Abstract
The interferon gamma-inducible protein 16 (IFI16) and its murine homologous protein p204 function in non-sequence specific dsDNA sensing; however, the exact dsDNA recognition mechanisms of IFI16/p204, which harbour two HIN domains, remain unclear. In the present study, we determined crystal structures of p204 HINa and HINb domains, which are highly similar to those of other PYHIN family proteins. Moreover, we obtained the crystal structure of p204 HINab domain in complex with dsDNA and provided insights into the dsDNA binding mode. p204 HINab binds dsDNA mainly through α2 helix of HINa and HINb, and the linker between them, revealing a similar HIN:DNA binding mode. Both HINa and HINb are vital for HINab recognition of dsDNA, as confirmed by fluorescence polarization assays. Furthermore, a HINa dimerization interface was observed in structures of p204 HINa and HINab:dsDNA complex, which is involved in binding dsDNA. The linker between HINa and HINb reveals dynamic flexibility in solution and changes its direction at ∼90° angle in comparison with crystal structure of HINab:dsDNA complex. These structural information provide insights into the mechanism of DNA recognition by different HIN domains, and shed light on the unique roles of two HIN domains in activating the IFI16/p204 signaling pathway.
INTRODUCTION
A number of innate immune receptors are involved in pathogen detection and host immune defense. Nucleic acids, both DNA and RNA, exhibit important pathogen-associated molecular patterns (1). Several families of intracellular nucleic acid receptors have been studied, including RIG-I-like receptors, cGAS, and the PYHIN family receptors (2–4). While the RIG-I family receptors primarily respond to dsRNA molecules (5), the cGAS and PYHIN family function as dsDNA receptors. The PYHIN family proteins, also known as hematopoietic interferon-inducible nuclear (HIN)-200 family, are a group of interferon-inducible genes. The human genome comprises four PYHIN family genes including AIM2, IFI16, PYHIN1 and MNDA. p202a, p202b, p203, p204, p205, p206 and AIM2 are PYHIN family members in murine. Except murine p202, human and murine PYHIN family members possess an N-terminal PYD domain and one or two C-terminal HIN (or HIN200) domains.
Early studies illustrated that PYHIN family genes are tumor inhibitory genes, while their mechanism of action remains poorly defined and is presumably related to its DNA binding activities. Several PYHIN family members can interact with nuclear proteins, and this action reveals their function in cell proliferation retardation. p204 can directly interact with UBF1, which is a ribosomal RNA-specific transcription factor that inhibits ribosomal RNA transcription (6). A recent study reported that another closely related murine PYHIN family member, p205, controls the expression of apoptosis-associated speck-like molecule containing CARD domain (ASC) mRNA to regulate inflammasome responses (7). Members of the PYHIN family are gaining immense attention due to the recent discovery of cytosolic DNA receptors and the initiation of innate immune responses related to infectious and autoimmune diseases. Among these, AIM2 is the most widely studied member of the PYHIN family (8,9). AIM2 forms an inflammasome in response to bacterial infection (10–12), viral infection (13) and cellular damage (14), and leads to downstream caspase-1 activation and IL-1β maturation. Although several studies have reported that IFI16 can also form inflammasomes with ASC, its role is better established in type I IFN pathways. IFI16 or its mouse homolog p204 is implicated in sensing double-stranded DNA (dsDNA) in the cytosol during DNA viral infection, and stimulates both type I IFN and proinflammatory responses (15,16). Moreover, IFI16 plays a crucial role in nuclear viral DNA sensing (15,17,18). IFI16 acts as a major sensor for lentivirus DNA in macrophages stimulating innate immune responses, which contributes to the early control of HIV viral replication (19). Mechanistically, IFI16 preferentially binds to stem-rich secondary structures in the ssDNA of HIV (19). In addition, IFI16 was reported to be required in the cGAS-STING DNA sensing pathway in macrophages during microbial infection (20,21). Furthermore, Iqbal et al. identified a nuclear protein histone H2B that interacts with IFI16. The H2B-IFI16-BRCA1 complex interacted with cGAS and STING in the cytoplasm. This complex leads to the phosphorylation of TBK1 and IRF3, nuclear translocation of pIRF3, and production of IFN-β. Eventually, such nuclear complexes mediate Kaposi's sarcoma-associated herpes virus (KSHV) and herpes simplex virus type-1 (HSV-1) genome recognition responses during viral infection (22). Recent studies have revealed that p204 is involved in the natural immune response. Unterholzner et al. demonstrated that p204 responds to DNA viruses by RNA interference (16). Later, Gray et al. observed that the PYHIN family was not necessary for the response to foreign DNA in mice depleted for the locus containing the PYHIN-encoding gene (23). Recently, Yi et al. reported that p204 is required for extracellular LPS/TLR4 during pathogen infection in macrophage-mediated innate immunity, and the PYD of p204 directly interacts with TLR4 to promote the dimerization of TLR4 (24). Chen et al. also demonstrated that p204 acts as a DNA sensor and is required for host defense against Staphylococcus aureus infection in p204-deficient mice (25). The aforementioned studies established a clear role of IFI16/p204 in innate immunity in response to various viral infections. Additionally, new evidence is accumulating regarding other functions of IFI16/p204, including cancer, regulation of EPC cell differentiation (26), and autoinflammatory diseases (27).
Recent structural characterization of PYHIN family proteins, particularly HIN:DNA interactions, has made significant progress in understanding its molecular recognition mechanism (Table 1). HIN domains can be further divided into three subtypes based on their sequence consensus. AIM2 HIN belongs to class C, whereas the first HIN domains of IFI16, p202 and p204 belong to class A, and their second HIN domains belong to class B (28). A canonical DNA-binding mode is observed in some HIN:DNA complex structures. These structures include both the human AIM2 HIN domain (PDB: 3RN2 and 3RN5) (29), murine AIM2 HIN domain (PDB: 4JBM) (30), and IFI16-HINb HIN domain (PDB: 3RNU) (29). In our previous study on DNA recognition by HIN domains, we identified a conserved DNA-binding mode for both human AIM2 HIN and IFI16 HINb, in which the concave surface formed by two OB folds of a HIN domain interacts with the phosphate backbone of dsDNA. The interaction between the HIN domain and DNA is sequence-independent and no base recognition is involved. Mechanistically, the positively charged residues (i.e. arginines and lysines) on the HIN surface interact with the negatively charged phosphate backbones of dsDNA. The electrostatic nature of such interactions is supported by high sensitivity to salt concentration in the solution (29). For the AIM2 inflammasome, structural studies have clarified the assembly mechanism around dsDNA and the activation of ASC inflammasome (31), whereas the signal mechanism of IFI16 remains elusive. The p202 protein is indeed a special member of the PYHIN family. Functionally, it is a negative regulator of p53-mediated apoptosis and AIM2 inflammasome activation (32,33). The latter function is attributed to its high affinity for binding to dsDNA (34). Structurally, its HINa domain binds to dsDNA in a noncanonical manner, as revealed from dsDNA complex structures by several groups (30,34,35). In this mode, a high positively charged surface at the back of the canonical DNA-binding linker region interacts with the phosphate backbone of ∼12 bp dsDNA; however, p202 HINb is a tetramer that lacks DNA-binding ability (35). Furthermore, IFI16 HINa:ssDNA interaction uses a different interface and the ssDNA ligand forms an imperfectly paired dsDNA in the crystal (PDB: 4QGU) (36).
Table 1.
Summary of published HIN domain structures of PYHIN family proteins
| Protein | Positions of Class A HIN | Positions of Class B HIN | Positions of Class C HIN | dsDNA | Resolution (Å) | PDB entry | Reference |
|---|---|---|---|---|---|---|---|
| hAIM2 | 144–343 | 20 mer | 2.55 | 3RN2* | (29) | ||
| hAIM2 | 144–343 | 19 mer | 2.50 | 3RN5 | (29) | ||
| mAIM2 | 158–349 | 13 mer | 2.22 | 4JBM* | (30) | ||
| IFI16 | 192–393 | 2.00 | 2OQ0* | (55) | |||
| IFI16 | 192–393 | 12 mer | 2.54 | 4QGU* | (36) | ||
| IFI16 | 571–766 | 2.35 | 3B6Y* | (55) | |||
| IFI16 | 571–766 | 1.80 | 3RLO | (29) | |||
| IFI16 | 571–766 | 2.25 | 3RLN | (29) | |||
| IFI16 | 571–766 | 16 mer | 2.50 | 3RNU* | (29) | ||
| p202 | 46–242 | 2.69 | 4JBJ* | (30) | |||
| p202 | 46–243 | 2.23 | 4L5Q | (35) | |||
| p202 | 46–242 | 14 mer | 2.96 | 4JBK | (30) | ||
| p202 | 46–243 | 20 mer | 1.87 | 4L5R* | (35) | ||
| p202 | 46–243 | 12 mer | 2.94 | 4L5S | (35) | ||
| p202 | 53–245 | 20 mer | 2.00 | 4LNQ | (34) | ||
| p202 | 244–445 | 3.40 | 4L5T* | (35) | |||
| p204 | 211–415 | 1.94 | 6OE9 | (56) | |||
hAIM2: human AIM2.
mAIM2: mouse AIM2.
