Summary
Rice (Oryza sativa) is the staple food for over half the world’s population. Drought stress imposes major constraints on rice yields. Intriguingly, labdane-related diterpenoid (LRD) phytoalexins in maize (Zea mays) affect drought tolerance, as indicated by the increased susceptibility of an insertion mutant of the class II diterpene cyclase ZmCPS2/An2 that initiates such biosynthesis. Rice also produces LRD phytoalexins, utilizing OsCPS2 and OsCPS4 to initiate a complex metabolic network.
For genetic studies of rice LRD biosynthesis the fast-growing Kitaake cultivar was selected for targeted mutagenesis via CRISPR/Cas9, with an initial focus on OsCPS2 and OsCPS4. The resulting cps2 and cps4 knock-out lines were further crossed to create a cps2×4 double-mutant. Both CPSs also were over-expressed.
Strikingly, all of the cv. Kitaake cps mutants exhibit significantly increased susceptibility to drought, which was associated with reduced stomatal closure that was evident even under well-watered conditions. However, CPS over-expression did not increase drought resistance, and cps mutants in other cultivars did not alter susceptibility to drought, although these also exhibited lesser effects on LRD production.
The results suggest that LRDs may act as a regulatory switch that triggers stomatal closure in rice, which might reflect the role of these openings in microbial entry.
Keywords: diterpenoids, rice, drought, stomata, phytoalexins
Introduction
Rice is the staple food for over half of the world’s peoples (Muthayya et al., 2014). However, in order to feed the increasing global population the current harvest of ~480 million tons will need to rise 15% within the next decade (Godfray et al., 2010). Among the various factors that constrain yield, drought stands out as one of the major stressors, with losses of over 25% often observed (Zhang et al., 2018). Accordingly, contributing factors to water use efficiency are of substantial interest (Gupta et al., 2020).
Coupled to such environmental stress is the enhanced susceptibility of the distressed plants to diseases caused by microbial pathogens (Bostock et al., 2014), which also contributes to reduced yields (Pandey et al., 2017). Among the various responses of plants to microbial infection is the production of an arsenal of antibiotic natural products (Bednarek & Osbourn, 2009). Such antimicrobial compounds whose production is strongly induced by infection are termed phytoalexins, while those constitutively present are termed phytoanticipins (VanEtten et al., 1994). Intriguingly, it has been proposed that diterpenoid phytoalexins may play a role in resistance to environmental stresses such as drought in cereal crop plants (Murphy & Zerbe, 2020).
In particular, it has been proposed that the maize kauralexins play a role in drought tolerance (Vaughan et al., 2015). The kauralexins are labdane-related diterpenoids (LRDs), which are defined by the activity of class II diterpene cyclases (Peters, 2010). These catalyze bicyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP), prototypically forming the eponymous labdadienyl/copalyl diphosphate (CPP). Such enzymes are then termed CPP synthases (CPSs). These most commonly produce what is termed the enantiomeric form of CPP (ent-CPP), as required for biosynthesis of the gibberellin A (GA) phytohormones in all vascular plants, although there are CPSs with such product outcome that are dedicated to more specialized metabolism (Zi et al., 2014). In maize there are two CPSs that produce ent-CPP, ZmCPS1/An1, which primarily functions in GA biosynthesis (Bensen et al., 1995), and ZmCPS2/An2, which seems to play a role in more specialized LRD metabolism (Harris et al., 2005).
A role for kauralexins in maize abiotic stress responses was suggested by the accumulation of these LRDs upon exposure to drought or high salinity and, particularly, the increased susceptibility of an an2 insertion mutant line to drought (Vaughan et al., 2015). Nonetheless, it has since been shown that maize also produces dolabralexins from ent-CPP in an An2 dependent manner, and these LRDs also accumulate in roots in response to various abiotic stresses, including drought (Mafu et al., 2018). Thus, only An2-dependent LRDs more generally can be assigned a role in resistance to abiotic stresses such as drought (Murphy & Zerbe, 2020). Moreover, the reduced plant biomass observed with an2 plants, particularly of the roots, leaves open the possibility that it is reduction in GA, rather than more specialized LRDs, that leads to the increased susceptibility to drought.
Rice has long been known to have extensive LRD metabolism (Peters, 2006). In rice GA metabolism is dependent on OsCPS1, as knocking out this gene leads to fully dwarfed plants (Sakamoto et al., 2004). By contrast, OsCPS2, which also leads to production of ent-CPP seems to be only involved in more specialized LRD biosynthesis (Otomo et al., 2004; Prisic et al., 2004). While OsCPS3 is a pseudogene, rice also contains OsCPS4, which leads to production of the stereochemically distinct syn-CPP (Otomo et al., 2004; Xu et al., 2004). Accordingly, more specialized LRD metabolism in rice can be divided into two branches (Figure 1), with those derived from syn-CPP (e.g., momilactones) dependent on OsCPS4, although the situation for those derived from ent-CPP (e.g., phytocassanes and oryzalexins A – F) is less clear. Indeed, although an OsCPS4 T-DNA insertion mutant line (cps4t) no longer produces momilactones (Xu et al., 2012), an OsCPS2 T-DNA insertion mutant line (cps2t) was found to produce similar amounts of phytocassanes as the relevant parental (wild-type) cultivar (cv.), albeit reductions in oryzalexins A – F levels were observed (Lu et al., 2018).
Figure 1.
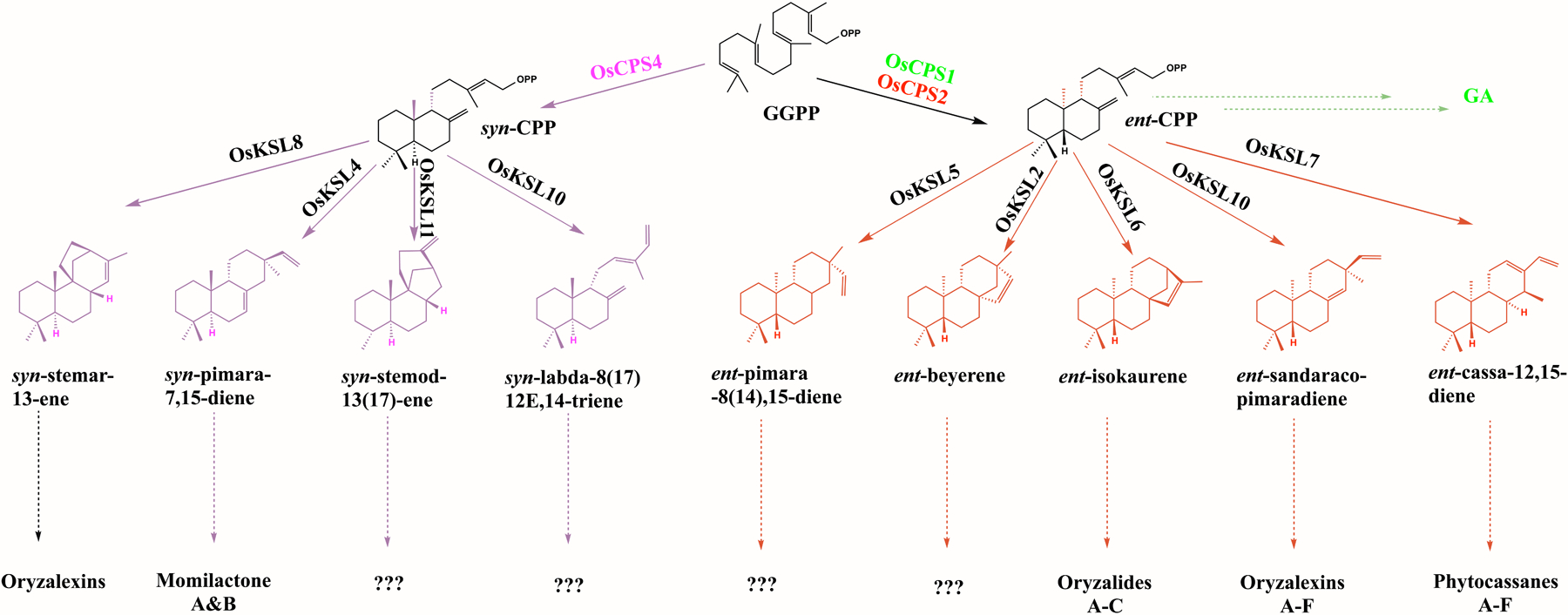
Rice labdane-related diterpenoid (LRD) metabolic network. Coloring indicates stereochemical or functional distinction. Dashed arrows indicate multiple enzymatic reactions leading to families of LRD derived from the shown diterpene olefins where known, while ‘???’ indicates presumed unknown derivatives (CPS, copalyl diphosphate synthase; KSL, kaurene synthase-like).
The known rice LRDs were originally isolated as phytoalexins and phytoanticipins, as well as allelochemicals (Schmelz et al., 2014). Most of the rice LRDs were isolated as phytoalexins against the fungal blast pathogen Magnaporthe oryzae, including the phytocassanes and oryzalexins A – F derived from ent-CPP, as well as oryzalexin S and momilactones A & B derived from syn-CPP (Peters, 2006). The remaining known rice LRDs are the oryzalides, oryzalexic acids and oryzadiones (termed here oryzalides & related LRDs), which are derived from ent-CPP but were isolated as phytoanticipins against the bacterial rice leaf blight pathogen Xanthomonas oryzae pathovar oryzae (Xoo) (Toyomasu, 2008). Interestingly, the momilactones also serve as allelochemicals (Kato-Noguchi & Peters, 2013). In addition, it has been further proposed that momilactones may enhance drought tolerance (Xuan et al., 2016). Beyond these known LRDs, it must be noted that rice produces others, derived from both ent- as well as syn- CPP, whose ultimate form and function remain uncertain (Figure 1).
