Abstract
The molecular evolution of the adaptive response at the host–pathogen interface has been frequently referred to as an ‘arms race’ between the host and bacterial pathogens. The innate immune system employs multiple strategies to starve microbes of metals. Pathogens, in turn, develop successful strategies to maintain access to bioavailable metal ions under conditions of extreme restriction of transition metals, or nutritional immunity. However, the processes by which evolution repurposes or re-engineers host and pathogen proteins to perform or refine new functions have been explored only recently. Here we review the molecular evolution of several human metalloproteins charged with restricting bacterial access to transition metals. These include the transition metal-chelating S100 proteins, natural resistance-associated macrophage protein-1 (NRAMP-1), transferrin, lactoferrin, and heme-binding proteins. We examine their coevolution with bacterial transition metal acquisition systems, involving siderophores and membrane-spanning metal importers, and the biological specificity of allosteric transcriptional regulatory proteins tasked with maintaining bacterial metallostasis. We also discuss the evolution of metallo-β-lactamases; this illustrates how rapid antibiotic-mediated evolution of a zinc metalloenzyme obligatorily occurs in the context of host-imposed nutritional immunity.
The Host–Pathogen Interface: A Struggle for Supremacy
An infection constitutes one of the major selective pressures that act on humans, and the host–pathogen interaction contributes to shape the genetic diversity of both organisms [1]. The general antagonistic paradigm suggests that, ultimately, the outcome of the encounter between host and bacteria is decided by the interplay between immune effectors that evolved to directly damage the microbial cell and countermeasures deployed by the pathogen to ‘blunt’ these host insults. However, this paradigm has been reconsidered in the context of the competition between host and pathogens for nutritional substrates, which enhances the impact of modest alterations in metabolism and nutrient sensing on the ultimate outcome of an infection [2]. Metal ions are unique examples of such shared nutrients since their essentiality pairs with their toxicity. Thus, bioavailability of every transition metal is tightly regulated in both bacteria and the eukaryotic host. Successful pathogens have evolved strategies to scavenge metal ions from host extracellular proteins that transport metal ions between tissues and organs [3], turning the human host into an accessible and rich reservoir of these nutrients. Consequently, host evolution is driven by a necessity to maintain the adapted bacterial population in check. This is illustrative of the Red Queen hypothesis, which claims that competing species continually evolve to maintain a level of fitness in a shared niche [3,4]. The inflammatory responses associated with innate immunity and further restriction of metal ions via metal-chelation is a process termed nutritional immunity (see Glossary) [5], which acts to prevent infections [3]. To preserve the physiological intracellular bioavailability of metal ions, pathogenic bacteria employ a systems-level response termed metallostasis that allows not only for metal scavenging, efflux, and intracellular sequestration, but also for ribosome remodeling and metabolic reprograming that occurs as a result of severe metal restriction.
Pathogen metallostasis and host innate immunity exert reciprocal selective pressures on one another that have shaped the proteins that function at this interface. On the host side, the core proteins of nutritional immunity can be grouped in three categories: (i) transporters that induce metal poisoning or starvation in intracellular pathogens (Figure 1A); (ii) multifunctional iron-transport proteins, and (iii) secreted multimetal-chelating immune proteins (Figure 1B). Bacterial systems-level responses against these host defensive proteins rely on transport systems specific to chemical speciation of metal nutrients in host tissues; tight regulation of metal acquisition systems across cell membranes is facilitated by the active transport of microorganism-derived metal-scavenging molecules, for example, siderophores and metallophores, and ultimately, a metabolism-level response to host-induced starvation (Figure 1). The highly complementary nature of this ‘all-hands-on-deck’ response enables pathogens to survive an ever-changing landscape of metal bioavailability within the infected host.
Figure 1. Schematic Illustration of Human Proteins Involved in Depletion of Essential Metal Ions during Nutritional Immunity (Both Panels, Left), and Bacterial Effectors of Metal Ion Acquisition and Metallostasis (Both Panels, Right).