*Used to compare with p204 HIN structures.
The domain organization of IFI16 is unique, as it is the only human PYHIN that has tandem HIN domains. Previously, we demonstrated that for individual HIN domains, AIM2 has a higher affinity for dsDNA than that of IFI16 HINb, whereas IFI16 HINa has the lowest affinity among the three HIN domains (16,29). Since both HIN domains of IFI16 bind to DNA, there could be several ways for HINab to bind dsDNA. Two linked HIN domains can potentially bind to two different DNA molecules, or only HINb binds to dsDNA, while HINa hangs out. The binding mode of the IFI16 HINab domain toward dsDNA remains unclear. A recent native stained electron micrographic study of IFI16:dsDNA complexes revealed that after binding dsDNA, IFI16 forms filaments on straight dsDNA with a width of 20–25 nm. It was further confirmed that both HIN domains bind to the same piece of dsDNA and do not open, bend, and crosslink dsDNA (37). Song et al. designed a series of linkers with different lengths and sequences between the IFI16 HINa and HINb domains, and then co-crystallized them with dsDNA; however, due to weak diffraction, the complex structure of IFI16 HINab:dsDNA was not obtained (38). Nevertheless, no detailed information is available on the binding of HINab to dsDNA during innate immune recognition.
To investigate the mechanism of HINab binding dsDNA, it is essential to solve the structure of HINab in complex with dsDNA. In the present study, fluorescence polarization assays were performed and the results indicated that the DNA-binding affinity of p204 HINab was higher than that of HINa and HINb, suggesting that HINa and HINb synergistically bind to dsDNA. Thereafter, we determined the crystal structures of p204 HINa and HINb with a highly similar conformation. The crystal structure of p204 HINab in complex with dsDNA was also determined, providing the first insight into the dsDNA-binding mode of tandem HIN domains. In the structure of p204 HINa and HINab:dsDNA complex, the HINa dimerization interface was observed and was also involved in binding to dsDNA. Thus, a structural view of tandem HIN domains of p204 binding to dsDNA not only fills the gap in understanding dsDNA recognition by this receptor, but also sheds light on the unique roles of the two HIN domains in activating the IFI16/p204 signaling pathway.
MATERIALS AND METHODS
Protein expression and purification
The coding region of the p204 gene (NM_008329.2) was amplified from the mice liver cDNA library (invitrogen). The HINa (residues 216–417), HINb (residues 427–619), and HINab (residues 216–619) domains of p204 were cloned into the pET30a vector with a TEV-cleavable N-terminal protein G β1 domain (GB1) tag or MBP tag (HINa with GB1 tag, HINb and HINab with MBP tag). The expression and purification of the three recombinant proteins were similar to those previously described (39). Briefly, transformed BL21 CodonPlus (DE3)-RIPL cells (Stratagene, Santa Clara, CA) were grown at 37°C and then induced with IPTG (0.2 mM) at 16°C for 4 h. Cells were lysed by sonication in buffer A (1 M NaCl, 20 mM Tris–HCl pH 8.0), 5 mM imidazole, DNase (Biomatik, Wilmington, DE, USA), and protease inhibitors (Roche Applied Science, Indianapolis, IN, USA). Next, the soluble protein was purified using a Hisprep IMAC column (GE Healthcare Bio-Sciences, Piscataway, NJ), and nonspecific nucleic acid contaminants were removed by 3 M ammonium sulfate precipitation. Thereafter, the protein pellet was redissolved and cleaved by the TEV protease. The protein samples were further purified using a second IMAC column and size-exclusion chromatography. Expression and purification of all mutant proteins followed a protocol similar to that used for wild-type proteins.
Crystallization, X-ray diffraction, and structure refinement
Purified p204 HINa and HINb proteins were concentrated to 20–50 mg/ml. The crystallization conditions were screened by the hanging-drop vapor-diffusion method with a liquid-handling robot. HINa was crystallized under the following conditions: 23% PEG4000, 0.2 M ammonium acetate, 100 mM sodium acetate pH 5.0, and an additional 20% ethylene glycol was used as a cryoprotectant solution. HINb was crystallized in 20% PEG300, 100 mM Bicine pH 8.5, and an additional 20% ethylene glycol was used as a cryoprotectant solution. X-ray diffraction data were collected at beamlines X12C, X29A and X25 of the National Light Source at the Brookhaven National Laboratory, and beamline SER-CAT 22ID and GM-CA 23ID of Advanced Photon Source (APS) at the Argonne National Laboratory. Data were processed using HKL-2000 (40) and XDS (41). The sequence identity was ∼56% between p204 HINa and IFI16 HINa, and 53% between p204 HINb and IFI16 HINb. All structures were solved by molecular replacement with Phaser (42) in the CCP4i GUI (43). IFI16 HINa (PDB: 2OQ0) and HINb (PDB: 3B6Y) were used as molecular replacement models in the initial search. The structure model was completed by manual building in Coot (44). Structure refinement was carried out using REFMAC5 (45) and PHENIX.refine (46). Structural figures were prepared using PyMol.
Crystallization and structural determination of p204 HINab:dsDNA complex
To obtain complex crystals of p204 HINab with dsDNA ligands, several different DNA lengths, blunt-ended or sticky-ended dsDNA were tested. These dsDNAs were added to the purified HINab protein sample at a molar ratio of 1.2:1 and were further concentrated to 20 mg/ml for crystallization screening. A 16 mer blunt-ended dsDNA with the sequence of 5′-ccatcagaaagagagc-3′ grew best crystals under the conditions of 4% PEG8000, 100 mM KCl, and 100 mM MES pH 6.5. An additional 15% ethylene glycol and 10% glycerol were used in the cryoprotectant solution. The crystals of HINab:dsDNA diffracted initially at ∼7 Å resolution, owing to the large unit cell dimensions (the length of c-axis = 784 Å, Table 2), which imposed a challenge to obtain higher resolution data. Thereafter, the crystals were dehydrated by briefly soaking in higher concentration of PEG8000 and the resolution was gradually improved to 4.5–5.0 Å. Data were processed using XDS (41). Correction for radiation damage was performed as previously described (47). The initial Wilson plot from the data shows a B-factor of up to 320 Å2. A correction of the B-factor and anisotropic B-factor (using PHENIX (46) Reflection File Editor) was applied to the data to reduce the Wilson B-factor to 120 Å2. Data quality and statistics are listed in Table 2. Molecular replacement was carried out with Phaser (42) using p204 HINa (PDB: 5YZP), HINb (PDB: 5YZW) and 16 mer dsDNA as search models. Structure refinement was performed with PHENIX (46,48,49), interspersed with manual rounds of model building in Coot (44). Initially, rigid body refinement was performed for individual HIN domains and two individual dsDNA fragments. Next, we applied the NCS and a secondary structure restraint for the individual coordinates and group B-factor refinements. The two high-resolution structures of p204 HINa (PDB: 5YZP) and p204 HINb (PDB: 5YZW) were used as reference models for restraints during the refinement cycles in PHENIX; however, the linkers between the HINa and HINb domains were manually rebuilt and refined according to the electron densities. Some atoms of the side chain of the linkers were removed during refinement due to the missing electron densities. The refinement results and model statistics are listed in Table 2.
Table 2.
