. reflections | 7029 | 19273 |
| R work/Rfree | 0.201/0.255 | 0.185/0.234 |
| No. atoms | ||
| Protein | 1366 | 1413 |
| DNA | / | 212 |
| Water | 86 | 169 |
| B-factors (Å2) | ||
| Protein | 38.0 | 43.9 |
| DNA | / | 63.4 |
| Water | 37.3 | 50.1 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.004 |
| Bond angles (°) | 0.514 | 0.596 |
| MolProbity score | 1.27 | 0.79 |
| Ramachandran plot | ||
| Favored | 98.2% | 98.3% |
| Allowed | 1.8% | 1.7% |
| Outliers | 0 | 0 |
*Values in parentheses are for the highest-resolution shell.
Fluorescence polarization analysis
Fluorescence polarization (FP) analysis was used to detect the RNA/DNA-binding affinities of RBM45RRM1–2 and its mutants. The FAM-labeled RNA or DNA at 100 nM was incubated with increasing amounts of RBM45RRM1–2 or its mutants in the binding buffer containing 10 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 1 mM EDTA and 5% (v/v) glycerol for 30 min at room temperature. The 535 nm fluorescence polarization measurements were carried out at 25°C on a SpectraMax Paradigm Multi-Mode detection platform (Molecular Devices, USA), with an excitation wavelength of 485 nm.
Gel filtration assays for binding of RBM45RRM1-2 with two-GAC-motif RNA
RBM45RRM1–2 (3 mg/ml) and RNA molecules that contained two GAC motifs at increasing distances (Supplementary Table S2) were mixed at a molar ratio of 1:1.2 in the GF buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl and 1 mM DTT) and incubated for 30 min on ice, and then loaded to a Superdex 200 Increase 10/300 GL (GE Healthcare) column equilibrated in the GF buffer. A 6-mer RNA (CGACGG) was used as a control.
Size exclusion chromatography with multi-angle light scattering (SEC-MALS)
A DAWN HELEOS-II (Wyatt Technology, Santa Barbara, CA, USA) multi-angle bright scattering detector and an Optilab T-rEX differential refractometer (Wyatt Technology) were used in-line with a Superdex 200 Increase 10/300 GL (GE Healthcare) column. RBM45RRM1–2 (3 mg/ml) and RNA containing two GAC motifs with different spaces were mixed at a molar ratio of 1:1.1 in the SEC-MALS buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl and 1 mM DTT) and incubated for 30 min on ice before each experiment. The samples were run at a flow rate of 0.4 ml/min in the SEC-MALS buffer at room temperature. The data were analyzed using ASTRA 6.1 software.
RESULTS
Structure of the N-terminal tandem RRM domains of human RBM45
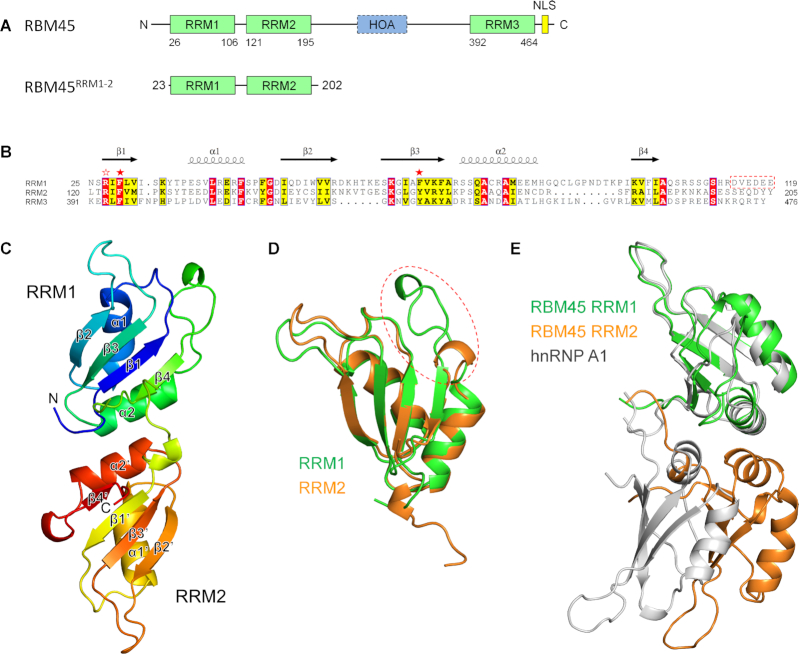
RBM45 contains two N-terminal tandem RRM domains (RRM1 and RRM2), a C-terminal RRM domain (RRM3) and an HOA domain between RRM2 and RRM3 (Figure 1A and B). In this study, we expressed, purified and crystallized the N-terminal tandem RRM domains (23–202, hereafter referred to as RBM45RRM1–2), which was free of nucleic acid contamination (Supplementary Figure S1), and solved the structure at a resolution of 2.5 Å (Figure 1C). The final model contained amino acid residues from Pro23 to Lys193, whereas the residues 109–111 in the interdomain linker region were disordered. Both RRM domains bore the typical RRM topology of β1–α1–β2–β3–α2–β4, as seen in other RRM structures. For clarification, the four β-strands and two helices in RRM2 will be named β1′–β4′ and α1′–α2′, respectively. In each domain, the four β-strands formed an antiparallel four-stranded β-sheet in the order β4–β1–β3–β2, with the two helices packed on one side of the sheet. Two RRM domains could be easily superimposed, with a Cα root-mean-square deviation (RMSD) of 0.82 Å for 53 residues, except for the α2–β4 loop regions, where the RRM1 was much longer than RRM2 (Figure 1B and D). The two domains were pseudo two-fold symmetric and interacted primarily through their α2 and α2′ helices and the interdomain loop, resulting in a pair of antiparallel potential RNA-binding sites on the same side (Figure 1C and Supplementary Figure S2A). The mutation of Ala175 in the inter-domain interface to arginine resulted in significant aggregation of RBM45RRM1–2 (Supplementary Figure S2B), suggesting that this interface was not just a result of crystal packing. The interaction between the two RRM domains was similar to those of hnRNP A1 (39,40) and hnRNP A2/B1 (41); however, they differed in detail. In hnRNP A1 and hnRNP A2/B1, the two RRM domains were not pseudo twofold symmetric; rather, the α2′ of RRM2 interacted with α2 and β4 of RRM1, resulting in a 10−20 Å shift in RRM2 relative to RBM45 (Figure 1E and Supplementary Figure S3). The residues involved in domain−domain interaction are not conserved between RBM45 and hnRNP A1 or hnRNP A2/B1 (Supplementary Figure S4).
