Abstract
The major clinical problem in human cancer is metastasis. Metastases are the cause of 90% of human cancer deaths. TAp63 is a critical suppressor of tumorigenesis and metastasis. ΔNp63 acts as a dominant-negative inhibitor to block the function of p53 and TAp63. Although several ubiquitin E3 ligases have been reported to regulate p63 stability, the mechanism of p63 regulation remains partially understood. Herein, we show that CHIP, an E3 ligase with a U-box domain, physically interacts with p63 and promotes p63 degradation. Notably, Hsp70 depletion by siRNA stabilizes TAp63 in H1299 cells and destabilizes ΔNp63 in SCC9 cells. Loss of Hsp70 results in a reduction in the TAp63-CHIP interaction in H1299 cells and an increase in the interaction between ΔNp63 and CHIP in SCC9 cells. Our results reveal that Hsp70 acts as a molecular switch to control CHIP-mediated ubiquitination and degradation of p63 isoforms. Furthermore, regulation of p63 by the Hsp70-CHIP axis contributes to the migration and invasion of tumor cells. Hence, our findings demonstrate that Hsp70 is a crucial regulator of CHIP-mediated ubiquitination and degradation of p63 isoforms and identify a new pathway for maintaining TAp63 or ΔNp63 stability in cancers.
INTRODUCTION
The major clinical problem in human cancer is metastasis (1). Metastases are the cause of 90% of human cancer deaths (1–3). TP63 is a homologue of the tumor suppressor p53 (4,5). The TP63 gene encodes two major protein isoforms, TAp63 and Delta Np63. TAp63 is a suppressor of tumorigenesis and metastasis (6–8). More aggressive, metastatic tumors lose TAp63 expression, suggesting that loss of TAp63 accelerates tumorigenesis and metastatic spread (1,5,9). Delta Np63 (ΔNp63 or DNp63) lacks the N- terminal transactivation domain found in TAp63 (6,7). ΔNp63 acts as a dominant-negative inhibitor to block the function of p53 and TAp63. ΔNp63 variants are also able to inactivate the transactivation function of p53, as well as the function of TA variants of p63 by directly competing with promoter regions and by incorporating into heterotetramers where they act in a dominant-negative manner (6-11). Due to these characteristics, TAp63 is often considered a tumor suppressor, while the ΔNp63 variant may function as an oncogene (6,12–14). The molecular mechanism underlying the regulation of TAp63 and ΔNp63 is still unknown.
CHIP (C-terminus of Hsc-70 interacting protein) is a highly conserved ubiquitin E3 ligase containing a U-box domain (15,16). CHIP contains an N-terminal tetratricopeptide repeat (TPR) domain responsible for its protein-protein interactions with Hsp70 and Hsp90, a central coiled-coiled domain responsible for its dimerization, and a C-terminal U-box domain responsible for its E-3 ligase function (17). CHIP constitutes an important link between the ubiquitin-proteasome system and the heat shock response pathway (15). E3 ligases often control the ubiquitination of multiple substrates (18,19); for example, CHIP acts as an E3 ligase for p53 (20), c-Myc (16), ErbB2 (21), EGFR (22), IRF-1 (23), PTEN (24), etc. The two opposite functions of CHIP, which promote or inhibit tumor growth, are intriguing and suggest a novel cell growth regulatory mechanism. It is possible that both functions are necessary at different stages of normal cell growth, and a balance of the two functions is maintained in normal cells. CHIP is underexpressed in breast, pancreatic, and gastric cancers (24–26), while it is overexpressed in gliomas and gallbladder carcinomas (27–30). Deletion of Chip in mice resulted in the development of apoptosis in multiple organs (31). However, the underlying mechanisms of this apoptosis remain unclear.
The heat-shock protein Hsp70 is one of the most highly evolutionarily conserved proteins (32). The N-terminal ATPase domain of Hsp70 can hydrolyze ATP, and the C-terminal substrate-binding domain can bind to unfolded polypeptides (33–36). These two functional domains are critical for the chaperone function of Hsp70. In humans, 13 Hsp70 homologs are expressed in distinct cellular compartments (32). Hsp70 can directly unfold misfolded proteins to maintain protein homeostasis. Knockout of Hsc70 in mice is lethal, and mice lacking the Hsp70–2 homolog (Hsp70.2) have a developmental defect in spermatogenesis (37). Hsp70.1/70.3 homozygous knockout mice show evidence of increased genomic instability. The HSC70/HSP70 proteins are acknowledged to have an oncogenic function; they are overexpressed in many invasive cancers, including bladder cancer, breast cancer, colorectal cancer, lung cancer, melanoma, and ovarian carcinomas (38). Depletion of Hsp70 induces cell death in lung cancer cells, but not in normal cells (39,40). However, the molecular mechanism of Hsp70 inhibition is not fully understood.
In this study, we identified that Hsp70 acts as a critical regulator to control CHIP-mediated p63 degradation. We found that CHIP physically interacts with p63. CHIP is overexpressed in several human cancer cell lines, and we found an inverse correlation between CHIP and p63 expression in these tumor samples. We present evidence that the depletion of Hsp70 by siRNA markedly increased TAp63 levels in H1299 non-small cell lung carcinoma cells and decreased levels of ΔNp63 in SCC9 squamous cell carcinoma cells. Importantly, loss of Hsp70 significantly decreased CHIP-TAp63 interactions in H1299 cells; by contrast, it increased CHIP-ΔNp63 interactions in SCC9 cells, indicating that Hsp70 is involved in CHIP-mediated p63 degradation. Regulation of p63 by the Hsp70-CHIP axis contributes to cancer cell migration and invasion. Moreover, we demonstrated that CHIP mediates p63 function in an Hsp70-dependent manner. Thus, our data reveal mechanistic insights into p63 regulation and may lead to the development of novel therapies for human cancers.
MATERIALS AND METHODS
Patient data and tumor bank
The patient's samples consisted of nine human prostate tumor tissues and two benign prostate neoplasm tissues. All samples were obtained from the Alberta Cancer Research Biobank (www.acrb.ca). The studies received research ethics approval from the Health Research Ethics Board – Biomedical Panel, University of Alberta.
Yeast two-hybrid screen
The yeast strain AH109 was co-transformed with the plasmid pAS2-1-CHIP and a mouse cDNA library in the pACT2 vector (Clontech). 6 × 107 transformants were screened, and twenty positive clones were isolated after two rounds of growth in the absence of histidine, adenine, and screening for β-galactosidase activity. Recovered plasmids from AH109 were used to co-transform Y190 yeast with either the full-length CHIP or CHIP mutants.
Reagents and antibodies
CHIP or CHIP mutants or Hsp70 or Hsp70 mutants were cloned into pcDNA3.1 or p3x Flag-CMV-10 (Sigma) or pET28a (Novagen). The Myc-MDM2 expressing plasmid was a kind gift from Dr. A.G. Jochemsen. Myc-Itch was obtained from Dr. Tony Pawson. Flag-Itch was obtained from Dr. A. Atfi. His-Ub is a kind gift from Dr. W. Gu. All PCR products were confirmed by sequencing. Doxycycline (Dox, D9891, Sigma), Cycloheximide (CHX, 01810, Sigma), Ver-155008 (SML0271, Sigma), and Hepatocyte growth factor (HGF, H9661, Sigma). Anti-p63 [p63 (ΔNp63 (N-16), sc-8609R, Santa Cruz Biotechnology; ab53039, Abcam; Poly 619002 (anti-p63 (ΔN), Poly 618902 (anti-TAp63), 938102 (anti-TAp63), BioLegend], anti-CHIP [H-231 and C10, Santa Cruz Biotechnology; EPR4447 (ab134064), Abcam], anti-Hsp70 (C92F3A5, Enzo Life Sciences), anti-Itch (ab109018, Abcam), anti-Mdm2 (2A10, Calbiochem; SMP14, BD Biosciences), anti-PRP19 (ab27692, Abcam), anti-p21 (F-5, Santa Cruz), anti-Myc (9E10, Roche), anti-Flag (M5, M2, Sigma), anti-HA (12CA5, Roche), anti-ubiquitin (550944, BD Biosciences), anti-actin (A2066, Sigma), anti-T7 tag (69522–4, Novagen) and anti-p53 (Pab1801, Santa Cruz Biotechnology) were used according to the manufacturer's instructions.
Cell culture and DNA transfection
Human H1299 (CRL-5803) and SCC9 cells (CRL-1629) were purchased from the American Type Culture Collection (ATCC). As indicated by ATCC, both cell lines’ identity is verified and shown to be free of contamination from other cell lines and microbes. SCC15, SCC25, FADU, A431, Sk-mel-2, Sk-mel-5, Sk-mel-8, M2, LOX-IMVI, MeWo, and Hs859T cells were provided and verified by Drs. M. Pasdar, A. Underhill and G. Chan (University of Alberta, Edmonton, AB, Canada). These cell lines were maintained in α-minimal essential medium (α-MEM, Gibco, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Gibco, Invitrogen). PCR routinely tested these cell lines for mycoplasma contamination by the following primers: Myco_fw1: 5′-ACACCATGGGAGCTGGTAAT-3′, Myco_rev1: 5′-CTTCATCGACTTTCAGACCCAAGGCA-3′. We verified that both H1299 cells and SCC9 cells are p53-deficient tumor cells, which expressed no p53 protein (41,42). The wild-type MEFs, TAp63−/- MEFs, and deltaNp63−/− MEFs were provided by Dr. E. Flores (43) and cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, Invitrogen) containing 10% FBS (Gibco, Invitrogen). H1299 cells and SCC9 cells were transfected by the calcium phosphate method described earlier (44), or using Lipofectamine 2000 (Invitrogen).
Gel filtration
As described previously (44), in brief, cell lysates (H1299 or SCC9 cells) were fractioned using FPLC protein purification system on Superose 6 column (GE Healthcare). The column was equilibrated with Tris buffer (50 mM Tris, pH7.5, 150 mM NaCl, 0.1% Triton X-100) and lysates (2 mg) were applied and eluted from the column with the same buffer. The flow rate was 0.4 ml/min, and 380 μl fractions were collected. The column was calibrated with BioRad gel filtration standards containing thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and Vitamin B12 (1.3 kDa).
siRNA experiments
For siRNA experiments, H1299 cells or SCC9 cells were transfected with the indicated siRNA constructs by using Lipofectamine 2000 (Invitrogen). We used the following sequences for these experiments: CHIP siRNA1: GGAGCAGGGCAATCGTCTG; CHIP siRNA2: AGGCCCTGGCCGACTGCCG. CHIP siRNA3: GAAGAAGCGCTGGAACAGC; CHIP siRNA4: ACCACGAGGGTGATGAGGA. The Itch target sequences were described earlier (45): AAGTGCTTCTCAGAATGAT and AACCACAACACACGAATTA. Hsp70-siRNA1: CTGGCCTTTCCAGGTGATC; Hsp70-siRNA2: GGACATCAGCCAGAACAAG. TAp63-siRNA: GCACACAGACAAATGAATT (13). ΔNp63-siRNA: CAATGCCCAGACTCAATTT (13).
Generation of knockdown cell lines
The 19-nt oligonucleotides derived from CHIP, Hsp70, Itch, TAp63 and ΔNp63 as described above, were cloned into the pSuper.neo vector, according to the manufacture's procedure (OligoEngine). We generated several knockdown cell lines for CHIP or Hsp70 or Itch or TAp63 or ΔNp63, the cells were transfected with the plasmids expressing CHIP-siRNAs, or Hsp70-siRNAs, or Itch-siRNA, or TAp63-siRNA, or ΔNp63-siRNA, selected in G418 (Invitrogen) for 2 weeks, and then independent stable clones were selected, and evaluated by western blotting.
Generation of H1299 and SCC9 with inducible pHsp70 expression
H1299 Tet-Off cells and SCC9 Tet-Off cells were generated by transfection of the pTet-Off plasmid and selected (G418, Geneticin) according to manufacturer's instruction (Clontech). The pTRE2puro-Hsp70 construct was transfected into the H1299 Tet-Off cells or the SCC9 Tet-Off cells and selected in the presence of 1.0 mg/ml puromycin. The expression of Hsp70 was screened in the presence or absence of doxycycline (Dox, 1.0 μg/ml). Clones were maintained in α-MEM medium containing 10% Tet system-approved FBS (Clontech).
