Abstract
Background
The degree to which infection with SARS-CoV-2 confers protection towards subsequent reinfection is not well described. In 2020, as part of Denmark's extensive, free-of-charge PCR-testing strategy, approximately 4 million individuals (69% of the population) underwent 10·6 million tests. Using these national PCR-test data from 2020, we estimated protection towards repeat infection with SARS-CoV-2.
Methods
In this population-level observational study, we collected individual-level data on patients who had been tested in Denmark in 2020 from the Danish Microbiology Database and analysed infection rates during the second surge of the COVID-19 epidemic, from Sept 1 to Dec 31, 2020, by comparison of infection rates between individuals with positive and negative PCR tests during the first surge (March to May, 2020). For the main analysis, we excluded people who tested positive for the first time between the two surges and those who died before the second surge. We did an alternative cohort analysis, in which we compared infection rates throughout the year between those with and without a previous confirmed infection at least 3 months earlier, irrespective of date. We also investigated whether differences were found by age group, sex, and time since infection in the alternative cohort analysis. We calculated rate ratios (RRs) adjusted for potential confounders and estimated protection against repeat infection as 1 – RR.
Findings
During the first surge (ie, before June, 2020), 533 381 people were tested, of whom 11 727 (2·20%) were PCR positive, and 525 339 were eligible for follow-up in the second surge, of whom 11 068 (2·11%) had tested positive during the first surge. Among eligible PCR-positive individuals from the first surge of the epidemic, 72 (0·65% [95% CI 0·51–0·82]) tested positive again during the second surge compared with 16 819 (3·27% [3·22–3·32]) of 514 271 who tested negative during the first surge (adjusted RR 0·195 [95% CI 0·155–0·246]). Protection against repeat infection was 80·5% (95% CI 75·4–84·5). The alternative cohort analysis gave similar estimates (adjusted RR 0·212 [0·179–0·251], estimated protection 78·8% [74·9–82·1]). In the alternative cohort analysis, among those aged 65 years and older, observed protection against repeat infection was 47·1% (95% CI 24·7–62·8). We found no difference in estimated protection against repeat infection by sex (male 78·4% [72·1–83·2] vs female 79·1% [73·9–83·3]) or evidence of waning protection over time (3–6 months of follow-up 79·3% [74·4–83·3] vs ≥7 months of follow-up 77·7% [70·9–82·9]).
Interpretation
Our findings could inform decisions on which groups should be vaccinated and advocate for vaccination of previously infected individuals because natural protection, especially among older people, cannot be relied on.
Funding
None.
Introduction
SARS-CoV-2, the cause of the COVID-19 epidemic, has resulted in over 117 million cases and over 2·6 million deaths worldwide as of March 7, 2021, as estimated by WHO. The presence or absence of protective immunity after infection with, or vaccination against, SARS-CoV-2 will affect transmission of the virus and severity of illness.1 The absence of pre-existing immunity to SARS-CoV-2 is thought to be responsible for the rapid spread of the virus globally and for the continuing pandemic. Therefore, greater understanding of the degree of protection against reinfection with SARS-CoV-2 is essential to refine appropriate intervention strategies.
Little is known about protection against SARS-CoV-2 repeat infections but two studies in the UK have found that immunity could last at least 5–6 months after infection.1, 2, 3 These data suggest that reinfection with SARS-CoV-2 is rare and occurs in less than 1% of individuals who previously tested positive for SARS-CoV-2. These findings are consistent with several single or small case studies, with only up to six patients, done in the USA, China, South Korea, and India, that found reinfection to occur within 26–142 days after the first infection, supported by genetic evidence and negative PCR test results in between the two infections.4, 5, 6, 7 The closely related viruses SARS-CoV and MERS-CoV induced immunity that typically lasted 2–3 years after infection.8, 9, 10
Research in context.
Evidence before this study
We searched PubMed and the bioRxiv and medRxiv preprint servers for publications between Sept 1, 2019, and Feb 14, 2021, without language restrictions, using the terms (“SARS-CoV-2” OR “COVID-19” OR “COVID” OR “coronavirus”) AND “reinfection” AND “immunity”. For preprint articles, we included the search term “human” and found 695 articles, of which 192 were published on the bioRxiv server and 503 on the medRxiv server. On PubMed, we found 112 peer-reviewed publications, and after filtering by human studies we identified 48 articles. Most of these peer-reviewed articles report that reinfection with SARS-CoV-2 is a rare event occurring in less than 1% of COVID-19 cases. Next, we repeated our PubMed search and changed the search term “reinfection” to “seroprevalence” to assess immunity to SARS-CoV-2 in the population at a given time. We identified 47 studies that reported a seroprevalence of SARS-CoV-2 based on serum antibody responses of 0·37–22·1%, highlighting the absence or low level of immunity in the population overall. When looking at natural immunity, we identified three cohort studies: one peer-reviewed article of reinfections in residents in two nursing care homes and two preprint articles, one that followed up 43 000 individuals in Qatar and found an estimated 95% protection against reinfection, and one of over 20 000 health-care workers in the UK that found an 83% lower risk of reinfection for at least 5 months after the first infection.
