Abstract
It is recognized that cell metabolism is tightly connected to other cellular processes such as regulation of gene expression. Metabolic pathways not only provide the precursor molecules necessary for gene expression, but they also provide ATP, the primary fuel driving gene expression. However, metabolic conditions are highly variable since nutrient uptake is not a uniform process. Thus, cells must continually calibrate gene expression to their changing metabolite and energy budgets. This review discusses recent advances in understanding the molecular and biophysical mechanisms that connect metabolism and gene regulation as cells navigate their growth, proliferation, and differentiation. Particular focus is given to these mechanisms in the context of organismal development.
Keywords: Gene regulation, glucose metabolism, ATP
Introduction
Studies of gene regulation and cell metabolism have typically focused on one process or the other, and yet these processes are mostly intertwined. Metabolism is driven by specific enzymatic products of gene expression. In turn, gene expression requires the continual synthesis of certain metabolites and ATP. Thus, the two processes are coupled and must coordinate with one another as cells navigate changing conditions of existence. Moreover, this coordination must be directed to particular cellular strategies such as growth, division, or differentiation according to the prevailing conditions.
The coupling of these two fundamental processes must deal with their individual dynamics. The timescale of metabolic supply-demand is much smaller than the timescale of gene expression (Figure 1A). The energy budget of cells is such that a minute-long lapse in ATP synthesis would totally deplete cellular ATP stores. Flux through the metabolic network (see Glossary) is very high, indeed. In contrast, the time taken for a gene to make an active protein product is longer by at least one order of magnitude. Thus, signaling between the two processes must somehow enable each to adapt and respond according to widely different timescales. How does it occur? This review places particular focus on work that addresses this question.
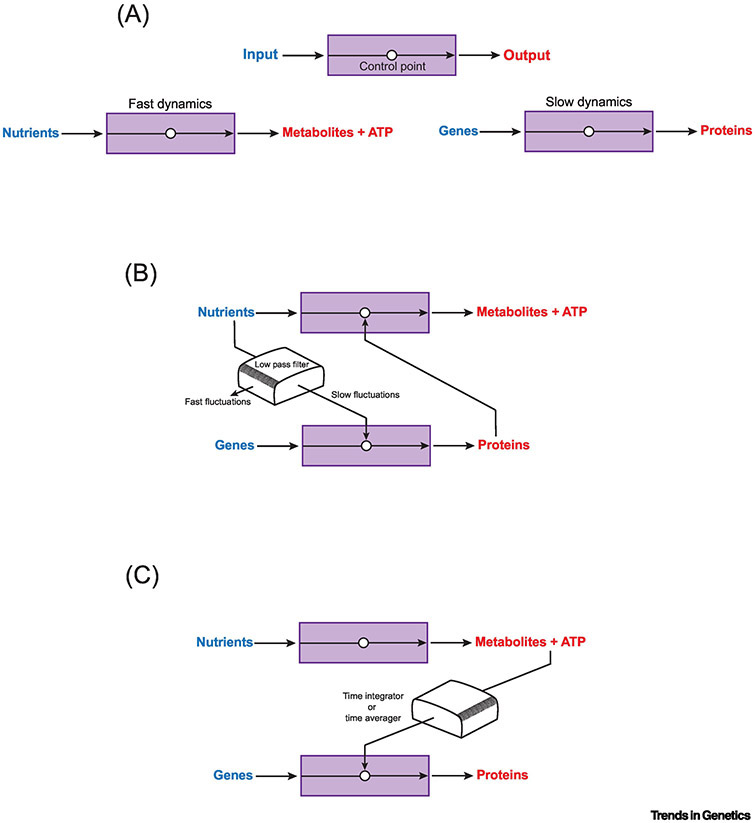
Figure 1. Coupling two processes with very different dynamics.
(A) A generic process can be represented as input into a "black box" to generate output. Control points regulate the process. Such a representation can be used to abstract metabolism (lower left) and gene regulation (lower right), which have very different black box dynamics. (B) The GRN coupling galactose metabolism and gene expression forms a low-pass filter that then synchronizes signals between the two processes. (C) An alternate mechanism to couple the two processes is for gene regulation to integrate or average the variable metabolic signals over time. The net result is to improve metabolic signal to noise and generate a synchronized response.
Low-Pass Filtering of Signals Between Metabolism and Gene Expression
Our earliest understanding of gene regulation was rooted in a metabolic outcome: the breakdown of lactose by E. coli. The pioneering work of Jacob, Monod, and many others provided key concepts that later were found to operate in eukaryotic gene regulation as well [1]. Gene regulation in the budding yeast, Saccharomyces cerevisiae, likewise, was pioneered by focusing on metabolic systems such as galactose metabolism [2, 3]. In these systems, gene regulation elicits an adaptive cellular response. A fluctuation in extracellular nutrient concentration induces a regulated change in nutrient uptake and metabolism until the concentration relaxes to a steady-state. The yeast galactose gene regulatory network (GRN) imparts adaptation to galactose metabolism, and additionally imparts bistability, when sensing the ratio of different extracellular metabolites [4, 5]. Since metabolism dynamics are so rapid compared to gene expression, what prevents GRNs such as the galactose GRN from regulating metabolic pathways chaotically? This question was explored with microfluidics to reveal dynamical features [6]. It was found that the galactose GRN is structured to behave like a low-pass filter. Such filters pass signals with low-frequency fluctuations and block signals with frequency fluctuations above a certain cutoff. The galactose GRN reliably responds to slow fluctuations (period length > 3 min) in external nutrients while ignoring faster fluctuations (period length < 1.5 min) [6]. This is one mechanism whereby metabolism and gene regulation can be reliably coupled (Figure 1B).
