Abstract
Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNAs characterized by a covalently closed-loop structure generated through a special type of alternative splicing termed back-splicing. Currently, an increasing body of evidence has demonstrated that 1) majority of circRNAs are evolutionarily conserved across species, stable, and resistant to RNase R degradation, and often exhibit cell-specific, and tissue-specific/developmental-stage-specific expression and can be largely independent of the expression levels of the linear host gene-encoded linear RNAs; 2) the biogenesis of circRNAs via back-splicing is different from the canonical splicing of linear RNAs; 3) circRNA biogenesis is regulated by specific cis-acting elements and trans-acting factors; 4) circRNAs regulate biological and pathological processes by sponging miRNAs, binding to RNA-binding protein (RBP), regulators of splicing and transcription, modifiers of parental gene expression, and regulators of protein translation or being translated into peptides in various diseases; 5) circRNAs have been identified for their enrichment and stability in exosomes and detected in body fluids such as human blood, saliva, and cerebrospinal fluids, suggesting that these exo-circRNAs have potential applications as disease biomarkers and novel therapeutic targets; 6) several circRNAs are regulated by oxidative stress and mediate reactive oxygen species (ROS) production as well as promote ROS-induced cellular death, cell apoptosis, and inflammation; 7) circRNAs have also emerged as important regulators in atherosclerotic cardiovascular disease, metabolic disease, and cancers; 8) the potential mechanisms of several circRNAs have been described in diseases, hinting at their potential applications as novel therapeutic targets. In this highlight, we summarized the current understandings of the biogenesis and functions of circRNAs and their roles in ROS regulation and vascular inflammation-associated with cardiovascular and metabolic disease.
Keywords: Circular RNAs (circRNAs), Vascular inflammation, Exosomes, Reactive oxygen species (ROS)
1. Introduction
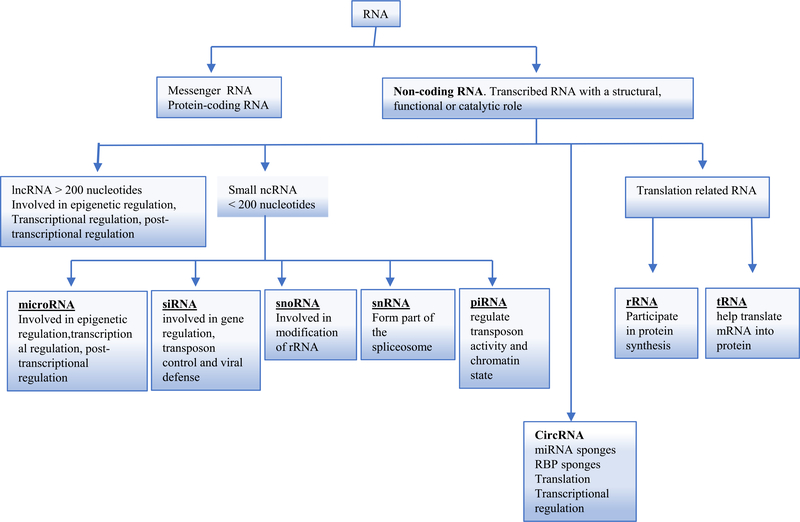
Cellular ribonucleic acids (RNAs) can be divided into two categories: coding and non-coding RNAs. Non-coding RNAs (ncRNAs) are functional RNAs that are transcribed from DNA, but are incapable of being translated into proteins. These ncRNAs can be classified into two groups based on their sizes, ncRNAs greater than 200 nucleotides are classified as long non-coding RNAs (lncRNAs), and ncRNAs less than 200 nucleotides are grouped as small ncRNAs. Small ncRNAs can be further classified into: microRNAs (miRNAs), small interfering RNAs (siRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), piwi-interacting RNAs (piRNAs), transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) (Esteller, 2011; Hombach & Kretz, 2016; Mattick & Makunin, 2006) (Fig. 1). In recent years, a large number of studies have shown that ncRNAs play fundamental regulatory roles in biological processes (Wei, Huang, Yang, & Kang, 2017). One of the significant examples in demonstrating the master gene functions of these ncRNAs is that we established the first mouse model of metabolically healthy obesity with miR155 deficiency in atherogenic apolipoprotein E deficient (ApoE−/−) background (Johnson et al., 2018; Virtue et al., 2017; Virtue, Wang, & Yang, 2012).
Fig. 1.
A classification of non-coding RNA and their biological functions. Abbreviations: ncRNA: non-coding RNA; circRNA: circular RNA; lncRNA: long non-coding RNA; miRNA: microRNA; siRNA: small interfering RNA; snoRNA: small nucleolar RNA; snRNA: small nuclear RNA; piRNA: piwi-interacting RNA; rRNA: ribosomal RNA; tRNA: transfer RNA.
In addition to lncRNAs and small ncRNAs, circular RNAs (circRNAs) represent a novel and large family of non-coding endogenous RNAs recently discovered in all eukaryotic cells. These ncRNAs arise from a particular alternative splicing (back-splicing) mechanism of precursor mRNAs (pre-mRNAs) (Chen, Huang, Wang, & Shan, 2015a). This back-splicing mechanism results in a covalently closed circular loop molecule that lack 5′ – 3′ ends and poly-adenylated tails (Salzman, 2016). Compared to the other types of ncRNAs, circRNAs are evolutionarily conserved among species, highly stable, and resistant to RNase R degradation (Table 1). These features provide circRNAs with many potential functions, such as (a) acting as miRNA sponges, (b) regulating the expression of parental genes, (c) regulating alternative splicing or translation, (d) acting as RNA-binding protein (RBP) sponges, and (e) being translated into peptides/proteins.
Table 1.
Three different types of non-coding RNAs and their features. Abbreviation: Pol II: polymerase II., pre-miRNAs: precursor miRNAs, PCR: polymerase chain reaction, RNA-FISH: RNA fluorescence in situ hybridization
| Feature | CircRNA | miRNA | lncRNA |
|---|---|---|---|
| Origin | Originated from pre-mRNA by pol II via back-splicing | Originate from mRNAs transcripts to form primary miRNA and then undergo editing by adenosine deaminases and undergo processing by RNA Pol II Drosha in the nucleus to form pre-miRNAs then exported to cytoplasm and cleaved by Dicer to form mature double-stranded miRNAs | Transcribed from various genomic regions by both RNA Pol II and III |
| Length | >200 nucleotides | 20 to 22 nucleotides | >200 nucleotides |
| Location | Nuclear and cytoplasm | Pre-miRNAs are found in the nucleus while mature miRNA found in the cytoplasm | Nuclear and cytoplasm |
| Degradation | Degraded by RNase L | Degraded by exoribonucleases such as small RNA degrading nuclease 1 (SDN1) and PNPase PNPT1 | Degraded by RNase P cleavage |
| Conservation | Evolutionary conserved | Evolutionarily conserved | poorly conserved |
| Stability | Highly stable | Highly stable | Unstable |
| miRNA sponges | Yes | No | No |
| Splicing regulation | Yes | Yes | Yes |
| Interaction with RBPs | Yes | Yes | Yes |
| Transcriptional regulation | Yes | Yes | Yes |
| Post-transcriptional regulation | Yes | Yes | Yes |
| Translational regulation | Yes | Yes | Yes |
| Methods of detection | circRNA seq, microarray, PCR, digital droplet PCR (ddPCR), RNA-FISH, and northern blot analysis | RNA-seq, microarray, PCR, and RNA-FISH | RNA-seq, microarray, PCR, and RNA-FISH |
CircRNAs were discovered more than four decades ago and were first identified in plant-based viruses via electron microscope in 1976 (Sanger, Klotz, Riesner, Gross, & Kleinschmidt, 1976). In 1979, circRNAs were identified in eukaryotic cells as endogenous RNA splicing products (Hsu & Coca-Prados, 1979). Then in 1986, circRNAs were identified in humans following hepatitis delta virus infection (Kos, Dijkema, Arnberg, van der Meide, & Schellekens, 1986). Therefore, circRNAs were typically considered as a functionless byproducts of aberrant RNA splicing and thus have not gained sufficient scientific attention. Two reports significantly transformed the field: the first study showed that RNA transcripts from many human genes were arranged in a non-canonical order, resulting in a type of circRNAs isoform (Salzman, Gawad, Wang, Lacayo, & Brown, 2012). The second study reported that circular transcripts of cerebellar degeneration-related protein 1 antisense RNA (CDR1as, ciRS7) could act as miRNA sponges for miR-7 (Memczak et al., 2013). These significant findings changed circRNAs into a new focal point of scientific research and rising stars in the ncRNA field. An excellent RNA database, exoRBase (http://www.exorbase.org/), has collected as many as 58,330 circRNAs, 15,501 lncRNAs, and 18,333mRNAs (Li et al., 2018a), suggesting that the numbers of circRNAs are even higher than that of mRNAs.
In recent years, the development and application of microarray techniques, high-throughput deep RNA sequencing (RNA-seq), and novel bioinformatics approaches have led to the discovery of many circRNAs (Broadbent et al., 2015; Danan, Schwartz, Edelheit, & Sorek, 2012; Fan et al., 2015; Lu et al., 2015; Yang, Duff, Graveley, Carmichael, & Chen, 2011) with some studies suggesting they may outnumber mRNAs (Li, Li, et al., 2018a). These recent advancements have demonstrated that circRNAs are incredibly abundant, relatively stable, diverse and conserved, and broadly expressed in eukaryotic cells (Jeck et al., 2013). Additionally, circRNAs exhibit cell-type, tissue-type or developmental stage-specific expression (Conn et al., 2015; Guo, Agarwal, Guo, & Bartel, 2014; Salzman et al., 2012; Salzman, Chen, Olsen, Wang, & Brown, 2013). Emerging evidence suggests that circRNAs are responsible for regulating complicated biological functions. For example, studies have shown a role for circRNAs as endogenous microRNA (miRNA) sponges (Hansen et al., 2013; Memczak et al., 2013; Wang et al., 2016a), transcriptional regulators of their encoding parental gene expressions (Li et al., 2017a; Zhang et al., 2013), and modulating alternative splicing (Ebbesen, Hansen, & Kjems, 2017). Furthermore, circRNAs can interact with RBPs, and act as scaffolds in the assembly of protein complexes (Du et al., 2016; Zhang et al., 2017; Zhang & Xin, 2018).
Recent studies have implicated circRNAs in physiological processes such as aging (Westholm et al., 2014) and insulin secretion (Xu, Guo, Li, & Yu, 2015). Additionally, circRNAs have been demonstrated to play a critical role in the pathogenesis of various human diseases, including atherosclerotic cardiovascular disorders (Burd et al., 2010; Holdt et al., 2016), diabetes (Zhao et al., 2017a), cancers (Bachmayr-Heyda et al., 2015; Chen et al., 2017a; Geng, Jiang, & Wu, 2018; Qin et al., 2016), Alzheimer’s disease (Lukiw, 2013; Zhao, Alexandrov, Jaber, & Lukiw, 2016), nervous system disorders (Bai et al., 2018), and osteoarthritis (Liu et al., 2016). In particular, we (Li et al., 2019a) and others reported that circRNAs are associated with cardiovascular and metabolic diseases; however, little is known about the exact role of circRNAs in vascular inflammation associated with cardiovascular and metabolic diseases.
Several studies reported that circRNAs can perform their biological functions inside the cell or exported by exosomes and taken up by neighboring (paracrine) or distant cells (endocrine) and affect many aspects of physiological and pathological conditions of the recipient cells (Bai, Lei, Huang, Jiang, & Zhou, 2019; Li et al., 2015a). Paracrine signaling has been proposed to promote cell-cell communication in various human cancers (Dou et al., 2016; Hon, Ab-Mutalib, Abdullah, Jamal, & Abu, 2019; Li, Zheng, et al., 2015a; Louis, Desoteux, & Coulouarn, 2019). However, the roles of exosomal circRNAs in propagating cardiovascular and metabolic inflammation have not been studied. Furthermore, the development of novel therapeutics targeting circRNAs may help mitigate vascular inflammation associated with cardiovascular and metabolic diseases.
2. CircRNA biogenesis
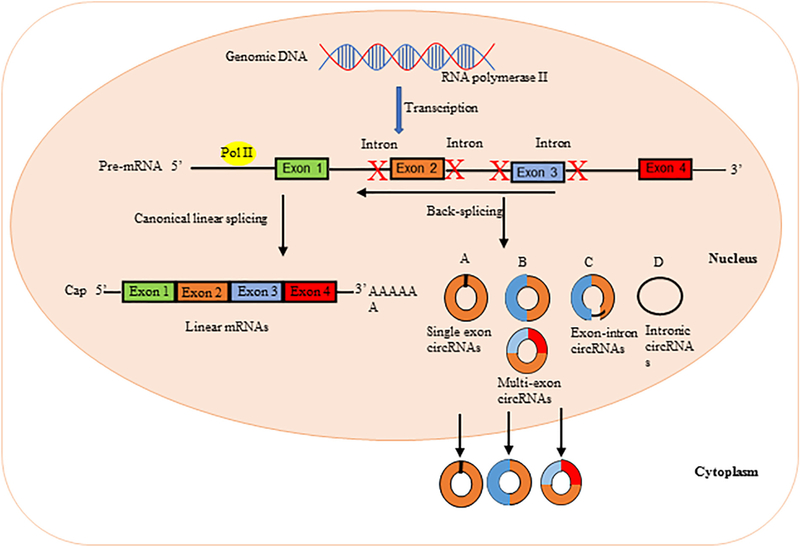
CircRNA biogenesis adds a modification step to the conventional generation of mRNAs. The precursor mRNA (pre-mRNA) splicing is catalyzed by the canonical spliceosome machinery to remove introns from the transcript and join exons leading to the formation of a linear mRNA with 5′–3′ polarity. However, circRNAs are generated from pre-mRNA by the action of RNA polymerase II (Chen, 2016). Most circRNAs do not follow the canonical 5′–3′ order, and are produced during back-splicing (Vicens & Westhof, 2014). These circRNAs are distinct from their linear RNA counterparts because they lack the 5′ – 3′ ends and poly-adenylated tail due to their closed covalent structure, which usually decides the fate of many RNA transcripts (Memczak et al., 2013). In general, circRNAs originated from exonic sequences, producing exonic circRNAs (ecircRNAs) (Jeck et al., 2013); or from intronic sequences, producing intronic circRNAs (ciRNAs) (Zhang et al., 2013). However, the exon-intron circRNAs (EIciRNAs) can be generated from intron-containing exons (Jeck & Sharpless, 2014). Exonic circRNAs are formed as a result of pre-mRNA splicing where the 3′ splice donor attaches to the 5′ splice acceptor forming an exonic circRNA (Jeck et al., 2013; Jeck & Sharpless, 2014). This type of splicing might occur with a single exon, but sometimes happens with multiple exons. However, if the introns between the exons are retained, the resulting circRNAs are referred to as exon-intron circRNAs (Li et al., 2015b). The intronic circRNAs are produced from intron lariat (Jeck et al., 2013; Li, Huang, et al., 2015b). The process of intronic circRNAs formation depends on GU-rich sequences near the 5’ splice site and C-rich sequences near the branch point. Finally, the lariat undergoes debranching and easily degraded while the mature intronic circRNA is generated (Barrett, Parker, Horn, Mata, & Salzman, 2017). These circRNAs are formed in the nucleus, and some of them are transported to the cytoplasm. The exonic circRNAs typically reside in the cytoplasm, but the intronic and exon intronic circRNAs remain in the nuclei (Li, Huang, et al., 2015b) (Fig. 2A).