Added value of this study
Through analysis of Danish population-level surveillance data with more than 10 million person-identifiable PCR test results in 2020, we estimated protective immunity to be approximately 80–83% in people younger than 65 years. We found no difference in immunity over the study period. Among those aged 65 years and older, immunity was estimated to be approximately 47%.
Implications of all the available evidence
Natural infection with SARS-CoV-2 led to observed protection against reinfection estimated to be approximately 80% after 6 months. However, the observed low natural immunity in people aged 65 years and older underlines the need to vaccinate previously infected individuals, in particular in this age group.
Denmark recorded its first positive SARS-CoV-2 case on Feb 27, 2020.11 Similar to other European countries, the epidemic was characterised by two infection surges (waves) in 2020, one in spring (March–May) and one in autumn–winter (September–December). Denmark has been doing high intensity testing for SARS-CoV-2 infection among its 5·8 million residents, with investment in test facilities done with the wider aim of keeping society open whenever possible. In addition to PCR tests done on symptomatic individuals by referral within the national health-care system, a parallel national testing system, primarily targeting non-symptomatic individuals, became widely available from May, 2020, known as TestCenter Denmark. As part of this parallel testing system, nationwide test stations offered free PCR testing to all residents aged 18 years and older, with subsequent expansion to everyone older than 2 years in September, 2020. The number of test stations has been gradually increasing over 2020, and by Dec 31, 2020, more than 10 million PCR tests had been done in Denmark on 3·96 million individuals in total. All tests are unequivocally person identifiable, which allows identification of individuals with more than one positive test at a national scale. The aim of our study was to estimate protection against repeat infections as measured by PCR positivity of SARS-CoV-2, including whether the estimated protection resulting from a first infection differed by age group, sex, or time period since infection.
Methods
Study design, data collection, and surveillance system
In this population-level observational study, we collected individual-level data from the Danish Microbiology Database (MiBa) for all individuals who had a PCR test for SARS-CoV-2 between Feb 26 and Dec 31, 2020. Electronic records of bookings and results are captured in the MiBa in a person-identifiable format and enriched with data (including age, sex, and vital status) from the civil registry system and other registries by the automated national surveillance system.12, 13 The surveillance system is hosted and maintained by Statens Serum Institut (SSI; Copenhagen, Denmark), the Danish National Institute for Infectious Disease Control and Prevention.
At each PCR-testing site that is part of TestCenter Denmark, throat swabs are collected and transported to central high-throughput laboratory facilities (one on the SSI campus or one that opened on Dec 7, 2020, in Aarhus, Denmark) and tested by PCR. Appointments for PCR tests are booked electronically or by telephone, with test results generally available within 48 h (positive, negative, or inconclusive) and communicated to the patient online via personal electronic health records (and their primary care physician). For those who were tested via referral as part of the national health-care testing system, patients attended hospitals or primary care centres and throat swabs were taken for PCR testing and transported to one of ten main clinical, public, microbiological laboratories. Depending on whether patients were tested as part of the national health-care system or TestCenter Denmark, different PCR platforms were used. The clinical microbiology laboratories applied a range of CE-marked commercial platforms or in-house assays that were all quality controlled according to clinical microbiology diagnostic standards. The TestCenter Denmark laboratory applied an RT-PCR assay with the E gene on SARS-CoV-2 as the target.14, 15
In December, 2020, rapid antigen testing also became freely available in separate dedicated public test stations (commercial antigen testing had been available to purchase before this date); these tests were not captured by surveillance and are not included in this analysis. The analyses we present in this Article are based on data extracted from the national surveillance system and covers all PCR tests, both those done within the national health-care system and in TestCenter Denmark.
Our analyses are based on existing Danish national COVID-19 surveillance data and did not require ethical approval. A copy of the protocol is available online.
Analysis of infections and reinfections during the second surge
For inclusion in this analysis, we selected all people in the country with a positive or negative PCR test from the first surge of the epidemic—ie, before June 1, 2020 (all inconclusive test results [approximately 1% of all tests] were removed from the dataset before analysis). We then followed up this cohort through the second surge of the epidemic—from Sept 1 to Dec 31, 2020—to see who contracted a (PCR-confirmed) SARS-CoV-2 infection during this period. The rates of infection during the second surge were compared across those with a positive or negative test from the first surge. Individuals who tested positive for the first time during the period June 1 to Aug 31 (ie, between the end of the first surge and the beginning of the second surge) were excluded from the analysis, as were people who died from any cause before Sept 1, 2020.
We calculated the rate of infection as the number of individuals with positive PCR tests during the second surge divided by the cumulative number of person-days at risk. We calculated the number of days at risk for each individual in the sample as the number of days from Sept 1, 2020, until the first positive test, or Dec 31, 2020, whichever came first. We censored follow-up time in the event of death. This non-informative censoring mechanism essentially assumed a similar infection rate would have been observed among those who died if they had survived, as was observed among the survivors with the same exposure status (whether previously infected or uninfected). We calculated the adjusted rate ratio (RR) and accompanying 95% CI using Poisson regression, adjusted for sex, age group (0–5, 6–14, 15–24, 25–34, 35–44, 45–54, 55–64, 65–74, 75–84, and ≥85 years), and test frequency (number of PCR tests done on each person in 2020 categorised as 1–2, 3–5, 6–10, and ≥11 tests) to control for potential confounding. Protection against repeat infections was calculated as 1 – adjusted RR, analogous to the method of estimating vaccine effectiveness from observational data.