While genetic evidence from two studies supports a role for the momilactones as allelochemicals, neither examined drought resistance, and these studies came to opposite conclusions regarding the relevance of these LRDs as phytoalexins against M. oryzae (Xu et al., 2012; Toyomasu et al., 2014). However, it is difficult to directly compare these studies, as they relied on insertion mutants derived from a variety of cultivars (cv.) – i.e., cv. Zhonghua 11, cv. Hwayoung and cv. Nipponbare (although these all are from the japonica subspecies). Similarly, although comparison of cps2t and cps4t has been reported (Lu et al., 2018), these also were found in distinct backgrounds (cv. Nipponbare and cv. Zhonghua 11, respectively). Accordingly, to enable genetic studies of rice LRD metabolism, it seemed prudent to utilize CRISPR/Cas9 genome editing to carry out targeted mutagenesis in a uniform background to allow more direct comparisons. For this purpose, the fast-growing cv. Kitaake was selected, with OsCPS2 and OsCPS4 targeted for initial studies, and the resulting cps2 and cps4 lines crossed to create a cps2 and cps4 double knock-out line (termed here cps2×4). Although the cps2 lines had a greater effect on LRD composition than observed with cps2t, the cps2 and cps4 lines still exhibited contrasting effects on susceptibility to Xoo, similar to what was previously reported with cps2t and cps4t (Lu et al., 2018), consistent with oryzalides & related LRDs serving as the relevant antibiotics. In addition, both cps2 and cps4, as well as cps2×4, were found to exhibit increased susceptibility to drought, which was traced to decreased stomatal closure in the mutants. The implications of these findings are then discussed.
Methods
Chemicals
Unless otherwise noted, all molecular biology reagents were purchased from Invitrogen (Carlsbad, CA, USA) and all other chemicals from Fischer Scientific (Waltham, MA, USA).
Plant materials
Rice plants (Oryza sativa L. ssp. japonica cv. Kitaake, Nipponbare or Zhonghua 11) were grown in standard growth chambers cycling between 12 h light at 28 °C and 12 h dark at 24 °C.
Construction of cps mutant lines
For guide RNA cloning, the intermediate vector pENTR-gRNA, engineered for two gRNAs was used, which contains two cloning sites (BtgZI and BsaI) following rice U6 promotors. For the BtgZI cloning site, the sense strand contains a 5’ 4-nt overhang of TGTT and the antisense strand contains a 5’ 4-nt AAAC overhang. For the BsaI cloning site, the sense strand contains a 5’ 4-nt overhang of GTGT and the antisense strand contains a 5’ 4-nt AAAC overhang. The gRNA sequences were synthesized (Supplemental Table S2), the first cloned into the BtgZI restriction site and the second was then inserted into the BsaI restriction site, followed by sequencing to verify the construct. These 2gRNA cassettes were then sub-cloned into the CRISPR vector pByo2-Cas9 using LR Clonase (i.e., the Invitrogen Gateway system). Escherichia coli strain XL1-Blue was used for molecular cloning of the Cas9/2gRNA constructs, which were then transformed into Agrobacterium tumefaciens strain EHA105 by electroporation. Immature embryo derived callus cells of rice (cv. Kitaake) were transformed were transformed using the Agrobacterium-mediated rice transformation method. Then, transformed calli and regenerated plantlets were selected on media containing hygromycin B.
For genotyping, genomic DNA (gDNA) was extracted from T0 transgenic rice seedling leaves using Plant DNAzol® Reagent (Invitrogen, USA). This gDNA was used for PCR amplification of relevant regions with specific primers flanking the target sites (Supplemental Table S2). The PCR amplicons were treated with ExoSAP-IT (Affymetrix, Santa Clara, CA) and then subjected to Sanger sequencing. The sequencing chromatograms were examined for patterns indicating diallelic plants. For diallelic mutations, the same genotyping method was used in the T1 generation transgenic rice. Homozygous mutant lines were selected and screened by PCR for the Cas9 gene. The lines used here are Cas9-free and homozygous for separate mutations that are expected to block translation of functional enzyme (see Supplemental Table S1 for the lesions found in each line).
Creation of a cps2 and cps4 double mutant line
The cps2–60 and cps4–11 mutant lines were crossed. Genotyping was carried out as described above, with every seedling screened for mutations in both genes. Seedlings that were diallelic for both OsCPS2 and OsCPS4 were selected and selfed. The resulting seedlings were again genotyped, and seeds harvested from those plants homozygous for the cps2–60 and cps4–11 mutations. These were designated cps2×4 double-mutants and used here.
Over-expression of OsCPS2 and OsCPS4
The open reading frame for OsCPS2 or OsCPS4 were amplified from cDNA by PCR using primers that also introduce unique KpnI and XbaI restriction sites at the 5′ and 3′ ends, respectively (Supplemental Table S2). The PCR fragment was cloned into the KpnI and XbaI restriction sites of the binary vector pBY02, such that expression is under control of the maize ubiquitin 1 gene promoter. These were verified by complete gene sequencing and the resulting pBY02-OsCPS2-OE and pBY02-OsCPS4-OE constructs were transformed into cv. Kitaake rice as described above. Over-expression of the relevant CPS was confirmed by semi-quantitative RT-PCR. Selection was carried out for three more generations to obtain non-segregating homozygous CPS2-OE and CPS4-OE lines.
Phytochemical analyses
Three-week old rice leaves were cut into 5 cm pieces and 0.1 g induced by floating on a solution of 0.5 mM CuCl2. After 72 h these induced leaves were frozen by submersion into liquid N2 and ground into fine power, which were extracted by shaking in 3 mL methanol for 72 h in a cold (4 °C) room. Each sample had 2.31 μg sclareol added as an internal standard. The methanol was filtered through a 0.2 μ nylon filter (F2513–2, Thermo) using a glass syringe, dried under a stream of N2 gas, and resuspended in 1 mL methanol for analysis.
Analyses were carried out via LC-MS/MS, with 15 μL samples injected onto a Supelco (Sigma-Aldrich, St. Louis, MO, USA) Ascentis C18 column (10 cm × 2.1 mm; 3 μ) using an Agilent Technologies 1100 Series HPLC system coupled to both a UV-Vis diode array detector and an Agilent Technologies Mass Selective Trap SL detector located in the Iowa State University W.M. Keck Metabolomics Research Laboratory. A binary gradient was used, consisting water with 0.1% acetic acid v/v (buffer A) and acetonitrile with 0.1% acetic acid v/v (buffer B). The solvent gradient elution was programmed as follows: initial, 40% buffer B; 0–13 min, a linear gradient from 40% buffer B to 85% buffer B; 13–14 min, a linear gradient from 85% buffer B to 100% buffer B; 14–14.5 min, a linear gradient from 100% buffer B to 40% buffer B. The known rice LRD phytoalexins were detected by MS/MS analysis, using an isolation selection window of ± 0.5 m/z. The isolated masses were reionized and the resulting mass spectra recorded.
The isolated parent ions and quantified secondary ions, along with corresponding retention time (RT), based on optimization with the available authentic standards, for the known rice LRD phytoalexins were as follows: momilactone A, m/z 315.2—271.2, RT 11.1 min; momilactone B, m/z 331.2—269.2, RT 9.0 min; oryzalexin A, m/z 303.2—285.2, RT 10.0 min; oryzalexin B, m/z 303.3—286.2, RT 9.7 min; oryzalexin C, m/z 301.2—283.2, RT 9.6 min; oryzalexin D, m/z 287.2—269.2 (147.2), RT 9.6 min; oryzalexin E, m/z 287.2—269.2, RT 11.3 min; oryzalexin S, m/z 287.2—269.2 (241.2), RT 9.7 min; phytocassane A, m/z 317.2—299.2, RT 7.6 min; phytocassane B, m/z 335.2—317.2, RT 7.3 min; phytocassane C, m/z 319.2—301.2, RT 6.8 min; phytocassane D, m/z 317.2—299.2, RT 9.7 min; phytocassane E, m/z 317.2—299.2, RT 8.3 min; phytocassane F, m/z 333.2—315.2, RT 7.3 min; sclareol, m/z 273.2—163.1, RT 13.6 min.
The relative amount of each LRD was determined with the 6300 Series Ion Trap LC/MS version 1.8 software package (Bruker), by comparison of total ion peak areas to that of the sclareol internal standard.
For root exudates phytochemical analyses were carried out as previously reported (Lu et al., 2018).
RNA isolation and qRT-PCR/RT-PCR
Total RNA was isolated using RNeasy® Plant Mini Kit (Qiangen, Carlsbad, CA, USA) according to the instructions of the manufacturer. To remove gDNA the Turbo DNA-freeTM kit (Thermo Scientific) was used. Reverse transcription (RT) was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). For qRT-PCR analysis a Step OnePlus thermocycler (in ISU DNA Facility) was employed, with the use of Power UpTM SYBR® Green Master Mix (Thermo Scientific), using actin (LOC_Os03g61970) for the reference gene, as previously described (Kitaoka et al., 2020).
Xoo infection assays
Bacterial leaf blight disease was assayed with three-week old rice seedlings. Xoo strain PXO99A was grown on TSA (1% wt/vol Tryptone, 1% sucrose, 0.1% glutamic acid, 1.5% agar, pH 6.6–6.8) plates at 28 °C for 2~3 days, scraped off, resuspended in sterile deionized water and washed twice. The suspension was then adjusted to an optical density of 0.5 at 600 nm, and this infiltrated into leaves with a needleless syringe. Each leaf was inoculated at five spots, with ten seedlings (one leaf each) infected for each line.
Three days after inoculation, leaf fragments containing the inoculated spots were harvested and ground in a sterile mortar with 2 mL sterile water per leaf, to create a bacterial seriflux. The seriflux was serially diluted ten-fold in sterile water up to nine times (i.e., 10−9) and 0.1 mL of each dilution spread on TSA plates in triplicate. These were incubated 2–3 days at 28 °C and the bacterial concentration in the seriflux calculated by colony counting and averaging over the three replicates.