Fig. 2A.
Schematic representation illustrating circRNA biogenesis. In the nucleus of eukaryotic cells, DNA is transcribed to form precursor mRNA (pre-mRNA), which contain coding exons and introns. Different from linear mRNAs, which are formed by canonical linear splicing and cutting away introns of the pre-mRNAs using small nuclear ribonucleoproteins (snRNPs), circular RNAs (circRNAs) are formed by back-splicing of the pre-mRNAs and circularization of the cut segment, where the 5’ end joins the 3’ end. (A) Single exon circRNAs: circular RNAs can be generated from a single exon; (B) Multi-exon circRNAs: circular RNAs can also be generated from two or more exons; (C) Exon-intron circRNAs: circular RNAs can contain intron(s) that have been retained between one or more circular exons; (D) Intronic circRNAs: introns can be excised from pre-mRNAs and circularize to give rise to circRNAs.
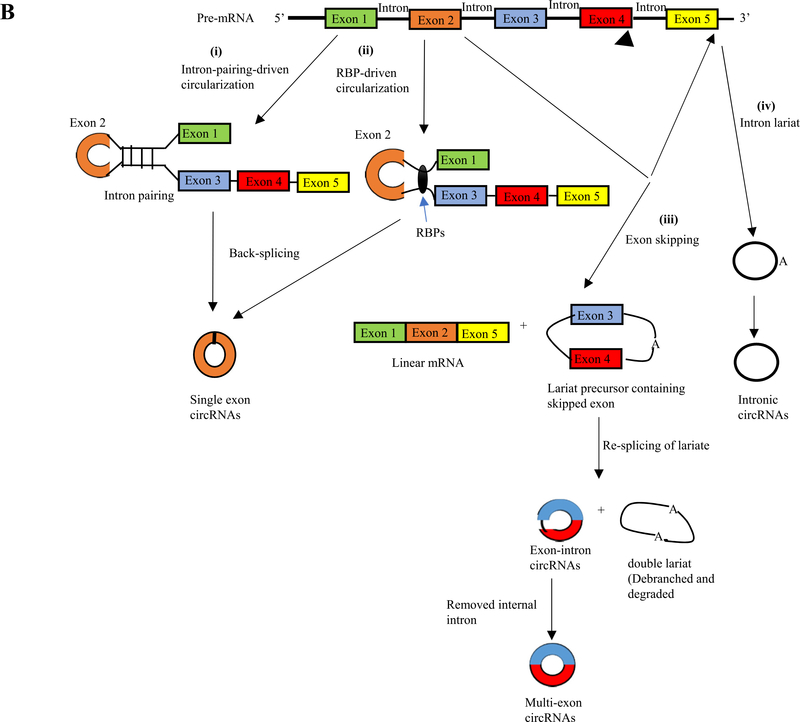
To date, four different hypothetical models of circRNA biogenesis have been proposed (Fig. 2B) including (i) intron-pairing-driven circularization; (ii) RNA binding protein (RBP)-driven circularization; (iii) exon-skipping; and (iv) intron lariat circularization (Barrett, Wang, & Salzman, 2015). The first model of circRNA biogenesis is intron pairing-driven circularization (Fig. 2Bi). In this model, two introns flanking the exon/exons of a pre-mRNAhave a structure capable of joining each other. The flanking introns approach each other creating a secondary conformation that makes the splice sites possible to carry on back-splicing and generate exonic circRNA. Adenosine deaminase 1 acting on RNA (ADAR1) is involved in the intron-pairing process of circRNA formation (Athanasiadis, Rich, & Maas, 2004). The second proposed model is the RBPs-driven circularization (Fig. 2Bii). In this model, RBPs bind to pre-mRNAs to connect the flanking introns. This process is induced by protein dimerization, which forms an RNA loop. Muscleblind like splicing regulator 1 (MBNL1) protein, is the most popular RBPs responsible for circRNA biogenesis (Chen & Yang, 2015). The third circRNA biogenesis model that can give rise to back-splicing is exon skipping (Fig. 2Biii). In this model, one or multiple exons of the mature mRNA will be missing. The lariat-driven circularization proceeds as the non-adjacent exons join producing linear mRNA and exon-intron or multiple exon circRNA transcript with lariat structure. Finally, the fourth proposed mechanism is the intron lariats (Fig. 2Biv) which can form intronic circRNAs (ciRNAs) (Kristensen et al., 2019). These three major subclasses of generated circRNAs are different in their location, formation, and biological function (Fig. 3).
Fig. 2B.
The proposed models of circRNA formation. i) Intron-pairing-driven circularization. Two complementary introns form a circular structure containing several introns and exons through a base-pairing connection. Finally, introns are removed to form exonic circRNAs (EcircRNAs). ii) RNA binding protein (RBP)-driven circularization. The binding of RBPs acts as a vehicle that binds two non-adjacent introns. Then circRNAs are generated after the removal of introns. iii) Exon skipping: the back-splicing process can take place because of exon skipping mechanism, which leads to lariat formation. This process can generate three different products: linear mRNA, an exonic or exonic-interonic circRNAs, and intron lariats iv) Intron lariat will generate intronic circRNAs.
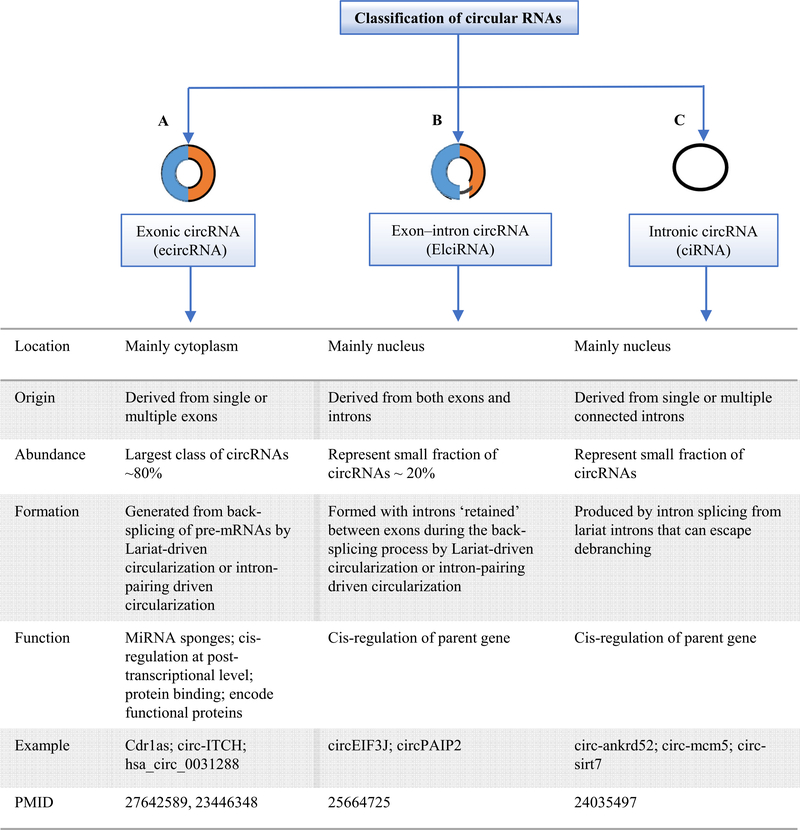
Fig. 3.
Three major subclasses of circRNAs. (A) Exonic circRNAs (ecircRNAs) consist of only exon(s) (usually less than five) and represent the most important group of circRNA class. EcircRNAs have cytoplasmatic location and may regulate microRNA and protein functions.(B) Exon-intron circRNAs (EIciRNAs) are composed of exons and retained introns. EIciRNAs have nuclear localization and have been found to be able to regulate gene transcription in cis and probably also in trans.(C) Intronic circRNAs (ciRNAs) are derived from intron lariats and are accumulated in the nucleus in which regulate gene transcription in cis.
3. Regulation of circRNA biogenesis
In general, the levels of steady-state circRNA expression in cells can be regulated by transcription regulation of circRNA-producing pre-mRNA or by circRNAs degradation. Emerging lines of evidence show that the transcriptional regulation of circRNAs involves both cis-regulatory elements and trans-acting factors that control the back-splicing machinery required for circRNA biogenesis. These factors include core spliceosome components, intronic complementary sequences (ICSs) flanking circle formation exons, and other regulatory RBPs. Both cis-elements and trans-factors are required to bring the downstream donor and upstream acceptor sites close together to promote back-splicing (Starke et al., 2015; Zhang et al., 2014a).
Previous studies reported that both canonical splice signals and spliceosome machinery are required for back-spliced circularization; and that back-splicing is usually coupled with canonical splicing (Ashwal-Fluss et al., 2014; Starke et al., 2015). It has been reported that the processing of circRNAs can be facilitated by either RNA pairing of reversely complementary sequences across their flanking introns or protein factors binding to pre-mRNAs to bridge flanking introns together (Ashwal-Fluss et al., 2014; Starke et al., 2015; Zhang, Wang, et al., 2014a). RNA pairing across flanking introns promotes exon circularization; and strong pairing capacity could dramatically increase the production of circRNAs. Two known RNA binding proteins, MBNL1 and ADAR1, are reported to play a significant role in circRNA biogenesis, where they bind to its own pre-mRNA and bridge two flanking introns close together leading to increased circRNA formation (Ashwal-Fluss et al., 2014; Ivanov et al., 2015). Notably, ADAR1 knockdown increased expression levels of some circRNAs, indicating a role of ADAR1 in the suppression of circRNA biogenesis (Chen et al., 2015b).
4. Properties of circRNAs
CircRNAs have several important properties including: 1) circRNAs are exist as endogenous ncRNA (Salzman et al., 2012); 2) circRNAs are widely expressed in eukaryotic cells (Jeck et al., 2013), and exhibit cell-specific and tissue-specific/developmental-stage-specific expression (Memczak et al., 2013; Xu,Wu, Han, Zhao, & Song, 2017). Despite the low levels of most circRNA expression, some are highly expressed than their corresponding linear mRNAs (Jeck et al., 2013; Memczak et al., 2013; Salzman et al., 2012). Previous studies reported that the expression of some circRNAs is more than 10-fold higher than the linear mRNA counterparts (Jeck et al., 2013). Also, other studies demonstrated that the expression of circRNA is not correlated with the expression of its linear mRNA as some circRNA is not detected even though the mRNA expression levels is too high (Nigro et al., 1991). Others demonstrated that circRNAs are highly expressed in the brain tissue (Rybak-Wolf et al., 2015). Different biological processes including epithelial-mesenchymal transition (EMT) (Conn et al., 2015), aging, and stress significantly changes the expression profile of circRNAs (Cortés-López et al., 2018; Fischer & Leung, 2017). Furthermore, RNA-seq analysis of human fetal and adult tissues including liver, kidney, heart, lung, stomach, and colon reported that up to 50% of circRNAs are tissue specific and their expression level is higher in fetal tissues than in adult tissues (Xu et al., 2017). Additionally, circRNAs are expressed at very low levels in human cancer cells (Bachmayr-Heyda et al., 2015). These studies strongly suggest that the expression levels of circRNA is a highly regulated process; 3) compared to linear RNAs, circRNAs form covalently closed-loop structures with neither 5′−3′ polarities nor poly-adenylated tails, resulting in reduced degradation by RNA exonuclease or RNase R, which makes them more stable than linear RNAs (Suzuki et al., 2006; Suzuki & Tsukahara, 2014); 4) The majority of the circRNAs including exonic circRNA are predominantly found in the cytoplasm (Jeck et al., 2013; Jeck & Sharpless, 2014; Memczak et al., 2013), however, few of them including the intron retained circRNAs such as intronic circRNA and exon-intron circRNA are exclusively located in the nucleus (Ebbesen et al., 2017; Zhang et al., 2013; Zhang, Wang, et al., 2014a). It was reported previously that some exonic circRNAs are also found in the nucleus (Errichelli et al., 2017) then exported to the cytoplasm. However, the mechanism by which circRNAs are exported from the nucleus to the cytoplasm remains unknown. As in the aforementioned, there are several models of circRNA biogenesis were hypothesized including lariat-driven circularization or exon skipping that generate intronic or exonic-intronic circRNAs and intron-pairing-driven circularization or direct back-splicing that generate exonic circRNA. Of note, previous studies demonstrated that intron-pairing driven circularization may occur more frequently than lariat-driven circularization (Jeck & Sharpless, 2014). Therefore, these studies indicates that exonic circRNAs represent the largest class (more than 80%) of total circRNAs and locate in the cytoplasm, while intronic circRNA represents only 20%(Chen, Huang, et al., 2015a) (Fig. 3).; 5) most of circRNAs are located in the cytoplasm and few are found in the nucleus (Memczak et al., 2013; Zhang et al., 2013); 6) circRNAs are highly conserved between species (Jeck et al., 2013; Salzman et al., 2012; Zhang, Wang, et al., 2014a); 7) most of circRNAs have microRNA response element (MRE), so they can interact with miRNAs and thus regulate target gene expressions (Hansen et al., 2013); and 8) vast majority of circRNAs can play roles in transcription and post-transcription regulation (Zhang et al., 2013).