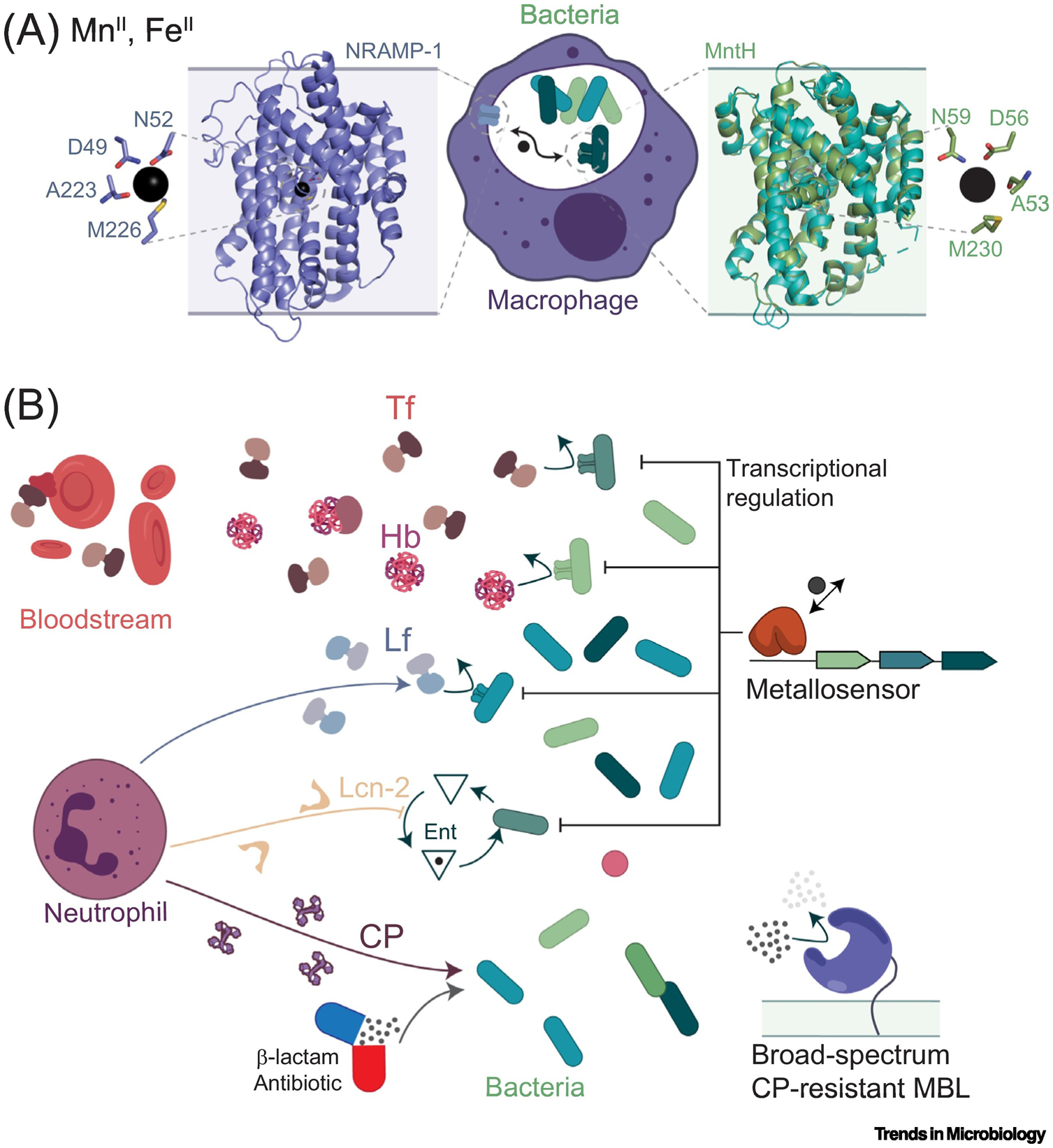
Proteins are shown in cartoon representation, metal ions as black spheres, and cell membranes as shaded rectangles. (A) Intracellular pathogens’ nutritional immunity proteins exemplified by the model human natural resistance-associated macrophage protein-1 (NRAMP-1) transporter DMT1 (Staphylococcus capitis DMT, PDB: 4WGW) and bacterial response exemplified by Deinococcus radiodurans MntH (PDB: 5KTE). Insets, MnII coordination chemistry. (B) Interplay between extracellular nutritional immunity proteins and bacterial response. Bacterial receptors for human serum transferrin (hTf), haptoglobin (Hb) and neutrophil-produced lactoferrin (Lf) drove the selection of variants with decreased affinity for the bacterial receptors or new immunomodulatory properties (curved arrows). Bacterial expression of metallophore production (exemplified by siderophore enterobactin or Ent) and import systems transcriptionally modulated by metallosensor proteins are counteracted by human neutrophil-produced lipocalin-2 (Lcn-2, also called siderocalin) and the multimetal-chelator calprotectin (CP). The latter, in addition to the use of β-lactam antibiotics, drove the selection for broad-spectrum metallo-β-lactamases (MBLs) from highly resistant pathogenic bacteria.
