Abstract
Metastasis is the leading cause of mortality from kidney cancer, and understanding the underlying mechanism of this event will provide better strategies for its management. Here we investigated the biological, functional, and clinical significance of lncTCL6 and its interacting miR-155 in clear cell renal cell carcinoma (ccRCC). We employed a comprehensive approach to investigate the lncTCL6-miR155-Src/Akt-mediated EMT pathway as a novel regulatory mechanism in ccRCC progression. Expression analyses revealed that lncTCL6 is downregulated in ccRCC compared to normal tissues. Overexpression of lncTCL6 in ccRCC cell lines impaired their oncogenic functions such as cell proliferation and migration/invasion and induced cell cycle arrest and apoptosis; conversely, depletion of lncTCL6 rescued these phenotypic effects. Furthermore, lncTCL6 directly interacted with miR-155. Unlike lncTCL6, miR-155 was overexpressed in ccRCC. Stable knockdown of miR-155 phenocopied the effects of lncTCL6 overexpression. Conversely, reconstitution of miR-155 and suppression of lncTCL6 in non-cancerous renal cell HK2 induced tumorigenic characteristics. Patients with higher expression of lncTCL6 and lower expression of miR-155 had better survival probability. When overexpressed, lncTCL6 recruited STAU1 and mediated decay of Src mRNA, followed by a marked downregulation of an integrated network of Src target genes involved in migration, invasion, and EMT. However, the interaction between miR-155 and lncTCL6 attenuated the regulatory role of lncTCL6 on Src-mediated EMT. In conclusion, this study is the first report documenting the lncTCL6-miR155-Src/Akt/EMT network as a novel regulatory mechanism in aggressive ccRCC and a promising therapeutic target to inhibit renal cancer.
Keywords: lncTCL6, miR155, Src, Akt, EMT, renal cancer
Introduction-
Each year 250,000 new cases of renal cancer are diagnosed worldwide, accounting for 2% of all cancers (1), and ranking highest in incidence of urological cancers. Clear cell renal cell carcinoma (ccRCC), is the most common subtype of renal cancer (2). Current therapies for renal cancer include conventional chemotherapy and radiation. Moreover, ccRCC metastasis is a significant obstacle for the substantial treatment of this disease (3). During metastasis, the tumor cells migrate from their original site to distant organs. The process of metastasis involves cell adhesion, invasion (4). Despite the significance of metastasis to ccRCC survival and morbidity, little is known about the cellular and molecular mechanisms mediating ccRCC metastasis. Few ccRCC metastatic biomarkers have been identified compared to other cancers. Therefore, identification of sensitive and specific biomarkers, as well as therapeutic targets for ccRCC metastasis urgently deserves attention.
The non-receptor tyrosine kinase Src is an important downstream effector of EGFR that is predominantly involved in renal cancer invasion (5,6). Its activation has been established as a poor prognostic factor in several types of cancers, and its contribution to the appearance of malignant phenotypes in renal cancer cells has also been reported (7). Accumulating evidence suggests that Src interacts with and stimulates the PI3K/Akt pathway in cancer cells (8,9).
Long non-coding RNA, defined as transcripts of more than 200 nucleotides that generally do not encode proteins, have been associated with diverse functions. To date, most work has been limited to understanding the function of lncRNAs located in the cell nucleus, and less is known about cytoplasmic lncRNAs and their mode of action. Cytoplasmic lncRNAs, act as ‘sponge’ for miRNA and are involved in post-transcriptional regulation affecting mRNA stability or accessibility to the translational machinery (10). lncRNATCL6 (lncTCL6) was first identified in T-cell leukemia located near the TCL1B protein-coding gene on chromosome 14q32.1. Recently, lncTCL6 was reported to be an independent predictor of ccRCC aggressiveness (11–15). Although the molecular function of lncTCL6 in renal cancer is unknown, it may modulate the EGFR/AKT pathway in placental tissue (16).
MicroRNAs (miRNAs) are a widely studied subclass of small ncRNAs, that post-transcriptionally regulate the expression of multiple genes through imperfect complementarity to their target mRNA transcripts (17). Recent studies have demonstrated the regulatory functions of miRNAs in ccRCC cell growth, apoptosis, migration, and invasion (18,19). miR-155 is localized within a genomic region known as B cell integration cluster, and plays important roles in immune response, aberrant cell proliferation and cancer (20). Overexpression of miR-155 has been found in colorectal carcinoma, breast cancer, lymphoma, and renal cancer (21–25). Further, understanding the mechanistic role of miR-155 in renal cancer metastasis is clinically important, as miR-155 has already been identified as a potential therapeutic target for anti-miRs (26).
Previous studies (27,28) have reported that miRNA-mRNA-lncRNA interaction play important roles in biological processes. However, none of these interaction/ networks have been shown to regulate the Src-Akt pathway that drives the metastasis in renal cancer. Therefore, further research is required to investigate miRNA-mRNA-lncRNA interactions in Src-Akt mediated ccRCC metastasis.
Here we report that lncTCL6 is significantly downregulated whereas miR-155–5p (hence forth referred as miR-155) is significantly upregulated in renal cancer tissues and cell lines. Low lncTCL6 and high miR-155 expression positively correlates with poor overall patient survival. In addition, we examined the biological and functional significance of these non-coding RNAs in ccRCC. We also identified that lncTCL6 plays a tumor-suppressive role in renal cancer and that its overexpression inhibits Src-Akt driven metastatic pathway via STAU1 mediated Src mRNA decay. For the first time, this study shows that lncTCL6 and miR-155 are potential biomarkers that regulate the Src-Akt mediated metastatic renal carcinoma.
Materials and Methods
Human Subjects and cells
Written informed consent was obtained from all patients, and the study was in accordance with institutional guidelines (Institutional review board approval no: 16–18555). Formalin-fixed, paraffin-embedded tissue blocks of radical nephrectomy specimens were obtained from the Pathology Department of the Veterans Affairs (VA) Medical Center of San Francisco (VAMC). All slides were reviewed by a board-certified pathologist for the identification of malignant and adjacent normal tissues. Tissues were micro dissected as described earlier (29).
All cell lines were acquired from the ATCC and were authenticated using DNA short -tandem repeat profiling by ATCC. Cells were confirmed mycoplasma free by using MycoFluor Mycoplasma detection kit (ThermoFisher Scientific, cat. # M7006). Normal renal epithelial cells HK-2 (ATCC number: CRL-2190) and renal cancer cell lines Caki-1 (ATCC # HTB-46), 786-O (ATCC # CRL-1932) were grown under recommended conditions. 786-O cells were cultured in RPMI media, Caki-1 in McCoy 5A (modified) media, each supplemented with 10% FBS and 1% penicillin/streptomycin. HK-2 cells were cultured in keratinocyte serum-free media (Gibco/Invitrogen) supplemented with prequalified human recombinant EGF 1–53 and bovine pituitary extract (BPE). All cell lines were maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Cells were passaged every 2–3 days and kept in culture for the duration of the experiments. Fresh batches of cells were used for repeat experiments. Cell line experiments were performed within 5 to 6 months of their procurement/resuscitation.
Establishing stable anti-miR-155 and lncTCL6 overexpressing cell lines
For lncTCL6 overexpression, cells were transfected with 10 μg of plasmid DNA, (TCL6 (NM_020550) Human Tagged ORF Clone), purchased from OriGene, Rockville, MD, USA using JetPRIME (Polyplus transfection, France). After transfection, cells (TCL6-OE, OE-control) were selected using G418 (1mg/ml) for 1–4 weeks. lncTCL6 expression levels were confirmed by qPCR using custom SYBR green probes. Two of the TCL6 overexpression clones- TCL6-OE-clone 1 and TCL6-OE-clone 2 were used for further studies.
The pLenti-III-miR-Off-hsa-miR-155-puro-GFP expression vector and the negative control vector pLenti-III-miR-Off-puro-GFP were purchased from Applied Biological Materials, Inc. (Richmond, CA). Caki-1 and 786-O cells were transfected with lentiviral packaging vectors (ABM, Richmond, CA) and lentiviral vectors expressing miR-Off-hsa-miR-155-puro-GFP or miR-Off-puro-GFP by JetPRIME (Polyplus transfection, France) according to the manufacturer’s protocol. After transfection, cells (anti-miR-155, miR-CON) were observed under a microscope to check for green fluorescence and then selected with 10 μg/mL puromycin for 1–4 weeks. Two of the anti-miR-155 clones- anti-miR-155-clone 1 and anti-miR-155-clone 2 were used for further studies.
TCL6-OE (clone 1) cells were used to carry out transfection experiments and related assays to understand the functional significance of si-TCL6 and siSTAU1 (Sigma Aldrich). The cells were transiently transfected with either 30nmol/L of siRNA or corresponding control using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to manufacturer’s protocol. All experiments were carried out for 72hrs.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from tissue samples and cell lines using a miRNeasy FFPE kit and miRNeasy mini kit (Qiagen,Valencia,CA), respectively according to the manufacturer’s instruction. RNA and miRNA were reverse transcribed into cDNA with the High capacity cDNA reverse transcription kit (Thermo Fisher). Quantitative real-time RT-PCR was performed in duplicate with QuantStudio 7 Flex-Real Time PCR System (Applied Biosystems) using TaqMan MicroRNA Assays and Gene Expression Assays, respectively, in accordance with the manufacturer’s instructions (Applied Biosystems). Human GAPDH, HPRT and RNU48 were used as endogenous controls for gene expression and miRNA, respectively. A QuantiFast SYBR Green PCR Kit was also utilized for gene expression analysis of miR-155/ lncTCL6 targets. The primers used for SYBR Green–based qPCR analyses are listed in Supplementary Table 1. Relative expression of RNA and miRNA were calculated using comparative Ct. The expression levels of miRNA/RNA were calculated as the amount of target miRNA/RNA relative to that of RNU48/ HPRT1/ GAPDH control to normalize the initial input of total RNA. Also, we used mRNA and miRNA expression data from The Cancer Genome Atlas (TCGA) now available at the genomic data commons (GDC) portal (https://portal.gdc.cancer.gov/).