Drought stress treatment and analysis
Surface-sterilized seeds were germinated on 1/2 MS (2.22 g Murashige Skoog, 15 g sucrose and 8 g agar per L) petri dishes (100 × 15 mm) for 7 days. A set of healthy and uniformly sized seedlings were then transferred into identical pots, each with an equivalent amount of soil, 2 seedlings per pot, and then grown for 18 days in trays (ten pots each). During this time, all trays were removed for watering and the pots shuffled to random locations within the trays daily, five hours into the light cycle. Subsequently, the plants were split into two treatment groups, one of which continued to be watered daily (control) and the other of which was placed into dry trays and no longer watered (drought stress). During this treatment, each pot was weighed and shuffled to random locations within the relevant trays daily, again five hours into the light cycle. Ten pots were grown for each line, with five then placed in each treatment group.
For stomatal analysis, three seedlings of each line were analyzed just before (0 day) and on the 3rd and 6th days during the drought stress treatment. Samples consisted of a 1 cm × 1 cm square excised from the 4th leaf (13 cm from the tip) and fixed in FAA (5% formalin, 5% acetic acid in 50% ethanol). After fixation, these samples were dehydrated through treatment with a graded ethanol series (70%, 85%, 95% and then 100%), with 2 changes at each concentration, for 1 hour each. Samples were critical point dried using a Denton Vacuum, Inc. Drying Apparatus, Model DCP-1 (Denton Vacuum, Moorestown, NJ).
The dried samples were mounted with the abaxial face up on aluminum stubs with double-sided tape and colloidal silver paint and sputter coated with palladium using Sputter Coater (Cressington, HR208). Images were captured using a scanning electron microscope at 10 KV (Hitachi SU-4800 field emission), using 11.3mm × 250 SE (M) for analysis of stomatal density and 11.3mm × 1.50K SE (U) for analysis of stomatal aperture status. Twenty images were captured for each sample.
Biomass analysis
For biomass analysis, plants were grown and treated with drought stress (with watered controls) as above. The plants were collected after 6 days, separated into root and shoot (arial) tissues, and weighed (fresh weight). These samples were dried in a 70 °C oven for three days, and then weighed again (dry weight).
Phytohormone analysis
For phytohormone analysis, plants were grown and treated with drought stress (with watered controls) as above. Leaf samples (50 mg) were collected, frozen in liquid N2, ground to a fine powder and then extracted with 0.5 mL of a 2:1 solution of 2-propanol and water with 0.2 % (v/v) HCl, containing 200 ppb of d6-ABA (abscisic acid), d4-SA (salicylic acid), d2-IAA (indole-acetic acid) and d5-JA (jasmonic acid) as internal standards. The homogenate was shaken at 300 rpm at 4 °C for 30 min, 1 mL of CHCl3 added, and the mixture then centrifuged at 13,000 g at 4 °C for 5 min. The organic solvent/extract was transferred to a new microtube, dried under N2 and resuspended in 100 μL methanol. The resuspension was filtered through a 0.1 μm nylon filter and then analyzed by LC-MS/MS at China Agriculture University, with quantification of ABA, IAA, SA and JA as previously described (Pan et al., 2010).
Results
Genetic manipulation of OsCPS2 and OsCPS4 in cv Kitaake
Kitaake has become a model rice cultivar based on its early flowering time, which enables a 9-week life cycle, as well as ease of propagation and transformation (Kim et al., 2013). Moreover, a high-quality genome sequence has been recently reported for cv. Kitaake (Jain et al., 2019), providing increased confidence in the precise targeting required for genome editing. In addition, as a member of the japonica subspecies (ssp.) Kitaake is expected to exhibit less redundancy in its LRD metabolism (Xu et al., 2007). Accordingly, this cultivar was selected for comprehensive genetic investigation of LRD metabolism, using a CRISPR/Cas9–2gRNA approach (Bi & Yang, 2017). Nevertheless, the constructs are designed to target sequences that are conserved in both of the two major subspecies of rice (i.e., ssp. indica as well as ssp. japonica), enabling facile utilization across different genetic backgrounds. Upon initiating these studies, it seemed prudent to re-investigate the effect of knocking-out OsCPS2 (OsKitaake02g36210) and OsCPS4 (OsKitaake04g09900) to determine if there are any distinct phenotypes in cv. Kitaake. For both genes, two distinct mutant alleles were identified by genotyping, and these plants selfed to create homozygous lines in which the CRISPR/Cas9–2gRNA gene-editing construct also has been segregated out (Supplemental Table S1). To highlight the utility of a common genetic background, a cps2×4 double knock-out line was created by crossing single mutant lines. All of the resulting mutant lines exhibited normal growth and development relative to wild-type (WT) cv. Kitaake (Supplemental Figure S1). For comparison, over-expression (OE) constructs for both OsCPS2 and OsCPS4 also were transformed into cv. Kitaake, and two lines for each (i.e., CPS2-OE and CPS4-OE) selected by semi-quantitative RT-PCR (Supplemental Figure S2).
Effect on LRD production
To investigate how these genetic manipulations of OsCPS2 and OsCPS4 affect LRD metabolism in cv. Kitaake, a previously developed LC-MS/MS method targeted at these phytochemicals was applied (Lu et al., 2018). However, this analysis was carried out with extracts from induced leaves rather than root exudates (Figure 2), as this yields greater amounts and numbers of the known LRDs. Nevertheless, of oryzalexins A – F, only oryzalexins D and E could be detected in cv. Kitaake. As expected, momilactones and oryzalexin S are no longer produced by either of the cv. Kitaake cps4 lines, nor the cps2×4 double-mutant. Some increased production of phytocassanes and oryzalexins D & E relative to WT cv. Kitaake was observed in the cps4–4 line, resembling the findings reported for the equivalent cv. Zhonghua 11 cps4t mutant (Lu et al., 2018). However, the other cv. Kitaake cps4–11 line did not exhibit such increases. Similar to the findings previously reported for the equivalent cv. Nipponbare cps2t mutant (Lu et al., 2018), oryzalexins D & E were not produced by the cv. Kitaake cps2 lines, although these were produced in increased amounts relative to WT by the cps2×4 double-mutant. Notably, in contrast to the lack of effect on phytocassane accumulation previously reported in the cv. Nipponbare cps2t mutant (Lu et al., 2018), these LRDs were produced at significantly lower levels relative to WT in the cv. Kitaake cps2 lines, and not at all by the cps2×4 double-mutant. In addition, the cv. Kitaake cps2 lines produced increased amounts of momilactones and oryzalexin S relative to WT, which contrasts to the findings reported with the cv. Nipponbare cps2t mutant (Lu et al., 2018). This presumably reflects their greater impact on production of the more prevalent phytocassanes, leading to reallocation of the GGPP precursor towards the alternate OsCPS4-dependent (i.e., syn-CPP derived), branch of rice LRD metabolism (see Figure 1). Nonetheless, overexpression of either OsCPS2 or OsCPS4 led to an overall increase in LRD production levels relative to WT cv. Kitaake. Surprisingly, rather than being limited to the corresponding branches of LRD metabolism, both CPS2-OE and CPS4-OE lines exhibited increased production of both momilactones and phytocassanes. These represent the most prevalent LRDs from each branch, suggesting that these overexpression lines somehow lead to increased allocation towards LRDs (i.e., increased production of GGPP).
Figure 2.
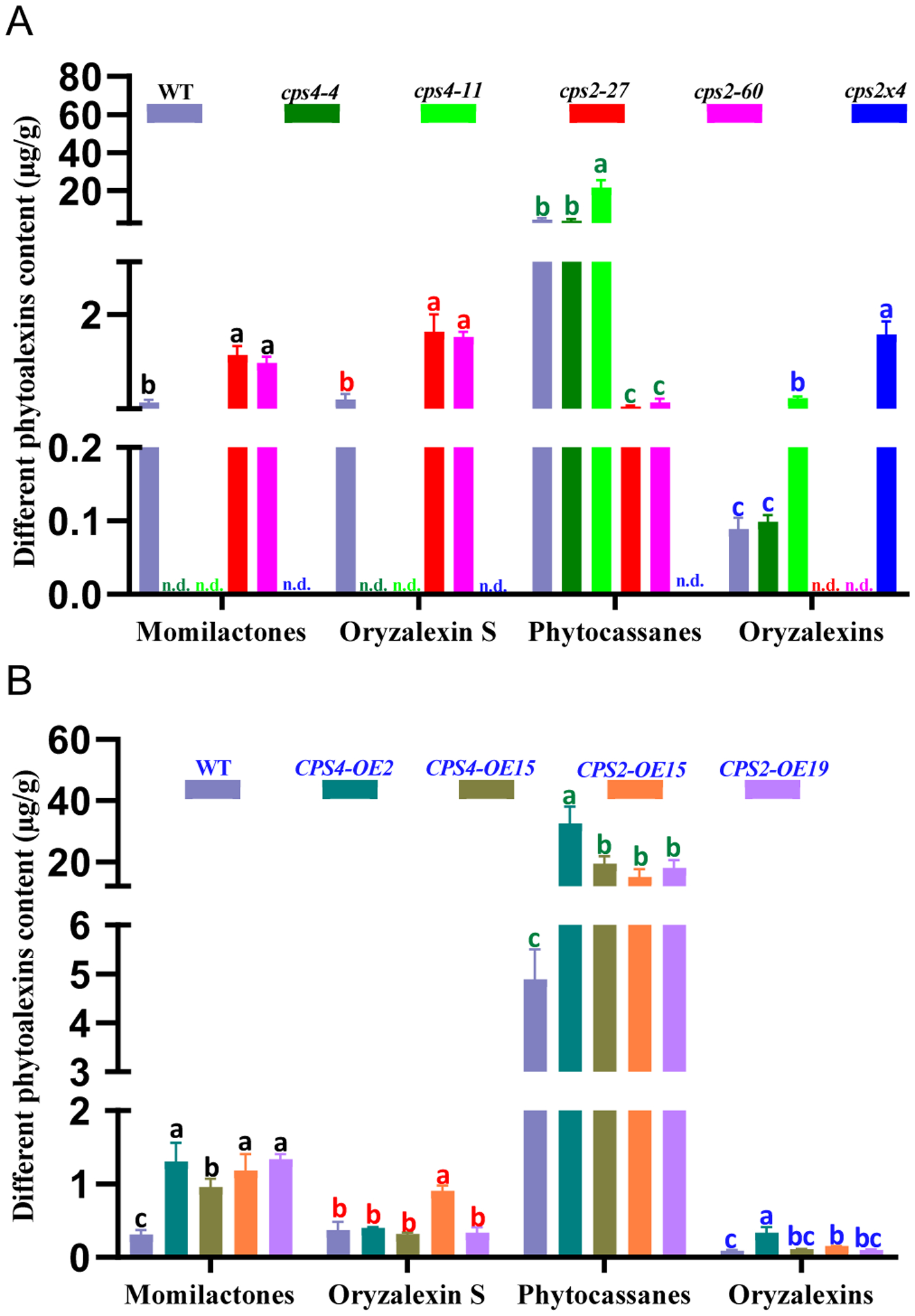
Effect of genetically manipulating CPSs in rice cultivar Kitaake on labdane-related diterpenoid (LRD) production. Histograms indicating LRD accumulation in induced leaf tissue (values reflect peak area for indicated LRD relative to that of the internal standard) from (A) wild-type (WT) and mutants (cps2, cps4 and cps2×4), or (B) WT and over-expression (OE) (CPS2-OE and CPS4-OE) plants assayed together (n = 3, error bars indicate standard deviation). Significant differences within each group of LRDs are indicated by different letters of the same color (P<0.01; using two-sided Fisher’s LSD). Three independent experiments were performed with similar results.