Recent advances in genome sequencing and gene synthesis have driven the development of powerful tools to assess protein evolution. These advances parallel a revolution in structural biology which has enabled the structure determination of large transmembrane protein complexes at atomic resolution, as well as developments in dynamics and thermodynamics approaches, to probe the molecular basis of specificity. Ancestral sequence reconstruction (ASR) might be the most important approach used to unveil how and when new functions appeared throughout evolution (Box 1). Furthermore, directed evolution assays and deep mutational scanning coupled to phylogenetic analyses allow for direct inference of selective pressures acting upon protein sequences (Box 1). In this review, we discuss the impact of evolutionary pressures responsible for the remarkable structural and functional diversity that continues to drive the ‘arms race’ over transition metal bioavailability in the infected host.
Box 1. Tools for Assessing Protein Evolution.
Identifying function-altering mutations in proteins is important because these can confer pathogen adaptability and evasion of the immune response of its host. A protein of interest can be compared with homologous proteins with different functions through horizontal analyses. Here, polymorphisms in residues important to catalysis are swapped between proteins, and functional shifts are experimentally assessed [120]. However, this approach usually fails because neutral substitutions occlude relevant ones, and epistatic interactions are not considered [120]. On the other hand, vertical analyses, like ASR, allow one to focus on substitutions that occurred when function changed. ASR is the calculation of ancient protein sequences using extant ones [121]. It is based on the phylogenetic analysis of a protein family obtained through multiple sequence alignments and a probabilistic model of evolution [120,121]. From this, ancestral protein sequences at every internal node of the phylogenetic tree can be inferred and artificially synthesized to evaluate changes in biological function during evolution. This has been beautifully illustrated by the Harms group for calgranulins and calprotectin [33]. By introducing mutations in the historical background where they occurred, ARS overcomes epistasis [120].
Multiple sequence alignments of extant proteins from evolutionarily divergent organisms allows us to quantitatively establish the presence of selective pressures acting upon a protein sequence. Positive selection is assessed by the estimation of the ratio between nonsynonymous (N) and synonymous (S) substitutions (dN:dS) across lineages [122]. Cross-analysis of dN:dS ratios between host-encoded and pathogen-encoded proteins is useful for identifying coevolving sites and causalities for natural selection [16].
Finally, artificial evolution methods uncover evolutionary pathways that lead to pathogen adaptation in hostile environments. All directed evolution approaches are based on expanding the sequence space of a given protein (through targeted or random mutagenesis), selecting for a desired phenotype, and allowing the genetic variants to replicate [123]. After multiple rounds of functional selection, it is possible to isolate a protein with new or optimized functionality [123]. In deep mutational scanning, the sequence space of a given protein is expanded in a high-throughput manner, as the functional consequences of every possible amino acid substitution are assessed for each position of a given protein. Necessarily, this requires a robust functional screening methodology [124]. In continuous evolution approaches, gene diversification, selection, and replication are carried out with minimal researcher intervention in continuous dilution systems [125]. Alternatively, the rapid evolution of a family of proteins, as the organism disseminates in new environments, can also be studied through molecular epidemiology, as in the case of β-lactamases [60].