Because people were prone to have more tests done if they thought they might be at increased risk of infection, there was a possibility of bias in the main analysis, especially if testing patterns differed among those with and without a previous positive test. For this reason, we did a sensitivity analysis in which we repeated the main analysis in a subgroup of people (nurses, doctors, social workers, and health-care assistants) who were tested frequently and routinely as part of their profession (details on professions were obtained through linkage with the registry on health authorisations from the Danish Patient Safety Authority).11
To determine the extent to which our findings depended on the gap between the first and second surge (ie, the minimum length of time allowed between repeat positive tests for an individual to be categorised as reinfected), we did two further sensitivity analyses, one in which the second surge began on Aug 1, 2020 (ie, 2 months after the end of the first surge), and one in which it began on Oct 1, 2020 (ie, 4 months after the end of the first surge). The period for the first surge was unchanged.
Alternative cohort analysis
Using an alternative analysis approach, we made full use of the available data to investigate rates of reinfection throughout the epidemic, not just during the second surge. Each individual with a PCR test result was followed up from the time of their first test, irrespective of the date and whether they had a positive or negative result, until Dec 31, 2020, or a new positive test at least 90 days later. If the initial test was negative, a subsequent positive test within the 90 days changed an individual's status from uninfected to previously infected. We compared the infection rate observed during follow-up when people were uninfected with the rate observed during follow-up of people who were previously infected. Those who tested positive during follow-up for the first time (ie, who had initially contributed time as an uninfected individual) remained in follow-up but, from the date of their first infection, contributed time as a previously infected individual. We estimated the adjusted RR using the same methods as for the main analysis, except the model was additionally adjusted for start month of follow-up (through inclusion of indicator variables) to minimise confounding due to variations in the underlying infection rate over time.
Variations by age, sex, and time since first infection
To assess whether the level of protection conferred by previous infection differed by sex or age, we expanded the alternative cohort analysis to include interaction terms with sex and age group (restricted to four age groups [0–34, 35–49, 50–64, ≥65 years] to avoid strata with few events). This expansion allowed us to calculate a protective effect estimate separately for each age group and by sex, and to test for evidence of effect modification using a likelihood ratio test. We used a similar approach to compare the level of protection against repeat infections as measured by positive PCR test before and after the first 6 months of follow-up. We did this analysis by splitting time at risk into two periods for those with more than 180 days of follow-up (individuals with <181 days of follow-up contributed with just a single period not exceeding 6 months), the first from 0 to 180 days, and the second from day 181 until the end of follow-up, whether through a positive test result, death, or study end. We included an interaction term with period in the model to allow for separate assessment of protection against repeat infections as measured by PCR positivity for the two periods. We also plotted a Kaplan-Meier curve of time until infection during follow-up.
We report proportions calculated using exact (Clopper-Pearson) 95% CIs. We did all analyses using SAS version 9.4 and generated graphs using Graphpad Prism (version 8.3.0).
Role of funding source
There was no funding source for this study.
Results
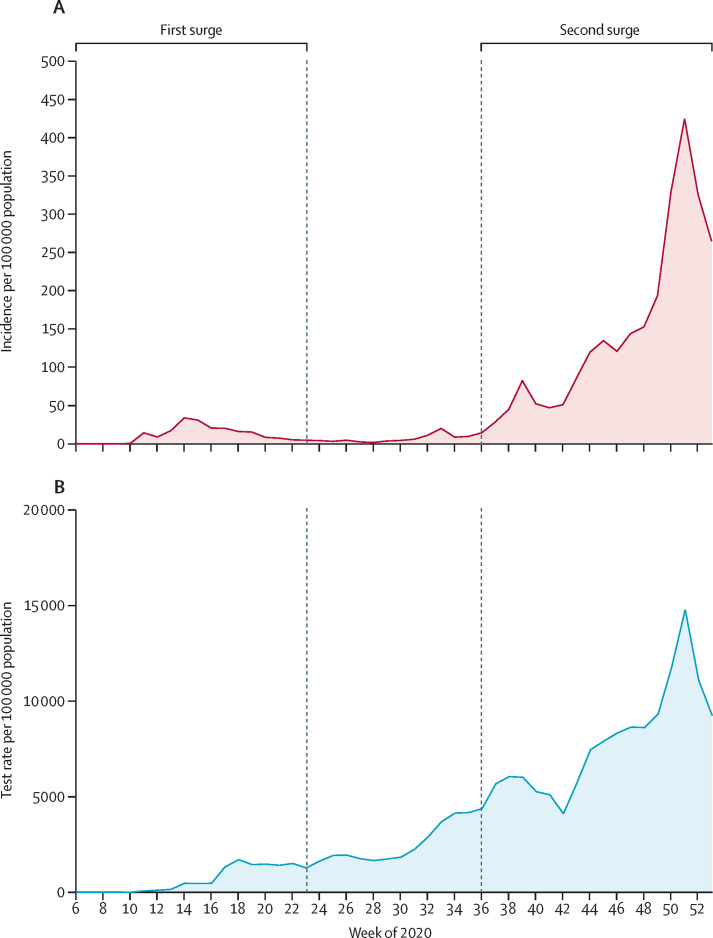
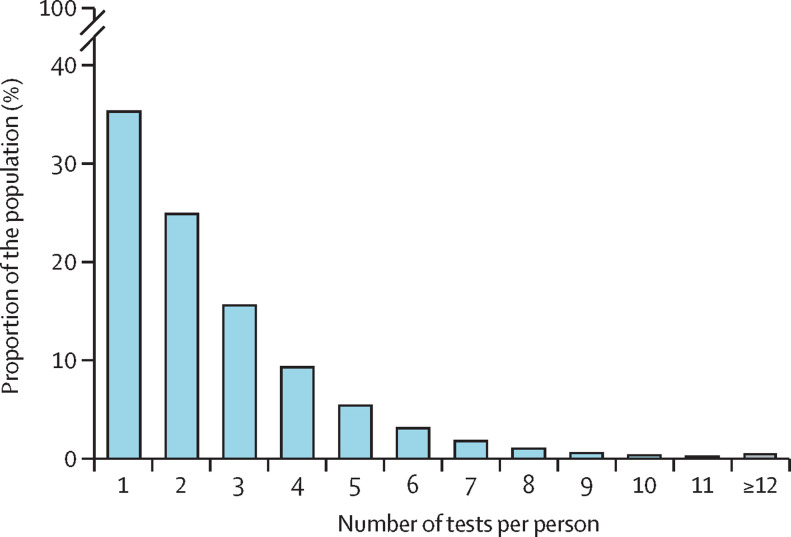