Figure 1.
The structure of RBM45RRM1–2. (A) The domains of human RBM45. (B) Sequence alignment of RRM1, RRM2 and RRM3. The identical residues and conserved residues are highlighted in the red and yellow background, respectively. The two conserved aromatic residues are denoted by red stars, the conserved arginine residues are denoted by a red blank star. (C) The overall structure of RBM45RRM1–2. (D) Superimposition of RRM1 and RRM2. The RRM1 and RRM2 are shown as green and orange cartoons, respectively. The α2−β4 loop is enclosed in a red dashed circle. (E) Superimposition of RBM45RRM1–2 and hnRNP A1. The RRM1 of RBM45 is superposed with the RRM1 of hnRNP A1 (PDB code: 1HA1). The RRM1 and RRM2 of RBM45 are colored green and orange, respectively, the hnRNP A1 is colored gray.
Both RRM1 and RRM2 recognize GAC-containing RNA
Previous studies have shown that the full-length RBM45 bound GACGAC (28), ACGC (29) and GGGACGGU (30) of RNA. To identify the RNA recognition sequences of RRM1 and RRM2 of RBM45, we synthesized a 5′-FAM-labeled 5′-GGACGG-3′ 6-mer RNA and analyzed its RBM45RRM1–2-binding affinity using the fluorescence polarization (FP) method. RBM45RRM1–2 bound the GGACGG RNA with a dissociation constant (KD) of 0.457 ± 0.053 μM, whereas the KD of the negative control—a 5′-FAM-labeled polyU 6-mer RNA—was 57.2 ± 10.2 μM (Figure 2A), suggesting that GGACGG was a preferred binding sequence for RBM45RRM1–2. We additionally analyzed the RBM45RRM1–2-binding affinity of a 5′-FAM-labeled 6-mer 5′-GGACGG-3′ single-stranded DNA (ssDNA) and discerned that RBM45RRM1–2 bound this ssDNA with a KD of 0.805 ± 0.108 μM (Supplementary Figure S5), slightly weaker relative to RNA, suggesting that RBM45RRM1–2 can also bind ssDNA.
Figure 2.
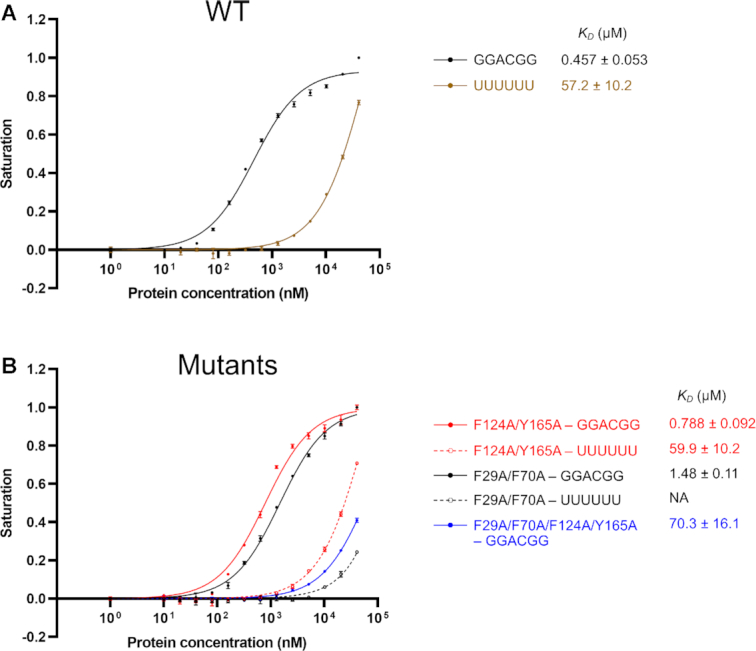
The RNA-binding affinities of RBM45RRM1–2 and its mutants. (A) The fluorescence polarization (FP) measures of binding affinities of RBM45RRM1–2 to GGACGG and UUUUUU RNA. (B) The FP measures of RNA-binding affinities of F124A/Y165A (RRM1 analog), F29/F70A (RRM2 analog) and F29/F70A/F124A/Y165A mutants of RBM45RRM1–2. The data shown here are the averages of three independent measurements with the same protein batch. The error bars indicate the standard deviations of three replicates.
Next, although we attempted to identify the recognition sequence of each single RRM domain, we failed to achieve an adequate amount of soluble RRM1 and RRM2 for the RNA-binding assay. Thus, we generated a double mutant where the two key aromatic amino acid residues in RRM2 (i.e. Phe124 and Tyr165) were mutated to alanine to mimic the RRM1; and a double mutant that the two key aromatic amino acid residues in RRM1 (Phe29 and Phe70) were mutated to alanine to mimic the RRM2. The F29A/F70A/F124A/Y165A quadruple mutant decreased the RNA-binding affinity approximately 150-fold (Figure 2B), indicating that mutations in these key residues virtually completely destroyed the RNA-binding ability of RBM45RRM1–2. This result allowed us to assess the RNA/DNA-binding affinities of the two single RRMs separately by using the above two double mutants. The RRM1 analog F124A/Y165A bound the GGACGG RNA 6-mer and ssDNA 6-mer with KDs of 0.788 ± 0.092 and 1.21 ± 0.09 μM, respectively, slightly weaker than the wild type (WT) RBM45RRM1–2, whereas the RRM2 analog F29A/F70A bound the GGACGG RNA 6-mer and ssDNA 6-mer with KDs of 1.48 ± 0.11 and 2.31 ± 0.19 μM, respectively, ∼3 times weaker than WT RBM45RRM1–2 (Figure 2B and Supplementary Figure S5). These results indicated that GGACGG was a preferred sequence binding both RRM1 and RRM2.
To determine the exact recognition sequence of RBM45RRM1–2 and each single RRM domain, we changed each residue of the 6-mer RNA to other bases and tested their RBM45RRM1–2-binding affinities. For WT RBM45RRM1–2, changing the positions 2 (G), 3 (A) or 4 (C) to any other bases resulted in 4−15-fold diminutions in RBM45RRM1–2-binding affinities, whereas changing the other positions did not significantly influence their affinities (Supplementary Table S1 and Figure S6), indicating that GAC was the critical recognition sequence of RBM45RRM1–2. For the F124A/Y165A mutant (RRM1 analog), the substitutions at positions 2, 3 or 4 resulted in 12−19-fold decreases in binding affinities, while the substitutions at other positions did not significantly influence the affinities (Supplementary Table S1 and Figure S7). For the F29A/F70A mutant (RRM2 analog), the substitutions at positions 2, 3 or 4 resulted in 13−20-fold decreases in binding affinities, while substitutions at other positions did not significantly influence affinities (Supplementary Table S1 and Figure S8). These results indicated that both RRM1 and RRM2 recognized the GAC sequence.