Effect on susceptibility to rice bacterial leaf blight
It was previously reported that, relative to the relevant parental/WT cultivar, the cv. Nipponbare cps2t mutant is more susceptible to the rice bacterial leaf blight pathogen Xoo, while the cv. Zhonghua 11 cps4t mutant is more resistant (Lu et al., 2018). However, the WT cultivars also exhibit significantly different susceptibility to Xoo, with cv. Zhonghua 11 exhibiting higher resistance than cv. Nipponbare or cv. Kitaake (Supplemental Figure S3). In addition, qRT-PCR analysis demonstrated that transcript levels of both OsCPS2 and OsCPS4 are increased in response to Xoo infection in cv. Kitaake (Supplemental Figure S4). To more directly compare the intriguing differential effect of knocking-out OsCPS2 versus OsCPS4, susceptibility of the cv. Kitaake mutants to infection by Xoo was investigated here. Consistent with the previously reported results, relative to WT cv. Kitaake the cps2 lines were significantly more susceptible to Xoo, with ~50% increases in bacterial numbers, while the cps4 lines exhibited increased resistance, with ~4-fold decreases in bacterial numbers, as measured by colony forming units (Figure 3A). Notably, the cps2×4 double-mutant is somewhat more susceptible to Xoo relative to WT cv. Kitaake than the cps2 lines, with ~3-fold increases in bacterial numbers (Figure 3B). Consistent with the observed overall increase in metabolic allocation towards LRDs with over-expression of either OsCPS2 or OsCPS4, all of the CPS2-OE and CPS4-OE lines are dramatically more resistant to Xoo relative to WT cv. Kitaake, with >10-fold (up to ~250-fold) decreases in bacterial numbers (Figure 3C).
Figure 3.
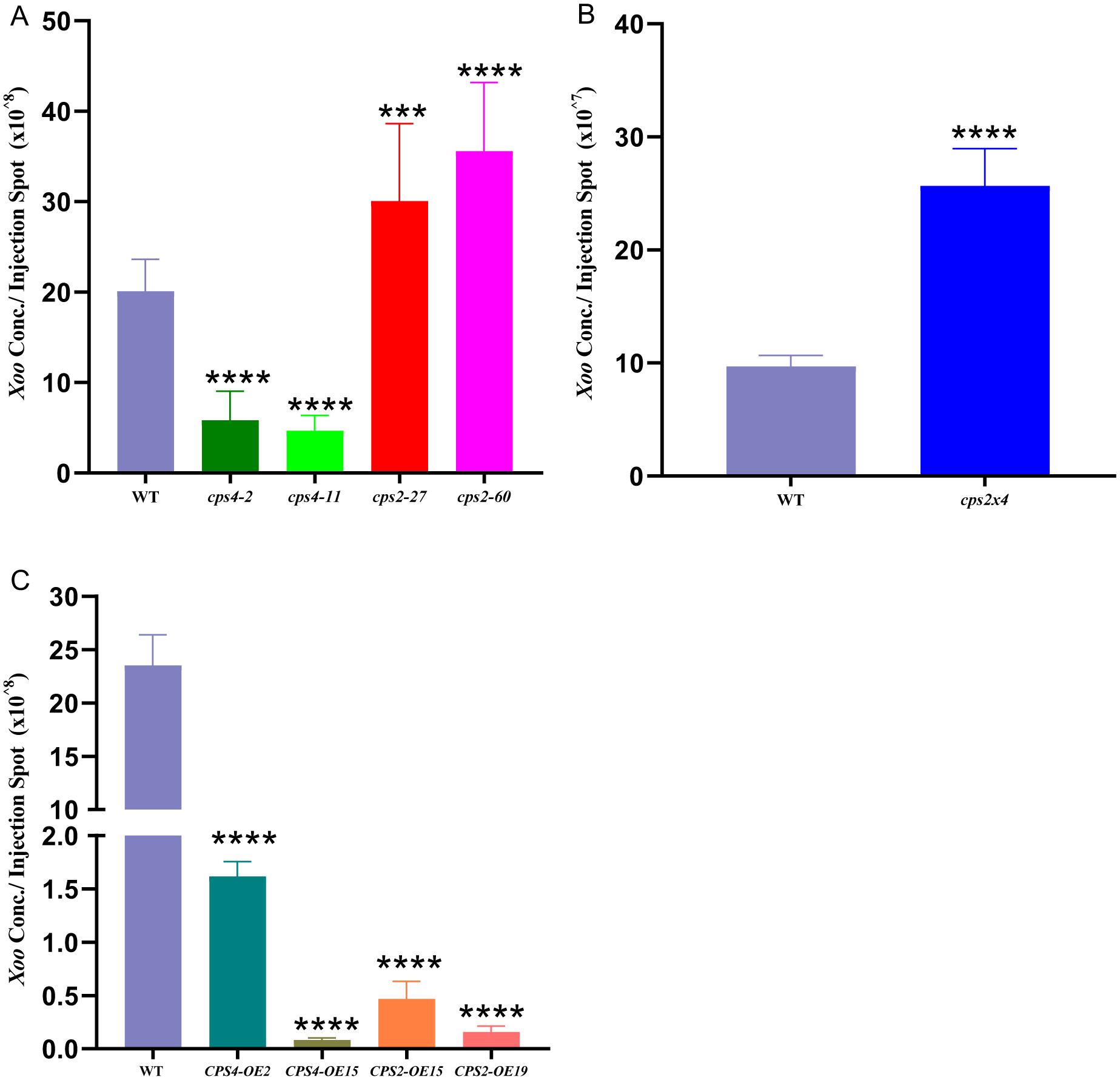
Effect of genetically manipulating CPSs in rice cultivar Kitaake on susceptibility to Xanthomonas oryzae pathovar oryzae (Xoo). Histograms indicating Xoo colony forming units found in infected leaves from (A) wild-type (WT), cps2 and cps4, or (B) WT and cps2×4, or (C) WT, CPS2-OE and CPS4-OE. The data are means ± SD of nine individual replicates, error bars indicate standard deviation. Asterisks represent significant differences derived from one-way ANOVA followed by Dunnett’s multiple comparisons tests for A and C, unpaired two tailed Student’s t-test for B to compare the different lines against the wildtype (***P < 0.001, ****P < 0.0001). Three independent experiments were performed with similar results.
Drought stress induces rice LRD biosynthesis
Based on the demonstrated role of ZmCPS2/An2 in maize drought tolerance (Vaughan et al., 2015), the role of the rice CPSs was investigated here. Intriguingly, qRT-PCR analysis demonstrated that both OsCPS2 and OsCPS4 transcript levels were increased by drought stress in cv. Kitaake, particularly in the roots (Figure 4A). This encouraged analysis of LRD biosynthesis in response to drought stress, which revealed LRD production in the roots, although not leaves, with increases observed relative to well-watered plants within three days of withholding water (Figure 4B).
Figure 4.