The capacity to do PCR testing for SARS-CoV-2 in Denmark increased rapidly over 2020, from the first tests in February up to the end of the year, when approximately 10% of the population was tested each week on average. During the first surge of the epidemic (ie, before June), 533 381 people were tested, of whom 11 727 (2·20%) were PCR positive. During the second surge (from Sept 1 to Dec 31, 2020), 3·48 million people were tested, of whom 150 159 (4·32%) tested positive (figure 1 ). By Dec 31, 2020, 3·96 million people—more than two-thirds of the population of 5·8 million people—had been tested at least once, of whom 2·55 million (64·4%) had been tested more than once (figure 2 ; appendix p 6).
Figure 1.
Weekly incidence of PCR-confirmed SARS-CoV-2 (A) and test rate (B) in Denmark over 2020
Data are presented per 100 000 population between Feb 3 (week 6) and Dec 31 (week 53), 2020.
Figure 2.
Number of tests per person
The total number of PCR tests done per person in Denmark in 2020 among the 3·96 million people who were tested at least once.
After excluding 610 people who tested positive for the first time between the first and second surges of the epidemic, and a further 7432 who died (from any cause) before the second surge (of whom 659 had tested positive for SARS-CoV-2 during the first surge; appendix p 2), 525 339 of those who were tested during the first surge of the epidemic remained in follow-up during the second surge. In this population, 11 068 (2·11%) individuals tested positive during the first surge, of whom 72 (0·65% [95% CI 0·51–0·82]) tested positive again during the second surge compared with 16 819 (3·27% [95% CI 3·22–3·32]) of 514 271 people who were negative during the first surge.
The daily rate of infection during the second surge was 5·35 positive tests per 100 000 people among those who had previously tested positive versus 27·06 per 100 000 people among those who previously tested negative (table 1 ). The adjusted RR of infection was 0·195 (95% CI 0·155–0·246) among those who previously tested positive compared with those who had previously only tested negative. The estimated protection against repeat infection after previous SARS-CoV-2 infection was 80·5% (95% CI 75·4–84·5; table 1).
Table 1.
Comparison of infection and reinfection rates before and after first SARS-CoV-2 infection in 2020 in Denmark
| Population | Confirmed new infection during follow-up | Person-days of follow-up | Infection rate*during follow-up | Adjusted rate ratio (95% CI)† | Estimated protection (95% CI) | ||
|---|---|---|---|---|---|---|---|
| Main analysis of reinfection during the second surge | |||||||
| Positive during first surge | 11 068 | 72 | 1 346 920 | 5·35 | 0·195 (0·155–0·246) | 80·5% (75·4–84·5) | |
| Negative during first surge | 514 271 | 16 819 | 62 151 056 | 27·06 | 1 (ref) | .. | |
| Alternative cohort analysis with reinfection at least 90 days after first infection‡ | |||||||
| Exposed periods | 28 875 | 138 | 2 447 924 | 5·64 | 0·212 (0·179–0·251) | 78·8% (74·9–82·1) | |
| Unexposed periods | 2 405 683 | 53 991 | 174 487 793 | 30·94 | 1 (ref) | .. | |
| Sensitivity analyses of reinfection during the second surge | |||||||
| In frequently tested nurses, doctors, social workers, and health-care assistants | |||||||
| Positive during first surge | 658 | 8 | 80 014 | 10·00 | 0·189 (0·094–0·379) | 81·1% (62·1–90·6) | |
| Negative during first surge | 14 946 | 934 | 1 798 184 | 51·94 | 1 (ref) | .. | |
| If the second surge was Aug 1 to Dec 31, 2020§ | |||||||
| Positive during first surge | 11 068 | 87 | 1 687 700 | 5·15 | 0·233 (0·189–0·287) | 76·7% (71·3–81·1) | |
| Negative during first surge | 514 562 | 17 110 | 78 098 000 | 21·91 | 1 (ref) | .. | |
| If the second surge was Oct 1 to Dec 31, 2020§ | |||||||
| Positive during first surge | 11 068 | 59 | 1 016 359 | 5·81 | 0·172 (0·133–0·222) | 82·8% (77·8–86·7) | |
| Negative during first surge | 513 025 | 15 573 | 46 739 367 | 33·32 | 1 (ref) | .. | |
Rate of infection per 100 000 person-days of follow-up.