Evolution of New Functions That Impact Nutritional Immunity in Human Host Proteins
The primordial origins of effector proteins that anchor nutritional immunity in humans are diverse. Some are ancestral metal-binding proteins that have more recently acquired novel molecular traits used to activate the immune response, while others have ancestral roles as damage-associated molecular patterns (DAMPs), with specific metal-binding properties only recently acquired, enhancing their antimicrobial activities. Although most proteins involved in nutritional immunity are soluble, and secreted into the extracellular space, the survival and replication of intracellular bacterial pathogens inside infected macrophages leverages a set of transporters normally found in the plasma membrane that are requisitioned to intoxicate these bacteria with ZnII and Cu or starve them for Fe, MnII, and possibly other metal ions [6] (Figure 1A). For example, the natural resistance–associated macrophage protein (NRAMP-1, SLC11) restricts MnII and other essential metals from intracellular pathogens, particularly those that replicate inside a membrane-enclosed compartment. Remarkably, NRAMP-1 shares the same molecular scaffold, metal coordination site, and metal specificity profiles with MntH, a cognate MnII importer and virulence determinant encoded by many human bacterial pathogens [7]. This suggests a common archaeal ancestor and highlights pathogen- and host-deployment of very similar tools to obtain these essential nutrients [8].
Interplay between Host Iron-Chelating Molecules and Bacterial Receptors
Upon infection, the innate immune system interfaces with systemic iron transport and increases the synthesis of several iron- and heme-binding proteins. One of these proteins is transferrin (Tf), a monomeric glycoprotein likely ubiquitous in metazoans, and namesake for the Tf protein superfamily. The exceptionally high affinity of Tf for FeIII, and its role in nutritional immunity in vertebrates, is well documented and widespread [9]. Despite its high affinity, Tf must release the iron upon endocytosis of a transferrin receptor (TfR)–Tf2 complex, the mechanism of which has only recently been explored [10,11] (Figure 2A). Although TfR is present only in vertebrates [12], an outer membrane-embedded transferrin receptor found in members of the families Neisseriaceae, Pasteurellaceae, and Moraxellaceae allows these bacteria to acquire FeIII from Tf in a process termed ‘iron piracy’ [13]. The bacterial transferrin receptor is