X-ray data collection and refinement table
| HINa | HINb | HINab:16 mer dsDNA | |
|---|---|---|---|
| Data set | TJ113–7 | TJ72–4 | JT4–12 |
| Data collection | |||
| Spacegroup | C2221 | P3121 | P6522 |
| Unit cell (a, b, c) (Å) | 57.93, 104.21, 90.42 | 99.72, 99.72, 107.05 | 101.39, 101.39, 783.61 |
| (α,β,γ) (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50–1.58 (1.68–1.58) | 50–2.00 (2.12–2.00) | 50–4.50 (4.67–4.50) |
| No of reflections (total/unique) | 287 766/37 551 | 447 016/41 767 | 195 843/15 339 |
| Redundancy | 7.66 (7.00) | 10.70 (10.85) | 12.80 (13.40) |
| Completeness (%) | 99.3 (97.7) | 99.6 (99.4) | 91.78 (82.36) |
| I/σ(I) | 20.45 (2.93) | 18.35 (2.31) | 11.80 (0.85) |
| Rmeas (%)a | 5.8 (71.0) | 7.4 (136.6) | 9.9 (320.1) |
| CC1/2 (%) | 99.9 (89.9) | 99.9 (84.1) | 99.9 (52.8) |
| Refinement | |||
| Resolution (Å) | 44.18–1.58 | 33.6–2.00 | 47.26–4.50 |
| No. of protein atoms | 1544 | 3096 | 9312 |
| No. of DNA atoms | 0 | 0 | 1312 |
| No. of solvent | 194 | 134 | 0 |
| No. of hetero-atoms | 16 | 0 | 0 |
| Wilson B-factor (Å2) | 22.5 | 44.1 | 112.1 |
| Average B-factor (Å2) (protein) | 30.97 | 65.44 | 162.29 |
| Average B-factor (DNA) (Å2) | 0 | 0 | 207.40 |
| RMSD bond lengths (Å) | 0.006 | 0.005 | 0.004 |
| RMSD bond angles (°) | 0.843 | 0.844 | 0.708 |
| R work (%)b | 18.36 | 20.45 | 25.97 |
| R free (%)c | 21.05 | 25.44 | 29.71 |
| Ramachandran plot favored/disallowed (%)d | 99.5/0 | 96.4/0 | 98.22/1.78 |
| PDB code | 5YZP | 5YZW | 5Z7D |
Values in the parentheses correspond to the last resolution shell.
a R meas = Σh(n/n – 1)1/2Σi|Ii(h) – <I(h)>|/ΣhΣiIi(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
b R work = Σh||Fobs(h)| – |Fcalc(h)||/Σh|Fobs(h)|, where Fobs(h) and Fcalc(h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
c R free is the R-value obtained for a test set of reflections consisting of a randomly selected 5% subset of the data set excluded from refinement.
dValues from Molprobity server (http://molprobity.biochem.duke.edu/).
Size exclusion chromatography assay
The apparent molecular masses of p204 HINab, HINa, HINb and mutant proteins were estimated using a 24 ml ENrich SEC 650 column (10/300 GL, Bio-Rad). Briefly, protein samples or molecular mass standards were applied to a 24 ml column at a flow rate of 0.6 ml/min and eluted with PBS. The standard proteins (Sigma-Aldrich, USA) used in this assay were β-amylase (200.0 kDa), albumin (66.0 kDa), and carbonic anhydrase (29.0 kDa).
Fluorescence polarization assay
5′-Fluorescein (FAM)- labeled (IDT, Coralville, IA, USA) ssDNA was dissolved in PBS and annealed with its reverse complement ssDNA by heating to 95°C and slowly cooling to room temperature. Fluorescence polarization (FP) assays were performed at room temperature. The wavelengths of fluorescence excitation and emission were 485 and 528 nm, respectively. Each well of a 384-well plate contained 20 nM fluorescent-labeled (5′-FAM) DNA probe and different concentrations of p204 HIN domain or mutant proteins (0, 3, 8, 16, 31, 48, 80, 120, 170, 230 and 300 μM) with a final volume of 80 μl in PBS. Three independent replicates were used. The binding curves were fitted according to a one-site binding model using GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). The ssDNA sequences used in the FP assay are listed in Supplementary Table S3.
Small angle X-ray scattering
Structural envelope of p204 HINab in solution was determined via small angle X-ray scattering (SAXS) data analysis. SAXS data were collected at beamline X9 of the National Synchrotron Light Source at the Brookhaven National Laboratory. SAXS was performed at different protein concentrations (1.00–3.80 mg/ml), and it was performed only once at a particular concentration. Purified p204 HINab samples with a concentration of 3.8 mg/ml were collected during 30 s exposures. Two-dimensional diffraction images were reduced to one dimension, and buffer scattering was subtracted. Supplementary Figure S2A displays the log of scattering intensities versus q and the Guinier plot. Data analysis of the radius of gyration (Rg) and Pr (pair-wise distance distribution function) calculations were performed using PRIMUS (50), and dummy residue models were produced using DAMMIN/DAMMIF (51,52) (Supplementary Figure S2B). Moreover, SAXS envelopes (Supplementary Figure S3C) were produced by pdb2vol of SITUS (http://situs.biomachina.org) and rendered by Chimera (53). Determined Rg and derived Pr from the SAXS experimental data are compared with those calculated by CRYSOL (54) from the crystal structure of HINab (Supplementary Table S1). Furthermore, SAXS data and models have been deposited in the SASBDB database (https://www.sasbdb.org/) under the accession number SASDK92. Data collection and scattering-derived parameters are listed in Supplementary Table S1.
RESULTS
The DNA-binding properties of p204 HIN domain
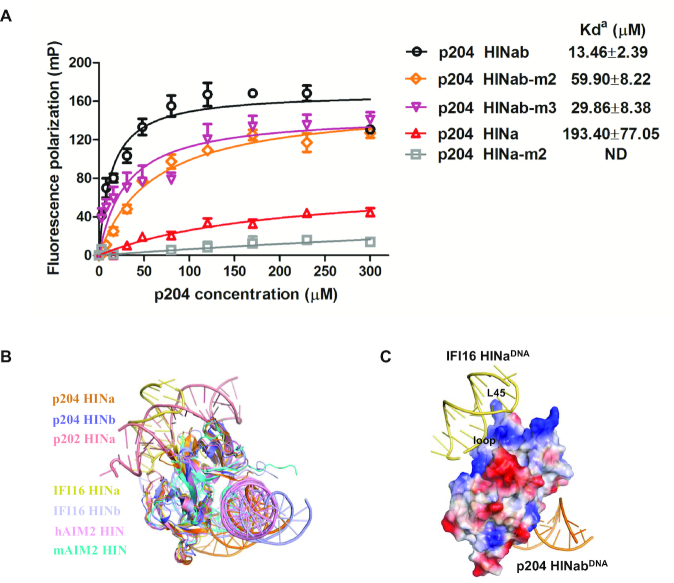
Previous studies have reported that the HIN domain of PYHIN family proteins can bind to DNA. To investigate the DNA-binding properties of the p204 HIN domain, we purified p204 HINab, HINa, and HINb to high purity (Supplementary Figure S1A). These proteins are all monomers in solution, as confirmed by size exclusion chromatography (Supplementary Figure S1B). The SAXS results indicate that p204 HINab also acts as a monomer in the absence of dsDNA (Supplementary Figure S2A–C and Table S1). The fluorescence polarization (FP) assay was performed to detect the binding affinities of p204 HINab on dsDNA with different lengths (Supplementary Table S2). As illustrated in Figure 1A, p204 HINab displayed binding affinity for 16 mer, 20 mer, and 30 mer dsDNA. The binding affinity of p204 HINab on 16 mer dsDNA is too low to be determined. The Kd value for the binding of p204 HINab to 20 mer dsDNA was determined to be 155.70 ± 37.51 μM and the binding affinity was increased to 13.46 ± 2.39 μM when binding to 30 mer dsDNA. Furthermore, we detected the DNA-binding affinity of p204 HINa and HINb for binding to 30 mer dsDNA. As illustrated in Figure 1B, the Kd value for the binding of p204 HINa and HINb to 30 mer dsDNA was 193.40 ± 77.05 and 157.00 ± 37.24 μM, respectively. These results indicated that the DNA-binding affinities of HINa and HINb were similar when considering the errors in the measurements; however, for human IFI16, Unterholzner et al. reported that the DNA-binding affinity of IFI16 HINb is higher than that of HINa (16). Furthermore, the DNA-binding affinity of p204 HINab is higher than that of HINa or HINb, suggesting that p204 HINa and HINb synergistically bind to dsDNA, which is consistent with the DNA-binding affinity of IFI16 HINab (16,29).
Figure 1.