5. Biological functions and mechanisms of circRNAs
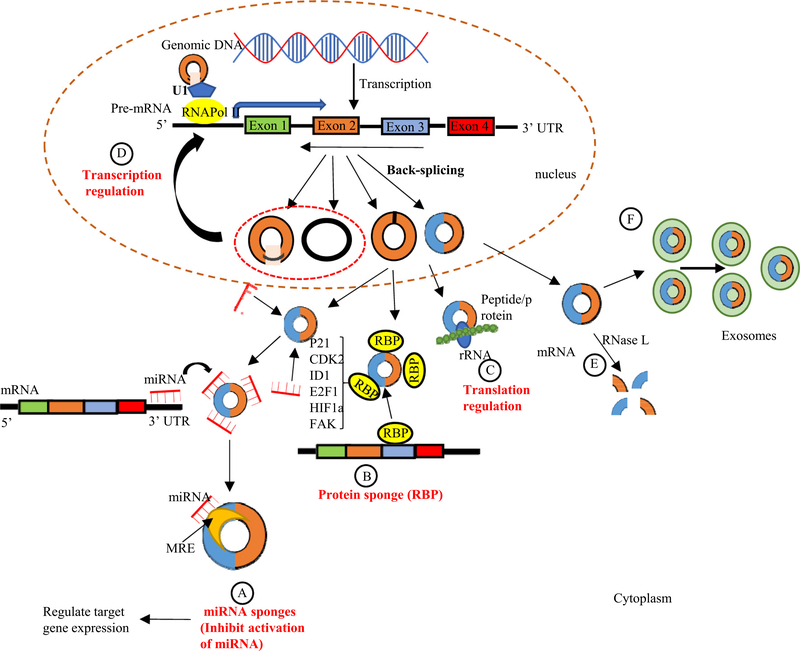
In recent years, the biological functions of circRNAs have become a hotspot of scientific research. A growing body of evidence shows that besides acting as miRNA sponges (Bartel, 2009), several other roles of circRNAs have been proposed (Fig. 4). CircRNAs may also interact with regulatory RBPs through their activities as protein sponges, decoys, scaffolds, and recruiters (Huang, Zheng, Wu, Chen, & Huang, 2020). By enhancing binding to RNA polymerase II, circRNAs located in the nucleus may modulate the transcription of their host genes, and regulate alternative splicing and transcription as well as translation (Conn et al., 2017).
Fig. 4.
Schematic representation of circRNA functions and degradation. (A)microRNA sponges: circRNAs can act as miRNA sponges by competing for miRNA binding sites (miRNA response elements (MREs)) and prevents miRNA from interacting with their target messenger RNA (mRNA) at 3’ untranslated region (UTR) leading to reduced the effect of miRNA-mediated regulatory activities.(B) Interaction with RNA binding proteins (RBPs): circRNAs may act as protein sponges, by directly binding to RBPs, and therefore retain them in the cytoplasm. These RBPs includes: cyclin-dependent kinase inhibitor 1 (p21), cyclin-dependent protein kinase 2 (CKD2), inhibitor of DNA binding 1 (ID1), E2F1, Hypoxia-inducible factor-1α (HIF1a), and preface focal adhesion kinase (FAK). (C) CircRNA can be translated with ribosome and encode peptides or proteins. (D) CircRNAs (e.g. EIciRNAs and ciRNAs) may interact with transcription complexes and enhance the expression of their parental genes. (E) The degradation of circRNAs. CircRNAs are globally degraded by RNase L in early cellular innate immune responses. (F) The elimination of circRNAs. circRNAs can be eliminated into the extracellular space by exosomes
5.1. CircRNAs can act as miRNA sponges
The majority of circRNAs are located in the cytoplasm, indicating that the circRNA is involved in post-transcriptional regulation and might function as miRNA sponges or competitive endogenous RNAs (ceRNAs) to regulate the expression of miRNA targets (Jeck et al., 2013). MicroRNAs can bind to the complementary sequences in the 3′-untranslated regions of mRNAs (Bartel, 2004) to facilitate mRNA degradation. Many circRNAs contain miRNA binding sites, also known as miRNA response elements (MREs), which allow them to sponge miRNAs and act as ceRNAs in order to inhibit miRNAs from negatively regulating their target mRNAs (Memczak et al., 2013). By binding to the 3’-untranslated region (UTR) of mRNAs, miRNAs can inhibit protein translation and facilitate mRNA degradation (Bartel, 2004) (Fig. 4A).
Since miRNAs have an inhibitory effect on their target genes, circRNA sponging leads to the upregulation of the miRNA target mRNAs and increases the expression of that gene products. Therefore, circRNAs indirectly regulate mRNAs translation. However, it is important to consider the stoichiometric relationship between the miRNA binding sites of the circRNA and the mRNA target sites of the miRNA. Of note, highly abundant circRNAs having many competing binding sites are more likely to have competing endogenous RNA function (Thomson & Dinger, 2016). For example, CDR1as, a circRNA that is highly expressed in the brain tissue, has more than 70 conserved seed-binding sites for miR-7, and miR-7 occupies many of these sites (Piwecka et al., 2017), which is involved in the regulation of several miR-7 target genes in the brain (Hansen et al., 2013; Memczak et al., 2013). Similarly, miR-671 has one complementary binding site to CDR1as, indicating that while miR-671 can mediate cleavage of CDR1as, CDR1as might regulate miR-7 levels and activity (Hansen et al., 2011; Hansen et al., 2013). Furthermore, the circRNA Sry acts as a miRNA sponge for miR-138, with at least 16 binding sites for miR-138 (Hansen et al., 2013). On the other hand, some circRNAs do not possess multiple binding sites for a specific miRNA, but they harbor many different types of miRNA binding sites. For instance, circRNA circITCH has multiple binding sites for miR-7, miR-17, and miR-214 leads an increase of E3-ubiquitinligase gene (INCH) expression (Li et al., 2015c). Even though, some identified circRNAs possess fewer miRNA binding sites than their co-linear mRNAs counterpart, but also exhibit their miRNA sponge function, indicating that the function of circRNAs as miRNA sponges is conserved across species. Recently, many studies have determined the sponge function of circRNAs; for example, circ_CHFR has been shown to promote oxidized low-density lipoprotein (oxLDL)-induced vascular smooth muscle cell proliferation, migration, and inflammation by interacting with miR-214–3p (Zhuang et al., 2020). In addition, homeodomain-interacting protein kinase 3 (HIPK3) circular RNA (circHIPK3) suppresses miR-30a-3p activity by sponging miR-30a-3p (Shan et al., 2017). These studies indicated that circRNAs might play a significant role as miRNA sponges in inflammatory cardiovascular diseases.
5.2. CircRNAs can act as RNA-binding protein (RBP) sponges
RBPs play a critical role in post-transcriptional regulatory processes associated with different biological activities, including cell proliferation, apoptosis, senescence, and cell responses to oxidative stress via post-transcriptional regulation (Brinegar & Cooper, 1647). Increasing evidence suggests that specific circRNAs can act as protein sponges by providing binding sites for specific RBPs to sequester and inhibit the biological activity of proteins through competitive inhibition (Zang, Lu, & Xu, 2020) (Fig. 4B) ). The interaction of circRNAs with the RBPs such as Elav like RNA binding protein 1 (HuR), KH-type splicing regulatory protein (KHSBP), tristetraprolin (TTP), heterogeneous nuclear ribonucleoprotein D, F-box family protein (AUF1), and other potent regulatory RBPs could indirectly affect the fate of their respective target mRNAs (Panda, Grammatikakis, Munk, Gorospe, & Abdelmohsen, 2017a). We and our collaborators recently reported that in a bone marrow (BM) transplantation mouse model with TTP deficient BM transplantation into low-density lipoprotein receptor deficient recipient mice (LDLR−/−), TTP deficient (TTP−/−) BM recipients display significantly higher systemic and multi-organ inflammation than TTP+/+ BM recipients, implying that the TTP-sponged circRNAs could enhance systemic inflammation (Saaoud et al., 2020). Previous studies showed that circFoxo3, the circular RNA variant of the Foxo3 gene, can interact with p21 and cyclin dependent kinase 2 (CDK2) to inhibit cell cycle progression. CircFoxo3 can also bind and inhibit senescence-associated proteins (ID1 and E2F transcription factor 1, E2F1) and stress-related proteins (hypoxia-inducible factor 1, HIF1a and focal adhesion kinase, FAK) in the cytoplasm to regulate cardiac senescence (Du et al., 2016; Du et al., 2017).
5.3. CircRNAs can regulate translation and alternative splicing
Besides acting as miRNA sponges and binding to RBPs, circRNAs have a potential role in regulating protein translation (Jeck & Sharpless, 2014) (Fig. 4C). Several circRNAs can bind open reading frames (ORFs) and be translated as protein fragments. It has been shown that protein/polypeptides can be coded by circRNAs both in vivo and in vitro only when the RNAs contain prokaryotic ribosome-binding sites, or internal ribosome entry site elements (IRES) (Chen & Sarnow, 1995; Perriman & Ares Jr., 1998). It has been confirmed that circ-ZNF609 can be translated into a protein functioning in myogenesis (Legnini et al., 2017). Recently, it has been reported that N6-methyladenosine (m6A) is the most common and abundant base methylation modification of RNAs and promotes the initiation of protein translation from circRNA in human cells. Many circRNAs contain the m6A motifs, and only one m6A is required to drive translation initiation (Yang et al., 2017a). Interestingly, a large number of circRNAs have the potential for translation, indicating that they may have a regulatory role in the stimulation of circRNA-derived proteins in cell responses to environmental stressors. Besides being translated themselves, there is emerging evidence that some circRNAs, including circPABPN1, can regulate translation of their associated linear mRNAs (Abdelmohsen et al., 2017). These studies provide a new direction for the functional studies of circRNAs.
Previous studies reported that ecircRNAs could play a role in alternative splicing as the circularization can compete with canonical splicing. Mannose-binding lectin 2 (MBL2) protein is a splicing factor, which can affect alternative MBL pre-mRNA splicing during the generation of MBL mRNA and circular MBL (circMBL). The second exon of the MBL gene contains sequences that form a circRNA transcript having conserved binding sites for MBL protein in the flanking intronic sequences. In an auto-regulatory manner, circMBL influences the selective splicing of the MBL mRNA. Additionally, MBL can interact with circMBL and its flanking introns to promote exon circularization. Therefore, the competition between back-splicing for circRNA generation and canonical splicing is evident from the concomitant reduction in circRNA and an increase in linear splicing (Ashwal-Fluss et al., 2014). These findings may hint that some circRNAs may play a role in controlling the expression of mRNA through binding to RBPs and affecting the canonical splicing.
5.4. CircRNAs act as gene expression regulators
There are competitions between the biosynthesis of circRNAs and linear RNAs, with the expression of linear RNAs being regulated by circRNAs. Recent studies showed that certain circRNAs such as intron (ciRNAs) and intron-exon (EIciRNAs) circRNAs can promote transcription of the parental genes. These circRNAs have few binding sites for miRNAs, suggesting that they may function differently (Zhang, Yang, & Chen, 2014b). Also, ciRNAs and EIciRNAs are widely localized and detected in the nuclei and act as transcriptional regulators by binding to U1 small nuclear ribonucleoproteins (U1 snRPN) and RNA polymerase II in the promotor region of genes, thus promoting the transcription of the parental gene (Ashwal-Fluss et al., 2014; Li, Huang, et al., 2015b) (Fig. 4D).
6. Degradation and elimination of circRNAs
Once generated, exonic circular RNAs progressively accumulate in the cytoplasm. CircRNAs are highly stable and have long half-lives because of their circular structure, which makes them naturally resistant to degradation by RNase exonucleases. Currently, very little is known about the mechanisms of how circRNAs are degraded in vivo. The first evidence of natural circRNAs degradation via endonuclease activity was found in vitro using RNase H and DIS3 homolog, exosome endoribonuclease and 3’−5’ exoribonuclease (Rrp44) (Zhao, Zhu, Limbo, & Russell, 2018). Also, RNase L has been identified globally to degrade circRNAs (Liu et al., 2019a) (Fig. 4E). Additionally, circRNA binding by miRNAs may initiate circRNA decay by Argonaute RNA-induced silencing complex (RISC) catalytic component 2 (Ago2)-mediated cleavage. For instance, circRNA CDR1as degradation was dependent on miR-671-mediated Ago2 cleavage (Hansen et al., 2011). Emerging studies have demonstrated that circRNAs are abundant and stable in exosomes. These exosomal circRNAs were identified and detected in human blood and urine (Li, Zheng, et al., 2015a; Memczak, Papavasileiou, Peters, & Rajewsky, 2015) (Fig. 4F). Furthermore, multiple circRNAs are secreted from cells into extracellular space by exosomes and further removed by the reticulo-endothelial system or eliminated by the kidney and liver (Choi & Lee, 2016; Lasda & Parker, 2016; Wang et al., 2019a). Recently, the RNA modification, N6-methyladenosines (m6A), as well as poly (I:C) stimulation were shown to mediate the activation of the endoribonuclease, RNase L, and ultimately the degradation of both mRNAs and circRNAs (Park et al., 2019).
