Abstract
We use single-molecule techniques to characterize the dynamics of prokaryotic DNA repair by non-homologous end-joining (NHEJ), a system comprised only of the dimeric Ku and Ligase D (LigD). The Ku homodimer alone forms a ∼2 s synapsis between blunt DNA ends that is increased to ∼18 s upon addition of LigD, in a manner dependent on the C-terminal arms of Ku. The synapsis lifetime increases drastically for 4 nt complementary DNA overhangs, independently of the C-terminal arms of Ku. These observations are in contrast to human Ku, which is unable to bridge either of the two DNA substrates. We also demonstrate that bacterial Ku binds the DNA ends in a cooperative manner for synapsis initiation and remains stably bound at DNA junctions for several hours after ligation is completed, indicating that a system for removal of the proteins is active in vivo. Together these experiments shed light on the dynamics of bacterial NHEJ in DNA end recognition and processing. We speculate on the evolutionary similarities between bacterial and eukaryotic NHEJ and discuss how an increased understanding of bacterial NHEJ can open the door for future antibiotic therapies targeting this mechanism.
INTRODUCTION
Non-homologous end joining (NHEJ) is a ligation process in which two free DNA ends, resulting from a double-strand break (DSB), are held in close proximity and then directly joined by core and accessory proteins without the need of a template (1–3). First discovered and characterized in Homo sapiens, eukaryotic NHEJ begins with threading of the heterodimeric, doughnut shaped Ku70/80 on each of the broken DNA ends. There it acts as a hub to directly or indirectly recruit downstream components, including the scaffolding proteins DNA-PKcs, PAXX and XLF, as well as the XRCC4-Ligase IV ligation complex (4). The requirements for the different components remain under debate as genetic data are obtained in backgrounds, where redundant pathways can obscure outcomes. Furthermore, biochemical data have traditionally required elevated protein concentrations to detect signals, conditions in which partial redundancies between components can again obscure outcomes. The downstream eukaryotic NHEJ proteins possess high mechanistic flexibility and have been reported to join a variety of different DNA substrates at about the same efficiency, thus manifesting the robustness and adaptability of the repair mechanism (5). Some results indicate that the DNA-PK holoenzyme, composed of dimeric Ku and DNA-PKcs, is the first complex capable of forming a ‘molecular synapsis’ between broken DNA ends (6–8). Importantly however, some ligation can occur in the absence of DNA-PKcs (9), a condition similar to the NHEJ systems found in prokaryotes or lower eukaryotes, such as Bacillus subtilis or yeast, which only possess Ku and use it to successfully carry out NHEJ (10). This leaves open the possibility that Ku may in fact be the central component in the formation of a molecular synapsis.
NHEJ has been demonstrated to be important for DSB repair in many, but not all, prokaryotic systems (11). In prokaryotic NHEJ, the homodimeric Ku and the ATP-dependent Ligase D (LigD) appear to be the only core factors involved in DSB repair (1,2). The fact that many bacterial Ku genes are known to be organized into operons also encoding LigD suggests that prokaryotic Ku and LigD form a species-specific NHEJ complex (11). This is further substantiated by the inability of eukaryotic Ku to stimulate prokaryotic LigD activity (2). The main features of the bacterial NHEJ machinery have previously been characterized for several different bacterial species, underscoring hallmarks of the repair process, such as protein binding events as well as their individual and joint activities on DNA (2,3,12–14). The proposed model for bacterial NHEJ is that Ku binds to the free ends, protects them from nuclease activity, and then recruits the multifunctional LigD that can process and ligate the free DNA ends (3). The multifunctional LigD is the main responsible protein for DNA-end processing, but it is anticipated that Ku modulates the order and extent of the resection by staying bound to the DNA post-recruitment of LigD (12). The prokaryotic Ku homologs display high similarity, and harbor the conserved core region of the human Ku70/80 heterodimer (13). Most bacterial Ku-homologs also have a C-terminal tail that varies in length and sequence between species. McGovern et al. demonstrated that the inner region of this tail in Ku from B. subtilis is responsible for recruiting LigD and further that the very end of the tail is used to control the threading of Ku on DNA ends and also to increase the concentration of Ku close to DNA by binding to the DNA backbone (15). The apparent minimalistic nature of the bacterial NHEJ process suggests that the two proteins involved possess multiple different functions in comparison to their homologs in eukaryots.
Little is known about the dynamics of the critical intermediate steps in bacterial NHEJ. Nevertheless the kinetics of the process is crucial to cell survival: cells that emerge from stationary phase or quiescence and initiate replication have no additional chromosome with which to carry out homologous recombination, yet must rapidly repair any DSBs that may emerge in the path of the replication fork. Single-molecule techniques enable the detection of transient intermediates that can aid in the understanding of such critical steps, which are difficult to assess in traditional bulk assays due to averaging effects. Here we use two complementary single-molecule techniques that are particularly well suited for interactions involving DNA ends to consecutively characterize the dynamics of the four main steps of NHEJ in B. subtilis, end-recognition by Ku, recruitment of LigD by Ku, ligation of the DSB, and disassembly of the complex post ligation. Molecular DNA forceps (8,16) reveal the dynamic nature of the synapsis between the two free DNA ends in the presence of proteins, while nanofluidics allows visualization of interactions between DNA and proteins by stretching the DNA in narrow channels (17). We show that Ku alone can form a weak synapsis on blunt-ended DNA that is stabilized when LigD is present in a manner that depends on the C-terminal arms of Ku. The stability of the synapsis is dramatically enhanced for 4 nt complementary overhangs. We also demonstrate that Ku, and potentially also LigD, remain bound to the site of repair for hours after the repair is completed, suggesting that there is an active system for removing the NHEJ components post ligation in vivo.
MATERIALS AND METHODS
Computational analysis
The structural model of the Ku homodimer from B. subtilis was generated following a comparative modeling protocol using the eukaryotic Ku70/80 heterodimer as a structural template. HHpred provided an initial sequence alignment between B. subtilis wild-type Ku (Kuwt) and the H. sapiens Ku70/80 heterodimer (PDB code: 1jey) sequences, sharing only 15% sequence identity (18). RosettaCM was used to build a structural model using the original protocol complemented with protocols for symmetry constraints (19) between the two B. subtilis Kuwt monomers and a set of distance restraints to improve the pairing of β-strands in the region spanning residues 45 to 80 (19,20). Locally, the sequence alignment was adjusted iteratively to match the large deletion existing between bacterial and eukaryotic sequences. Evolutionary rates were mapped at the surface of the model using the rate4site algorithm and a multiple sequence alignment of B. subtilis Kuwt homologs obtained after a blast search on UniprotDB, followed by a re-alignment of full-length homolog sequences by MAFFT selecting the top 150 first homologs after redundancy reduction <95% sequence identity (21,22). A model of B. subtilis Ligase D (LigD) was obtained using the same comparative modeling protocol as for Kuwt using PDB templates 6NHX for the 1–310 region and 5DMU for the 320–600 region both sharing 25% and 29% sequence identity with LigD, respectively. Covariation analyses were performed using the contact prediction server RaptorX and using as query the full Kuwt sequence and either full LigD sequence or the polymerase domain only (region 320–611) (23). The generated co-alignments contained 2575 and 1000 sequences for Kuwt versus the polymerase domain of LigD in the phylogenic or genomic modes, respectively, while they contained 2900 and 1265 sequences when the full-length sequence of LigD was used.
Protein expression and purification
The codon optimized DNA sequences for Kuwt, encoded by the ykoV gene in B. subtilis and the C-terminal truncated variant Kucore, corresponding to M1 to K253 of the full-length protein, were cloned into separate pET His1a plasmids under control of the T7-promoter and with kanamycin resistance as a selection marker. A His6-tag was added to the 5′ ends of the Kuwt and Kucore open reading frames (Supplementary methods). All construct design and cloning procedures were performed by the Protein Expertise Platform at Umeå University (Umeå, Sweden). Plasmid pSMG217, encoding LigD with a His6-tag on the C-terminal, was gratefully received from François Lecointe (Micalis Institute, Paris, France).
Protein expression
Plasmid constructs encoding Kuwt, Kucore and LigD were transformed into Escherichia coli BL21(DE3) (Invitrogen) and transformants were grown over-night on LB-agar plates supplemented with 50 μg/ml kanamycin. O/n-cultures were started by inoculating a single colony in LB medium (Merck) supplemented with 50 μg/ml kanamycin, followed by incubation at 37°C, 200 rpm. The o/n-cultures were diluted in 3 liters of fresh LB-medium supplemented with 50 μg/ml kanamycin and incubated at 37°C, 200 rpm until OD600 = 0.5, whereafter the temperature was lowered to 30°C for Kuwt and Kucore and 16°C for LigD. Protein expression was induced at OD600 = 0.7 by adding 0.5 mM IPTG (Thermofisher Scientific), followed by incubation for 3 h for Kuwt and Kucore and 18 h for LigD. The cells were harvested by centrifugation in a swinging bucket rotor centrifuge (30 min, 4°C, 4200 g) and stored at –80°C until further handling.