Adjusted for sex, age group, and test frequency, and, for the alternative cohort analysis only, start month of follow-up.
Exposed periods are periods of follow-up time contributed by individuals with previous infection and unexposed periods are contributed by individuals without a previous infection.
For the sensitivity analyses exploring 2 months and 4 months of separation between the two surges, surge one was unchanged.
Test frequency during the second surge was a little higher among those who did not have a positive test result during the first surge than among those who tested positive during the first surge (appendix p 3). In a sensitivity analysis, we restricted the sample to the 15 604 frequently tested nurses, doctors, social workers, and health-care assistants that were present in the sample. They had a median of 10 tests (IQR 9–12) done each in 2020, and 658 (4·2%) tested positive during the first surge (table 1). Eight (1·2%) of 658 who tested positive in the first surge also tested positive during the second surge. By contrast, among those who remained uninfected during the first surge, 934 (6·2%) of 14 946 tested positive during the second surge. The adjusted RR was 0·189 (95% CI 0·094–0·379) and the estimated protection against reinfection was 81·1% (95% CI 62·1–90·6). Moving the date on which the second surge began in two sensitivity analyses, and thereby revising the definition of reinfection in our study by changing the gap between the first and second positive test, only slightly affected the estimated protection against repeat infection (table 1).
2 432 509 individuals were included in the alternative cohort analysis, with 28 875 (1·19%) individuals contributing exposed time periods and 2 405 683 (98·90%) contributing unexposed time periods, with 2049 contributing to both unexposed and exposed time periods, with a total of 138 reinfections. No individual tested positive more than twice. The results from this alternative cohort analysis were very similar to those of the main analysis (table 1), even though it was based on more events because of the additional follow-up time afforded by the analytical approach, which also allows for analysis of reinfection throughout the calendar year. The estimated protection against repeat infection in this analysis was 78·8% (95% CI 74·9–82·1).
Also in the alternative cohort analysis, we found little evidence that the degree of protection against repeat infection as measured by PCR positivity conferred by previous infection varied by age group below age 65 years. However, protection against repeat infection among those aged 65 years and older was lower than among younger age groups (table 2 ). We found no evidence of differences in the estimates of protection against repeat infection by sex, nor did we find any evidence that protection against repeat infection was waning after 6 months of follow-up (table 2; appendix p 7).
Table 2.
Protection against reinfection with SARS-CoV-2 by sex, age group, and time since first infection, in the alternative cohort analysis
|
Number of infections during follow-up |
Infection rate* |
Adjusted rate ratio (95% CI)† | Estimated protection (95% CI) | p value‡ | ||||
|---|---|---|---|---|---|---|---|---|
| Exposed individuals | Unexposed individuals | Exposed individuals | Unexposed individuals | |||||
| Overall | 138 | 53 991 | 5·64 | 30·94 | 0·212 (0·179–0·251) | 78·8% (74·9–82·1) | .. | |
| Sex | ||||||||
| Female | 78 | 30 225 | 5·68 | 30·87 | 0·209 (0·167–0·261) | 79·1% (73·9–83·3) | 0·84 | |
| Male | 60 | 23 766 | 5·59 | 31·03 | 0·216 (0·168–0·279) | 78·4% (72·1–83·2) | .. | |
| Age group, years | ||||||||
| 0–34 | 49 | 26 829 | 5·92 | 38·13 | 0·173 (0·131–0·229) | 82·7% (77·1–86·9) | <0·0001 | |
| 35–49 | 32 | 12 071 | 5·16 | 31·92 | 0·199 (0·141–0·282) | 80·1% (71·8–85·9) | .. | |
| 50–64 | 26 | 10 111 | 4·25 | 27·42 | 0·187 (0·127–0·274) | 81·3% (72·6–87·3) | .. | |
| ≥65 | 31 | 4980 | 8·01 | 16·92 | 0·529 (0·372–0·753) | 47·1% (24·7–62·8) | .. | |
| Time in follow-up, months | ||||||||
| 3–6 | 84 | 37 357 | 5·57 | 27·28 | 0·207 (0·167–0·256) | 79·3% (74·4–83·3) | 0·67 | |
| ≥7 | 54 | 16 634 | 2·66 | 14·48 | 0·223 (0·171–0·291) | 77·7% (70·9–82·9) | .. | |
Rate of infection per 100 000 person-days of follow-up.
Adjusted for sex, age group, test frequency, and start month of follow-up.
p value from likelihood ratio tests comparing models with and without interaction terms to capture evidence of effect heterogeneity across subgroups.
