Abstract
N 6-Methyladenosine (m6A) is the most abundant modification within diverse RNAs including mRNAs and lncRNAs and is regulated by a reversible process with important biological functions. Human YTH domain family 2 (YTHDF2) selectively recognized m6A-RNAs to regulate degradation. However, the possible regulation of YTHDF2 by protein post-translational modification remains unknown. Here, we show that YTHDF2 is SUMOylated in vivo and in vitro at the major site of K571, which can be induced by hypoxia while reduced by oxidative stress and SUMOylation inhibitors. SUMOylation of YTHDF2 has little impact on its ubiquitination and localization, but significantly increases its binding affinity of m6A-modified mRNAs and subsequently results in deregulated gene expressions which accounts for cancer progression. Moreover, Disease-free survival analysis of patients with lung adenocarcinoma derived from TCGA dataset reveals that higher expression of YTHDF2 together with higher expression of SUMO1 predicts poor prognosis. Our works uncover a new regulatory mechanism for YTHDF2 recognition of m6A-RNAs and highlight the importance of YTHDF2 SUMOylation in post-transcriptional gene expression regulation and cancer progression.
INTRODUCTION
Over 140 types of chemical modification of RNAs are found in recent decades. N6-Methyladenosine (m6A), is one of the most abundant internal modifications especially in mRNAs and lncRNAs (1), which mediates post-transcriptional gene expression regulation. The m6A modification is dynamically deposited by the m6A methyltransferase complex (‘writers’) containing METTL3, METTL14 and WTAP (2), while reversed by m6A demethylases FTO and AlkBH5 (‘erasers’) (3,4). Transcriptome analyses have revealed that m6A modification is enriched within a RR(m6A)CH motif (where R refers to G or A and H refers to U, A or C) and mainly located in 5′ untranslated regions (UTRs) and 3′UTRs adjacent to stop codons of mammalian mRNAs (5,6). The reversible nature of m6A modification and the reliable ways to detect this modification transcriptome-wide have attracted researchers’ interest to discover its biological roles. Until now, the m6A modification has been shown to regulate mRNA processing, translation, and degradation (7) and to be involved in many cellular features such as stemness (8), immune response (9–11), viral infections (12), oocyte maturation (13), spermatogenesis (14), and cancer (15).
The m6A readers, YTH (YT521-B homology) domain proteins including YTHDC1 that is mainly located to the nucleus and YTHDF1, YTHDF2, YTHDF3 and YTHDC2 mainly located in the cytoplasm, can selectively recognize and bind to m6A motif within the consensus RR(m6A)CH sequence (5,16–18). The cytoplasmic m6A readers play critical roles in accelerating metabolism of m6A-modified mRNAs. YTHDF1 and YTHDF3 binding to m6A motif enhances mRNA translation (19,20), while YTHDF2 recognizes m6A motif to facilitate the degradation of m6A-modified transcripts by recruiting the CCR4-NOT deadenylase (20,21). The C-terminal of YTHDF2 is responsible for binding to m6A motif while the N-terminal can recruit deadenylase (21). Moreover, the crystal structure of YTH domain in YTHDF2 at 2.1 Å resolutions has been solved to show that two residues W432 and W486 are very important for recognition of m6A (18). However, the functional regulation of the specific m6A readers remains unexplored, for examples, whether and how post-translational modifications (PTMs) of YTHDF2 modulates its functions.
The SUMOylation pathway introduces, in a reversible and dynamic manner, post-translational modifications (PTMs) in the form of SUMO to protein substrates. SUMOylation is catalyzed by the dimeric E1 SAE1/UBA2, the single E2 Ubc9 and E3 ligases, whereas it can be removed by Sentrin-specific Proteases (SENPs) (22). It regulates a large variety of cellular processes by altering the activity, stability, localization and protein-protein interaction of substrates, in response to environmental stimuli and cellular stresses (23–28). Recently, we firstly reported that SUMOylation is involved in regulation of m6A modification levels, in which the m6A-RNA methyltransferase METTL3 is SUMOylated to repress its methyltransferase activity (29).
In this study, we found that YTHDF2 was modified by SUMO1 at the major site K571 in vitro and in vivo. SUMOylation of YTHDF2 increased its binding with transcriptome-wide m6A modified mRNAs, and subsequently changed the gene expression profiles, which resulted in promoting proliferation, migration, colony formation and tumor growth of lung cancer cell H1299. These results suggested that SUMOylation of YTHDF2 was a novel molecular mechanism underlying regulation of m6A-modified RNAs recognition and degradation.
MATERIALS AND METHODS
Antibodies and reagents
Antibodies against YTHDF2 (24744-1-AP), GST (#66001-1-Ig), β-Actin (#6008-1-Ig) and His-tag (66005-1-Ig) were purchased from Proteintech™. Antibodies against Flag (M2, #F1804) and HA (16B12, #MMS-101P) were obtained from Sigma. Antibodies against m6A (17-3-4-1, #ab208577), GAPDH (#ab37168), SUMO1 (Y299, #ab32058), GFP (#ab183734) and SENP1 (EPR3844, #ab108981) were from Abcam. Antibody against Myc (#2278) was from Cell Signaling Technology. Antibody against Lamin B1 (8D1, #sc-56,144) was from Santa Cruz Biotech. Protein G Plus/Protein A agarose suspension (#IP05) was purchased from Calbiochem. Ni2+-NTA agarose beads were obtained from Qiagen. Polybrene (hexadimethrine bromide, # H9268) and puromycin (#P8833) were from Sigma.
Plasmids and short interfering RNAs
Human YTHDF2 was amplified by PCR using KOD-plus Kit (TOYOBO) and subcloned into the vectors pEF-5HA, pEGX-4T-1 and pCMV-Myc, respectively. The fragment of YTHDF2 C-terminal corresponding to amino acid region from E384 to the end K579 was constructed into pEF-5HA. Point mutations of YTHDF2 were introduced by using KOD-plus-mutagenesis Kit (TOYOBO) according to the manufacturer's protocol. The construct pCMV-Myc-YTHDF2△568–579 was constructed by deleting the amino acid region from 568 to 579 in pCMV-Myc-YTHDF2-WT plasmid. The BG-PLAC2 fragment was amplified from the pTBG-PLAC2 plasimd kindly provided by Dr LG Wu (21), which contained two m6A motifs: (5′-CATCGCAAGAAGAGAAGCACAGAAGGGGCAGGAGAGACACTCAGAGGCACTTCCGCTCTTGCCCAGGACATTTTCCCAGCCACACCTTTGCCCAAGCCGTGCCCCCTGCCTGGAGCACTTTTCAACCTCTTCTCT-3′). The constructs pCW-BG-PLAC2 and pcDNA3.1-BG-PLAC2-12 × -MS2 were constructed by inserting the BG-PLAC2 fragment into the vectors pCW57.1 (with BamHI and NheI) and pcDNA3.1-12X-MS2 (with NotI and XhoI), respectively. The pEGFP-C3-SON was constructed by inserting an 84-nt fragment derived from the ORF of SON gene containing three m6A motifs (5′-AACACCATGGACTCCCAGATGTTAGCGTCTAGCACCATGGACTCCCAGATGTTAGCAACTAGCTCCATGGACTCCCAGATGTTA-3′) into the pEGFP-C3 vector (with XhoI and SacII). The shRNA sequence 5′-GCAGACTTGCAGTTTAAGTAT-3′ targeting YTHDF2 3′-UTR (sh-YTHDF2) was cloned into the Lentiviral vector pLKO.1 (Sigma). The YTHDF2 cDNA was amplified with the primer containing KOZAK sequence (GCCACC) and Flag-tag sequence (GATTACAAGGATGACGATAAG), and then subcloned into the Lentiviral vector CD513B (System Biosciences).
Cell culture and transfection
HEK-293T, 293FT, HeLa and H1299 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone) containing 10% fetal bovine serum (Biowest) and 1% penicillin and streptomycin (Invitrogen). All cells were cultured in 5% CO2-humidified incubator at 37°C. Cell transfection was performed using Lipofectamine 2000 (Invitrogen).
SUMOylation assays
SUMOylation of YTHDF2 was confirmed by three different methods. The SUMOylation analysis by Ni2+-NTA pull down was performed as previously described (23,25–27). The SUMOylation analysis by Co-immunoprecipitation (Co-IP) under denaturing conditions was used to detect both exogenous and endogenous SUMOylation of YTHDF2 following the published protocol (30), with minor changes as our previous publication described (29). Briefly, cells were lysed with lysis buffer S (20 mM sodium phosphate pH 7.4, 150 mM NaCl, 1% SDS, 1% Triton, 0.5% sodium deoxycholate, 5 mM EDTA, 5 mM EGTA, 20 mM N-ethylmaleimide (NEM) 5mM DTT and Protease inhibitor cocktail), boiled for 10 min, and then sonicated until the lysate became fluid. The lysates clarified by centrifugation at 16 000g for 10 min at 4°C were finally diluted 1:10 with lysis buffer S without SDS, and then incubated with the anti-SUMO1 or anti-YTHDF2 antibodies and Protein A/G beads overnight at 4°C. Beads were washed five times with high-salt buffer, boiled for 10 min in SDS sample loading buffer, and followed by Western blotting analysis.
The SUMOylation analysis by bacterial reconstitution system was performed as previously described (23,26,31). Briefly, the GST-tagged recombinant YTHDF2-WT plasmid was transformed into BL21(DE3) Escherichia coli cells alone or together with pE1E2S1 and were induced at 16°C with 1 mM IPTG for 12 h. The pellets were lysed in PBS-L solution (50 mM NaH2PO4 pH7.2, 150 mM NaCl, 1 mM PMSF, 1 mM DTT, 1 mM EDTA, 0.1% (v/v) Triton X-100) and then sonicated for 10 min on ice. After centrifuging at 17 000g for 20 min, the supernatants were mixed with GST-Sefinose Resin (GE healthcare) for 4 h at 4°C. Then the beads were washed three times with PBS-L solution and eluted with GSH buffer (50 mM Tris pH 8.0, 20 mM GSH). The purified protein was detected by Western blotting.
Immunofluorescence staining
Cells were seeded on the glass cover slips and treated under different conditions for indicated time, and then fixed with 4% paraformaldehyde. After permeablization by 0.1% Titon X-100 for 30 min, cells were blocked by 5% BSA for 1 h, and then incubated with anti-Flag or Anti-YTHDF2 antibody diluted (1:100) overnight. Subsequently, fluorescent dye-conjugated secondary antibody was applied for 2 h away from light and the nucleus was stained by DAPI for 30 min. Finally, the immunofluorescence images were recorded by a laser scanning confocal microscopy.
qRT-PCR
RNAs were extracted by TRIZOL reagent (Invitrogen) and then treated with DNase I (Fermentas) to degrade genomic DNA. Reverse transcription was performed using the PrimeScript RT-PCR Kit (#RR037A, TAKARA) according to the manufacturer's instructions. Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (#4309155, Applied Biosystems) to analyze the RNA abundance of BG-PLAC2. Primers used for real-time PCR were listed below:
BG-PLAC2 Forward: 5′TGAGGAGAAGTCTGCGGTCAC 3′
BG-PLAC2 Reverse: 5′ GGACTCGAAGAACCTCTGGGT 3′
RNA immunoprecipitation assay (RIP)
The RNA immunoprecipitation assay (RIP) was performed as previously described (23,26). The cells transfected with indicated plasmids were lysed with RIP lysis buffer [150 mM NaCl, 50 mM Tris–HCl pH 7.4, 1% NP40, 1 mM dithiothreitol, 100 U/ml RNase inhibitor (Fermentas), 400 μM VRC (New England BioLabs) and Protease inhibitor cocktail (Roche)] for 30 min on ice, then centrifuged at 15 000g for 20 min to clear the lysate. One-tenth of the lysates was used as Input, and other lysates were incubated with protein A/G agarose beads and antibodies at 4°C overnight. The beads were washed three times with RIP buffer and the bound RNAs was isolated using Trizol (Sigma) following instructions, and then reversely transcribed using the PrimeScript RT-PCR Kit (#RR037A, TAKARA). The immunoprecipitated RNAs of BG-PLAC2 associated with YTHDF2 were measured by q-PCR analysis and m6A dot plot analysis. The enrichment of BG-PLAC2 associate with YTHDF2 was normalized by the input abundance of BG-PLAC2.