Protein purification
All subsequent steps were carried out at 4°C. The Kuwt and Kucore cell pellets were resuspended in 50 ml pre-chilled Buffer A (50 mM Tris–HCl pH 8, 1 M NaCl) and the LigD pellet in Buffer B (50 mM Tris–HCl pH 8, 0.2 M NaCl). A cOmplete mini protease inhibitor tablet (Roche) was added before disrupting the cells by sonication for 10 min (5 s sonication + 10 s pause) at an amplitude of 70%. The homogenized cell suspensions were centrifuged (1 h, 4°C, 18 500 g) and the supernatants were passed through a 0.22 μm Nalgene filter (Thermofisher Scientific). Imidazole (Merck) was added to a final concentration of 20 mM before loading onto a 5 ml HisTrap FF Ni-NTA column (GE Healthcare). An Äkta purifier (GE Healthcare) was used to perform a gradient elution over 20 column volumes with a gradual increase of imidazole starting from 20 mM to 1 M in Buffer A for Kuwt and Kucore and Buffer B for LigD. The Kuwt and Kucore proteins were loaded on a size exclusion column (Superdex 16/600 200 pg, GE Healthcare), pre-equilibrated with Buffer A, followed by an isocratic elution. The column was then re-equilibrated with Buffer D (100 mM Tris–HCl pH 8, 0.4 M NaCl, 2 mM DTT) and the purified protein was once again eluted in the same way. The pooled fractions containing LigD from the Ni-NTA purification were diluted with 50 mM Tris–HCl, pH 8, to obtain a final concentration of 75 mM NaCl. The diluted protein was loaded onto a 5 ml Hitrap Heparin HP column (GE Healthcare), pre-equilibrated with Buffer C (50 mM Tris–HCl pH 8, 75 mM NaCl) and a gradient elution was performed over 15 column volumes by the proportion of NaCl to a final concentration of 1 M. The eluted protein was loaded on a size exclusion column (Superdex 16/600 200 pg, GE Healthcare), pre-equilibrated with Buffer D, followed by an isocratic elution. The purified proteins were finally concentrated using an Amicon Ultra Centrifugal Filter (Merck), followed by the addition of glycerol to obtain a final buffer composition of 50 mM Tris–HCl pH 8, 0.2 M NaCl, 1 mM DTT, 50% glycerol. The proteins were stored at –20°C.
The human Ku70/80, PAXX and DNA-PKcs proteins were gratefully received from the research group of Tom Blundell (Cambridge) and prepared as described elsewhere (24,25).
Fluorescent labelling of Ku
A plasmid construct was designed, where a His6-tag and a DNA sequence encoding the SNAP-tag was added on the N-terminal of the full-length Kuwt sequence (Supplementary methods). The fusion protein was expressed and purified as described above, except for the final size exclusion chromatography, which was done with Buffer B supplemented with 2 mM DTT. The fusion protein was functionalized with an Alexa-647 fluorophore by mixing 5 μM of protein and 10 μM SNAP-Surface® Alexa Fluor® 647 (NEB) in a total volume of 2 ml followed by incubation at room temperature for 3 h. The labelled protein was separated from non-reacted substrate by size exclusion chromatography using a Superdex 16/600 200 pg column (GE Healthcare), pre-equilibrated with Buffer B supplemented with 1 mM DTT. The degree-of-labelling was spectrophotometrically determined to 71% using a correction factor of 0.03 to account for the absorbance of Alexa-647 at 280 nm. The labelled protein (KuAF647) was stored at –80°C.
Ligation activity assay
100 ng of phosphorylated 1000 bp DNA (NoLimits, Thermofisher Scientific) was mixed with Ku at increasing monomer concentrations ranging from 0.2 to 1 μM in Buffer L (50 mM Tris–HCl pH 8, 30 mM NaCl, 5 mM MgCl2) and incubated for 30 min at 37°C. LigD was subsequently added at a final concentration of 0.1 μM in a total volume of 18 μl, followed by incubation at 37°C. The ligation reaction was quenched after 2 h by adding 2 μl of stop-buffer (5 mg/ml Proteinase K, 2% SDS, 0.1 M EDTA), incubated at 37°C for 30 min for complete protein digestion and loaded on a 0.8% agarose gel. The gel electrophoresis was run at 80 V for 90 min in 1× TAE buffer, post-stained with SYBR Gold (Invitrogen) and imaged using a ChemiDoc MP Imaging System (BioRad).
Electrophoretic mobility shift assay (EMSA)
Two pairs of complementary primer sequences, of which one sequence in each pair was labelled with 6-fluorescein amidite (6-FAM) on the 5′ end, were purchased from IdtDNA and ATDBio (Supplementary methods). The primers were combined to form one blunt-ended and one sticky-ended DNA substrate with a 4 nt overhang, which were subsequently loaded on a preparative 6% polyacrylamide gels (19:1 acrylamide: bisacrylamide) for further purification. The gels were run at 15 V/cm for 50 min in 0.5× TBE, followed by cutting out a thin gel-slice containing the DNA substrate. The gel-slice was fragmented in 10 mM Tris–HCl pH 7.4, 1 mM EDTA by mechanical stress and the DNA was allowed to diffuse into the soluble phase. Gel fragments were separated by centrifugation from the soluble phase containing the pure DNA substrate.
Protein was mixed at different concentrations (indicated in each figure) with 25 nM purified substrate DNA in 50 mM Tris–HCl pH 8, 30 mM NaCl, 1 mM EDTA and 10% glycerol in a total reaction volume of 10 μl. The samples were incubated at room temperature for 20 min before loaded on a 6% native polyacrylamide gel (19:1 acrylamide: bisacrylamide, 0.5× TBE). The electrophoresis was run at 15 V/cm for 40 min in 0.5× TBE and the gel was imaged using a ChemiDoc MP Imaging System (Bio-Rad) to visualize the fluorescently labelled DNA.
Single-molecule forceps
The juncture DNA was engineered as described elsewhere except that the leash was 610 bp instead of 690 bp (8). The DNA construct sequences are listed in Supplementary methods. Briefly, two pieces of 1500 bp DNA originating from Chromid-95 plasmid were connected by a 610 bp DNA leash, two ends of the juncture DNA were ligated to 1 kb biotin- and dig-labeled DNA through XbaI and SacI, allowing it to be further linked to streptavidin-coated magnetic beads and an anti-dig-coated glass surface, respectively. The juncture DNA process gives two DNA tips with length of 58 bp, that could be cleaved by Xmal/SmaI to generate 4 nt complementary overhangs or blunt ends, respectively.
The method to engineer the juncture DNA with 4 nt non-complementary overhangs was further modified from the above. The backbone was the same as the juncture DNA described above except the leash was 690 bp, and the length of the two DNA tips were 47 bp and 41 bp, respectively. The two DNA tips contained Nb.BbvCI (NEB) nicking endonuclease cleavage sites at the ends that generate 9 nt overhangs by Nb.BbvCI digestion. The two 9 nt overhangs, 5′GCTCTTACA and 5′GCCTCATAT, were ligated to their complementary oligos 4bp-overhang-1, 5′Pho-GGCCTGTAAGAGC and 4bp-overhang-2, 5′Pho-CCGGATATGAGGC with 1:100 molar ratio at room temperature for 3 h by DNA ligase (NEB).
All single-molecule forceps experiments were conducted at 34°C in buffer RB (20 mM K·HEPES pH 7.8, 100 mM KCl, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 0.05% Tween 20 and 0.5 mg/ml BSA). Bacterial NHEJ components were used at a concentration of 5 nM for all three proteins. The human NHEJ components were used at a concentration of 10 nM for PAXX and 1 nM for DNA-PKcs.
Single-molecule magnetic trapping data giving DNA extension as a function of time under automatic force-modulation cycles was obtained using the Picotwist software suite (Picotwist S.A.R.L). The typical modulation cycle was ∼120 s at 0.01 pN and ∼120 s at a higher pulling force of 1.4 pN. Time-traces were analyzed to extract the fraction of traction cycles for which, upon increasing the force, the DNA extension is initially observed to be shorter than the maximum extension, but then recovers maximum extension during that same high-force portion of the traction cycle (referred to as frequency of end-binding events). We also extracted all amplitudes of the end-binding events. Finally, for all those end-binding events, in which end-binding ruptured within the same traction cycle, the duration of the end-binding event was determined. Synapsis lifetimes were only analyzed for end-binding events, for which the change in DNA extension observed upon rupture was consistent with specific synapsis (i.e. within 3 standard deviations of the expected length of the amplitude at a specific pulling force). A complete summary of parameters for all single-molecule forceps experiments is provided in Supplementary Tables S1–S3.