Kinase Signals Between Metabolism and Gene Expression
There are several well-studied mechanisms by which cells use kinases to transduce their metabolic state and adjust gene expression. One such mechanism in mammalian cells integrates growth factor signals and amino acid status via the mTOR transduction pathway (for recent review see [7]). mTOR regulates the coordinate expression or nuclear localization of several transcription factors that individually are responsive to internal or external cues such as stress. In doing so, mTOR coordinates the activation of these factors to achieve an integrated and directed outcome on gene expression.
Insulin and growth factors also signal via the protein kinase AKT to phosphorylate some members of the FOXO transcription factor family, leading to their nuclear export and destruction [8]. Within the context of cells in developing organisms, AKT influences not only metabolic responses but also fate choices. For mouse neural and hematopoietic stem cells, FOXO factors maintain cell quiescence by suppression of cell cycle genes [9, 10]. With growth factor signaling, this repression is relieved, and cells proliferate and self-renew. In human embryonic stem cells, FOXO1 appears to regulate the expression of two crucial transcription factors for the stemness program, OCT4 and SOX2 [11].
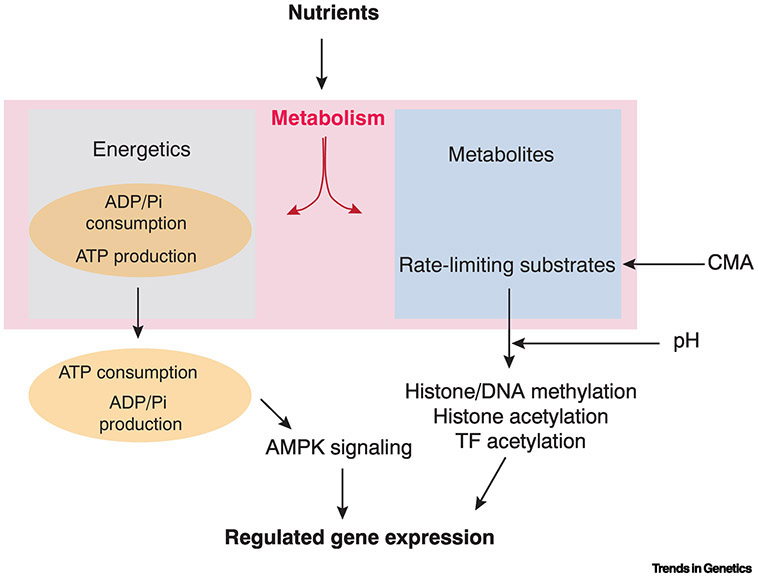
Another means for cells to sense and adjust their metabolic state is by monitoring ATP levels. Gene expression can be regulated by the ratio of ATP and AMP, which is mediated by the protein kinase AMPK (for recent review see [12]). When AMP levels are raised, AMPK is activated and it phosphorylates proteins such as the general transcription regulator p300 [13]. It also phosphorylates selective transcription factors such as members of the FOXO family [14]. The net result is a sustained increase in catabolic activities as a means to replenish energy stores (Figure 2). However, it should be noted that phosphorylation of proteins by ATP is generally not limited by ATP concentration but by kinase activity [15]. Thus, phosphorylation of transcription factors is not a general transducer linking ATP status to overall gene regulation.
Figure 2. Roles for metabolism in gene regulation.
Scheme to categorize the distinct outcomes of nutrient uptake, which generates energy and metabolites. Energy is expended by hydrolysis of ATP into ADP and inorganic phosphate (Pi), and this flux controls the rate at which biochemical processes such as transcription, translation, and molecular degradation occur. Certain signaling pathways such as AMPK actively sense ATP status and regulate specific genes in response. Generation of certain metabolites regulates gene expression because these metabolic substrates can act as rate-limiting factors in modulating epigenetic and protein modifications. Other metabolic processes can tune these modifications. Examples include chaperone-mediated autophagy (CMA), an alternate protein degradation mechanism from the proteasome, and efflux of lactate from cells, leading to higher intracellular pH.
One organelle where gene expression does directly sense ATP concentration is the mitochondrion. When yeast cells undergo respiration, mitochondria generate ATP, which causes an upregulation of mitochondrial gene transcription [16]. This effect can be reconstituted in vitro, where various mitochondria gene promoters have unique sensitivity (Km) to ATP concentration. The mitochondrial RNA polymerase is thought to impart this ATP dependence on transcription.
Metabolites as Signals to Transcription Factors
Some transcription factors are post-translationally modified by acetylation and methylation. Methyltransferases catalyze methylation using the donor metabolite S-adenosyl-methionine (SAM-e). This modifies not only proteins but also DNA at specific genomic loci. Histone acetyltransferases catalyze the transfer of an acetyl group from the central metabolite acetyl-Coenzyme A (acetyl-CoA) onto lysines in various proteins including histones. Although removal of acetyl groups from proteins is not obligatorily dependent on a metabolite, Sirtuin deacetylases are activated by NAD+, linking this energy metabolite to transcription control [17]. Crucially, the rates of acetylation and methylation are sensitive to the concentration of metabolite substrates such as NAD+, SAM-e, and acetyl-CoA [18]. All of these metabolites are generated as part of the glucose metabolic network (Figure 3). Hence, the dynamical behavior of glucose metabolism can globally or locally regulate transcription via modification of transcription factors and chromatin. Since acetyl and methyl modifications of proteins tend to have long lifetimes, they either integrate or average all of the fluctuations in glucose metabolism over that long timescale. The end-result is that transcription regulation is only sensitive to long-term changes in glucose metabolism (Figure 1C).
Figure 3. An overview of glucose metabolism.