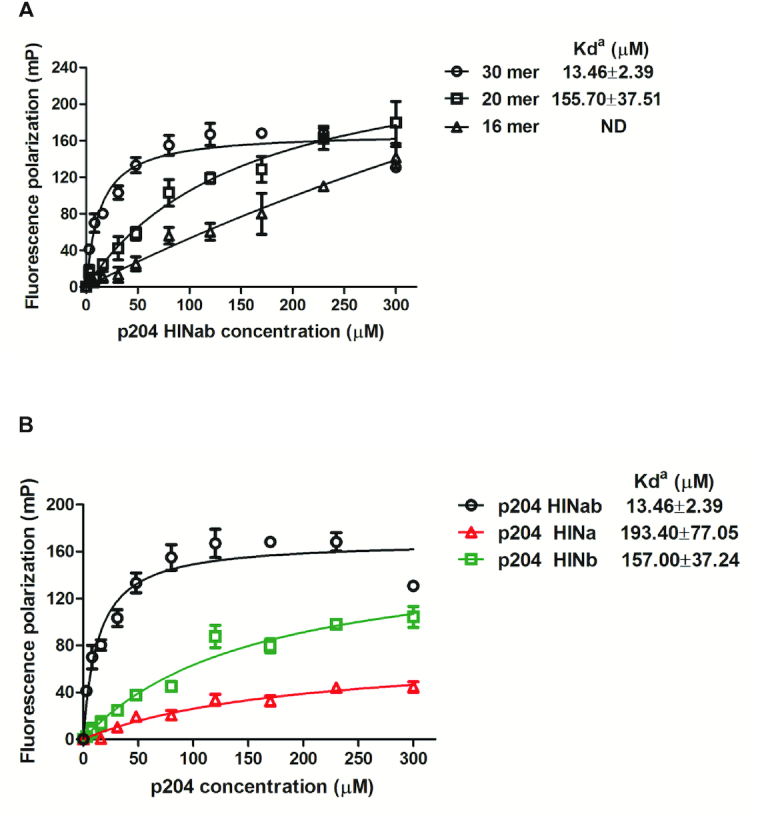
The DNA-binding properties of p204 HIN domain. (A) Fluorescence polarization (FP) assay of p204 HINab binding to FAM-labeled 16 mer, 20 mer and 30 mer dsDNA. The apparent Kd values (Kda) are shown for each dsDNAs. ND means not determined. (B) FP assay of p204 HIN domain binding to FAM-labeled 30 mer dsDNA. The apparent Kd values (Kda) are shown for p204 HINab, HINa and HINb domain.
Structures of p204 HINa and HINb domain
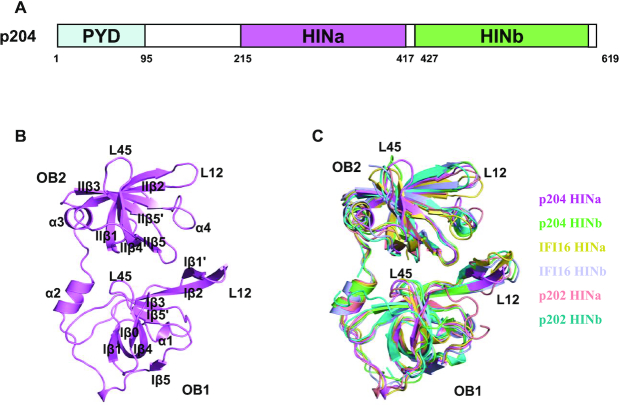
The p204 protein has a tripartite structure comprising three well-defined domains, the PYD domain, and two HIN domains (HINa and HINb domain) (Figure 2A). We determined the crystal structures of the p204 HINa domain at 1.58 Å resolution and HINb domain at 2.0 Å resolution. The diffraction data collection and refinement statistics are presented in Table 2. Moreover, p204 HINa crystallized in the C2221 space group with one molecule in the asymmetric unit (Figure 2B) and HINb crystallized in the P3121 space group with two molecules in the asymmetric unit (Supplementary Figure S3A). Two HINb molecules adopt a similar conformation with an RMSD of 0.6 Å (align 188 Cα) (Supplementary Figure S3B and Table S3). Furthermore, the structural alignment of p204 HINa and HINb results in RMSDs of 1.24 Å (HINb chain A, align 180 Cα) and 1.25 Å (HINb chain B, align 183 Cα) (Supplementary Figure S3B and Table S3). The overall topologies of both HIN domains are highly similar, containing two classical OB folds (OB1 and OB2) joined by a linker containing two α-helices. Each OB fold comprises a β-barrel formed by five twisted β strands (β1–β5).
Figure 2.
Structures of p204 HINa and HINb domains. (A) Domain organization of p204 protein. p204 has an N-terminal PYD domain, a ∼120 residues linker region, followed by a consecutive HINa and HINb domains. The residue numbers at the predicted domain boundaries were labeled. (B) Crystal structure of p204 HINa domain. HINa is shown in violet with the secondary structures labeled. (C) Structural superposition of HIN domains of p204, IFI16 and p202. Structures of p204 HINa and HINb are shown in violet and lime. Structures of IFI16 HINa (PDB: 2OQ0) and HINb (PDB: 3B6Y) are shown in yellow orange and light blue. Structures of p202 HINa (PDB: 4JBJ) and HINb (PDB: 4L5T) are shown in salmon and cyan. Loops L12 and L45 in OB1 and OB2 folds are labeled.
The known HIN domains of the PYHIN family proteins present a highly similar topology. Structural superposition revealed that the HIN domains of p204 were similar to those of other HIN domains, including murine p202 HINa (PDB: 4JBJ) (30) and HINb (PDB: 4L5T) (35) as well as human IFI16 HINa (PDB: 2OQ0) and HINb (PDB: 3B6Y), with RMSDs in the range of 0.99 (align 183 Cα) to 1.72 (align 166 Cα) Å (Supplementary Table S3). Slight differences were found in L12 (loop between Iβ1′ and Iβ2 or loop between IIβ1 and IIβ2) and L45 (loop between Iβ4 and Iβ5′ or loop between IIβ4 and IIβ5) in both OB folds (Figure 2C), with slightly different surface charges (Supplementary Figure S3C), indicating the presence of distinct DNA-binding surfaces. The surface charge slightly differed between p204 HINa and HINb. The region where DNA is bound by other HIN domain proteins acts as the linker between OB1 and OB2, or between L12 and L45 on OB folds. The positive potential of p204 HINa was mainly concentrated on the OB linker, L45 of OB1, L12 and L45 of OB2. The positive potential of HINb is mainly concentrated on the OB linker, L12, of the two OB folds.
Complex structure of p204 HINab:dsDNA
To obtain p204 HINab:dsDNA complex crystals, numerous DNA ligands with varying lengths, sequences, and ends were tested. We obtained single crystals of p204 HINab complexed with 16 mer blunt-ended dsDNA with a sequence of 5′-ccatcagaaagagagc-3′ (Supplementary Figure S4A). Despite their large sizes, most of the crystals are poorly diffracted X-rays. After screening hundreds of crystals, a 4.50 Å dataset was obtained in the P6522 space group. The structure was determined by molecular replacement (MR) using the p204 HINa and HINb structures reported here. The Fo-Fc map clearly indicated the presence of dsDNA in the crystal (Supplementary Figure S4B). In the second round of MR, two pieces of 16 mer dsDNA were placed into the density. The electron density of the dsDNA in the crystal was continuous. After initial refinement, the density of the linker region between p204 HINa and HINb became clear and was successfully built manually. The final structural refinement adopted NCS and secondary structure restraints for the individual coordinates and group B-factor refinements in PHENIX (46,48,49). The final model was validated using Molprobity (Table 2).
The final model had three copies of p204 HINab and two copies of 16 mer dsDNA. The orientation of the three HINab molecules formed a C-ring shaped structure for binding the dsDNA ligand (Figure 3A and Supplementary Figure S4C). Two copies of dsDNA were slightly bent in the asymmetric unit, and three p204 HINab molecules bind to the major groove of dsDNA in a similar mode. After superposition of the structures of HINa:dsDNA and HINb:dsDNA in the p204 HINab:dsDNA complex, all HINa molecules bind to dsDNA via the α2 helix and the C-terminal loop of OB2, whereas all HINb molecules bind to dsDNA via the α2 helix and the N-terminal loop of OB1 (Figure 3B). The conformation of three dsDNA molecules bound to HINb revealed a large shift. No obvious conformational changes in HINa and HINb domains were observed with or without bound dsDNA (Figure 3C). The long flexible linker between HINa and HINb may impact the large shift of dsDNA bound HINb.
Figure 3.