Single-molecule nanofluidics
KuAF647 was used together with LigD to covalently join 10 000 bp phosphorylated DNA fragments (NoLimits, Thermofisher Scientific). 0.5 μM KuAF647 was incubated with 1 μg of DNA in Buffer L for 30 min at 37°C followed by the addition of LigD to a final concentration of 0.1 μM. The ligation reaction was carried out at 37°C for 90 min and the DNA intercalating dye YOYO-1 was subsequently added at a ratio of 1 dye molecule per 5 bp of DNA. The DNA was diluted 20 times and the buffer condition was adjusted to match Buffer S (10 mM Tris–HCl pH 7.8, 10 mM NaCl, 0.06% (w/w) SDS and 5 mM DTT), which was used as a standard buffer for all nanofluidics experiments, before loading on the nanofluidic chip. The in situ digestion of the DNA bound proteins was done using a previously described method (26), where Proteinase K was added to the DNA–protein complex, confined in a 300 × 130 nm2 reaction chamber, while recording the respective emissions from YOYO-1 and Alexa-647 for two min. The emission depletion of Alexa-647 was compared to that of reference complexes that were only subjected to photobleaching.
For the non-covalent DNA bridging experiments, Ku was mixed with 4 μM sticky-ended (12 nt complementary overhangs) λ-phage DNA (48 502 bp, Roche) or blunt-ended T7-DNA (39936 bp, York-bio) at a concentration of 33 and 40 nM respectively (corresponding to a ratio of 100 homodimers per DNA end) in 10 mM Tris–HCl pH 7.8, 10 mM NaCl and incubated at 37°C for 1 h. For the PciI-digested λ-phage DNA (4 nt complementary overhangs), 33 nM Kuwt was mixed with DNA at 4 μM. YOYO-1 was added at a 1:5 ratio and the sample was diluted to obtain the correct stretching conditions according to Buffer S. The YOYO-1 labelled molecules were confined in 100 × 100 nm2 or 100 × 150 nm2 nanofluidic channels and visualized on an inverted fluorescence microscope (Zeiss AxioObserver.Z1) equipped with a 63x oil immersion objective (NA = 1.46, Zeiss), a 1.6× optovar magnification changer, a Colibri 7 LED light source (Zeiss) and an iXon EMCCD camera (Andor). For single-channel imaging, blue light (469/38 nm) was used to excite the sample and the subsequent emission was passed through a single band pass filter (530/30 nm) before reaching the detector. For dual-channel imaging blue and red light (631/33 nm) was alternately used to excite YOYO-1 and Alexa-647, respectively. The emitted light was passed through a 90 HE multi bandpass emission filter (Zeiss) before reaching the detector. An LED trigger box was used to shorten the recording time. Up to 100 frames were recorded with an exposure time of 100 ms for the single-channel imaging. For the dual-channel imaging an exposure time of 20 ms was used for YOYO-1 and 300 ms for Alexa-647. The collected single-channel images were analyzed using a custom-written Matlab-based software to convert image stacks to kymographs, from which the mean extension of each molecule was calculated (27). The dual-color images were analyzed using the open source image processing program Fiji (28) and Matlab. Kymographs were generated for each channel and the relative distance between each fluorescent dot was determined using a custom written Matlab script.
Surface plasmon resonance (SPR)
Apparent binding affinity constants were determined using surface plasmon resonance on a Biacore X100 instrument (GE Healthcare). The complementary primer sequences (Supplementary methods), of which one was functionalized with biotin on the 5′ end, were combined and diluted in HBS-P buffer (10 mM HEPES pH 7.4, 0.15 M NaCl, 0.005% Surfactant P20). The DNA substrate was immobilized in one of two flow channels on a streptavidin coated sensor chip SA (GE Healthcare). Kuwt and Kucore were diluted in HBS-P buffer and their association to DNA was recorded by injecting protein at a flow rate of 30 μl/min for 180 s. The dissociation phase was observed for 600 s by flowing HBS-P buffer followed by surface regeneration using 0.2 M NaOH. Triplicates were run for each concentration and the responses were background corrected by subtracting the signal from the non-modified flow channel. The resulting sensograms were fitted to the standard kinetic interaction model provided by the Biacore system for interaction of an analyte with a heterogeneous ligand.
Photobleaching experiments
KuAF647 and λ-phage DNA were mixed in 10 mM Tris–HCl pH 7.8, 10 mM NaCl and 5 mM DTT and incubated for 1 h at 37°C. The final concentration of λ-DNA was 4 μM and of KuAF647 was either 16.5 or 33 nM (corresponding to a ratio of 50 and 100 homodimers per DNA end, respectively). Control samples with T7 DNA were also prepared with similar DNA and protein concentrations. KuAF647 was also incubated with a 50 bp double-stranded DNA substrate (10 nM DNA and 120 nM Ku) that was used as a control. The DNA was labeled with YOYO-1 (one YOYO-1 molecule per 5 basepairs). β-Mercaptoethanol (Sigma, 2% v/v) was added to minimize photo-nicking during imaging.
DNA-Ku complexes were stretched on activated microscope coverslips following published protocols (29–31) The glass coverslips were silanized by incubating them in a mixture of Allyltrimethoxysilane (ATMS, 95%, Sigma-Aldrich), (3-aminopropyl)triethoxysilane (APTES, ≥ 98%, Sigma-Aldrich) and Acetone (Sigma-Aldrich) in a 1:1:100 ratio for at least 30 min at room temperature. Next, the coverslips were cleaned with acetone and water and dried with nitrogen. A charged coverslip was placed on a Menzel-Gläser glass slide (Thermo Scientific) and 3.7 μl of the DNA sample was pipetted onto the edge of the coverslip. The capillary force between the activated coverslip and glass slide pulled the solution under and the DNA molecules were stretched and aligned on the surface of the glass. The edges were sealed with nail polish to prevent drying.
DNA-Ku complexes stretched on glass coverslips were imaged using ELYRA PS.1/LSM 780 inverted microscope (Zeiss) equipped with a Plan-Apochromat 63x oil immersion objective (NA = 1.4, Zeiss), an Argon-laser 458, 488, 514 nm (35 mW) and an Andor iXon 885 EMCCD camera. Images were acquired as 1000 frame time-series with 30 millisecond exposure for each frame. All imaging was carried out at the Center for Cellular Imaging at Gothenburg University.
Photobleaching steps analysis (PBSA) was performed in Python using the quickPBSA package (32). The trace analysis algorithm in quickPBSA involves trace extraction for regions of interest (ROI), preliminary step detection for each trace, and iterative refinement using Bayesian marginal posterior (33). The localization of ROIs in the microscopy image stacks was carried out in Fiji (28) using the plugin Spot Intensity Analysis Ver. 0.3 (Vale Lab, UCSF) and the coordinates of the ROIs were passed on to quickPBSA for trace extraction. After trace extraction, all traces were filtered using a Butterworth lowpass filter (implemented in Python using SciPy) to improve signal-to-noise ratio and step identification. The number of fluorophores identified in the first frame of image stacks were used to estimate the number of protein dimers in ROIs and are plotted as histograms.
RESULTS
Bacillus subtilis Kuwt and the C-terminally truncated derivative Kucore both generate stable DNA end synapsis
The ‘protein ring’ in the Ku dimer is conserved among all kingdoms of life. It has previously been reported that all prokaryotic Ku proteins comprise a central core-domain that displays a substantial level of homology with the eukaryotic Ku70 protein subunit (13,15). A 3D model of the homodimeric Kuwt from B. subtilis (Figure 1A) was computed by comparative modeling using the structure of the heterodimeric eukaryotic Ku70/80 complex as template and optimizing the model to account for the large sequence variations that exist between the bacterial and eukaryotic homologs (only 15% sequence identity). Despite the sequence divergence, the homodimeric model of Kuwt predicted a 20 Å hole in the complex core center through which the DNA can fit (34). An evolutionary analysis of bacterial Ku homologs was performed and the resulting residues were mapped to the surface of the Kuwt model, emphasizing a crown of conserved amino-acids surrounding the hole, including a significant number of positively charged residues (Supplementary Figure S1). This supports the notion that the ability of Ku to thread onto DNA ends is an evolutionarily conserved feature. With respect to Kuwt, Kucore refers to a C-terminal truncation that lacks the last helix colored in red in Figure 1A and the downstream 35 C-terminal disordered residues that have been proposed to be responsible for the interaction with LigD (15).