7. Approaches studying circRNAs
Since the initial discovery of circRNAs, various biochemical tools have been discovered and identified to detect and validate the existence of circRNAs and their localization, biogenesis, biological functions, disease implication, interacting molecules, and therapeutic potential. Furthermore, online resources, bioinformatics and statistical approaches have been developed to quantify the expression of circRNAs and identify new circRNAs with high confidence (Li, Yang, & Chen, 2018b) (Table 2A). First, high-throughput circRNA sequencing (circ-seq) analysis has enabled the identification of thousands of circRNAs. The unique feature of this technique is that circRNAs are enriched with RNase R digestion to eliminate most linear transcripts and keep circRNAs intact (Jeck et al., 2013; Salzman et al., 2013). Second, microarray high-throughput analysis is another useful technique to detect the expression levels of specific circRNAs. This method typically requires pre-treatment of the RNA samples with RNase R to degrade linear transcripts and enhance circRNA detection. The disadvantages of this technique include: 1) microarray platforms incorporate only a limited number of known circRNAs, 2) circRNAs identification is based only on the junction sequences and circRNAswith shared junction sequences cannot be distinguished, and 3)microarray cannot inform on the internal sequence of a given circRNA (Li et al., 2016a; Qu et al., 2015). A third technology used to study circRNA is reverse transcription-polymerase chain reaction (RT-PCR) has been used to validate the high-throughput circ-seq and microarray analysis. This technique requires the use of divergent primers spanning the circRNA junction, and the extracted circRNAs must be enriched with RNase R followed by RT-qPCR analysis, but often this step can be avoided if the circRNA is abundant, as divergent primers do not amplify linear RNA (Panda, Abdelmohsen, & Gorospe, 2017b). Fourth, digital droplet PCR (ddPCR) analysis is used to quantify circRNA copy number (Hindson et al., 2011). Compared to the conventional RT-qPCR, ddPCR is more accurate, provides absolute numbers, and measures low-abundance RNAs. However, it requires special instrumentation, software, and proprietary reagents (Quan, Sauzade, & Brouzes, 2018). Fifth, northern blot analysis is another technique used to investigate circRNAs size, isoforms, processing, sequence, and abundance, and also to distinguish between a circRNA and its linear counterpart (Pamudurti et al., 2017). Sixth, RNA fluorescence in situ hybridization (RNA-FISH) coupled with high-resolution microscopy is a commonly used technique to examine the abundance and localization of RNA molecules. RNA-FISH probe targets circRNA junctions to detect and quantify several circRNAs and determine the co-localization of circRNAs with proteins (Itzkovitz & van Oudenaarden, 2011; Jeck & Sharpless, 2014).
Table 2A.
Approaches to study circRNAs.
| Method | Criteria | Unique feature | PMID |
|---|---|---|---|
| High-throughput circRNA sequencing (Circ-seq) analysis | Allows identification and annotation, high-throughput screening, and profiling under different conditions | Pre-treatment with RNase R is required to eliminate most linear transcripts while circRNAs are left intact Circ-seq analysis detects circRNA junctions. |
28903484 27739534 |
| CircRNA microarrays analysis | A useful tools for high-throughput analysis and multiple comparisons of the expression levels of specific circRNAs | Pre-treatment with RNase R is required to reduce the presence of linear RNAs and enhance circRNA detection and quantification | 26451160 26484292 |
| Reverse transcription-polymerase chain reaction (RT-PCR) | Used to validate the high-throughput screening Used to assess the relative abundance of the circRNAs Amplify speculative back-spliced junction (BSJ) sites but, this can amplify the linear RNA containing the same BSJ sequence locus and detects both circular and linear RNAs because of their sequence overlap. |
Require the use of divergent primers that are designed to amplify and detect the BSJ, which can be designed using online resources such as CircInteractome Divergent primers will not amplify linear RNA Require the use of the RNA exonuclease enrichment strategy to digest all linear RNAs but does not digest circRNAs |
16682442 |
| Digital droplet PCR (ddPCR) analysis | A relatively new technique which used to quantify and measure the absolute levels of circRNAs | ddPCR is more accurate, provides absolute numbers, and measures low-abundance of circRNAs than RT-PCR | 22035192 |
| Northern blotting | Detect the abundances, sizes, isoforms, processing, and sequence of circRNAs. Can be employed to distinguish between a circRNA and its linear counterpart. The NB probes were labelled with the radioisotope 32P or non-radioactive probes labelled with digoxigenin. |
This method is not suitable for high-throughput screening. A short probes can be employed that span the splice junction. |
25921068 |
| RNA fluorescence in situ hybridization (RNA-FISH) | Provide a tool for circRNAs visualization, subcellular localization, and quantification. | Require the use of RNA-FISH probes targeting circRNA back-spliced junctions to avoid recognition of the linear RNA counterpart. RNA FISH combined with RNase R treatment to impair linear RNA signals. |
25664725 27050392 |
| Online circRNA resources and databases | Allow the researchers to access the online circRNA databases. Examples of online databases (See table 1B) |
||
| Overexpression of circRNAs | Gain of function analysis | This strategy used to design expression vectors for the ectopic expression of circRNAs and overexpress the brain-specific CDR1as and the testis-specific circSry | 29174924 26450910 28344080 |
| Depletion of circRNAs (knockdown) | Loss of function analysis Can be achieved by using circ siRNA/shRNA and CRISPR/Cas9-mediated genome editing The CircInteractome tool provides multiple options for circRNA siRNA |
Designed specifically to target the circRNA junction without affecting the linear mRNA counterpart | 21964070 18521077 25854182 27986464 |
Furthermore, several online circRNA resources and public databases have been developed to provide key bioinformatics knowledge about circRNAs (Table 2B) including exoRBase (http://www.exorbase.org/) which was mentioned earlier in this review. CircIntractome can be used to design divergent primers and siRNA (Dudekula et al., 2016). CircBase can provide information about circRNA identity and tissue specificity based on RNA-seq data and identify potential interacting factors such as miRNAs and RBPs (Memczak et al., 2013).
Table 2B.
Databases and online resources for circRNA research.
| Database | Website | Note | PMID |
|---|---|---|---|
| CircBase | http://www.circbase.org/ | Provides online data sets of circRNAs expression levels and sequence | 25234927 |
| CircInteractome | http://circinteractome.nia.nih.gov | Identification of RBPs and miRNAs binding sites on human circRNAs, design of divergent primers and siRNAs | 26669964 |
| circRNADb | http://reprod.njmu.edu.cn/circrnadb | Provides information on the circRNAs such as genomic information, genomic sequence, exon splicing, IRES, and open reading frames (ORFs) | 27725737 |
| exoRBase | http://www.exoRBase.org | Identify circRNAs in human blood exosomes. The first release of exoRBase contains 58,330 circRNAs | 30053265 |
| BBBomics | http://bioinformaticstools.mayo.edu/bbbomics/ | CircRNA database specific for blood-brain barrier | 26973449 |
| CSCD | http://gb.whu.edu.cn/CSCD | A database for cancer-specific circRNAs that provides information on circRNAs expression, interacting miRNAs and RBPs | 29036403 |
| TSCD | http://gb.whu.edu.cn/TSCD | Provides a comprehensive characterization of tissue-specific circRNAs in the human and mouse genomes | 27543790 |
| CIRCpedia | http://www.picb.ac.cn/rnomics/circpedia | CircRNAs expressed in different species, cell types, and tissues | 30172046 |
| CircR2Disease | http://bioinfo.snnu.edu.cn/CircR2Disease/ | Provides a comprehensive resource for circRNA deregulation in different diseases | 29741596 |
| TRcirc | http://www.licpathway.net/TRCirc | Provides a resource for transcriptional regulation information of circRNAs | 30184150 |
| CircView | http://gb.whu.edu.cn/CircView | Allow users to visualize circRNAs and view the regulatory elements, such as microRNA response elements and RNA-binding protein binding sites | 29106456 |
Since circRNAs have the ability to regulate gene expression, it exist as potential therapeutic tool for various diseases (Li, Yang, & Chen, 2018b). Currently, molecular tools for circRNA manipulation are under investigation. Furthermore, the manipulation of circRNA expression levels is another strategy to understand the impact of its expression levels in disease progression. CircRNA overexpression (gain of function) and knockdown (loss of function) approaches have been successfully used to study circRNAs. CircRNA gain of function approaches to overexpress circRNAs have been used to design expression vectors containing the circRNA sequence and delivered by viral vector systems such as plasmid transfection and AAV vector (Hansen et al., 2013; Meganck et al., 2018) which already used in clinical trials (Jessup et al., 2011). The limitation of this technique is the formation of concatemers if the RNA polymerase cannot recognize the transcription terminator site (TTS) (Barrett & Salzman, 2016). Therefore, northern blot is required to exclude concatemer expression. Non-viral strategies are also developed to produce exogenous circRNAs in vitro in eukaryotic cells (Hansen et al., 2013; Wesselhoeft, Kowalski, & Anderson, 2018).
In addition, circRNA loss-of-function experiment is usually used to knockdown the circRNAs expression. Small interfering RNA (siRNA) and small hairpin RNA (shRNA) designed specifically to target the circRNA junction (back-splice site) without affecting the linear mRNA counterpart are used to successfully knockdown circRNA expression (Chen et al., 2017b). However, the limitation of this technique is that the siRNA may target the linear mRNA or other circRNAs with the same junction sequence (Holdt et al., 2016; Wang, Long, et al., 2016a). Therefore, the negative effect of siRNA-mediated knockdown on the parental gene expression should always be examined and ruled out. Also, CRISPR-Cas9 genome editing technology can be used to delete intronic complementary regions and potentially generate circRNA knockouts (Abudayyeh et al., 2017; Cox et al., 2017). This tool was used to remove the circRNA CDR1as locus from the mouse genome to generate CDR1as-loss-of-function mutant mice (Piwecka et al., 2017). Moreover, the CRISPR/Cas9 genome editing was used to remove the intronic complement sequence (ICS) of the circGCN1L1-flanking introns to suppress its expression without affecting the linear mRNA (Zhang et al., 2016). In general, manipulation tools for circRNA are a considerable gain in uncover circRNA function in diseases, and they have perfect prospects for being progressing into circRNA-based therapeutic strategies in the future.
8. Exosomal circRNAs
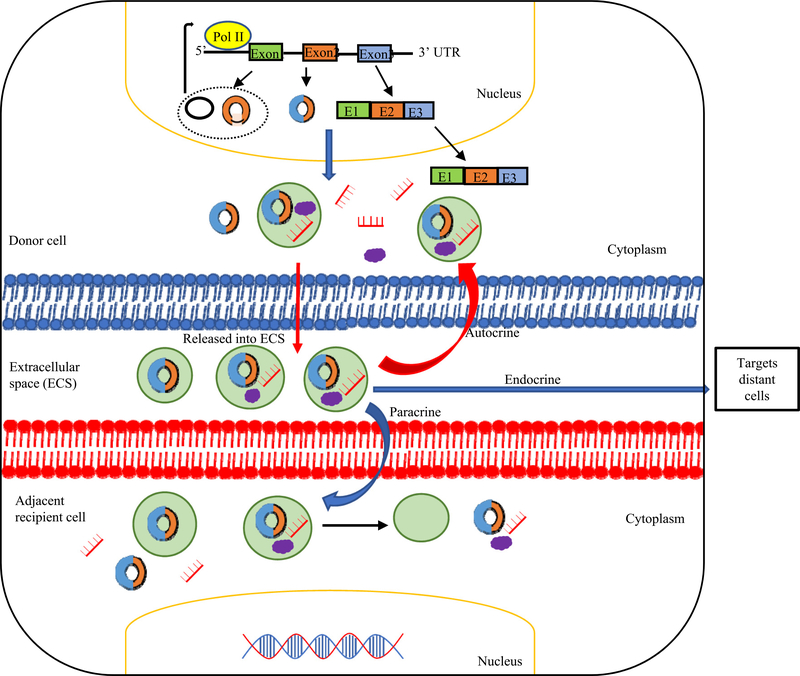
CircRNAs have a high degree of stability and resistance to exonuclease degradation; therefore, they may be accumulated in the cells if not controlled by cellular mechanism (Conn et al., 2015). One of the most important cellular mechanisms is the excretion of circRNAs from cells through extracellular vesicles such as exosomes. Exosomes are membrane-bound vesicles of endocytic origin secreted by most cell types (Xu et al., 2018; Yang et al., 2017b) and contain cellular components such as proteins, lipids, mRNAs, and miRNA (Valadi et al., 2007) and participate in cell-to-cell communication, and transfer genetic information (Bai et al., 2019). Recently, circRNAs have been detected in exosomes indicating that cells may use exosomes to transport circRNAs to communicate to other cells (Lasda & Parker, 2016). In 2015, Li et al. reported, for the first time, that exosomes contain abundant circRNAs (Lener et al., 2015). These circRNAs within the exosomes (referred to as exo-circRNAs) are stable and circulate in the serum after RNase R treatment. The presence of exo-circRNAs has been confirmed in a variety of cells. It has been reported that numerous exo-circRNAs could be released into extracellular space and detected in the blood, which is used as potential biomarkers in cancers. In addition, exo-circRNAs could enter the cell again (autocrine effect), taken by adjacent cell (paracrine effect), or enter a distant cell (endocrine effect) (Fig. 5) and release their content (Milman, Ginini, & Gil, 2019). However, the functions of exosomal circRNAs are not fully clear. By delivering circRNAs, exosomes play a key role in regulating signaling transduction between neighboring or distant cells and transfer biological activities to recipient cells (Li, Zheng, et al., 2015a; Théry, Zitvogel, & Amigorena, 2002).
Fig. 5.
Generation of exosomal circRNAs (exo-circRNAs). circRNAs can be loaded into exosomes, released by donor cells into extracellular space, and enter the cells again (Autocrine effect) or targets nearby cells (Paracrine effect) or targets a distant cells (Endocrine effect) through endocytosis, thus modulating gene expression in recipient cells. Some exo-circRNAs are not bind to miRNAs in exosomes, they are able to sponge specific in target cells leading to target gene activation; or exo-circRNAs are bind to miRNAs in exosome. After entering target cells, miRNAs are released and target genes can be silenced.
Furthermore, circRNAs have been reported to be able to bind to miRNAs, which are also shown to be abundant in exosomes (exomiRNAs) (Li, Zheng, et al., 2015a). CircHIPK3 has been reported to be transferred from cardiomyocytes into cardiac microvascular endothelial cells by exosomes and protect endothelial cells from oxidative damage and vascular dysfunction in vitro under oxidative stress conditions by sponging miR-29 (Wang et al., 2019b). Additionally, exosomal ncRNAs are involved in cardiovascular diseases and their potential use as disease biomarkers (Jaquenod De Giusti, Santalla, & Das, 2019). However, the roles of exo-circRNAs in accelerating and propagating cardiovascular and metabolic inflammation still require further study.