Glucose is transported into cells where it is catabolized by glycolysis to yield pyruvate. Pyruvate is then either converted into lactate (which can be excreted from the cell) or transported into mitochondria and metabolized into acetyl-CoA, fueling the tricarboxylic acid (TCA) cycle. NADH and FADH2, produced through glycolysis and the TCA cycle, are used by the mitochondrial electron transport chain for generating an electrochemical proton gradient, which drives oxidative phosphorylation (OxPhos) for ATP production. Since mitochondrial acetyl-CoA cannot enter the cytosol, citrate made in the TCA cycle can enter the cytosol and be converted to acetyl-CoA. This is then a substrate for protein acetylation. Glucose can also enter the pentose phosphate pathway via glucose-6-phosphate. This ultimately produces nucleotides, which in addition to being nucleic acid precursors, also generate S-adenosyl-methionine (SAM-e), a substrate for protein methylation. Note that shunting glucose to form cytosolic acetyl-CoA and SAM-e comes at the expense of oxidative phosphorylation.
One such change is the magnitude of glucose metabolism. Mammals undergo a 24-hour cycle of metabolic activity, reflecting the solar cycle. Levels of NAD+ fluctuate with a 24-hour period as a consequence. Two NAD+ dependent proteins, the mouse sirtuin SIRT1 and the ADP-ribosyltransferase PARP, bind to two transcription factors in the core circadian regulatory network [19-21]. When they bind the CLOCK/BMAL complex, they affect DNA binding of the complex and its ability to activate target gene transcription. In addition, the complex recruits SIRT1 to target genes leading to their selective deacetylation [19, 20]. Thus, NAD+ plays a critical role in the functioning of the mouse circadian clock.
Change in the mode of glucose metabolism is another means to regulate transcription. High levels of glucose consumption by aerobic glycolysis are known to increase the concentration of cytosolic acetyl-CoA, since metabolites are shunted out of the TCA cycle to the cytosol where they generate acetyl-CoA (Figure 3). However, ATP yield is low from glycolysis. In contrast, oxidative phosphorylation efficiently generates ATP but keeps acetyl-CoA in the mitochondria (Figure 3). When cells change from one mode to the other, this can have profound effects on gene regulation. One example is seen when neural crest cells are specified in vertebrate embryos. When they initiate differentiation, neural crest cells increase expression of glycolytic enzymes and engage in aerobic glycolysis rather than oxidative phosphorylation [22]. In chick embryos, this change promotes the association of two transcription factors, YAP and TEAD, with one another. YAP and TEAD are downstream effectors of Hippo signaling, in which YAP/TEAD complexes activate gene transcription [23]. However, in neural crest cells, YAP/TEAD complexes form due to glycolysis, leading to transcriptional activation of several genes associated with an epithelial-to-mesenchymal (EMT) transition [22]. The EMT enables neural crest cells to delaminate from the neural tube and migrate throughout the developing embryo [24].
Signaling from glycolysis to EMT has positive feedback, whereby the EMT transcriptional program can in turn activate glycolysis. This has been observed in human breast cancer cells. EMT is a feature of cancer cells as they progress towards metastasis [24]. Human breast cancer cells generate complexes of the transcription factor SNAIL and DNA methyltransferase DNMT1, which then methylate and silence the E-cadherin gene, resulting in cell delamination from epithelia [25]. The complex also methylates the gene encoding fructose 1,6-biphosphatase (FBP1) [26]. This silences FBP1 expression and thereby activates aerobic glycolysis over a long timescale. Persistent aerobic glycolysis is a common feature of cancer cells [27].
Aerobic glycolysis can also affect transcription factor modification in unexpected ways. Cells of the vertebrate tail bud are actively glycolytic, causing export of lactic acid and protons from the cells [28] (Figure 3). This leads to an increase in intracellular pH, which promotes non-enzymatic acetylation of the protein β-catenin, an intracellular transducer of Wnt signaling. Acetylation is sufficient for β-catenin to activate transcription of mesoderm-specific Wnt target genes, and the determination of paraxial mesoderm [28]. In conclusion, there are diverse ways in which long-term changes in glucose metabolism can alter transcription programming by transcription factors (Figure 2).
Metabolites as Signals to Chromatin
Acetyl and methyl modifications of histones can be so longlasting that epigenetic “memory” is imparted to cells. Hence, these modifications are another means by which rapid fluctuations in glucose metabolism are integrated or averaged over long timescales (Figure 1C). Hovever, in this case, regulation by metabolism can be exerted on a genome-wide scale. This has particular relevance for cell differentiation. A shift from aerobic glycolysis to oxidative phosphorylation is observed in mouse embryonic and adult stem cells during cell differentiation toward specific fates [29-32]. Glycolysis occurs in pluripotent human embryonic stem cells, stimulating the acetylation of histones in chromatin [33]. It is thought that enhanced histone acetylation maintains cells in their stem state. When the cells switch to oxidative phosphorylation, the synthesis of cytosolic acetyl-CoA drops, and histone acetylation levels also go down, leading to reprogramming of transcription towards lineage restriction. However, this relationship is the opposite in human neural stem cells. Neural differentiation depends on increasing the level of cytosolic acetyl-CoA to elevate the levels of histone acetylation in neural-specific genes [34]. Even though neural stem cells shift from glycolysis to oxidative phosphorylation as they differentiate, this is accompanied by a regulated efflux of citrate from mitochondria that is then used to generate the high levels of cytosolic acetyl-CoA [35] (Figure 3).
Embryonic stem cell transitions are also regulated by a form of metabolism called chaperone-mediated autophagy (CMA) [36, 37]. CMA degrades isocitrate dehydrogenase and reduces the level of α-ketoglutarate (Figure 3), down-regulating DNA and histone demethylases due to their requirement for α-ketoglutarate as a cofactor [37]. However, in mouse ES cells, the pluripotency factors OCT4 and SOX2 inhibit CMA, thereby maintaining a hypomethylated chromatin state in pluripotency. ES cell differentiation triggers the onset of CMA and inhibition of the demethylases, shifting the balance of chromatin to a more-methylated state (Figure 2). In summary, metabolism acts to modulate longlasting chromatin states via methylation and acetylation.