Expression and recombinant protein preparation
All GST or His-tagged recombinant proteins were expressed in the Escherichia coli strain BL21 (DE3, Novagen), treated with isopropyl-β-d-thiogalactoside to induce fusion protein expression, and purified using glutathione Sepharose 4B (Amersham) for GST-fusion proteins or using Ni2+-NTA agarose (Qiagen) for His-fusion proteins.
Immunoprecipitation and measurement of TAp63 and ΔNp63 half-life
Cells were lysed in 50 mM Tris–HCl (pH 8.0), 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, protease inhibitor tablet (Roche), and immunoprecipitated with indicated antibodies. The immune complexes were collected with protein A-agarose beads and washed four times with the same lysis buffer. The immunoprecipitates were analyzed by immunoblotting. To measure the half-live of TAp63 or ΔNp63 proteins, H1299 or SCC9 cells with depleted Hsp70, or CHIP or Hsp70 and CHIP by siRNA were treated with 25 μg/ml cycloheximide (CHX) to inhibit de novo protein synthesis; protein levels were monitored by immunoblotting with the p63-specific antibodies at the indicated time points. The relative amounts of TAp63 or ΔNp63 proteins were determined by densitometry and normalized using actin.
In vitro ubiquitination assay
The in vitro ubiquitination assay was performed as described previously with some modifications (41,44). CHIP-mediated ubiquitination reactions were carried out by adding E1 (20–40 ng, Calbiochem), E2 (UbcH5b, 100 ng, Calbiochem), ubiquitin or His-ubiquitin (3–5 μg, Sigma), His-TAp63 or His-ΔNp63 (0.2–0.5 μg) or His-Hsp70 (0.2–0.4 μg) in ubiquitination buffer (50 mM Tris–HCl [pH 7.4], 2 mM ATP, 5 mM MgCl, 2 mM DTT) to a final volume of 30 μl. The reactions were incubated at 30°C for 1.0–1.5 h. The reactions were stopped with 2× SDS loading buffer, resolved by SDS-PAGE gels, and analyzed by western blotting. Or the mixtures were immunoprecipitated with the indicated antibodies and analyzed by western blotting.
In vivo ubiquitination assay
Cells were transfected with expressing plasmids encoding TAp63, or ΔNp63, or Myc-Itch or Flag-CHIP, and HA-tagged ubiquitin. After 30 h, cells were harvested, lysed, and immunoprecipitated with indicated antibodies. Immune complexes recovered with protein A-Sepharose were washed four times with radioimmunoprecipitation assay buffer [RIPA buffer: 50 mM Tris–HCl (pH7.6), 150 mM NaCl, 1.0% (v/v) NP-40, 0.5% (w/v) Sodium Deoxycholate, 1.0 mM EDTA, 0.5 mM EGTA, 1.0% (w/v) SDS and protease inhibitor tablet (Roche)], separated on 10% SDS-PAGE, and analyzed by immunoblotting as described previously (44).
His-ubiquitin pull-down assay
As described previously (46), H1299 cells were transfected with His-tagged ubiquitin and the indicated expression plasmids. Thirty hours after transfection, the cells were lysed in buffer A (6 M guanidine–HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris–HCl (pH 8.0); 10 mM imidazole at pH 8.0). Approximately 500 μg of cell lysates were added to 50 μl of equilibrated Ni-NTA agarose and were allowed to incubate for 3 h at room temperature. Beads were then washed once with Buffer A, followed by two washes with Buffer A/TI (1 vol of Buffer A, 3 vol of Buffer TI [25 mM Tris–Cl, 20 mM imidazole at pH 6.8]), and one wash with Buffer TI; all of the washes used 1 ml of the buffer. After extensive washing, the precipitates were boiled with SDS loading buffer and then subjected to SDS-PAGE followed by immunoblotting analysis.
Chromatin Immunoprecipitation assay
As previously described (41,44). Briefly, we fixed and prepared lysates from 3 × 107 H1299 cells. Cells were sonicated to an average fragment size of 500 bp. A 100-μl aliquot of sonicated chromatin (3 × 106 cell equivalents) was used per immunoprecipitation. Chromatin was diluted in 1 ml of 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl [pH 8] containing 2 μg of sheared salmon sperm DNA and precleared by mixing with protein A-Sepharose 4 fast flow beads for 2 h at 4°C. Anti-TAp63 antibodies (Poly618902) or Rabbit IgG were used for immunoprecipitation. After overnight incubation at 4°C, immune complexes were collected with protein A-Sepharose beads in the presence of 2 μg of salmon sperm DNA and 45 μl of yeast tRNA for 1 h, and washed sequentially for 10 min once in TSEI (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl [pH 8.0] and 150 mM NaCl), 4 times in TSEII (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl [pH 8.0], 500 mM NaCl), once in buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris–HCl [pH 8.0]) and three times in TE. Samples were extracted twice in 250 μl of 1% SDS and 0.1 M NaHCO3 and heated at 65°C overnight to reverse crosslinks. DNA was purified by extraction with phenol-chloroform, precipitated with ethanol, and resuspended in 50 μl TE. ChIP-enriched DNA was quantitated by real-time PCR with primers amplifying the Hsp70 promoter. The following primer pairs were used for Hsp70 ChIP: (i) Hsp70 promoter region, forward, 5′-ccacatacctcaggcttaaacc-3′; reverse, 5′- cggttgtgcagtttgatattgag-3′. (ii) Hsp70-control, forward, 5′-accccatcatcagcggac-3′; reverse, 5′-tccttgagtcccaacagtcc-3′.
Luciferase reporter gene assay
H1299 cells were co-transfected with a luciferase reporter construct (p21-Luc) containing the p53 binding site from the p21WAF1 promoter and TAp63 or ΔNp63 alone, or in combination with CHIP, MDM2, or Myc-Itch. A β-galactosidase reporter construct, pCMV-β-gal (Promega) was included in all the transfection reactions. Luciferase activity was measured 2 days post-transfection using an LB9507 luminometer and the luciferase assay reagent (Promega); values were normalized to β-galactosidase activity. Alternatively, a luciferase reporter construct (Hsp70-Luc) containing the p63 binding site from the Hsp70 promoter or Hsp70 mutant reporter construct was transfected into the H1299 Tet-OFF cells or the SCC9 Tet-OFF cells. Luciferase activity was measured in the absence or presence of doxycycline (Dox) at the indicated time points.
Apoptosis assay
As described previously (44,47), for Annexin V staining (BD, Biosciences), these cells were transfected with indicated expression plasmids. The cells were then trypsinized, washed, and resuspended in PBS containing 25 μg/ml Annexin-V-FITC and 50 μg/ml 7-AAD (7-amino-actinomycin D) prior to FACS analysis. The data were analyzed using FlowJo software (TreeStar Inc).
GST pull-down assay
The purified GST-fusion proteins (2–5 μg) were added to unlabeled cell extracts, and incubated in non-ionic lysis buffer (100 mM NaCl, 20 mM Tris-HCl [pH 8.0], 0.5% NP-40, 1 mM EDTA, 1 mM EGTA) on ice for 1 h. The mixtures were centrifuged for 15 min at 4°C to remove precipitating proteins, and the clarified lysates were mixed with glutathione–Sepharose beads previously washed in lysis buffer and incubated for 1 h at 4°C with slow shaking. The bound beads were then washed four times with the same lysis buffer, heated at 95°C in SDS-loading buffer to release bound proteins, and analyzed by SDS-PAGE. Proteins were analyzed by immunoblotting.
Cell cycle analysis
H1299 cells were transfected with indicated expression plasmids. After 40 h, the cells were washed, fixed with 70% ethanol, treated with100 μg/ml RNase A and labeled with 50 μg/ml propidium iodide (PI) for 3 h at 4°C and analyzed by flow cytometry (Becton Dickinson). The data were analyzed using FlowJo software (TreeStar Inc).
Migration and invasion assay
As described previously (47), the effect of Hsp70/CHIP on the migratory and invasive capability of the neoplastic cells was measured by Transwell assay. For the migration assay, 2 × 104 cells were seeded into the insert in 24-well Transwell units. For the invasion assay, 2 × 104 cells were seeded into the insert with precoated-Matrigel matrix (BD, Biosciences) in 24-well Transwell units. After a 24 h incubation period, the inside of each insert was swabbed and stained. The cells that had passed through the filter into bottom wells were fixed in methanol for 20 min, stained with 0.1% crystal violet for 15 min and photographed with a photomicroscope (Nikon), and quantified by counting using the ImageJ plugin ‘cell counter’ and expressed as a percentage of the control group, with control values setting at 100%.
Scattering assay
As described previously (48), 2500 cells were seeded in six-well plates and allowed to settle for 32 h. Cells were incubated in the indicated chemicals, and scattering was monitored using a phase-contrast ×20 objective of an inverted microscope (Zeiss, Germany) coupled to a CCD camera (Roper Scientific, France), capturing pictures every 30 min for 16 h. For knockdown experiments, H1299 cells were transfected with siRNA 24 h after seeding, washed 5 h later, and incubated with 10 ng/ml HGF (Sigma) for 48 h, as indicated. Phase-contrast images were taken, and lysates were harvested for immunoblotting analysis to verify knockdown after scattering. For each condition, >1000 cells from three independent experiments were counted and tagged as ‘scattered' or ‘non-scattered' in the ImageJ to calculate the proportion of scattered cells. Scattered cells were defined as cells growing in colonies of four cells or less or cells that were touching only one other cell.
Statistical analysis
Statistical significance was analyzed by a two-tailed Student's t-test and expressed as a P-value using GraphPad Prism8 software. A P-value <0.05 was considered significant. Pearson correlations were determined using OriginPro 2019 software; significance (two-tailed test) was considered if P <0.05 (44).
RESULTS
CHIP is associated with p63 and Hsp70 in cells
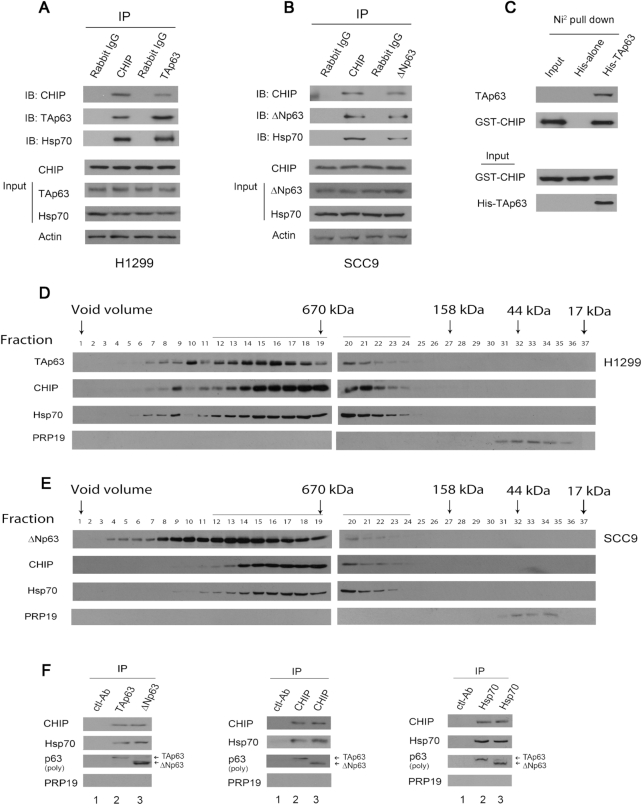
We conducted a yeast two-hybrid screen, and p63 was identified as a CHIP interacting protein. Given that CHIP binds to Hsp70 (15), we sought to determine whether endogenous CHIP, Hsp70 and p63 interact under more physiological conditions. As shown in Figure 1A, TAp63 was present in the anti-CHIP but not in the control rabbit IgG immunoprecipitates. Hsp70 coimmunoprecipitated with CHIP and TAp63 in H1299 cells. Similarly, ΔNp63 coimmunoprecipitated with CHIP and Hsp70 in SCC9 cells (Figure 1B). Next, we performed an in vitro Ni2+ pull-down assay and revealed that GST-CHIP binds to His-TAp63 but not to His alone, implying that this interaction is indeed direct; similar to the interaction of CHIP with PTEN (one of the substrates of CHIP), this interaction is not mediated by the chaperone complex in vitro (Figure 1C). To determine whether p63-CHIP, p63-Hsp70 or p63-CHIP-Hsp70 complexes exist in cells, H1299 cells (with high levels of TAp63 expression) and SCC9 cells (with high levels of expression of ΔNp63, unpublished data) were fractionated by gel filtration chromatography (44). The fractions were analyzed by western blotting with the corresponding antibodies. As shown in Figure 1D and E, one major peak was observed in fractions 12–19 (1.6 MDa-670 kDa), although the elution patterns of ΔNp63, CHIP and Hsp70 covered a more extensive range of fractions than that of TAp63. Additionally, a smaller peak was observed in fractions 20–24 (670–158 kDa). Co-elution of TAp63/or ΔNp63, with CHIP, or Hsp70 is not conclusive evidence that these proteins exist in the same multiprotein complex in cells. To address this issue, the fractions (co-eluted proteins) were pooled, subjected to immunoprecipitation with antibodies specific for TAp63/ΔNp63, CHIP or Hsp70, and analyzed by western blotting. These data confirmed that TAp63 and ΔNp63 associate with CHIP and Hsp70 in cells (Figure 1F).