Currently, a database named exoRBase (http://www.exorbase.org/) is mainly a collection of all long RNA species derived from RNA-seq data analyses of human blood exosomes. It studied the RNA expression profiles in normal individuals and patients with different diseases. The first release of exoRBase contains 58,330 circRNAs, 15,501 lncRNAs and 18,333 mRNAs from 87 blood exosomal RNA-seq datasets. Compared to healthy individuals, patients with coronary heart disease (CHD) significantly upregulated 2,425 exo-circRNAs and downregulated 485 exo-circRNAs (Li, Li, et al., 2018a). These finding indicated that exo-circRNAs play a role in the development of the disease and can be used as disease biomarkers.
9. CircRNAs as potential diagnostic biomarkers
In the early stages of a disease, the characterization of biomarkers is considered a very promising strategy in the diagnosis and prevention of the disease. Since circRNAs are highly abundant, stable, and have tissue-specific expression, previous reports highlighted their potential as disease biomarkers. In addition to this, it has been shown that circRNAs are packaged and released in exosomes that provide additional protection to it, and increasing amounts were found in exosomes when compared to the cells (Li, Zheng, et al., 2015a). Previous studies have demonstrated that the circRNAs hsa_circ_002059 and hsa_circ_104916 were dramatically downregulated in the plasma and gastric cancer tissues and significantly correlated with tumor metastasis, age, and sex, suggesting the potential of these circRNAs as a stable diagnostic biomarker for gastric cancer (Li et al., 2015d; Li et al., 2017b). Additionally, circ-ITCH can be used to diagnose esophageal cancer (Li, Zhang, et al., 2015c), whereas hsa_circ_0005075 can be used as a potential biomarker of hepatocellular carcinoma (Qin et al., 2016). Although several circRNAs have been examined and suggested to exert important functions in the pathological process of various cardiovascular diseases (CVDs) (Fan et al., 2017), however, very few circRNA biomarker candidates for CVDs have been reported. The expression of circRNAs was investigated in peripheral blood of patients with coronary heart disease (CHD) and results suggested that circRNA hsa_circ_0124644 could be used as a diagnostic biomarker of CHD (Memczak et al., 2015; Zhao et al., 2017b). Another study reported that circulating circRNA hsa_circ_0021001 was decreased in peripheral blood of patients with intracranial aneurysms (IA), and was considered to be a potential diagnostic marker for IA (Teng et al., 2017). Furthermore, bioinformatics analysis of peripheral blood circRNA expression profiles in hypertensive patients identified circRNA hsa-circ-0005870 as a novel biomarker for diagnosis of hypertension (Wu, Jin, & Cai, 2017). Another study identified circRNA MICRA as a novel prognostic biomarker for patients with heart failure after myocardial infarction (Salgado-Somoza, Zhang, Vausort, & Devaux, 2017). CircRNAs can also be used as diagnostic biomarkers in central nervous system (CNS). These circRNAs can cross the blood-brain barrier (BBB), enter into the blood and cerebrospinal fluid (CSF), and thus be used as novel biomarkers (Lu & Xu, 2016).
10. Regulation of oxidative stress and reactive oxygen species (ROS) production by circRNA
Oxidative stress is characterized by an imbalance between oxidants and antioxidants and a disruption of redox signaling (Sies, 2015). It is generally accepted that oxidative stress can lead to cell and tissue injury having a fundamental role in vascular dysfunction and inflammation (Siti, Kamisah, & Kamsiah, 2015; Steven et al., 2019). Under physiological conditions, ROS control vascular function by modulating various redox-sensitive signaling pathways. However, in vascular disorders, oxidative stress triggers endothelial dysfunction and inflammation, affecting several cells in the vascular wall including endothelial cells and vascular smooth muscle cells (Steven et al., 2019; Sun et al., 2020). Recently, we reported that mitochondrial ROS, uncoupled from ATP synthesis, determine endothelial cell activation status for both physiological recruitment of patrolling cells (at low ROS levels) and pathological recruitment of inflammatory cells (at high ROS levels) (Cheng et al., 2017; Li et al., 2013; Li et al., 2016b; Li et al., 2017c; Nanayakkara, Wang, & Yang, 2019). Several studies demonstrated that several circRNAs are regulated by oxidative stress and mediate ROS production as well as promote ROS induced cellular death, cell apoptosis, and inflammation (Cheng, Cao, Xue, Xia, & Xu, 2019; Li et al., 2020a; Liang et al., 2020). A previous study showed that sodium/calcium exchanger 1 (ncx1) circular RNA (circNCX1) was upregulated in response to ROS (H2O2) in cardiomyocytes by acting as a miR-133a-3p sponge to release the pro-apoptotic gene cell death inducing p53 target 1 (CDIP1) and promote cardiomyocyte apoptosis (Li et al., 2018c). However, H2O2 stimulation resulted in the downregulation of circHIPK3 expression in human osteoblast whereas circHIPK3 overexpression alleviated H2O2-induced cell viability reduction, cell death and apoptosis (Liang et al., 2020). Furthermore, autophagy-related circular RNA (circACR) is downregulated by high glucose (HG) irritation, and its overexpression attenuated HG-aroused SchwannRSC96 cell apoptosis, autophagy, and ROS generation (Liu, Chen, Yao, & Kang, 2019) (Table 3). Interestingly, circHIPK3 can be transferred by exosomes released from hypoxia-pretreated cardiomyocytes into cardiac microvascular endothelial cells, and protect endothelial cells from oxidative damage and vascular dysfunction in vitro under oxidative stress conditions by sponging miR-29a (Wang, Zhao, et al., 2019b). These studies indicated that circRNAs play a critical role in vascular pathology especially in disease conditions wherein oxidative stress plays a crucial role, such as endothelial dysfunction, vascular inflammation, diabetes mellitus, and atherosclerosis (Fuschi, Maimone, Gaetano, & Martelli, 2019).
Table 3.
Circular RNAs regulates oxidative stress and reactive oxygen species (ROS). ROS including hydrogen peroxide (H2O2) has been shown to upregulate or downregulate circRNAs expression which either promote or attenuate the effect of ROS on the cell viability and apoptosis. Also, high glucose has been shown to affect circRNA expression and then regulate ROS production by acting as miRNAs sponge. Abbreviations: H2O2: Hydrogen peroxide; HG: High glucose; ROS: Reactive oxygen species; CircNCX1: Sodium/calcium exchanger 1 (ncx1) circular RNA; CircPRKCI: Protein kinase C iota type (PRKCI) circular RNA; CircHIPK3: Homeodomain-interacting protein kinase 3 (HIPK3) circular RNA; CircLRP6: Low-density lipoprotein receptor-related protein 6 (LRP6) circular RNA; CircACR: Autophagy-related circular RNA (ACR); HMGB1: High mobility group box 1; TLR4: Toll like receptor 4; NF-κB: Nuclear factor kappa-light-chain-enhancer of activated B cells; PI3K: Phosphoinositide 3-kinase; AKT: Protein kinase B; mTOR: mammalian target of rapamycin
| Stimulus | CircRNA | Expression of circRNAs | Function | Target | PMID |
|---|---|---|---|---|---|
| H2O2 | CircNCX1 | Upregulated by H2O2 | Promote cardiomyocyte apoptosis by acting as an endogenous miR-133a-3p sponge and suppress the activity of pro-apoptotic gene cell death-inducing protein (CDIP1) | miR-133a-3p | 30613267 |
| H2O2 | CircRNA-4099 | Upregulated by H2O2 | Overexpression of circRNA-4099 aggravated H2O2-induced cell apoptosis, ROS production and cell fibrosis | miR-706 and trigger keap1/Nrf2 and p38MAPK | 31479678 |
| H2O2 | CircPRKCI | Downregulated by H2O2 | Overexpression of circPRKC1 attenuated H2O2-induced neuronal cell death and apoptosis | miR-545 and miR-589 | 31053300 |
| H2O2 | CircHIPK3 | Downregulated by H2O2 | Overexpression of circHIPK3 in human osteoblasts alleviated H2O2-induced viability reduction, cell death and apoptosis | miR-124 | 31955154 |
| HG | CircLRP6 | Upregulated by HG | Regulate HG-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells by upregulating HMGB1 and activating TLR4/NF-κB signaling | miR-205 | 31087368 |
| HG | CircACR | Downregulated by HG | Overexpression of circACR relieved HG-aroused SchwannRSC96 cell apoptosis, autophagy and ROS generation | miR-145–3p and inhibit PI3K/AKT/mTOR pathway | 31886589 |
11. Association of innate immune response with circRNA
The activation of the innate immune system results in enhanced responsiveness to subsequent triggers, which is termed trained immunity or innate immune memory (Netea & van der Meer, 2017). Not only the innate immune cells such as (monocytes, macrophages, and NK cells), but also traditional non-immune cells such as endothelial cells and smooth muscle cells have this memory function (Hamada, Torre, Drancourt, & Ghigo, 2018; Shao et al., 2020a). As discussed above, we recently proposed a new working model that endothelial cells are conditional innate immune cells (Wang, Zhao, et al., 2019b). Challenged cells with exogenous or endogenous insults results in metabolic reprogramming and epigenetic modifications (Lu et al., 2019), which is phenomenon seen in trained immunity of innate immune cells. Furthermore, when cells are exposed to a subsequent non-specific immune stimulus, they respond more strongly (secondary response) than to the primary insult (Netea, Quintin, & van der Meer, 2011; Zhong, Yang, Feng, & Yu, 2020). The metabolic reprogramming in trained immunity is characterized by increased glycolysis, increased acetyl-CoA generation, and increased cholesterol (mevalonate) pathway synthesis (Penkov, Mitroulis, Hajishengallis, & Chavakis, 2019). Initiation of innate immune memory through trained immunity is the likely mechanism behind the non-specific protective effects of certain vaccines (Benn, Netea, Selin, & Aaby, 2013), while increased inflammatory responsiveness of the cells due to trained immunity could also play a central role in inflammatory diseases (Bekkering, Joosten, van der Meer, Netea, & Riksen, 2013). We recently reported that pro-atherogenic lysophosphatidylcholine (LPC) activates human aortic endothelial cells (HAECs) (Li et al., 2018d; Li et al., 2018e; Li, Fang, et al., 2016b) and upregulates trained immunity pathways (TIPs) via analyzing our RNA-Seq data and histone 3 lysine 14 acetylation (H3K14ac)-chromatin immunoprecipitation (CHIP)-Seq data, both performed on HAEC treated with LPC (Lu et al., 2019). CircRNAs have been shown to play a critical role in glycolysis through sponging miRNAs, which represses the expression of several enzymes and transporters of glycolysis pathway (Yu et al., 2019). Some experimental studies reported that circMAT2B (Li et al., 2019b) and circ-PRMT5 (Ding, Guo, Deng, & Li, 2020) promote glycolysis, while knockdown of circAKT3 (Xu, Jiang, Wu, & Zhang, 2020), circCUX1 (Li et al., 2019c), and circDENND4C (Ren et al., 2019) inhibits glycolysis in cancer cells.
Microbial lipopolysaccharides (LPS) induce Toll-like receptor (TLR) pathways and NF-κ activation leading to modulation of genes involved in innate and adaptive immunity (Rosadini & Kagan, 2017). A previous study reported that the activation of TLR in mouse macrophages regulate the expression of circRasGEF1B. Additionally, circRasGEF1B could modulate the expression of intercellular adhesion molecule 1 (ICAM-1) as part of the LPS response, and that knockdown of this circRNA leads to a reduction in ICAM-1expression levels in vitro (Ng et al., 2016). Furthermore, innate immune response and trained immunity contribute to the development of atherosclerotic cardiovascular diseases (Hansson & Hermansson, 2011).
12. Regulation of vascular inflammation by circRNA
Endothelial dysfunction (Etwebi, Landesberg, Preston, Eguchi, & Scalia, 2018; He et al., 2016; Shao et al., 2014) and vascular inflammation are associated with several pathological processes such as inflammatory and ischemic cardiovascular disease as well as metabolic disorders (Carmeliet, 2003; Puro, Kohmoto, Fujita, Gardner, & Padovani-Claudio, 2016).We recently proposed a new working model that endothelial cells are conditional innate immune cells (Wang, Zhao, et al., 2019b), which are equipped with receptors including Toll-like receptors and inflammasome/caspase-1 (Li et al., 2016c; Li et al., 2017d; Shen et al., 2010; Wang et al., 2016b; Yang, Yin, & Wang, 2008; Yin et al., 2013) for danger/pathogen-associated molecular patterns (DAMPs/PAMPs) for sensing various DAMPs and conditional DAMPs such as pro-atherogenic lysophospholipids (Li et al., 2016d; Shao et al., 2017; Wang et al., 2016c), hyperlipidemia (Yin et al., 2015), hypoxia (Fu et al., 2017), uremic toxins (Ferrer et al., 2016; Monroy et al., 2015; Sun et al., 2018), LPS (Sha et al., 2015; Virtue et al., 2017), pro-inflammatory cytokines such as interleukin-7 (IL-17) (Mai et al., 2016) and anti-inflammatory cytokines including interleukin-35 (IL-35) (Li et al., 2012; Li, Fang, Yang, Wang, & Yang, 2017; Li, Shao, et al., 2018e) and interleukin-10 (IL-10) (Li et al., 2020b) as well as having innate immune memory (trained immunity) functions (Lu et al., 2019; Shao et al., 2020b; Zhong et al., 2020). Thus, targeting vascular endothelial dysfunction and inflammation has enormous therapeutic potential in the prevention and treatment of vascular complications. Several lines of evidence indicate that circRNAs are expressed in different cardiovascular diseases (Holdt et al., 2016; Wang, Long, et al., 2016a; Zheng et al., 2016) that are usually associated with endothelial dysfunction, vascular smooth muscle cell proliferation and migration, and vascular inflammation (Eelen, de Zeeuw, Simons, & Carmeliet, 2015; Sena, Pereira, & Seiça, 1832).