A Biophysical Perspective on Metabolic Signaling to Chromatin
Eukaryotes, with their larger genomes, are thought to dynamically package DNA into different chromatin states in order to modulate accessibility of DNA to transcription factors and RNA polymerases. Most consider the expenditure of energy in this process as a byproduct of consuming acetyl-CoA, SAM-e, and other metabolites to modify chromatin conformation. However, a thermodynamic argument has recently been made to explain why eukaryotic gene regulation is coupled to cell energetics [38]. Transcription of eukaryotic genes, particularly ones involved in development, are often very sensitive to the input of upstream transcription factors [39]. Gene expression responds sharply to a small change in transcription factor concentration. In the absence of energy expenditure, with transcription factors binding at thermodynamic equilibrium, there are constraints on how switch-like transcription initiation can occur [38]. However, if energy is expended to maintain binding away from thermodynamic equilibrium, then transcription initiation can become remarkably sensitive to transcription factors. Although this argument is grounded in purely physical modeling, we know that energy is expended to actively alter chromatin accessibility by means of histone modification and nucleosome displacement.
The early Drosophila embryo is being exploited to experimentally test this model. Transcription of the hunchback gene is regulated by the transcription factor Bicoid, whose concentration varies as a spatially extended gradient across the embryo [39]. hunchback is also subject to opening of its chromatin accessibility by Bicoid and the pioneer factor Zelda [40, 41], since both factors are linked to histone acetylation [42, 43]. Live imaging of hunchback transcription has allowed for measurement of dynamical features of its transcription, including the rate of polymerase loading and the transcriptional onset time [44]. In wild type and mutant embryos, these measured features can only be explained if Bicoid and Zelda actively alter chromatin accessibility for transcription factor binding [44]. Crucially, transcription responds very sharply to a small change in Bicoid concentration [39]. This observation is consistent with a biophysical explanation for how histone acetylation might allow transcription to become remarkably sensitive to transcription factor levels.
Gene Regulation During Development - An Energetics Perspective
Metabolism not only supports production of the metabolite precursors of protein and nucleic acid, but it also supports energy conversion to carry out their syntheses (Figure 2). Moreover, energy is expended to maintain order in a cell: to assemble and disassemble molecular machines, generate forces, and change the activity of molecular pathways over time and space. The balance between energy uptake and energy expenditure can be measured as the heat dissipation from a cell or organism to its surroundings. Since cells are too small to make precise measurements of heat, focus on energetics and gene regulation has primarily been in the context of developing organisms.
Heat dissipation measurement has been made for a variety of developing embryos, and it ranges from 60 to 170 nJ/sec per embryo [45-47]. Since the chemical potential of ATP is the primary energetic fuel, this rate of heat dissipation corresponds to hydrolysis of an equivalent of 25 - 75 μM ATP/sec [48], or the complete turnover of cellular ATP in approximately 1 minute [46].
As embryonic development proceeds, heat dissipation slowly increases by 40 to 70%, reflecting increasing metabolic rate [45-47]. The major biosynthetic processes of gene expression, RNA and protein synthesis, account for less than 10% of the energy expended, suggesting that the energy budget is mostly devoted to other biochemical processes [47]. Indeed, small oscillations in heat dissipation are detected due to the biochemical activity of the cell cycle oscillator circuit within proliferating cells during embryogenesis [46].
The energy budget of a cell then is composed of many competing processes that expend energy by consuming ATP, balanced with the rate of energy uptake in the form of nutrients (Figure 2). If energy uptake changes for whatever reason, then energy expenditure must adjust to prevent the total depletion of ADP or ATP within minutes [15]. Since ATP concentration is invariant to changing nutrient conditions [49], it is thought that energy expenditure adjusts to the rate of ATP synthesis [50, 51]. This was nicely demonstrated for Drosophila larvae whose heat dissipation was monitored as the animals grew [52]. Genetic ablation of the genes encoding insulin-like peptides causes cells to reduce their uptake of circulating sugars by ~40%. There is a corresponding 30% decrease in energy expenditure by the body, and as a result, the animals grow and develop more slowly [52].
Like other biochemical processes, gene expression adjusts with the energy budget. If energy expenditure for other biochemical processes goes up, then polymerases and ribosomes have less available ATP to synthesize gene products. Or if energy uptake goes up, polymerases and ribosomes have more available ATP to make their products. Short-term fluctuations in the energy budget are likely integrated or averaged by the longer timescales of gene expression, but long-term changes in the energy budget do affect gene expression (Figure 1C). Long-term changes do not necessarily affect the steady-state concentration of gene products like mRNA and protein [53], but they do affect gene expression dynamics. If less ATP is available for polymerases, ribosomes and proteosomes, then changes in protein output take a longer period of time to complete [54-56].