As the GAC motif is a major N6-methyladenosine (m6A) RNA modification site (42,43), we next analyzed the binding affinity of RBM45RRM1–2 with a methylated RNA, 5′-GG(m6A)CGG-3′. The FP assays showed that the WT RBM45RRM1–2, as well as the F124A/Y165A and the F29A/F70A mutants, bound methylated RNA with similar affinities of unmethylated RNA (Supplementary Figure S9), which suggested that neither RRM1 nor RRM2 of RBM45 possesses selectivity for methylation of RNA.
Structure of RBM45RRM1-2 in complex with ssDNA
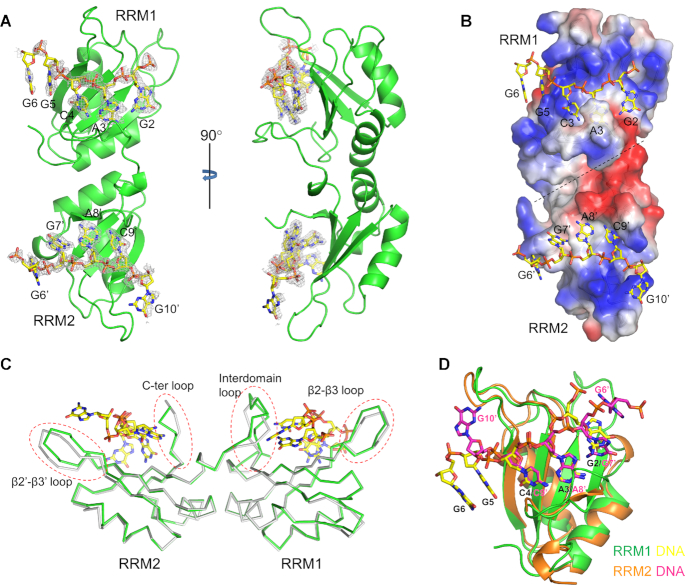
To investigate the mechanism underlying the RNA recognition of RBM45, we attempted to co-crystallize RBM45RRM1–2 with RNA or ssDNA in different sequences and lengths. Ultimately, we obtained crystals of RBM45RRM1–2 in complex with an 11-nt 5′-CGACGGGACGC-3′ (the GAC motif residues are underlined) ssDNA, which contained two GAC motifs, and solved the structure at a resolution of 1.8 Å. Clear electron densities of the GAC motifs were observed at the potential RNA-binding sites of RRM1 and RRM2. The final model contained RBM45 residues Pro23–Asn194 and two ssDNA fragments, with GACGG bound to RRM1 and GGACG bound to RRM2 (Figure 3A). For clarification, we numbered the nucleotides according to the sequence of the 11-nt ssDNA, the RRM1-bound DNA from G2 to G6, and the RRM2-bound DNA from G6′ to G10′. Although an 11-nt ssDNA containing two GAC motifs was used for co-crystallization, only two 5-nt ssDNA segments, each of which contained one GAC motif, were found in the structure. As expected, the two ssDNAs bound in an antiparallel manner to the positively charged patches of RBM45RRM1–2 (Figure 3B). The distance between each end of the two DNAs did not allow a direct linking, suggesting that the two short DNA strands were not a result of a disorder. The two RRM domains of the same molecule most likely bound different DNA molecules. Interestingly, the 5′ ends of the RRM1- and RRM2-bound DNAs were close to the 3′ ends of the RRM1- and RRM2-bound DNAs in the symmetrical molecule, respectively (Supplementary Figure S10), suggesting that the ssDNA that contained two GAC motifs might mediate crystal packing.
Figure 3.
Structure of RBM45RRM1–2−ssDNA complex. (A) The overall structure of the RBM45RRM1–2−ssDNA complex. RBM45 is shown as green cartoon, the ssDNAs are shown as yellow sticks. The simulated annealing omit map of ssDNA (Fo − Fc contoured at 2.5 σ) are shown as gray meshes. (B) The surface electrostatic potential of RBM45RRM1–2. The ssDNA bind to the positively charged regions. (C) Structural changes of RBM45RRM1–2 by DNA-binding. The RBM45RRM1–2 in complex and apo structures are shown as green and gray ribbons, respectively. The DNA is shown as yellow sticks. The changed regions are enclosed in red dashed circles. (D) Superimposition of RRM1 and RRM2. The RRM1 and RRM2 are shown as green and orange cartoons, respectively. The ssDNA bound RRM1 and RRM2 are shown as yellow and magenta sticks.
The interaction between the two RRM domains in the complexed structure was identical to that in the protein alone structure. RBM45RRM1–2 in two structures could be superimposed with a Cα RMSD of 0.33 Å for 123 residues. Two structures differed principally in some loop regions, including the β2−β3 loop, the interdomain loop, the β2′−β3′ loop and the C-terminal loop, which were all involved in ssDNA binding (Figure 3C). The interdomain loop, which was partially disordered in the protein alone structure, interacted with the RRM1-bound DNA and was entirely built into the complexed structure. Both the ssDNA strands in the structure adopted extended conformations. The GAC motifs bound to two RRM domains could be superimposed well; in particular, the adenines and cytosines that bound to two domains bore exactly the same conformation (Figure 3D).
Structural details of nucleic acid recognition
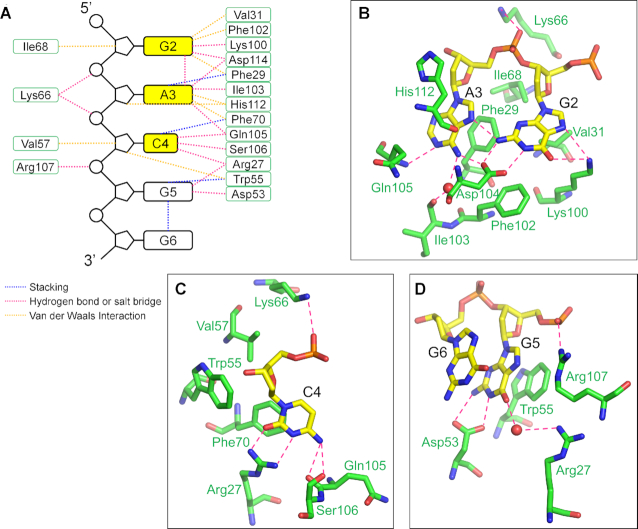
Both the backbone atoms and the base moieties of the ssDNA were involved in the protein−DNA interaction (Figures 4A and 5A). Specifically, in RRM1, the N1 and N2 of G2 formed two hydrogen bonds with the side chain of Asp114, whereas the O6 and N7 formed two hydrogen bonds with the side chain of Lys100. The purine ring of A3 was sandwiched between the conserved aromatic residue Phe29 in β1 and His112 in the interdomain loop. The N1 formed a hydrogen bond with the main-chain amino of Gln105, and the N6 hydrogen-bonded with the side chain of Asp114 directly and with the main-chain carbonyl of Ile103 through a water molecule (Figure 4A and B). The pyrimidine ring of C4 was stacked with the conserved aromatic residue Phe70 in β3; its O2 and N3 formed two hydrogen bonds with the guanidine group of Arg27, and its N4 formed hydrogen bonds with the main-chain carbonyl groups of Gln105 and Ser106 (Figure 4A and C). The purine ring of G5 was stacked with Trp55, and the N1 and N2 formed two hydrogen bonds with the side chain of Asp53; its O6 formed a hydrogen bond with the side-chain amino of Arg27 through a water molecule. G6 did not directly contact protein but was stacked with G5 (Figure 4A and D).