composed of two proteins, transferrin-binding proteins A and B (TbpA, TbpB), and each binds Tf at well defined, yet distinct interfaces [14,15]. This interaction with TbpA/B has likely driven Tf evolution, as suggested by the identification of positive selection sites in primate transferrins, located primarily in the Fe-binding C-lobe that binds TbpA, thus inhibiting that interaction, and blunting Fe piracy by the invading microbe [15] (Figure 2C). These positively selected sites are not expected to interfere with the binding of Tf to TfR, which primarily involves the N-lobe (Figure 2A); as a result, these Tf variants are expected to have little or no impact on systemic iron distribution in the host. This highlights an underlying evolutionary advantage of the bilobed architecture of Tf which accommodates a simultaneous evolutionary response to two orthogonal selective pressures, one derived from the host and the other away from the bacterial receptor (Figure 2C).
Figure 2. Selection of Human Iron Transport Proteins and Bacterial Uptake Systems.
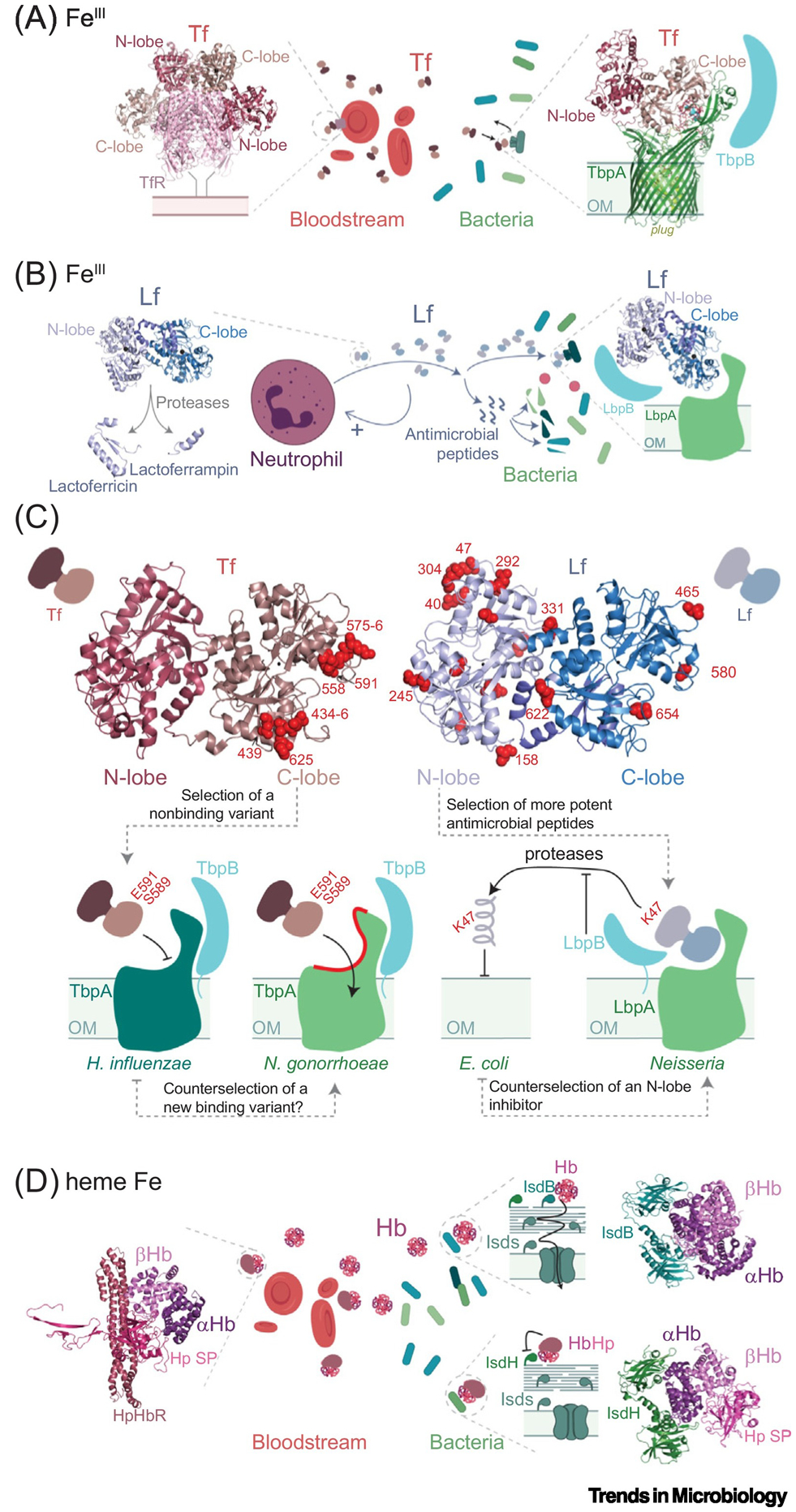
Proteins are shown in cartoon representation and cell membranes as shaded rectangles. (A) Human transferrin (Tf, PBD: 1SUV) interacts through distinct interfaces (N-lobe or C-lobe) with its cognate receptor in erythrocytes (red cells) and the Neisseria receptor TsbA (shown in complex with Tf, PBD: 3V89) and TsbB. (B) neutrophil-produced lactoferrin (Lf, PDB: 1B0l) with the antimicrobial peptides produced by proteolysis, and bacterial response exemplified by LbpAB [16]. (C) Above: positively selected sites across the primate clade represented as red spheres on human serum transferrin (hTf, PDB: 3V83) [15] and human lactoferrin (hLf, PDB:1B01) [16]. Below: schematic representation of the interaction of these proteins with bacterial Tf receptors in iron piracy, highlighting the location of key selected sites on both proteins that either prevent binding to the bacterial receptor (Tf) or convey antimicrobial activity against certain pathogens (Lf). The dashed arrows between bacterial species indicate an increased relative fitness. (D) Hemoglobin (Hb, in complex with haptoglobin, PDB: 4WJG) and bacterial response exemplified by Staphylococcus aureus receptors (IsdH, PDB: 6TB2 and IsdB, PDB: 5VMM), where the HbHp complex inhibits the bacterial receptor. Abbreviations: E. coli, Escherichia coli; H. influenzae, Haemophilus influenzae; N. gonorrhoeae, Neisseria gonorrhoeae; OM, outer membrane of Gram-negative bacteria.