Discussion
We used a large national surveillance dataset of individually referable PCR test results to estimate the degree to which previous infection with SARS-CoV-2 results in protection against repeat infection. We found protection in the population to be 80% or higher in those younger than 65 years, but to be approximately 47% in those aged 65 years and older. We did not see signs of waning protection against repeat infection within the year 2020.
Our estimates for overall protection after previous infection with SARS-CoV-2 of 77–83% are in line with several other cohort studies from the UK, Qatar, and the USA that reported reinfection to be rare and occurring in fewer than 1% of all COVID-19 cases.2, 3, 16, 17 How long protection against repeat infection lasts after previous SARS-CoV-2 infection remains unknown because too little time has elapsed since the beginning of the pandemic, but one study of more than 20 000 health-care workers in the UK found that the risk of reinfection with SARS-CoV-2 was reduced by 83% for at least 5 months after primary infection.3 Another study of 12 541 health-care workers in the UK showed 89% protection lasting at least 6 months.2 A study from Qatar screening 43 000 people by PCR suggested that protection against repeat infection occurred for 95% of individuals who tested positive, lasting for at least 7 months.17 Previous studies have found that antibodies to other coronaviruses wane over time and allow for reinfection in the long term; however, the exact longevity of antibody responses after coronavirus infection is still uncertain. For circulating human coronavirus, the estimated period of protective immunity was 11 months.18 For MERS-CoV, antibodies were decreasing after approximately 5 months while immunity lasted up to 3 years, and for SARS-CoV, up to 2 years.8, 9, 10, 19 Estimates of seroprevalence of IgG antibodies against SARS-CoV-2 are variable depending on the laboratory methods used, selected cohort, geographical location, and ethnicity of participants and their socioeconomic background.20 In our study in which we classified time between infection and reinfection into two major time periods, late and early, we did not observe an effect that would indicate waning protection against repeat infections during our study period.
In addition to epidemiological studies, longitudinal serological and other immunological studies are needed to provide information on mechanisms of immunity against SARS-CoV-2 and its duration. An observational study from the USA that included 3·2 million people who had antibody tests for SARS-CoV-2 examined their subsequent PCR-test patterns.16 3 months after the index date of their serological test, PCR tests were positive for individuals with a negative SARS-CoV-2 antibody test at least ten times more often than for those who had a positive antibody test.16 Many studies have examined adaptive immunity after SARS-CoV-2 infection.1 In a longitudinal study of immunological memory to SARS-CoV-2, about 95% of individuals retained immunity for up to 8 months after infection based on measurements of antibodies, memory B cells, and CD4 and CD8 T cells.21 Although concentrations of antibodies against both SARS-CoV-2 spike and receptor binding domain decreased moderately over the 8-month study period, the number of memory B cells increased and memory CD4 and CD8 T cells had a half-life of 3–5 months. Thus, the different types of immunological memory as part of the adaptive immune system were active but had distinct kinetics, and measurements of circulating antibodies did not appear to predict T-cell memory.
We estimated relatively low protection against reinfection in people aged 65 years or older compared with younger individuals. Those aged 65 years and older had less than 50% protection against repeat SARS-CoV-2 infections after the first infection. However, another study group, who used a different study design, found a high degree of protection against reinfection among older people.22 Our finding that older people were more likely than younger people to test positive again if they had already tested positive could be explained by natural age-related changes in the immune system of older adults, also referred to as immune senescence. These changes affect both the innate and adaptive immune system and coordination of immune responses, and hence result in older people being more susceptible to emerging infectious diseases, such as SARS-CoV, MERS-CoV, and other viruses.23, 24, 25 Coordination of SARS-CoV-2-specific CD4 and CD8 T-cell responses have been found to be disrupted in individuals aged 65 years and older but not in younger individuals.26 Additionally, scarcity of naive T cells was associated with ageing and worse COVID-19 outcomes.26 In light of this evidence, our analysis highlights the need to protect older people against reinfection with SARS-CoV-2 by vaccination, physical distancing measures, and personal protective equipment, such as facemasks, regardless of previous infection status.
The reinfection potential of health-care workers is of particular interest because of their high risk of exposure to the virus and frequent tests regardless of clinical signs and symptoms. In our sensitivity analysis of health-care workers, we found similar results as in our main analysis. Several seroprevalence studies of health-care workers have found that the risk of infection with SARS-CoV-2 is higher in this group than in the general population.20, 27, 28, 29 In one study from Iran, the seroprevalence for IgG was almost 20%.27 A seroprevalence study among health-care workers in Denmark found that the risk of infection was 1·38 times higher in front-line health-care workers working in COVID-19 wards than in other health-care workers in the hospital.30 In our study, we found that the infection rate among health-care professionals was around twice that in the general population.
A main strength of our study is the size and completeness of our dataset, which is based on the entire population of Denmark and includes every individual that has been tested for SARS-CoV-2 between Feb 26 and Dec 31, 2020. We took advantage of the fact that Denmark has a large testing capacity, offering free testing within the population without needing a referral, and regardless of age, whether an individual is symptomatic or asymptomatic, or whether they suspect infection or not. This framework enabled us to study differences within age groups. As described, test facilities became more easily accessible over the course of the study period and the number of tests done per week increased by up to ten times in the second surge compared within the first surge. We do not think the change in the overall number of tests done has affected our analysis; in fact, the change might have made our analysis more complete because those who had a positive test in the first surge would probably not have been restricted in their access to testing during the second surge.