Figure 4.
The interactions between RRM1 and ssDNA. (A) A schematic drawing of RRM1−ssDNA interaction. The DNA bases, deoxyriboses, and phosphates are represented as black boxes, pentagons and circles, respectively. The recognition bases are highlighted in yellow. The amino acid residues are represented as green boxes. The magenta, blue, and orange dashed lines denote hydrogen bonds or salt bridges, stacking and Van der Waals interactions, respectively. (B−D) Detail views of interactions between DNA and RRM1 of RBM45. The involved RBM45 and DNA residues are shown as green and yellow sticks, respectively. The hydrogen bonds and salt bridges are indicated by magenta dashed lines.
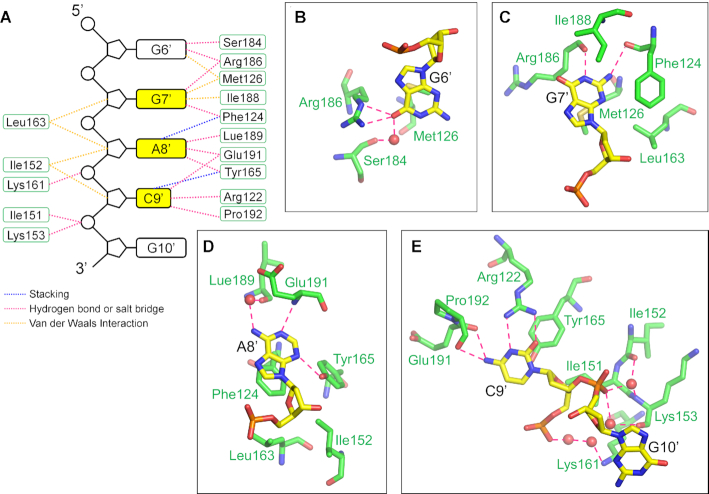
Figure 5.
The interactions between RRM2 and ssDNA. (A) A schematic drawing of RRM2−ssDNA interaction. The DNA bases, deoxyriboses and phosphates are represented as black boxes, pentagons and circles, respectively. The recognition bases are highlighted in yellow. The amino acid residues are represented as green boxes. The magenta, blue and orange dashed lines denote hydrogen bonds or salt bridges, stacking and Van der Waals interactions, respectively. (B−E) Detail views of interactions between DNA and RRM2 of RBM45. The involved RBM45 and DNA residues are shown as green and yellow sticks, respectively. The hydrogen bonds and salt bridges are indicated by magenta dashed lines.
In RRM2, the base moiety of G6′ contacted the side chain of Met126, whereas the O6 hydrogen-bonded with the side chain of Arg186 and with the main-chain carbonyl of Ser184 through a water molecule. The N7 hydrogen-bonded with the side chain of Arg186 through a water molecule (Figure 5A and B). The purine ring of G7′ in the GAC motif was sandwiched between two hydrophobic side chains of Met126 and Ile188, and its N1 and N2 formed hydrogen bonds with the main-chain carbonyl groups of Arg186 and Phe124, respectively (Figure 5A and C). The purine ring of A8′ was stacked with the conserved aromatic residue Phe124, whereas its N1 and N3 formed hydrogen bonds with the main-chain amino group of Glu191 and the hydroxyl group of Tyr165, respectively. Its N6 hydrogen-bonded with the main-chain carbonyl of Leu189 through a water molecule (Figure 5A and D). The pyrimidine ring of C9′ was stacked with the conserved aromatic residue Tyr165. The O2 and N3 formed two hydrogen bonds with the guanidine group of Arg122, and N4 formed hydrogen bonds with the main-chain carbonyl groups of Glu191 and Pro192 (Figure 5A and E).
Binding assays of the interaction surface
To validate our structural findings, we generated several mutants and analyzed their binding to the GGACGG 6-mer RNA using the FP method. As we failed to obtain an adequate amount of soluble single RRM1 or RRM2, the F124A/Y165A and F29A/F70A mutants were used to mimic the single RRM1 domain and single RRM2 domain, respectively. The point mutants of key residues in RRM1 and RRM2 were generated based on the F124A/Y165A and F29/F70A mutants, respectively. For RRM1, the FP assays showed that the mutants of the residues interacted with G2, K100A and D114A, bound RNA with KD values of 4.59 and 1.13 μM, respectively. The former resulted in an approximately 5-fold reduction in RNA-binding affinity, while the latter did not significantly affect the affinity. Mutation of the aromatic residue that stacked with A3 (F29A) bound RNA with a KD of 7.38 μM, attenuating the RNA-binding affinity approximately 9-fold. Mutation of the aromatic residue stacked with C4 (F70A) bound RNA with a KD of 16.8 μM, reducing the RNA-binding affinity more than 20-fold. Mutation of the arginine that interacted with C4 by two hydrogen bonds (R27A) reduced the RNA-binding affinity approximately 18-fold, with a KD of 14.4 μM. For residues that interacted with G5, the W55A and D53A mutants bound RNA with KD values of 3.54 and 1.99 μM, reducing the binding affinities 4.5- and 2.5-fold, respectively (Figure 6A). These results indicated that Phe29, Phe70 and Arg27 were critical for the RNA-binding of RRM1 and that Lys100 and Trp55 also played important roles in RNA-binding.
Figure 6.