Cell viability and colony formation assay
Cell viability of established stable and their respective control cells as well as cells transiently transfected with siRNAs and miR-155 mimic was determined at 24, 48, and 72 hours using a CellTiter 96 Aqueous Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer’s instructions. This is a colorimetric assay which measures the activity of reductase enzymes. Briefly cells were seeded at a density of 1.2×103 for Caki-1 and 1×103 for 786-O per well in flat bottomed 96-well plates. At indicated times, CellTiter 96 Aqueous One reagent was added to each well and absorbance was measured at 490nm using SpectraMAX 190 (Molecular Device, Sunnyvale, CA).
For colony-forming assays, stable Caki-1 and 796-O anti-miR-155 / TCL6-OE cells and their respective control cells were plated at a low density onto 10cm dishes (1000 cells/plate). These cells were allowed to grow until visible colonies were formed.
Similarly for cells with transient miR-155 overexpression and suppression of lncTCL6, 72 hrs post-transfection, cells were counted and seeded at low density. Following 8 days (786-O) or 12 days (Caki-1) of cell adherence, colonies of cells were fixed with Geimsa stain and further stained with crystal violet reagent. The colonies were counted and recorded.
Cell-cycle and Apoptosis analysis
Stable cells (anti-miR-155 / lnc-TCL6-OE and negative controls) and transiently transfected cells were harvested using accutase (Corning, NY) and washed with cold PBS.
For cell cycle, PBS-washed cells were fixed in cold 70% ethanol overnight at −20°C. Fixed cells were further washed with PBS, stained with PI/RNase Staining Buffer (BD Pharmingen, San Diego, CA) and incubated for 30 minutes at room temperature in the dark. 10,000 gated events/sample were counted and analyzed using BD FACS Verse (BD Pharmingen, San Diego, CA).
For apoptosis, cold PBS-washed cells were resuspended in 1× binding buffer and stained with Annexin V-FITC and 7AAD viability dye as per manufacturer’s instructions (Annexin V-FITC/7AAD kit, Beckman Coulter). Cells were then incubated for 30 minutes at room temperature in the dark. 10,000 gated events/sample were counted and analyzed using BD FACSVerse.
Migration and Invasion assay
Culture inserts of 8-μm pore size (Transwell; Costar, Corning, NY) were placed into the wells of 24-well culture plates and used for migration and invasion assay. For cell invasion chambers were coated with Matrigel (BD Biosciences, San Jose; 100ug/well). Cells (0.5×105 cells/ml for migration and 1×105 cells/ml for invasion) were seeded in serum-free medium in the top chamber inserts and cells could invade into the bottom chambers containing 500ul of culture medium with 10% FBS. For cells with transient miR-155 overexpression and suppression of lncTCL6, 72 hrs post-transfection, cells were counted and seeded for migration and invasion assay.
After 48hrs of incubation at 37°c with 5%CO2 for migration and after 72hrs of incubation at 37°c with 5%CO2 for invasion, cells that migrated or invaded through pores were fixed and then stained with 0.5% crystal violet, later cells were photographed using a light microscope. Crystal violet was then solubilized with methanol and quantified at 540nm by a kinetic microplate reader (Spectra MAX 190; Molecular Devices, Sunnyvale, CA). Percent migration or invasion was analyzed based on the absorbance values.
Dual-Luciferase reporter assay
Luciferase reporter vectors were constructed by ligation with annealed custom oligonucleotides (Supplementary Table 1) containing the putative target binding and off-target sites of lncTCL6 into pmiR-GLO reporter vector (Promega, catalog no. E1330). These plasmids with putative target binding site (wild type) and off-target site (mutant) were used for further experiments. For reporter assay, Caki-1 and 786-O cells were transiently transfected with wild-type or mutant vector DNA (1 ng) along with miRVana miRNA mimic (miR-155 mimic) and miRNA mimic negative control#1 (Thermo Fisher Scientific, 50nmol/L each) using JetPrime transfection reagent (Polyplus-transfection, Illkirch, France). The luciferase activity was measured 72 hours after transfection using a Dual-Luciferase Reporter Assay System (Promega) as per manufacturer’s instructions. Relative luciferase activity was calculated by normalizing to Renilla luminescence.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) was performed to investigate the binding of lncTCL6 and Src mRNAs to STAU1 protein. Cells overexpressing lncTCL6 were harvested, fixed and an imprint RIP kit was used as per manufacturer’s instructions (Sigma-Aldrich). STAU1 and IgG (control) antibodies were used for immunoprecipitation. The RIP RNA fraction was reverse transcribed to cDNA using High capacity cDNA reverse transcription kit (Thermo Fisher). Final analysis was performed using RT-qPCR and shown as fold enrichment of lncTCL6, Src to STAU1 antibody with respect to IgG.
Western blot analysis
Cells were lysed with RIPA buffer (Thermo Scientific) plus Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Protein concentration was determined using BCA Protein Assay (Thermo Fisher Scientific). Total proteins (30–50μg) were separated by NuPAGE 4%–12% Bis-Tris Protein Gels (Invitrogen) and subsequently transferred onto nitrocellulose membrane using wet transfer method. Prior to incubation with 1:1,000-fold diluted primary antibodies overnight at 4°C, blots were blocked in Odyssey blocking buffer (LI-COR) for an hour. The primary antibodies used are listed in Supplementary Table 2. After washing the membranes, either goat anti-rabbit IgG (H+L) 800 W or goat anti-mouse IgG (H+L) 680RD was applied for 60 minutes at room temperature (1:20,000, LI-COR Biosciences). Membranes were again washed with PBS containing Tween 20. Blots were scanned using an Odyssey Infrared Imaging System Scan and quantification was carried out with the LI-COR Odyssey scanner and software (LI-COR Biosciences).
Immunofluorescence
Stable cells overexpressing lncTCL6 and anti-miR-155 along with their respective controls were fixed in 4% paraformaldehyde for 15 minutes. Prior to overnight incubation with 1:100-fold diluted primary antibody, cells were blocked with blocking buffer (1× PBS/5% normal goat serum/0.3% Triton X-100) for 1 hour. After washing with PBS, cells were treated with 1:100-fold diluted secondary antibody for 2 hours and counterstained with 0.5 μg/mL of 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes. Cells were then mounted using Prolong Gold Antifade reagent and images were captured using Zeiss microscope (model: Axio Imager.D2). The primary antibodies used are listed in Supplementary Table 2.
In vivo study
For in vivo studies, 0.67 × 107 cells (Caki-1 cells expressing control miR and anti-miR-155-clone 1) were injected into nude mice subcutaneously. Caliper measurements were recorded once a week and tumor growth were calculated using formula [(x)2*y}/2 where x, width<y, length. Once palpable tumors developed (average volume = 80 mm3), and tumor growth was followed for 35 days. Experiment was terminated on 35th day, tumors were excised and snap frozen for future use. All animal care was in accordance with the institutional guidelines (IACUC approval no.: 16–004).
Statistical analysis
Statistical analyses were conducted with GraphPad Prism 8. All quantified data represent the average of three or more independent experiments performed at different times or as indicated. Error bars show SEM of independent experiments. All tests were performed two-tailed and P values <0.05 were considered statistically significant. MiRNA/mRNA sequencing data were analyzed using Mann–Whitney U test. Youden index was calculated to generate a cut-off value to determine miR-155 high or low expression groups with the SFVAMC samples. The endpoints were defined from time of surgery until time of the death, or last follow-up. This cut-off value was used for Kaplan–Meier and uni-/multivariate analyses. Kaplan–Meier analyses were used for overall survival curves and P values were calculated with the log-rank test. Uni-/multivariate analyses were computed by Cox proportional hazards model. The Spearman r correlation was computed to compare the correlation between miR-155, lncTCL6 and Src in the TCGA cohort.
Results
lncTCL6 is suppressed in renal tumors and cancer cell lines
For initial screening we analyzed the expression of lncTCL6 in the KIRC-TCGA dataset (Fig. 1A). We also analyzed the relative expression of lncTCL6 in SFVAMC cohort of human renal cancer clinical specimens by real-time PCR. Micro dissected tissues (n=67) and matched adjacent normal regions were used for this analysis. In agreement with previous reports the levels of lncTCL6 were statistically downregulated in cancer tissues as compared to their normal counterparts (Fig. 1B). We also investigated lncTCL6 levels in renal cancer cell lines 786-O, Caki-1 and compared them with HK2 (Fig. 1C). The levels of lncTCL6 were significantly low in 786-O and Caki-1 suggesting that lncTCL6 may function as a tumor suppressor gene.
Figure 1. lncTCL6 is a downregulated and miR-155 is upregulated in RCC and lncTCL6 is direct target of miR-155.