YTHDF2-bound m6A RNA detection by Co-immunoprecipitation (Co-IP)
The binding of YTHDF2 with endogenous m6A RNAs was tested by Co-immunoprecipitation as previous reports (32,33) with minor changes. Cells transfected with indicated plasmids were UV-crosslinked before collected. Then the cell pellet was re-suspended with lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% NP-40, 100 U/ml RNase inhibitor and Protease inhibitor cocktail). YTHDF2 was immunoprecipitated with anti-HA antibody. The immunoprecipitation complex was washed twice with high-salt buffer (50 mM Tris–HCl pH 7.4, 300 mM NaCl), followed by two additional washes with low-salt buffer (50 mM Tris–HCl pH 7.4, 150mM NaCl). The amount of YTHDF2-bound m6A RNAs were detected by Western blot analysis with anti-m6A antibody.
MeRIP-Seq
MeRIP-Seq was performed by Cloudseq Biotech Inc. (Shanghai, China) according to the published procedure (6,29) with minor modifications. Briefly, m6A RNA immunoprecipitation was performed with the GenSeqTM m6A RNA IP Kit (GenSeq, China) by following the manufacturer's instructions. Both the input sample without immunoprecipitation and the m6A IP samples were used for library generation with NEBNext® Ultra II Directional RNA Library Prep Kit (New England Biolabs). The library quality was evaluated with BioAnalyzer 2100 system (Agilent Technologies). Library sequencing was performed on the Illumina Hiseq instrument with 150 bp paired-end reads.
RIP-Seq
RIP-Seq and subsequent bioinformatics analysis were all done by Cloud-Seq Biotech (Shanghai, China). Cells were lysed in an ice-cold lysis buffer. Then, RNA immunoprecipitation (RIP) was performed with the GenSeqTM RIP Kit (GenSeq, China). RNA extracted using Trizol by following manufacturer's instruction (Thermo Fisher Scientific). rRNAs were removed from the immunoprecipitated RNA and input RNA samples by using Ribo-Zero™ rRNA Removal Kit (Illumina, San Diego, CA, USA). RNA libraries were constructed by using rRNA-depleted RNAs with TruSeq Stranded Total RNA Library Prep Kit (Illumina) according to the manufacturer's instructions. Libraries were controlled for quality and quantified using the BioAnalyzer 2100 system (Agilent Technologies). Library sequencing was performed on the Illumina Hiseq instrument with 150 bp paired-end reads according to the manufacturer's instructions.
m6A dot-blotting assay
The m6A dot blot assay was conducted as previously described (29) with minor changes. Briefly, total RNAs were extracted by TRIZOL reagent (Invitrogen) and the mRNAs were isolated by using GenEluteTM mRNA Miniprep Kit (Sigma). The concentration and purity of mRNAs pulled down by YTHDF2 were measured by NanoDrop 2000. Then, the mRNAs were denatured at 95°C for 5 min and chilled on ice immediately after denaturation. RNA samples were dropped onto the Hybond-N+ membrane (GE Healthcare). After UV cross-linking, the membrane was washed in 1xPBS-T buffer, blocked with 5% non-fat milk in PBST and incubated with anti-m6A antibody overnight at 4°C. After incubating with secondary antibody, the membrane was visualized with enhanced chemiluminescence. Methylene blue staining was used to verify the equal amount of RNAs.
Sequencing data analysis
Paired-end reads were harvested from Illumina HiSeq 4000 sequencer. After 3′ adaptor-trimming and low quality reads removing by cutadapt software (v1.9.3) (34), the high quality trimmed reads were aligned to the human reference genome (GRCh37/hg19) by hisat2 software (v2.0.4) (35). For RIP-seq and RNA-seq, guided by the Ensembl gtf gene annotation file, cuffdiff software (36) (v2.2.1, part of cufflinks) was used to get the transcript level and gene level FPKM. In RIP-seq, fold change and P-value were calculated based on FPKM IP/Input, differentially binding mRNAs were identified. The RIP targets were defined as genes with FPKM ≥0.5, IP/Input ≥1.5. In RNA-seq, the fold change and P-value were calculated based on FPKM, differentially expressed mRNA were identified for an enrichment score of 1.5 and P value <0.05. For MeRIP-seq, Methylated sites on RNAs (peaks) were identified by MACS software (37). Differentially methylated sites were identified by diffReps (38). The cutoff threshold for FDR of 5.5 × 10−7 and fold enrichment score of 2 were used to obtain differentially methylated sites in MeRIP-seq.
Cell proliferation assay
Cell proliferation capability was measured by Cell Counting Kit-8 assay. Stable cell lines were seeded at a density of 1 × 103 cells/well on 96-well plates. The cellular activity was tested by adding 10 μL of CCK8 reagents into each well and reading the absorbance at a wavelength of 450 nm every day.
Wound healing assay
For the wound healing assay, stable H1299 cells were seeded in 12-well plates for 24 h to get confluent monolayer. Cells were scratched and continued to culture in serum-free DMEM for migration. Photos were taken as indicated time until the wound was healed. The healing percent was calculated by ImageJ
Soft agar colony formation assay
The procedure of soft agar colony formation assay was adapted from the previous reports (25). 2 ml of 0.6% base agar gel (Amresco) with 10% FBS was placed in six-well plates, and then followed by layering 1 × 103 cells in 2 ml of 0.35% agar gel with 10% FBS on the surface. The photographs of the cell colonies developed in soft agar were taken at an indicated day after 0.05% crystal violet staining, and the numbers of colonies were counted. Triplicate repeats were performed.
3D culture assays
3D culture assays was performed as described before (39). Briefly, 2 × 103 cells diluted in 7.5 μl of medium mixed with 7.5 μl of 3D culture matrix (Millipore) into the inner well of μ-slides (IBIDI). After concretion, the cell culture medium was added on the surface. Microscopy images were taken after 4 days.
Xenograft tumor model
The experiment of xenograft tumour model was conducted according to the previous research (25). Stable H1299 cell lines diluted into 100μl were injected subcutaneously into 5-week-old nude mice (n = 5) at the final concentration of 3 × 106 cells. Mice were sacrificed 4 weeks later, and tumors were weighed and photographed. All animal studies were conducted with the approval and guidance of Shanghai Jiao Tong University Medical Animal Ethics Committees.
Statistical analysis
Data are presented as mean±SD or±SEM for qPCR, cell proliferation, wound healing assay, mouse xenograft model and soft agar colony assay. Statistical comparisons were performed by t-tests (two tailed) or Wilcoxon and Mann–Whitney test as indicated in the article. All statistical analyses were performed with GraphPad Prism 5. A value of P < 0.05 was considered statistically significant and P-value < 0.05 was marked with (*), < 0.01 with (**) or < 0.001 with (***).
RESULTS
YTHDF2 is SUMOylated in vitro and in vivo
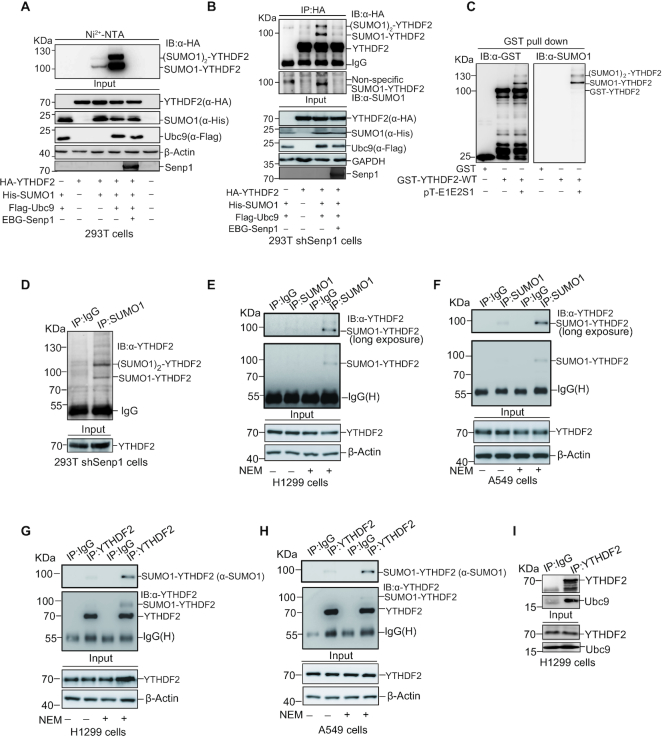
To identify whether YTHDF2 is SUMOylated in cells, HA-YTHDF2 and His-tagged SUMO1, SUMO2 or SUMO3 were transiently transfected into HEK-293T cells, respectively. His-SUMO conjugated YTHDF2 was pulled down by Ni2+-NTA resin and Western blotting showed that YTHDF2 was modified strongly by SUMO1 but weakly by SUMO2 or SUMO3 (Supplementary Figure S1A). YTHDF2 was modified by SUMO1 with a size of Mr ∼95–100 kDa (the expected normal size of YTHDF2 is 70 kDa), which is YTHDF2 covalently conjugated with one molecule of SUMO1 (SUMO1-YTHDF2). YTHDF2 modified with two molecules of SUMO1 [(SUMO1)2-YTHDF2] in a size of Mr ∼120 kDa was also weakly observed (Supplementary Figure S1A, lane 3). Thus, we focused on SUMO1 modification in the following studies.