Knowledge of a first positive test could potentially affect the behaviour of an individual, resulting in differential misclassification. Individuals with a previous positive PCR test might engage in more high-risk activities (eg, not wearing a facemask) because of assumed immunity, and therefore be more likely to test positive a second time. By contrast, and probably more likely, such individuals might be less likely to have a second PCR test because they might believe themselves to be immune. Such behaviour would result in an overestimation of the protective effect of previous infection. We addressed this potential overestimation in two ways: by adjusting analyses for the number of tests done and through the sensitivity analysis of health-care workers. The results of this analysis corroborated the results of the main analysis. The different approaches we adopted to the analysis of the data did not change the overall findings, and neither did changing the defined time period between the first and second surges. In fact, the increase in the period between surges resulted in a slight increase in observed protection against repeat infection, suggesting that the criteria for reinfection became more specific because fewer recrudescent infections were misclassified as reinfections. Therefore, we believe we can draw representative conclusions about protection against repeat infection in the population.
One of the limitations of our study is that we could not correlate symptoms with protection against repeat infection because detailed clinical parameters are not typically recorded unless the patient was admitted to hospital due to severe COVID-19 symptoms. Our dataset includes test results from people with few or no symptoms that might have resulted in a comparatively lower immune response than if we had only included individuals with moderate or severe symptoms. However, had we included only individuals with moderate or severe infections, our findings would then have been generalisable only to individuals with symptomatic infections. Also, misclassification of reinfections might have occurred if detectable virus RNA lingered for more than 3 months in some patients. However, this potential bias is unlikely to have affected our results substantially because we addressed this potential misclassification in the analysis by altering the defined time period between the pandemic surges. Some misclassifications by PCR tests might have occurred; however, the test used is believed to be highly accurate, with a sensitivity of 97·1% and specificity of 99·98%.31 Therefore, we would only expect approximately two false positive results for every 10 000 tests in uninfected people and approximately three false negative results for every 100 tests in people who are infected. Our findings are only very slightly affected when taking account of test accuracy (data not shown). Finally, new variants of SARS-CoV-2 with the 484K or 501Y receptor binding area substitutions have recently appeared in Denmark,32, 33, 34 with some variants known to be more transmissable then the original.35, 36 During the study period, such variants were not yet established in Denmark; although into 2021 this pattern is changing.37 More prospective and longitudinal cohort studies coupled with molecular surveillance are needed to characterise antibody titres and waning of protection against repeat infections and the effect of antigenic shifts or drifts of the virus on immunity.
In summary, we found that protection against repeat SARS-CoV-2 infection is robust and detectable in the majority of individuals, protecting 80% or more of the naturally infected population who are younger than 65 years against reinfections within the observation period. However, we observed that individuals aged 65 years and older had less than 50% protection against repeat SARS-CoV-2 infection. Because the older age group is more prone to a serious clinical course of illness, this finding highlights the need to implement protective measures for the older population in the form of effective vaccines and enhanced physical distancing and infection control, even in those known to be previously infected. Furthermore, our data indicate that vaccination of previously infected individuals should be done because natural protection cannot be relied on.
Data sharing
De-identified participant-level data are available for access to members of the scientific and medical community for non-commercial use only. Applications should be submitted to Forskerservice at The Danish Health Data Authority, where they will be reviewed on the basis of relevance and scientific merit. Data are available now, with no defined end date.
Acknowledgments
Acknowledgments
We thank the staff in test sites, Departments of Clinical Microbiology, the virology laboratory, and TestCenter Denmark at SSI, and the staff of the Data Integration and Analysis Secretariat at SSI. DM was funded by the European Centers for Disease Control and Prevention programme European Programme for Public Health Microbiology Training. DM also thanks her supervisors for their support and scientific input.