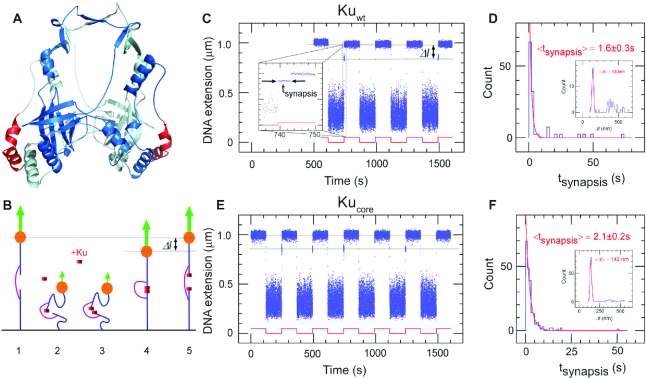
Figure 1.
The core structure of bacterial Ku forms short-lived synapsis of blunt DNA ends. (A) 3D model of the bacterial Kuwt homodimer, computed by comparative modelling to the eukaryotic Ku70/80 heterodimer. The red regions highlight the protruding C-terminal arms, which are deleted in the Kucore construct (downstream C-terminal tail of 35 residues predicted as disordered is not represented). (B) Schematic showing the molecular forceps experiment where 1.5 kb (blue) dsDNA segments with blunt ends are joined through a 600 bp dsDNA ‘bridge’ segment (magenta) anchored 59 bases from each end. The blue dsDNA segments are attached to a glass surface (black) and tethered to a magnetic bead (orange), respectively. When extended by a 1.4 pN force (green arrow) by a magnet and in the absence of interacting proteins (1) the DNA construct will have a maximal extension of ∼1 μm. (2) When the force is lowered to the femtoNewton range the ends can meet (3) and for instance be held together by Ku. This can be seen when (4) the DNA extension does not recover to 1 μm when the force is returned to its initial value: in this case the extension of the construct will be shorter by roughly 140 nm. (5) The construct will return to its initial extension upon disruption of the synapse. (C) Representative time-trace obtained upon force modulation (red) in the presence of Kuwt. Inset shows an expanded view of a rupture event of a single end-to-end synapsis. Δl corresponds to the change in DNA extension upon rupture, and tsynapsis the duration of the synaptic event. Events are indexed by coordinate pair (Δl, tsynapsis). (D) Lifetime distribution for end-specific events is fit to a single-exponential distribution (red), yielding a lifetime of 1.6 ± 0.3 s (SEM, n = 51). End-specific events are identified (inset) as having a Δl value within three standard deviations of the mean expected amplitude change given bridge mechanics (Gaussian fit in red, <Δl> = 140 ± 12 nm, SD, n = 99). (E, F), As for C and D but for Kucore. The mean lifetime is derived from a single-exponential fit giving a value of 2.1 ± 0.2 s (SEM, n = 212). Gaussian fit parameters for amplitude distribution are <Δl> = 142 ± 11 nm SD, n = 251).
The first step of prokaryotic NHEJ is the identification of the broken ends by Ku (11). We assessed the ability of Kuwt to form DNA end-to-end synapsis and the dynamics of the formed complex using single-molecule forceps (described elsewhere (8) and summarized in the Methods section and in Figure 1B). Blunt-ended DNA forceps were incubated with 5 nM Kuwt at a force of ∼0.01 pN to allow the synapsis formation and subsequently extended at 1.4 pN by magnetic tweezers. The resulting time-traces for Kuwt show that it sustained frequent synapsis between DNA ends (Figure 1C) with a mean synapsis life-time of 1.6 ± 0.3 s (Figure 1D). Similar results were obtained with Kucore (2.1 ± 0.2 s, Figure 1E, F). The lifetime of the synapsis was only weakly dependent on the force at which the complex was ruptured, changing by less than a factor of two for a nearly twentyfold increase in force, from 0.35 pN to 6 pN (Supplementary Tabe S2). These results establish Kuwt from B. subtilis as a mediator of DNA end-joining and imply that the core of the homodimer sustains this function without the aid of its C-terminal arms. This is in contrast to human Ku70/80 that cannot bridge DNA on its own (Supplementary Figure S2, (35)).
To elucidate whether the capability of Ku to bridge DNA is affected by the physical nature of the DNA ends, the same experiment was repeated using DNA forceps with 4 nt complementary overhangs on the 5′ end. In the absence of any protein, no interactions were observed between the ends of this construct (Supplementary Figure S3). For both Kuwt (Figure 2A) and Kucore (Figure 2B) synaptic interactions were readily observed, and we found that a majority of the synapses formed for this construct could not be torn apart at a stretching force of 1.4 pN within 1 h. We also performed the same experiment for 4 nt non-complementary overhangs, where we observed a synapsis life-time similar to that of blunt ends (Supplementary Figure S4). These results suggest that Ku-mediated synapsis permits the pairing of two complementary DNA ends and that the pairing, in turn, stabilizes the synapsis. Human Ku70/80 did not generate specific synapsis on its own on the complementary overhangs (Supplementary Figure S5). To obtain a synaptic life-time similar to bacterial Ku on blunt ends for human NHEJ we had to combine DNA-PK and PAXX (Supplementary Figure S6A–C, (8)). Interestingly, these human complexes were insensitive to the nature of the DNA ends (Supplementary Figure S6D-F), in stark contrast to bacterial Ku.
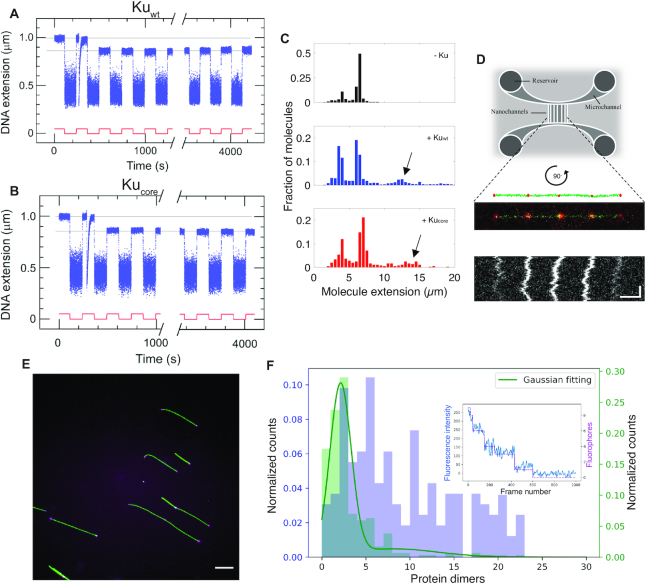
Figure 2.
The core structure of Ku forms long-lived synapsis of sticky DNA ends. Representative time-trace of molecular forceps experiment where a sticky-ended DNA construct with 4 nt complementary overhangs interacts with (A) Kuwt and (B) Kucore. A highly stable synapsis forms, which is not disrupted within 1 h of force-cycling at 1.4 pN. (C) Extension histogram of λ-DNA (black, top, N = 407), λ-DNA + Kuwt (blue, center, N = 1145) and λ-DNA + Kucore (red, bottom, N = 349) in 100 × 150 nm2 nanofluidic channels. Upon addition of protein at a ratio of 100 homodimers per DNA end, circularization and concatemerization of DNA is observed for both Ku variants. The peaks at ∼4 μm and ∼7 μm correspond to single circular and linear λ-DNA molecules, respectively. Longer extensions (indicated by arrows) correspond to concatemers of λ-DNA molecules. (D) Top: A schematic of the nanofluidic chip design, where the vertically aligned nanochannels (100 × 150 nm2) span the two horizontal microchannels. The samples are loaded in the circular reservoirs. Center: A schematic of four λ-DNA molecules (green) joined together by KuAF647 (red) confined in a nanochannel is shown together with the fluorescence microscopy image of a corresponding DNA–protein complex. Bottom: A kymograph shows the positions of the fluorescent KuAF647 over time. Horizontal and vertical scale-bars correspond to 5 μm and 5 s, respectively. (E) Representative fluorescence image of YOYO-1 labelled λ-DNA (green) bound with KuAF647 (magenta) stretched on glass cover slips. Scale-bar corresponds to 10 μm. (F) Normalized histograms for the number of KuAF647 protein dimers bound to λ-DNA (4 μM) at a protein concentration of 33 nM (purple) and a 50 bp blunt-ended DNA substrate (10 nM) in the presence of 120 nM KuAF647 (green). The histograms were constructed from the number of homodimers at different loci on DNA stretched on glass coverslips. The total number of fluorophores on all bound dimers were counted using photobleaching steps analysis. The number of fluorophores was normalized by the protein labeling efficiency (71%) to derive the number of protein dimers. The histogram corresponding to 50 bp DNA was fitted to a Gaussian mixture model (solid green line), from which the total number of homodimers was estimated to be 2.13 ± 1.21. Inset: a representative photobleaching curve (blue) over 1000 frames along with step-detection and enumeration of the number of fluorophores (magenta) A total of 9 fluorophores are observed for this particular λ-DNA-KuAF647 complex.