(A) Expression levels of TCL6 in KIRC-TCGA cohort (normal =72 and tumor = 518), p value calculated by Mann-Whitney test. (B) Relative TCL6 expression in ccRCC tissue and matched adjacent normal regions as assessed by qRT-PCR (SFVAMC cohort, n=67), p value calculated by Mann-Whitney test. (C) Relative expression levels of TCL6 in Caki-1, 786-O compared to normal non-malignant renal cell line HK2 (N=3). (D) Expression levels of miR-155 in the KIRC-TCGA cohort (normal =71 and tumor = 544), p value calculated by Mann-Whitney test. (E) Relative miR-155 expression in ccRCC tissue and matched adjacent normal regions as assessed by qRT-PCR (SFVAMC cohort, n=83), p value calculated by Mann-Whitney test. (F) Relative expression levels of miR-155 in Caki-1, 786-O compared to normal non-malignant renal cell line HK2 (N=3) (G) Correlation of miR-155 expression with lncTCL6 expression in patients from TCGA cohort. p-value calculated using the Spearman test of correlation. (H) Complimentary binding sites for miR-155 in TCL6 gene. (I) and (J) Luciferase assays showing decreased reporter activity after co-transfection of either wild-type target sequence compared with mutant target site (control) with miR-155 in Caki-1 and 786-O cells (N=3).
We checked the expression of miR-155 in RCC samples from TCGA. We found that miR-155 was significantly upregulated in cancer tissues as compared to normal (Fig. 1D). The expression of miR-155 in the SFVAMC cohort of human renal cancer clinical specimens (n=83) was significantly higher as compared to matched adjacent normal regions, (p<0.0001) (Fig. 1E). The miR-155 levels were also high in 786-O, Caki-1 as compared to HK2 (Fig. 1F) suggesting that miR-155 may function as an oncomir.
We performed correlation analysis between miR-155 and lncTCL6 in cancer tissues from TCGA data cohort. Though we found a weak negative correlation between miR-155 expression and lncTCL6 levels, it was statistically significant (Fig. 1G). Hence, we checked if there are any miR-155 binding sites within lncTCL6 gene. Computational algorithms predicted potential target sites within lncTCL6 with complementary binding sites for the seed sequence of miR-155 (Fig. 1H). To check whether a direct interaction is involved between miR-155 and lncTCL6, we performed luciferase reporter assays. We found that co-transfection of miR-155 along with the lncTCL6 wild type binding site caused a significant decrease in luciferase activity compared to control (Fig. 1I and 1J). These results suggest that miR-155 directly targets lncTCL6.
lncTCL6 gain of function exerts tumor suppressor effects in RCC
Uncontrolled growth of cells and survival are common characteristics of cancer cells that undergo metastatic dissemination. To assess the tumor suppressive role of lncTCL6, we stably overexpressed lncTCL6 in both renal cancer cell lines (Fig. 2A, Supplementary Fig. S1A) as well as performed phenotype rescue experiments in these cells using siRNA for TCL6. We found that post si-TCL6 transfection the levels of lncTCL6 reduced significantly in both the cell lines (Fig. 2B). We performed functional assays with these lncTCL6-OE cells and in TCL6-OE cells transfected with si-TCL6. A significant decrease in cell viability was observed over time in Caki-1, 786-O TCL6 overexpressing cells (Fig. 2C) whereas there was significant increase in proliferation in cells with suppressed expression of lncTCL6 (Fig. 2D). The lncTCL6 overexpressing clones had low colony formation ability as the number of foci decreased when compared to control cells. Contrary to this, the si-TCL6 cells showed increased colonies as compared to cells transfected with control siRNA (Fig. 2E, Supplementary Fig. S1B)
Figure 2. lncTCL6 is tumor suppressor with an anti-metastatic role in RCC.
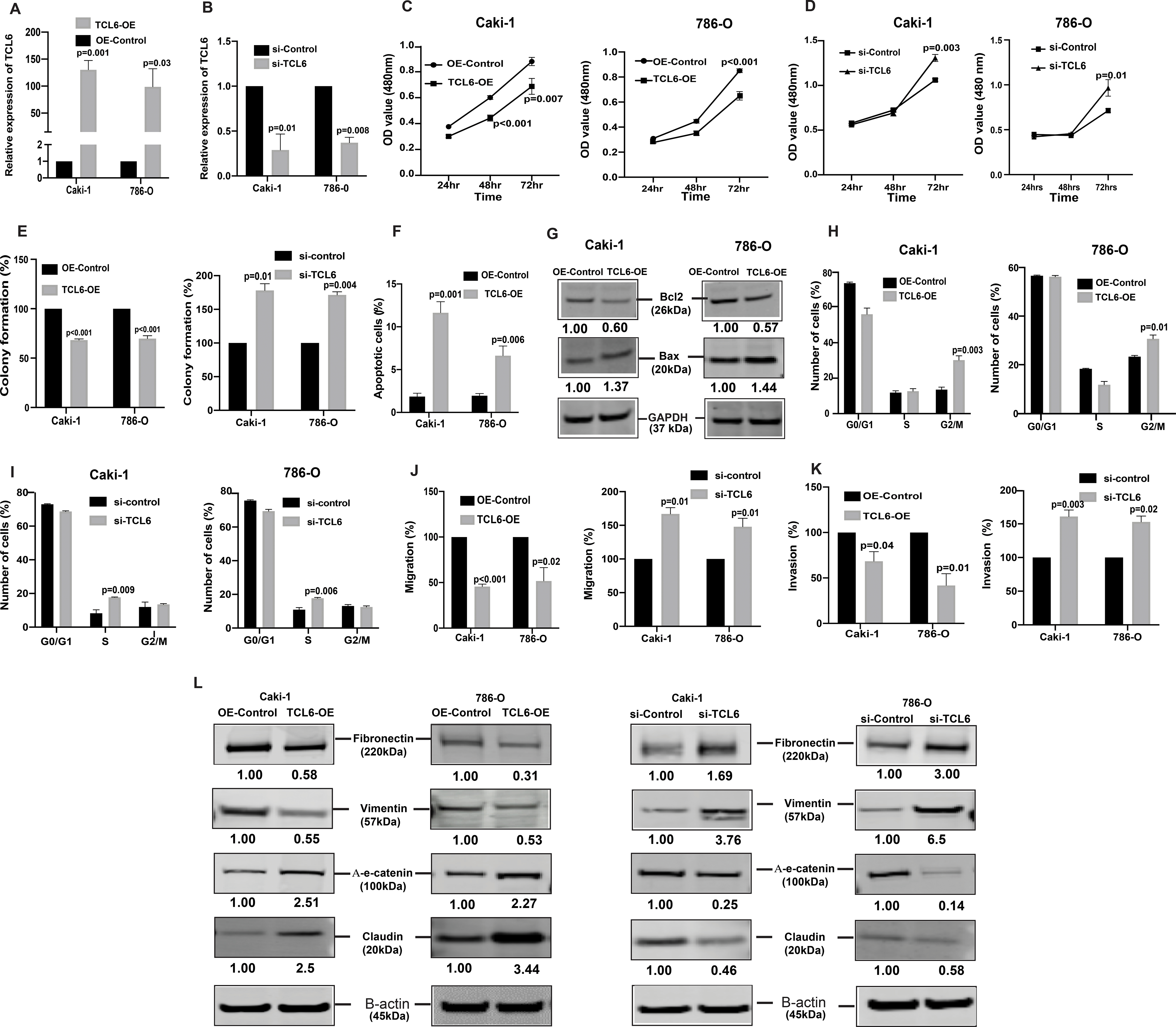
To evaluate the functional significance of TCL6 in renal cancer, TCL6 gene was overexpressed in Caki-1/786-O cell lines by transfection followed by selection of cells stably overexpressing TCL6 stably (TCL6-OE). For comparison, cells transfected with control plasmid (OE-control) were used. Later these TCL6-OE cells were used to perform rescue experiments (A) Relative expression levels of TCL6 in stable TCL6-OE cells as compared to control cells. (B) Relative expression levels of TCL6 in cells transfected with siTCL6 as compared to control cells. (C) Proliferation of TCL6-OE stable Caki-1(left), 786-O(right) cells by MTS assay (D) Proliferation of TCL6 suppressed Caki-1(left), 786-O (right) cells by MTS assay (E) Colony formation assay (graphical representation) in TCL6-OE-stable cells (left) and cells transfected with siTCL6 compared to control cells (right). (F) Apoptosis assay represented as a graph to show total apoptosis (early +late) in stable TCL6-OE cells. (G) Immunoblots showing apoptotic proteins in TCL6-OE Caki-1 and 786-O cells compared to control cells with GAPDH as endogenous control. (H) Cell cycle analysis showing significant increase in G2/M phase in TCL6-OE Caki-1 and 786-O cells compared to control cells. (I) Cell cycle analysis showing significant increase in S phase in Caki-1 and 786-O cells with suppressed TCL6 expression compared to control cells. Transwell migration assays (J) and invasion assays (K) in TCL6-OE and si-TCL6 treated Caki-1 and 786-O cells as compared to their respective controls. (L) Immunoblot assay showing EMT related proteins in Caki-1 and 786-O cells stably expressing TCL6 and transient transfected siTCL6 as compared to control cells, B-actin as endogenous control. The numbers below the blot represent relative protein level, which was determined from band intensity using Image studio software of LI-COR and normalized relative to cells stably expressing control plasmid or treated with si-control.
Graphs are mean of three independent experiments (N=3). P value was calculated by Student t test. Bar = mean ± standard error mean (SEM).
lncTCL6 overexpression triggers G2/M arrest and induces apoptosis in renal cancer cells
FACS (Fluorescence activated cell sorting) analysis for apoptosis was performed using Annexin-V-FITC-AAD dye. In both cell lines, the percentage of total apoptotic cells (early + late apoptosis) was significantly increased (1.9% to 11.6% for Caki-1 and 2% to 6.6% for 786-O) in response to lncTCL6 overexpression with a corresponding decrease in the viable cell population (Fig. 2F). These results indicate a tumor suppressive role for lncTCL6 in renal cancer. This was further supported by an increase in BAX and decrease in BCl2 in lncTCL6-OE cells as detected by immunoblot analysis (Fig. 2G). FACS analysis revealed that lncTCL6 gain of function led to a significant increase in the number of Caki-1 cells in the G2/M phase of the cell cycle (13.6% to 30.2%). Similar results were observed in 786-O cells with an increase in the G2/M population (23.3% to 30.7%) (Fig. 2H). Thus, suggesting that overexpression of lncTCL6 triggers G2/M arrest. On contrary the cells with decreased TCL6 levels showed increased cells in S-phase of the cell cycle (Fig 2I) supporting our earlier observation that lncTCL6 suppression promotes cell growth.