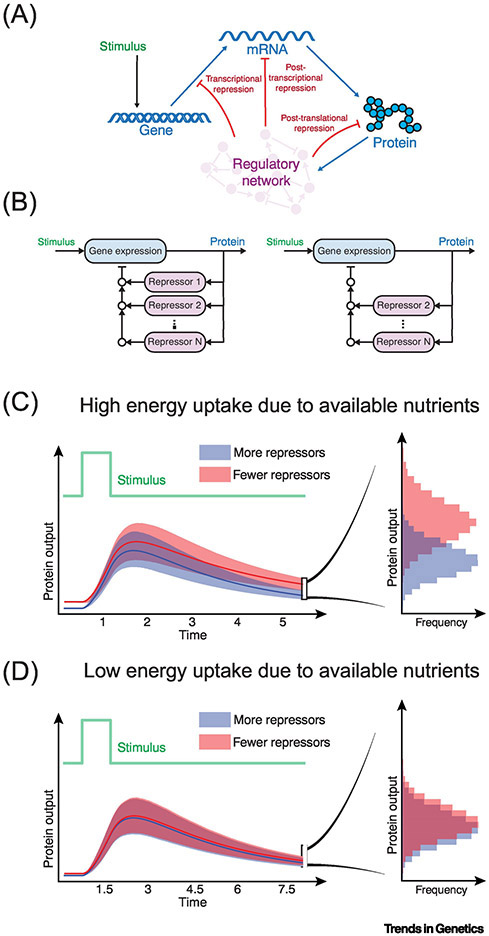
This variability in gene expression dynamics has unexpected effects on the role of repressors of gene expression, as suggested by a generalized theoretical treatment of gene expression dynamics [56]. Repressors are frequently employed to bring gene expression down to a resting steady state, acting after a controlled stimulus or uncontrolled environmental stimulus triggers expression (Figure 4A). To efficiently and rapidly return expression levels to base-line, often there are multiple repressors acting on the same gene at the same time (Figure 4B). A cell’s energy budget has a profound influence on the need for multiple repressors. If energy expenditure is restricted owing to less nutrient uptake by a cell, then not all of the repressors are needed to bring down expression within the expanded timeframe [56]. This makes the repressors become wholly redundant with one another (Figure 4C,D). Modeling predicts that this phenomenon should hold for repressors of transcription, mRNA stability, and protein synthesis/stability [56]. For example, a transcription repressor becomes redundant not only with other transcription repressors but also post-transcriptional and post-translational repressors.
Figure 4. Gene regulation and energy uptake.
(A) A stimulus activates gene expression, followed by repression of gene expression at different steps, thereby reducing protein output down to a resting steady-state. (B) Multiple repressors can act on gene expression, and the repressors can be more (left) or fewer (right) in number. (C,D) When the stimulus is transient, expression dynamics are pulse-like owing to repression. Shown are the average dynamics of protein output when there are more (blue line) or fewer (red line) repressors. Shaded areas show the distribution of curves for protein output from many cells. The histograms to the right, plot the frequency distribution of protein output at a defined time as indicated. In (C), energy uptake in the system is high owing to nutrient availability, allowing the gene expression machinery to expend ATP at high rates. Fewer repressors lead to higher protein output as would be intuitively predicted. In (D), energy uptake is low owing to limited nutrient availability, causing the gene expression machinery to expend ATP at lower rates. In this scenario, fewer repressors do not result in higher protein output. The repressors become redundant with each other. An implication of this is that multi-layered repression enables gene expression to uniformly operate under a wide variety of metabolic conditions. This might be a driving force in how gene regulation has evolved in organisms.
Experiments support this surprising prediction. MicroRNAs are repressors of gene expression, and they act post-transcriptionally. One such microRNA in Drosophila, called miR-7, represses expression of the yan gene [57]. It acts in parallel with a transcription repressor of yan and a post-translational repressor that destabilizes Yan protein by ubiquitination [58]. Although miR-7 acts in parallel with the other repressors, its loss causes cells to take longer for Yan protein levels to relax to a basal steady state. However, if glucose uptake by cells is restricted, then loss of miR-7 has no effect on the time it takes for Yan protein to relax to its basal state [56]. A more indirect assay is consistent with this observation. Defective cell differentiation caused by loss of miR-7 repression of yan is not observed if glucose uptake by cells is restricted. Likewise, loss of the post-translational repression of yan has less impact on cell differentiation if glucose uptake is restricted. Similar observations are seen with loss of transcription repressors of other target genes in other tissues [56]. The phenomenon is so profound that loss of the entire family of microRNAs has almost no impact on Drosophila development when glucose uptake is restricted [56].
A similar phenomenon also appears to occur in the nematode Caenorhabditis elegans. When the L2 stage of larval development is artificially lengthened by pheromone/hormone treatment, the developmental phenotypes of mutation in the let-7 family are suppressed [59]. This microRNA family represses expression of the target gene hbl-1 [60]. In conclusion, the energy budget dictates how rapidly cells can modulate gene expression, and limited energy uptake makes repressors less essential for proper modulation of expression.
Temperature Considered
Although mammals are endothermic, the vast majority of species on the planet are ectotherms, whose body temperatures rely on their environment. Temperature affects chemical reactions in complex ways, and yet the Arrhenius equation (k = Ae−Ea/RT) has been a powerful tool to predict the temperature dependence of biochemical reaction rates [61]. Indeed, it has been useful to describe the resting metabolic rates observed in microbes, plants, and animals [62]. Remarkably, this description of metabolism can be used to describe developmental processes such as the growth rate of vertebrate and invertebrate animals [63]. However, recent evidence suggests that developmental rates of animals deviate from the idealized Arrhenius Law, and this deviation is possibly due to non-idealized behavior of metabolic enzymes such as glyceraldehyde-3-phosphate dehydrogenase [64].
It is somewhat surprising that development follows the Arrhenius Law, since huge numbers of biochemical reactions occur simultaneously in cells and tissues throughout the developing body. The thousands of temperature-dependent reactions associated with gene expression must occur in parallel, ensuring that processes unfold in synchrony. One possible solution to this problem is that the rate-limiting steps controlling gene expression across the body have similar temperature dependencies. Fundamental cell metabolism might be such a common rate-limiting step for gene expression [63]. An alternative solution is that development has built-in checkpoints to resynchronize gene expression events that are slightly diverged due to temperature. One such checkpoint has been described for the Drosophila embryo. Microfluidics was used to incubate a single embryo at two different temperatures such that one half of the embryo was continually exposed to one or the other temperature [65]. Although nuclear division rates in the two halves were highly asynchronous, the subsequent transcription and translation of embryonic genes became completely synchronized in both halves [65]. Synchronization requires the action of endogenous small interfering RNAs (endo-siRNAs) in the embryo [66]. Because endo-siRNAs repress gene expression, it is possible that they attenuate the accelerated gene expression that is occurring in the warmer half and thereby allow the slower half to catch up.