To further validate YTHDF2 is modified by SUMO1, firstly we transfected HA-YTHDF2 alone or together with His-tagged SUMO1, a SUMO-conjugating enzyme E2 Flag-Ubc9 and a deSUMO enzyme Senp1 (Sentrin/SUMO-specific protease 1) into 293T cells. Similarly, His-SUMO1 conjugated YTHDF2 was pulled down by Ni2+-NTA resin precipitation and then immunoblotted with anti-HA antibody. SUMOylated YTHDF2 was significantly increased by Ubc9, but this was greatly weakened by co-transfection with Senp1 (Figure 1A, lanes 3, 4). Secondly, SUMOylation of YTHDF2 was confirmed by the method of immunoprecipitation (IP). HA-YTHDF2 with or without His-SUMO1 and Flag-Ubc9 were co-transfected into the 293T-shSENP1 cells which endogenous SENP1 was knocked down by a specific shRNA for SENP1 (Supplementary Figure S1B). Cell lysates were immuneprecipited with anti-HA antibody to pull down exogenous HA-YTHDF2, and then immunoblotted with anti-HA and anti-SUMO1 antibodies, showing two specific shifted bands of SUMOylated-YTHDF2 (Figure 1B). Thirdly, to examine whether YTHDF2 is SUMOylated in vitro, we performed an E. coli-based prokaryotic SUMOylation assay (23,26,31). Plasmids GST-YTHDF2 and pE1E2S1 (co-expressing two enzymes E1, E2 and SUMO1) were co-transformed into BL21(DE3) competent cells to reconstitute SUMOylation system. After GST pull-down of GST-YTHDF2, immunoblotting with anti-SUMO1 and anti-GST antibodies were performed to verify that GST-YTHDF2 was SUMOylated as two up-shifted bands with sizes of 110 and 130 kDa (Figure 1C). Finally, to examine the key point that YTHDF2 is also endogenously modified by SUMO1, we performed the SUMOylation assay by IP with anti-SUMO1 antibody in 293T-shSENP1 (Figure 1D), H1299 (Figure 1E), and A549 (Figure 1F) cells to pull down the SUMOylated-proteins, and subsequently immunoblotted with anti-YTHDF2 antibody to show that YTHDF2 was moderately modified by endogenous SUMO1 with one major band in a size of Mr ∼95–100 kDa. Moreover, this result was confirmed by reciprocal IP with anti-YTHDF2 antibody and immunoblotted with anti-SUMO1 antibody, showing that endogenous YTHDF2 was naturally modified by SUMO1 with one major band in H1299 (Figure 1G) and A549 (Figure 1H) cells when N-Ethylmaleimide (NEM) was added to the lysis buffer for stabilization of SUMO-conjugates. As Ubc9 is the only E2 for SUMO modification, YTHDF2 may interact with Ubc9. To validate this, lysates from HEK293T cells transfected with Myc-YTHDF2, Flag-Ubc9 or both were used for co-IP with anti-Myc antibody. The following immunoblotting results showed that YTHDF2 indeed interacted with Ubc9 (Supplementary Figure S1C). More convincingly, the interaction between endogenous YTHDF2 and Ubc9 in H1299 cells were detected by IP with anti-YTHDF2 antibody, and followed by western blotting with anti-Ubc9 antibody (Figure 1I). All above results proved that YTHDF2 was SUMOylated by SUMO1 both in vitro and in vivo.
Figure 1.
YTHDF2 is SUMOylated in vitro and in vivo. (A, B) YTHDF2 is modified by SUMO1 and this can be removed by Senp1. (A) HA-YTHDF2 and His-SUMO1/Flag-Ubc9 were transfected with or without EBG-Senp1 into HEK-293T cells as indicated. SUMO1 conjugated proteins were pull-down by Ni2+-NTA resin, and were immunoblotted with anti-HA antibody. Total cell lysates were immunoblotted with indicated antibodies. (B) Senp1 was stably knocked down by shRNA in HEK-293T cells and the cells were transfected with indicated plasmids. Cell lysates were immunoprecipitated (IP) with anti-HA antibody, and then immunoblotted with anti-SUMO1 and anti-HA antibodies, respectively. (C) SUMOylation of YTHDF2 is validated in the E. coli based in vitro system. The construct pGEX-4T-1-YTHDF2 was transformed into E. coli BL21 (DE3) with or without pE1E2SUMO1. After expression induction, bacteria were lysed and incubated with GST resin, and eluates were immunoblotted with anti-GST and anti-SUMO1 antibodies. (D) YTHDF2 is endogenously SUMOylated. Lysates from HEK-293T shSENP1 cells were used for IP with normal IgG or anti-SUMO1 antibody, and then immunoblotted with anti-YTHDF2 antibody. (E–H) Endogenous YTHDF2 is naturally modified by SUMO1. H1299 and A549 cells were lysed for IP with normal IgG or anti-SUMO1 antibody, and then immunoblotted with anti-YTHDF2 antibody (E, F), or IP with normal IgG or anti-YTHDF2 antibody, and then immunoblotted with anti-YTHDF2 or anti-SUMO1 antibody (G, H). The cell lysis buffer was added with or without N-Ethylmaleimide (NEM, 20 mM) for stabilization of SUMO conjugates. (I) Ubc9 interacts with YTHDF2. H1299 cells were lysed for IP with anti-YTHDF2 antibody, and then immunoblotted with anti-Ubc9 antibody.
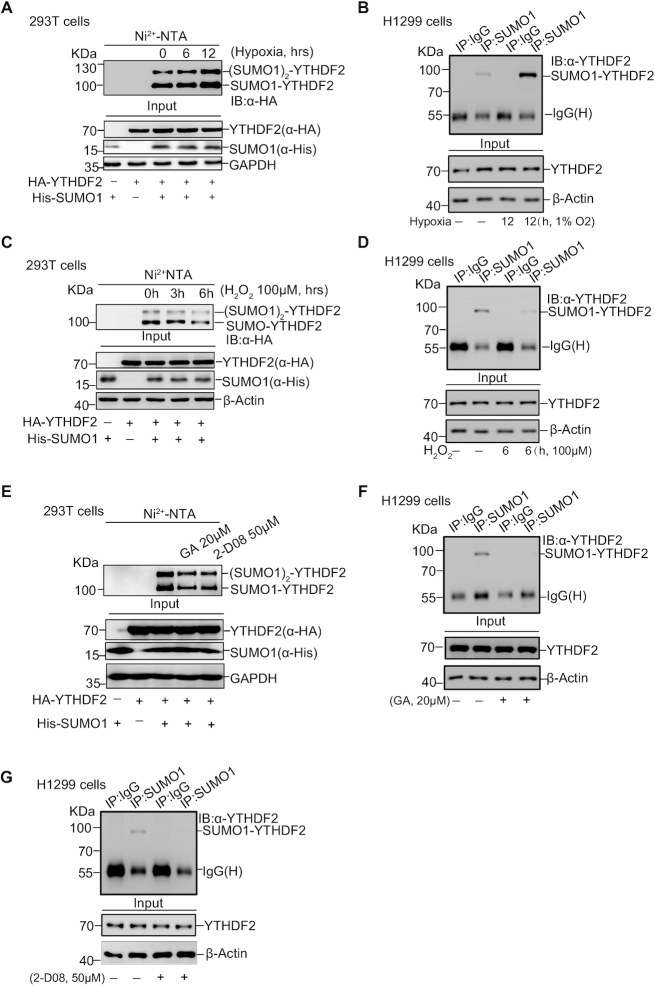
SUMOylation of YTHDF2 is regulated by hypoxia, oxidative stress and SUMO inhibitors
We wondered whether SUMOylation of YTHDF2 can be induced upon certain cellular stresses. 293T cells transfected with His-SUMO1 and HA-YTHDF2 were treated with hypoxia (1% O2) or hydrogen peroxide (100 μM H2O2) for indicated time before harvested for Ni2+-NTA pull down assay. We found that SUMOylation of YTHDF2 was increased under hypoxia (Figure 2A) whereas decreased by oxidative stress (Figure 2C) without obvious protein abundance turbulence, thus indicating that SUMOyaltion may control YTHDF2 functions in response to certain cellular stresses such as hypoxia. These results were also confirmed by the IP method for SUMOylation detection of endogenous YTHDF2 in H1299 cells under the same treatment of hypoxia (Figure 2B) or hydrogen peroxide (Figure 2D). As it has been reported that heat shock stress affects the function of YTHDF2 (40), we also checked SUMOylation of YTHDF2 under heat shock for the indicated time, showing little change after treatment (Supplementary Figure S1D). Due to the importance of SUMOylation in regulating diverse biologic functions, several novel small-molecule inhibitors targeting SUMOylation have been developed. We chose Ginkgolic acid (GA), a SUMO-activating enzyme E1 inhibitor (41,42) and 2-D08, a SUMO-conjugating enzyme E2 UBC9 inhibitor (43,44) for following studies. GA inhibits SAE1 (SUMOylation E1) by blocking the formation of SUMO E1-SUMO intermediate (41) while 2-D08 can prevent transferring of SUMO from the UBC9-SUMO thioester to substrates (44). After treatment for the indicated concentrations, the Ni2+-NTA pull down assay showed that SUMOylation of YTHDF2 was gradually decreased with increasing concentrations of GA and 2-D08 (Supplementary Figure S1E, F). Based on these, HA-YTHDF2 alone or together with His-tagged SUMO1 were transiently transfected into 293T cells and then separately treated with GA (20 μM) or 2-D08 (50 μM). The results showed both GA and 2-D08 could inhibit YTHDF2 SUMOylation (Figure 2E). Moreover, by using the method of IP with anti-SUMO1 antibody and immunoblotting with anti-YTHDF2 antibody, we found that SUMOylation of endogenous YTHDF2 in H1299 cells was effectively inhibited by both GA and 2-D08 (Figure 2F, G). In short, above results illustrate that SUMOylation of YTHDF2 is increased upon hypoxia whereas repressed by H2O2 and SUMOylation inhibitors.
Figure 2.
SUMOylation of YTHDF2 is regulated by hypoxia, oxidative stress and SUMO inhibitors. (A, B) Hypoxia upregulates SUMOylation of YTHDF2. HEK-293T cells were transfected with HA-YTHDF2 and His-SUMO1 and treated with hypoxia (1% O2) for the indicated time. SUMO1 conjugated proteins were enriched by Ni2+-NTA resin, and then immunoblotted with anti-HA antibody (A). SUMOylation of endogenous YTHDF2 under hypoxia treatment for 12 hours was detected by IP with anti-SUMO1 and immunoblotted with anti-YTHDF2 antibody in H1299 cells (B). (C, D) Hydrogen peroxide (H2O2) downregulates SUMOylation of YTHDF2. HEK-293T cells were transfected with HA-YTHDF2 and His-SUMO1, and treated with H2O2 (100 μM) as indicated time. The Ni2+-NTA resin pull down assay was performed to detect YTHDF2 SUMOylation (C). H1299 cells treated H2O2 (100 μM) for 6 h were lysed for detection of endogenous YTHDF2 SUMOylation by using the IP/WB method as Figure 2B (D). (E–G) SUMOylation inhibitors (Ginkgolic acid and 2-D08) decrease YTHDF2 SUMOylation. HEK-293T cells were transfected with HA-YTHDF2 and His-SUMO1, and treated with Ginkgolic acid (GA, 20 μM) or 2-D08 (50 μM) for 12 or 24 h, respectively. SUMOylation of YTHDF2 was detected by Ni2+-NTA resin pull-down assay (E). SUMOylation of endogenous YTHDF2 in H1299 cells treated with 20 μM GA for 12 h (F) or 50 μM 2-D08 for 24 h (G) was detected by IP with anti-SUMO1 and immunoblotting with anti-YTHDF2 antibody.