Overexpression of lncTCL6 reduces invasiveness of renal cancer cell lines
To determine whether lncTCL6 has a role in ccRCC metastasis, we performed in vitro chemotactic transwell migration and invasion assays. lncTCL6-OE had anti-migratory and anti-invasive effects on renal cancer cell lines. Less absorbance was observed at 560nm for lncTCL6 overexpressing cells compared to control cells in the migration assay (100 vs 45.6% for Caki-1 and 100 vs 51.9% for 786-O, Fig. 2J) and overexpression of lncTCL6 also significantly decreased the cell invasiveness (100 vs 55 % for Caki-1 and 100 vs 41.7% for 786-O, Fig. 2K). Similar results were obtained from clone 2 of lncTCL6-OE cells (Supplementary Fig. S1C and S1D). However there was reversal of phenotype when TCL6 levels were suppressed in these cells (Fig. 2J and 2K).
Since change in migration and invasion are directly associated with Epithelial-to-Mesenchymal transition (EMT), we also looked at EMT markers. Our results showed an increase in epithelial markers like α-E-catenin, claudin and decrease in mesenchymal markers like fibronectin and vimentin at protein levels in TCL6 overexpressing cells. But there was an opposite effect on levels of EMT proteins in the cells with suppressed TCL6 levels (Fig. 2L). We found similar effect over the mRNA levels for these EMT markers in cells with TCL6 overexpression and suppression (Supplementary Fig. S1E - S1H). Taken together, lncTCL6 suppression contributes to increased migration, invasion and EMT that are hallmarks of metastatic dissemination causing renal cancer cells to become more aggressive.
miR-155 suppression mimics lncTCL6 overexpression in renal cancer
To assess the oncogenic role of miR-155, we stably suppressed miR-155 in both renal cancer cell lines (Caki-1, 786-O, two clones each) (Supplementary Fig. S2A and S2B) and performed functional assays. Similar to lncTCL6 overexpression, miR-155 knockdown in both the cell lines showed reduced cell viability (Fig. 3A) and decreased colony formation (Fig. 3B, Supplementary Fig. S2C). To further verify the role of miR-155 in renal cancer cells, xenograft tumor formation assay was performed in vivo. miR-155 inhibited or control cells (Caki-1, clone 1) were injected into nude mice. Inhibition of miR-155 by anti-miR-155 in Caki-1 cells significantly decreased the capability of this metastatic cell line to form tumors in vivo. We found that tumors in the anti-miR-155 group grew more slowly and tumor size (1837 vs. 363.4 mm3) was smaller than that of control group (Fig. 3C and 3D). These results demonstrate that miR-155 acts as an oncogene and promotes aggressive tumor growth in vivo.
Figure 3. Knockdown of miR-155 functionally phenocopies the effect of lnRNATCL6 overexpression.
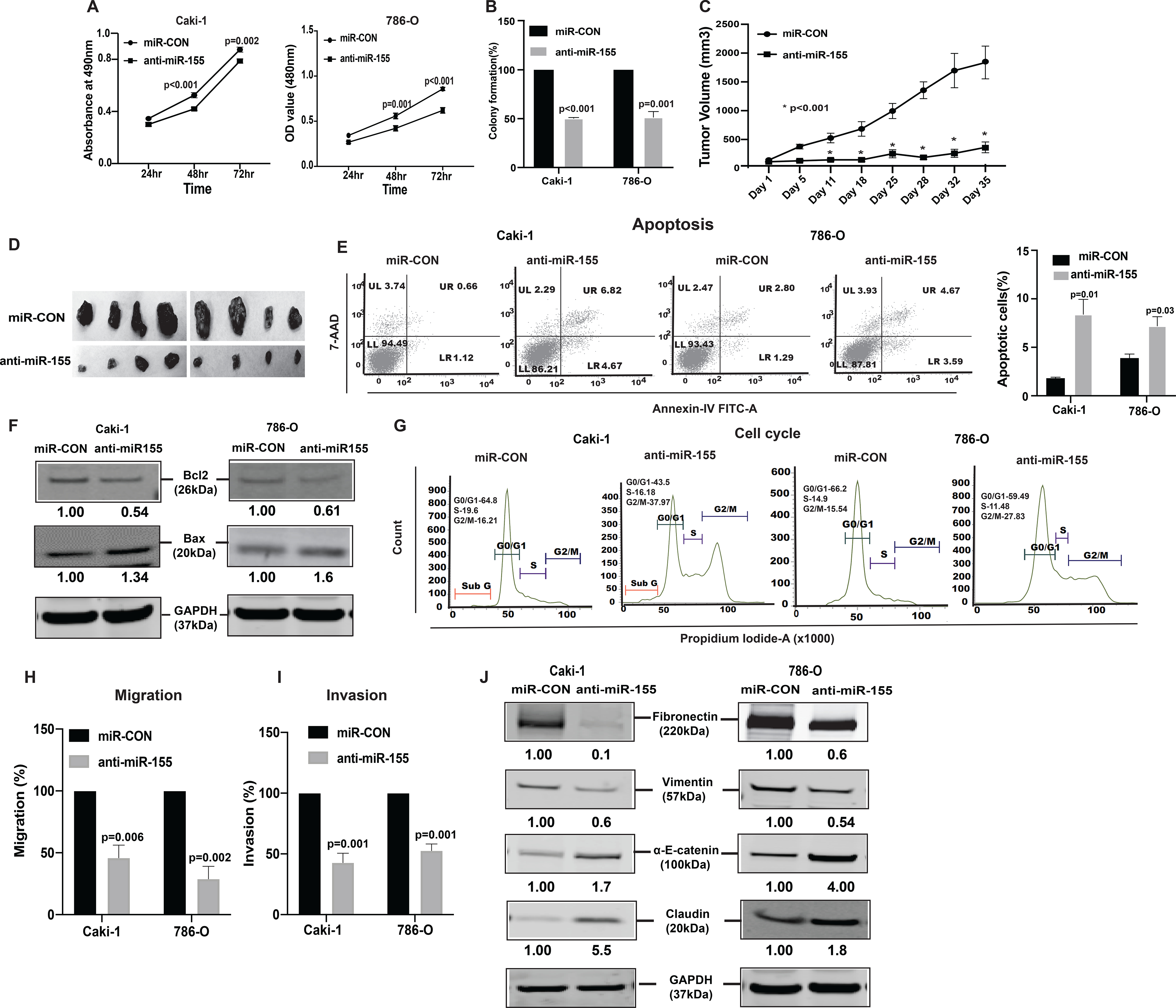
(A) Proliferation of anti-miR-155-stable Caki-1(left), 786-O(right) cells by MTS assay. (B) Colony formation assay (graphical representation) in anti-miR-155-stable Caki-1 and 786-O cells compared to controls. (C) Tumor volumes of xenograft tumors from mice injected with anti-miR-155-stable Caki-1 cells and control cells at indicated time points. Data represent the mean of each group ± SE. (D) Images of extracted tumors from miR-CON and anti-miR-155-stable groups at Day 35 are shown. (E) FACS apoptosis analyses of anti-miR-155-stable Caki-1 and 786-O cells compared to control. Graph represents total apoptosis (early + late). (F) Immunoblots showing apoptotic proteins in anti-miR-155-stable Caki-1 and 786-O cells compared to control cells with GAPDH as endogenous control. (G) Cell cycle analysis showing significant increase in G2-M phase of anti-miR-155-stable Caki-1 and 786-O cells. Caki-1 and 786-O cells stably expressing anti-miR-155 were subjected to transwell assay to evaluate chemotactic migration (H) and invasion (I) as seen in graphical representation for both Caki-1 and 786-O cells. (J) Immunoblot assay showing EMT related proteins in Caki-1 and 786-O cells stably expressing anti-miR-155 plasmid as compared to control cells, GAPDH as endogenous control. The numbers below the blot represent relative protein level, which was determined from band intensity using Image studio software of LI-COR and normalized relative to cells stably expressing control plasmid Graphs are mean of three independent experiments, (N=3). P value was calculated by Student t test. Bar = mean ± standard error mean (SEM).
FACS analysis showed an increase in apoptotic cells (Fig. 3E). This was supported by an increase in BAX and a decrease in BCl2 protein, markers of proliferation and apoptosis (Fig. 3F). Also, the cells with stable miR-155 knockdown were arrested at the G2/M phase of the cell cycle like cells with lncTCL6 overexpression (Fig. 3G).Transwell migration and invasion assays indicated that miR-155 suppression significantly reduced the migration (Fig. 3H) and invasion (Fig. 3I) of both cell lines. The anti-miR-155 clone 2 cells also showed a similar effect on the migratory and invasive features of RCC cells (Supplementary Fig. S2D and S2E).