Organisms can actually exploit the temperature dependence of biochemical reactions over short timescales to elicit gene expression changes over much longer timescales. The plant Arabidopsis thaliana does this for a developmental process called vernalization [67]. The transcription factor NTL8 accumulates in meristem cells when plants are exposed to cold temperatures. Accumulation occurs because NTL8 protein is less diluted by cell growth when the temperature is cold; cells metabolize and grow more slowly at colder temperatures. The accumulated NTL8 activates a long-term transcriptional program leading to vernalization [67]. Thus, temperature-dependent metabolism can be used as a thermosensor for gene regulation of processes that occur much later in life.
Concluding Remarks
The fields of metabolism and gene regulation are vast, and even the intersection between the two includes a considerable library of research. This review has touched upon some of the more recent advances plus it has discussed some aspects that might be considered non-traditional by some. One of the take-home messages is the importance of dynamics. The timescale of metabolic supply-demand is very different from the timescale of gene expression. The energy budget of cells is such that a minute-long lapse in ATP production will completely deplete cellular ATP stores, while gene expression takes much longer. Thus, signaling between the two processes tends to be primarily from metabolism to gene expression and not the other way around. Metabolism is primarily regulated by post-translational mechanisms, owing to their typically rapid dynamics. Since global gene expression appears to consume only a small fraction of the energy budget of a cell, the signaling from metabolic processes to gene regulation predominantly goes through transcription. Rapid adjustment of gene expression might be less necessary if it is not a major consumer of cellular energy. Another take-home message is that metabolism is not simply about making metabolites but is also about energy conversion. Both are required for gene expression, and balancing the metabolite flux with energy flux is crucial for meeting the demands of producing proteins from genes.
What might we expect in the future (see Outstanding Questions)? The field has greatly benefited from the diversity of expertise practiced by its participants; biochemistry (of course), but also biophysics, molecular genetics, systems biology, and even applied mathematics. The future will continue to broaden this diversity and provide us with an expanded perspective of a complex and fascinating problem!
Outstanding Questions.
How does the circadian rhythmicity of metabolism affect the dynamics of gene expression, not only of cycling genes but genes that are constitutively expressed?
How do cells in different regions of the body experiencing metabolic variation manage to synchronize their gene expression for the purposes of coordinated regulation?
Since the mode of glucose metabolism regulates epigenetics in cells of the early embryo, how do oocytes and spermatocytes epigenetically reprogram given their different modes of metabolism?
Does the energy expended for nucleosome displacement and modification drive transcription factor activity away from thermodynamic equilibrium and imbue them with properties different from bacteria gene regulation?
Synthetic gene regulatory circuits have been made that can generate unique and desired outcomes. Can synthetic metabolic circuits be made to accomplish similar outcomes for gene expression?
Feedback repression provides gene regulation with the ability to adapt to variable metabolic conditions. Are there other types of regulatory circuits (i.e. feedforward loops) that enable the same thing?
Highlights.
Gene expression may only account for 10% of a cell’s energy budget, which undergoes complete ATP turnover every minute.
Multi-layered feedback repression enables gene expression to adapt to variable metabolic rates.
Energy expenditure via nucleosome displacement and modification regulates TF accessibility and ultrasensitivity.
Aerobic glycolysis versus oxidative phosphorylation have distinct effects on epigenetic modifications.
Acknowledgements
I would like to thank the NIH for financial support (R35GM118144). I would like to thank my colleagues in the NSF-Simons Center for Quantitative Biology for stimulating discussions and reading drafts of the manuscript.
Glossary
- Arrhenius equation
The rate constant k of a chemical reaction depends on an exponent of absolute temperature T, the gas constant R, and Ea, the activation energy of the reaction. Proposed in 1889 by Svante Arrhenius, the formula is k = Ae−Ea/RT. It is seen as an empirical relationship useful for calculating the rate of chemical reactions and the energy of activation.
- Bicoid
This zinc-finger transcription factor is translated from mRNA specifically located at the anterior pole of the Drosophila one-cell embryo. The protein passively diffuses to form a concentration gradient from anterior-to-posterior poles. The protein enters the nuclei of the syncitial embryo, where it binds DNA and activates transcription of select genes. Expression of these genes initiates the division of the embryo into distinct body parts and forms the anterior-to-posterior axis of the fly.
- β-Catenin
This protein serves two important functions in epithelial cells. A pool of it is localized to the adherens junction where it binds the intracellular domain of cadherin proteins. It is required for adherens junction integrity. A cytosolic pool is highly unstable unless the cell receives a Wnt signal. The signal stabilizes cytosolic β-catenin, allowing it to enter the nucleus and associate with the DNA-binding transcription factor TCF. This heterodimer is able to activate transcription of target genes. Acetylation of β-catenin enhances its activation potency.
- CMA
Chaperone-mediated autophagy is a proteolytic system that contributes to degradation of specific intracellular proteins. CMA substrate proteins are targeted to lysosomes and translocated into the lysosomal lumen through the action of chaperones located at both sides of the membrane and a translocation complex. CMA substrates contain a five-amino acid motif that is recognized by a heat shock protein in the cytoplasm.
- EMT
Epithelial to Mesenchymal transition is a cell behavior limited to cells within epithelia. These cells lose adhesion with other cells in the epithelium and undergo migratory behaviors typical of mesenchyme. While EMT is a natural process that occurs in response to developmental or physiological cues, it is also observed in tumor cells. There, they become migratory and leave the site of the tumor, potentially becoming metastatic.
- Endo-siRNAs
Small interfering RNAs (siRNAs) are 21-nucleotide single-stranded RNAs. They associate with Argonaute proteins to form RISC, which cleaves target mRNAs if they contain sequence complementary to the siRNA sequence. This is known as RNAi and is typically applied in biomedical research. However, cells naturally make RNA that either folds or anneals into a long double-stranded conformation, thereby becoming processed by Dicer to form endogenous siRNAs (endo-siRNAs). The functions of these range from heterochromatin formation, transposon silencing, to post-transcriptional repression of cellular gene expression.