Lactoferrin (Lf) is another paradigmatic member in the Tf superfamily; it arose from a duplication of a primordial Tf gene in the ancestor of eutherian mammals (Figure 2B). Lf is a crucial component of the mucosal innate immune response within the granules of polymorphonuclear leukocytes, and its N-lobe has evolved new immunomodulatory properties that are independent of Fe binding (Figure 2B). The N-lobe is highly cationic and capable of disrupting the microbial membrane, while also harboring two regions that, when processed by host proteases, liberate peptides that exhibit potent antimicrobial activity (Figure 2B). Indeed, most of the residues undergoing positive selection in primates localize to the N-lobe, particularly in those regions that are precursors to antimicrobial peptides [16] (Figure 2C). Additionally, some sites in Lf under positive selection in the infected host negate a physical interaction with a bacterial inhibitory protein, for example, Streptococcus pneumoniae PspA or the iron piracy receptor Neisseria LbpB which specifically extracts Fe from the Lf N-lobe (Figure 2C). Overall, the evolution of the Tf superfamily in humans exemplifies how gene duplication and subfunctionalization dramatically alter the landscape of host–microbe conflict [4].
The observation that Fe-binding proteins constitute an important reservoir of genetic resistance to infectious disease is not exclusive to the Tf protein family, as recurrent positive selection on primate α- and β-globins restricts hemoglobin (Hb) binding and Fe acquisition by pathogenic Staphylococcus aureus in another form of iron piracy [17] (Figure 2D). Here, the human acute-phase protein haptoglobin (Hp) restricts the availability of Hb-bound Fe to the pathogen by physically blocking one of the two staphylococcal Hb-binding protein-mediated heme extraction pathways, by the iron-regulated surface determinant IsdH [18] (Figure 2D). Remarkably, Hp prevents bacterial heme extraction not by direct interaction with IsdH but by restricting the conformational ensemble of Hb, thus weakening the receptor interaction [19]. It is not yet known if the second staphylococcal Hb receptor IsdB appeared as a result of a duplication event in pathogenic Staphylococcus spp. to evade Hp-mediated inhibition of Fe uptake (Figure 2D), but it is known that IsdB is found exclusively in staphylococci capable of infecting deep tissues and causing invasive disease [17].
The Origins of the Antimicrobial Activity of Calprotectin
The innate immune system has also evolved modern proinflammatory proteins deployed to restrict a broader variety of transition metal ions beyond FeIII. These proteins are members of the S100 superfamily of calcium-binding proteins found only in vertebrates and in their closest phy-logenetically related group [20] (Figure 3A,B). While S100 proteins have multiple intracellular functions [21], most are secreted into the extracellular milieu and bind pattern-recognition receptors (PRRs) which generally results in the initiation and upregulation of an inflammatory response, for example, the receptor for advanced glycation end products (RAGE) and Toll-like receptor 4 (TLR4) [21]. These receptors are well-known for their recognition of DAMPs or pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS). In contrast to what is considered an ancestral proinflammatory response characteristic of nearly all S100 subgroups, the calgranulin subgroup, composed of S100A7, S100A8, S100A9, and S100A12 in humans, has recently evolved a unique antibacterial activity defined by transition metal sequestration, while also retaining proinflammatory activity [20,22–24] (Figure 3B).
Figure 3. (A) Model of Action for Neutrophil-Produced Multimetal-Chelating Calprotectin (CP, PDB: 4GGF).