K571 is the major SUMOylation site of YTHDF2
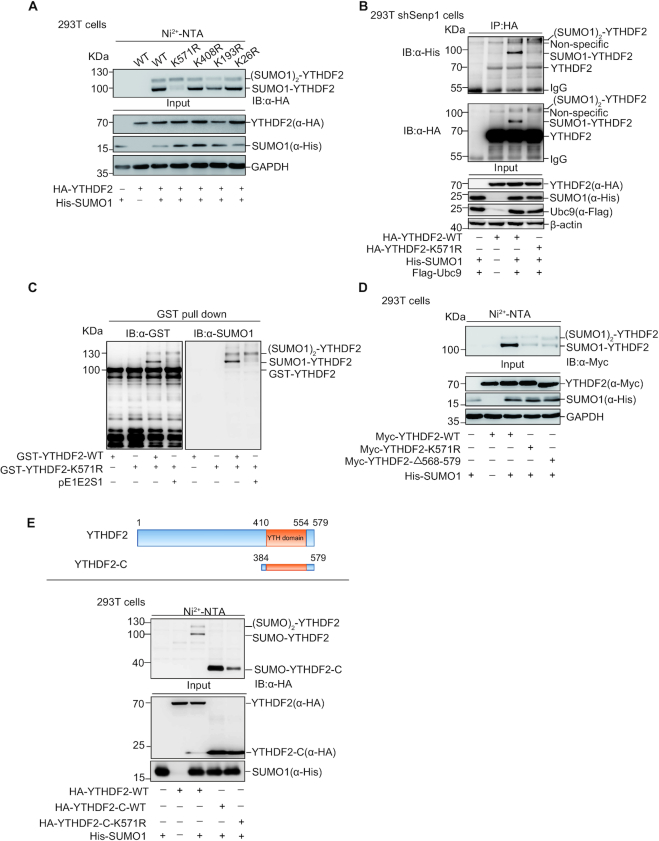
To identify the major SUMOylation sites of YTHDF2, we used the SUMOplot program (http://www.abcepta.com/sumoplot) to predict putative SUMOylation sites (Supplementary Figure S1G). Among those candidates seven lysines (Ks) with high-ranking including K571, K408, K193 and K26 were individually mutated to arginine (R) for preliminary screening of SUMOylation sites. The SUMOylation assays by Ni2+-NTA pull down showed that the mutation of K571R abolished the main band of SUMOylated-YTHDF2 with a size of ∼100 kDa (Figure 3A), indicating that SUMOylation of YTHDF2 mainly occurred at K571. To confirm this result, we performed IP and prokaryotic SUMOylation assays. 293T-shSENP1 cells transfected with HA-YTHDF2-WT or -K571R together with His-SUMO1 and Flag-Ubc9 were lysed for IP SUMOylation assay with anti-HA antibody and followed by immunoblotting analysis with anti-His and anti-HA antibodies, showing that SUMOylation of YTHDF2-K571R was greatly reduced compared to that of YTHDF2-WT (Figure 3B). Next, pE1E2S1 and pGST-YTHDF2-WT or -K571R were co-transformed into E.coli BL21 cells for prokaryotic SUMOylation assay, showing that the mainly specific SUMOylated-YTHDF2 band disappeared in GST-YTHDF2-K571R (Figure 3C). We also generated the construct of Myc-YTHDF2-△568–579, in which the fragment from aa 568 to the end 579 of YTHDF2 was deleted. The Ni2+-NTA pull down assay showed that the band of SUMOylated YTHDF2 with a size of molecular weight about 100 kDa almost disappeared when the amino acid fragment containing K571 were deleted (Figure 3D). The N-terminal of YTHDF2 is responsible for recruiting a deadenylase complex and the C-terminal is for recognizing m6A motif (21). Moreover, the truncated form of YTHDF2 C-terminal (aa 384–579) was used for the Ni2+-NTA pulldown SUMOylation assay. The result showed the truncated form YTHDF2-C-WT with a size of ∼20 KDa had only one SUMO band (∼40 kDa), which indicated that the other SUMOylation site might locate at the N-terminal (Figure 3E). Compared with YTHDF2-C-WT, YTHDF2-C-K571R greatly reduced the 40-kDa SUMOylated band, confirming K571 was a major SUMOylation site. However, above results suggested that there might be two SUMOylation sites which one is prominent while the other is subordinate, and K571 was the main SUMOylation site of YTHDF2. Based on the prediction of the SUMOplot program, we mutated more other possible sites including K452, K401 and K286 and performed Ni2+-NTA pulldown SUMOylation assay, but unfortunately we had not found the other SUMOylation site in this strategy (Supplementary Figure S1H). Taken together, we proved that K571 was the major SUMOylation site of YTHDF2.
Figure 3.
YTHDF2 is mainly SUMOylated at K571. (A) Point mutants of YTHDF2 with His-SUMO1 were transfected into HEK-293T as indicated. SUMO1 conjugated proteins enriched by Ni2+-NTA resin, and eluates were immunoblotted with anti-HA antibody. (B) HEK-293T shSenp1 cells were transfected as indicated constructs. Cell lysates were IP with anti-HA antibody, and then immunoblotted with anti-HA and anti-His antibodies. (C) Mutation of K571R impairs YTHDF2 SUMOylation in the E.coli SUMOylation system. The construct pGEX-4T-1-YTHDF2 or pGEX-4T-1-YTHDF2-K571R was transformed into E. coli BL21 (DE3) with or without pE1E2SUMO1. Bacteria lysates were incubated with GST resin, and then eluates were immunoblotted with anti-GST and anti-SUMO1 antibodies. (D) Deletion of C-terminus (aa 569–579) abolishes YTHDF2 SUMOylation, as like mutant YTHDF2-K571R. His-SUMO1 with Myc-YTHDF2, Myc-YTHDF2-K571R, or Myc-YTHDF2-Δ568–579 was transfected into HEK-293T cells. SUMO1 conjugated proteins were enriched by Ni2+-NTA resin, and eluates were immunoblotted with anti-HA antibody. (E) The truncated C-terminal form, mutant K571R and full length of YTHDF2 with His-SUMO1 were transfected into HEK-293T cells. The Ni2+-NTA resin pull down assay was performed to detect SUMOylation of YTHDF2.
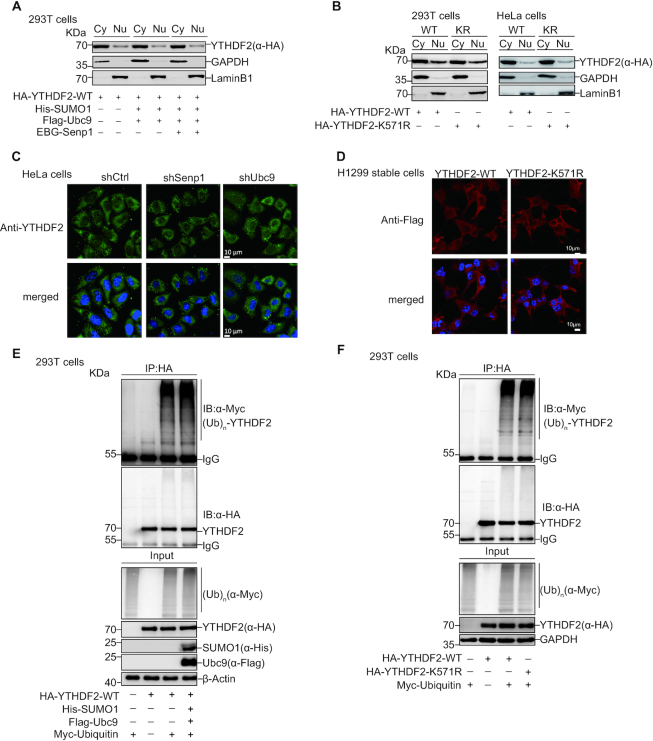
SUMOylation of YTHDF2 does not affect its localization and ubiquitination
SUMOylation takes part in many cellular processes and signaling pathways by altering the activity, stability, localization and protein–protein interaction of targeted proteins (23–27). For more comprehensive understanding of YTHDF2 SUMOylation, we sought to assess the potential effect of YTHDF2 SUMOylation on its subcellular localization. 293T cells transfected HA-YTHDF2 with or without SUMO1/Ubc9 and SENP1 were extracted for the nuclear/cytosolic fractionation assays. YTHDF2 mainly resided in the cytosol as previous reported (40) and did not relocate under different levels of SUMOylation (Figure 4A). We also collected 293T and HeLa cells transfected with HA-YTHDF2-WT or HA-YTHDF2-K571R for separating cytoplasmic and nuclear protein fractions. Similarly, there were no different sub-cellular locations between WT and K571R (Figure 4B). Moreover, we checked the subcellular localization of YTHDF2 with immunofluorescence assay in HeLa cells by knocking down SENP1 or Ubc9. The majority of YTHDF2 was present in cytosol and did not relocate into nucleus (Figure 4C). The same result was validated in H1299 stable cells which Flag-tagged YTHDF2-WT or YTHDF2-K571R was re-expressed by the lentiviral expressing system in shYTHDF2 cells, in which endogenous YTHDF2 was silenced by a short hairpin RNA targeting YTHDF2 3′UTR (Figure 4D).
Figure 4.
SUMOylation of YTHDF2 does not affect its localization and ubiquitination. (A–D) SUMOylation of YTHDF2 does not affect its localization. (A, B) HEK-293T or HeLa cells were transfected with indicated constructs. Nuclear and cytosolic fractions were extracted by the nuclear/cytosol fractionation kit and immunoblotted with anti-HA, anti-GADPH and anti-LaminB1 antibodies (A-B). Stable cells HeLa-shControl, HeLa-shUbc9 or HeLa-shSenp1 were immunostained with anti-YTHDF2 antibodies and DAPI. Immunofluorescence pictures were taken. Scale bar, 10 μm (C). Stable cells H1299-shYTHDF2-WT, or -K571R were immunostained with anti-Flag antibodies or DAPI. Immunofluorescence pictures were taken. Scale bar, 10 μm (D). (E, F) SUMOylation of YTHDF2 does not affect its ubiquitination. HEK-293T cells were transfected with indicated plasmids. Cell lysates were used for IP with anti-HA antibody, and then immunoblotted with anti-Myc and anti-HA antibodies. Total cell lysates were immunoblotted with antibodies as indicated.
Since YTHDF2 SUMOylation was induced by hypoxia while repressed by H2O2, we detected the localizations of YTHDF2 after these two treatments by nuclear/cytosolic fractionation assays. 293T cells transfected with HA-YTHDF2 with or without SUMO1/Ubc9 under treatment of hypoxia or H2O2, were separated into cytosolic and nuclear fraction. No significant relocation change of YTHDF2 was observed (Supplementary Figure S2A-B). The same result was confirmed by utilization of GA or 2-D08 inhibiting SUMOylation (Supplementary Figure S2C, D). We also compared the distribution of HA-YTHDF2-WT and HA-YTHDF2-K571R in the same way as above. Consistently, there was no significant change of nuclear/cytosolic locations of YTHDF2 between WT and K571R (Supplementary Figure S2E, H). Furthermore, we displayed the distribution of YTHDF2 in stable cell lines HeLa-shCtrl, HeLa-shSENP1 and HeLa-shUbc9 (Supplementary Figure S3A), and H1299 expressing Flag-YTHDF2-WT and -K571R (Supplementary Figure S3B) with different treatments previously mentioned, by immunofluorescence staining. Similarly, we also did not find any different change in relocation of YTHDF2. All these results supported the conclusion that YTHDF2 SUMOylation is weakly connected with its localization.
To check whether SUMOylation of YTHDF2 influences its ubiquitination, we transfected HA-YTHDF2-WT and Myc-ubiquitin with or without SUMO1/Ubc9 into 293T cells and found that ubiquitination of YTHDF2 was not affected by its SUMOylation status (Figure 4E). We also validated that ubiquitination was comparable between YTHDF2-WT and YTHDF2-K571R (Figure 4F). To figure out whether YTHDF2 ubiquitination might be influenced by hypoxia or H2O2, we transfected HA-YTHDF2-WT and Myc-Ubiquitin with or without SUMO1/Ubc9 into 293T cells, and then treated with hypoxia and H2O2. There was no significant influence on YTHDF2 ubiquitination (Supplementary Figure S4A, B). Next, we compared the levels of YTHDF2 ubiquitination with treatment of SUMOylation inhibitors GA and 2-D08. Similarly, no remarkable difference was displayed between the treated group or non-treated group (Supplementary Figure S4C, D). Also, the same results were confirmed by comparing ubiquitination between YTHDF2-WT and YTHDF2-K571R under these treatments (Supplementary Figure S5A–D). In brief, these results suggested that SUMOylation of YTHDF2 does not affect its ubiquitination.