Contributors
SE and KM conceived the idea for the study and provided methodological input. DM and CHH provided further input into the methodology and study design. CHH did the statistical analyses and SMG verified the underlying data. CHH, DM, and SE wrote the first draft of the manuscript. SMG and her team at SSI developed and maintained the data management systems for surveillance. All authors had full access to the data and contributed to interpreting the data and writing of the manuscript. CHH and SMG accessed and verified the underlying data for the study. All authors approved the final version and had final responsibility for the decision to submit for publication.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lumley SF, O'Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall V, Foulkes S, Charlett A, et al. Do antibody positive healthcare workers have lower SARS-CoV-2 infection rates than antibody negative healthcare workers? Large multi-centre prospective cohort study (the SIREN study), England June to Nov 2020. medRxiv. 2021 doi: 10.1101/2021.01.13.21249642. published online Jan 15. (preprint). [DOI] [Google Scholar]
- 4.Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To KK-W, Hung IF-N, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. published online Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J-S, Kim SY, Kim TS, et al. Evidence of severe acute respiratory syndrome coronavirus 2 reinfection after recovery from mild coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1421. published online Nov 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta V, Bhoyar RC, Jain A, et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1451. published online Sept 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe PG, Perera RAPM, Park WB, et al. MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg Infect Dis. 2017;23:1079–1084. doi: 10.3201/eid2307.170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L-P, Wang N-C, Chang Y-H, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne DC, Iblan I, Rha B, et al. Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg Infect Dis. 2016;22:1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reilev M, Kristensen KB, Pottegård A, et al. Characteristics and predictors of hospitalization and death in the first 11 122 cases with a positive RT-PCR test for SARS-CoV-2 in Denmark: a nationwide cohort. Int J Epidemiol. 2020;49:1468–1481. doi: 10.1093/ije/dyaa140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, Pedersen L, Sørensen HT. The Danish Civil Registration System as a tool in epidemiology. Eur J Epidemiol. 2014;29:541–549. doi: 10.1007/s10654-014-9930-3. [DOI] [PubMed] [Google Scholar]
- 13.Voldstedlund M, Haarh M, Mølbak K. The Danish Microbiology Database (MiBa) 2010 to 2013. Euro Surveill. 2014;19 doi: 10.2807/1560-7917.es2014.19.1.20667. [DOI] [PubMed] [Google Scholar]
- 14.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyngse FP, MØlbak K, Frank KT, Nielsen C, Skov RL, Kirkeby CT. Association between SARS-CoV-2 transmission risk, viral load, and age: a nationwide study in Danish households. medRxiv. 2021 doi: 10.1101/2021.02.28.21252608. published online March 5. (preprint). [DOI] [Google Scholar]
- 16.Harvey RA, Rassen JA, Kabelac CA, et al. Association of SARS-CoV-2 seropositive antibody test with risk of future infection. JAMA Intern Med. 2021 doi: 10.1001/jamainternmed.2021.0366. published online Feb 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Raddad LJ, Chemaitelly H, Coyle P, et al. SARS-CoV-2 reinfection in a cohort of 43,000 antibody-positive individuals followed for up to 35 weeks. medRxiv. 2021 doi: 10.1101/2021.01.15.21249731. published online Jan 15. (preprint). [DOI] [Google Scholar]
- 18.Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao W-C, Liu W, Zhang P-H, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 20.Rostami A, Sepidarkish M, Leeflang MMG, et al. SARS-CoV-2 seroprevalence worldwide: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;27:331–340. doi: 10.1016/j.cmi.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeffery-Smith A, Iyanger N, Williams SV, et al. Antibodies to SARS-CoV-2 protect against re-infection during outbreaks in care homes, September and October 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.5.2100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goronzy JJ, Li G, Yu M, Weyand CM. Signaling pathways in aged T cells - a reflection of T cell differentiation, cell senescence and host environment. Semin Immunol. 2012;24:365–372. doi: 10.1016/j.smim.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solana R, Pawelec G, Tarazona R. Aging and innate immunity. Immunity. 2006;24:491–494. doi: 10.1016/j.immuni.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Tan W, Lu Y, Zhang J, et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.24.20042382. published online March 26. (preprint). [DOI] [Google Scholar]
- 26.Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poustchi H, Darvishian M, Mohammadi Z, et al. SARS-CoV-2 antibody seroprevalence in the general population and high-risk occupational groups across 18 cities in Iran: a population-based cross-sectional study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30858-6. published online Dec 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudberg A-S, Havervall S, Månberg A, et al. SARS-CoV-2 exposure, symptoms and seroprevalence in healthcare workers in Sweden. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galanis P, Vraka I, Fragkou D, Bilali A, Kaitelidou D. Seroprevalence of SARS-CoV-2 antibodies and associated factors in healthcare workers: a systematic review and meta-analysis. J Hosp Infect. 2021;108:120–134. doi: 10.1016/j.jhin.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iversen K, Bundgaard H, Hasselbalch RB, et al. Risk of COVID-19 in health-care workers in Denmark: an observational cohort study. Lancet Infect Dis. 2020;20:1401–1408. doi: 10.1016/S1473-3099(20)30589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorentzen HF, Schmidt SA, Sandholdt H, Benfield T. Estimation of the diagnostic accuracy of real-time reverse transcription quantitative polymerase chain reaction for SARS-CoV-2 using re-analysis of published data. Dan Med J. 2020;67:67. [PubMed] [Google Scholar]
- 32.Toovey OTR, Harvey KN, Bird PW, Tang JWW. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J Infect. 2021 doi: 10.1016/j.jinf.2021.01.025. published online Feb 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang JW, Toovey OTR, Harvey KN, Hui DDS. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect. 2021 doi: 10.1016/j.jinf.2021.01.007. published online Jan 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang JW, Tambyah PA, Hui DS. Emergence of a new SARS-CoV-2 variant in the UK. J Infect. 2020 doi: 10.1016/j.jinf.2020.12.024. published online Dec 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.European Centre for Disease Prevention and Control Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. Dec 20, 2020. https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf
- 36.WHO . World Health Organization; Dec 31, 2020. Emergencies preparedness, response: SARS-CoV-2 variants.https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/ [Google Scholar]
- 37.Bager P, Wohlfahrt J, Fonager J, et al. Increased risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark. SSRN. 2021 doi: 10.1016/S1473-3099(21)00290-5. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3792894 published online March 2. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified participant-level data are available for access to members of the scientific and medical community for non-commercial use only. Applications should be submitted to Forskerservice at The Danish Health Data Authority, where they will be reviewed on the basis of relevance and scientific merit. Data are available now, with no defined end date.