Figure 1.
CHIP is associated with endogenous p63 and Hsp70 in cancer cells. (A) H1299 cells were immunoprecipitated with anti-CHIP (H-231) or anti-TAp63 (Poly 618092) antibodies as indicated and immunoblotted with the indicated antibodies. Direct western blots for CHIP (H-231), TAp63 (Poly 618092) and Hsp70 (C92F3A5) are shown in the lower panels. (B) Similar to (A), except that SCC9 cells and an anti-ΔNp63 antibody (Poly 619002) were used. (C) In vitro interactions of CHIP and TAp63 were evaluated in Ni2+ pull-down assays. GST-CHIP or His-TAp63 fusion proteins were purified from E. coli. The ability of GST-CHIP to bind to His-TAp63 was analyzed by immunoblotting with an antibody against GST (GST-CHIP). (D) Gel filtration of H1299 lysates. H1299 cell lysates were fractioned on a Superose 6 column. Serial fractions were collected, and proteins were concentrated and analyzed for the presence of TAp63, CHIP, Hsp70, and PRP19 (control). (E) Similar to (D), except that SCC9 cell lysates were fractioned. (F) The fractions 12–21 from H1299 cells (D), or SCC9 cells (E) were pooled, immunoprecipitated, and analyzed by western blotting with CHIP-specific (H-231), TAp63-specific (Poly 618902), DNp63-specific (poly 619002) and PRP19-specific (ab27692) antibodies, as indicated. Lane 1 fractions are mixed from E and D (fractions 12–21); lane 2 fractions are from D (fractions 12–21), lane 3 fractions are from E (fractions 12–21). All experiments were performed in triplicate.
The TP63 gene contains a primary promoter (P1) upstream of the coding sequence and an alternative promoter (P2) located in intron 3 (Supplementary Figure S1A). P1 encodes p53-like isoforms that contain the N-terminal acidic transactivation domain and are designated TAp63, while P2 generates N-terminal truncated isoforms that lack this transactivation domain and are designated delta Np63 (ΔNp63 or DNp63). To identify the domain of TAp63 that interacts with CHIP, five Myc-tagged TAp63 constructs were generated with various TAp63 mutations (Supplementary Figure S1B). H1299 cells were co-transfected with plasmids expressing T7-TAp63 or one of the Myc-TAp63 mutants and Flag-CHIP. After 24 h of transfection, the cells were lysed, and the cell extracts were mixed with the indicated antibodies. As shown in Supplementary Figure S1C, CHIP was present with constructs 541C, 363C and 108C, which were co-immunoprecipitated; these constructs share the C-terminal SAM-domain and post-SAM domain (also called: transactivation-inhibition domain (TIM), 7). However, co-immunoprecipitation does not occur with N362 or N540, which does not contain the SAM-domain and the post-SAM domain, indicating that both SAM domain and post-SAM domain are required for CHIP binding. We then generated a wild-type GST-CHIP construct and various GST-CHIP mutant constructs and performed in vitro GST pull-down assays (Supplementary Figure S1D). We observed that the TPR domain of CHIP is required for binding to TAp63 in vitro (Supplementary Figure S1E).
CHIP/Hsp70 expression modulates the clinical characteristics of human cancers
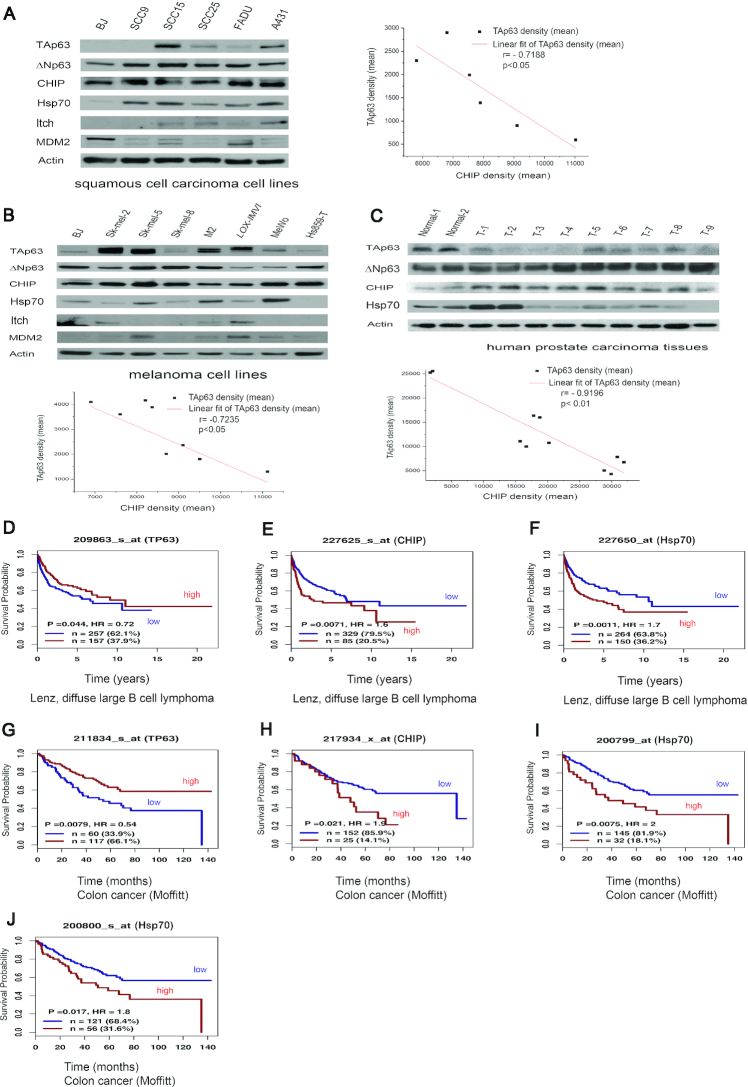
We next examined the association of CHIP, Hsp70, and p63 expression with the clinical characteristics of human cancers. CHIP and ΔNp63 were found to be overexpressed in human squamous cell carcinoma cell lines (Figure 2A) and in human prostate cancer tissues (Figure 2C), but not in human melanoma cell lines (Figure 2B). We identified an inverse correlation between elevated CHIP expression and low or undetectable TAp63 levels (P < 0.01, Pearson correlation test). A similar inverse correlation between CHIP and ΔNp63 expression was found (Supplementary Figure S2A-2C, P < 0.01, Pearson correlation test). Three human prostate cancer tissue samples (T2, T3, T4) had low levels of TAp63; however, one tumor sample (T5) did not display an inverse correlation between CHIP and TAp63 expression. This relationship is not entirely reciprocal, because some samples had decreased CHIP levels, suggesting that additional factors could be involved.
Figure 2.
CHIP is overexpressed in cancer cells and human prostate tumors. CHIP and Hsp70 are associated with low overall survival in human cancers. (A) CHIP is overexpressed in squamous cell carcinoma cell lines. Cell extracts were prepared and immunoblotted with TAp63-specific (Poly 618902), ΔNp63-specific (Poly 619002), CHIP-specific (H-231), Hsp70-specific (C92F3A5), Itch-specific (ab109018), MDM2-specific (SMP14) antibodies. The inverse correlation between TAp63 and CHIP protein levels was tested with a Pearson correlation using OriginPro 2019 software. Pearson correlation: –0.7188; significance (two-tailed test): P < 0.05. (B) Similar to (A), except that melanoma cell lines were used. The inverse correlation between TAp63 and CHIP protein levels was tested with a Pearson correlation. Pearson correlation: –0.85895; significance (two-tailed test): P < 0.05. (C) Whole lysates were harvested from nine invasive prostate carcinoma tissue samples and two benign prostate neoplasm samples as indicated and analyzed by western blotting with the antibodies against TAp63 (Poly 618902), ΔNp63 (Poly 619002), CHIP (H-231), and Hsp70 (C92F3A5). T denotes tumor tissues. The inverse correlation between TAp63 and CHIP protein levels was tested with a Pearson correlation. Pearson correlation: –0.9196; significance (two-tailed test): P < 0.01. An antibody to actin was used as a loading control. (D–F). Kaplan–Meier survival curves generated from www.genomicscape.com revealed that patients with high levels of CHIP and Hsp70 with lower levels of TP63 had a significantly lower overall survival in the Lenz, diffuse large B cell lymphoma. (G–J). Kaplan–Meier survival curves generated from www.genomicscape.com revealed that patients with high levels of CHIP and Hsp70 with low levels of TP63 had a significantly lower overall survival in human colon cancers. Please note the number: 209863 (D), 227625 (E), 227650 (F), 211834 (G), 217934 (H), 200799 (I), and 200800 (J) represented different ‘Affymetrix ID’ number.
To assess the effect of CHIP, Hsp70 and TP63 on overall survival (OS) in human cancers, Kaplan–Meier survival analysis was performed. We used an online database (www.genomicscape.com) to assess the correlation of CHIP, Hsp70 and TP63 mRNA expression with overall survival in human cancers. As shown in Figure 2D–F, high levels of CHIP and Hsp70, and low levels of TP63 expression were strongly correlated with worse survival in the Lenz, diffuse large B cell lymphoma (www.genomicscape.com). Similar findings were observed in human colon cancers, and the differences were significant (P <0.01) (Figure 2G–J). In addition, the literature on p63 is complicated, because some data do not distinguish between the TA and ΔN isoforms (due to a lack of specific antibodies or the use of TAp63- or ΔNp63-specific primers). Taken together, our results and the OS data substantiate the oncogenic properties of CHIP and Hsp70 in several human cancers.
CHIP promotes degradation of p63
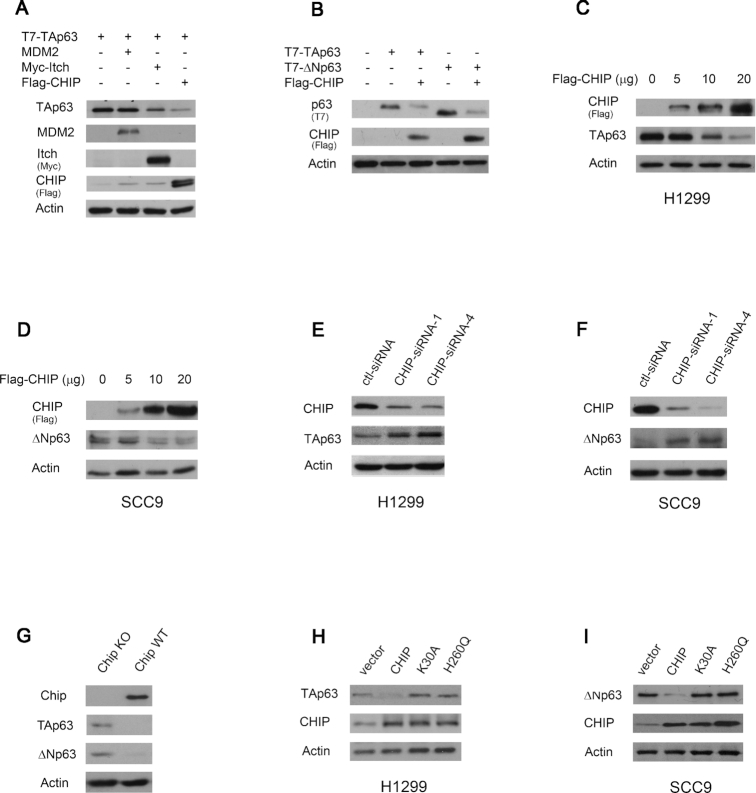
CHIP is an ubiquitin E3 ligase that selectively ubiquitinates and degrades proteins. To date, only a few E3 ligases, such as Itch, Nedd4 and WWP1 have been reported to ubiquitinate p63 proteins (45,49,50). As shown in Figure 3A, the levels of TAp63 were markedly reduced by co-expression of CHIP or Itch, but not by co-expression of MDM2. Similar data were obtained in cells co-expressing ΔNp63 and CHIP (Figure 3B). We then evaluated the ability of CHIP to regulate the endogenous TAp63 or ΔNp63 protein levels in H1299 cells and SCC9 cells, respectively. Transient overexpression of CHIP decreased the expression of TAp63 in H1299 cells and ΔNp63 in SCC9 cells (Figure 3C and D). To determine whether CHIP is critical in regulating the levels of TAp63 in H1299 cells and ΔNp63 in SCC9 cells, endogenous CHIP was subjected to siRNA-mediated knockdown. As shown in Figure 3E and F, CHIP-siRNA-1 and CHIP-siRNA-4 effectively inhibited CHIP expression in both cell lines, while a scrambled control siRNA had no effect. Notably, the decrease in the CHIP level was accompanied by an increase in the TAp63 protein (H1299) or ΔNp63 protein (SCC9) level (Figure 3E and F). As predicted, the basal levels of endogenous TAp63 and ΔNp63 proteins were also much higher in Chip-knockout mouse embryonic fibroblasts (MEFs) than in parental wild-type (wt) MEFs (Figure 3G). Next, we investigated whether CHIP mediates TAp63 or ΔNp63 degradation through the ubiquitin-proteasome pathway. Plasmids expressing CHIP or Itch and TAp63 or ΔNp63 were co-transfected into H1299 or SCC9 cells in the presence or absence of MG132, a proteasome inhibitor (44). Similar to transfection with the Itch plasmid (6,45), addition of MG132 significantly increased TAp63 or ΔNp63 levels in the presence of exogenous CHIP compared with those in untreated cells, suggesting that CHIP promotes TAp63 or ΔNp63 degradation via the ubiquitin-proteasome pathway (Supplementary Figure S3A and S3B). The TPR domain of CHIP is required for binding to TAp63 (Supplementary Figure S1E). The K30A mutation, a point mutation in the TPR domain of CHIP, was reported to disrupt the binding of the TPR domain to Hsp70. CHIP (H260Q), a U box mutant, lost its E3 ligase activity (51). As shown in Figure 3H, neither point mutant degraded TAp63 in H1299 cells. Similar results were obtained in SCC9 cells (Figure 3I), indicating that the binding activity and E3 ligase activity of CHIP are required for CHIP-mediated p63 degradation.