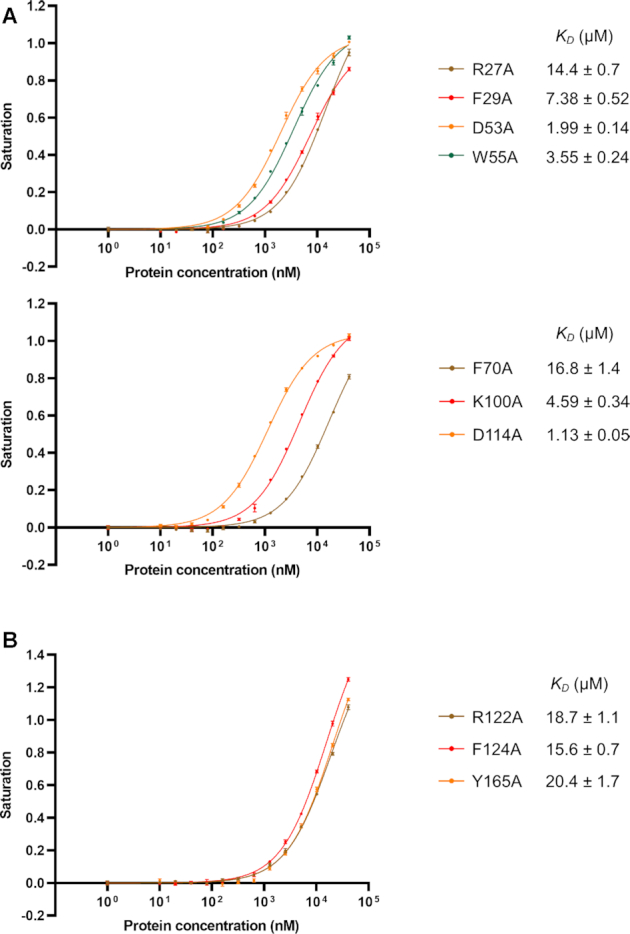
RNA-binding affinities of RBM45RRM1–2 mutants. (A) The FP assays of mutants of key residues in RRM1. The mutants were generated based on the F124A/Y165A mutant (RRM1 analog). Only mutated residues in the RRM1 domain are indicated in each plot. (B) The FP assays of mutants of key residues in RRM2. The mutants were generated based on the F29A/F70A mutant (RRM2 analog). Only mutated residues in the RRM2 domain are indicated in the plot. The data shown here are the averages of three independent measurements with the same protein batch. The error bars indicate the standard deviations of three replicates.
For RRM2, the FP results showed that mutation of the aromatic residue stacked with A8 (F124A) bound RNA with a KD of 15.6 μM, which was approximately 10-fold weaker than the RRM2 analog. Mutation of the aromatic residue stacked with C9 (Y165A) reduced the RNA-binding affinity approximately 14-fold, with a KD of 20.4 μM. Mutation of Arg122, which formed two hydrogen bonds with C9, to alanine reduced the RNA-binding affinity approximately 12-fold, with a KD of 18.7 μM (Figure 6B). These results indicated that Phe124, Tyr165 and Arg122 were critical for the RNA-binding of RRM2.
Binding of RBM45RRM1-2 with RNA containing two GAC motifs
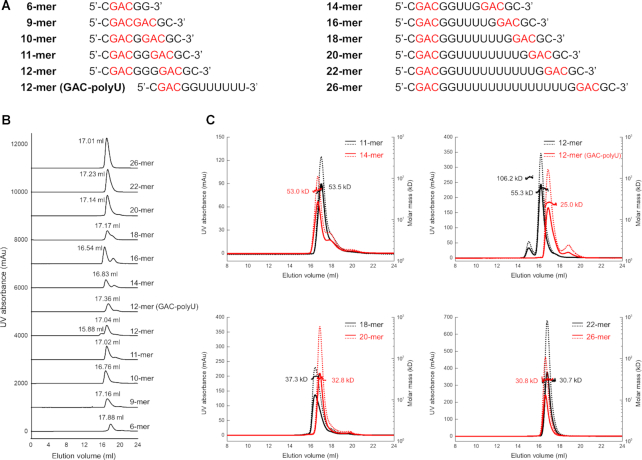
To further study the RNA-binding property of RBM45RRM1–2, we analyzed the binding of RBM45RRM1–2 with a set of longer RNA molecules containing two GAC motifs at increasing distances (Figure 7A) using the gel filtration method (Figure 7B). Some of them were selected for further analysis by the size exclusion chromatography with multi-angle light scattering (SEC-MALS) method to determine the accurate molecular weights (Figure 7C). The gel filtration retention volumes of the protein−RNA complexes with 2-GAC-motif RNA with 0–2 nt spaces (9-mer, 10-mer and 11-mer RNA) were significantly smaller than that of the 6-mer one-GAC-motif RNA, which could only form one protein−one RNA (1P−1R) or 1P−2R complex, implying these RNAs formed larger complexes with RBM45RRM1–2. The SEC-MALS results showed that the molecular weight of the RBM45RRM1–2−11-mer RNA was about 53.5 kDa, close to the theoretical molecular weight of two proteins and two RNAs (53.2 kDa), suggesting that they most likely formed a 2P−2R complex. Interestingly, when the distance was increased to 3 nt (12-mer RNA), in addition to the main peak with a retention volume close to that of the RBM45RRM1–2−11-mer RNA complex, there was a small peak with a significantly smaller retention volume. The SEC-MALS result showed that the molecular weights of the main and small peaks were approximately 55.3 and 106.2 kDa, respectively, consistent with the theoretical molecular weights of 2P−2R (53.9 kDa) and 4P−4R (107.8 kDa) complexes, respectively. These results suggested that the 12-mer RNA can form a higher-order assembly with RBM45RRM1–2 in addition to the 2P−2R complex. To verify whether this binding property was sequence-specific, we performed the same experiments with a 12-mer RNA containing one GAC motif and a 3′ polyU sequence and found that the high molecular weight peak was disappeared, and the main peak was moved to a larger retention volume with a molecular weight of about 25.0 kDa, close to the theoretical molecular weights of a 1P−1R complex (26.8 kDa). These results suggested that the assembly of RBM45RRM1–2−12-mer RNA was sequence-specific. However, when the distance between the two GAC motifs was further increased, the peak of the higher-order assembly was not observed. The gel filtration and the SEC-MALS results suggested that the RNA containing two GAC motif with 5 nt or 7 nt spaces (14-mer or 16-mer RNA) formed a 2P−2R complex as the 11-mer RNA did. The binding of a 9-nt-spaced 2-GAC-motif RNA (18-mer RNA) resulted in a broader gel filtration peak with a larger retention volume than that of 16-mer RNA and a molecular weight of approximately 37.3 kDa, which suggested a mixture of 2P−2R (theoretical molecular weight 57.6 kDa) and 1P−1R (theoretical molecular weight 28.8 kDa) complexes. The retention volumes of gel filtration peaks of 11-nt-, 13-nt- and 17-nt-spaced 2-GAC-motif RNA−RBM45RRM1–2 samples were all larger than that of 16-mer RNA, with molecular weights of approximately 32.8, 30.7, and 30.8 kDa, respectively, close to the theoretical molecular weights of 1P−1R complexes of RBM45RRM1–2 with 20-mer (29.4 kDa), 22-mer (30.0 kDa) and 26-mer (31.2 kDa) RNA, suggesting that these RNAs all formed 1P−1R complexes with RBM45RRM1–2. These results implied that RBM45RRM1–2 can form 1P-1R, 2P-2R or 4–4R complexes with two-GAC-motifs RNA dependent on the distances between the two motifs.