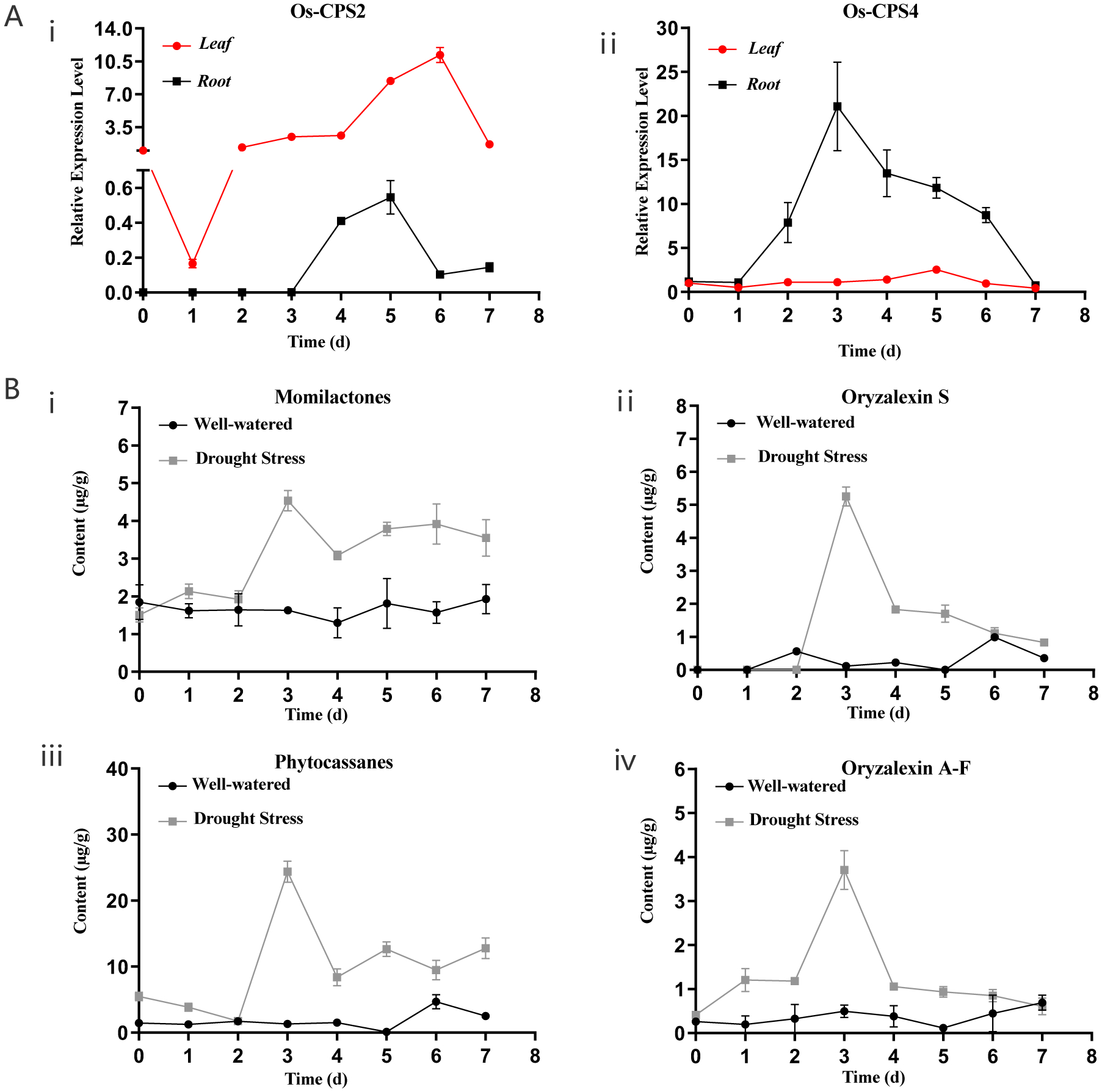
Drought stress induces LRD biosynthesis in rice cultivar Kitaake. Graphs indicating relative levels over time following withholding of water for (A) mRNA in root (black) and leaves (red) for (i) OsCPS2 or (ii) OsCPS4; or (B) in roots of (i) momilactones, (ii) oryzalexin S, (iii) phytocassanes, or (iv) oryzalexins (n = 3, error bars indicate standard deviation). Three independent experiments were performed with similar results.
CPS knock-out lines exhibit increased susceptibility to drought
Strikingly, upon withholding water plants from the cv. Kitaake cps mutant lines exhibited an increased rate of wilting, reflecting reduced leaf water content, relative to WT (Figure 5A–C). However, there was no significant difference in biomass, either following such drought treatment or with normal watering (Supplemental Figure S5). In addition, despite the reduced leaf water content, the cv. Kitaake cps mutant lines exhibited increased transpiration during drought stress treatment, as simple weighing of the pots in which the plants were growing revealed increased rates of water loss relative to WT, which was evident as early as two days after water was withheld (Figure 5D&E), with no significant differences observed in the watered control pots (Supplemental Figure S6). By contrast, the OsCPS2- and OsCPS4- OE lines did not show any change in susceptibility to drought (Supplemental Figure S7).
Figure 5.
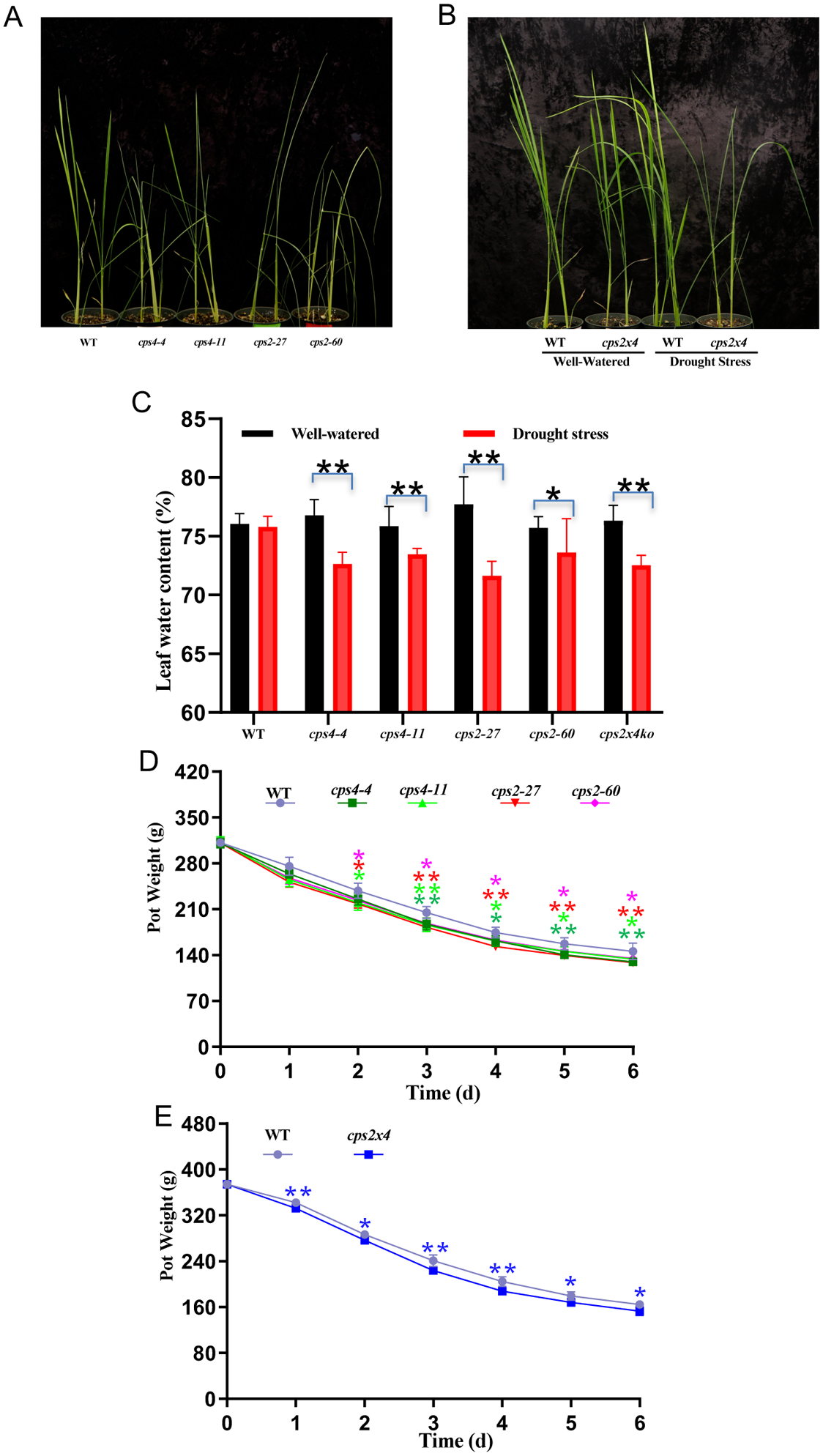
Effect of rice cultivar Kitaake cps mutants on drought resistance. (A and B) Representative pictures of (A) wild-type (WT), cps2 and cps4 mutant plants after drought stress treatment (withholding of water for 6 days) and (B) WT and cps2×4 plants following drought stress treatment as well as controls (continued watering). (C) Analysis of leaf water content following drought stress treatment (n = 6, error bars show standard deviation; *P < 0.05 and **P < 0.01, calculated using unpaired two-tailed Student’s t-test). Four independent experiments were performed with similar results. (D and E) Analysis of water loss during drought stress treatment from pots with (D) WT, cps2 or cps4 plants or (E) WT and cps2×4 plants (n = 5, error bars indicate standard deviation). Asterisks represent significant differences derived from one-way ANOVA followed by Dunnett’s multiple comparisons tests to compare the different lines against WT at each time point (*P < 0.05 and **P < 0.01; colors correlated to that for each cps mutant line). Four independent experiments were performed with similar results.
A role for OsCPS2 and OsCPS4 in Rice Stomatal Closure
The rate of transpiration largely depends on stomata (Nilson and Assmann, 2007). Thus, the stomatal density and aperture status (Figure 6A) of WT cv. Kitaake and the cps mutants were examined via scanning electron microscopy at several time points during the drought stress treatment (0, 3 and 6 days after withholding water). There were no significant differences in stomatal density between WT and the cps mutants before withholding water or even after three days of drought stress, at which point differences in transpiration rate were already evident. Although the cps mutants exhibited small increases in stomatal density after six days of drought stress (Supplemental Figure S8), this was likely a response to the decreased water content of their leaves as moderate water deficits have previously been shown to induce stomatal formation in grasses (Xu & Zhou, 2008). Strikingly, all of the cps mutants exhibited greater numbers of open stomata, with significantly more (~4-fold) observed to be fully open even prior to the withholding of water (Figure 6B). In addition, while drought stress treatment induced more stomatal closure in WT cv. Kitaake, this was not observed with the cps mutants (Figure 6, c.f., B, C and D).