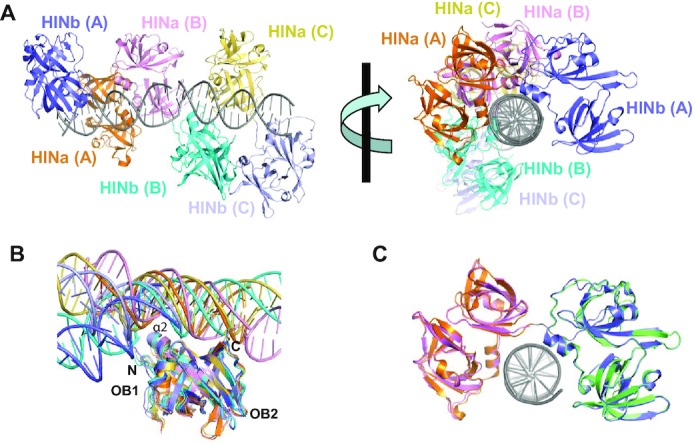
Crystal structure of p204 HINab:dsDNA complex. (A) Overall structure of p204 HINab:dsDNA complex. There are three p204 HINab molecules binding to two copies of dsDNA. Molecule A is shown in orange (HINa) and slate (HINb). Molecule B is shown in pink (HINa) and cyan (HINb). Molecule C is shown in yellow orange (HINa) and light blue (HINb). Two copies of dsDNA are shown in gray. (B) Similar DNA-binding mode of p204 HINa and HINb. Structures of HINa:dsDNA and HINb:dsDNA in p204 HINab:dsDNA complex were aligned, the position of dsDNA are conserved with slight bending. The DNA-binding surface formed by α2, C-termini of HINa, and N-termini of HINb, were showed in the relevant position. dsDNAs are shown in the same colour as the bound HINa or HINb. (C) Structural superposition of p204 HINa (violent), HINb (lime) and HINab:dsDNA.
The HINa dimerization interface of p204
Interestingly, a similar HINa dimerization interface was observed in both p204 HINa and HINab:dsDNA structures (Figure 4A and B). The butterfly-shaped dimer is formed by two p204 HINa molecules with 180° rotational symmetry. This interface comprises primarily of the α2–α3 helices and Iβ3, the N-terminus of β1, and the linker between IIβ2 and IIβ3. The surface accessible surface area of the p204 HINa dimer is ∼850 Å2. In order to explore the influence of HINa dimerization interface for recognizing DNA, we constructed mutant HINab-m1 and HINa-m1 to disrupt the dimerization interface of HINa (Supplementary Figure S5A, B and Table S4). HINab-m1 and HINa-m1 were purified with high purity, and both were monomers in the solution (Supplementary Figure S6A–D). As illustrated in Figure 4C, the DNA-binding affinity of HINab-m1 and HINa-m1 slightly decreased compared with that of wild-type HINab and HINa, indicating the HINa dimerization interface attributes to, but does not dominate DNA binding. Presumably, the dimerization interface was not completely abrogated, and there may be other residues involved in the dimeric interfaces. Moreover, a similar HINa dimer interface was observed in the previously reported IFI16 HINa:ssDNA complex structure (36). Structural superposition of p204 HINa dimer and IFI16 HINa dimer in the DNA-binding mode revealed that the conformation of the p204 HINa dimer was more compact (Figure 4D). Comparing the structures of p204 HINa dimer in the presence and absence of dsDNA, the HINa dimer was shifted after binding to dsDNA, which was closer to dsDNA (Figure 4E), suggesting that the p204 HINa dimer may change its conformation upon binding to dsDNA.
Figure 4.
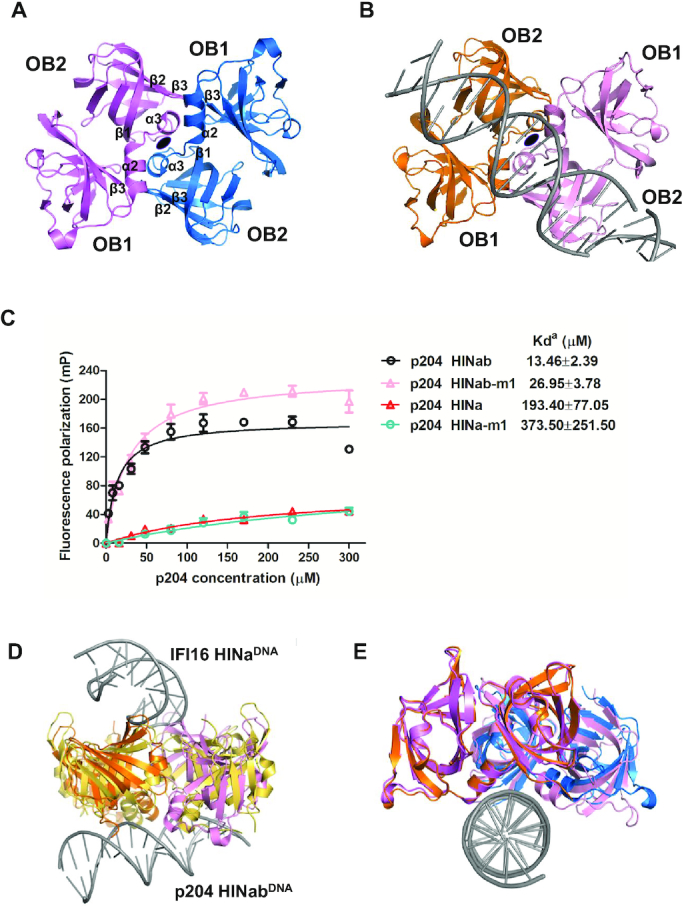
The dimeric interface of p204 HINa domain. (A) HINa dimer in p204 HINa crystal. Two HINa molecules are related by a 2-fold rotational asymmetry and coloured differently. The secondary structure components involved in the dimeric interface were labeled. (B) HINa dimer in p204 HINab:dsDNA complex crystal. (C) FP assay of the dimeric interface of p204 HINab and HINa binding to 30 mer dsDNA. The apparent Kd values (Kda) are shown for p204 HINab, HINa and mutants. (D) Structural comparison of HINa dimer from p204 HINab:DNA and IFI16 HINa:DNA complexes. HINa dimer from IFI16 HINa: DNA complex is shown in yellow orange. HINa dimer from p204 HINab:DNA is shown in orange and pink. (E) Structural comparison of HINa dimer from p204 HINa and HINab:dsDNA complex. HINa dimer from p204 HINa structure is shown in violet and slate. HINa dimer from p204 HINab:dsDNA complex is shown in orange and pink.
The dsDNA-binding mode of p204 HINab
The DNA-binding modes of p204 HINa and HINb were highly similar in the PYHIN family. The OB1–OB2 linker containing α2–α3 helices faces DNA, which has been reported in previous structures including both human and murine AIM2 HIN domains, and the IFI16 HINb domain (29). To investigate the DNA-binding affinity of HINa and HINb on the p204 HINab domain, we introduced four mutants, including HINab-m2 (mutation on HINa DNA-binding interface), HINab-m3 (mutation on HINb DNA-binding interface), HINab-m4 (mutation on HINa dimerization and HINb DNA-binding interface), and HINab-m5 (mutation on both HINa and HINb DNA-binding interface) (Supplementary Figure S5C and Table S4). HINab-m3 with low purity could be obtained due to degradation (Supplementary Figure S6A); however, HINab-m4 and HINab-m5 could not be obtained due to severe degradation and precipitation (Supplementary Figure S6A). All HINab mutants were monomers in solution (Supplementary Figure S6C). The DNA-binding affinities of wild-type and mutant p204 HIINab (HINab-m2 and m3) were measured by the FP assay. As illustrated in Figure 5A, the mutants HINab-m2 and m3 had lower affinities for 30 mer dsDNA. Furthermore, mutant HINa-m2 (mutation on HINa DNA-binding interface) with high purity was introduced to detect the DNA-binding ability of the p204 HINa domain (Supplementary Figure S6B). HINa-m2 is a monomer in solution (Supplementary Figure S6D). Figure 5A illustrates that HINa-m2 possesses a diminished DNA-binding affinity comparable to that of wild-type HINa. These results indicated that both HINa and HINb play vital roles in p204 sensing DNA.
Figure 5.
The DNA-binding mode of HIN domains in PYHIN family. (A) FP assays of the DNA-binding interface of p204 HINab and HINa binding to 30 mer dsDNA. The apparent Kd values (Kda) are shown for p204 HINab, HINa and mutants. ND means not determined. (B) Structural comparison of HIN domains in complex with DNA. Structures of p204 HINa:DNA (orange) and HINb: DNA (slate) are from HINab:dsDNA complex. Structures of p202 HINa (PDB: 4L5R), human AIM2 HIN (PDB: 3RN2), murine AIM2 HIN (PDB: 4JBM), IFI16 HINa (PDB: 4QGU) and IFI16 HINb (PDB: 3RNU) are shown in salmon, pink, cyan, yellow orange and light blue, respectively. (C) Positive surface electrostatics of p204 HINa facing the DNA from the complexc of IFI16 HINa:DNA. It was set to 20% transparency. The loop between α3 and IIβ1, and L45 of OB2 on p204 HINa are labeled.