SUMOylation increases the binding affinity of YTHDF2 with m6A-modified RNAs
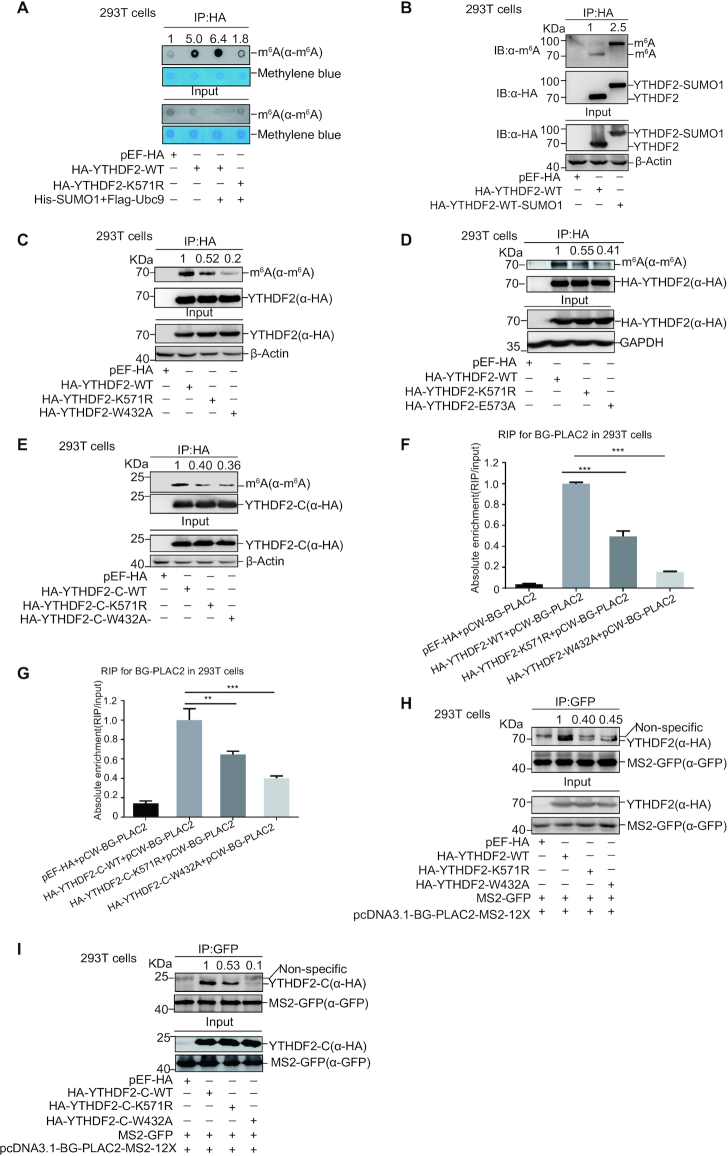
Since SUMOylation regulates SUMO-targeted enzymes activity (29), we wondered whether SUMOylation of YTHDF2 influences its function on metabolism of m6A RNAs. To explore this hypothesis, 293T cells transfected HA-YTHDF2-WT or -K571R together with or without SUMO1/Ubc9 were lysed for IP with anti-HA antibody, and then RNAs pulled down by HA-YTHDF2-WT or -K571R were extracted for dot-blotting assay with anti-m6A antibody, which could detect the abundance of m6A-modified RNAs bound by YTHDF2. As expectedly, m6A-modified RNAs were greatly pulled down by YTHDF2-WT when compared to that by the control vector pEF-HA; and the pull-down m6A RNAs were increased by co-transfection of SUMO1/Ubc9 with YTHDF2-WT but not with YTHDF2-K571R (Figure 5A). This result suggested that SUMOylation of YTHDF2 increased its binding affinity with m6A-modified RNAs. Next, we generated a HA-YTHDF2-SUMO1 fusion protein expression plasmid, in which the C-end of YTHDF2 was linked with SUMO1 (aa 2–96) to mimic SUMOylated-YTHDF2 protein. The RIP assay was performed according to the protocols published before (32,33). UV-crosslinked cells were lysed for immunoprecipitation with anti-HA antibody, and followed by immunoblotting with the m6A antibody, showing that the m6A-modified RNAs bound by HA-YTHDF2-SUMO1 was much more than that by HA-YTHDF2 (Figure 5B). To check whether the reduced binding of mutant K571R to m6A-modified RNAs can be rescued by fusion of SUMO1, we also generated a HA-YTHDF2-K571R-SUMO1 fusion protein expression plasmid to mimic SUMOylated-YTHDF2 protein. Indeed, the abundance of m6A-modified RNAs pulled down by HA-YTHDF2-K571R-SUMO1 was much more compared to that by HA-YTHDF2-K571R (Supplementary Figure S6A). Since two residues W432 and W486 of YTHDF2 protein are important for specific recognition of m6A motif, and mutations of W432A and W486A markedly decreased the binding affinity of YTHDF2 to m6A-modified RNAs (18), so one mutant W432A was chose as a control in this study. 293T cells transfected with HA-YTHDF2-WT, -K571R or -W432A were UV-crosslinked, and then lysed for IP with HA antibody. Subsequently, the result of immunoblotting with anti-m6A antibody showed that m6A-modified RNAs bound by YTHDF2-K571R or -W432A were less than those by YTHDF2-WT (Figure 5C). Moreover, the binding affinity of m6A modified-RNAs to the double mutant K571R/W432A was less than that of WT, and slight weaker than that of the single mutant K571R or W432A (Supplementary Figure S6B).
Figure 5.
SUMOylation enhances YTHDF2 binding with m6A-modified RNAs. (A) Enrichment of m6A-RNAs bound by YTHDF2 in dot-blot assay. HA-YTHDF2-WT or -K571R with/without His-SUMO1/Flag-Ubc9 was transfected into HEK-293T cells. RNP Complexes were pull down by IP with anti-HA antibody, and then immunoblotted with anti-m6A antibody or stained by methylene blue (upper panels). The abundance of m6A RNAs from cells was detected by dot-blotting with anti-m6A antibody, and equal loading of RNAs was verified by methylene blue staining (lower panels). (B–E) The abundance of m6A bound by YTHDF2 was detected by IP method. HEK-293T cells were transfected with HA-YTHDF2-WT or HA-YTHDF2-WT-SUMO1 fusion protein (B), HA-YTHDF2-WT, -K571R or -W432A (C), HA-YTHDF2-WT, -K571R or -E573A (D), and truncated form HA-YTHDF2-C-WT, -K571R or -W432A (E). Cells were UV-crosslinked before harvested. Lysates were used for IP by anti-HA antibody, and then immunoblotted with antibodies against HA or m6A. (F-G) The reporter pCW-BG-PLAC2 with full-length HA-YTHDF2-WT, -K571R or -W432A (F), and truncated form HA-YTHDF2-C-WT, -K571R or -W432A (G) were transfected into 293T cells. Cell lysates were used for IP with anti-HA antibody, and then the RNP complexes were extracted with Trizol for qRT-PCR of PLAC2. The relative recruitment was calculated by normalizing to the Input. (H-I) MS2-GFP and pcDNA3.1-BG-PLAC2-12X-MS2 with full-length HA-YTHDF2-WT, -K571R or -W432A (H), and truncated form HA-YTHDF2-C-WT, -K571R or -W432A (I) were transfected into HEK-293T cells. After UV-crosslinked, cells were lysed for IP with anti-GFP antibody, and followed by immunoblotting with anti-HA antibody. The relative fold of YTHDF2 bound by MS2-GFP indirectly was analyzed by ImageJ.
To exclude possible impact of other modification rather than dependency on SUMO conjugation, E573 in the SUMO consensus motif was mutated to alanine to get HA-YTHDF2-E573A. We transfected HA-YTHDF2-WT, -K571R or -E573A into 293T cells and performed a RIP assay. The m6A-RNAs bound by YTHDF2-E573A were truly decreased compared those by YTHDF2-WT (Figure 5D). Furthermore, we employed the truncated form YTHDF2-C(384–579), which can recognize m6A-RNAs but not induce degradation of m6A transcripts (16,21), to repeat the same above experiment as the full-length YTHDF2, and confirmed that YTHDF2-C-K571R or -W432A indeed decreased its binding affinity with m6A-modified RNAs (Figure 5E). In addition, whether the other SUMOylation sites located at the N-terminal of YTHDF2 participate in this process need further investigation.
A long non-coding RNA PLAC2 as YTHDF2-targeted RNA with abundant m6A motif reported previously (16) was selected for further validation. The pCW-BG-PLAC2 was constructed by inserting BG-PLAC2 (21) containing a rabbit BG coding region and a fragment with two m6A motifs from LncRNA PLAC2 into the vector pCW57 (Supplementary Figure S6C). The designed RIP assay (Supplementary Figure S6D) was performed by transfection of pCW-BG-PLAC2 together with YTHDF2-WT, -K571R or -W432A into 293T cells. The follow-up qRT-PCR analysis showed that the enrichment of PLAC2 RNA by YTHDF2-K571R was less than that by YTHDF2-WT, although YTHDF2-W432A as a positive control enriched much less (Figure 5F). Moreover, we observed the similar patterns of PLAC2 RNA recruitment by the truncated forms of YTHDF2-C-WT, -K571R or -W432A in the similar experiments (Figure 5G). Next, the fragment of BG-PLAC2 was cloned into the vector pcDNA3.1-12 × MS2 to generate the construct pcDNA3.1-BG-PLAC2-12 × MS2. Since MS2-GFP fusion protein recognizes the 12 × MS2 binding sites and YTHDF2 can bind to m6A motifs in BG-PLAC2 (Supplementary Figure S6E), thus we used this system to support our above hypothesis. 293T cells transfected with MS2-GFP, pcDNA3.1-BG-PLAC2-12 × -MS2 and HA-YTHDF2-WT, -K571R or -W432A, respectively, were lysed for RIP assay with anti-GFP antibody. The follow-up Western-blotting analysis with anti-HA antibody showed that YTHDF2-K571R pulled down by BG-PLAC2 was much less than YTHDF2-WT (Figure 5H), suggesting that the mutation of K571R weakened its binding affinity with m6A motifs located at the LncRNA PLAC2 transcript. The same results were repeated with the truncated forms of YTHDF2-C (Figure 5I).
Since YTHDF2 SUMOylation was regulated under certain conditions, we checked the binding affinity of m6A-modified RNAs with YTHDF2 under different treatments, as shown in Supplementary Figure S7. H1299 cells treated with hypoxia, H2O2, GA, or 2-D08 were lysed for RIP assay with anti-YTHDF2 antibody or normal IgG, and then the abundance of m6A-RNA bound by YTHDF2 was detected by the dot-blot method. The results revealed the binding affinity of YTHDF2 toward m6A-RNA was increased under hypoxia treatment (Supplementary Figure S7A), whereas the amount of m6A-RNA bound by YTHDF2 was decreased when treated with H2O2, GA, or 2-D08 (Supplementary Figure S7B-D). We also repeated these experiments in stable H1299 cells, which YTHDF2 was re-expressed in a H1299-shYTHDF2 cell line (Supplementary Figure S8A), and gotten the same conclusion (Supplementary Figure S7E–H). Taken together, all above results demonstrated that SUMOylation of YTHDF2 at K571 increases the binding affinity with m6A-modified RNAs.