- GRN
A gene regulatory network is a collection of genes within a single genome whose constituent molecules (DNA, RNA, protein) interact with each other in defined and specific ways. One network typically has a distinct function for the organism, i.e., a GRN that programs cells to differentiate into skeletal muscle. Some genes in a GRN only serve to regulate other genes in the network, either by repression or activation. GRNs are often represented as graph networks from a modeling perspective.
- Hunchback gene
A Drosophila gene that is first expressed in the early syncitial embryo stage. Its transcription is directly activated by Bicoid through binding of the protein to the promoter. Activation is strictly dependent on Bicoid concentration, with a sharp threshold concentration, below which no hunchback transcription occurs. This ensures that the gene is only expressed in the anterior half of the embryo. The gene encodes a transcription factor whose function is to regulate genes that subdivide the embryo into 14 segments.
- Metabolic network
Cellular metabolism is the sum of reactions involving the conversion of a carbon source, usually glucose, into the metabolites used to synthesize macromolecules. This can be represented as a metabolic network of reactions. ATP and ADP are central cofactors that connect parts of the metabolic network with each another and with networks representing other processes such as RNA and protein synthesis. Conversion of ATP to ADP is key to maintaining all of the networks out of equilibrium and thus driving biosynthesis
- MicroRNAs
These 21 nucleotide single stranded RNAs associate with Argonaute proteins and are similar to siRNAs. Unlike siRNAs, microRNAs are processed from primary transcripts that locally fold into short stem-loop structures whereupon they are cleaved by Drosha and Dicer. Base-pairing between a microRNA and its target mRNAs usually occurs in the mRNA 3’UTR and is partially complementary. Binding elicits a weak increase in mRNA turnover rate and weak decrease in translation efficiency. There are over 2,000 microRNA species in humans and they likely regulate over half of the protein-coding transcriptome. They are found in virtually all plants and animals
- Vernalization
A plant’s flowering process is induced by exposure of the plant or its seeds to cold temperature. For Arabidopsis, some varieties have delayed flowering without vernalization. Vernalization of the meristem section of the Arabidopsis plant is sufficient to confer competence to respond to floral inductive cues. This competence can be retained for as long as 300 days
- Wnt
A family of secreted proteins exclusively found in animals. These proteins act as extracellular ligands for cell-surface receptors on other cells. Bound receptors trigger a signal transduction cascade within the cytoplasm, resulting in stabilization of the protein ß-catenin and its translocation into the nucleus. There, it associates with a protein of TCF family, and the complex acts as a sequence specific transcription factor to regulate gene expression. Frequently, these target genes mediate transitions in cell fate for the receiving cells.
- Yan
A Drosophila gene that encodes a zinc finger transcription repressor. It is a central gene target for receptor tyrosine kinase (RTK) signals that multipotent cells receive at many stages of development and in many tissue types. RTK signaling triggers MAPK to phosphorylate Yan protein, leading to its ubiquitination and rapid destruction. This de-represses Yan target genes whose function is to lineage-restrict cells. In addition, the yan gene is subject to inhibition by other repressors such as the microRNA miR-7 and transcription factor Pointed.
- Zelda
A zinc finger protein that acts as a pioneer factor, increasing enhancer accessibility for transcription factors to bind DNA throughout the genome of the early Drosophila embryo. Zelda mRNA is deposited in the Drosophila egg, and is translated upon fertilization where it becomes uniformly distributed throughout the embryo. Zelda binding to specific loci in the genome is associated with histone depletion and open chromatin accessibility as determined by ATAC-seq. This primes genes located in these regions for transcription and they are among the first zygotic genes to be expressed in the embryo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beckwith J (2011) The operon as paradigm: normal science and the beginning of biological complexity. J Mol Biol 409 (1), 7–13. [DOI] [PubMed] [Google Scholar]
- 2.Klar AJ and Halvorson HO (1974) Studies on the positive regulatory gene, GAL4, in regulation of galactose catabolic enzymes in Saccharomyces cerevisiae. Mol Gen Genet 135 (3), 203–12. [DOI] [PubMed] [Google Scholar]
- 3.Johnston SA and Hopper JE (1982) Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A 79 (22), 6971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante-Chong R et al. (2015) Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc Natl Acad Sci U S A 112 (5), 1636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venturelli OS et al. (2015) Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol 13 (1), e1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett MR et al. (2008) Metabolic gene regulation in a dynamically changing environment. Nature 454 (7208), 1119–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu GY and Sabatini DM (2020) mTOR at the nexus of nutrition, growth, ageing and disease. Nature Reviews Molecular Cell Biology 21 (4), 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafalski VA et al. (2012) Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci 125 (Pt 23), 5597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik JH et al. (2009) FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5 (5), 540–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renault VM et al. (2009) FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5 (5), 527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X et al. (2011) FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol 13 (9), 1092–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzig S and Shaw RJ (2018) AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19 (2), 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W et al. (2001) Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J Biol Chem 276 (42), 38341–4. [DOI] [PubMed] [Google Scholar]
- 14.Greer EL et al. (2007) The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 282 (41), 30107–19. [DOI] [PubMed] [Google Scholar]
- 15.Locasale JW and Cantley LC (2011) Metabolic flux and the regulation of mammalian cell growth. Cell Metab 14 (4), 443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amiott EA and Jaehning JA (2006) Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol Cell 22 (3), 329–38. [DOI] [PubMed] [Google Scholar]
- 17.Sauve AA and Youn DY (2012) Sirtuins: NAD(+)-dependent deacetylase mechanism and regulation. Curr Opin Chem Biol 16 (5-6), 535–43. [DOI] [PubMed] [Google Scholar]
- 18.Reid MA et al. (2017) The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 19 (11), 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asher G et al. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134 (2), 317–28. [DOI] [PubMed] [Google Scholar]