Figure 6.
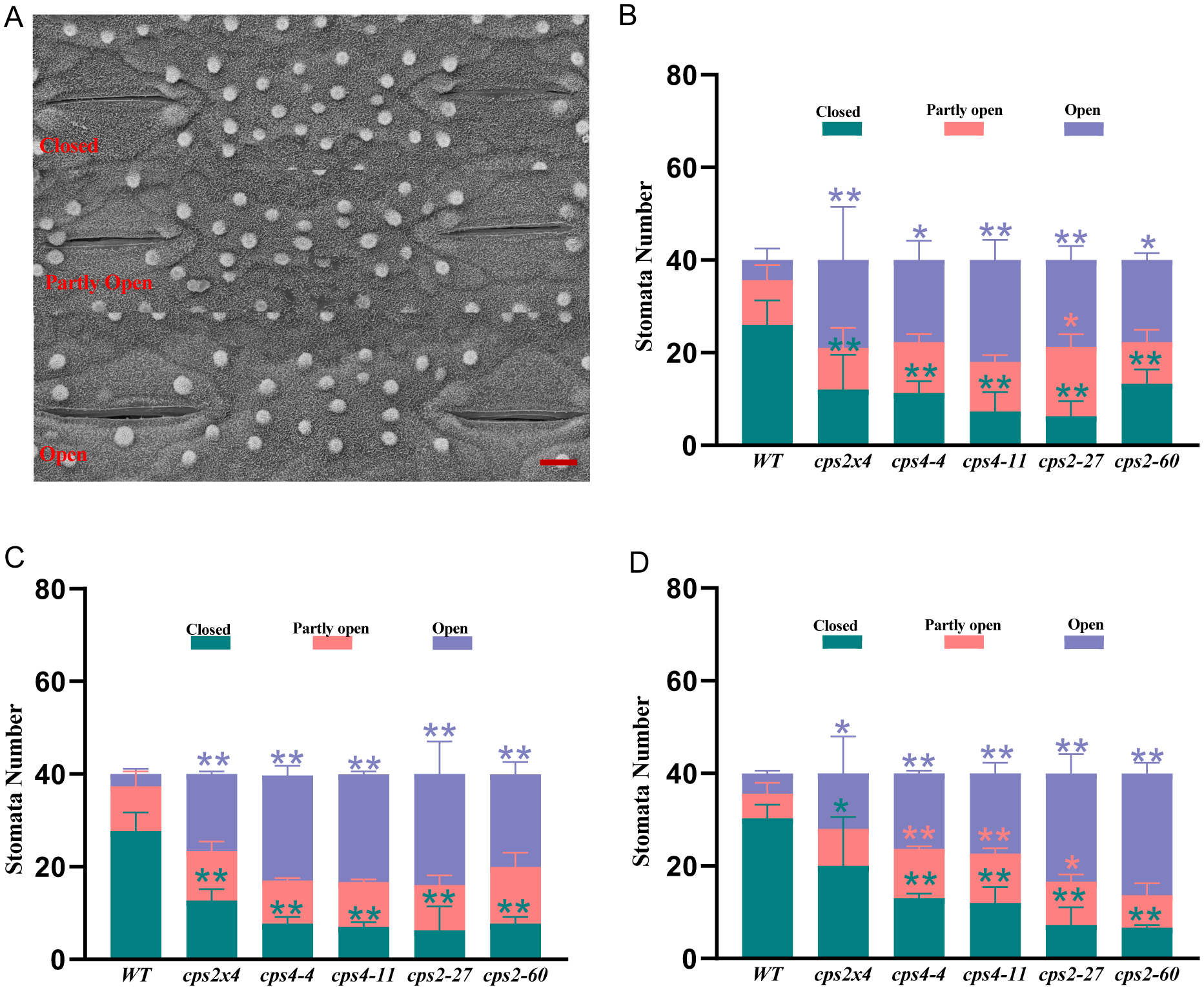
Effect of rice cultivar Kitaake cps mutants on stomata aperture. (A) SEM image representing closed, partially open and open stomata (bar = 6 μ). (B – D) Histograms indicating the number of stomata falling into each of these aperture statuses for wild-type (WT), cps2, cps4 and cps2×4 plants grown together (B) before, and (C) three or (D) six days after, withholding water (n = 3, error bars indicate standard deviation). Significant differences within each aperture status are indicated by different asterisks of the same color, which were derived from one-way ANOVA followed by Dunnett’s multiple comparisons tests to compare the different lines against WT at each time point (*P < 0.05 and **P < 0.01; colors correlated to aperture status). Two independent experiments were performed with similar results.
LRDs act independently of ABA on stomatal closure
The role of abscisic acid (ABA) in triggering stomatal closure is well known (Murata et al., 2015). Thus, phytohormone analysis was carried out with WT cv. Kitaake and the cps mutants at the same time points during the drought stress treatment as for stomatal analysis (i.e., 0, 3 and 5 days after withholding water). Notably, all lines exhibited significant increases in ABA level during the drought stress treatment (Figure 7). This indicates that the defect in stomatal closure in response to drought stress is either downstream or independent of ABA. Indeed, the even larger increases in ABA levels observed in the cps mutants relative to WT indicates appropriate induction of this hormone in response to their earlier wilting. Moreover, the lack of uniform difference in ABA levels before the onset of drought stress contrasts with the observed increase in open stomata even under well-watered conditions for the cps mutants, suggesting the more specific hypothesis that the OsCPS2 and OsCPS4 dependent LRDs act independently of ABA to induce stomatal closure. Given the lack of uniform difference in levels of the other analyzed phytohormones (Supplemental Figure S9), it appears that the observed defect in stomatal closure cannot be attributed to these either (i.e., jasmonates, salicylic acid or indole-acetic acid).
Figure 7.
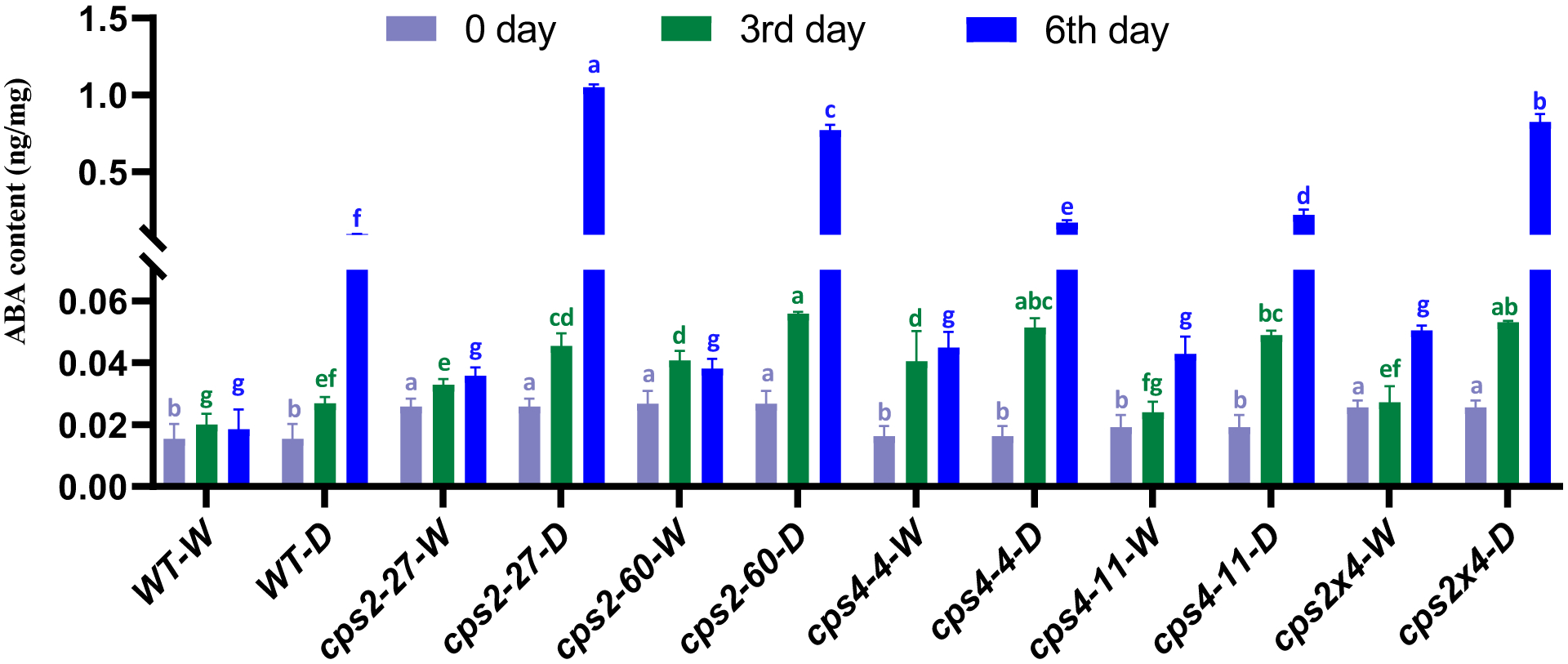
Effect of rice cultivar Kitaake cps mutants on abscisic acid (ABA) levels in response to drought stress treatment either before, three or six days after withholding water (n = 6, error bars indicate standard deviation). Significant differences within each time point are indicated by different letters of the same color (P<0.01; using two-sided Fisher’s LSD).