Figure 3.
CHIP decreases the levels of TAp63 and ΔNp63. (A) The expression of CHIP decreases the levels of TAp63 in cells. Plasmids expressing T7-TAp63, or in combination with MDM2, Myc-Itch, or Flag-CHIP, were transfected into H1299 cells and analyzed by immunoblotting with T7 tag-specific (69522-4), MDM2-specific (SMP14), Myc tag-specific (9E10), Flag tag-specific (M2) antibodies. (B) H1299 cells were transfected with T7-TAp63 or T7-ΔNp63, or together with Flag-CHIP, and analyzed by immunoblotting. (C) H1299 cells were transfected with increasing amounts of the Flag-CHIP expression plasmid. The levels of expression of Flag-CHIP and endogenous TAp63 protein were determined by immunoblotting with Flag-specific (M2) and TAp63-specific (Poly 618902) antibodies. An antibody against actin was used as a loading control. (D) Similar to the (C), except that SCC9 cells were transfected. The protein levels of endogenous ΔNp63 and CHIP expression were detected by western blotting with Flag-specific (M2) for CHIP and ΔNp63-specific (Poly 619002) antibodies. (E) H1299 cells were transfected with CHIP-siRNA-1 or CHIP-siRNA-4 and analyzed by immunoblotting with CHIP-specific (H-231) and TAp63-specific (Poly 618902) antibodies. (F) Similar to (E), except that SCC9 cells were used. The protein levels of endogenous ΔNp63 and CHIP were detected by western blotting with CHIP-specific (H-231) and ΔNp63-specific (Poly 619002) antibodies. (G) Cell extracts were prepared from Chip−/−and Chip+/+ MEFs. The amount of endogenous Chip, TAp63, and ΔNp63 proteins were detected by western blotting with Chip-specific (EPR4447), TAp63-specific (938102), ΔNp63-specific (N-16) antibodies. (H) H1299 cells were transfected with CHIP, or CHIP mutants (K30A or H260Q), or empty vector, and analyzed by western blotting with TAp63-specific (Poly 618902), CHIP-specific (H-231) antibodies. (I) Similar to (H), except that SCC9 cells and ΔNp63-specific (Poly 619002) antibody were used. All experiments were performed in triplicate.
Hsp70 acts as a switch for CHIP-mediated TAp63 and ΔNp63 degradation
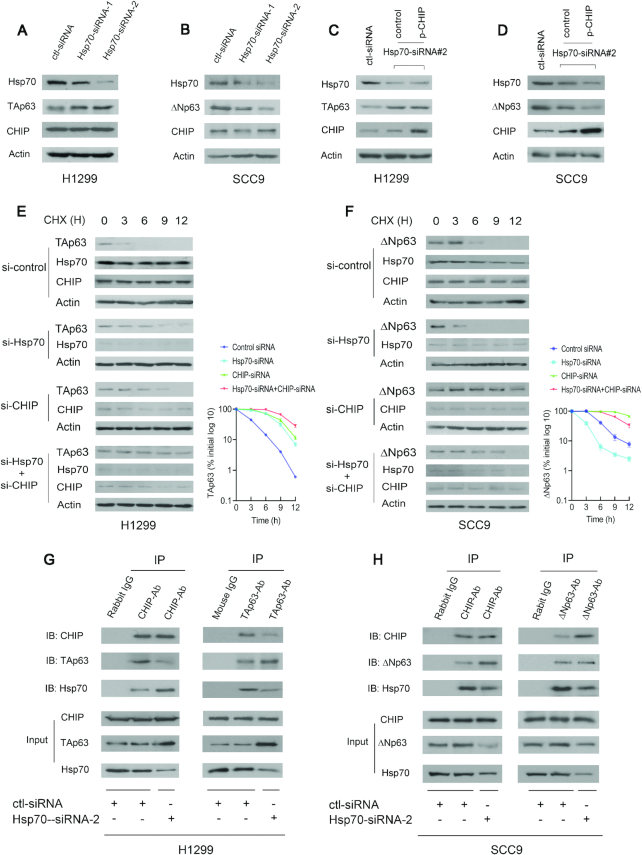
Molecular chaperones, such as Hsp70, regulate p53 stability (20). We sought to determine whether Hsp70 affects the function of CHIP in regulating the TAp63 level in H1299 cells and the ΔNp63 level in SCC9 cells. In H1299 cells, TAp63 protein levels were increased when Hsp70 was depleted by siRNA; in contrast, in SCC9 cells, the levels of ΔNp63 were greatly decreased when Hsp70 was depleted (Figure 4A and B). Overexpression of CHIP did not affect TAp63 protein levels when Hsp70 was depleted, suggesting that CHIP is unable to degrade TAp63 in Hsp70-depleted H1299 cells (Figure 4C). In contrast, reintroduction of CHIP further decreased ΔNp63 protein levels in Hsp70 depleted SCC9 cells (Figure 4D). To further explore whether CHIP regulates the stability of endogenous TAp63 or ΔNp63 proteins, H1299 cells or SCC-9 cells transfected with Hsp70-siRNA, CHIP-siRNA, both Hsp70-siRNA and CHIP-siRNA, or control-siRNA were treated with cycloheximide (CHX) to inhibit de novo protein synthesis. The half-life of TAp63 was maintained at ∼3 h in H1299 cells treated with control-siRNA (Figure 4E, upper image), and the half-life of ΔNp63 was maintained at 6 h in SCC-9 cells treated with siRNA-control (Figure 4F, upper image). The half-life of endogenous TAp63 was increased to ∼6 h in H1299 cells treated with Hsp70-siRNA or CHIP-siRNA (Figure 4E, middle image). The half-life of ΔNp63 was decreased to 3 h in SCC9 cells treated with Hsp70-siRNA; but was increased to more than 9 h in SCC-9 cells treated with CHIP-siRNA (Figure 4F, middle image). Co-depletion of CHIP and Hsp70 increased the TAp63 half-life to 9 h in H1299 cells (Figure 4E, lower image). However, the half-life of ΔNp63 in SCC9 cells with depletion of both CHIP and Hsp70 remained at 6 h (Figure 4F, lower image). These data demonstrated that Hsp70 destabilizes the TAp63 protein and stabilizes the ΔNp63 protein.
Figure 4.
Hsp70 is a molecular switch that regulates CHIP-mediated p63 degradation. (A) H1299 cells were transfected with Hsp70-siRNA-1 or Hsp70-siRNA-2 and analyzed by western blotting with Hsp70-specific (C92F3A5), TAp63-specific (Poly 618902), CHIP-specific (H-231) antibodies. Actin expression was examined as a loading control. (B) Similar to (A), except that SCC9 cells and ΔNp63-specific (Poly 619002) antibody were used. (C) H1299 cells were transfected with Hsp70-siRNA2 or control-siRNA. 30 h later, the cells were further transfected with the CHIP expression plasmid and analyzed by western blotting using the indicated antibodies. (D) Similar to (C), except that SCC9 cells and ΔNp63-specific (Poly 619002) antibody were used. (E) H1299 cells were transfected with control-siRNA, or Hsp70-siRNA2 (si-Hsp70), or CHIP-siRNA4 (si-CHIP), or both Hsp70-siRNA2 and CHIP-siRNA4, and followed by treatment with cycloheximide (CHX) (20 μg/ml). The half-life of endogenous TAp63 was measured by western blotting with TAp63-specific (Poly 618902) antibody at the indicated time-points. The protein levels of TAp63 expression were determined by densitometry and plotted (right image). Errors bars indicate the SEM (n = 3). (F) Similar to (E), except that SCC9 cells were used. The half-life of endogenous ΔNp63 was measured by western blotting with ΔNp63-specific (Poly 619002) antibody. (G) Cell extracts prepared from H1299 cells treated with control-siRNA or Hsp70-siRNA-2 were immunoprecipitated with anti-CHIP (H-231) or anti-TAp63 (Poly 618902) antibodies and immunoblotted with anti-CHIP (C10), anti-TAp63 (938102) and anti-Hsp70 (C92F3A5) antibodies. Direct western blots for CHIP, TAp63 and Hsp70 are shown in the lower panels. (H) Similar to (G), except that SCC9 cells and anti-ΔNp63-specific (Poly 619002) antibody were used as indicated.
The main difference between TAp63 and ΔNp63 is their N-terminus and the alternative promoter (P2) for ΔNp63; thus, Hsp70 plays opposite roles since CHIP binds to both p63 isoforms. To further determine the mechanism by which p63 is regulated by Hsp70, we examined whether Hsp70 regulates TAp63 and ΔNp63 stability by modulating the CHIP-TAp63 or the CHIP-ΔNp63 interaction. H1299 cells or SCC9 cells treated with or without Hsp70-siRNA were immunoprecipitated with the indicated antibodies and analyzed by immunoblotting. Ablation of Hsp70 by siRNA decreased the CHIP-TAp63 interaction and increased the CHIP-Hsp70 interaction, resulting in increased TAp63 protein levels in H1299 cells (Figure 4G). By contrast, depletion of Hsp70 increased the CHIP-ΔNp63 interaction and reduced the CHIP-Hsp70 interaction, leading to a decreased level of ΔNp63 in SCC9 cells (Figure 4H). Thus, Hsp70 controls p63 stabilization through regulation of the CHIP-p63 interaction in different cancer cells.
Hsp70 is involved in response to various stressors, such as heat, UV, γ-irradiation, and chemicals that cause DNA damage (52–54). After DNA damage or exposure to other forms of cellular stress, Hsp70 levels increase. Several mechanisms control the Hsp70 heat shock response. However, the mechanisms of Hsp70-mediated responses to various stressors are largely unknown. For the first time, we demonstrated that DNA sequences within the promoter of Hsp70 contain functional TAp63 DNA binding sites and are efficiently transactivated by TAp63 (Supplementary Figure S4A–C). Notably, we observed that Hsp70 expression was very low in TAp63−/− MEFs compared to wt MEFs (Supplementary Figure S4D). The protein levels of Hsp70 were not significantly changed in ΔNp63−/− MEFs (Supplementary Figure S4E). The p53-MDM2 feedback loop is widely recognized (Supplementary Figure S4F). Similar to the relationship between p53 and MDM2, TAp63−/− MEFs express very low levels of Hsp70. TAp63 transactivates Hsp70; upregulated Hsp70, together with CHIP, then mediates p63 degradation (Supplementary Figure S4G).