Figure 7.
The binding assays of RBM45RRM1–2 with RNA containing two GAC motifs with different spaces. (A) The RNA sequences used for gel filtration and SEC-MALS assays. The GAC motifs are highlighted in red. (B) The gel filtration results of RBM45RRM1–2 interacted with RNA containing two GAC motif at increasing distances. The assays were performed using a Superdex 200 Increase 10/30 GL column. The 280 nm absorbance of each experiment is shown. The RNA used for each assay is denoted on the right side of each curve. The retention volume of each peak is denoted near the peak. The 6-mer RNA containing one GAC motif was used as a control. (C) The SEC-MALS results of RBM45RRM1–2−RNA complexes. Curves in different colors represent different samples. The RNA used in experiments are denoted on the upper right of each plot. The 280 nm and 260 nm absorbances are shown as solid and dashed curves, respectively. The molecular weight values measured by SEC-MALS are denoted.
DISCUSSION
In the present study, we identified the RRM1 and RRM2 domains of RBM45 as both recognizing the GAC sequence of RNA or ssDNA, which suggested that RBM45 prefers to bind RNA/ssDNA with multiple GAC motifs. The recognition sequence of RBM45 has been investigated by both high-throughput (28,29) and biochemical (30) methods. An RNAcompete (44) study showed that full-length RBM45 recognized GACGAC sequence (28), a duplex of GAC motif, whereas an RNA Bind-n-Seq (45) study reported another sequence recognized by full-length RBM45, ACGC (29). Our finding agreed with the former result and differ slightly from the latter. The RNA-binding of RBM45 was also analyzed by the biolayer interferometry (BLI) method, which showed that RBM45 bound to a GGGACGGU sequence (30). This result was consistent with our finding because the RNA they used contained a GAC motif. In this work, we identified the accurate and condensed recognition sequence of RRM1 and RRM2 of RBM45, which will help identify the physiologic targets of RBM45. In addition, as it was reported that RBM45 was involved in DNA damage response by interacting with FUS (27), the ssDNA-binding ability of RBM45 suggested that RBM45 might also directly interact with ssDNA during the FUS-related DNA damage response.
Our structural results revealed that the recognition primarily involved two aromatic residues and an arginine residue in each domain. The two aromatic residues were conserved in RRMs and played critical roles in RNA-binding (25). The aromatic residue in β1 (Phe29 in RRM1 and Phe124 in RRM2) was stacked with the adenine of the GAC motif, while the aromatic residue in β2 (Phe70 in RRM1 and Tyr165 in RRM2) was stacked with the cytosine of the GAC motif. Whereas the stacking interactions between the conserved aromatic residues and bases, as well as the electrostatic interactions between basic amino acids and the backbone phosphates, provide the fundamental binding power for nucleotides, the hydrogen-bond interactions between the atoms of the bases and the side-chain or main-chain atoms of the protein determine the RNA/DNA-binding specificity. In both RRM1 and RRM2 of RBM45, the hydrogen bonds of the two adenine bases principally involved the main-chain atoms, and the guanidine group of the arginine (Arg27 in RRM1 and Arg122 in RRM2) formed two hydrogen bonds with the cytosine. Our binding assays showed that the four aromatic residues and two arginine residues were critical for RNA-binding, confirming our structural results.
In addition to the dinucleotide that binds at these two positions, the RRM domain usually binds to additional nucleotides (25,46). In RRM1 of RBM45, the bases of two guanines, G2 and G5, were found to directly bind RRM1. Our binding assays showed that the binding of G2 was sequence-specific. Mutation of Lys100, which formed two hydrogen bonds with the base of G2, significantly reduced the RNA-binding affinity, supporting the specificity of G2. G2 also formed two hydrogen bonds with Asp114 in the interdomain linker; however, the mutation of Asp114 did not significantly affect binding. One possible explanation for this apparent contradiction might be that the interdomain linker was rich in acidic residues (Figure 1B) and that the other acidic residues might therefore have rescued the effect of the Asp114 mutation. The binding of G5 was not sequence-specific, and the mutation of Asp53, which formed hydrogen bonds with G5, did not significantly affect the binding. In RRM2, the bases of G6′ and G7′ directly bound protein, although the binding of G6′ was not sequence-specific. In contradistinction, the binding of G7′ was sequence-specific, with the specificity determined by hydrogen bonds between the base and the two main-chain atoms in β1′ and β4′. The three key residues in each RRM and most of the residues that are involved in human RBM45RRM1–2−DNA interaction are conserved among RBM45s from human to Drosophila (Supplementary Figure S11), suggesting that the recognition mechanisms of RBM45 might be conserved across a wide range of organisms.