Similarly, suppression of miR-155 also resulted in upregulation of α-E-catenin, Claudin and simultaneous decrease in vimentin, fibronectin expression both at protein levels (Fig. 3J) and mRNA (Supplementary Fig. S2F and S2G) levels.
miR-155 overexpression exerts tumorigenic effects in non-malignant HK-2 cells
In a reciprocal approach, we overexpressed miR-155 and suppressed lncTCL6 expression in normal immortalized renal epithelial cell line (Human keratinocyte −2, HK2) using miRVana miRNA mimic and siRNA TCL6. miR-155 overexpression and TCL6 downregulation was confirmed by RT-PCR (Fig. 4A, 4B). We performed all functional assays using these miR-155 overexpressing and TCL6 suppressed cells. Our results suggest that both the over-expression of miR-155 and suppression of TCL6 increased proliferation (Fig. 4C, 4D) and colony formation (Fig. 4D). Cell cycle analysis showed a significant increase in S-phase (Fig. 4E). We also observed an increase in motility and invasiveness of these non-malignant epithelial cells (Fig. 4F, 4G). This phenotypic effect was supported by increased expression of mesenchymal markers and a decrease in epithelial markers both at the mRNA (Supplementary Fig.S3A and S3B) and protein levels in both miR-155 overexpressing and TCL6 suppressed HK2 cells (Fig. 4H).
Figure 4. miR-155 overexpression and TCL6 suppression increases invasiveness and proliferation of normal immortalized renal epithelial cell line, HK2.
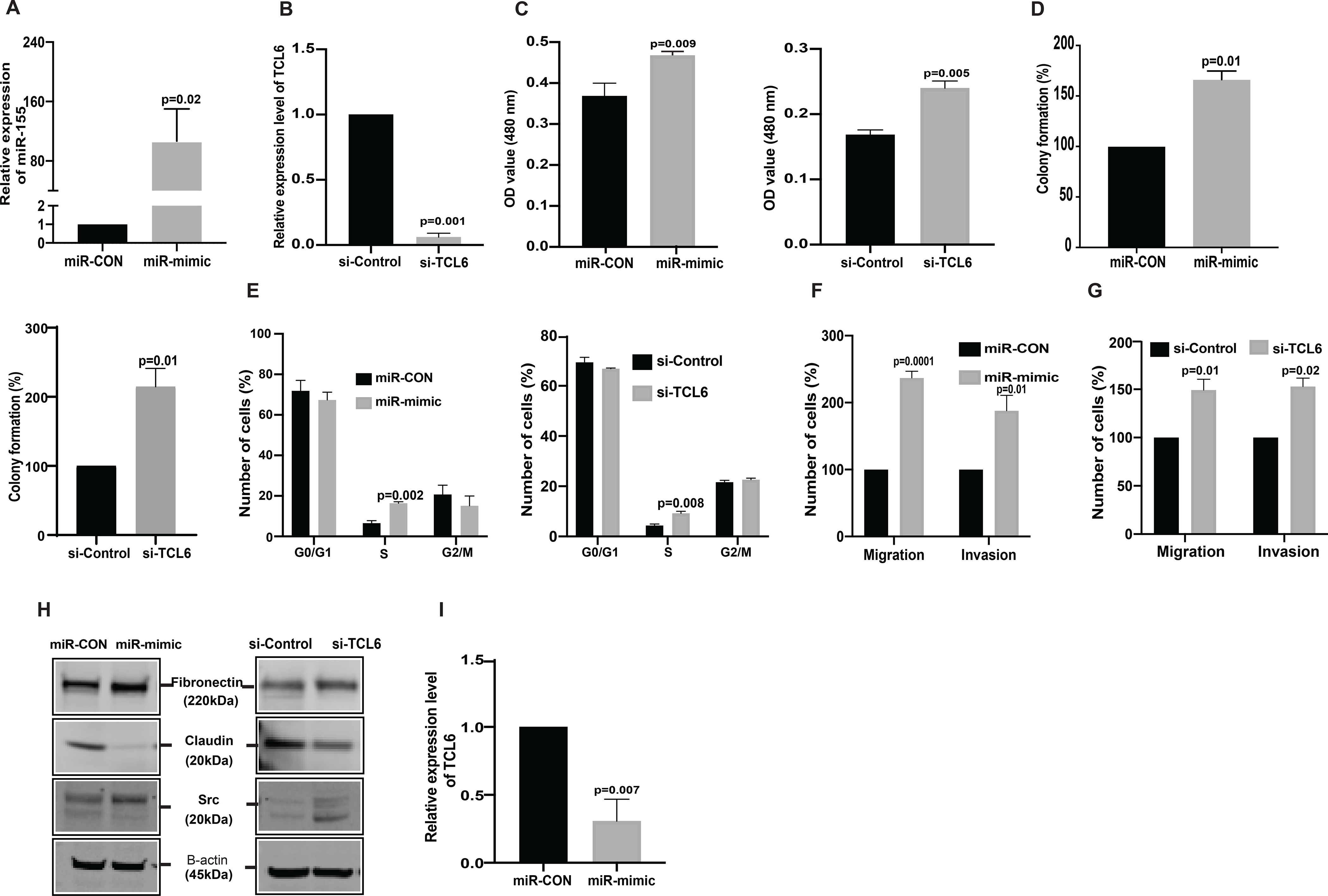
HK2 was treated with miRVANA miR-155 mimic /control mimic (miR-CON) as well as si-TCL6 and corresponding control followed by functional assays (performed 72 hours post-transfection). (A) Relative miR-155 expression as assessed by real-time PCR. (B) Relative expression of TCL6 cells treated with siTCL6 (C) Cell viability in HK2 cells after miR-CON/miR-155 mimic transfections and after si-control/si-TCL6 transfections as assessed by MTS assay at 72 hr time-point. (D) Colony formation assay shows increase in clonogenicity of HK2 cells post miR-155 overexpression and post TCL6 suppression as compared to control (E) Cell cycle analysis showing significant increase in S-phase for the HK2 cells transfected with miR-155 mimic as well as the HK2 cells transfected with si-TCL6 as compared to cells transfected with control (F) Transwell migration assays and invasion assays in miR-CON/miR-155 mimic transfected cells (G) Transwell migration assays and invasion assays in si-Control/si-TCL6 transfected cells (H) Relative expression of EMT markers (Fibronectin, Claudin, Src) in miR-155 overexpressing and TCL6 suppressed HK2 cells as compared to control cells. (I) TCL6 expression in miR-155 overexpressing HK2 cells as compared to control cells.
The bar graph (mean ± standard error mean (SEM) represent the data from three technical replicates,N=3 P value was calculated by Student t test.
Having demonstrated that lncTCL6 is a direct target of miR-155, and that miR-155 has pro-metastatic effects in renal cancer, we sought further evidence linking the pro-oncogenic functions of miR-155 to lncTCL6 inhibition. We performed RT-qPCR with miR-155 depleted and overexpressed cells. We found that over-expression of miR-155 levels led to decreased expression of lncTCL6 as compared to control cells (Fig 4I). Also, suppression of miR-155 in turn led to increased lncTCL6 expression in both clone 1 and clone 2 of anti-miR-155 treated Caki-1 and 786-O cells (Supplementary Fig S3C and S3D).
These results further confirm the biological role of lncTCL6 and miR-155 in renal cancer as their suppression and overexpression respectively induces the tumorigenic attributes in HK2 cells.
lncTCL6 attenuates renal cancer progression through STAU1 mediated decay of Src mRNA
We next examined if lncTCL6 is involved in post-transcriptional regulation affecting EMT and metastasis in RCC. According to iLoc-LncRNA and lncLocator in silico tools (30,31) lncTCL6 is localized mostly in the cytoplasm (Supplementary Fig. S4A, B). It is known that cytoplasmic lncRNAs can interact with proteins that in turn bind to target mRNA and regulate their stability. One such regulatory protein is STAU1, a double stranded RNA binding protein, that binds within the translationally active target mRNAs. Thus we looked to see if lncTCL6 interacts with STAU1 protein using a RNA-protein complex prediction tool, RPI-Pred (32), that predicted possible interaction between lncTCL6 and STAU1 protein (Supplementary Fig. S4C).
From the gene co-expression network, we checked the relationship between lncTCL6 and downstream effector mRNAs. According to the network, lncTCL6 was correlated with Src expression levels (Fig. 5A). Also, Src expression is high in renal cancer (33) and in metastatic cell lines as compared to primary cells (34). In lncTCL6 overexpressing Caki-1 and 786-O cells, both the mRNA (Fig. 5B) was significantly decreased as compared to control cells. Similar results were seen in TCL6 and miR-155 suppressed cells (Fig. 5C, 5D). These cells also showed concomitant decrease in protein levels of Src as compared to the control cells (Fig 5E – 5G). Further we performed RNA immunoprecipitation assay using STAU1 antibody. LncTCL6 overexpressing cells were harvested and STAU1 protein was immunoprecipitated together with bound RNA. These experiments revealed that higher levels of lncTCL6 and Src mRNA were co-precipitated with STAU1 compared to IgG (Fig. 5H). In order to confirm the involvement of STAU1 in Src mRNA degradation we checked the effect of STAU1 suppression on Src protein and mRNA levels. We found that decrease in STAU1 levels (Fig. 5I), decreases Src levels both at RNA and protein levels (Fig. 5J, 5K). Also, decrease in STAU1 levels led to increase mesenchymal markers and concomitant decrease in epithelial markers (Fig. 5K).
Figure 5. LncTCL6 degrades Src mRNA by STAU1 mediated RNA degradation, and thereby attenuates Src-Akt mediated EMT transition in renal carcinoma.