SUMOylation of YTHDF2 promotes cancer progression
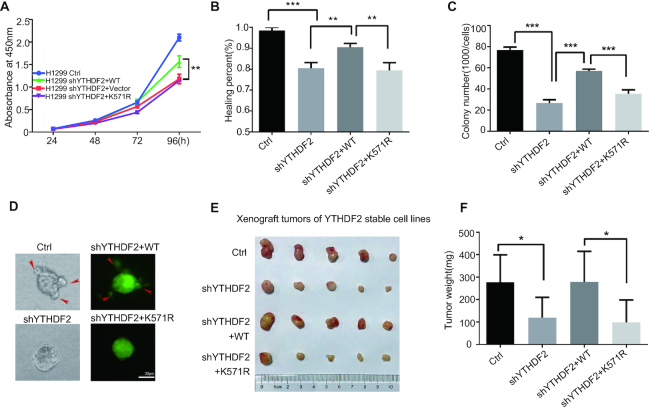
Since YTHDF2 is one of the main m6A readers and SUMOylation could enhance its binding affinity with m6A-modified RNAs, we wandered whether SUMOylation of YTHDF2 is involved in cancer progression. To this end, we generated stable H1299 cell lines, in which endogenous YTHDF2 was silenced by a short hairpin RNA targeting YTHDF2 3′UTR (shYTHDF2), and then YTHDF2-WT and YTHDF2-K571R were re-introduced by the Lentiviral expressing system. The expression levels of endogenous YTHDF2 and re-expressed YTHDF2-WT or YTHDF2-K571R were assessed by Western-blot analysis (Supplementary Figure S8A). Knockdown of YTHDF2 significantly inhibited cell proliferation (Figure 6A), cell migration (Figure 6B and S8B), and soft-agar colony formation ability (Figure 6C and Supplementary Figure S8C), which were rescued by re-expression of Flag-tagged YTHDF2-WT but not YTHDF2-K571R. To test the invasive ability of stable cells, we used the method of 3D growth cell cultures on extracellular matrix to mimic the in vivo conditions (39). Stable H1299 cells re-expressing YTHDF2-WT, as like control cells, their colonies showed elongated and scattered morphology, whereas both sh-YTHDF2 cells and re-expressing shYTHDF2-K571R cells grew more tightly and seemed like smooth spheroid (Figure 6D). This indicated that YTHDF2 SUMOylation is required for the invasive ability of tumor cells. Furthermore, we investigated whether YTHDF2 SUMOylation affects xenograft tumour growth in vivo. Four stable H1299 cell lines (control, sh-YTHDF2, shYTHDF2-WT and shYTHDF2-K571R) were injected into the flanks of nude mice subcutaneously. Being consistent with above results, knockdown of YTHDF2 suppressed tumor growth in mice as reflected by the decrease of tumor size and tumor weight compared with the control group, which was recovered by re-expression of YTHDF2-WT but not by YTHDF2-K571R (Figure 6E, F). We further detected SUMOylated-YTHDF2 in the xenograft tumors from nude mice by IP method, showing that the SUMOylation level of YTHDF2 were decreased in K571R group compared to that in WT group (Supplementary Figure S8D). All these results suggested that SUMOylation of YTHDF2 promoted cancer progression.
Figure 6.
SUMOylation of YTHDF2 promotes cancer progression. (A) Mutation K571R of YTHDF2 inhibits cell proliferation of H1299 cells as determined by CCK8 assays. Stable H1299 cell lines were seeded and then cell proliferation was measured by CCK8 values as indicated time. (B) Mutation K571R of YTHDF2 inhibits cell migration of H1299 cells as verified by wound healing assay. Images were captured at indicated time. The healing percentage was calculated by image J and was presented by histogram. Error bars ±SD represent of three independent experiments. (C) SUMOylation of YTHDF2 promotes soft-agar colony formation. 1000 of each stable H1299 cell line were seeded per well in 2 ml of medium containing 10% FBS with 0.35% soft agarose and layered over a 0.6% solidified agarose. Photographs were taken 21 days after seeding. The number of colonies was calculated. Each value represents as mean±SEM of three independent experiments with triplicates. (D) Mutation K571R of YTHDF2 downregulates the invasive ability of stable H1299 cells by 3D culture growth assay. Photos were taken at 5 days post seeding. The images (left panels) were taken under the white light, and the green signals indicated the expression of GFP in the plasmid pCD513B-Flag-YTHDF2. (E, F) SUMOylation of YTHDF2 promotes xenograft tumor growth in vivo. 3 × 106 of each stable H1299 cell line was injected subcutaneously into male BALB/c nude mice individually. Mice were sacrificed 35 days later, and tumors were dissected (E) and weighted (F, n = 5). Two-tailed Student's t-tests were used for statistical analysis, and differences with P values of <0.05 (*), < 0.01 (**) or < 0.001 (***) were considered significant.
SUMOylation of YTHDF2 influences transcriptome-wide mRNA levels and gene expressions
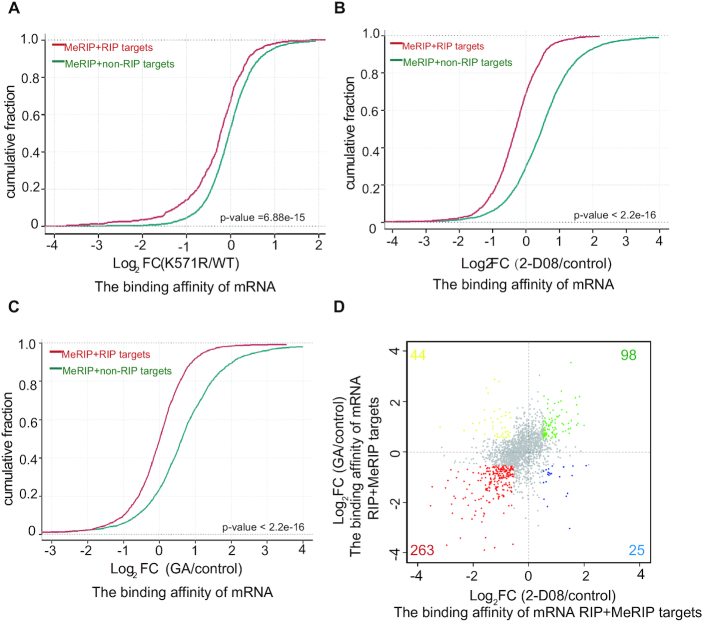
To explore the underlying mechanism of YTHDF2 SUMOylation in regulation of cancer progression, we performed RNA immunoprecipitation throughput sequencing (RIP-seq) and antibody-based methylated RNA immunoprecipitation throughput sequencing (MeRIP-seq) in the stable cell lines, H1299-shYTHDF2 re-expressing YTHDF2-WT or YTHDF2-K571R, to map target transcripts of YTHDF2 and m6A marked transcripts, respectively. A significant enrichment of m6A modifications in Flag-YTHDF2-bound RNAs (RIP) was observed (Supplementary Figure S9A, Extended Data Supplementary Tables S1, S9, S10, S11), as similar to that reported in previous report (16). RIP targets were defined as genes with FPKM ≥0.5 and IP/input ≥1.5-fold. Combined analysis of RIP-seq and MeRIP-seq in YTHDF2-WT stable cells, we got the m6A modified transcripts in RIP-seq. Then we categorized these transcripts into two groups: m6A-modified and non-RIP targets (MeRIP+non-RIP targets), m6A-modified and RIP targets (MeRIP+RIP targets). As expected, MeRIP + RIP targets showed a globally and significantly lower binding affinities in the mutant YTHDF2-K571R (Figure 7A), which was consistent well with our above biochemical results (Figure 5). Furthermore, to exclude the possibility structural change of K571R leading to different binding affinity, we treated H1299-YTHDF2-WT stable cells with SUMOylation inhibitors (GA and 2-D08) and then performed RIP-seq. With the m6A-sequencing data obtained from MeRIP-seq, the RIP targets were also divided into two groups: m6A-modified and non-RIP targets (MeRIP+non-RIP targets), m6A-modified and RIP targets (MeRIP + RIP targets). The results displayed that the binding affinities to m6A-motified mRNAs of YTHDF2 were significantly decreased by the treatment with SUMO inhibitors 2-D08 and GA (Figure 7B, C). There were 992 genes changed in the RIP targets of both GA and 2-D08 treated groups, among which 575 (57.9%) genes were down-regulated (Supplementary Figure S9B, Extended Data Supplementary Tables S5, S6). Consistently, there were 61% (263/430) genes down-regulated in the MeRIP+RIP targets in both 2-D08 and GA treatment (Figure 7D), which illustrated the decreased binding capability of YTHDF2-WT to m6A-mRNAs was dependent on SUMOylation inhibition. As YTHDF2 binds and subsequently mediates degradation of m6A-modified mRNAs, so we wondered whether SUMOylation of YTHDF2 affects the mRNA levels by altering the binding affinity. MeRIP-seq analysis showed that the abundance of m6A modification in the SUMO-site mutant YTHDF2-K571R group was increased around the stop codons (Supplementary Figure S9C, Extended Data Supplementary Tables S1 and S9) where the mRNA m6A modification is highly enriched as reported (6,7) and the presence of YTHDF2 destabilizes the m6A-modified mRNAs. This also implied that SUMOylation of YTHDF2 especially decreased the total m6A abundance at the stop codons, which might lead to degradation of mRNAs.
Figure 7.
SUMOylation of YTHDF2 increases its binding with m6A-mRNAs. (A) Cumulative frequency of the binding affinity with log2-fold change in MeRIP + non-RIP targets (green) and MeRIP+RIP-targets (red) between YTHDF2-WT and YTHDF2-K571R groups (Extended Data Supplementary Tables S1, S2). P values were calculated with a Kolmogorov-Smirnov test. (B, C) Cumulative distribution of mRNA log2-fold changes between 2-D08 (treated with 2-D08) and control (Extended Data Supplementary Tables S1 and S3) (B), GA (treated with GA) and control (Extended Data Supplementary Tables S1 and S4) (C) for MeRIP+non-RIP targets (green) and MeRIP+RIP-targets (red). P values were calculated with a Kolmogorov–Smirnov test. (D) Distribution of genes with a significant change in both 2-D08 treated group and GA treated group compared to control for RIP+MeRIP targets in stable H1299 shYTHD2-WT cells (Extended Data Tables S1, S5 and S6).
To validate the hypothesis, an 84-nt fragment from SON mRNA containing three GAC motifs, which are easily methylated and recognized by YTHDF2 to mediate degradation by the CCR4-NOT deadenylase complex (16,21), was inserted into the vector pEGFP-C3 to generate the construct pEGFP-C3-GFP-SON (Supplementary Figure S10A). As expectedly, YTHDF2 can recognize m6As in the SON fragment and consequentially lead to degradation of GFP-SON mRNA and further low protein expression, thus we used the GFP-SON system to assess the different degrading ability of YTHDF2-WT and -K571R. The protein levels of GFP-SON were less in the transfected with HA-YTHDF2-WT than that of the HA-YTHDF2-K571R or HA-YTHDF2-W432A in either 293T-shYTHDF2 (Supplementary Figure S10B, upper panels) or 293T-shSENP1 (Supplementary Figure S10C, upper panels) cells. Moreover, overexpression of SUMO1/Ubc9 together with YTHDF2-WT but not YTHDF2-K571R significantly decreased the protein level of GFP-SON (Supplementary Figure S10D, upper panels). However, as negative controls, YTHDF2-WT, mutants YTHDF2-K571R and -W432A had no any effects on the GFP expression when the SON fragment was absent (Figures S10B-D, lower panels). All these data suggested that the SUMOylation of YTHDF2 promoted the degradation of m6A-modified mRNAs by increasing its binding affinity.