- 20.Nakahata Y et al. (2008) The NAD+−dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134 (2), 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asher G et al. (2010) Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142 (6), 943–53. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya D et al. (2020) Metabolic Reprogramming Promotes Neural Crest Migration via Yap/Tead Signaling. Dev Cell 53 (2), 199–211 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moya IM and Halder G (2019) Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 20 (4), 211–226. [DOI] [PubMed] [Google Scholar]
- 24.Kerosuo L and Bronner-Fraser M (2012) What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Semin Cell Dev Biol 23 (3), 320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong C et al. (2012) G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 122 (4), 1469–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong C et al. (2013) Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23 (3), 316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberti MV and Locasale JW (2016) The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 41 (3), 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oginuma M et al. (2020) Intracellular pH controls WNT downstream of glycolysis in amniote embryos. Nature 584 (7819), 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondoh H et al. (2007) A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal 9 (3), 293–9. [DOI] [PubMed] [Google Scholar]
- 30.Chung S et al. (2008) Developmental restructuring of the creatine kinase system integrates mitochondrial energetics with stem cell cardiogenesis. Ann N Y Acad Sci 1147, 254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folmes CD et al. (2011) Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14 (2), 264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takubo K et al. (2013) Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12 (1), 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moussaieff A et al. (2015) Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21 (3), 392–402. [DOI] [PubMed] [Google Scholar]
- 34.Zheng X et al. (2016) Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W et al. (2019) TIGAR promotes neural stem cell differentiation through acetyl-CoA-mediated histone acetylation. Cell Death Dis 10 (3), 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuervo AM and Dice JF (1996) A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273 (5274), 501–3. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y et al. (2020) Chaperone-mediated autophagy regulates the pluripotency of embryonic stem cells. Science 369 (6502), 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Estrada J et al. (2016) Information Integration and Energy Expenditure in Gene Regulation. Cell 166 (1), 234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregor T et al. (2007) Probing the limits to positional information. Cell 130 (1), 153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z et al. (2014) Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev 28 (6), 608–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannon CE et al. (2017) Concentration dependent chromatin states induced by the bicoid morphogen gradient. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu D et al. (2004) The co-activator CREB-binding protein participates in enhancer-dependent activities of bicoid. J Biol Chem 279 (47), 48725–33. [DOI] [PubMed] [Google Scholar]
- 43.Li XY et al. (2014) Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3, doi: 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eck E et al. (2020) Quantitative dissection of transcription in development yields evidence for transcription factor-driven chromatin accessibility. bioRxiv 10.1101/2020.01.27.922054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagano Y and Ode KL (2014) Temperature-independent energy expenditure in early development of the African clawed frog Xenopus laevis. Phys Biol 11 (4), 046008. [DOI] [PubMed] [Google Scholar]
- 46.Rodenfels J et al. (2019) Heat Oscillations Driven by the Embryonic Cell Cycle Reveal the Energetic Costs of Signaling. Dev Cell 48 (5), 646–658 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song Y et al. (2019) Energy budget of Drosophila embryogenesis. Curr Biol 29 (12), R566–R567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alberty RA and Goldberg RN (1992) Standard thermodynamic formation properties for the adenosine 5'-triphosphate series. Biochemistry 31 (43), 10610–5. [DOI] [PubMed] [Google Scholar]
- 49.Brown GC (1992) Control of respiration and ATP synthesis in mammalian mitochondria and cells. Biochem J 284 ( Pt 1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atkinson DE (1977) Cellular Energy Metabolism and Its Regulation, Academic Press. [Google Scholar]
- 51.Milo R and Phillips R (2015) Cell Biology by the Numbers, First edn., Garland Science. [Google Scholar]
- 52.Zhang H et al. (2009) Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci U S A 106 (46), 19617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brauer MJ et al. (2005) Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell 16 (5), 2503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerosa L et al. (2013) Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol Syst Biol 9, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papagiannakis A et al. (2017) Autonomous Metabolic Oscillations Robustly Gate the Early and Late Cell Cycle. Mol Cell 65 (2), 285–295. [DOI] [PubMed] [Google Scholar]
- 56.Cassidy JJ et al. (2019) Repressive Gene Regulation Synchronizes Development with Cellular Metabolism. Cell 178 (4), 980–992 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X and Carthew RW (2005) A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123 (7), 1267–77. [DOI] [PubMed] [Google Scholar]
- 58.Silver SJ and Rebay I (2005) Signaling circuitries in development: insights from the retinal determination gene network. Development 132 (1), 3–13. [DOI] [PubMed] [Google Scholar]
- 59.Ilbay O and Ambros V (2019) Pheromones and Nutritional Signals Regulate the Developmental Reliance on let-7 Family MicroRNAs in C. elegans. Curr Biol 29 (11), 1735–1745 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abbott AL et al. (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 9 (3), 403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolfe J and Bagnall DJ (1980) Arrhenius Plots Curves or Straight Lines? Ann. Bot 45, 485–488. [Google Scholar]
- 62.Gillooly JF et al. (2001) Effects of size and temperature on metabolic rate. Science 293 (5538), 2248–51. [DOI] [PubMed] [Google Scholar]
- 63.Gillooly JF et al. (2002) Effects of size and temperature on developmental time. Nature 417 (6884), 70–73. [DOI] [PubMed] [Google Scholar]
- 64.Crapse J et al. (2020) Evaluating the simple Arrhenius equation for the temperature dependence of complex developmental processes. bioRxiv 10.1101/2020.07.17.208777. [DOI] [Google Scholar]
- 65.Lucchetta EM et al. (2005) Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434 (7037), 1134–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucchetta EM et al. (2009) The endo-siRNA pathway is essential for robust development of the Drosophila embryo. PLoS One 4 (10), e7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y et al. (2020) Temperature-dependent growth contributes to long-term cold sensing. Nature 583 (7818), 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]