Hsp70 plays a critical role in regulating CHIP-mediated p63 ubiquitination
To investigate the mechanisms of CHIP-mediated degradation of TAp63 and ΔNp63, we transfected plasmids expressing T7-TAp63, Myc-Itch, Flag-CHIP or Flag-CHIPΔU (deleted U-box), and HA-ubiquitin into H1299 cells. The cell extracts were immunoprecipitated with a T7-specific antibody and immunoblotted with an anti-HA antibody. As shown in Figure 5A, immunoprecipitated TAp63 (or proteins associated with TAp63) was heavily ubiquitinated in the presence of Itch or CHIP (Figure 5A, upper image). Consistently, CHIPΔU was unable to promote TAp63 ubiquitination, suggesting that the U-box of CHIP is essential for its E3 ligase activity. Similar results were obtained for ΔNp63 in SCC9 cells (Figure 5B). We next investigated whether Hsp70 may directly regulate p63 ubiquitination. Ubiquitination of TAp63 was significantly decreased in H1299 cells with Hsp70 depletion by siRNA compared to cells treated with control-siRNA, and this decrease was accompanied by an increase in the TAp63 protein level (Figure 5C). In contrast, depletion of Hsp70 significantly increased ubiquitination of ΔNp63 and decreased the protein levels of ΔNp63 (Figure 5D). Ubiquitination of TAp63 was markedly decreased in Chip−/− MEFs compared with wild-type MEFs (Supplementary Figure S5A). Re-introduction of Chip expression in Chip−/- MEFs restored TAp63 ubiquitination, indicating that TAp63 is a target of CHIP in cells (Supplementary Figure S5B). Similar data were obtained for ΔNp63 ubiquitination in Chip−/− MEFs (Supplementary Figure S5C and D). To provide direct evidence that the modified TAp63 species corresponded with the ubiquitin conjugated forms, we co-expressed His-tagged ubiquitin with T7-TAp63, Flag-CHIP or Myc-Itch in H1299 cells and isolated His-ubiquitin conjugated proteins under denaturing conditions. After extensive washing, ubiquitinated proteins were eluted and analyzed by western blotting using a TAp63-specific antibody. We confirmed that TAp63 was heavily ubiquitinated in the presence of CHIP or Itch (Supplementary Figure S5E).
Figure 5.
Hsp70 regulates CHIP-mediated p63 ubiquitination. (A) CHIP promotes TAp63 ubiquitination. H1299 cells were transfected with plasmids expressing HA-ubiquitin (Ub), T7-TAp63, Myc-Itch, Flag-CHIP, or Flag-CHIPΔU, as indicated. Cell extracts were immunoprecipitated with a T7-specific (69522-4) antibody against TAp63, and analyzed by immunoblotting with antibodies to HA (12CA5). Direct western blots for TAp63 (T7), Itch (Myc) and CHIP (Flag, M2) are shown in the bottom panels. (B) Similar to (A), except that the T7-ΔNp63 construct was used. (C) H1299 cells were transfected with Hsp70-siRNA-2 or control-siRNA constructs. 40 h later, the cells were further transfected with a plasmid expressing HA-Ub. Lysates were immunoprecipitated with a TAp63-specific (Poly 618902) antibody and analyzed by western blotting with an HA-specific (12CA5) antibody to detect ubiquitinated TAp63. Western blots for TAp63, Hsp70, CHIP, and actin are shown in the lower panel. (D) Similar to (C), except that SCC9 cells and an ΔNp63-specific (Poly 619002) antibody were used. (E) H1299 cells were transfected with plasmids expressing CHIP, HA-Ub, or empty vector. Thirty hours later, the cells were treated with Ver-155008 (25 μM), an inhibitor of Hsp70, and immunoprecipitated with a TAp63-specific (Poly 618902) antibody and analyzed by western blotting with an HA-specific (12CA5) antibody to detect ubiquitinated TAp63. The western blots for TAp63, Hsp70, CHIP and actin are shown in the lower panel. (F) Similar to (E), except that SCC9 cells and an ΔNp63-specific (Poly 619002) antibody were used. (G) CHIP promotes TAp63 and ΔNp63 ubiquitination in vitro. Western blot of the in vitro ubiquitination reaction probed with a Ub-specific antibody (BD, Biosciences) to detect ubiquitinated p63. Direct Western blots for CHIP (anti-GST) and p63 (anti-His) are shown in the lower panel. (H) Western blot analysis of a coupled in vitro ubiquitination-immunoprecipitation. After the in vitro ubiquitination, the samples were immunoprecipitated with a TAp63-specific (Poly 618902) antibody and analyzed by western blotting with a ubiquitin-specific antibody (550944, upper image). Direct western blots for TAp63 (Poly 618902), CHIP (H-231), and Hsp70 (C92F3A5) are shown in the lower panels. (I) Similar to (H), except that the reaction was immunoprecipitated with an ΔNp63-specific (Poly 619002) antibody as indicated. All experiments were performed in triplicate.
Ver-155008 is a small molecule inhibitor of Hsp70 (55–57). We next investigated whether the inhibitory effect of Ver-155008 on Hsp70 affected CHIP-mediated p63 ubiquitination. Plasmids expressing CHIP and HA-Ub were co-transfected into H1299 cells. The transfected cells were treated with increasing amounts of Ver-155008 (5–10 μM), immunoprecipitated with TAp63-specific antibodies, and analyzed by western blotting with an anti-HA antibody to detect ubiquitinated TAp63. As shown in Figure 5E, ubiquitination of TAp63 was significantly decreased in the presence of Ver-155008. As expected, TAp63 protein levels were increased. Consistently, the levels of Hsp70 expression were unchanged after Ver-155008 treatment. Ubiquitination of ΔNp63 was markedly increased, accompanied by decreased protein levels of ΔNp63 in SCC9 cells (Figure 5F). To determine whether TAp63 or ΔNp63 could be a substrate for CHIP-dependent ubiquitination in vitro, we performed an in vitro ubiquitination assay (41,44). Affinity purified GST-CHIP or GST alone, along with His-TAp63 or His-ΔNp63, was added to bacterial extracts containing recombinant E1 and E2 (UbcH5b). As shown in Figure 5G, CHIP promoted the ubiquitination of TAp63 and ΔNp63 in vitro. We next determined whether CHIP mediated p63 ubiquitination is regulated by Hsp70 in vitro. CHIP, TAp63, and ubiquitin co-incubated with His-Hsp70 or His-protein alone were immunoprecipitated with TAp63-specific antibodies, analyzed by western blotting with Ub-specific antibodies to detect ubiquitinated TAp63 protein. Addition of Hsp70 increased the ubiquitination of TAp63 but did not further reduce TAp63 protein levels, suggesting that the decrease in the level of TAp63 mediated by CHIP had peaked in this in vitro system (Figure 5H). In contrast, Hsp70 reduced ΔNp63 ubiquitination and resulted in increased levels of ΔNp63 (Figure 5I). The TAp63 and ΔNp63 immunoblots revealed that CHIP promotes multi-ubiquitination of p63 in vitro (Figure 5H and I, lower images). Together, our results revealed that Hsp70 plays a key role in regulating CHIP-mediated p63 ubiquitination.
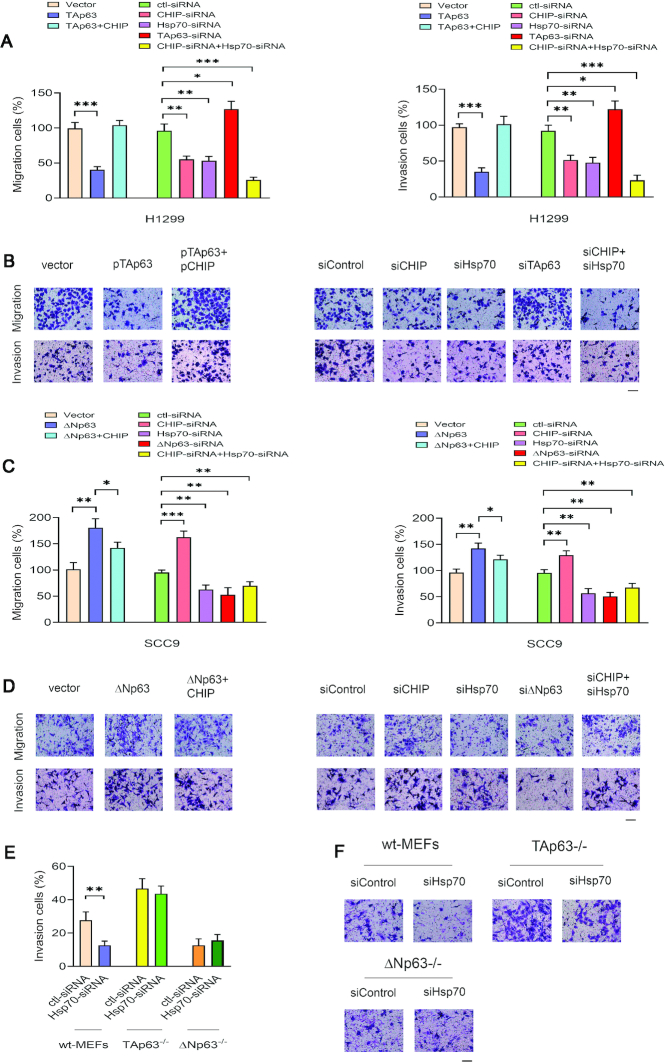
Regulation of p63 by Hsp70-CHIP axis contributes to cancer cell migration and invasion
To determine the role of TAp63 in cell scattering, a cell scattering assay was performed (48). As shown in Supplementary Figure S6A (upper panel), scattering was significantly enhanced in cells with TAp63 depletion by siRNA compared to cells treated with control-siRNA, suggesting the involvement of TAp63 in regulating cell scattering. Hepatocyte growth factor (HGF) was reported as a scatter factor for its ability to induce the spread of cancer cells (58). HGF induced scattering of H1299 cells; however, cell scattering was only slightly increased in TAp63-depleted cells with HGF treatment compared to cells transfected with control-siRNA treated with HGF (Supplementary Figure S6A, lower panel). We investigated whether CHIP affects the tumor-suppressive properties of TAp63. Endogenous CHIP was depleted in H1299 cells by two CHIP-siRNAs, which targeted different regions of the CHIP gene. The scattering was significantly reduced in cells with CHIP depletion by two CHIP-siRNAs compared to cells treated with control-siRNA (Supplementary Figure S6B, upper panel). Similar results with HGF treatment were observed in H1299 cells with CHIP-knockdown (Supplementary Figure S6B, lower panel). The scattered cells were counted and plotted (Supplementary Figure S6A and S6B, right panel). Depletion of CHIP by siRNA in H1299 cells resulted in a significant increase of TAp63 protein level. These data were reproducible with two independent CHIP-siRNAs and with TAp63 depletion by siRNA, indicating that the reduction in cell scattering is in a TAp63-dependent manner (Supplementary Figure S6C).
Metastasis is a multi-step process that includes the migration and invasion of cancer cells, and it is associated with 90% of cancer deaths (1–3). TAp63 is a suppressor of metastasis (6–8). We next determined whether Hsp70 affects tumor dissemination by regulating CHIP-mediated p63 degradation. We examined whether overexpression of CHIP or depletion of CHIP, or depletion of Hsp70 by siRNA affects the migration and invasion of cancer cells. Plasmids expressing empty vector, TAp63 alone, or TAp63 and CHIP were transfected into H1299 cells. We performed migration and invasion assays in vitro (47,59). Expression of TAp63 pronouncedly decreased the ability of H1299 cells to migrate; co-expression of TAp63 and CHIP significantly increased the migration of these cells (Figure 6A, left). Furthermore, overexpression of TAp63 decreased the invasion of these cells, and co-expression of CHIP and TAp63 increased the invasive ability of these cells (Figure 6A, right). To determine the biological significance of CHIP or Hsp70 targeting TAp63 or ΔNp63 in cancer cells, we examined the migration and invasion of H1299 cells with CHIP and/or Hsp70 depletion. As shown in Figure 6A, a reduction in CHIP or Hsp70 significantly decreased the migration and invasion of cells by two-fold relative to those of H1299 control cells (vector alone); in contrast, depletion of TAp63 by siRNA produced the opposite effects. Representative images are shown (Figure 6B). Consistent with previous studies (12,13,60), expression of ΔNp63 significantly increased the migration and invasion of SCC9 cells, also, co-expression of ΔNp63 and CHIP in SCC9 cells reduced their migration and invasion relative to those of cells expressing ΔNp63 alone (Figure 6C, left). Depletion of CHIP by siRNA increased the migration and invasion of these cells because of the increased ΔNp63 protein level, and depletion of ΔNp63 by siRNA significantly decreased the migration and invasive capacities of these cells relative to those of SCC9 cells treated with control-siRNA (Figure 6C, right). As expected, Hsp70 depletion resulted in significantly decreased levels of cancer cell migration and invasion. Representative images are shown in Figure 6D. Our results demonstrate that inhibition of cancer cell migration and invasion by Hsp70 depletion (accompanied by high levels of TAp63 in H1299 cells and low levels of ΔNp63 in SCC9 cells) is dependent on the cell type or context.