The formation of synapsis in the presence of Ku was further investigated using nanofluidic channels. Kuwt or Kucore was mixed with λ-DNA (48.5 kb) with 12 nt complementary 5′-overhangs at a ratio of 100 homodimers per DNA end (33 nM Ku and 4 μM DNA bp). The samples were confined in 100 × 150 nm2 nanofluidic channels and visualized by fluorescence microscopy. Importantly, since the DNA can be stretched in nanochannels without any attached handles, the DNA is extended simply due to the confinement, and thereby intermolecular DNA complexes can be readily investigated (36). The presence of either Kuwt or Kucore significantly increased the fraction of λ-DNA concatemers (Figure 2C), and circular λ-DNA (see (27), Supplementary Figure S7 and Supplementary methods) formed by intramolecular hybridization of the overhangs. Furthermore, imaging of fluorescently labeled KuAF647 was used to confirm the presence of the protein at the interface of the bridged DNA molecules. The equally spaced ‘dots’ show that KuAF647 was indeed present at the synapsis at an average distance of 6.8 μm, which correlates well with one λ-DNA molecule extended to ∼40% of its contour length (Figure 2D, additional examples in Supplementary Figure S8). The variation in emission intensity from the dots suggests that the number of protein units at each synapsis varies. We also observed dots at the ends of the molecules, and these were of similar emission intensity as the internal dots, suggesting that the number of protein units bound were similar, whether at synapses or at DNA ends. Concatemer formation was also observed for Kuwt when incubated with λ-DNA, digested with PciI to generate 4 nt complementary overhangs (Supplementary Figure S9). Hence, consistent with the single-molecule forceps data, Ku promotes annealing of complementary DNA ends.
To determine the number of proteins at each Ku locus we turned to single-fluorophore bleaching experiments. Figure 2E, F show results for λ-DNA incubated with 33 nM KuAF647. Taking into account the 71% labeling efficiency and that each KuAF647 homodimer can accommodate two labels, we observed that the number of Ku's per locus varied significantly. We observed some loci with two to three Ku homodimers bound, while other loci were occupied by up to 20 dimers. As a control, we used a 50 bp DNA substrate, where we know that up to three Ku homodimers can bind (see EMSA experiments below). 83% of all DNA–protein complexes on the 50 bp substrate that were photobleached were observed to be bound with an average number of homodimers corresponding to 2.13 ± 1.21. Importantly, these results suggest that as few as two homodimers were enough to form a stable synapsis. Results at a lower Ku concentration and for the blunt ended T7-DNA were similar to λ-DNA with KuAF647 at 33 nM (Supplementary Figure S10).
For blunt-ended T7-DNA (39.9 kb), no stable concatemers were observed in the nanofluidics experiments in the presence of either of the Ku variants. This is in agreement with the short-lived synapsis detected with the single-molecule forceps and with previous studies using TEM (15). However, for both Ku variants, we detected complexes in the nanofluidic channels in which two T7-DNA molecules were arranged with a region of overlap between their ends, giving rise to a local increase in fluorescence emission from the YOYO-1 labelled DNA at the center of the extended DNA–protein complex (Supplementary Figure S11). Similar overlapping complexes were occasionally also observed for sticky-ended λ-DNA-Ku complexes (not shown). This may suggest interaction, independent of the protruding C-terminal arms of Kuwt, between the exterior of the Ku core structure at the DNA ends and the sugar-phosphate backbone of the DNA.
The binding of multiple Ku homodimers to DNA is cooperative
The results in Figure 2D-F suggest that the number of proteins at each synapsis varies. To characterize this further, we investigated the DNA binding of bacterial Kuwt and Kucore to a blunt-ended 50 bp DNA substrate and a 50 bp substrate with a 4 nt overhang using EMSA (Supplementary Figure S12). The distinct ladder pattern indicates that complexes of increasing sizes were formed. No notable difference was observed for the different DNA substrates. For Kuwt it appears that up to three homodimers could be accommodated on the 50 bp substrate DNA, which agrees with the photobleaching experiments above. This suggests an approximative footprint of 15 bp, consistent with structural data for human Ku (37). For Kucore, up to four homodimers could be loaded onto the 50 bp DNA substrate, yielding a smaller estimated footprint of approximately 11 bp. Most probably, the 35-residue disordered extension at the C-terminal arms in the Kuwt homodimer obstructs further loading of protein dimers onto the DNA molecule, either through repulsive forces or steric hindrance. From the EMSA gels it also appears that the proportion of complexes with one bound homodimer did not increase as rapidly as the proportion of complexes with two bound homodimers. This suggests that the Ku-homodimers thread on a single DNA molecule in a cooperative manner, similar to the human Ku70/80 heterodimer (38). This is consistent with surface plasmon resonance (SPR) measurements, where a single-binding-site model was not adequate to describe the binding of Kuwt and Kucore to DNA. Instead, the obtained sensograms were fitted to a multisite model, which yielded two apparent equilibrium constants KD1 = 6.7 nM and KD2 = 9.4 pM for Kuwt (χ2 = 4.83). The corresponding values for Kucore are 9.3 nM and 330 pM, respectively (χ2 = 1.83, Supplementary Figure S13). Hence, while the first binding affinity was similar, the second was 30-fold stronger for Kuwt than for Kucore. The SPR data also yielded the kon and koff for the process, which demonstrated that a large fraction of the difference between the binding constants for the second Ku homodimer could be explained by a ∼20 times lower dissociation rate for Kuwt compared to Kucore (Supplementary Table S4). The deconvolved curves representing the binding of each protein unit to the DNA substrate displayed a typical 1:1 binding behavior for the first interacting protein and a sigmoidal binding pattern for the second protein, suggesting cooperative binding (Supplementary Figure S13C, D). These results show that the protruding C-terminal arms are not involved in the stabilization of the first Ku homodimer onto the DNA end, but that they are involved in the binding of the second protein unit to the DNA substrate.
LigD interacts strongly with the C-terminal arms of two Kuwt homodimers
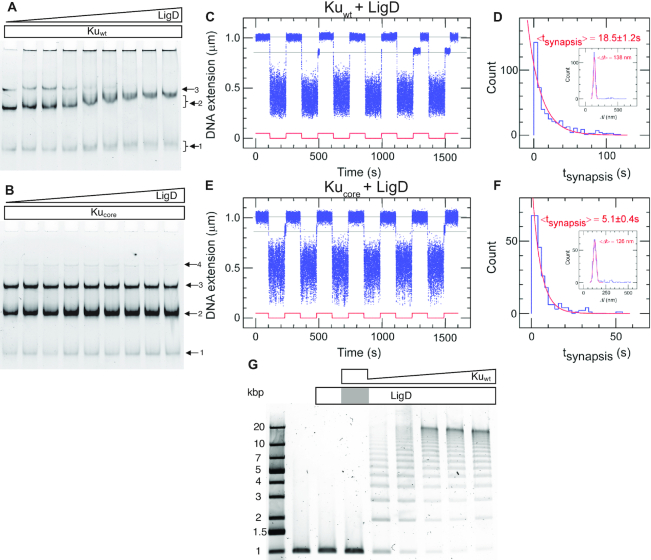
After binding and bridging of the DSB by Ku, the next step in the NHEJ process is the recruitment of LigD by Ku. It has previously been reported that there is a weak DNA-independent interaction between Kuwt and LigD and no detectable interaction between Kucore and LigD in solution (15). We combined each Ku variant (0.2 μM) with the 50 bp blunt-ended DNA substrate and titrated LigD at concentrations ranging from 0 to 0.8 μM. The resulting products were separated by EMSA. For Kuwt (Figure 3A) three bands appeared, corresponding to one, two and three Kuwt homodimers loaded on the DNA substrate, respectively. As the LigD concentration increased, a super-shift of the complex with two Kuwt homodimers was observed. This unsmeared and shifted 2-Kuwt band indicates a stable association of LigD to the Kuwt-bound DNA. A super-shift was also observed for the complex with only one Kuwt homodimer bound. The smeared nature and smaller magnitude of this band indicate a transient association of LigD, suggesting that the ligase prefers two Kuwt homodimers to stably ‘dock’ onto DNA. The size of the LigD-bound 3-Kuwt population was most probably too large to enter the gel and was hence stuck in the wells, which explains the reduction of the 3-Kuwt population at high LigD concentrations. The fact that the 3-Kuwt band started to disappear approximately at the same LigD concentration at which the 2-Kuwt band started to shift, suggests that LigD was attracted at approximately the same efficiency to the 3-Kuwt population as the 2-Kuwt population. For Kucore (Figure 3B) there is no detectable super-shift at increasing concentrations of LigD, manifesting the importance of the C-terminal arms for recruiting LigD to Ku-bound DNA in B. subtilis. This is in contrast to the Pseudomonas LigD, which is stimulated by Ku, truncated by up to 64 amino-acids at the C-terminal (39). The presence of unresolved DNA in the wells for Kuwt, but not for Kucore, further demonstrates the ability of LigD to form large DNA–protein complexes when interacting with Kuwt, in agreement with TEM imaging by McGovern et al. (15).