Biotic and abiotic stress resistance varies with genetic background
While OsCPS2 and OsCPS4 clearly appear to play a role in drought resistance via regulation of stomatal closure in cv. Kitaake, the available cv. Nipponbare cps2t and cv. Zhonghua 11 cps4t mutants were found to be no more susceptible to drought stress than their parental/WT cultivars (Supplemental Figure S10). However, it also was found that cv. Kitaake is significantly more susceptible to drought than either cv. Nipponbare or cv. Zhonghua 11 (Figure 8A), with wilting occurring three days earlier. In addition, comparison of induced LRD accumulation found that cv. Zhonghua 11 produces significantly more LRDs than either cv. Nipponbare or cv. Kitaake (Figure 8B), although it is interesting that the cv. Nipponbare cps2t mutant makes similar amounts of the more prevalent phytocassanes as WT cv. Kitaake (Supplemental Figure S11). Notably, their relative susceptibility to Xoo infection was inversely related to induced LRD accumulation, with cv. Zhonghua 11 exhibiting significantly increased resistance relative to cv. Nipponbare and cv. Kitaake (Supplemental Figure S3). In addition, it was further observed that Xoo infection had differential effect on induction of OsCPS2 versus OsCPS4 in these three cultivars, with greater induction of OsCPS2 but little to none of OsCPS4 in cv. Zhonghua 11, while the converse is found in cv. Nipponbare (i.e., induction of OsCPS4 but not OsCPS2), and both are induced in cv. Kitaake (Supplemental Figure S12), which matches the more effective nature of the OsCPS2-dependent than OsCPS4-dependent LRDs that was verified here (Figure 3).
Figure 8.
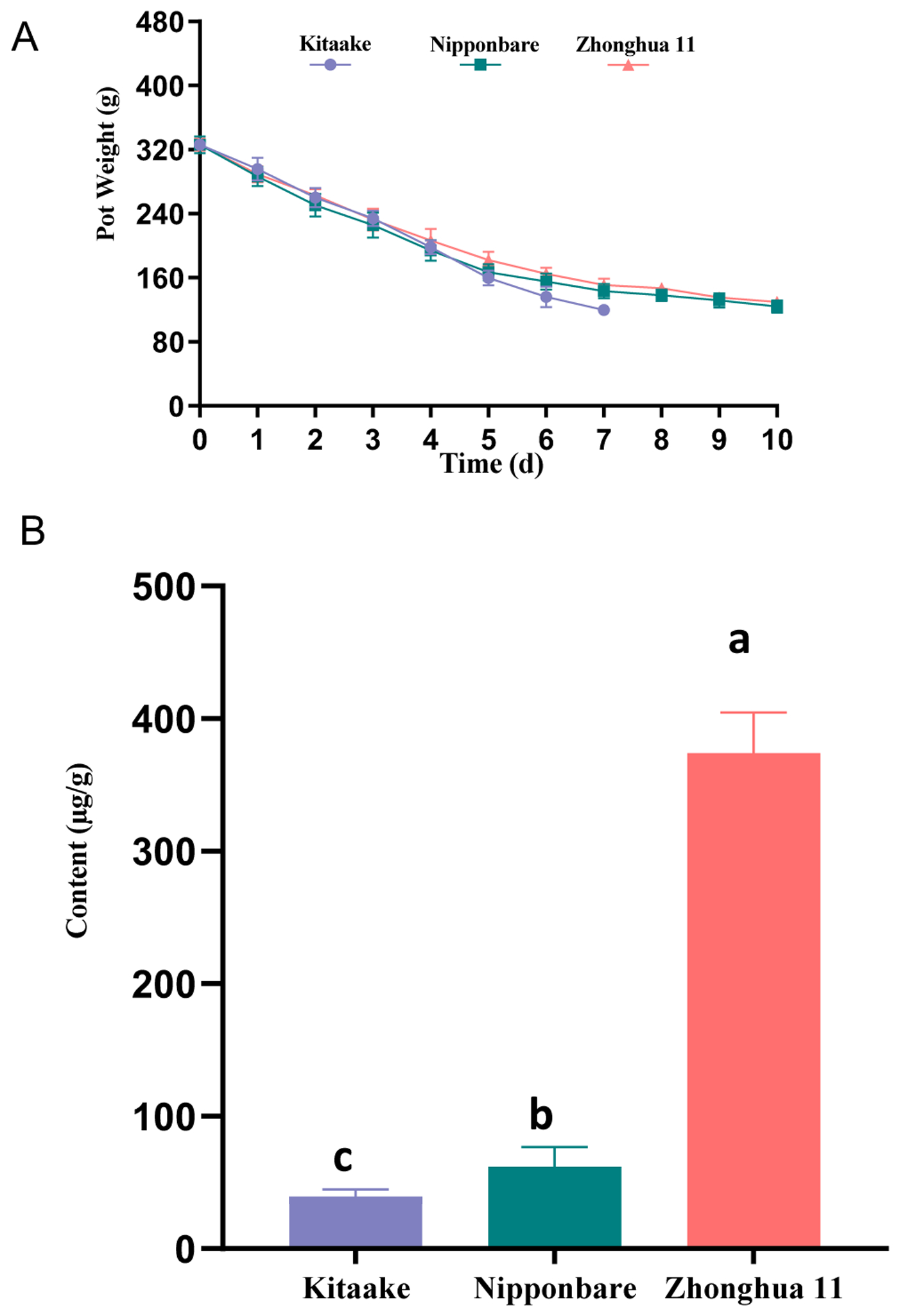
Variation between rice cultivars (cv.). Comparison of cv. Kitaake, cv. Nipponbare and cv. Zhonghua 11. (A) Graph indicating pot weights over time following withholding of water (n = 5, error bars indicate standard deviation). Four independent experiments were performed with similar results. (B) Histogram indicating labdane-related diterpenoid (LRD) levels in root exudates, with values from peak area for all known LRDs relative to that of the internal standard (n = 3, error bars indicate standard deviation). Significant differences are indicated by different letters (p<0.01; using Fisher’s LSD test). Three independent experiments were performed with similar results.
Discussion
Upon selecting cv. Kitaake for comprehensive investigation of rice LRD metabolism it seemed prudent to re-investigate the effect of knocking-out OsCPS2 and OsCPS4. Indeed, relative to what was previously reported for the cv. Nipponbare cps2t mutant (Lu et al., 2018), the cv. Kitaake cps2 mutant lines exhibited a greater impact on ent-CPP derived LRD production. In particular, while both significantly reduce production of oryzalexins A – F relative to their respective parental/WT cultivar, only in the cv. Kitaake cps2 lines is production of the more prevalent phytocassanes also significantly reduced (Figure 2A). By contrast, the cv. Kitaake cps2×4 double-mutant produces significantly more oryzalexins. Given that all of these cps2 mutants also exhibit the same increased susceptibility to Xoo (Figure 3, A and B), this result supports the hypothesis that it is the ent-CPP derived oryzalides & related LRDs that act as effective phytoanticipins against Xoo (i.e., rather than phytocassanes and/or oryzalexins A – F). However, it remains possible that other ent-CPP derived LRDs might serve as antibiotics against Xoo (i.e., the unknown elaborated natural products derived from ent-pimara8(14),15-diene and/or ent-beyerene; see Figure 1), instead of, or in addition to, the oryzalides & related LRDs.
Given this hypothesis, it might be considered surprising that both OsCPS2- and OsCPS4- OE lines exhibit increased resistance to Xoo (Figure 3C). However, all of these lines exhibit increased production of both syn- and ent- CPP derived LRDs (Figure 2B). Speculatively, this might stem from increased production of GGPP, potentially as a response to the metabolic drain imposed by CPS over-expression on other GGPP-derived essential metabolites such carotenoids, phytol, etc., enabling increased production of all LRDs, including the ent-CPP derived more effective antibiotics against Xoo.
Perhaps most intriguing is the observed increased susceptibility to drought for all the cv. Kitaake cps mutants (Figure 5). While a similar effect was reported for the maize an2 mutant, those results were complicated by the observed reduction in root biomass. By contrast, the rice cv. Kitaake cps mutants do not exhibit any consistent effect on biomass. Instead, these were found to exhibit a defect in stomatal closure, even in the absence of drought stress (Figure 6). Given that OsCPS2 and OsCPS4 serve in more specialized metabolism, it appears that the resulting LRD natural products promote stomatal closure. Based on the appropriate induction of ABA in response to drought stress and lack of uniform increase under well-watered conditions where significantly higher proportions of stomata are open in the cps mutants than WT, it appears that the LRD natural products act independently of the ABA signaling pathway.
Notably, based on the equivalent ratios of stomatal aperture status observed with the cps2×4 double-mutant, as well as lack of effect on drought resistance in the CPS OE lines, the LRDs seem to operate as a regulatory switch (i.e., simply requiring the presence of sufficient amounts to exceed a set threshold). This hypothesis may pertain to the observation that the effect of knocking-out the CPSs on drought resistance appears to be specific to cv. Kitaake. In particular, cv. Zhonghua 11 naturally produces much higher amounts of LRDs (Figure 7B), which then might exceed such a threshold even in the absence of OsCPS4, leading to the lack of observed effect for the cv. Zhonghua 11 cps4t mutant on drought resistance. Similarly, although cv. Nipponbare does not make significantly more LRDs than cv. Kitaake, the cv. Nipponbare cps2t mutant has a much smaller effect on production of phytocassanes, the most prevalent ent-CPP derived LRDs, than observed with the cv. Kitaake cps2 lines. Indeed, the cv. Nipponbare cps2t mutant makes equivalent amounts of phytocassanes as WT cv. Kitaake (Supplemental Figure S9), which is then presumably sufficient to meet the threshold. Accordingly, the potential role for LRDs to serve as a regulatory switch promoting stomatal closure may be more widespread in rice.