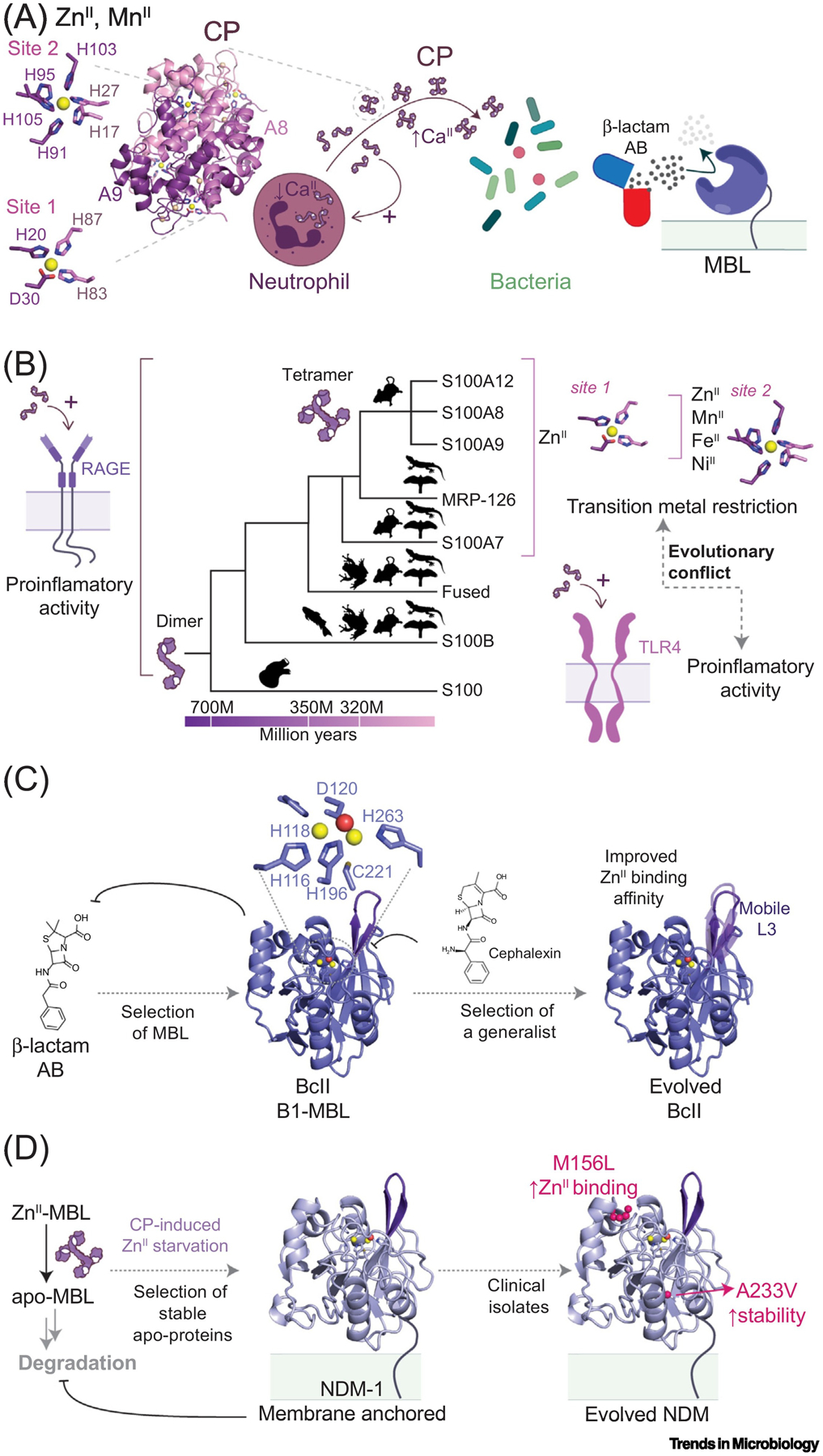
Dimeric CP is secreted to the extracellular milieu upon activation of neutrophils where high CaII levels induce heterotetramerization [30], which enhances transition metal affinity and resistance to extracellular proteases. Details of the transition metal coordination site 1 (ZnII-specific) and site 2 are shown to the right. Left: β-lactam antibiotics and bacterial response exemplified with metallo-β-lactamases (MBLs), which are sensitive to ZnII chelation by CP. (B) S100 protein evolution across vertebrates. They arose in the last common ancestor of vertebrates (represented by the fish, frog, mouse, bird, and lizard silhouettes) and urochordates around 700 million years ago, while calgranulins appeared in the ancestor of amniotes (320 million years ago). In mammals, the calgranulin clade expanded via gene duplication events (top) [20,23]. Illustration of conserved interaction of most S100 proteins with the receptor for advanced glycation end products (RAGE) (left) and calgranulin-specific interaction with TLR4, exemplified by heterodimeric CP and homodimeric S100A8 and S100A9 (bottom, right) [21,23]. Illustration of nutritional immunity activity restricted to calgranulins, with heterotetrameric CP eliciting broad-spectrum metal restriction attributed to a metal-agnostic binding site 2 that is unique in the S100 superfamily (top, right). (C) and (D): Evolution of MBLs. Proteins are shown in cartoon representation and the ZnII ions as yellow spheres and coordinated water molecule as a red sphere. (C) The extensive use of β-lactam antibiotics acted on bacteria as a selective pressure that resulted in the expansion of MBLs in bacterial populations, such as BcII (PDB: 4NQ4). BcII cannot hydrolyze carbapenem and is sensitive to ZnII depletion, as apo-MBLs are readily degraded by proteases in the periplasm. Further selection exerted by carbapenem antibiotics and nutritional immunity results in variants with increased antibiotic binding site flexibility, which enhances substrate promiscuity, and increased ZnII binding affinity [61,68]. (D) In NDM-1 (PDB: 5ZGZ), membrane anchoring prevents proteolysis in the periplasm of the apo-form and renders the protein highly resistant to nutritional immunity-imposed ZnII restriction [71,72]. Further mutations from clinical isolates (two are shown) that occur from the active site of the protein enhance resistance toward host-imposed ZnII deprivation [74].