When structures of HINa:dsDNA and HINb:dsDNA from p204 HINab:dsDNA complex are superposed, the relative position of dsDNA is in a similar position, although DNA undergoes slight bending (Figure 3B). Structural superposition of HIN:DNA complexes revealed that p204 HINa and HINb, human and murine AIM2 HIN (PDB: 3RN2 and 4JBM) and IFI16 HINb (PDB: 3RNU) have a similar DNA-binding mode (Figure 5B). They bind DNA via the OB1–OB2 linker containing α2–α3 helices. The corresponding DNA-binding surface of IFI16 HINa on p204 HINa also presents positive potential, indicating that p204 HINa is likely to bind DNA by the surface formed by the loop between α3 and IIβ1, and L45 of OB2 (Figure 5C). Sequence alignment of HIN domains indicated that the known DNA-binding residues were highly conserved (Figure 6). The DNA-binding residues of p202 HINa and IFI16 HINa were mainly located on L12 and L45 of OB2. The residues of human and murine AIM2 HIN and IFI16 HINb were mainly located on α2–α3 helices, IIβ4 and IIβ5, whereas some conserved residues are also found in p204 HINa and HINb domains.
Figure 6.
Sequence alignment of HIN domains. These HIN domains are from representative human and murine PYHIN family proteins. The secondary structure components were labeled on the top of the alignment. Highly conserved residues are highlighted in yellow. Positively charged residues in L12 and L45 are coloured in red. The residues involved in binding DNA are in black boxes. The red dotted boxes show the residues speculated to be involved in DNA-binding in p204 HINa and HINb domains.
HIN domain linkers between HINa and HINb are dynamically flexible
Fitting of the crystal structures into the SAXS envelope (Figure 7A) indicates that the linker (14 residues, 413–426) between HINa and HINb domains differs from that in the p204 HINab:dsDNA complex (Figure 7B). By superimposing on the HINa domain, the HINb domain swings up >90° toward the dsDNA (Figure 7C). Conformation changes of the linker between HINa and HINb indicate that the linker is flexible. Both HINa and HINb domains are bound to dsDNA through electrostatic charge surfaces in the solution; thus, the linker is forced to change its conformation upon DNA binding. Moreover, three copies of p204 HINab in complex with two dsDNA molecules revealed different linker conformations (hinge angles), as illustrated in Supplementary Figure S7A. Yin et al. observed that p202 HINab formed a tetramer but displayed the ‘heterogenous’ conformations, that is, the linkers (11 residues, 243–253) between HIN1 and HIN2 present different conformations in negative stain (electron microscopy) images (35). The linker in p204 HINab (14 residues) (Supplementary Figure S7A) is longer than that of p202 HINab (11 residues) (Supplementary Figure S7B), whereas the dsDNA in IFI16 HIN domains is wrapped by a long ‘linker’ mixed with loops and helices (residues 394–514), as illustrated in Supplementary Figure S7C (Supplementary Table S5).
Figure 7.
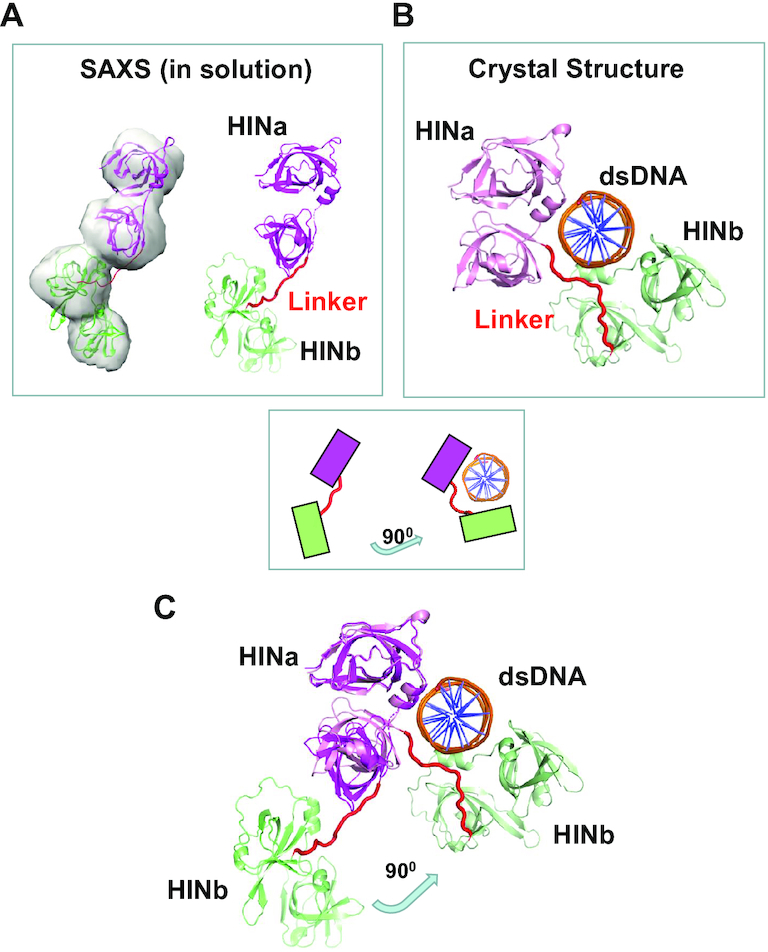
The dynamic flexible linker between HINa and HINb of p204. (A) HIN domain linker of p204 in the solution by SAXS analysis. (B) HIN domain linker in the crystal structure of p204 HINab:dsDNA complex. (C) Conformational changes of HIN domain linker in p204 with and without dsDNA. The superposition of SAXS model (dsDNA-free) and crystal structure (dsDNA-bound) of p204 shows HINb domain swing 90° toward binding to dsDNA (for clarifing view, one of the two dsDNAs is omitted from the figures).
DISCUSSION
PYHIN family receptors belong to an important type of cytosolic DNA sensor that recognizes self as well as pathogenic double-stranded (ds) and several single-stranded (ss) DNAs, although their downstream signaling is diverse. Therefore, understanding the molecular mechanism of DNA ligand recognition and assembly of large protein:nucleic acid is a crucial step. Several single HIN domains and ds/ss DNA complex structures are available. The complex structures of human AIM2 HIN:dsDNA (PDB: 3RN2), murine AIM2 HIN:dsDNA (PDB: 4JBM) and IFI16 HINb:DNA (PDB: 3RNU) present a similar DNA-binding mode; however, p202 HINa and IFI16 HINa bind DNA in two different modes. In the present study, the DNA-binding modes of p204 HINa and HINb were highly similar. Furthermore, this binding mode is shared by HIN domains from human and murine AIM2 (PDB: 3RN2 and 4JBM) and IFI16 HINb domain (PDB: 3RNU). In this binding mode, the HIN domain binds to DNA mainly via a linker connecting two OB folds, whereas p202 HINa binds to DNA mainly via loops of OB1 and OB2 folds. Additionally, IFI16 HINa binds to DNA through IIβ4 and the loop between IIβ4 and IIβ5′. The most significant structural differences among HIN domains are the charge distributions on their surfaces, resulting in a slightly different DNA-binding mode. We revealed that p204 HINab presents the canonical dsDNA-binding mode of the PYHIN family HIN domains, which is not related to its downstream signaling pathways; however, the diverse biological activities of PYHIN family members are determined by the PYD signaling domain.