Given the hypothesis that OsCPS2- and OsCPS4-dependent LRDs serve a regulatory role in promoting stomatal closure, which is observed even in the absence of drought stress, this implies a separate function for the observed induction of LRD biosynthesis (Figure 4). Indeed, such induction is only observed in roots, with no increased accumulation of LRDs in the leaves. As suggested for the root accumulation of LRDs observed in maize, such selective induction is consistent with the optimal defense theory (Vaughan et al., 2015). In particular, this postulates that such production of antibiotics in roots under drought stress conditions reflects the vital role of these organs in obtaining the limiting factor (i.e., water).
In conclusion, the results reported here indicate that LRDs play a role in triggering stomatal closure in rice, providing insight into water use efficiency in this critically important crop plant. Given that stomata provide opening for microbial entry, it can be speculated that stomatal closure in response to phytoalexins might have evolved as a mechanism to deny pathogens an opportunity to gain access to the plant interior. Regardless of such speculation, these results demonstrate another important role for LRD natural products in rice.
Supplementary Material
Supplemental Figure S1. Rice cultivar Kitaake cps mutants exhibit normal growth.
Supplemental Figure S2. Selection of rice cultivar Kitaake CPS over-expression (OE) lines.
Supplemental Figure S3. Variation between rice cultivars in susceptibility to Xanthomonas oryzae pathovar oryzae (Xoo).
Supplemental Figure S4. Induction of rice CPSs by Xanthomonas oryzae pathovar oryzae (Xoo) infection in rice cultivar Kitaake.
Supplemental Figure S5. Rice cultivar Kitaake cps mutants do not affect biomass.
Supplemental Figure S6. Equivalent water loss for rice cultivar Kitaake cps mutants under well-watered conditions.
Supplemental Figure S7. Rice CPS over-expression (OE) does not affect drought resistance.
Supplemental Figure S8. Rice cultivar Kitaake cps mutants do not affect stomata density.
Supplemental Figure S9. Rice cultivar Kitaake cps mutants do not uniformly affect other phytohormone levels in response to drought stress treatment.
Supplemental Figure S10. Rice cultivar (cv.) Nipponbare cps2t and cv. Zhonghua 11 cps4t mutants do not affect drought resistance.
Supplemental Figure 11. Contrasting effect of rice cultivar (cv.) Kitaake (Kit) cps2 and cv. Nipponbare (Ni) cps2t relative to their respective wild-type (WT) on phytocassanes level in root exudates.
Supplemental Figure S12. Induction of rice CPSs by Xanthomonas oryzae pathovar oryzae (Xoo) infection varies between cultivars.
Supplemental Table S1. Genotypes from CRISPR/Cas9 mutagenesis.
Supplemental Table S2. Primers used in this study.
Acknowledgements
This work was supported by grants from the NIH (GM131885 to R.J.P.), and USDA-NIFA (2020-67013-32557 to R.J.P. and B.Y.), as well as a fellowship from the International Postdoctoral Exchange Fellowship Program (20170057 to J.Z.).
References
- Bednarek P, Osbourn A. 2009. Plant-microbe interactions: chemical diversity in plant defense. Science 324: 746–748. [DOI] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. 1995. Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Yang B. 2017. Gene Editing With TALEN and CRISPR/Cas in Rice. Prog Mol Biol Transl Sci 149: 81–98. [DOI] [PubMed] [Google Scholar]
- Bostock RM, Pye MF, Roubtsova TV. 2014. Predisposition in plant disease: exploiting the nexus in abiotic and biotic stress perception and response. Annu Rev Phytopathol 52: 517–549. [DOI] [PubMed] [Google Scholar]
- Gupta A, Rico-Medina A, Cano-Delgado AI. 2020. The physiology of plant responses to drought. Science 368: 266–269. [DOI] [PubMed] [Google Scholar]
- Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ. 2005. The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol. Biol 59: 881–894. [DOI] [PubMed] [Google Scholar]
- Jain R, Jenkins J, Shu S, Chern M, Martin JA, Copetti D, Duong PQ, Pham NT, Kudrna DA, Talag J, et al. 2019. Genome sequence of the model rice variety KitaakeX. BMC Genomics 20: 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Noguchi H, Peters RJ. 2013. The role of momilactones in rice allelopathy. J. Chem. Ecol 39: 175–185. [DOI] [PubMed] [Google Scholar]
- Kim SL, Choi M, Jung KH, An G. 2013. Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J Exp Bot 64: 4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka N, Zhang J, Oyagbenro RK, Brown B, Wu Y, Yang B, Li Z, Peters RJ. 2020. Interdependent evolution of biosynthetic gene clusters for momilactone production in rice. Plant Cell DOI: 10.1093/plcell/koaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang J, Brown B, Li R, Rodriguez-Romero J, Berasategui A, Liu B, Xu M, Luo D, Pan Z, et al. 2018. Inferring Roles in Defense from Metabolic Allocation of Rice Diterpenoids. Plant Cell 30: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafu S, Ding Y, Murphy KM, Yaacoobi O, Addison JB, Wang Q, Shen Z, Briggs SP, Bohlmann J, Castro-Falcon G, et al. 2018. Discovery, Biosynthesis and Stress-Related Accumulation of Dolabradiene-Derived Defenses in Maize. Plant Physiol 176: 2677–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Mori IC, Munemasa S. 2015. Diverse stomatal signaling and the signal integration mechanism. Annu Rev Plant Biol 66: 369–392. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Zerbe P. 2020. Specialized diterpenoid metabolism in monocot crops: Biosynthesis and chemical diversity. Phytochemistry 172: 112289. [DOI] [PubMed] [Google Scholar]
- Muthayya S, Sugimoto JD, Montgomery S, Maberly GF. 2014. An overview of global rice production, supply, trade, and consumption. Ann N Y Acad Sci 1324: 7–14. [DOI] [PubMed] [Google Scholar]
- Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. 2004. Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J. 39: 886–893. [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. 2010. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992. [DOI] [PubMed] [Google Scholar]
- Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M. 2017. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front Plant Sci 8: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ. 2006. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317. [DOI] [PubMed] [Google Scholar]
- Peters RJ. 2010. Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep 27: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ. 2004. Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136(4): 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. 2004. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134: 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ. 2014. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79: 659–678. [DOI] [PubMed] [Google Scholar]
- Toyomasu T 2008. Recent Advances Regarding Diterpene Cyclase Genes in Higher Plants and Fungi. Biosci. Biotechnol. Biochem 72: 1168–1175. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Usui M, Sugawara C, Otomo K, Hirose Y, Miyao A, Hirochika H, Okada K, Shimizu T, Koga J, et al. 2014. Reverse-genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiol Plant 150: 55–62. [DOI] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE. 1994. Two classes of plant antibiotics: phytoalexins versus ‘phytoanticipins’. Plant Cell 6: 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Christensen S, Schmelz EA, Huffaker A, McAuslane HJ, Alborn HT, Romero M, Allen LH, Teal PE. 2015. Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 38: 2195–2207. [DOI] [PubMed] [Google Scholar]
- Xu M, Galhano R, Wiemann P, Bueno E, Tiernan M, Wu W, Chung IM, Gershenzon J, Tudzynski B, Sesma A, et al. 2012. Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol. 193: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ. 2004. Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39: 309–318. [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya N, Coates RM, Peters RJ. 2007. Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68: 312–326. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhou G. 2008. Responses of leaf stomatal density to water status and its relationship with photosynthesis in a grass. J Exp Bot 59: 3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan TD, Minh TN, Anh LH, Khanh TD. 2016. Allelopathic momilactones A and B are implied in rice drought and salinity tolerance, not weed resistance. Argon. Sustain. Dev 36: 52. [Google Scholar]
- Zhang J, Zhang S, Cheng M, Jiang H, Zhang X, Peng C, Lu X, Zhang M, Jin J. 2018. Effect of Drought on Agronomic Traits of Rice and Wheat: A Meta-Analysis. Int J Environ Res Public Health 15: 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ. 2014. To Gibberellins and Beyond! Surveying the Evolution of (Di)Terpenoid Metabolism. Annu. Rev. Plant Biol 65: 259–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Rice cultivar Kitaake cps mutants exhibit normal growth.
Supplemental Figure S2. Selection of rice cultivar Kitaake CPS over-expression (OE) lines.
Supplemental Figure S3. Variation between rice cultivars in susceptibility to Xanthomonas oryzae pathovar oryzae (Xoo).
Supplemental Figure S4. Induction of rice CPSs by Xanthomonas oryzae pathovar oryzae (Xoo) infection in rice cultivar Kitaake.
Supplemental Figure S5. Rice cultivar Kitaake cps mutants do not affect biomass.
Supplemental Figure S6. Equivalent water loss for rice cultivar Kitaake cps mutants under well-watered conditions.
Supplemental Figure S7. Rice CPS over-expression (OE) does not affect drought resistance.
Supplemental Figure S8. Rice cultivar Kitaake cps mutants do not affect stomata density.
Supplemental Figure S9. Rice cultivar Kitaake cps mutants do not uniformly affect other phytohormone levels in response to drought stress treatment.
Supplemental Figure S10. Rice cultivar (cv.) Nipponbare cps2t and cv. Zhonghua 11 cps4t mutants do not affect drought resistance.
Supplemental Figure 11. Contrasting effect of rice cultivar (cv.) Kitaake (Kit) cps2 and cv. Nipponbare (Ni) cps2t relative to their respective wild-type (WT) on phytocassanes level in root exudates.
Supplemental Figure S12. Induction of rice CPSs by Xanthomonas oryzae pathovar oryzae (Xoo) infection varies between cultivars.
Supplemental Table S1. Genotypes from CRISPR/Cas9 mutagenesis.
Supplemental Table S2. Primers used in this study.