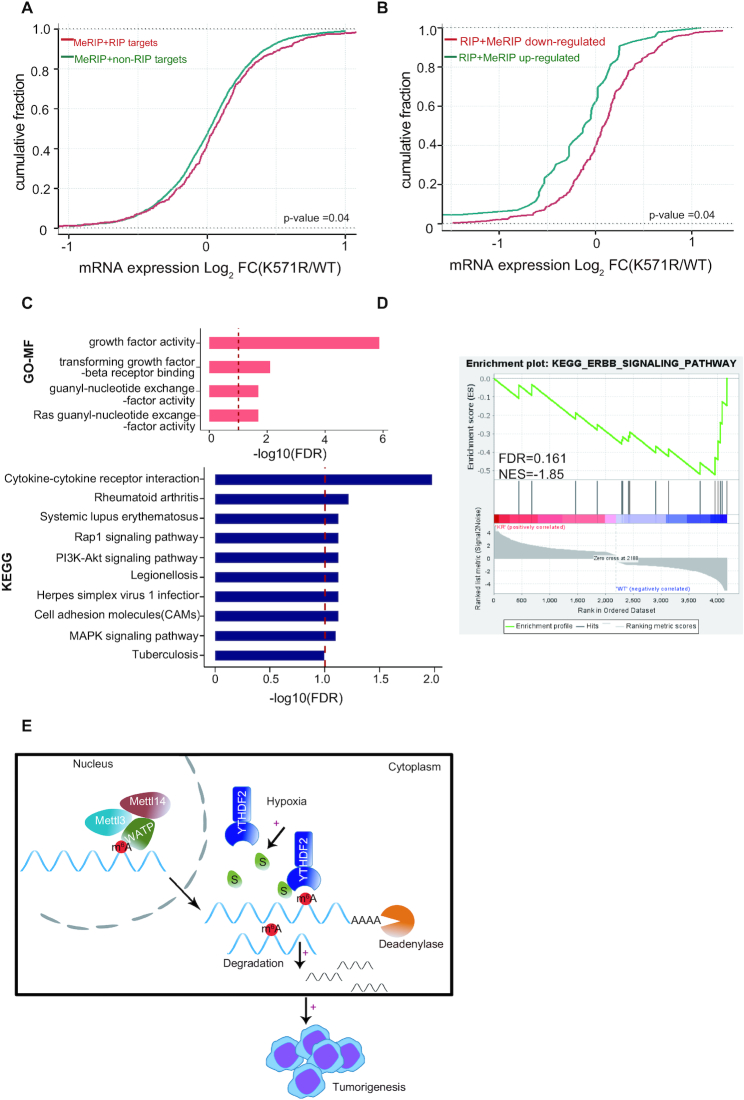
Next, we applied mRNA sequencing to assess transcriptome-wide mRNA levels. The increase of mRNA reads for MeRIP+RIP targets was observed in YTHDF2-K571R compared to those in YTHDF2-WT (Figure 8A). Furthermore, to verify the relationship between the binding ability and mRNA levels, we divided MeRIP+RIP targets into the low affinity group (MeRIP+RIP down-regulated, Fold change ≤ −1.5) and the high affinity group (MeRIP+RIP up-regulated, Fold change ≥ +1.5) according to the binding capability in YTHDF2-K571R versus YTHDF2-WT. As shown in Figure 8B, the mRNA levels in the MeRIP + RIP down-regulated group were much higher than those in the MeRIP + RIP up-regulated group, which revealed that SUMOylation of YTHDF2 preferentially increased the binding affinity and sequentially mediated degradation of certain mRNAs.
Figure 8.
SUMOylation of YTHDF2 influences transcriptome-wide mRNA levels and gene expressions. (A, B) SUMOylation of YTHDF2 alters gene expression profile in H1299 stable cell lines. Cumulative frequency of mRNA with log2-fold change in MeRIP+non-RIP targets (green) and MeRIP+RIP targets (red) between YTHDF2-WT and YTHDF2-K571R groups (Extended Data Supplementary Tables S1, S2 and S7) (A). Cumulative frequency of mRNA with log2-fold change in MeRIP+RIP down-regulated targets (red) and MeRIP + RIP up-regulated targets (green) groups between YTHDF2-WT and YTHDF2-K571R (Extended Data Supplementary Tables S1, S2 and S7) (B). P values were calculated using a Kolmogorov–Smirnov test. (C) Gene Ontology (GO) enrichment analysis of molecular function and KEGG analysis for alterative genes between YTHDF2-WT and YTHDF2-K571R stable cell lines (Extended Data Table S8) which were conducted by clusterProfiler package (68,69) in R platform. The vertical red bar indicated FDR 10%. (D) GSEA of differently expressed mRNAs in RNA-seq compared between YTHDF2-WT and -K571R in stable H1299 cell lines. NES, normalized enrichment score; FDR, false discovery rate. Note that FDR <0.25 was considered significant (Extended Data Table S8). (E) A model for SUMOylation of YTHDF2 promoting cancer progression. SUMOylation of YTHDF2 induced by tumor microenvironmental hypoxia enhanced its binding affinity towards m6A-RNAs, and subsequently altered gene expression profiles thus to promote cancer progression.
To identify the functional pathways which are associated with YTHDF2 SUMOylation, we adopted the Gene ontology, KEGG and Gene Set Enrichment Analysis (GSEA) for the deregulated mRNAs in between H1299-YTHDF2-WT and H1299-YTHDF2-K571R cells. Gene Ontology analysis revealed that the deregulated mRNAs were enriched in growth factor activity, transforming growth factor beta receptor binding, guanyl-nucleotide exchange factor activity and Ras guanyl-nucleotide exchange factor activity (Figure 8C, upper panel). GSEA also showed that genes highly expressed in the WT group were enriched in ERBB-signaling pathway (Figure 8D) which had close relationship with lung cancer and played roles in tumor progression (45). In addition, the enrichment of cytokine-cytokine receptor interaction, cell adhesion molecules (CAMs) were observed in the altered mRNAs (Figure 8C, lower panel) which are associated with cancer growth and metastasis (46–48). On the basis of above functional enrichment analyses, we found that the deregulated gene expressions were enriched for pathways associated with cancer progression, which could explain that YTHDF2 is involved in several cancers (49–51) and SUMOylation of YTHDF2 promoted cancer progression (Figure 6 and Supplementary Figure S8)
Lastly, to detect whether YTHDF2 or SUMOylated-YTHDF2 is correlated with the survival rate of lung adenocarcinoma, we subdivided YTHDF2 into two groups according to the upper and lower quartiles. Clearly, there was no significant difference of disease survival rate (P = 0.379; Figure S11A) between these two groups in TCGA database. The high or low expression levels of SUMO1 were unrelated to patient survival (P = 0.091; Figure S11B). But to our surprise, when combining analysis of YTHDF2 with SUMO1, we found that patients with higher YTHDF2 expression had higher disease-free survival rates in ones with high SUMO1 expression than with low SUMO1 expression (P = 0.022; Figure S11C); whereas the disease-free survival of patients with lower YTHDF2 expression was not influenced by the SUMO1 expression levels (P = 0.461; Figure S11D). Taken together, our findings suggested that SUMOylation of YTHDF2 participated in cancer progression by affecting the mRNA levels and gene expressions in several key pathways.
In conclusion, we depicted a novel mechanism for YTHDF2 in regulation of cancer progression (Figure 8E). SUMOylation of YTHDF2 increased its binding affinity towards m6A-modified RNAs and subsequently affected gene expressions, thereby promoted cancer progression. Especially, microenvironmental hypoxia in tumors could induce YTHDF2 SUMOylation and by means of this mechanism promoted tumorigenesis and malignant transformation. Our study suggests that modulation of YTHDF2 SUMOylation has potential clinical implications.
DISCUSSION
As one of the most potent modifications in RNA, m6A occurs at different RNA types mainly including mRNAs and LncRNAs and thereby takes part in many biological processes, especially in cancer progression (20,52–55). YTHDF2, one of the major readers, recognizes m6A modified RNAs and thereby induces their degradatin (16). Until now, studies have mainly focused on the function of YTHDF2 (49,56,57), however the regulatory mechanism of YTHDF2 itself had not yet been reported. YTHDF2 consists of a C-terminal YTH domain responsible for binding m6A and a P/Q/N-rich N-terminal region for recruiting CNOT1 to degrade m6A-modified RNA (21). We found YTHFD2 was modified by SUMO1 at two sites: the K571 was dominate (Figure 3) and the C-terminal truncation (aa 384–579) of YTHDF2 can be SUMOylated only in one site which suggested another site might locate on the N-terminal (aa 1–383). Most importantly, microenvironmental hypoxia enhanced YTHDF2 SUMOylation (Figure 2A), indicating this modification is connected to tumorigenesis.
SUMOylation affects many aspects of target proteins such as activity, stability, localization and protein-protein interactions (23–27). We did not observe that SUMOylation of YTHDF2 changed its cellular localization and affected ubiquitination in physiological conditions (Figure 4), even under treatment with hypoxia, hydrogen peroxide or SUMO inhibitors (Supplementary Figures S2–S5). Nevertheless, as the main SUMO-site K571 of YTHDF2 located near the YTH domain which specifically binds to m6A-modified RNAs, it might influence the binding capability of YTHDF2 toward m6A-RNAs. Indeed, we found that SUMOylation of YTHDF2 at K571 increased its binding activity with m6A-RNAs by using different experimental approaches (Figure 5). The analysis of RIP-seq and MeRIP-seq in H1299-shYTHDF2 cells re-expressing YTHDF2-WT and YTHDF2-K571R exactly confirmed this conclusion (Figure 7A). MeRIP+RIP targets showed globally and significantly lower binding affinities in the mutant YTHDF2-K571R when compared with YTHDF2-WT. Compared to the control group, the binding capacities of YTHDF2 to RIP targets in treated group with either 2-D08 or GA were decreased, especially to MeRIP + RIP targets (Figure 7B and C). Besides, the down-regulated genes were highly overlapped in 2-D08 and GA treated group (Figure 7D, Supplementary Figure S9B). All these results suggested SUMOylation of YTHDF2 indeed increased its binding affinity towards m6A-RNAs and the possible mechanism might be the crystal structure change upon SUMOylation, which need further exploration but be not included in this study. Consequently, SUMOylated YTHDF2 promoted more m6A-RNAs degradation. Combined analysis of RNA-seq, RIP-seq and MeRIP-seq showed that the mRNA levels were up-regulated in shYTHDF2 stable cells re-expressing YTHDF2-K571R compared with those in re-expressing YTHDF2-WT (Figure 8A-B). Moreover, higher abundance of m6A modification was rich in YTHDF2-K571R cells especially around the stop codons (Supplementary Figure S9C). Therefore, we proved that SUMOylation of YTHDF2 promoted degradation of certain mRNAs by increasing its binding affinity with m6A.
Many studies have reported YTHDF2 linked to tumorigenesis (58–61) and dysregulation of the SUMO pathway appeared in a variety of malignancies such as brain, lung, liver, pancreas, lymphoma and multiple myeloma (62–67). In this study, we investigated the relationship of YTHDF2 SUMOylation and lung cancer. The association between the expression of YTHDF2 and disease free survival (DFS) of patients with lung adenocarcinoma in TCGA data-base implied YTHDF2 might not affect disease free survival. However, by stratified analysis of SUMO1 in high expression YTHDF2 group, we found the patients with higher abundance of SUMO1 seemed to be associated with poor prognosis (Supplementary Figure S11), which suggested SUMOylation of YTHDF2 might promote cancer progress. More importantly, we found that the SUMO-site mutant YTHDF2-K571R repressed cancer progression in H1299 cells compared with YTHDF2-WT group (Figure 6). In line with this, the RNA-seq data displayed the differently expressed genes between K571R and WT in H1299 stable cell lines mainly enriched in several tumor-associated pathways such as growth factor activity, transforming growth factor beta receptor binding, guanyl-nucleotide exchange factor activity, Ras, guanyl-nucleotide exchange factor activity and ERBB-signaling pathway (Figure 8C-D).