Figure 3.
The C-terminal arms of Ku are required for recruitment of LigD and full stabilization of the pre-ligation complex. EMSA gel of a 50 bp blunt-ended DNA substrate incubated with 0.2 μM (A) Kuwt and (B) Kucore and an increasing concentration of LigD (0–0.8 μM, 0.1 μM increments). The arrows indicate the number of Ku homodimers loaded on the DNA substrate. (C) Time-trace of molecular forceps experiment (blue), where a blunt-ended DNA substrate is used together with Kuwt (5 nM) and LigD (5 nM). End-to-end synapses were detected upon repeated force cycling (red). (D) Lifetime distribution of the specific end-binding events is fit to a single-exponential, giving a mean lifetime of 18.5 ± 1.2 s (SEM, n = 459). End-specific events are identified (inset) as having a Δl value within three standard deviations of the mean expected amplitude change given bridge mechanics (Gaussian fit in red, <Δl> = 138 ± 15 nm, SD, n = 486). (E, F) as C and D but for Kucore, giving a mean synapsis lifetime of 5.1 ± 0.4s (SEM, n = 270). End-specific events are identified (inset) as having a Δl value within 3 standard deviations of the mean expected amplitude change given bridge mechanics (Gaussian fit in red, <Δl> = 126 ± 16 nm, SD, n = 305). (G) Ligation of a 1000 bp blunt-ended DNA substrate with a constant amount of LigD (0.1 μM) and increasing concentration of Kuwt (0–1 μM, 0.2 μM increments). The grey area in the LigD bar indicates the absence of LigD in the corresponding sample, where only Ku is added at a concentration of 1 μM.
We next returned to the single-molecule DNA forceps to probe the dynamics of the synaptic complexes in presence of both Ku and LigD. The time-traces for Kuwt (Figure 3C) and Kucore (Figure 3E) in the presence of LigD show that synapses were formed with life-times of 18.5 ± 1.2 s (Figure 3D) and 5.1 ± 0.4 s (Figure 3F), respectively. LigD on its own could not generate synapsis on DNA with blunt ends (Supplementary Figure S14). The substantial increase in synapsis life-time for Kuwt in the presence of LigD shows that the ligase plays a central role in stabilizing the bridge between two Ku-bound DNA molecules, at least for blunt ends. The notable difference in synaptic stability observed in this experiment for Kuwt compared to Kucore further supports that the protruding C-terminal arms are important for the recruitment of LigD to the Ku-bound DNA ends. At the same time, the moderate increase in Kucore synaptic lifetime when LigD was added indicates that LigD has other weaker means of interacting with DNA-bound Ku than only through the C-terminal arms. We furthermore note that another metric can appear relevant here: the ‘synaptic efficiency’ which reflects the percentage of traction cycles, which resulted in a specific interaction event as defined by having the correct amplitude in the extension-change signal. It is a reflection of the equilibrium statistics of the system. Although this metric is subject to caution as it can vary with specific activity of a protein sample, it appears to increase notably from ∼5% in the presence of only Kuwt to ∼40% in the presence of LigD (Supplementary Table S1). We also note that the synaptic efficiency showed only a modest increase to ∼14% when LigD was combined with Kucore. It should be noted that it is unlikely that these variations reflect changes in association rates of the complexes as the increases in efficiency appear by and large to follow, and could well be explained by, the increases in synaptic lifetime.
After establishing the importance of the C-terminal tail for the interaction with Kuwt, we wanted to investigate where on LigD the tail binds. The C-terminal arm of Kuwt possesses a highly conserved surface patch, which could be responsible for the Ku–LigD interaction. A closer covariation analysis based on the ComplexContact server (23) (Supplementary Figure S15) between Kuwt and the polymerase domain of LigD revealed that a region of the C-terminal helix of Kuwt (residues 260–265), absent in the Kucore construct, constitutes some of the highest covariation signals with the Pol-domain of LigD (in the regions spanning residues 526–531 and 547–551). The strength of the signal was mild (between 0.3 and 0.4) corresponding to ∼40% chance that the prediction was correct, which was strengthened by biochemical evidence that the C-terminal arms of Kuwt, and in particular the region between residues 256 and 277, are important for LigD recruitment (15). Further support to this observation is the fact that the interaction between Ku and LigD has been shown to occur via the POL domain of LigD, at least in mycobacteria (40). This, along with the preferential loading of LigD onto DNA bearing two Kuwt homodimers, suggests that one LigD protein may be responsible for bridging and ultimately ligating two free DNA ends. The identification of this patch on LigD is analogous to the Ku-binding motifs identified for several human NHEJ partners (41).
The ligation activity is limited by the availability of Ku
When LigD is bound at the synapsis, the next step is the actual ligation of the two DNA ends. It is known that the NHEJ ligation activity is stimulated by the presence of Ku, and the protruding C-terminal arms of Kuwt play a crucial role in recruiting LigD to the repair site (15,42). However, the ligation efficiency has been reported to stagnate at elevated concentrations of LigD, suggesting that higher concentrations of the ligase may pose an inhibitory effect on the ligation process (15). To further examine this phenomenon, a ligation assay was performed using 1000 bp blunt-ended DNA, where Kuwt was titrated at concentrations ranging from 0.2 to 1 μM monomers, while keeping the LigD concentration constant at 0.1 μM. The resulting ligation product was separated on an agarose gel (Figure 3G). Upon increasing the concentration of Kuwt longer concatemers were formed by efficient ligation, while the ligation activity was completely lost in the presence of Kucore (Supplementary Figure S16). This shows that the ligation activity is strictly controlled by the availability of Kuwt. The lowest concentration of Kuwt in the titration series was set to 0.2 μM, roughly corresponding to 11 homodimers per each 1000 bp DNA molecule. Despite the excessive amounts of Kuwt relative to the substrate DNA, an unligated portion of DNA was observed, consistent with the proposition that Ku depletes during the course of ligation and that multiple Ku dimers bind to a single DNA molecule in a cooperative manner.
B. subtilis Ku threads onto DNA and remains immobile at the junction of two ligated DNA molecules
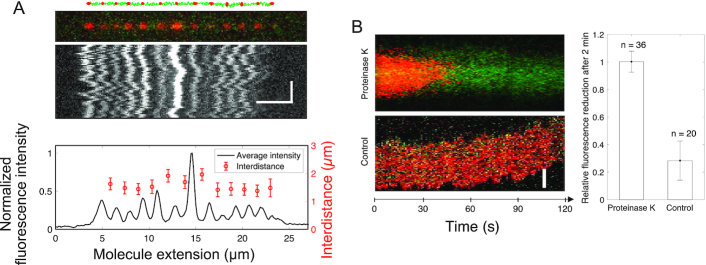
The results from the EMSA showed that Ku acts stoichiometrically at the synapses, suggesting that the protein remains bound after ligation is completed and is not recycled. In order to study the fate of the protein after ligation is completed, KuAF647 was used with LigD to covalently join 10 000 bp blunt-ended DNA molecules. The resulting ligated DNA molecules were stretched in 100 × 100 nm2 nanofluidic channels and imaged using fluorescence microscopy up to 4 h after the ligation was completed (Figure 4A, additional examples in Supplementary Figure S17). Upon completion of ligation, the fluorescent KuAF647 homodimer was bound statically to the DNA, equally spaced with a distance corresponding to 1.6 μm, in agreement with a 10 000 bp DNA stretched to approximately 46% of its contour. The emission intensity profile suggests that varying numbers of KuAF647 homodimers had been threaded on the DNA ends prior to ligation and that they remained bound at the ligation site after the ligation had occurred. This agrees with the fact that Ku acts stoichiometrically during the ligation (Figure 3G), suggesting that another protein is at play for removing the Ku post-ligation in vivo. To further confirm covalent attachment of the DNA ends, we exposed the nanoconfined DNA–protein complex (Figure 4B) to Proteinase K in situ. This is possible in a nanofluidic device, where the solution surrounding the nanoconfined DNA can be actively exchanged in real time (26). The hypothesis was that if the DNA-ends were ligated, the DNA concatemers would remain intact when the proteins are digested and washed away. The rapid reduction of fluorescence emission from KuAF647 in combination with the stable emission from the DNA molecule, which has a constant extension throughout the entire procedure (Supplementary Figure S18), demonstrates that Proteinase K removed KuAF647 (and most likely LigD) by digestion, while leaving the ligated DNA intact. Complete depletion of KuAF647 fluorescence emission was observed within 2 min for all DNA–protein complexes subjected to Proteinase K treatment (n = 36), whereas non-treated reference complexes showed a reduction of only 28 ± 14% in the same time (n = 20), a reduction attributed to photobleaching of the fluorophore.
Figure 4.