Structural superposition of HIN:DNA complexes revealed that the conformations of IFI16 HINa and p202 HINa are more similar to p204 HINa than to p204 HINb. After observing the DNA-binding mode in the p204 HINab:dsDNA complex structure, we found that p204 HINa may have a second DNA-binding interface that is similar to the DNA-binding surface of IFI16 HINa. The second DNA-binding surface of p204 HINa is mainly formed by L12 and L45 of OB2 fold, whereas the DNA-binding surface of IFI16 HINa is formed by IIβ4 and L45 of OB2 (36). Moreover, IFI16 HINa has a third DNA-binding surface formed by α1, the linker between α1 and Iβ4, and L45 of OB1. The second DNA-binding surfaces of other HIN domains were also observed in multiple asymmetry units (Supplementary Figure S8). Human AIM2 has a second DNA-binding interface formed by the loop between Iβ1/Iβ1′ and Iβ4, as well as has a third DNA-binding surface formed by L12 of OB2. The second DNA interface of p202 HINa is formed by the linker between Iβ3, Iβ4, and α3. The second DNA-binding surface was not observed in the structures of murine AIM2 HIN:DNA (PDB: 4JBM) and IFI16 HINb:DNA complex (PDB: 3RNU). Furthermore, various DNA-binding interfaces provide a structural basis for explaining the formation of wide DNA filaments. For example, the width of full-length IFI16:dsDNA complexes was consistently between 20 and 25 nm (∼200–250 Å), as observed by the negatively stained electron microscope (37); however, the width of IFI16 HINb binding to one dsDNA molecule was ∼80 Å. The width of more IFI16 HINb:DNA complex in multiple asymmetry units is approximately 22 nm, which is consistent with the width of 20–25 nm. Additionally, IFI16 HINa binds to other DNA molecules via a different DNA-binding surface to stabilize the filaments. Furthermore, the presence of the PYD domain may limit further expansion of IFI16:dsDNA complex filaments.
In this study, we determined the complex structure of p204 HINab:dsDNA, which revealed that three HINab molecules form a C-ring shaped structure for binding dsDNA. In the absence of dsDNA, p204 HINab is a monomer in solution, as confirmed via size exclusion chromatography and SAXS assay. Moreover, p204 HINa and HINb synergistically bind to dsDNA, and the DNA-binding affinity of HINab is higher than that of HINa and HINb. Although p204 HINa is a monomer in solution, it presents a dimer in the crystal structure of the HINa and HINab:dsDNA complex. The dimerization interface of HINa is involved in binding dsDNA confirmed by FP assay. Furthermore, p204 is the homolog of IFI16 and the identity of its full-length sequence is 36.95%; thus, it may have highly similar topological structure. The full length of IFI16 has an extended conformation, indicating that the full length of p204 also presents an extended structure before recognizing dsDNA. Acting as the homotypic protein–protein interaction domain, the N-terminal PYD domain of p204 interacts with other PYD domains and drives filament assembly in a mode similar to that of IFI16 after binding to DNA (37). Because the linker between HINa and HINb is longer in IFI16 than that in p204, perhaps the conformation of IFI16 HINab is more elongated than that of p204 HINab in the solution. Morrone et al. reported that more than 15 mer dsDNA is required for binding IFI16 by the FP assay (37). In accordance with our results, presumably the minimum length of dsDNA to bind HINab is approximately 16 mer and the minimum length to induce p204 HINab dimerization is at least 20 mer (observed from the HINab:dsDNA complex). Based on our results and previous reports, we propose a working model of p204 and IFI16 that recognizes dsDNA to activate downstream signaling pathways (Figure 8). In the resting state, the tripartite protein IFI16/p204 presented an extended conformation in the cytoplasm. Once recognizing dsDNA from invading virus, IFI16/p204 binds to dsDNA by its C-terminal HINab domain, in which HINa and HINb synergistically bind to dsDNA. Subsequently, more HINab molecules were arranged to form a C-ring shaped structure to bind long dsDNA. Simultaneously, the binding of dsDNA stabilizes the dimerization of the HINa domain, resulting in the close proximity of the adjacent N-terminal PYD domain, which aggregates to activate STING. Activated STING leads to the phosphorylation of TBK1 and IRF3 and induces the production of IFNβ and other proinflammatory cytokines to defend against the virus.
Figure 8.
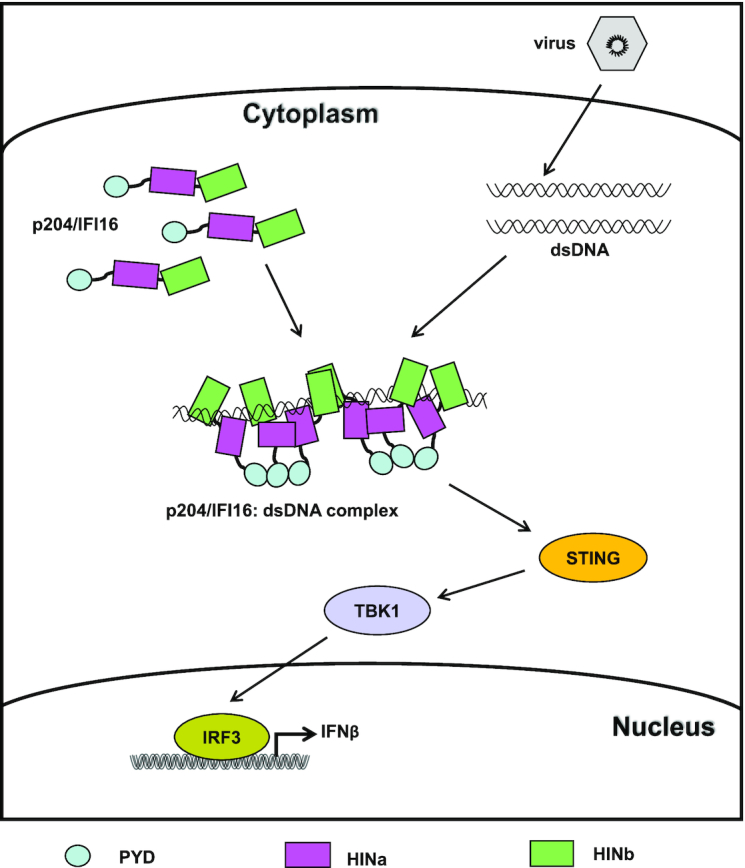
A working model of IFI16/p204 recognizing dsDNA to active downstream signaling pathway. In the absence of DNA, IFI16/p204 presents an extended conformation in the cytoplasm. Once recognizing dsDNA from invading virus, IFI16/p204 HINab domain synergistically binds to dsDNA. More HINab molecules bind to the long dsDNA and form a C-ring shaped structure around dsDNA. The binding of dsDNA stabilizes the dimerization of HINa domain, resulting in the adjacent N-terminal PYD domain are closer and aggregate to activate STING. Activated STING leads to the phosphorylation of TBK1 and IRF3 and induces the production of IFNβ and other proinflammatory cytokines to defend against the virus infection.
In summary, our studies on p204 provide the first crystal structure of the HINab domain in complex with dsDNA, revealing a similar nature of dsDNA recognition by HIN domains. This structure further revealed that dsDNA binding to the HINab domain induced slight conformational changes in the hinge between HINa and HINb. Most interestingly, a uniform HINa dimerization mode was identified. The structure of the p204 HINab:dsDNA complex, together with p204 HINa and HINb structures, completes the molecular description of p204 involved in recognizing dsDNA. These results markedly advance our understanding of the mechanism by which IFI16/p204 receptors sense dsDNA by their tandem HIN domain.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5YZP (p204 HINa), 5YZW (p204 HINb), 5Z7D (p204 HINab: dsDNA complex).
Supplementary Material
ACKNOWLEDGEMENTS
X-ray diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamline at the APS, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. SAXS data were collected at beamline X9 of the National Synchrotron Light Source at the Brookhaven National Laboratory. We would like to thank Editage (www.editage.com) for English language editing.
Contributor Information
Xiaojiao Fan, Department of Obstetrics and Gynecology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230001, P.R. China.
Jiansheng Jiang, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Dan Zhao, Hefei National Laboratory for Physical Sciences at Microscale, the CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027 China.
Feng Chen, Hefei National Laboratory for Physical Sciences at Microscale, the CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027 China.
Huan Ma, Hefei National Laboratory for Physical Sciences at Microscale, the CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027 China.
Patrick Smith, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Leonie Unterholzner, Division of Biomedical and Life Sciences, Faculty of Health and Medicine, Lancaster University, Lancaster LA1 4YQ, UK.
Tsan Sam Xiao, Department of Pathology, Case Western Reserve University, Cleveland, OH 44106, USA.