Figure 6.
The Hsp70-CHIP axis regulates cancer cell migration and invasion. (A) H1299 cells were transfected with plasmids expressing empty vector, TAp63, and TAp63 together with CHIP, or the depletion of CHIP or Hsp70 or both CHIP and Hsp70 or TAp63 by siRNA. Transwell migration (uncoated) and invasion (Matrigel matrix-coated) assays were performed. The numbers of migration cells (left panel) or invasion cells (right panel) were quantified using ImageJ software. All experiments were performed in triplicate. The P-values were calculated by the Student's t-test. (B) Digital photomicrographs of migration cells or invasion cells. All images are representative of three independent experiments at a magnification of 200×. (C) Similar to the (A), except that SCC9 cells were used. (D) Representative images of SCC9 cells are presented as indicated. (E) Wild-type (wt) MEFs, or TAp63−/− MEFs, or ΔNp63−/− MEFs with depleted Hsp70 were subjected to Transwell invasion assays. The percentage of invasion cells was quantified from three independent experiments. (F) Representative images in MEFs cells are presented as indicated. Scale bar: 50 μM. *P < 0.05. **P < 0.01. ***P < 0.001.
TAp63−/− MEFs exhibited increased invasive ability compared to wt-MEFs (Figure 6E). Notably, Hsp70 ablation by siRNA significantly reduced the invasive ability of wt-MEFs; however, no change was observed in TAp63−/− MEFs and ΔNp63−/− MEFs, confirming that Hsp70 is involved in cancer cell invasion in a p63-dependent manner. Representative photographs of invaded MEFs are presented in Figure 6F.
Hsc70/Hsp70 proteins are considered to have an oncogenic function; however, the role of CHIP in cancers remains controversial. We next investigated the ability of CHIP or Hsp70 to immortalize primary mouse embryonic fibroblasts (MEFs). We analyzed the transforming ability of CHIP and Hsp70 by evaluating the capacity for anchorage-independent growth. CHIP and Hsp70 significantly increased the oncogenic capability of oncogenic ras in wt-MEFs (Supplementary Figure S6D). The data indicated that CHIP and Hsp70 could cooperate with the activated ras gene to transform MEFs. p53 suppressed focus formation and that more foci were formed in p53−/− MEFs than wild-type MEFs (44). Similarly, the ectopic expression of CHIP or Hsp70 alone significantly increased transformation in TAp63−/− MEFS compared to control cells (Supplementary Figure S6E, upper panel). However, co-expression of CHIP or Hsp70 with ras did not cause a further increase of foci formation in TAp63−/− MEFs (Supplementary Figure S6E, lower panel). Together, these data indicate that the transforming ability of CHIP or Hsp70 to cooperate with ras is mainly TAp63-dependent. The effects of CHIP or Hsp70, or in combination with ras on the transformation of MEFs, are summarized in Supplementary Figure S6F.
Hsp70 interferes with the transactivation activity of the TAp63 protein and TAp63-mediated apoptosis regulation in a cell type-dependent manner
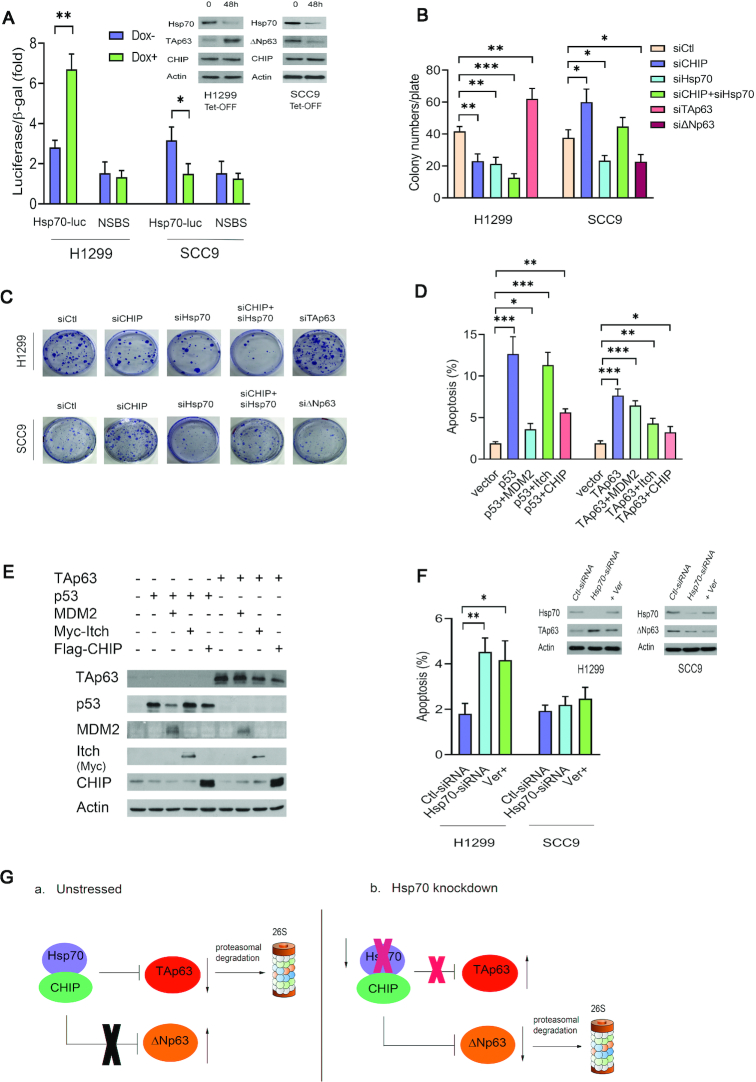
TAp63 and its isoforms are able to transactivate p63-responsive genes related to tumor suppression, such as p21 and Bax (7). Although ΔNp63 lacks the N-terminal transactivation domain, it is able to transactivate other targets through the function of a second transactivation domain (7,9–11). p63 can bind to p53 response elements (p53REs). However, the transcriptional activity of p63 is increased by at least two-fold when it is bound to p63 response elements (p63REs), which contain different DNA base pairs at the 5th and 16th positions compared to p53REs (61,62). Recent functional experiments have demonstrated that TAp63 specifically transactivates Dicer, Sharp-1 and Maspin, factors involved in suppressing metastasis (8,63,64). These studies strengthen the link between TAp63 and its tumor suppressor function. To determine the functional consequences of the interaction of CHIP with TAp63 or ΔNp63, we examined the effect of Itch, or CHIP expression on TAp63 mediated transcriptional activity. H1299 cells were co-transfected with a luciferase reporter construct (p21-Luc) containing the p53 binding site in the p21WAF1 promoter and with TAp63 or ΔNp63 with or without the E3 ligases Itch or CHIP (44). We chose the p21-Luc reporter because we used wild-type p53 as a control in these experiments. Similar to Itch, CHIP repressed TAp63 mediated transactivation activity (Supplementary Figure S7A). ΔNp63 appeared to activate p21-Luc reporter expression less efficiently than did TAp63. We examined the effect of overexpression of TAp63 or ΔNp63 and Itch or CHIP on cell proliferation. Cell proliferation ability was evaluated with a colony formation assay in vitro. As shown in Supplementary Figure S7B, cell proliferation was significantly inhibited by TAp63 and restored by co-expression of CHIP and TAp63, or co-expression of Itch and TAp63. However, proliferation was greatly enhanced by ΔNp63 and attenuated by co-expression of CHIP and ΔNp63 or co-expression of Itch and ΔNp63. Colony numbers were counted and plotted (Supplementary Figure S7C). A portion of each cell extract was lysed and analyzed by western blotting (Supplementary Figure S7D). We next transfected the Hsp70-Luc reporter into H1299 Tet-OFF cells or SCC9 Tet-OFF cells, as indicated. The reduction in Hsp70 expression was accompanied by high levels of TAp63 and increased luciferase activity of Hsp70-Luc, but not of the NSBS (nonspecific binding site) mutant-Luc reporter construct in the presence of doxycycline (Dox) in H1299 cells; however, in the presence of Dox in SCC9 cells, the reduction in Hsp70 expression resulted in a decrease in ΔNp63 levels, which led to a reduction in Hsp70-Luc activity (Figure 7A). Consistent with this finding, we observed that ΔNp63 protein levels were significantly decreased when Hsp70 levels were reduced in the presence of Dox in SCC9 cells (Figure 7A, top right image).
Figure 7.
Hsp70 is critical for the CHIP-mediated p63 function. (A) H1299 Tet-OFF-Hsp70 cells or SCC9 Tet-OFF-Hsp70 cells were transfected with an Hsp70-Luc reporter construct or a control reporter with a nonspecific binding site (NSBS) and treated in the presence or absence of doxycycline (1.0 μg/ml) for 48 h. Luciferase activity was subsequently measured. The protein levels of Hsp70, TAp63, or ΔNp63, CHIP expression were detected by western blotting with Hsp70-specific (C92F3A5), TAp63-specific (Poly 618902), or ΔNp63-specific (Poly 619002), and CHIP-specific (H-231) antibodies. (B) H1299 cells and SCC9 cells were transfected with CHIP-siRNA (siCHIP), Hsp70-siRNA (siHsp70), both siCHIP and siHsp70, TAp63-siRNA (siTAp63), or ΔNp63 (siΔNp63) as indicated. Colonies that were resistant to G418 were enumerated 15 days after drug selection. Data are means, and all experiments were performed in triplicate. (C) Representative images in H1299 cells and SCC9 cells are presented as indicated. (D) H1299 cells were transfected with plasmids expressing p53 or TAp63 alone, or in combination with MDM2 or with Itch or with CHIP. The inhibitory effect of MDM2, Itch, and CHIP on p53 or TAp63-dependent apoptosis was determined by annexin V staining and flow cytometry. Error bars indicate the SEM (n = 3). (E) The protein levels of p53, TAp63, MDM2, Itch and CHIP expression were visualized by western blotting with p53-specific (Pab1801), MDM2-specific (2A10), anti-Myc (9E10, for Itch), anti-Flag (M5, for CHIP) antibodies. (F) H1299 cells or SCC9 cells were treated with Hsp70-siRNA, or Ver-155008, a specific inhibitor of Hsp70. The apoptosis assays were performed. The protein levels of Hsp70, TAp63, or ΔNp63 were analyzed by western blotting with Hsp70-specific (C92F3A5), TAp63-specific (Poly 618902), and ΔNp63-specific (Poly 619002) antibodies. An antibody against actin was used as a loading control. All experiments were performed in triplicate. (G). The model we propose is shown in panel (A) In unstressed conditions, Hsp70 and CHIP form a complex. Hsp70 is required for CHIP-mediated TAp63 degradation. Hsp70 inhibits CHIP-mediated DNp63 degradation. (B) In the Hsp70 knockdown, CHIP is unable to degrade TAp63. Loss of Hsp70 leads to an increase the CHIP-ΔNp63 interaction and results in decreased levels of ΔNp63. *P < 0.05. **P < 0.01, ***P < 0.001.
To explore the roles of CHIP and Hsp70 in a more physiological setting, endogenous CHIP or Hsp70 was depleted with siRNA in H1299 or SCC9 cells. Consistently, the colony numbers were significantly reduced as a result of CHIP or Hsp70 depletion in H1299 cells. The colony numbers were further decreased by co-depletion of CHIP and Hsp70 in H1299 cells, suggesting that CHIP and Hsp70 are necessary for H1299 cell growth (Figure 7B). In contrast, in SCC9 cells, knockdown of CHIP significantly increased the colony numbers; however, depletion of Hsp70 decreased the colony numbers due to the low levels of ΔNp63. Representative colony formation assay results are presented in Figure 7C. Next, we investigated whether CHIP overexpression inhibits TAp63-dependent apoptosis. H1299 cells were transfected with p53 or TAp63 expression construct alone, or in combination with MDM2, or with Itch or with CHIP, as indicated. Annexin V staining was used to determine whether transient CHIP overexpression could rescue cells from TAp63-dependent cell death. Here, p53 and MDM2 were selected as positive controls (41). p53 and Itch were chosen as negative controls (6,45). As shown in Figure 7D, expression of TAp63 alone resulted in apoptosis, while apoptosis was largely prevented by co-expression of TAp63 and CHIP or TAp63 and Itch. Western blotting was used to visualize the ectopic expression levels of p53 or TAp63 and the E3-ligases, as indicated (Figure 7E).