Ku remains stably bound to DNA post ligation. (A) Schematic of 10 000 bp blunt-ended DNA fragments (green) ligated by LigD, where KuAF647 (red) is stably bound to the ends and junctions post ligation, together with the corresponding fluorescence microscopy image where the DNA–protein complex has been stretched in a nanofluidic channel (100 × 100 nm2). The fluorescence intensity plot (black) displays variation in the emission from the equally spaced KuAF647 (red), normalized to the highest emission intensity observed. The error bars correspond to the standard deviation. Horizontal and vertical scale-bars correspond to 5 μm and 2 s, respectively. (B) Kymograph showing the emission from KuAF647 (red) and YOYO-1 labelled DNA (green) with time upon addition of Proteinase K in situ (top) in nanochannels with a dimension of 300 × 130 nm2 that allow for active addition of protein solution to the confined DNA–protein complex. The minimal level of photobleaching of KuAF647 is displayed in the control kymograph (bottom). The histogram reports the average relative reduction in fluorescence emission from KuAF647 after 2 min with or without Proteinase K (n = 36 and n = 20, respectively). Scale-bar corresponds to 5 μm.
DISCUSSION
Our work provides new functional and kinetic insights into prokaryotic NHEJ, involving only the two proteins Ku and LigD. We go through the bacterial NHEJ reaction step by step and dissect the dynamics of the process using suitable single molecule techniques. Based on our findings, we propose a model for the bacterial NHEJ process, where Ku loads cooperatively onto DNA ends and forms DNA bridges, in a manner that does not depend on the C-terminal arms. The C-terminal arms of Kuwt are however very important for the recruitment of LigD onto DNA, that subsequently leads to a stabilization of the synapsis and efficient ligation. The interaction between Ku and LigD corresponds to the binding of NHEJ partners to Ku in the eukaryotic system via the so-called Ku-binding motifs (41,43) and we identified a potential patch for this interaction on LigD. Interestingly, prokaryotic Ku cannot spontaneously release from repaired DNA, indicating an as-yet-unidentified signaling mechanism is at work in prokaryotes to actively remove this component from DNA. A similar mechanism has been proposed also for the human system (44) where removal of the protein from DNA after ligation was found to be related to the polyubiquitylation of the K48 residue of Ku80 (45). Furthermore, it has recently been proposed that the recruitment of p97 ATPase and its co-factor Ufd1 will cause extraction of the ring-shaped Ku70/80 heterodimer from the DNA (44).
Our experiments are particularly powerful in determining the kinetics and dynamics of synapsis formation, which is of great importance, in particular in bacteria that duplicate very fast. We conclude, for the first time, that Ku alone is sufficient to generate a weak end-synapsis. Furthermore, we observe that the simple bacterial NHEJ system is very sensitive to the nature of the DNA ends. Although blunt-end synapsis lasts for 2 seconds, synapsis of DNA ends with 4 nt complementary overhangs is extremely long-lived, in fact too long-lived for us to measure in the molecular forceps assay. Base-pairing worth ΔG in free energy should increase the lifetime of blunt DNA end-synapsis by a Boltzmann factor exp(ΔG/kBT). Thus, a 2 s mean synapsis lifetime would be extended to 4 × 104 s by a 4 nt overhang contributing to lower the free energy of the complex by ΔG ∼10 kBT. From this we conclude that in bacterial NHEJ the DNA ends are allowed to come into sufficiently close contact as to be able to anneal if complementary sequences are present. As a result, in the bacterial NHEJ system there is little mechanistic flexibility: if bacterial Ku finds complementary DNA ends it will simply hold them together and enact ligation without sampling other ends. Experiments with non-complementary overhangs confirm that base-pairing is required to form the long-lived synapsis, but a synapsis with a similar life-time as for blunt ends is formed also with 4 nt non-complementary overhangs.
Our experiments also highlight the interest in studying simple prokaryotic systems to guide current and future work on the more complex eukaryotic systems. In the human system the lifetime of DNA end-synapsis mediated by DNA-PK and PAXX is essentially independent of whether the DNA ends are blunt (∼2 s) or complementary (∼3 s). This indicates that the human system has evolved to preclude annealing of complementary DNA ends at its early stages, allowing it to remain less deterministic as to which ends will be ligated together. Our observations are consistent with those from the Loparo laboratory (46) indicating that initial pairing in the human system as permitted by the presence of DNA-PK and PAXX does not allow the DNA ends to get as close to each other as when the downstream components XLF, XRCC4 and Lig IV are present. This allows the human system to maintain extensive mechanistic flexibility, as would be expected given the numerous roles of human NHEJ, ranging from repair of exogenous DNA breaks to the endogenous breaks which are a fundamental aspect of V(D)J recombination and generation of a diverse immune repertoire. Our experiments provide additional evolutionary perspectives to guide future work on NHEJ, from studying the yeast system, which again does not possess DNA-PKcs, as well as ciliates which do possess DNA-PKcs and use it to purge their micronuclear genome of transposons.
In addition to reporting significant similarities and differences between prokaryotic and eukaryotic NHEJ, our study also reveals important molecular details for future development of antibiotics that target bacterial NHEJ. One option would be a drug that acts on the interface between either two Ku homodimers or between one Ku homodimer and LigD, to inhibit repair of DSBs. A drug inhibiting NHEJ could then, for example, be used together with a DNA damaging drug to increase the efficiency of the latter. Such combination therapies are possible future treatments against the ever-increasing health concern of bacteria becoming resistant to common antibiotics (3).
To conclude, we present the first single molecule study of bacterial NHEJ, where we characterize the dynamics of synapsis formation by bacterial Ku and discover that the homodimeric protein is able of promoting hybridization of short complementary overhangs. The synapsis is stabilized by LigD and post-ligation Ku remains bound, suggesting another system is available in vivo for removing Ku. This system is yet to be identified.
DATA AVAILABILITY
Experimental data and computer programs for performing all data analyses described herein are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Istvan Horvath and Ranjeet Kumar, Chalmers University of Technology, Gothenburg, Sweden, for their support in SPR experiments and protein purification, Mikael Lindberg, Protein Expertise Platform, Umeå, Sweden, for producing the bacterial Ku constructs, François Lecointe, Micalis Institute, Paris, France, for sharing the construct of LigD, Tom Blundell and Shikang Liang, University of Cambridge, Cambridge, UK, for sharing Ku70/80, PAXX and DNA-PKcs, the research team of Tobias Ambjörnsson, Lund University, Lund, Sweden, for developing the Matlab based analysis tool for the nanofluidic experiments, the Centre for Cellular Imaging at the University of Gothenburg and the National Microscopy Infrastructure, NMI (VR-RFI 2019-00022) for assisting in microscopy for photobleaching experiments and Myfab Chalmers for providing the instruments needed to fabricate the nanofluidic devices.
Notes
Present address: Jing L. Wang, School of Medicine, Stanford University, Stanford, USA.
Contributor Information
Robin Öz, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg SE 41296, Sweden.
Jing L Wang, Institut Jacques Monod, Université de Paris, CNRS, UMR7592, Paris, France; Ecole Normale Supérieure, IBENS, CNRS, INSERM, PSL Research University, Paris 75005 France.
Raphael Guerois, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Université Paris-Saclay, Gif-sur-Yvette 91198, France.
Gaurav Goyal, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg SE 41296, Sweden.
Sriram KK, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg SE 41296, Sweden.
Virginie Ropars, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Université Paris-Saclay, Gif-sur-Yvette 91198, France.
Rajhans Sharma, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg SE 41296, Sweden.
Firat Koca, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg SE 41296, Sweden.
Jean-Baptiste Charbonnier, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Université Paris-Saclay, Gif-sur-Yvette 91198, France.
Mauro Modesti, Cancer Research Center of Marseille, CNRS, Inserm, Institut Paoli-Calmettes, Aix-Marseille Université, Marseille 13009, France; Equipe Labélisée, Ligue Nationale Contre le Cancer, Paris 75013, France.
Terence R Strick, Institut Jacques Monod, Université de Paris, CNRS, UMR7592, Paris, France; Ecole Normale Supérieure, IBENS, CNRS, INSERM, PSL Research University, Paris 75005 France; Equipe Labélisée, Ligue Nationale Contre le Cancer, Paris 75013, France.
Fredrik Westerlund, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg SE 41296, Sweden.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council [2015-5062 to F.W.]; Stiftelsen Olle Engqvist Byggmästare and the European Research Council, in the form of an ERC consolidator grant [nanoDNArepair, no. 866238]; work on this topic in the Strick Lab is supported by the Equipes Labellisées Programme of the French National League Against Cancer; M.M. is supported by the French National Research Agency, the French National Cancer Institute and the French National League Against Cancer. JBC is supported by the French National Research Agency [ANR-20-CE11-0026] and the National Infrastructure FRISBI [ANR-10-INBS-0005]. Funding for open access charge: Swedish Research Council [2015-5062].