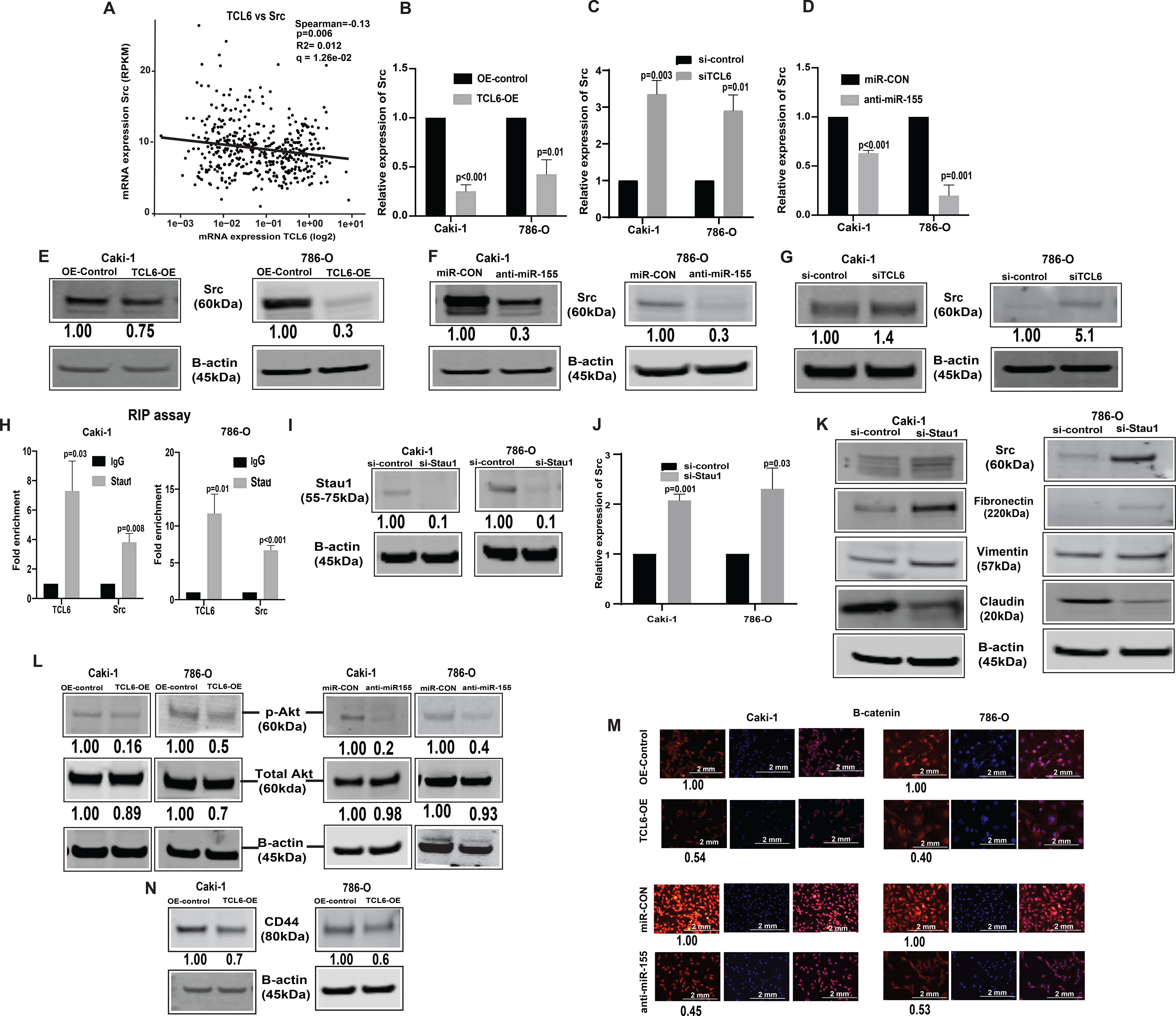
(A) Negative correlation between TCL6 and Src expression in KIRC-TCGA dataset. Spearman test was used to analyze the correlation. Relative expression of Src (mRNA) in cells with TCL6 overexpression (B), in TCL6-OE cells transfected with si-TCL6 (C) and in cells with stable miR-155 inhibition (D) as compared to their respective control cells. Immunoblot of Src in Caki-1 and 786-O cells stably overexpressing TCL6 (E), cells expressing anti-miR-155 and (F) cells transfected with si-TCL6 (G) as compared to respective control cells. (H) RNA-immunoprecipitation using STAU1 antibody shows enrichment of TCL6 and Src as compared to control antibody (IgG) in TCL6 over-expressing cells. (I) STAU1 expression in TCL6-OE cells transfected with siSTAU. (J) Src mRNA expression in TCL6-OE cells transfected with siSTAU (K) Immunoblot showing expression of Src and EMT markers (Fibronectin, Vimentin, Claudin) in TCL6-OE cells transfected with siSTAU (L) Immunoblot showing suppression of phosphorylated Akt in cells stably expressing TCL6 and anti-miR-155 as compared to respective controls. (M) Immunostaining of p-®-catenin (red) counterstained with DAPI (blue) in Caki-1 and 786-O cells expressing anti-miR-155 stably and overexpressing TCL6 as compared to respective control cells, scale bar: 2mm (right bottom). (N) Immunoblot showing suppression of metastatic marker CD44 in cells stably expressing TCL6 as compared to respective controls. The numbers below the blot represent relative protein level, which was determined from band intensity using Image studio software of LI-COR and normalized relative to cells stably expressing control plasmid or treated with si-control.
The bar graphs represent mean ± standard error mean (SEM) from three set of experiments, (N=3). P value was calculated by Student t test. Immunoblot and immunofluorescence staining was performed for three and two biological replicates respectively..
Overexpression of lncTCL6 and silencing of miR-155 attenuates EMT and metastasis associated genes
Since Src is a known regulator of the Akt mediated EMT pathway we checked the levels of Akt, b-catenin in both miR-155 suppressed and lncTCL6 overexpressed cells. We found that there is a significant decrease in expression of Akt, b -catenin in both lncTCL6-OE and anti-miR-155 stable cells as compared to their respective controls (Fig. 5L and 5M). Molecular studies have shown that high CD44 expression is correlated with EMT and metastasis, so we looked at its expression in TCL6-OE renal cancer cells. We found a significant decrease in CD44 levels in lncTCL6-OE cells (Fig. 5N), indicating EMT-metastasis in renal carcinoma is associated with the Src-Akt pathway which in turn is regulated by miR-155 through lncTCL6 downregulation (Supplementary Fig. S5).
Clinical utility of lncTCL6 and miR-155 in ccRCC
In view of the observed widespread lncTCL6 downregulation in renal cancer clinical specimens, we evaluated the potential clinical significance of lncTCL6 expression. Clinical demographics of the study cohort are summarized in Supplementary Table 3. We examined the correlation between lncTCL6 expression and different grades of renal carcinoma in both SFVAMC and TCGA cohorts and found that lower levels of lncTCL6 are correlated with higher grades of cancer (Fig. 6A and 6B, respectively). In contrast higher levels of miR-155 expression correlated with higher grades of renal carcinoma in both the cohorts (Fig. 6C and 6D).
Figure 6. Clinical significance of lncTCL6 and miR-155 in RCC.