As the RNA-binding specificity of a single RRM domain is usually limited, RBPs often contain multiple RRM domains for higher specificity as well as higher affinity (47). However, the three-dimensional (3D) arrangements of domains are diverse in multiple-RRM RBPs with known structures. The two tandem RRM domains in TDP-43 (48), PABP1 (49), HuD (50), HuR (51), Sxl (52) and Hrp1 (53) create continuous RNA-binding surfaces, although their 3D arrangements are not identical (Supplementary Figure S12A). Contrarily, the two tandem RRMs in hnRNP A1 (54), hnRNP A2/B1 (41) and PTB (55) form separate and approximately antiparallel RNA-binding sites (Supplementary Figure S12B). Also, there are many RBPs, such as nucleolin (56) and CUGBP1 (57), containing tandem RRM domains without stable interaction. Although the arrangement of RRM1 and RRM2 of RBM45 differs from those of any known tandem RRM structures, it is similar to hnRNP A1 and hnRNP A2/B1, which result in a pair of antiparallel RNA-binding sites. While the RBPs that contain multiple RRMs with continuous RNA-binding surfaces usually bind adjacent recognition elements in one RNA strand, RBPs containing separate RNA-binding sites can bind separated recognition elements in both the same RNA strand (Supplementary Figure S13A) and different RNA strands (Supplementary Figure S13B). In this work, we tested the binding of RBM45RRM1–2 with RNA molecules that contained two GAC motifs with increasing distances and found that when the distance between the two GAC motifs was longer than 9 nt, RBM45RRM1–2 formed a 1:1 complex with RNA. When the distance was short (0−7 nt), RBM45RRM1–2 tended to form a 2:2 complex with RNA, implying that the two RRM domains bound two GAC motifs from two RNA molecules respectively (Supplementary Figure S13C). We also observed a 4:4 complex when the distance was 3 nt, which suggested that RBM45RRM1–2 could form a higher-order assembly with this RNA through a protein−RNA network (Supplementary Figure S13D). These results implied that the binding of one RBM45RRM1–2 with two GAC motifs of the same RNA molecule will happen only when the distance between two motifs is long enough. While the distance is short, the two GAC motifs will bind RRM domains from different RRM45RRM1–2 molecules, leaving two unbound RRM domains to recruit other RNA molecules mediating RNA-RNA interactions. It should be noted that because RNA molecules generally form secondary structures, the spatial distance between two motifs may be significantly different from their distance in the sequence, therefore, our in vitro results with short RNA can not be simply applied to transcripts in vivo.
Although the association between the cytoplasmic aggregation of RBM45 and neurodegenerative diseases has been well established, no physiological target of RBM45 has been reported to date. To date, the only reported RBM45 target is a viral RNA segment identified in vitro, which contained a GAC sequence (30), consistent with our finding. Our structural results and binding assays suggested that the targets of RBM45 should contain separate multiple GAC motifs. Given that the recognition sequence of the RRM1 or RRM2 contains only three bases, the RNA-binding specificity of a single domain should be relatively low. Multiple domains usually significantly improve the binding specificity (47), however, our binding assays showed that RBM45RRM1–2 can bind to RNA with two GAC motifs spaced over a wide range of distances, therefore, the improvement in binding specificity by dual domains of RBM45 should be limited. RNA sequences that match the recognition feature of RBM45RRM1–2 may occur every few hundred nucleotide residues, therefore most transcripts are expected to carry such sequences. However, this does not mean that RBM45 can bind to most transcripts, because the accessibility of RBP is significantly affected by the secondary structure of RNA (58,59). Our structural results suggested that the RRM1 and RRM2 of RBM45 could only bind to a GAC motif in extended conformation. The prediction of the targets of RBM45 requires not only the sequence information but also the secondary structure information, which is largely lacking and highly dependent on bioinformatical predictions at present. Therefore, although our study has figured out a sequence characteristic of the RNA recognized by RBM45RRM1–2, the identification of RBM45 targets is still a challenging work.
The C-terminal RRM of RBM45, RRM3, also contains the two conserved aromatic residues and the arginine that interacts with cytosine (Figure 1B), implying that it may also possess RNA-binding affinity. The HOA domain between RRM2 and RRM3 was reported to mediate the oligomerization of RBM45 (20) and interact with other ALS-related RBPs such as FUS (27). Therefore, the full-length RBM45, which might carry three RNA-binding modules and a protein-protein interaction domain, may form more complex RNA-protein networks. Our structural and biochemical results suggest a model encompassing the physiologic and pathologic functions of RBM45. Under normal conditions, RBM45 functions as an RNA-binding protein that binds RNA that contains separate multiple GAC motifs. While under stress, RBM45 binds multiple RNA molecules and forms complex protein-RNA networks with the participation of other proteins, such as TDP-43 and FUS, thus leading to aggregation.
DATA AVAILABILITY
The atomic coordinates and structure factors of RBM45RRM1–2 and RBM45RRM1–2−ssDNA have been deposited to the Protein Data Bank (PDB) with accession numbers 7CSX and 7CSZ, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at beamlines BL17U and BL18U of the Shanghai Synchrotron Radiation Facility (SSRF) for assistance during data collection. We thank Xiuhai Wang, Xu Li and the staff of the National Training Center for Laboratory Techniques of Life Science, University of Science and Technology of China, for their help during SEC-MALS experiments. We thank the staff for providing technical support with using the facility of the Institute of Health Sciences & Technology, Anhui University.
Contributor Information
Xiaolei Chen, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
Zhongmei Yang, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
Wenfeng Wang, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
Kaiyue Qian, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; Key Laboratory of Human Microenvironment and Precision Medicine of Anhui Higher Education Institutes, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
Mingjie Liu, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; Key Laboratory of Human Microenvironment and Precision Medicine of Anhui Higher Education Institutes, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
Junchao Wang, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; Key Laboratory of Human Microenvironment and Precision Medicine of Anhui Higher Education Institutes, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
Mingzhu Wang, Institutes of Physical Science and Information Technology, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China; Key Laboratory of Human Microenvironment and Precision Medicine of Anhui Higher Education Institutes, Anhui University, 111 Jiulong Road, Hefei 230601, Anhui, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31470719 to M.W.]. Funding for open access charge: Natural Science Foundation of China [31470719 to M.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005; 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 2. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 3. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glisovic T., Bachorik J.L., Yong J., Dreyfuss G.. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008; 582:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh G., Pratt G., Yeo G.W., Moore M.J.. The Clothes Make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015; 84:325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nussbacher J.K., Tabet R., Yeo G.W., Lagier-Tourenne C.. Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron. 2019; 102:294–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M.et al.. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006; 314:130–133. [DOI] [PubMed] [Google Scholar]
- 8. Kwiatkowski T.J. Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T.et al.. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009; 323:1205–1208. [DOI] [PubMed] [Google Scholar]