Conflict of interest statement. None declared.
REFERENCES
- 1. de Vega M. The minimal bacillus subtilis nonhomologous end joining repair machinery. PLoS One. 2013; 8:e64232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weller G.R., Kysela B., Roy R., Tonkin L.M., Scanlan E., Della M., Devine S.K., Day J.P., Wilkinson A., di Fagagna F.D.et al.. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002; 297:1686–1689. [DOI] [PubMed] [Google Scholar]
- 3. Shuman S., Glickman M.S.. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007; 5:852–861. [DOI] [PubMed] [Google Scholar]
- 4. Chaplin A.K., Blundell T.L.. Structural biology of multicomponent assemblies in DNA double-strand-break repair through non-homologous end joining. Curr. Opin. Struct. Biol. 2020; 61:9–16. [DOI] [PubMed] [Google Scholar]
- 5. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yaneva M., Kowalewski T., Lieber M.R.. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997; 16:5098–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeFazio L.G., Stansel R.M., Griffith J.D., Chu G.. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002; 21:3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J.L., Duboc C., Wu Q., Ochi T., Liang S.K., Tsutakawa S.E., Lees-Miller S.P., Nadal M., Tainer J.A., Blundell T.L.et al.. Dissection of DNA double-strand-break repair using novel single-molecule forceps. Nat. Struct. Mol. Biol. 2018; 25:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang H.H., Watanabe G., Gerodimos C.A., Ochi T., Blundell T.L., Jackson S.P., Lieber M.R.. Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. J. Biol. Chem. 2016; 291:24377–24389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emerson C.H., Bertuch A.A.. Consider the workhorse: nonhomologous end-joining in budding yeast. Biochem. Cell Biol. 2016; 94:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bowater R., Doherty A.J.. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLos Genet. 2006; 2:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Della M., Palmbos P.L., Tseng H.-M., Tonkin L.M., Daley J.M., Topper L.M., Pitcher R.S., Tomkinson A.E., Wilson T.E., Doherty A.J.. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004; 306:683–685. [DOI] [PubMed] [Google Scholar]
- 13. Aravind L., Koonin E.V.. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001; 11:1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doherty A.J., Jackson S.P., Weller G.R.. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001; 500:186–188. [DOI] [PubMed] [Google Scholar]
- 15. McGovern S., Baconnais S., Roblin P., Nicolas P., Drevet P., Simonson H., Pietrement O., Charbonnier J.B., Le Cam E., Noirot P.et al.. C-terminal region of bacterial Ku controls DNA bridging, DNA threading and recruitment of DNA ligase D for double strand breaks repair. Nucleic Acids Res. 2016; 44:4785–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kostrz D., Wayment-Steele H.K., Wang J.L., Follenfant M., Pande V.S., Strick T.R., Gosse C.. A modular DNA scaffold to study protein–protein interactions at single-molecule resolution. Nat. Nanotechnol. 2019; 14:988–993. [DOI] [PubMed] [Google Scholar]
- 17. Frykholm K., Nyberg L.K., Westerlund F.. Exploring DNA-protein interactions on the single DNA molecule level using nanofluidic tools. Integr. Biol. 2017; 9:650–661. [DOI] [PubMed] [Google Scholar]
- 18. Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2004; 21:951–960. [DOI] [PubMed] [Google Scholar]
- 19. DiMaio F., Leaver-Fay A., Bradley P., Baker D., Andre I.. Modeling symmetric macromolecular structures in Rosetta3. PLoS One. 2011; 6:e20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song Y.F., DiMaio F., Wang R.Y.R., Kim D., Miles C., Brunette T.J., Thompson J., Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013; 21:1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pupko T., Bell R.E., Mayrose I., Glaser F., Ben-Tal N.. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002; 18:S71–S77. [DOI] [PubMed] [Google Scholar]
- 23. Zeng H., Wang S., Zhou T.M., Zhao F.F., Li X.F., Wu Q., Xu J.B.. ComplexContact: a web server for inter-protein contact prediction using deep learning. Nucleic Acids Res. 2018; 46:W432–W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V.et al.. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015; 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sibanda B.L., Chirgadze D.Y., Blundell T.L.. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2009; 463:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Öz R., Sriram K., Westerlund F.. A nanofluidic device for real-time visualization of DNA–protein interactions on the single DNA molecule level. Nanoscale. 2019; 11:2071–2078. [DOI] [PubMed] [Google Scholar]
- 27. Frykholm K., Nyberg L.K., Lagerstedt E., Noble C., Fritzsche J., Karami N., Ambjornsson T., Sandegren L., Westerlund F.. Fast size-determination of intact bacterial plasmids using nanofluidic channels. Lab Chip. 2015; 15:2739–2743. [DOI] [PubMed] [Google Scholar]
- 28. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allemand J.F., Bensimon D., Jullien L., Bensimon A., Croquette V.. pH-dependent specific binding and combing of DNA. Biophys. J. 1997; 73:2064–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Labit H., Goldar A., Guilbaud G., Douarche C., Hyrien O., Marheineke K.. A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. BioTechniques. 2008; 45:649–658. [DOI] [PubMed] [Google Scholar]
- 31. Singh V., Johansson P., Torchinsky D., Lin Y.-L., Öz R., Ebenstein Y., Hammarsten O., Westerlund F.. Quantifying DNA damage induced by ionizing radiation and hyperthermia using single DNA molecule imaging. Translational Oncology. 2020; 13:100822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hummert J., Yserentant K., Fink T., Euchner J., Herten D.-P.. Photobleaching step analysis for robust determination of protein complex stoichiometries. 2020; bioRxiv doi:26 August 2020, preprint: not peer reviewed 10.1101/2020.08.26.268086. [DOI] [PMC free article] [PubMed]
- 33. Tsekouras K., Custer T.C., Jashnsaz H., Walter N.G., Pressé S.. A novel method to accurately locate and count large numbers of steps by photobleaching. Mol. Biol. Cell. 2016; 27:3601–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watson J.D., Crick F.H.C.. Molecular structure of nucleic acids - a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. [DOI] [PubMed] [Google Scholar]
- 35. Wang J.L., Duboc C., Wu Q., Ochi T., Liang S., Tsutakawa S.E., Lees-Miller S.P., Nadal M., Tainer J.A., Blundell T.L.et al.. Dissection of DNA double-strand-break repair using novel single-molecule forceps. Nat. Struct. Mol. Biol. 2018; 25:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang K., Humbert N., Kk S., Lequeu T., Lin Y.-L., Mely Y., Westerlund F.. Annealing of ssDNA and compaction of dsDNA by the HIV-1 nucleocapsid and Gag proteins visualized using nanofluidic channels. Q. Rev. Biophys. 2019; 52:e2. [DOI] [PubMed] [Google Scholar]
- 37. Walker J.R., Corpina R.A., Goldberg J.. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001; 412:607–614. [DOI] [PubMed] [Google Scholar]
- 38. Ma Y., Lieber M.R.. DNA length-dependent cooperative interactions in the binding of Ku to DNA. Biochemistry. 2001; 40:9638–9646. [DOI] [PubMed] [Google Scholar]
- 39. Zhu H., Shuman S.. Gap filling activities of Pseudomonas DNA ligase D (LigD) polymerase and functional interactions of LigD with the DNA end-binding Ku protein. J. Biol. Chem. 2010; 285:4815–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pitcher R.S., Tonkin L.M., Green A.J., Doherty A.J.. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J. Mol. Biol. 2005; 351:531–544. [DOI] [PubMed] [Google Scholar]
- 41. Nemoz C., Ropars V., Frit P., Gontier A., Drevet P., Yu J., Guerois R., Pitois A., Comte A., Delteil C.et al.. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat. Struct. Mol. Biol. 2018; 25:971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Ory A., Zafra O., de Vega M.. Efficient processing of abasic sites by bacterial nonhomologous end-joining Ku proteins. Nucleic Acids Res. 2014; 42:13082–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frit P., Ropars V., Modesti M., Charbonnier J.B., Calsou P.. Plugged into the Ku-DNA hub: The NHEJ network. Prog. Biophys. Mol. Biol. 2019; 147:62–76. [DOI] [PubMed] [Google Scholar]
- 44. van den Boom J., Wolf M., Weimann L., Schulze N., Li F., Kaschani F., Riemer A., Zierhut C., Kaiser M., Iliakis G.et al.. VCP/p97 Extracts sterically trapped Ku70/80 rings from DNA in double-strand break repair. Mol. Cell. 2016; 64:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Postow L., Ghenoiu C., Woo E.M., Krutchinsky A.N., Chait B.T., Funabiki H.. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008; 182:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Graham T.G.W., Walter J.C., Loparo J.J. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell. 2016; 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Experimental data and computer programs for performing all data analyses described herein are available from the corresponding author upon reasonable request.