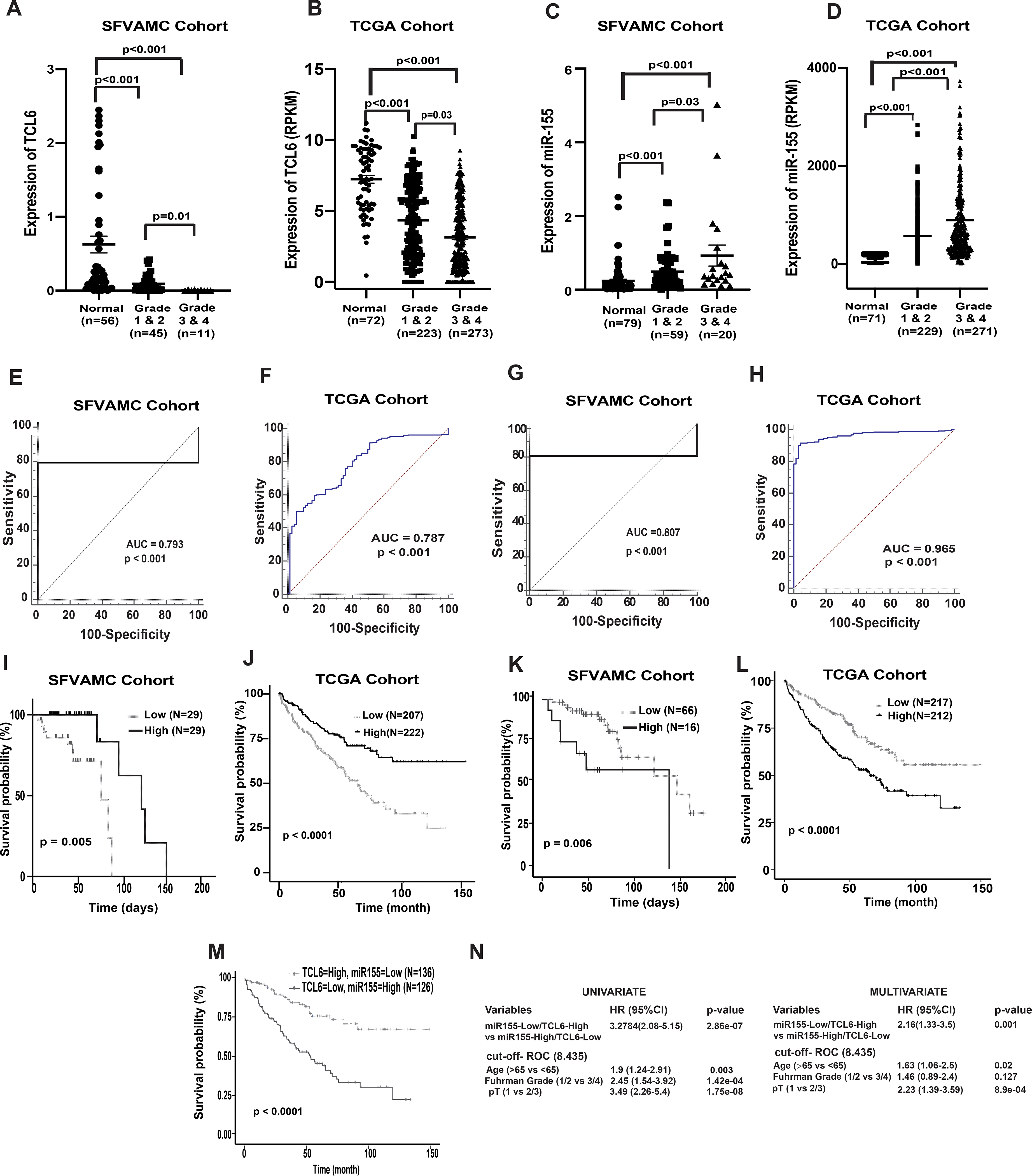
(A) Expression levels of lnc TCL6 across different grades in SFVAMC cohort [normal (n=56), grade 1 & 2 (n=45) and grade 3 & 4 (n=11)], p value calculated by Mann-Whitney test. (B) Expression levels of lncTCL6 across different grades in KIRC-TCGA cohort [normal (n= 72), grade 1 & 2 (n=223) and grade 3 & 4 (n=273)], p value calculated by Mann-Whitney test. (C) Expression levels of miR-155 among different grades in SFVAMC cohort [normal (n=79), grade 1 & 2 (n=59) and grade 3 & 4 (n=20)], p value calculated by Mann-Whitney test. (D) Expression levels of miR-155 among different grades in KIRC-TCGA cohort [normal (n= 71), grade 1 & 2 (n=229) and grade 3 & 4 (n=271)], p value calculated by Mann-Whitney test. (E) Receiver operating curve (ROC) analysis showing ability of lncTCL6 to differentiate between malignant and non-malignant SFVAMC samples. (F) Receiver operating curve (ROC) analysis showing ability of lncTCL6 to differentiate between malignant and non-malignant TCGA samples. (G) Receiver operating curve (ROC) analysis showing ability of miR-155 to differentiate between malignant and non-malignant SFVAMC samples. (H) Receiver operating curve (ROC) analysis showing ability of miR-155 to differentiate between malignant and non-malignant TCGA samples. (I) Overall survival of renal cancer patients (SFVAMC cohort) with high lncTCL6 expression (n=29) compared to low expression (n=29) performed by Kaplan- Meier analysis, p-value calculated by log-rank test. (J) Overall survival of renal cancer patients (KIRC TCGA cohort) with high lncTCL6 expression (n=222) compared to low expression (n=207) performed by Kaplan- Meier analysis, p-value calculated by log-rank test. (K) Overall survival of renal cancer patients (SFVAMC cohort) with high miR-155 expression (n=16) compared to low miR155 expression (n=66) performed by Kaplan- Meier analysis, p-value calculated by log-rank test. (L) Overall survival of renal cancer patients (KIRC TCGA cohort) with high miR-155 expression (n=253) compared to low miR155 expression (n=253) performed by Kaplan- Meier analysis, p-value calculated by log-rank test. (M) Overall survival of renal cancer patients (KIRC TCGA cohort) with high miR-155 and low TCL6 expression (n=126) compared to low miR155 and high TCL6 expression (n=136) performed by Kaplan- Meier analysis, p-value calculated by log-rank test. (N) Logistic regression model showing high TCL6 and low miR-155 expression could be a unique risk factor for patient survival.
Further, we determined the capability of lncTCL6 and miR-155 to act as a diagnostic biomarker for renal cancer by performing ROC analyses with SFVAMC cohort and TCGA cohort (Fig. 6E–6F and 6G–6H, respectively). ROC analyses showed that both lncTCL6 and miR-155 expression is a significant parameter to discriminate between normal and tumor tissues.
Further we checked the relationship between expression of lncTCL6, miR-155 with overall survival of patients with renal cancer in both cohorts. We stratified both the SFVAMC and TCGA cohorts based on lncTCL6 and miR-155 expression (high vs low) using median and Youden index (Supplementary Fig. S6) and performed Kaplan-Meier (KM) survival analysis. KM analysis showed the survival probability was significantly higher in patients with high lncTCL6 expression compared to those with low expression in both cohorts (Fig. 6I and 6J). These results indicate that downregulation of lncTCL6 expression is associated with poor survival outcome in renal cancer. Also, for both cohorts, miR-155 expression showed an opposite trend, the survival probability for patients with low miR-155 expression was higher compared to those with higher expression (Fig. 6K and 6L).
Since both lncTCL6 and miR-155 are significantly linked to the overall survival of the patients with RCC, we stratified the TCGA data in two groups one with high lncTCL6 - low miR-155 expression and the other with low lncTCL6 - high miR-155 expression using the earlier cut-off values and performed KM analysis. We found that the high lncTCL6 - low miR-155 expression group had a higher overall survival of patient as compared to the other group (Fig. 6M). Further, univariate and multivariate analyses using these groups revealed that low lncTCL6 - high miR-155 expression is associated with poor prognosis, indicating that this interaction between lncTCL6 and miR-155 has potential to be an independent prognostic biomarker for RCC (Fig. 6N). Thus, the interaction between lncTCL6 and miR-155 may be a clinically important parameter with prognostic and diagnostic significance in ccRCC.
Discussion
Histological subtypes of RCC are highly heterogeneous in their biology and therapeutic outcomes with clear-cell RCC (ccRCC) being the most common (70–80%) and one of the most aggressive subtypes (2). Currently there are not enough predictive biomarkers and have limited understanding of the signaling pathways prevalent in RCC progression and metastasis. Thus, new biomarkers for diagnosis and targeted therapy are urgently needed to improve patient care and treatment. LncRNAs and miRNAs drive cancer phenotypes and are promising targets for effective diagnosis and therapeutic intervention in the fight against cancer (35–38). The validation of lncRNAs as clinical biomarkers and their role in molecular mechanisms driving ccRCC metastasis are yet to be elucidated. In this study we show that lncTCL6-miR-155-Src/Akt/EMT network is a novel regulatory mechanism in ccRCC progression and metastasis.
We observed lncTCL6 to be significantly downregulated in two study cohorts (SFVAMC and TCGA), which is associated with poor prognosis. Recent work on lncTCL6 by other investigators using other RCC cohorts confirm our findings, of it being a tumor-suppressor and its decreased expression linked to poor overall survival (13,15,39). In addition, the levels of lncTCL6 were inversely associated with advanced tumor grade and shorter overall survival. Our findings revealed that lncTCL6 expression can distinguish between cancerous/non-cancerous tissues in RCC and can be used as a diagnostic ccRCC biomarker. Functionally, overexpression of TCL6 significantly decreased ccRCC cell proliferation, colony formation, cell migration, invasion and induced G2/M arrest and apoptosis in vitro. And suppression of lncTCL6 in these cells led to reversal to cancer phenotype.
On the other hand, miR-155 expression was significantly upregulated in ccRCC tissues compared to corresponding non-tumor tissues. This agrees with previous renal cancer studies (22,23). The miR-155 depleted cells showed slower tumor growth as well as low tumor volume in vivo, highlighting the potential of miR-155 suppression as a ccRCC therapeutic modality. Previous studies have shown that miR-155 enhances malignant tumor phenotypes by promoting cell proliferation. There are reports of miR-155 accelerating cell invasion in gastric and renal carcinoma cells (40,41). Similar to these results we also found that suppression of miR-155 decreased motility and invasiveness in RCC cells. Furthermore, we also confirmed the oncogenicity of miR-155 and tumor suppressive nature of lncTCL6. Overexpression of miR-155 and suppression of lncTCL6 led to increased proliferation, colony formation, migration, and invasion in non-malignant HK2 cells. Thus, the present study has expanded our current knowledge about the role of miR-155 and lnc TCL6 in ccRCC aggressiveness. This study also shows a novel regulatory pathway of Src-Akt induced EMT triggered via miR-155 targeting lncTCL6.
Further, we found a weak inverse correlation between miR-155 and lncTCL6 in TCGA renal cancer tissue cohort. miR-155 has a complementary binding site on lncTCL6. Luciferase reporter assay revealed that miR-155 could bind to the wild type target sequence but not a mutated sequence. Furthermore, miR-155 overexpression and suppression led to decreased and increased expression of TCL6 in vitro respectively. This shows that lncTCL6 is a direct target of miR-155 in ccRCC. It is true that miRNAs or lncRNAs may affect a series of genes and further in-depth studies are required to confirm this association. However in our study restoration of lncTCL6 attenuated the effects of miR-155 on RCC cell migration and invasion whereas suppression of TCL6 in these cells reversed the behavior of cells. This indicates that exploiting lncTCL6 and miR-155 interaction may be a promising therapeutic approach to inhibit ccRCC progression and metastasis.