CircRNAs participate in cardiovascular disease via miRNA sponging and thus regulating their target genes to maintain homeostasis (Su & Lv, 2020). Some of these circRNAs (pro-inflammatory circRNAs) have been shown to promote endothelial dysfunction, vascular inflammation, and cardiovascular disease (Table 4A). Of note, circRNA from lncRNA ANRIL (antisense ncRNA in the INK4 locus) was positively correlated with INK4/ARF (Cdkn2a–Cdkn2b genes encoding three potent tumor suppressors, namely p16Ink4a, p19Arf and p15Ink4b) expression and atherosclerosis risk (Burd et al., 2010). Circ-Sirt1 binds to miR-132/212 and controls NF-kappaB activation to mediate inflammatory phenotypic switching of vascular smooth muscle cells (VSMCs) which play a fundamental role in neointimal formation and atherosclerosis (Kong et al., 2019). Also, circRNA CDR1as could regulate the miR-7 on its target gene expression and promote myocardial infarction via promoting cell apoptosis (Geng et al., 2016). In 2017, Du et al. discovered that circRNA circ-Foxo3 promotes cellular senescence and cardiac cell death (Du et al., 2017). Meanwhile, other circRNAs (anti-inflammatory circRNAs) have been shown to attenuate endothelial dysfunction and vascular inflammation (Table 4B). For instance, circRNA circ_0003204 was significantly upregulated in oxLDL-activated endothelial cells, and its silencing promoted endothelial cell proliferation and angiogenesis (Li, Ma, & Yu, 2017f). However, more pro-inflammatory circRNAs associated with cardiovascular diseases have been identified and characterized than the anti-inflammatory circRNAs. In general, circRNAs are potentially involved in vascular dysfunction/inflammation; however, we recently reported that pro-atherogenic LPC induced circRNAs may contribute to homeostasis in LPS-induced HAEC activation (Li, Sun, et al., 2019a). Of note, the relevance of circRNAs to vascular inflammation and its molecular mechanism in cardiovascular diseases remains poorly characterized, and a better understanding of circRNA involvement in vascular-related inflammation and cardiovascular diseases will form a basis for the development of these circRNAs as biomarkers in the cardiovascular system. Additionally, a better understanding of circRNA may lead to discovery and the development of therapeutic agents as well as better patient prognosis.
Table 4A.
Summary of identified Pro-inflammatory circRNAs. Different circRNAs that promotes vascular inflammation and cardiovascular diseases. Abbreviations: AS: Atherosclerosis; AIS: Acute ischemic stroke; CHD: coronary heart disease; HUVECs: human umbilical vein endothelial cells; VEC: vascular endothelial cells; VSMCs: vascular smooth muscle cells; Mø: macrophages; PBMC: peripheral blood mononuclear cells; Ox-LDL: oxidized low- density lipoprotein; LPS: lipopolysaccharides; Circ-RELL1: circular RNA-receptor expressed in lymphoid tissues-like protein 1 (RELL1); CircANRIL: circular antisense non-coding RNA at the INK4 locus (cANRIL); CircRNA HECTD1: circular RNA HECT domain E3 ubiquitin protein ligase 1.
| CircRNAsUp | Expression of circRNAs | Cells | Function | Mechanism/target | Pathways | PMID |
|---|---|---|---|---|---|---|
| circ-RELL1 | Upregulated by ox-LDL | HUVECs | Increased endothelial inflammation in ox-LDL-stimulated HUVECs | miR-6873–3p | miR-6873–3p/ MyD88/NF-κB pathway | 32113679 |
| RNA-ZNF609 | Upregulated in high glucose conditions, hypoxia stress and in diabetic retina | Retinal ECs | Aggravated oxidative stress/hypoxic stress-induced HUVEC apoptosis increased endothelial dysfunction | miR-615–5p | cZNF609/miR-615–5p/MEF2A | 28824721 |
| hsa_circ_0068087 | Upregulated in high glucose conditions | HUVECs | Accelerate TLR4/NF-κB/NLRP3 inflammasome-mediated inflammation and EC dysfunction in the high glucose condition | miR-197 | TLR4/NF-κB/NLRP3 inflammasome | 31108165 |
| CircHIPK3 | Upregulated in diabetic retina | Retinal VECs | Increased retinal VECs dysfunction/inflammation | miR-30a-3p | circHIPK3-miR-30a-3p-VEGFC/WNT2/FZD4 pathway | 28860123 |
| CircANRIL | Overexpression in AS model | VECs | Increased VECs inflammation and AS | - | - | 28683453 |
| Circ_0003575 | Upregulated by ox-LDL | HUVECs | Attenuated EC proliferation and angiogenesis | - | - | 28946214 |
| CircRNA-0044073 | Overexpression in AS | VSMCs/VECs/blood cells | Promoted the proliferation of VSMCs and HUVECs Promotes inflammation in AS |
miR-107 | JAK/STAT signaling pathway | 30864721 |
| Circ_CHFR | Upregulated by ox-LDL | VSMCs, Plasma | Promoted the ox-LDL-induced cell proliferation, migration, and inflammation | miR-214–3p | Wnt3/β-catenin signal pathway | 32271446 |
| CircWDR77 | Upregulated in high glucose conditions | VSMCs | Induced VSMCs proliferation and migration | miR-124 | CircWDR77-miR-124-FGF2 pathway | 29042195 |
| Circ_Lrp6 | AS | VSMCs | Induced VSMCs proliferation and migration, and neointima hyperplasia; proatherogenic | miR-145 | - | 30582454, 30763218 |
| hsa_circ_0029589 (circCHFR) | Upregulated by ox-LDL and in AS | VSMCs | Promotes VSMCs migration and proliferation | miR-370 | miR-370/FOXO1/ Cyclin D1 pathway | 31048182 |
Table 4B.
Anti-inflammatory circRNAs. Different circRNAs that attenuates vascular inflammation and cardiovascular diseases. Abbreviation: HUVEC: human umbilical vein endothelial cells; AS: atherosclerosis; VSMCs: vascular smooth muscle cells; Mø: macrophages; HAECs: human aortic endothelial cells; Ox-LDL: oxidized low-density lipoprotein; TNF-α: tumor necrosis factor-alpha;
| CircRNA name | Expression of circRNAs | Cells/tissue | Function | Mechanism/Target | Pathways | PMID |
|---|---|---|---|---|---|---|
| hsa_circRNA-00 54633 | Upregulated in high glucose conditions | HUVEC | Decreased the high glucose-induced endothelial cell dysfunction, including proliferation, migration and promote angiopoiesis | microRNA-218 | microRNA-218/heme oxygenase-1 axes | 29693114 |
| CircANRIL | Downregulated in AS | VSMCs/Mø | Atheroprotection against human AS | Binds to PES1, thereby impairing pre-rRNA processing and ribosome biogenesis | - | 27539542 |
| Circ_0003204 | Upregulated by ox-LDL and in cerebral AS | HAECs | Inhibits proliferation, migration and tube formation of ECs in AS | miR-370 | miR-370–3p/TGFβR2/phosph-SMAD3 pathway | 31900142 |
| Circ-Sirt1 | Downregulated by TNF-α stimulation and in neointima hyperplasia | VSMCs | Inhibits inflammatory phenotypic switching of VSMCs | miR-132/212 | circ-Sirt1/miR-132/212/NF-κB pathway | 30820544 |
13. Conclusion
CircRNAs are now a noteworthy area in the field of RNA. Several studies have demonstrated that circRNAs are an abundant, diverse, stable, and conserved class of RNA molecules, representing a new type of regulatory ncRNA. Nevertheless, their regulation and biological roles are not yet clearly understood, as well as their degradation, localization, and their involvement in disease pathogenesis. Recent studies have demonstrated that circRNAs can act as miRNAs sponges, RBP sponges, regulate transcription, or affect gene expression, and a growing body of evidence suggests that there might be other functions remaining to be revealed. Recently, a circRNA database has been constructed to provide tissue-specific circRNA expression profiles and circRNA-miRNA-gene regulatory networks. Considering the studies mentioned above, circRNAs might be useful therapeutic agents. Controlling the expression of circRNAs in specific tissues and cells of the body might yield greatly reduced endothelial dysfunction and vascular inflammation associated with cardiovascular and metabolic diseases. The studies mentioned above provide novel insights on the roles of circRNAs in regulating ROS, vascular inflammation associated with cardiovascular and metabolic diseases.
Acknowledgments
FS carried out the primary literature search and drafted the manuscript. Others provided material input and helped revising the manuscript. XY supervised the study, data analysis, and manuscript writing. All authors read and approved the final manuscript.
Source of Funding
Our research activities are supported by grants from the National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (XY and HW). The content in this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
References
- Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. (2017). Identification of hur target circular rnas uncovers suppression of pabpn1 translation by circpabpn1. RNA Biology 14, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. (2017). Rna targeting with crispr-cas13. Nature. 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. (2014). Circrna biogenesis competes with pre-mrna splicing. Molecular Cell 56, 55–66. [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, & Maas S (2004). Widespread a-to-i rna editing of alu-containing mrnas in the human transcriptome. PLoS Biology 2, e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. (2015). Correlation of circular rna abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific Reports 5, 8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Lei K, Huang F, Jiang Z, & Zhou X (2019). Exo-circrnas: A new paradigm for anticancer therapy. Molecular Cancer 18, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, et al. (2018). Circular rna dlgap4 ameliorates ischemic stroke outcomes by targeting mir-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 38, 32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Parker KR, Horn C, Mata M, & Salzman J (2017). Cirs-7 exonic sequence is embedded in a long non-coding rna locus. PLoS Genetics 13, e1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, & Salzman J (2016). Circular rnas: Analysis, expression and potential functions. Development (Cambridge, England) 143, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, & Salzman J (2015). Circular rna biogenesis can proceed through an exon-containing lariat precursor. eLife. 4, e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004). Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009). Micrornas: Target recognition and regulatory functions. Cell. 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S, Joosten LA, van der Meer JW, Netea MG, & Riksen NP (2013). Trained innate immunity and atherosclerosis. Current Opinion in Lipidology 24, 487–492. [DOI] [PubMed] [Google Scholar]
- Benn CS, Netea MG, Selin LK, & Aaby P (2013). A small jab - a big effect: Nonspecific immunomodulation by vaccines. Trends in Immunology 34, 431–439. [DOI] [PubMed] [Google Scholar]
- Brinegar AE, & Cooper TA (1647). Roles for rna-binding proteins in development and disease. Brain Research 2016, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, & Sabeti PC (2015). Strand-specific rna sequencing in plasmodium falciparum malaria identifies developmentally regulated long non-coding rna and circular rna. BMC Genomics 16, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, & Sharpless NE (2010). Expression of linear and novel circular forms of an ink4/arf-associated non-coding rna correlates with atherosclerosis risk. PLoS Genetics 6, e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P (2003). Angiogenesis in health and disease. Nature Medicine 9, 653–660. [DOI] [PubMed] [Google Scholar]
- Chen CY, & Sarnow P (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular rnas. Science (New York, N.Y.) 268, 415–417. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. (2017b). Circular rna profile identifies circpvt1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Letters 388, 208–219. [DOI] [PubMed] [Google Scholar]
- Chen L, Huang C, Wang X, & Shan G (2015a). Circular rnas in eukaryotic cells. Current Genomics 16, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, et al. (2017a). Circrna_100290 plays a role in oral cancer by functioning as a sponge of the mir-29 family. Oncogene. 36, 4551–4561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen LL (2016). The biogenesis and emerging roles of circular rnas. Nature reviews. Molecular and Cellular Biology 17, 205–211. [DOI] [PubMed] [Google Scholar]
- Chen LL, & Yang L (2015). Regulation of circrna biogenesis. RNA Biology 12, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Xiang JF, Zhu S, Chen S, Yin QF, Zhang XO, et al. (2015b). Adar1 is required for differentiation and neural induction by regulating microrna processing in a catalytically independent manner. Cell Research 25, 459–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Nanayakkara G, Shao Y, Cueto R, Wang L, Yang WY, et al. (2017). Mitochondrial proton leak plays a critical role in pathogenesis of cardiovascular diseases. Advances in Experimental Medicine and Biology 982, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Cao X, Xue L, Xia L, & Xu Y (2019). Circprkci-mir-545/589-e2f7 axis dysregulation mediates hydrogen peroxide-induced neuronal cell injury. Biochemical and Biophysical Research Communications 514, 428–435. [DOI] [PubMed] [Google Scholar]
- Choi H, & Lee DS (2016). Illuminating the physiology of extracellular vesicles. Stem Cell Research & Therapy 7, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. (2015). The rna binding protein quaking regulates formation of circrnas. Cell. 160, 1125–1134. [DOI] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, et al. (2017). A circrna from sepallata3 regulates splicing of its cognate mrna through r-loop formation. Nature plants. 3, 17053. [DOI] [PubMed] [Google Scholar]
- Cortés-López M, Gruner MR, Cooper DA, Gruner HN, Voda AI, van der Linden AM, et al. (2018). Global accumulation of circrnas during aging in caenorhabditis elegans. BMC Genomics 19, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, et al. (2017). Rna editing with crispr-cas13. Science (New York, N.Y.) 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan M, Schwartz S, Edelheit S, & Sorek R (2012). Transcriptome-wide discovery of circular rnas in archaea. Nucleic Acids Research 40, 3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Guo L, Deng Z, & Li P (2020). Circ-prmt5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting mir-188–5p/hk2 axis. Annals of Hepatology 19, 269–279. [DOI] [PubMed] [Google Scholar]
- Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. (2016). Circular rnas are down-regulated in kras mutant colon cancer cells and can be transferred to exosomes. Scientific Reports 6, 37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. (2017). Foxo3 circular rna promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European Heart Journal 38, 1402–1412. [DOI] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, & Yang BB (2016). Foxo3 circular rna retards cell cycle progression via forming ternary complexes with p21 and cdk2. Nucleic Acids Research 44, 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, & Gorospe M (2016). Circinteractome: A web tool for exploring circular rnas and their interacting proteins and micrornas. RNA Biology 13, 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen KK, Hansen TB, & Kjems J (2017). Insights into circular rna biology. RNA Biology 14, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eelen G, de Zeeuw P, Simons M, & Carmeliet P (2015). Endothelial cell metabolism in normal and diseased vasculature. Circulation Research 116, 1231–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. (2017). Fus affects circular rna expression in murine embryonic stem cell-derived motor neurons. Nature Communications 8, 14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M (2011). Non-coding rnas in human disease. Nature Reviews. Genetics 12, 861–874. [DOI] [PubMed] [Google Scholar]