Tengchuan Jin, Department of Obstetrics and Gynecology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230001, P.R. China; Hefei National Laboratory for Physical Sciences at Microscale, the CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027 China; CAS Center for Excellence in Molecular Cell Science, Shanghai, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Fund for Young Scholars [31800639]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB29030104]; National Natural Science Foundation of China [31870731, 31971129]; Fundamental Research Funds for the Central Universities and the 100 Talents Program of the Chinese Academy of Sciences; National Institutes of Health [R01GM127609]; intramural research program of National Institute of Allergy and Infectious Diseases. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences [XDB29030104].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wu J., Chen Z.J.. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014; 32:461–488. [DOI] [PubMed] [Google Scholar]
- 2. Unterholzner L. The interferon response to intracellular DNA: why so many receptors?. Immunobiology. 2013; 218:1312–1321. [DOI] [PubMed] [Google Scholar]
- 3. Hornung V. SnapShot: nucleic acid immune sensors, part 2. Immunity. 2014; 41:1066–1066. [DOI] [PubMed] [Google Scholar]
- 4. Xiao T.S. The nucleic acid-sensing inflammasomes. Immunol. Rev. 2015; 265:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoneyama M., Onomoto K., Jogi M., Akaboshi T., Fujita T.. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015; 32:48–53. [DOI] [PubMed] [Google Scholar]
- 6. Liu C.J., Wang H., Lengyel P.. The interferon-inducible nucleolar p204 protein binds the ribosomal RNA-specific UBF1 transcription factor and inhibits ribosomal RNA transcription. EMBO J. 1999; 18:2845–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh S., Wallerath C., Covarrubias S., Hornung V., Carpenter S., Fitzgerald K.A.. The PYHIN protein p205 regulates the inflammasome by controlling asc expression. J. Immunol. 2017; 199:3249–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., Alnemri E.S.. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009; 458:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., Fitzgerald K.A.. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009; 458:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernandes-Alnemri T., Yu J.W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E.et al.. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 2010; 11:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S., Bauernfeind F., Ablasser A., Hartmann G., Fitzgerald K.A., Latz E., Hornung V.. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 2010; 40:1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sauer J.D., Witte C.E., Zemansky J., Hanson B., Lauer P., Portnoy D.A.. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010; 7:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., Vanaja S.K., Monks B.G., Ganesan S., Latz E.et al.. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 2010; 11:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu B., Jin C., Li H.B., Tong J., Ouyang X., Cetinbas N.M., Zhu S., Strowig T., Lam F.C., Zhao C.et al.. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016; 354:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kerur N., Veettil M.V., Sharma-Walia N., Bottero V., Sadagopan S., Otageri P., Chandran B.. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011; 9:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S.et al.. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010; 11:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Orzalli M.H., Conwell S.E., Berrios C., DeCaprio J.A., Knipe D.M.. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4492–E4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diner B.A., Lum K.K., Cristea I.M.. The emerging role of nuclear viral DNA sensors. J. Biol. Chem. 2015; 290:26412–26421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jakobsen M.R., Bak R.O., Andersen A., Berg R.K., Jensen S.B., Tengchuan J., Laustsen A., Hansen K., Ostergaard L., Fitzgerald K.A.et al.. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4571–E4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Almine J.F., O’Hare C.A., Dunphy G., Haga I.R., Naik R.J., Atrih A., Connolly D.J., Taylor J., Kelsall I.R., Bowie A.G.et al.. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 2017; 8:14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jonsson K.L., Laustsen A., Krapp C., Skipper K.A., Thavachelvam K., Hotter D., Egedal J.H., Kjolby M., Mohammadi P., Prabakaran T.et al.. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 2017; 8:14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iqbal J., Ansari M.A., Kumar B., Dutta D., Roy A., Chikoti L., Pisano G., Dutta S., Vahedi S., Veettil M.V.et al.. Histone H2B-IFI16 recognition of nuclear herpesviral genome induces cytoplasmic interferon-beta responses. PLoS Pathog. 2016; 12:e1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray E.E., Winship D., Snyder J.M., Child S.J., Geballe A.P., Stetson D.B.. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity. 2016; 45:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yi Y.S., Jian J., Gonzalez-Gugel E., Shi Y.X., Tian Q., Fu W., Hettinghouse A., Song W., Liu R., He M.et al.. p204 is required for canonical lipopolysaccharide-induced TLR4 signaling in mice. EBioMedicine. 2018; 29:78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen W., Yu S.X., Zhou F.H., Zhang X.J., Gao W.Y., Li K.Y., Liu Z.Z., Han W.Y., Yang Y.J.. DNA Sensor IFI204 contributes to host defense against staphylococcus aureus infection in mice. Front. Immunol. 2019; 10:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J., Zhang X., Zhao Z., Li X., Wang X., Chen M., Song B., Ii M., Shen Z.. Regulatory roles of interferon-inducible protein 204 on differentiation and vasculogenic activity of endothelial progenitor cells. Stem Cell Res. Ther. 2016; 7:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choubey D., Panchanathan R.. IFI16, an amplifier of DNA-damage response: Role in cellular senescence and aging-associated inflammatory diseases. Ageing Res. Rev. 2016; 28:27–36. [DOI] [PubMed] [Google Scholar]
- 28. Cridland J.A., Curley E.Z., Wykes M.N., Schroder K., Sweet M.J., Roberts T.L., Ragan M.A., Kassahn K.S., Stacey K.J.. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol. Biol. 2012; 12:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin T., Perry A., Jiang J., Smith P., Curry J.A., Unterholzner L., Jiang Z., Horvath G., Rathinam V.A., Johnstone R.W.et al.. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012; 36:561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ru H., Ni X., Zhao L., Crowley C., Ding W., Hung L.W., Shaw N., Cheng G., Liu Z.J.. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013; 23:855–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang B., Yin Q.. AIM2 inflammasome activation and regulation: a structural perspective. J. Struct. Biol. 2017; 200:279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xin H., D'Souza S., Jorgensen T.N., Vaughan A.T., Lengyel P., Kotzin B.L., Choubey D.. Increased expression of Ifi202, an IFN-activatable gene, in B6.Nba2 lupus susceptible mice inhibits p53-mediated apoptosis. J. Immunol. 2006; 176:5863–5870. [DOI] [PubMed] [Google Scholar]
- 33. Roberts T.L., Idris A., Dunn J.A., Kelly G.M., Burnton C.M., Hodgson S., Hardy L.L., Garceau V., Sweet M.J., Ross I.L.et al.. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009; 323:1057–1060. [DOI] [PubMed] [Google Scholar]
- 34. Li H., Wang J., Wang J., Cao L.S., Wang Z.X., Wu J.W.. Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta Crystallogr. F Struct. Biol. Commun. 2014; 70:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yin Q., Sester D.P., Tian Y., Hsiao Y.S., Lu A., Cridland J.A., Sagulenko V., Thygesen S.J., Choubey D., Hornung V.et al.. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013; 4:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ni X., Ru H., Ma F., Zhao L., Shaw N., Feng Y., Ding W., Gong W., Wang Q., Ouyang S.et al.. New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16. J. Mol. Cell Biol. 2016; 8:51–61. [DOI] [PubMed] [Google Scholar]
- 37. Morrone S.R., Wang T., Constantoulakis L.M., Hooy R.M., Delannoy M.J., Sohn J.. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E62–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z., Zheng X., Wang Y., Song H.. Bacterial expression of the HINab domain of IFI16: purification, characterization of DNA binding activity, and co-crystallization with viral dsDNA. Protein Expr. Purif. 2014; 102:13–19. [DOI] [PubMed] [Google Scholar]
- 39. Jin T., Perry A., Smith P., Jiang J., Xiao T.S.. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J. Biol. Chem. 2013; 288:13225–13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 41. Kabsch W. Xds. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Potterton E., Briggs P., Turkenburg M., Dodson E.. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D, Biol. Crystallogr. 2003; 59:1131–1137. [DOI] [PubMed] [Google Scholar]
- 44. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 46. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diederichs K., Junk M.. Post-processing intensity measurements at favourable dose values. J. Appl. Crystallogr. 2009; 42:48–57. [Google Scholar]
- 48. Schroder G.F., Levitt M., Brunger A.T.. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010; 464:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brunger A.T., Adams P.D., Fromme P., Fromme R., Levitt M., Schroder G.F.. Improving the accuracy of macromolecular structure refinement at 7 A resolution. Structure. 2012; 20:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003; 36:1277–1282. [Google Scholar]
- 51. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Franke D., Svergun D.I.. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009; 42:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 54. Svergun D., Barberato C., Koch M.H.J.. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995; 28:768–773. [Google Scholar]
- 55. Liao J.C., Lam R., Brazda V., Duan S., Ravichandran M., Ma J., Xiao T., Tempel W., Zuo X., Wang Y.X.et al.. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure. 2011; 19:418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tian Y., Yin Q.. Structural analysis of the HIN1 domain of interferon-inducible protein 204. Acta Crystallogr. F-Struct. Biol. Commun. 2019; 75:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5YZP (p204 HINa), 5YZW (p204 HINb), 5Z7D (p204 HINab: dsDNA complex).