Emerging reports also suggest that both the expression and activation of Akt and Src is associated with the appearance of malignant phenotypes and reduced survival in renal carcinoma(7,42). Src signaling has been demonstrated to play a pleiotropic role in mediating various malignant functions. From bioinformatic tools it is apparent that lncTCL6 can interact with Src mRNA and STAU1 protein. Also, Src is negatively correlated with lncTCL6 expression in TCGA dataset and, over-expression of lncTCL6 reduces Src mRNA levels. It is known that lncRNA exerts its biological effects mainly by binding to RNA binding proteins (RBP). For example, lncRNA TINCR physically interacts with STAU1 protein to regulate SMD (10,43) STAU1 is a part of a highly conserved family of double-stranded RNA-binding proteins implicated in mRNA transport, stability and translation (44,45). In this study, RNA immunoprecipitation assay using STAU1 antibody showed specific association of lncTCL6 with Src. Also, downregulation of STAU1 protein in TCL6 overexpressing cells increased the expression of Src both at mRNA and protein level. From these results we hypothesize that lncTCL6 recruits STAU1 protein to Src mRNAs and mediates its decay. Inhibition of Src is known to decrease AKT and β-catenin phosphorylation, thus further attenuating ccRCC epithelial to mesenchymal transition (46). In our study, we found similar effects of Src decay on Akt mediated EMT progression.
In conclusion, we demonstrate for the first time that miR-155 directly interacts with lncTCL6 in ccRCC resulting in activation of the Src-Akt pathway to promote ccRCC metastasis. Further, over-expression of lncTCL6 inhibits cell growth and epithelial to mesenchymal progression by STAU-1 mediated decay of Src mRNA. Modulation of their expression by either repressing miR-155 or overexpressing lncTCL6 provides a novel therapeutic approach to regulate lncTCL6-miR 155-Src-Akt-EMT network driven metastasis in ccRCC.
Supplementary Material
Statement of significance.
This study’s investigation of noncoding RNA interactions in renal cell carcinoma identify miRNA-155-lncRNA TCL6-mediated regulation of the Src-Akt-EMT network as a novel mechanism of disease progression and metastasis.
Acknowledgements
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript.
Grant support
This study was supported by the Department of Veterans Affairs VA Merit Review 101 BX001123, Senior Research Career Scientist Award (Rajvir Dahiya, IK6-BX004473), and the National Institutes of Health / National Cancer Institute RO1CA199694, RO1CA196848.
Footnotes
Conflict of interest
There are no conflicts of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9–29 [DOI] [PubMed] [Google Scholar]
- 2.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res 2012;22:2089–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Zhao X, Qi J, Yang C, Cheng H, Ren Y, et al. Eight proteins play critical roles in RCC with bone metastasis via mitochondrial dysfunction. Clin Exp Metastasis 2015;32:605–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006;127:679–95 [DOI] [PubMed] [Google Scholar]
- 5.Bai L, Yang JC, Ok JH, Mack PC, Kung HJ, Evans CP. Simultaneous targeting of Src kinase and receptor tyrosine kinase results in synergistic inhibition of renal cell carcinoma proliferation and migration. Int J Cancer 2012;130:2693–702 [DOI] [PubMed] [Google Scholar]
- 6.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res 2011;71:2611–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonezawa Y, Nagashima Y, Sato H, Virgona N, Fukumoto K, Shirai S, et al. Contribution of the Src family of kinases to the appearance of malignant phenotypes in renal cancer cells. Mol Carcinog 2005;43:188–97 [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Xu X, Luo J, Wang H, Zhou S. Rapamycin Enhances the Anti-Cancer Effect of Dasatinib by Suppressing Src/PI3K/mTOR Pathway in NSCLC Cells. PLoS One 2015;10:e0129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke L, Xiang Y, Guo X, Lu J, Xia W, Yu Y, et al. c-Src activation promotes nasopharyngeal carcinoma metastasis by inducing the epithelial-mesenchymal transition via PI3K/Akt signaling pathway: a new and promising target for NPC. Oncotarget 2016;7:28340–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011;470:284–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Ma T, Li Q, Wang S, Sun W, Li W, et al. Identification of a lncRNAassociated competing endogenous RNAregulated network in clear cell renal cell carcinoma. Mol Med Rep 2019;20:485–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou W, Tang Q, Bi F. [Comprehensive analysis of the aberrantly expressed profiles of lncRNAs, miRNAs and the regulation network of the associated ceRNAs in clear cell renal cell carcinoma]. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 2019;36:267–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K, Lu XF, Luo PC, Zhang J. Identification of Six Potentially Long Noncoding RNAs as Biomarkers Involved Competitive Endogenous RNA in Clear Cell Renal Cell Carcinoma. Biomed Res Int 2018;2018:9303486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Zhang C, He W, Gou X. Construction and comprehensive analysis of dysregulated long non-coding RNA-associated competing endogenous RNA network in clear cell renal cell carcinoma. J Cell Biochem 2018 [DOI] [PubMed] [Google Scholar]
- 15.Su H, Sun T, Wang H, Shi G, Zhang H, Sun F, et al. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget; Vol 8, No 4 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu LP, Gong YB. LncRNA-TCL6 promotes early abortion and inhibits placenta implantation via the EGFR pathway. Eur Rev Med Pharmacol Sci 2018;22:7105–12 [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu X, et al. miR-141 is a key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res 2014;20:2617–30 [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Shen D, Li H, Zhang Y, Lv X, Huang Q, et al. MicroRNA-185 inhibits cell proliferation and induces cell apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma. Urol Oncol 2015;33:169 e1–11 [DOI] [PubMed] [Google Scholar]
- 20.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol 2009;28:264–84 [DOI] [PubMed] [Google Scholar]
- 21.Li T, Yang J, Lv X, Liu K, Gao C, Xing Y, et al. miR-155 regulates the proliferation and cell cycle of colorectal carcinoma cells by targeting E2F2. Biotechnol Lett 2014;36:1743–52 [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Ma X, Yao Y, Li H, Fan Y, Zhang Y, et al. miR-155 regulates the proliferation and invasion of clear cell renal cell carcinoma cells by targeting E2F2. Oncotarget 2016;7:20324–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Tian D, Zhang B, Zhang Y, Yan D, Wu S. Overexpression of miR-155 in clear-cell renal cell carcinoma and its oncogenic effect through targeting FOXO3a. Exp Ther Med 2017;13:2286–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Chen T, Zhong Z, Wang Y, Li Y, Zhao X. microRNA-155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in renal cancer cells. Mol Med Rep 2012;5:949–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lokeshwar SD, Talukder A, Yates TJ, Hennig MJP, Garcia-Roig M, Lahorewala SS, et al. Molecular Characterization of Renal Cell Carcinoma: A Potential Three-MicroRNA Prognostic Signature. Cancer Epidemiology Biomarkers & Prevention 2018;27:464. [DOI] [PubMed] [Google Scholar]
- 26.Witten L, Slack FJ. miR-155 as a novel clinical target for hematological malignancies. Carcinogenesis 2019 [DOI] [PubMed] [Google Scholar]
- 27.Wu Q, Guo L, Jiang F, Li L, Li Z, Chen F. Analysis of the miRNA–mRNA–lncRNA networks in ER+ and ER− breast cancer cell lines. Journal of Cellular and Molecular Medicine 2015;19:2874–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Dong R, Diao S, Du J, Fan Z, Wang F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Research & Therapy 2017;8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni P, Dasgupta P, Bhat NS, Shahryari V, Shiina M, Hashimoto Y, et al. Elevated miR-182–5p Associates with Renal Cancer Cell Mitotic Arrest through Diminished. Mol Cancer Res 2018;16:1750–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D, Chen W, et al. iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics 2018;34:4196–204 [DOI] [PubMed] [Google Scholar]
- 31.Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 2018;34:2185–94 [DOI] [PubMed] [Google Scholar]
- 32.Suresh V, Liu L, Adjeroh D, Zhou X. RPI-Pred: predicting ncRNA-protein interaction using sequence and structural information. Nucleic Acids Res 2015;43:1370–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo J, Podolak J, Degnin C, Chen Y, Robinson KW, Pukay M, et al. Expression of VHL and SRC in metastatic renal cell carcinoma (mRCC). Journal of Clinical Oncology 2016;34:e16133–e [Google Scholar]
- 34.Suwaki N, Vanhecke E, Atkins K, Graf M, Swabey K, Huang P, et al. A HIF-regulated VHL-PTP1B-Src signaling axis identifies a therapeutic target in Renal Cell Carcinoma. Science translational medicine 2011;3:85ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol 2018;15:541–63 [DOI] [PubMed] [Google Scholar]
- 36.Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, et al. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene 2018;37:5037–53 [DOI] [PubMed] [Google Scholar]
- 37.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673–7 [DOI] [PubMed] [Google Scholar]
- 38.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005;353:1768–71 [DOI] [PubMed] [Google Scholar]
- 39.Yang FY, Wang Y, Wu JG, Song SL, Huang G, Xi WM, et al. Analysis of long non-coding RNA expression profiles in clear cell renal cell carcinoma. Oncol Lett 2017;14:2757–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y, et al. MicroRNA-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor-beta receptor 2. Cancer Sci 2018;109:618–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei RJ, Zhang CH, Yang WZ. MiR-155 affects renal carcinoma cell proliferation, invasion and apoptosis through regulating GSK-3beta/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2017;21:5034–41 [DOI] [PubMed] [Google Scholar]
- 42.Roelants C, Giacosa S, Pillet C, Bussat R, Champelovier P, Bastien O, et al. Combined inhibition of PI3K and Src kinases demonstrates synergistic therapeutic efficacy in clear-cell renal carcinoma. Oncotarget 2018;9:30066–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013;493:231–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. The EMBO Journal 2007;26:2670–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, et al. Staufen1 senses overall transcript secondary structure to regulate translation. Nature Structural & Molecular Biology 2014;21:26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan H, Meng X, Guo W, Cai P, Li W, Li Q, et al. Transmembrane-Bound IL-15–Promoted Epithelial-Mesenchymal Transition in Renal Cancer Cells Requires the Src-Dependent Akt/GSK-3β/β-Catenin Pathway 1 2. Neoplasia 2015;23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