Depletion of Hsp70 or repression of Hsp70 by Ver-155008 greatly enhanced apoptosis in H1299 cells, but no change was observed in SCC9 cells, confirming that Hsp70 is a key regulator that controls p63 function (Figure 7F). Induction of cell cycle arrest by TAp63 is vital for its tumor-suppression function. To determine whether CHIP can inhibit TAp63-induced G1 arrest, H1299 cells were transfected with plasmids expressing p53 or TAp63 alone, or in combination with these E3-ligases, as indicated. Cell cycle analysis by propidium iodide (PI) staining and flow cytometry was performed (41,44). The relative proportion of cells in each phase (G1, S and G2/M) of the cell cycle was determined by flow cytometry (Supplementary Figure S7E, left panel). An increase in the G1/S ratio is considered as an indicator of G1 arrest (41,44). Similar to overexpression of p53, overexpression of TAp63 increased the percentage of cells in G1 phase and decreased in the percentage of cells in S phase, resulting in an increase in the G1/S ratio from 1.6 to 2.6. Notably, co-expression of these E3 ligases with TAp63 showed a significant decrease in the G1/S ratio, indicating that like Itch or MDM2, CHIP significantly decreased TAp63-induced G1 arrest (Supplementary Figure S7E, right panel). Consistent with this result, increased G1 arrest due to increased endogenous TAp63 expression was observed in H1299 cells with depletion of CHIP or Hsp70 (Supplementary Figure S7F, left panel). CHIP or Hsp70 depletion by siRNA showed a significant increase in the G1/S ratio, indicating that the G1 arrest occurred in a TAp63-dependent manner (Supplementary Figure S7F, right panel). We proposed a model to elucidate the role of Hsp70 in the regulation of the CHIP-p63 axis in unstressed cells and Hsp70 knockdown cells (Figure 7G). Taken together, our findings reveal that Hsp70 is involved in the regulation of CHIP-mediated p63 transcriptional activation, growth suppression, and cell cycle checkpoint control.
DISCUSSION
TAp63, a homolog of the tumor suppressor p53, transactivates many p53 target genes leading to apoptosis or cell cycle arrest (5,11). Delta Np63 (ΔNp63/DNp63) lacks the N-terminal transactivation domain, which is found in TAp63. Although ΔNp63 cannot induce apoptosis, it can act in a dominant-negative manner to block the function of p53, TAp73 and TAp63 (7). ΔNp63 is overexpressed in many human cancers, including squamous cell carcinomas. TAp63 is generally considered to be a tumor suppressor, and ΔNp63 an oncogene. However, the molecular mechanism of p63 regulation remains unknown. We demonstrated, for the first time, that the chaperone protein Hsp70 acts as an integral molecular switch for controlling CHIP-mediated p63 ubiquitination and degradation.
CHIP is a critical negative regulator of TAp63 and ΔNp63 through the ubiquitin-proteasome system. CHIP, p63, and Hsp70 form a complex in cells. CHIP physically interacts with TAp63 and ΔNp63. The U-box of CHIP is required for its E3 ubiquitin ligase activity towards TAp63 and ΔNp63. Elevated protein levels of CHIP were observed in human squamous cell carcinoma cell lines and human prostate cancer patient tissues. Most strikingly, there was an inverse correlation between high levels of CHIP and low levels of TAp63 in these cancer cells and human tumor samples. Based on the online database (www.genomicscape.com), high CHIP and Hsp70 expression levels and low levels of TP63 expression were associated with a significantly decreased probability of overall survival in different types of human cancers.
CHIP is a particularly interesting E3 ligase involved in linking the ubiquitin-proteasome system to the heat-shock response system. It has been defined as having both oncogenic and tumor-suppressing functions. Our findings revealed that CHIP might have an oncogenic role via interaction with and negative regulation of TAp63. Hsp70 proteins are primarily considered to have oncogenic functions and are overexpressed in many invasive carcinomas (38). Although wild-type p53 forms transient complexes with Hsp70 (20), when CHIP is overexpressed, p63-Hsp70 complexes may become more stable as p53 is locked into the CHIP-mediated degradation pathway. The regulatory pathways through which Hsp70 acts as a switch to differentially modulate the degradation of p63 isoforms are complex. It is possible that when Hsp70 is depleted, TAp63 becomes inaccessible to CHIP, and ΔNp63 is more accessible to CHIP in different cell types. It is also possible that the two isoforms of p63 might have differential susceptibilities to folding/unfolding by Hsp70, which then modulates their susceptibility to degradation. Ver-155008 specifically interacts with the ATPase binding domain of Hsp70 and inhibits its activity (55–58). By targeting Hsp70, Ver-155008 inhibits cancer cell proliferation and induces apoptosis (55–58). The protein levels of Hsp70 were unchanged in several cancer cells (e.g. H1299, A549, H1975) after Ver-155008 treatment. However, Hsp70 protein levels were upregulated in PC12 cells treated with Ver-155008 (65–67). The change in Hsp70 protein levels in response to Ver-155008 treatment may be cell type or context-dependent. We found that the TAp63 protein level increased and that the ubiquitination of TAp63 was significantly reduced in H1299 cells after Ver-15008 treatment. In contrast, after treatment with Ver-155008, the ubiquitination of ΔNp63 increased significantly, while the protein level ΔNp63 was decreased in SCC9 cells. The complexity of these regulatory pathways needs to be further investigated.
One critical question raised is how TAp63 (a tumor suppressor) and ΔNp63 (an oncogene) are regulated by CHIP. The two apparently opposing functions of p63—promotion and inhibition of tumorigenesis—are intriguing and may be mediated by a novel cell growth regulatory mechanism in cells. A major discovery from our study is that Hsp70 acts as a fine-tuning switch that controls the CHIP-mediated degradation of TAp63 and ΔNp63. The effects of Hsp70 on regulating the CHIP-mediated function of p63 are multiple and included the following. (1) Depletion of Hsp70 by siRNA (or repression by Ver-155008, a specific inhibitor of Hsp70) resulted in decreased levels of TAp63 ubiquitination and increased levels of TAp63 protein in H1299 cells; in contrast, Hsp70 depletion resulted in increased levels of ΔNp63 ubiquitination and reduced levels of ΔNp63 protein in SCC9 cells. (2) Depletion of Hsp70 influenced the CHIP-p63 interaction in a cell type-dependent manner. (3) Consistent with those findings, Hsp70 overexpression increased TAp63 ubiquitination and decreased TAp63 protein levels; in contrast, ectopic Hsp70 expression reduced ΔNp63 ubiquitination and stabilized the ΔNp63 protein. (4) Control of p63 regulation by Hsp70 contributed to cancer cell migration and invasion. (5) Hsp70 was involved in CHIP-mediated functions of p63, including apoptosis and cell cycle arrest.
An autoregulatory feedback loop involving p53 and MDM2 is well established (68–70). Similar to MDM2 expression, Hsp70 expression is deficient in TAp63−/−MEFs but not in ΔNp63−/−MEFs. Loss of Hsp70 increased TAp63 levels. TAp63 transactivated Hsp70. Hsp70, in turn, promoted CHIP-mediated TAp63 degradation in a cell type-dependent manner. In contrast, the reduction in Hsp70 expression increased CHIP-ΔNp63 interactions and markedly reduced the levels of ΔNp63 in SCC9 cells. TAp63 significantly increased the reporter activity of Hsp70-Luc, but not the Hsp70mut-Luc reporter construct. In contrast, ΔNp63 activated the Hsp70-Luc reporter much less efficiently than did TAp63, suggesting that TAp63 and Hsp70 form an autoregulatory feedback loop. In summary, our findings provide an insight into the mechanism of p63 regulation in cells. Our data demonstrate that Hsp70 is a critical regulator of p63, which could be therapeutically targeted in human cancer. Thus, the outcome of this study has significant implications for improving cancer therapy.
DATA AVAILABILITY
All other relevant data supporting the findings of this study are available within the article and supplementary information files.
Supplementary Material
ACKNOWLEDGEMENTS
The Hsp70-siRNA constructs were generated by Hadeel Alyenbaawi in the lab. We thank the University of Alberta for research equipment and technical support for this research. We thank the Flow Core Facility at the University of Alberta for technical support.
Contributor Information
H Helena Wu, 370 Heritage Medical Research Center, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
Benfan Wang, 370 Heritage Medical Research Center, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
Stephen R Armstrong, 370 Heritage Medical Research Center, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
Yasser Abuetabh, 370 Heritage Medical Research Center, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
Sarah Leng, 370 Heritage Medical Research Center, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
Wilson H Y Roa, Department of Oncology, Cross Cancer Institute, 11560 University Ave., University of Alberta, Edmonton, Alberta T6G 1Z2, Canada.
Azeddine Atfi, Laboratory of Cell Signaling and Carcinogenesis, INSERM UMRS938, 184 Rue du Faubourg St-Antoine, 75571 Paris, France.
Adriano Marchese, Department of Pharmacology, Stritch School of Medicine, Loyola University Chicago, 2160 S. First Ave., Maywood, IL 60153, USA.
Beverly Wilson, Department of Pediatrics, University of Alberta, 11405 - 87 Ave., Edmonton, Alberta T6G 1C9, Canada.
Consolato Sergi, Department of Laboratory Medicine and Pathology (5B4. 09), University of Alberta, Edmonton, AB T6G 2B7, Canada.
Elsa R Flores, Department of Molecular Oncology, H. Lee Moffitt Cancer Center, 12902 Magnolia Drive, Tampa, FL 33612, USA.
David D Eisenstat, Department of Oncology, Cross Cancer Institute, 11560 University Ave., University of Alberta, Edmonton, Alberta T6G 1Z2, Canada; Department of Pediatrics, University of Alberta, 11405 - 87 Ave., Edmonton, Alberta T6G 1C9, Canada.