In general, our studies demonstrated the SUMOylation of YTHDF2 increased its binding affinity to m6A-modified RNAs, thus leading to degradation of certain RNAs and finally promoted tumor cell growth and migration. Especially, the microenvironmental hypoxia could induce YTHDF2 SUMOylation, which might accelerate cancer progression.
DATA AVAILABILITY
All the raw data and processed files have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE164917.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: G.H. and X.Z. performed most of the experiments; L.L., Q.Y., X.L. C.H., R.L., R.C. and Y.W. helped with all experiments; J.Y. and B.J. supervised this project and analyzed data; J.Y., X.Z. and G.H. wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Guofang Hou, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Department of Oncology, Shanghai 9th People's Hospital, Shanghai Jiao Tong University School of Medicine, 280 Mohe Road, Shanghai 201999, China.
Xian Zhao, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Lian Li, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Qianqian Yang, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Xiaojia Liu, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Caihu Huang, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Runhui Lu, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Ran Chen, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Yanli Wang, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Bin Jiang, Department of Oncology, Shanghai 9th People's Hospital, Shanghai Jiao Tong University School of Medicine, 280 Mohe Road, Shanghai 201999, China.
Jianxiu Yu, State Key Laboratory of Oncogenes and Related Genes, Department of Biochemistry and Molecular Cell Biology & Shanghai Key Laboratory of Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81630075, 31671345, 81721004, 81871862, 81972585] and China’s National Key R&D Programme (NKP) [2019YFE0110600]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Desrosiers R., Friderici K., Rottman F.. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X.et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G.et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H.et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M.et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 6. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 8. Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S.et al.. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- 9. Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U.et al.. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019; 566:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H., Hu X., Huang M., Liu J., Gu Y., Ma L., Zhou Q., Cao X.. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 2019; 10:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winkler R., Gillis E., Lasman L., Safra M., Geula S., Soyris C., Nachshon A., Tai-Schmiedel J., Friedman N., Le-Trilling V.T.K.et al.. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019; 20:173–182. [DOI] [PubMed] [Google Scholar]
- 12. Dang W., Xie Y., Cao P., Xin S., Wang J., Li S., Li Y., Lu J.. N(6)-methyladenosine and viral infection. Front Microbiol. 2019; 10:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasowitz S.D., Ma J., Anderson S.J., Leu N.A., Xu Y., Gregory B.D., Schultz R.M., Wang P.J.. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLos Genet. 2018; 14:e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., Zheng H., Klungland A., Yan W.. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E325–E333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun T., Wu R., Ming L.. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019; 112:108613. [DOI] [PubMed] [Google Scholar]
- 16. Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D.et al.. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014; 8:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J.. Structural basis for the discriminative recognition of N6-Methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 2015; 290:24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., Tian Y., Li J., He C., Xu Y.. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014; 24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L.. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016; 7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Y., He Y., Wang X., Liang Z., He G., Zhang P., Zhu H., Xu N., Liang S.. Protein SUMOylation modification and its associations with disease. Open Biol. 2017; 7:170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu C., Chen C., Huang J., Zhang H., Zhao X., Deng R., Dou J., Jin H., Chen R., Xu M.et al.. SUMOylation at K707 of DGCR8 controls direct function of primary microRNA. Nucleic Acids Res. 2015; 43:7945–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan H., Deng R., Zhao X., Chen R., Hou G., Zhang H., Wang Y., Xu M., Jiang B., Yu J.. SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis. Mol. Cancer. 2017; 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J., Yan J., Zhang J., Zhu S., Wang Y., Shi T., Zhu C., Chen C., Liu X., Cheng J.et al.. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 2012; 3:911. [DOI] [PubMed] [Google Scholar]
- 26. Chen C., Zhu C., Huang J., Zhao X., Deng R., Zhang H., Dou J., Chen Q., Xu M., Yuan H.et al.. SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat. Commun. 2015; 6:8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qu Y., Chen Q., Lai X., Zhu C., Chen C., Zhao X., Deng R., Xu M., Yuan H., Wang Y.et al.. SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1. Mol. Cancer. 2014; 13:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo C., Henley J.M.. Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life. 2014; 66:71–77. [DOI] [PubMed] [Google Scholar]
- 29. Du Y., Hou G., Zhang H., Dou J., He J., Guo Y., Li L., Chen R., Wang Y., Deng R.et al.. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018; 46:5195–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barysch S.V., Dittner C., Flotho A., Becker J., Melchior F.. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 2014; 9:896–909. [DOI] [PubMed] [Google Scholar]
- 31. Uchimura Y., Nakamura M., Sugasawa K., Nakao M., Saitoh H.. Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Anal. Biochem. 2004; 331:204–206. [DOI] [PubMed] [Google Scholar]
- 32. Alarcon C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F.. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015; 519:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alarcon C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F.. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015; 162:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 35. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L.. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013; 31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J.. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013; 8:e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao X., Wang Y., Deng R., Zhang H., Dou J., Yuan H., Hou G., Du Y., Chen Q., Yu J.. miR186 suppresses prostate cancer progression by targeting Twist1. Oncotarget. 2016; 7:33136–33151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B.. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fukuda I., Ito A., Hirai G., Nishimura S., Kawasaki H., Saitoh H., Kimura K., Sodeoka M., Yoshida M.. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem. Biol. 2009; 16:133–140. [DOI] [PubMed] [Google Scholar]
- 42. Liu K., Wang X., Li D., Xu D., Li D., Lv Z., Zhao D., Chu W.F., Wang X.F.. Ginkgolic acid, a SUMO-1 inhibitor, inhibits the progression of oral squamous cell carcinoma by alleviating SUMOylation of SMAD4. Mol Ther Oncolytics. 2020; 16:86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou P., Chen X., Li M., Tan J., Zhang Y., Yuan W., Zhou J., Wang G.. 2-D08 as a SUMOylation inhibitor induced ROS accumulation mediates apoptosis of acute myeloid leukemia cells possibly through the deSUMOylation of NOX2. Biochem. Biophys. Res. Commun. 2019; 513:1063–1069. [DOI] [PubMed] [Google Scholar]
- 44. Kim Y.S., Keyser S.G., Schneekloth J.S. Jr. Synthesis of 2′,3′,4′-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg. Med. Chem. Lett. 2014; 24:1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roskoski R. Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014; 79:34–74. [DOI] [PubMed] [Google Scholar]
- 46. Saiki I. Cell adhesion molecules and cancer metastasis. Jpn. J. Pharmacol. 1997; 75:215–242. [DOI] [PubMed] [Google Scholar]
- 47. Apte R.N., Voronov E.. Interleukin-1–a major pleiotropic cytokine in tumor-host interactions. Semin. Cancer Biol. 2002; 12:277–290. [DOI] [PubMed] [Google Scholar]
- 48. Apte R.N., Dotan S., Elkabets M., White M.R., Reich E., Carmi Y., Song X., Dvozkin T., Krelin Y., Voronov E.. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006; 25:387–408. [DOI] [PubMed] [Google Scholar]
- 49. Hou J., Zhang H., Liu J., Zhao Z., Wang J., Lu Z., Hu B., Zhou J., Zhao Z., Feng M.et al.. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer. 2019; 18:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H., Zuo H., Liu J., Wen F., Gao Y., Zhu X., Liu B., Xiao F., Wang W., Huang G.et al.. Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018; 28:1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paris J., Morgan M., Campos J., Spencer G.J., Shmakova A., Ivanova I., Mapperley C., Lawson H., Wotherspoon D.A., Sepulveda C.et al.. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019; 25:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bartosovic M., Molares H.C., Gregorova P., Hrossova D., Kudla G., Vanacova S.. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017; 45:11356–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rosa-Mercado N.A., Withers J.B., Steitz J.A.. Settling the m(6)A debate: methylation of mature mRNA is not dynamic but accelerates turnover. Genes Dev. 2017; 31:957–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coots R.A., Liu X.M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.B.. m(6)A facilitates eIF4F-Independent mRNA Translation. Mol. Cell. 2017; 68:504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millan-Zambrano G., Robson S.C., Aspris D., Migliori V., Bannister A.J., Han N.et al.. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017; 552:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li M., Zhao X., Wang W., Shi H., Pan Q., Lu Z., Perez S.P., Suganthan R., He C., Bjoras M.et al.. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018; 19:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ivanova I., Much C., Di Giacomo M., Azzi C., Morgan M., Moreira P.N., Monahan J., Carrieri C., Enright A.J., O’Carroll D.. The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell. 2017; 67:1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M.et al.. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018; 67:2254–2270. [DOI] [PubMed] [Google Scholar]
- 59. Sheng H., Li Z., Su S., Sun W., Zhang X., Li L., Li J., Liu S., Lu B., Zhang S.et al.. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2019; 41:541–550. [DOI] [PubMed] [Google Scholar]
- 60. Li Z., Qian P., Shao W., Shi H., He X.C., Gogol M., Yu Z., Wang Y., Qi M., Zhu Y.et al.. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018; 28:904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhong L., Liao D., Zhang M., Zeng C., Li X., Zhang R., Ma H., Kang T.. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019; 442:252–261. [DOI] [PubMed] [Google Scholar]
- 62. Yang W., Wang L., Roehn G., Pearlstein R.D., Ali-Osman F., Pan H., Goldbrunner R., Krantz M., Harms C., Paschen W.. Small ubiquitin-like modifier 1–3 conjugation [corrected] is activated in human astrocytic brain tumors and is required for glioblastoma cell survival. Cancer Sci. 2013; 104:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li H., Niu H., Peng Y., Wang J., He P.. Ubc9 promotes invasion and metastasis of lung cancer cells. Oncol. Rep. 2013; 29:1588–1594. [DOI] [PubMed] [Google Scholar]
- 64. Guo W.H., Yuan L.H., Xiao Z.H., Liu D., Zhang J.X.. Overexpression of SUMO-1 in hepatocellular carcinoma: a latent target for diagnosis and therapy of hepatoma. J. Cancer Res. Clin. Oncol. 2011; 137:533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chien W., Lee K.L., Ding L.W., Wuensche P., Kato H., Doan N.B., Poellinger L., Said J.W., Koeffler H.P.. PIAS4 is an activator of hypoxia signalling via VHL suppression during growth of pancreatic cancer cells. Br. J. Cancer. 2013; 109:1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hoellein A., Fallahi M., Schoeffmann S., Steidle S., Schaub F.X., Rudelius M., Laitinen I., Nilsson L., Goga A., Peschel C.et al.. Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood. 2014; 124:2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Driscoll J.J., Pelluru D., Lefkimmiatis K., Fulciniti M., Prabhala R.H., Greipp P.R., Barlogie B., Tai Y.T., Anderson K.C., Shaughnessy J.D. Jret al.. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010; 115:2827–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zou J.B., Chai H.B., Zhang X.F., Guo D.Y., Tai J., Wang Y., Liang Y.L., Wang F., Cheng J.X., Wang J.et al.. Reconstruction of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in cerebral infarction. Sci. Rep. 2019; 9:12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw data and processed files have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE164917.