- Etwebi Z, Landesberg G, Preston K, Eguchi S, & Scalia R (2018). Mechanistic role of the calcium-dependent protease calpain in the endothelial dysfunction induced by mpo (myeloperoxidase). Hypertension (Dallas, Tex. : 1979) 71, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Weng X, Zhao Y, Chen W, Gan T, & Xu D (2017). Circular rnas in cardiovascular disease: An overview. BioMed Research International 2017, 5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, et al. (2015). Single-cell rna-seq transcriptome analysis of linear and circular rnas in mouse preimplantation embryos. Genome Biology 16, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer LM, Monroy AM, Lopez-Pastrana J, Nanayakkara G, Cueto R, Li YF, et al. (2016). Caspase-1 plays a critical role in accelerating chronic kidney disease-promoted neointimal hyperplasia in the carotid artery. Journal of Cardiovascular Translational Research 9, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JW, & Leung AK (2017). Circrnas: A regulator of cellular stress. Critical Reviews in Biochemistry and Molecular Biology 52, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Vadalia N, Xue ER, Johnson C, Wang L, Yang WY, et al. (2017). Thrombus leukocytes exhibit more endothelial cell-specific angiogenic markers than peripheral blood leukocytes do in acute coronary syndrome patients, suggesting a possibility of trans-differentiation: A comprehensive database mining study. Journal of Hematology & Oncology 10, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuschi P, Maimone B, Gaetano C, & Martelli F (2019). Noncoding rnas in the vascular system response to oxidative stress. Antioxidants & Redox Signaling 30, 992–1010. [DOI] [PubMed] [Google Scholar]
- Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. (2016). The circular rna cdr1as promotes myocardial infarction by mediating the regulation of mir-7a on its target genes expression. PLoS One 11, e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Jiang J, & Wu C (2018). Function and clinical significance of circrnas in solid tumors. Journal of Hematology & Oncology 11, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, & Bartel DP (2014). Expanded identification and characterization of mammalian circular rnas. Genome Biology 15, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada A, Torre C, Drancourt M, & Ghigo E (2018). Trained immunity carried by non-immune cells. Frontiers in Microbiology 9, 3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. (2013). Natural rna circles function as efficient microrna sponges. Nature. 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. (2011). Mirna-dependent gene silencing involving ago2-mediated cleavage of a circular antisense rna. The EMBO journal. 30, 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson GK, & Hermansson A (2011). The immune system in atherosclerosis. Nature Immunology 12, 204–212. [DOI] [PubMed] [Google Scholar]
- He H, Guo F, Li Y, Saaoud F, Kimmis BD, Sandhu J, et al. (2016). Adiporedoxin suppresses endothelial activation via inhibiting mapk and nf-κb signaling. Scientific Reports 6, 38975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. (2011). High-throughput droplet digital pcr system for absolute quantitation of DNA copy number. Analytical Chemistry 83, 8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. (2016). Circular non-coding rna anril modulates ribosomal rna maturation and atherosclerosis in humans. Nature Communications 7, 12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach S, & Kretz M (2016). Non-coding rnas: Classification, biology and functioning. Adv. Exp. Med. Biol 937, 3–17. [DOI] [PubMed] [Google Scholar]
- Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, & Abu N (2019). Extracellular vesicle-derived circular rnas confers chemoresistance in colorectal cancer. Scientific Reports 9, 16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, & Coca-Prados M (1979). Electron microscopic evidence for the circular form of rna in the cytoplasm of eukaryotic cells. Nature. 280, 339–340. [DOI] [PubMed] [Google Scholar]
- Huang A, Zheng H, Wu Z, Chen M, & Huang Y (2020). Circular rna-protein interactions: Functions, mechanisms, and identification. Theranostics 10, 3503–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, & van Oudenaarden A (2011). Validating transcripts with probes and imaging technology. Nature Methods 8, S12–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. (2015). Analysis of intron sequences reveals hallmarks of circular rna biogenesis in animals. Cell Reports 10, 170–177. [DOI] [PubMed] [Google Scholar]
- Jaquenod De Giusti C, Santalla M, & Das S (2019). Exosomal non-coding rnas (exoncrnas) in cardiovascular health. Journal of Molecular and Cellular Cardiology 137, 143–151. [DOI] [PubMed] [Google Scholar]
- Jeck WR, & Sharpless NE (2014). Detecting and characterizing circular rnas. Nature Biotechnology 32, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. (2013). Circular rnas are abundant, conserved, and associated with alu repeats. RNA (New York, N.Y.) 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, et al. (2011). Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (cupid): A phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum ca2 +-atpase in patients with advanced heart failure. Circulation 124, 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drummer C, Virtue A, Gao T, Wu S, Hernandez M, et al. (2018). Increased expression of resistin in microrna-155-deficient white adipose tissues may be a possible driver of metabolically healthy obesity transition to classical obesity. Frontiers in Physiology 9, 1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong P, Yu Y, Wang L, Dou YQ, Zhang XH, Cui Y, et al. (2019). Circ-sirt1 controls nf-κb activation via sequence-specific interaction and enhancement of sirt1 expression by binding to mir-132/212 in vascular smooth muscle cells. Nucleic Acids Research 47, 3580–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A, Dijkema R, Arnberg AC, van derMeide PH, & Schellekens H (1986). The hepatitis delta (delta) virus possesses a circular rna. Nature. 323, 558–560. [DOI] [PubMed] [Google Scholar]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, & Kjems J (2019). The biogenesis, biology and characterization of circular rnas. Nature Reviews. Genetics 20, 675–691. [DOI] [PubMed] [Google Scholar]
- Lasda E, & Parker R (2016). Circular rnas co-precipitate with extracellular vesicles: A possible mechanism for circrna clearance. PLoS One 11, e0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. (2017). Circ-znf609 is a circular rna that can be translated and functions in myogenesis. Molecular Cell 66 (22–37.e29). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener T, Gimona M, Aigner L, Börger V, Buzas E, Camussi G, et al. (2015). Applying extracellular vesicles based therapeutics in clinical trials - an isev position paper. Journal of Extracellular Vesicles 4, 30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Sun Y, Drummer Ct LY, Yu D, Zhou Y, et al. (2019a). Increasing upstream chromatin long-range interactions may favor induction of circular rnas in lysopc-activated human aortic endothelial cells. Frontiers in Physiology 10, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Ma L, & Yu B (2017f). Circular rna hsa_circ_0003575 regulates oxldl induced vascular endothelial cells proliferation and angiogenesis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 95, 1514–1519. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. (2015c). Circular rna itch has inhibitory effect on escc by suppressing the wnt/β-catenin pathway. Oncotarget. 6, 6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hao X, Wang H, Liu Z, He Y, Pu M, et al. (2016a). Circular rna expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology 40, 1334–1344. [DOI] [PubMed] [Google Scholar]
- Li H, Yang F, Hu A, Wang X, Fang E, Chen Y, et al. (2019c). Therapeutic targeting of circ-cux1/ewsr1/maz axis inhibits glycolysis and neuroblastoma progression. EMBO Molecular Medicine 11, e10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, et al. (2017b). Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. OncoTargets Therapy 10, 3521–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. (2018c). A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting mir-133a-3p. Theranostics 8, 5855–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. (2015d). Using circular rna as a novel type of biomarker in the screening of gastric cancer. Clinica Chimica Acta 444, 132–136. [DOI] [PubMed] [Google Scholar]
- Li Q, Pan X, Zhu D, Deng Z, Jiang R, &Wang X (2019b). Circular rnamat2b promotes glycolysis and malignancy of hepatocellular carcinoma through the mir-338–3p/pkm2 axis under hypoxic stress. Hepatology (Baltimore, Md.) 70, 1298–1316. [DOI] [PubMed] [Google Scholar]
- Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. (2018a). Exorbase: A database of circrna, lncrna and mrna in human blood exosomes. Nucleic Acids Research 46, D106–d112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Li Y, Kuo YM, Andrews AJ, Nanayakkara G, et al. (2016b). Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arteriosclerosis, Thrombosis, and Vascular Biology 36, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Mai J, Choi ET, Wang H, & Yang XF (2013). Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. Journal of Hematology & Oncology 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Sun Y, Shao Y, Yang WY, Jiang X, et al. (2020b). Anti-inflammatory cytokines il-35 and il-10 block atherogenic lysophosphatidylcholine-induced, mitochondrial ros-mediated innate immune activation, but spare innate immune memory signature in endothelial cells. Redox Biology 28, 101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Yang WY, Chan K, Lavallee M, Xu K, et al. (2017c). Mitochondrial ros, uncoupled from atp synthesis, determine endothelial activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells. Canadian Journal of Physiology and Pharmacology 95, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Yang WY, Wang H, & Yang X (2017). Il-35, as a newly proposed homeostasis-associated molecular pattern, plays three major functions including anti-inflammatory initiator, effector, and blocker in cardiovascular diseases. Cytokine. 122:154076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mai J, Virtue A, Yin Y, Gong R, Sha X, et al. (2012). Il-35 is a novel responsive anti-inflammatory cytokine–a new system of categorizing anti-inflammatory cytokines. PLoS One 7, e33628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shao Y, Sha X, Fang P, Kuo YM, Andrews AJ, et al. (2018e). Il-35 (interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of h3k14 (histone 3 lysine 14). Arteriosclerosis, Thrombosis, and Vascular Biology 38, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang L, Fang P, Sun Y, Jiang X, Wang H, et al. (2018d). Lysophospholipids induce innate immune transdifferentiation of endothelial cells, resulting in prolonged endothelial activation. The Journal of biological chemistry. 293, 11033–11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang L, & Chen LL (2018b). The biogenesis, functions, and challenges of circular rnas. Molecular Cell 71, 428–442. [DOI] [PubMed] [Google Scholar]
- Li Y, Gao X, Wang Z, Liu W, Xu F, Hu Y, et al. (2020a). Circular rna 4099 aggravates hydrogen peroxide-induced injury by down-regulatingmicrorna-706 in l02 cells. Life Sciences 241, 116826. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. (2015a). Circular rna is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Research 25, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Huang X, Li X, Gong R, Yin Y, Nelson J, et al. (2016c). Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Front Biosci (Landmark Ed) 21, 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Li RS, Samuel SB, Cueto R, Li XY, Wang H, et al. (2016d). Lysophospholipids and their g protein-coupled receptors in atherosclerosis. Front Biosci (Landmark Ed) 21, 70–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Nanayakkara G, Sun Y, Li X, Wang L, Cueto R, et al. (2017d). Analyses of caspase-1-regulated transcriptomes in various tissues lead to identification of novel il-1beta-, il-18- and sirtuin-1-independent pathways. Journal of Hematology & Oncology 10, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M,Wang X, et al. (2015b). Exon-intron circular rnas regulate transcription in the nucleus. Nature Structural & Molecular Biology 22, 256–264. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. (2017a). Corrigendum: Exon-intron circular rnas regulate transcription in the nucleus. Nature Structural & Molecular Biology 24, 194. [DOI] [PubMed] [Google Scholar]
- Liang J, Shen YC, Zhang XY, Chen C, Zhao H, & Hu J (2020). Circular rna hipk3 downregulation mediates hydrogen peroxide-induced cytotoxicity in human osteoblasts. Aging. 12, 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. (2019a). Structure and degradation of circular rnas regulate pkr activation in innate immunity. Cell 177 (865–880. e821). [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. (2016). Circular rna related to the chondrocyte ecm regulates mmp13 expression by functioning as a mir-136 “sponge” in human cartilage degradation. Scientific Reports 6, 22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen X, Yao J, & Kang J (2019). Circular rna acr relieves high glucose-aroused rsc96 cell apoptosis and autophagy via declining microrna-145–3p. Journal of Cellular Biochemistry. 10.1002/jcb.29568. [DOI] [PubMed] [Google Scholar]
- Louis C, Desoteux M, & Coulouarn C (2019). Exosomal circrnas: New players in the field of cholangiocarcinoma. Clinical Science (London, England : 1979) 133, 2239–2244. [DOI] [PubMed] [Google Scholar]
- Lu D, & Xu AD (2016). Mini review: Circular rnas as potential clinical biomarkers for disorders in the central nervous system. Frontiers in Genetics 7, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, et al. (2015). Transcriptome-wide investigation of circular rnas in rice. RNA (New York, N.Y.) 21, 2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sun Y, Drummer C, Nanayakkara GK, Shao Y, Saaoud F, et al. (2019). Increased acetylation of h3k14 in the genomic regions that encode trained immunity enzymes in lysophosphatidylcholine-activated human aortic endothelial cells - novel qualification markers for chronic disease risk factors and conditional damps. Redox Biology 24, 101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ (2013). Circular rna (circrna) in alzheimer’s disease (ad). Frontiers in Genetics 4, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J, Nanayakkara G, Lopez-Pastrana J, Li X, Li YF, Wang X, et al. (2016). Interleukin-17a promotes aortic endothelial cell activation via transcriptionally and post-translationally activating p38 mitogen-activated protein kinase (mapk) pathway. The Journal of Biological Chemistry 291, 4939–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, & Makunin IV (2006). Non-coding rna. Human Molecular Genetics 15, 17–29 (15 Spec No 1:R17–29). [DOI] [PubMed] [Google Scholar]
- Meganck RM, Borchardt EK, Castellanos Rivera RM, Scalabrino ML, Wilusz JE, Marzluff WF, et al. (2018). Tissue-dependent expression and translation of circular rnas with recombinant aav vectors in vivo. Molecular Therapy–Nucleic Acids 13, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. (2013). Circular rnas are a large class of animal rnas with regulatory potency. Nature. 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Memczak S, Papavasileiou P, Peters O, & Rajewsky N (2015). Identification and characterization of circular rnas as a new class of putative biomarkers in human blood. PLoS One 10, e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Ginini L, & Gil Z (2019). Exosomes and their role in tumorigenesis and anticancer drug resistance. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 45, 1–12. [DOI] [PubMed] [Google Scholar]
- Monroy MA, Fang J, Li S, Ferrer L, Birkenbach MP, Lee IJ, et al. (2015). Chronic kidney disease alters vascular smooth muscle cell phenotype. Front Biosci (Landmark Ed) 20, 784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanayakkara GK, Wang H, & Yang X (2019). Proton leak regulates mitochondrial reactive oxygen species generation in endothelial cell activation and inflammation - a novel concept. Archives of Biochemistry and Biophysics 662, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Quintin J, & van der Meer JW (2011). Trained immunity: A memory for innate host defense. Cell Host & Microbe 9, 355–361. [DOI] [PubMed] [Google Scholar]
- Netea MG, & van der Meer JW (2017). Trained immunity: An ancient way of remembering. Cell Host & Microbe 21, 297–300. [DOI] [PubMed] [Google Scholar]
- Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, & Ea CK (2016). Inducible rasgef1b circular rna is a positive regulator of icam-1 in the tlr4/lps pathway. RNA Biology 13, 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. (1991). Scrambled exons. Cell. 64, 607–613. [DOI] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. (2017). Translation of circrnas. Molecular Cell 66 (9–21.e27). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Abdelmohsen K, & Gorospe M (2017b). Rt-qpcr detection of senescence-associated circular rnas. Methods in molecular biology (Clifton, N.J.) 1534, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Grammatikakis I, Munk R, Gorospe M, & Abdelmohsen K (2017a). Emerging roles and context of circular rnas. Wiley Interdiscip Rev RNA. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. (2019). Endoribonucleolytic cleavage of m(6)a-containing rnas by rnase p/mrp complex. Molecular Cell 74 (494–507.e498). [DOI] [PubMed] [Google Scholar]
- Penkov S, Mitroulis I, Hajishengallis G, & Chavakis T (2019). Immunometabolic crosstalk: An ancestral principle of trained immunity? Trends in Immunology 40, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, & Ares M Jr. (1998). Circular mrna can direct translation of extremely long repeating-sequence proteins in vivo. RNA (New York, N.Y.) 4, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. (2017). Loss of a mammalian circular rna locus causes mirna deregulation and affects brain function. Science (New York, N.Y.) 357. [DOI] [PubMed] [Google Scholar]
- Puro DG, Kohmoto R, Fujita Y, Gardner TW, & Padovani-Claudio DA (2016). Bioelectric impact of pathological angiogenesis on vascular function. Proceedings of the National Academy of Sciences of the United States of America 113, 9934–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, et al. (2016). Hsa_circ_0001649: A circular rna and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers Section A Disease Markers 16, 161–169. [DOI] [PubMed] [Google Scholar]
- Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, et al. (2015). Microarray expression profile of circular rnas in human pancreatic ductal adenocarcinoma. Genomics data. 5, 385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan PL, Sauzade M, & Brouzes E (2018). Dpcr: A technology review. Sensors (Basel, Switzerland) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Liu J, Feng Y, Li Z, He L, Li L, et al. (2019). Knockdown of circdennd4c inhibits glycolysis, migration and invasion by up-regulating mir-200b/c in breast cancer under hypoxia. Journal of Experimental & Clinical Cancer Research 38, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosadini CV, & Kagan JC (2017). Early innate immune responses to bacterial lps. Current Opinion in Immunology 44, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. (2015). Circular rnas in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell 58, 870–885. [DOI] [PubMed] [Google Scholar]
- Saaoud F, Wang J, Iwanowycz S, Wang Y, Altomare D, Shao Y, et al. (2020). Bone marrow deficiency of mrna decaying protein tristetraprolin increases inflammation and mitochondrial ros but reduces hepatic lipoprotein production in ldlr knockout mice. Redox Biology 101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Somoza A, Zhang L, Vausort M, & Devaux Y (2017). The circular rna micra for risk stratification after myocardial infarction. International journal of cardiology. Heart & vasculature. 17, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J (2016). Circular rna expression: Its potential regulation and function. Trends in Genetics 32, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, & Brown PO (2013). Cell-type specific features of circular rna expression. PLoS Genetics 9, e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, & Brown PO (2012). Circular rnas are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger HL, Klotz G, Riesner D, Gross HJ, & Kleinschmidt AK (1976). Viroids are single-stranded covalently closed circular rna molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America 73, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena CM, Pereira AM, & Seiça R (1832). Endothelial dysfunction - a major mediator of diabetic vascular disease. Biochimica et Biophysica Acta 2013, 2216–2231. [DOI] [PubMed] [Google Scholar]
- Sha X, Meng S, Li X, Xi H, Maddaloni M, Pascual DW, et al. (2015). Interleukin-35 inhibits endothelial cell activation by suppressing mapk-ap-1 pathway. The Journal of Biological Chemistry 290, 19307–19318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. (2017). Circular noncoding rna hipk3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 136, 1629–1642. [DOI] [PubMed] [Google Scholar]
- Shao Y, Cheng Z, Li X, Chernaya V, Wang H, & Yang X-F (2014). Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction-a novel mechanism for maintaining vascular function. Journal of Hematology & Oncology 7, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Nanayakkara G, Cheng J, Cueto R, Yang WY, Park JY, et al. (2017). Lysophospholipids and their receptors serve as conditional damps and damp receptors in tissue oxidative and inflammatory injury. Antioxidants & Redox Signaling 28 (10), 973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Saredy J, Yang W, Sun Y, Lu Y, Saaoud F, … Yang X (2020b). Vascular endothelial cells and innate immunity. Arteriosclerosis, Thrombosis, and Vascular Biology 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Saredy J, Yang WY, Sun Y, Lu Y, Saaoud F, et al. (2020a). Vascular endothelial cells and innate immunity. Arteriosclerosis, Thrombosis, and Vascular Biology 40, e138–e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Yin Y, Mai J, Xiong X, Pansuria M, Liu J, et al. (2010). Caspase-1 recognizes extended cleavage sites in its natural substrates. Atherosclerosis. 210, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H (2015). Oxidative stress: A concept in redox biology and medicine. Redox Biology 4, 180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siti HN, Kamisah Y, & Kamsiah J (2015). The role of oxidative stress, antioxidants and vascular inflammation in cardiovascular disease (a review). Vascular Pharmacology 71, 40–56. [DOI] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. (2015). Exon circularization requires canonical splice signals. Cell Reports 10, 103–111. [DOI] [PubMed] [Google Scholar]
- Steven S, Frenis K, Oelze M, Kalinovic S, Kuntic M, Bayo Jimenez MT, et al. (2019). Vascular inflammation and oxidative stress: Major triggers for cardiovascular disease. Oxidative Medicine and Cellular Longevity 2019, 7092151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, & Lv X (2020). Revealing new landscape of cardiovascular disease through circular rna-mirna-mrna axis. Genomics. 112, 1680–1685. [DOI] [PubMed] [Google Scholar]
- Sun Y, Johnson C, Zhou J, Wang L, Li YF, Lu Y, et al. (2018). Uremic toxins are conditional danger- or homeostasis-associated molecular patterns. Front Biosci (Landmark Ed) 23, 348–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lu Y, Saredy J, Wang X, Drummer Iv C, Shao Y, et al. (2020). Ros systems are a new integrated network for sensing homeostasis and alarming stresses in organelle metabolic processes. Redox Biology 101696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, & Tsukahara T (2014). A viewof pre-mrna splicing from rnase r resistant rnas. International Journal of Molecular Sciences 15, 9331–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, & Mayeda A (2006). Characterization of rnase r-digested cellular rna source that consists of lariat and circular rnas from pre-mrna splicing. Nucleic Acids Research 34, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Chen Y, Chen H, He X, Wang J, Peng Y, et al. (2017). Circular rna hsa_circ_0021001 in peripheral blood: A potential novel biomarker in the screening of intracranial aneurysm. Oncotarget 8, 107125–107133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Zitvogel L, & Amigorena S (2002). Exosomes: Composition, biogenesis and function. Nature Reviews. Immunology 2, 569–579. [DOI] [PubMed] [Google Scholar]
- Thomson DW, & Dinger ME (2016). Endogenous microrna sponges: Evidence and controversy. Nature Reviews. Genetics 17, 272–283. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, & Lötvall JO (2007). Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nature Cell Biology 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Vicens Q, & Westhof E (2014). Biogenesis of circular rnas. Cell. 159, 13–14. [DOI] [PubMed] [Google Scholar]
- Virtue A, Johnson C, Lopez-Pastrana J, Shao Y, Fu H, Li X, et al. (2017). Microrna-155 deficiency leads to decreased atherosclerosis, increased white adipose tissue obesity, and non-alcoholic fatty liver disease: A novel mouse model of obesity paradox. The Journal of Biological Chemistry 292, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue A, Wang H, & Yang XF (2012). Micrornas and toll-like receptor/interleukin-1 receptor signaling. Journal of Hematology & Oncology 5, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. (2016a). A circular rna protects the heart from pathological hypertrophy and heart failure by targeting mir-223. European Heart Journal 37, 2602–2611. [DOI] [PubMed] [Google Scholar]
- Wang L, Fu H, Nanayakkara G, Li Y, Shao Y, Johnson C, et al. (2016b). Novel extracellular and nuclear caspase-1 and inflammasomes propagate inflammation and regulate gene expression: A comprehensive database mining study. Journal of Hematology & Oncology 9, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y-F, Nanayakkara G, Shao Y, Liang B, Cole L, et al. (2016c). Lysophospholipid receptors, as novel conditional danger receptors and homeostatic receptors modulate inflammation—novel paradigm and therapeutic potential. Journal of Cardiovascular Translational Research 9, 343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. (2019a). Exosomal circrnas: Biogenesis, effect and application in human diseases. Molecular Cancer 18, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao R, Liu W, Wang Z, Rong J, Long X, et al. (2019b). Exosomal circhipk3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the mir-29a/igf-1 pathway. Oxidative Medicine and Cellular Longevity 2019, 7954657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JW, Huang K, Yang C, & Kang CS (2017). Non-coding rnas as regulators in epigenetics (review). Oncology Reports 37, 3–9. [DOI] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, & Anderson DG (2018). Engineering circular rna for potent and stable translation in eukaryotic cells. Nature Communications 9, 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. (2014). Genome-wide analysis of drosophila circular rnas reveals their structural and sequence properties and age-dependent neural accumulation. Cell Reports 9, 1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Jin L, & Cai J (2017). Profiling and bioinformatics analyses reveal differential circular rna expression in hypertensive patients. Clinical and experimental hypertension (New York, N.Y. : 1993) 39, 454–459. [DOI] [PubMed] [Google Scholar]
- Xu H, Guo S, Li W, & Yu P (2015). The circular rna cdr1as, via mir-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific Reports 5, 12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Yang WY, Nanayakkara GK, Shao Y, Yang F, Hu W, et al. (2018). Gata3, hdac6, and bcl6 regulate foxp3+treg plasticity and determine treg conversion into either novel antigen-presenting cell-like treg or th1-treg. Frontiers in Immunology 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wu J, Han P, Zhao Z, & Song X (2017). Circular rna expression profiles and features in human tissues: A study using rna-seq data. BMC Genomics 18, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jiang T, Wu C, & Zhang Y (2020). Circakt3 inhibits glycolysis balance in lung cancer cells by regulating mir-516b-5p/stat3 to inhibit cisplatin sensitivity. Biotechnology Letters 42, 1123–1135. [DOI] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, & Chen LL (2011). Genomewide characterization of non-polyadenylated rnas. Genome Biology 12, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Nanayakkara GK, Drummer C, Sun Y, Johnson C, Cueto R, et al. (2017b). Low-intensity ultrasound-induced anti-inflammatory effects are mediated by several new mechanisms including gene induction, immunosuppressor cell promotion, and enhancement of exosome biogenesis and docking. Frontiers in Physiology 8, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Yin Y, &Wang H (2008). Vascular inflammation and atherogenesis are activated via receptors for pamps and suppressed by regulatory t cells. Drug Discov Today Ther Strateg. 5, 125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. (2017a). Extensive translation of circular rnas driven by n(6)-methyladenosine. Cell Research 27, 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Li X, Sha X, Xi H, Li YF, Shao Y, et al. (2015). Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arteriosclerosis, Thrombosis, and Vascular Biology 35, 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Pastrana JL, Li X, Huang X, Mallilankaraman K, Choi ET, et al. (2013). Inflammasomes: Sensors of metabolic stresses for vascular inflammation. Front Biosci (Landmark Ed) 18, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. (2019). Circrnas in cancer metabolism: A review. Journal of Hematology & Oncology 12, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang J, Lu D, & Xu A (2020). The interaction of circrnas and rna binding proteins: An important part of circrna maintenance and function. Journal of Neuroscience Research 98, 87–97. [DOI] [PubMed] [Google Scholar]
- Zhang M, & Xin Y (2018). Circular rnas: A new frontier for cancer diagnosis and therapy. Journal of Hematology & Oncology 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, & Yang L (2014a). Complementary sequence-mediated exon circularization. Cell. 159, 134–147. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, et al. (2017). Circular rnas: Emerging cancer biomarkers and targets. Journal of Experimental & Clinical Cancer Research 36, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, et al. (2016). The biogenesis of nascent circular rnas. Cell Reports 15, 611–624. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang L, & Chen LL (2014b). Life without a tail: New formats of long noncoding rnas. The international journal of biochemistry & cell biology. 54, 338–349. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. (2013). Circular intronic long noncoding rnas. Molecular Cell 51, 792–806. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhu M, Limbo O, & Russell P (2018). Rnase h eliminates r-loops that disrupt DNA replication but is nonessential for efficient dsb repair. EMBO Reports 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Alexandrov PN, Jaber V, & Lukiw WJ (2016). Deficiency in the ubiquitin conjugating enzyme ube2a in alzheimer’s disease (ad) is linked to deficits in a natural circular mirna-7 sponge (circrna; cirs-7). Genes 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, et al. (2017b). Peripheral blood circular rna hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Scientific Reports 7, 39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Li X, Jian D, Hao P, Rao L, & Li M (2017a). Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetologica 54, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. (2016). Circular rna profiling reveals an abundant circhipk3 that regulates cell growth by sponging multiple mirnas. Nature Communications 7, 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Yang X, Feng Y, & Yu J (2020). Trained immunity: An underlying driver of inflammatory atherosclerosis. Frontiers in Immunology 11, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JB, Li T, Hu XM, Ning M, Gao WQ, Lang YH, et al. (2020). Circ_chfr expedites cell growth, migration and inflammation in ox-ldl-treated human vascular smooth muscle cells via the mir-214–3p/wnt3/β-catenin pathway. European Review for Medical and Pharmacological Sciences 24, 3282–3292. [DOI] [PubMed] [Google Scholar]