Roger P Leng, 370 Heritage Medical Research Center, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Women & Children's Health Research Institute (WCHRI); Canadian Institutes of Health Research (CIHR) (to R.P.L.); Saudi Arabia (to Y.A.); D.D.E. holds the Muriel & Ada Hole Kids with Cancer Society Chair in Pediatric Oncology, University of Alberta. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Steeg P.S. Targeting metastasis. Nat. Rev. Cancer. 2016; 16:201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta G.P., Massague J.. Cancer metastasis: building a framework. Cell. 2006; 127:679–695. [DOI] [PubMed] [Google Scholar]
- 3. Welch D.R., Hurst D.R.. Defining the hallmarks of metastasis. Cancer Res. 2009; 79:3011–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dötsch V., Andrews N.C., Caput D., McKeon F.. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998; 2:305–316. [DOI] [PubMed] [Google Scholar]
- 5. Yang A., McKeon F.. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell. Biol. 2000; 1:199–207. [DOI] [PubMed] [Google Scholar]
- 6. Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011; 18:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Armstrong S.R., Wu H., Wang B., Abuetabh Y., Leng R.. The regulation of tumor suppressor p63 by the ubiquitin-proteasome system. Int. J. Mol. Sci. 2016; 17:E2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Su X., Chakravarti D., Cho M.S., Liu L., Gi Y.J., Lin Y.L., Leung M.L., El-Naggar A., Creighton C.J., Suraokar M.B.et al.. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010; 467:986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flores E.R., Sengupta S., Miller J.B., Newman J.J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T.. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005; 7:363–372. [DOI] [PubMed] [Google Scholar]
- 10. Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A.. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999; 398:708–713. [DOI] [PubMed] [Google Scholar]
- 11. Flores E.R., Tsai K.Y., Crowley D., Sengupta S., Yang A., McKeon F., Jack T.. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002; 416:560–564. [DOI] [PubMed] [Google Scholar]
- 12. Keyes W.M., Pecoraro M., Aranda V., Vernersson-Lindahl E., Li W., Vogel H., Guo X., Garcia E.L., Michurina T.V., Enikolopov G.et al.. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011; 8:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang X., Lu H., Yan B., Romano R., Bian Y., Friedman J., Duggal P., Allen C., Chuang R., Ehsanian R.et al.. ΔNp63 versatilely regulates a broad NF-κB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011; 71:3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gatti V., Fierro C., Annicchiarico-Petruzzelli M., Melino G., Peschiaroli A.. ΔNp63 in squamous cell carcinoma: defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 2019; 13:981–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qian S.B., McDonough H., Boellmann F., Cyr D.M., Patterson C.. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006; 440:551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paul I., Ahmed S.F., Bhowmik A., Deb S., Ghosh M.K.. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene. 2013; 32:1284–1295. [DOI] [PubMed] [Google Scholar]
- 17. Nikolay R., Wiederkehr T., Rist W., Kramer G., Mayer M.P., Bukau B.. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 2004; 279:2673–2678. [DOI] [PubMed] [Google Scholar]
- 18. Koepp D.M., Harper J.W., Elledge S.J.. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999; 97:431–434. [DOI] [PubMed] [Google Scholar]
- 19. Ang X.L., Harper W.J.. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005; 24:2860–2870. [DOI] [PubMed] [Google Scholar]
- 20. Esser C., Scheffner M., Höhfeld J.. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J. Biol. Chem. 2005; 280:27443–27448. [DOI] [PubMed] [Google Scholar]
- 21. Zhou P., Fernandes N., Dodge I.L., Reddi A.L., Rao N., Safran H., DiPetrillo T.A., Wazer D.E., Band V., Band H.. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 2003; 278:13829–13837. [DOI] [PubMed] [Google Scholar]
- 22. Wang T., Yang J., Xu J., Li J., Cao Z., Zhou L., You L., Shu H., Lu Z., Li H.et al.. CHIP is a novel tumor suppressor in pancreatic cancer through targeting EGFR. Oncotarget. 2014; 5:1969–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Narayan V., Pion E., Landré V., Müller P., Ball K.L.. Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase CHIP. J. Biol. Chem. 2011; 286:607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahmed S.F., Deb S., Paul I., Chatterjee A., Mandal T., Chatterjee U., Ghosh M.K.. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 2012; 287:15996–16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jang K.W., Lee K.H., Kim S.H., Jin T., Choi E.Y., Jeon H.J., Kim E., Han Y.S., Chung J.H.. Ubiquitin ligase CHIP induces TRAF2 proteasomal degradation and NF-kappaB inactivation to regulate breast cancer cell invasion. J. Cell Biochem. 2011; 112:3612–3620. [DOI] [PubMed] [Google Scholar]
- 26. Gan L., Liu D.B., Lu H.F., Long G.X., Mei Q., Hu G.Y., Qiu H., Hu G.Q.. Decreased expression of the carboxyl terminus of heat shock cognate 70 interacting protein in human gastric cancer and its clinical significance. Oncol. Rep. 2012; 28:1392–1398. [DOI] [PubMed] [Google Scholar]
- 27. Wen J., Luo K.J., Hu Y., Yang H., Fu J.H.. Metastatic lymph node CHIP expression is a potential prognostic marker for resected esophageal squamous cell carcinoma patients. Ann. Surg. Oncol. 2013; 20:1668–1675. [DOI] [PubMed] [Google Scholar]
- 28. Liang Z.L., Kim M., Huang S.M., Lee H.J., Kim J.M.. Expression of carboxyl terminus of Hsp70-interacting protein (CHIP) indicates poor prognosis in human gallbladder carcinoma. Oncol. Lett. 2013; 5:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang L., Liu L., He X., Shen Y., Liu X., Wei J., Yu F., Tian J.. CHIP promotes thyroid cancer proliferation via activation of the MAPK and AKT pathways. Biochem. Biophys. Res. Commun. 2016; 477:356–362. [DOI] [PubMed] [Google Scholar]
- 30. Li F., Xie P., Fan Y., Zhang H., Zheng L., Gu D., Patterson C., Li H.. C terminus of Hsc70-interacting protein promotes smooth muscle cell proliferation and survival through ubiquitin-mediated degradation of FoxO1. J. Biol. Chem. 2009; 284:20090–20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai Q., Zhang C., Wu Y., McDonough H., Whaley R.A., Godfrey V., Li H.H., Madamanchi N., Xu W., Neckers L.et al.. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003; 22:5446–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B.. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019; 20:665–680. [DOI] [PubMed] [Google Scholar]
- 33. Flaherty K.M., DeLuca-Flaherty C., McKay D.B.. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990; 346:623–628. [DOI] [PubMed] [Google Scholar]
- 34. Mayer M.P., Schröder H., Rüdiger S., Paal K., Laufen T., Bukau B.. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 2000; 7:586–593. [DOI] [PubMed] [Google Scholar]
- 35. Kampinga H.H., Craig E.A.. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010; 11:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kityk R., Vogel M., Schlecht R., Bukau B., Mayer M.P.. Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 2015; 6:8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gong J., Weng D., Eguchi T., Murshid A., Sherman M.Y., Song B., Calderwood S.K.. Targeting the hsp70 gene delays mammary tumor initiation and inhibits tumor cell metastasis. Oncogene. 2015; 34:5460–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murphy M.E. The HSP70 family and cancer. Carcinogenesis. 2013; 34:1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nylandsted J., Rohde M., Brand K., Bastholm L., Elling F., Jäättelä M.. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frese S., Schaper M., Kuster J., Miescher D., Jäättelä M., Buehler T., Schmid R.A.. Cell death induced by down-regulation of heat shock protein 70 in lung cancer cell lines is p53-independent and does not require DNA cleavage. J. Thorac. Cardiovasc. Surg. 2003; 126:748–754. [DOI] [PubMed] [Google Scholar]
- 41. Leng R., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S.. Pirh2, a p53 induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003; 112:779–791. [DOI] [PubMed] [Google Scholar]
- 42. John L.S.S., Sauter E.R., Herlyn M., Litwin S., Adler-Storthz K.. Endogenous p53 gene status predicts the response of human squamous cell carcinomas to wild-type p53. Cancer Gene Ther. 2000; 7:749–756. [DOI] [PubMed] [Google Scholar]
- 43. Chakravarti D., Su X., Cho M.S., Bui N.H.B., Coarfa C., Venkatanarayan A., Benham A.L., González R.E.F., Alana J., Xiao W.et al.. Induced multipotency in adult keratinocytes through down-regulation of ΔNp63 or DGCR8. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E572–E581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu H., Pomeroy S.L., Ferreira M., Teider N., Mariani J., Nakayama K.I., Hatakeyama S., Tron V.A., Saltibus L.S., Spyracopoulos L.et al.. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med. 2011; 17:347–356. [DOI] [PubMed] [Google Scholar]
- 45. Rossi M., Aqeilan R.I., Neale M., Candi E., Salomoni P., Knight R.A., Croce C.M., Melino G.. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Du C., Wu H., Leng R.. UBE4B targets phosphorylated p53 at serines 15 and 392 for degradation. Oncotarget. 2016; 7:2823–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang B., Wu H., Chai C., Pichiorri F., Lewis J., Eisenstat D., Pomeroy S., Leng R.. MicroRNA-1301 suppresses tumor cell migration and invasion by targeting the p53/UBE4B pathway in multiple human cancer cells. Cancer Lett. 2017; 401:20–32. [DOI] [PubMed] [Google Scholar]
- 48. Muller P.A., Trinidad A.G., Timpson P., Morton J.P., Zanivan S., van den Berghe P.V., Nixon C., Karim S.A., Caswell P.T., Noll J.E.et al.. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2013; 32:1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bakkers J., Camacho-Carvajal J.M., Nowak M., Kramer C., Danger B., Hammerschmidt M.. Destabilization of DeltaNp63alpha by Nedd4-mediated ubiquitination and Ubc9-mediated sumoylation, and its implications on dorsoventral patterning of the zebrafish embryo. Cell Cycle. 2005; 4:790–800. [DOI] [PubMed] [Google Scholar]
- 50. Li Y., Zhou Z., Chen C.. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008; 15:1941–1951. [DOI] [PubMed] [Google Scholar]
- 51. Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K.I.. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001; 276:33111–33120. [DOI] [PubMed] [Google Scholar]
- 52. Sierra-Rivera E., Voorhees G.J., Freeman M.L.. Gamma irradiation increases Hsp70 in Chinese hamster ovary cells. Radiat. Res. 1993; 135:40–45. [DOI] [PubMed] [Google Scholar]
- 53. Calini V., Urani C., Camatini M.. Overexpression of HSP70 is induced by ionizing radiation in C3H 10T1/2 cells and protects from DNA damage. Toxicol. In Vitro. 2003; 17:561–566. [DOI] [PubMed] [Google Scholar]
- 54. Schilling D., Kühnel A., Konrad S., Tetzlaff F., Bayer C., Yaglom J., Multhoff G.. Sensitizing tumor cells to radiation by targeting the heat shock response. Cancer Lett. 2015; 360:294–301. [DOI] [PubMed] [Google Scholar]
- 55. Massey A.J., Williamson D.S., Browne H., Murray J.B., Dokurno P., Shaw T., Macias A.T., Daniels Z., Geoffroy S., Dopson M.et al.. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 2010; 66:535–545. [DOI] [PubMed] [Google Scholar]
- 56. Schlecht R., Scholz S.R., Dahmen H., Wegener A., Sirrenberg C., Musil D., Bomke J., Eggenweiler H.M., Mayer M.P., Bukau B.. Functional analysis of Hsp70 inhibitors. PLoS One. 2013; 8:e78443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumar S., Stokes J. 3rd, Singh U.P., Gunn K.S., Acharya A., Manne U., Mishra M.. Targeting Hsp70: a possible therapy for cancer. Cancer Lett. 2016; 374:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weidner K.M., Arakaki N., Hartmann G., Vandekerckhove J., Weingart S., Rieder H., Fonatsch C., Tsubouchi H., Hishida T., Daikuhara Y.et al.. Evidence for the identity of human scatter factor and human hepatocyte growth factor. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:7001–7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chai C., Wu H., Wang B., Eisenstat D.D., Leng R.. MicroRNA-498 promotes proliferation and migration by targeting the tumor suppressor PTEN in breast cancer cells. Carcinogenesis. 2018; 39:1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhuang Z., Xie N., Hu J., Yu P., Wang C., Hu X., Han X., Hou J., Huang H., Liu X.. Interplay between ΔNp63 and miR-138-5p regulates growth, metastasis and stemness of oral squamous cell carcinoma. Oncotarget. 2017; 8:21954–21973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ortt K., Sinha S.. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 2006; 580:4544–4550. [DOI] [PubMed] [Google Scholar]
- 62. Perez C.A., Ott J., Mays D.J., Pietenpol J.A.. p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm. Oncogene. 2007; 26:7363–7370. [DOI] [PubMed] [Google Scholar]
- 63. Zou Z., Anisowicz A., Hendrix M.J., Thor A., Neveu M., Sheng S., Rafidi K., Seftor E., Sager R.. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994; 263:526–529. [DOI] [PubMed] [Google Scholar]
- 64. Tan E.H., Morton J.P., Timpson P., Tucci P., Melino G., Flores E.R., Sansom O.J., Vousden K.H., Muller P.A.J.. Functions of TAp63 and p53 in restraining the development of metastatic cancer. Oncogene. 2014; 33:3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prince T., Ackerman A., Cavanaugh A., Schreiter B., Juengst B., Andolino C., Danella J., Chernin M., Williams H.. Dual targeting of HSP70 does not induce the heat shock response and synergistically reduces cell viability in muscle invasive bladder cancer. Oncotarget. 2018; 9:32702–32717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu F., Lin D., Jiang W., Meng L., Xu Y., Wang C., Wang X., He H., Xu D., Zhu Y.. HSP70 inhibitor VER155008 suppresses pheochromocytoma cell and xenograft growth by inhibition of PI3K/AKT/mTOR and MEK/ERK pathways. Int. J. Clin. Exp. Pathol. 2019; 12:2585–2594. [PMC free article] [PubMed] [Google Scholar]
- 67. Wen W., Liu W., Shao Y., Chen L.. VER-155008, a small molecule inhibitor of HSP70 with potent anti-cancer activity on lung cancer cell lines. Exp. Biol. Med. 2014; 239:638–645. [DOI] [PubMed] [Google Scholar]
- 68. Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997; 88:323–331. [DOI] [PubMed] [Google Scholar]
- 69. Kruse J.P., Gu W.. Modes of p53 regulation. Cell. 2009; 137:609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hafner A., Bulyk M.L., Jambhekar A., Lahav G.. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019; 20:199–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other relevant data supporting the findings of this study are available within the article and supplementary information files.