- 9. Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P.et al.. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009; 323:1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ling S.C., Polymenidou M., Cleveland D.W.. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013; 79:416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Es M.A., Hardiman O., Chio A., Al-Chalabi A., Pasterkamp R.J., Veldink J.H., van den Berg L.H.. Amyotrophic lateral sclerosis. Lancet. 2017; 390:2084–2098. [DOI] [PubMed] [Google Scholar]
- 12. Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 13. Elbaum-Garfinkle S. Matter over mind: liquid phase separation and neurodegeneration. J. Biol. Chem. 2019; 294:7160–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng C., Trojanowski J.Q., Lee V.M.. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 2020; 16:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhine K., Vidaurre V., Myong S.. RNA Droplets. Annu. Rev. Biophys. 2020; 49:247–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang H., Ji X., Li P., Liu C., Lou J., Wang Z., Wen W., Xiao Y., Zhang M., Zhu X.. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci. 2020; 63:953–985. [DOI] [PubMed] [Google Scholar]
- 17. Tamada H., Sakashita E., Shimazaki K., Ueno E., Hamamoto T., Kagawa Y., Endo H.. cDNA cloning and characterization of Drb1, a new member of RRM-type neural RNA-binding protein. Biochem. Biophys. Res. Commun. 2002; 297:96–104. [DOI] [PubMed] [Google Scholar]
- 18. Cooper-Knock J., Robins H., Niedermoser I., Wyles M., Heath P.R., Higginbottom A., Walsh T., Kazoka M., Ince P.G., Hautbergue G.M.et al.. Targeted genetic screen in amyotrophic lateral sclerosis reveals novel genetic variants with synergistic effect on clinical phenotype. Front. Mol. Neurosci. 2017; 10:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mashiko T., Sakashita E., Kasashima K., Tominaga K., Kuroiwa K., Nozaki Y., Matsuura T., Hamamoto T., Endo H.. Developmentally regulated RNA-binding protein 1 (Drb1)/RNA-binding motif protein 45 (RBM45), a nuclear-cytoplasmic trafficking protein, forms TAR DNA-binding protein 43 (TDP-43)-mediated cytoplasmic aggregates. J. Biol. Chem. 2016; 291:14996–15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y., Collins M., Geiser R., Bakkar N., Riascos D., Bowser R.. RBM45 homo-oligomerization mediates association with ALS-linked proteins and stress granules. Sci. Rep. 2015; 5:14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y., Collins M., An J., Geiser R., Tegeler T., Tsantilas K., Garcia K., Pirrotte P., Bowser R.. Immunoprecipitation and mass spectrometry defines an extensive RBM45 protein-protein interaction network. Brain Res. 2016; 1647:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collins M., Li Y., Bowser R.. RBM45 associates with nuclear stress bodies and forms nuclear inclusions during chronic cellular stress and in neurodegenerative diseases. Acta Neuropathol. Commun. 2020; 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins M., Riascos D., Kovalik T., An J., Krupa K., Krupa K., Hood B.L., Conrads T.P., Renton A.E., Traynor B.J.et al.. The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol. 2012; 124:717–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uversky V.N. The roles of intrinsic disorder-based liquid-liquid phase transitions in the “Dr. Jekyll-Mr. Hyde” behavior of proteins involved in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Autophagy. 2017; 13:2115–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maris C., Dominguez C., Allain F.H.. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005; 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- 26. Clery A., Blatter M., Allain F.H.. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008; 18:290–298. [DOI] [PubMed] [Google Scholar]
- 27. Gong J., Huang M., Wang F., Ma X., Liu H., Tu Y., Xing L., Zhu X., Zheng H., Fang J.et al.. RBM45 competes with HDAC1 for binding to FUS in response to DNA damage. Nucleic Acids Res. 2017; 45:12862–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A.et al.. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dominguez D., Freese P., Alexis M.S., Su A., Hochman M., Palden T., Bazile C., Lambert N.J., Van Nostrand E.L., Pratt G.A.et al.. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell. 2018; 70:854–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J., Ganaie S.S., Cheng F., Xu P., Ning K., Wang X., Kleiboeker S., Cheng S., Qiu J.. RNA binding motif protein RBM45 regulates expression of the 11-kilodalton protein of parvovirus B19 through binding to novel intron splicing enhancers. mBio. 2020; 11:e00192-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q.-S., Zhang K.-H., Cui Y., Wang Z.-J., Pan Q.-Y., Liu K., Sun B., Zhou H., Li M.-J., Xu Q.et al.. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018; 29:68. [Google Scholar]
- 32. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 33. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qian K., Li M., Wang J., Zhang M., Wang M.. Structural basis for mRNA recognition by human RBM38. Biochem. J. 2020; 477:161–172. [DOI] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shamoo Y., Krueger U., Rice L.M., Williams K.R., Steitz T.A.. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat. Struct. Biol. 1997; 4:215–222. [DOI] [PubMed] [Google Scholar]
- 40. Xu R.M., Jokhan L., Cheng X., Mayeda A., Krainer A.R.. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997; 5:559–570. [DOI] [PubMed] [Google Scholar]
- 41. Wu B., Su S., Patil D.P., Liu H., Gan J., Jaffrey S.R., Ma J.. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018; 9:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei C.M., Moss B.. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977; 16:1672–1676. [DOI] [PubMed] [Google Scholar]
- 43. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 44. Ray D., Kazan H., Chan E.T., Pena Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., Hughes T.R.. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009; 27:667–670. [DOI] [PubMed] [Google Scholar]
- 45. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Auweter S.D., Oberstrass F.C., Allain F.H.. Sequence-specific binding of single-stranded RNA: is there a code for recognition?. Nucleic Acids Res. 2006; 34:4943–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lukavsky P.J., Daujotyte D., Tollervey J.R., Ule J., Stuani C., Buratti E., Baralle F.E., Damberger F.F., Allain F.H.. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 2013; 20:1443–1449. [DOI] [PubMed] [Google Scholar]
- 49. Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K.. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999; 98:835–845. [DOI] [PubMed] [Google Scholar]
- 50. Wang X., Tanaka Hall T.M.. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001; 8:141–145. [DOI] [PubMed] [Google Scholar]
- 51. Wang H., Zeng F., Liu Q., Liu H., Liu Z., Niu L., Teng M., Li X.. The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr. D. Biol. Crystallogr. 2013; 69:373–380. [DOI] [PubMed] [Google Scholar]
- 52. Handa N., Nureki O., Kurimoto K., Kim I., Sakamoto H., Shimura Y., Muto Y., Yokoyama S.. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999; 398:579–585. [DOI] [PubMed] [Google Scholar]
- 53. Perez-Canadillas J.M. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006; 25:3167–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding J., Hayashi M.K., Zhang Y., Manche L., Krainer A.R., Xu R.M.. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999; 13:1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L.et al.. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005; 309:2054–2057. [DOI] [PubMed] [Google Scholar]
- 56. Allain F.H., Bouvet P., Dieckmann T., Feigon J.. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000; 19:6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Teplova M., Song J., Gaw H.Y., Teplov A., Patel D.J.. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010; 18:1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Taliaferro J.M., Lambert N.J., Sudmant P.H., Dominguez D., Merkin J.J., Alexis M.S., Bazile C.A., Burge C.B.. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol. Cell. 2016; 64:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bevilacqua P.C., Ritchey L.E., Su Z., Assmann S.M.. Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 2016; 50:235–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of RBM45RRM1–2 and RBM45RRM1–2−ssDNA have been deposited to the Protein Data Bank (PDB) with accession numbers 7CSX and 7CSZ, respectively.