The four mammalian calgranulins likely arose as a result of gene duplication from an ancestral calgranulin; a fifth member of the calgranulin subfamily, sauropsid calgranulin (MPR126), is found only in birds and reptiles [20,23]. All five calgranulins harbor a conserved tetrahedral ZnII coordination site, characterized by a low- to sub-nanomolar affinity, and exert some level of antimicrobial activity towards certain pathogens [25–28]. By contrast, the heterodimeric complex of S100A8 and S100A9 (named calprotectin, CP), and particularly in its CaII-induced S100A82/S100A92 tetrameric form, exerts potent antibacterial activity against a broader spectrum of pathogens [24,29,30]. CP is unique among calgranulins as it harbors an additional transition-metal coordination site 2 at the heterodimer interface, consisting of six histidines from S100A8 and S100A9 subunits (Figure 3A). Site 2 is largely metal-agnostic and conveys nanomolar or greater affinity for all late 3d-block divalent metal ions, ZnII, MnII, FeII, NiII, CoII, and CuII (Figure 3B). This allows CP to further restrict the growth of bacterial strains that harbor mechanisms to evade ZnII-specific metal starvation induced by the other calgranulins [29,31,32]. Indeed, a recent ASR study (Box 1) reveals that only the ancestral CP conserves the hexa-histidine site and exhibits superior antibacterial activity, while the ancestral calgranulins show proinflammatory effects, albeit weaker than those of modern calgranulins [33].
Thus, the dual function of calgranulins requires two seemingly orthogonal characteristics: one as a short lived proinflammatory cytokine and the other as a protease-resistant transition metal-chelating protein. This apparent evolutionary conflict is elegantly resolved by the fact that S100A8 and S100A9 are each capable of forming both homodimers and the heterodimer CP. The antibacterial function is predominantly achieved by the CaII-bound heterotetrametric form of CP that is strongly resistant to degradation by proteases and binds TLR4 poorly [34,35]; by contrast, the relatively low stability of the S100A8 and S100A9 homo-oligomers enhances the temporal nature of an optimized proinflammatory response; indeed, this may have evolved through a single nucleotide polymorphism from a proteolysis-resistant, weakly proinflammatory ancestral calgranulin [33]. Emerging evidence suggests that CP and host-resident pathogens continue to coevolve with the discovery of a physical association growth-inhibition mechanism in Borrelia burgdorferi that is dependent on the ancestral ZnII site, but independent of both metal restriction and the proinflammatory response [36]. On the bacterial side, a TonB-dependent transporter in Neisseria seems to have evolved to specifically capture human CP-bound ZnII and transport the ion to the periplasm, overcoming ZnII starvation [37,38]. In the case of Neisseria gonorrhoeae this is a specific adaptation to the human host as the bacterial receptor shows higher affinity for ZnII bound to the hexa-histidine site of human CP than to mouse CP [37]. Furthermore, and as discussed later, metal chelation by CP induces evolutionary pressure on metallo-β-lactamases, which depend on ZnII coordination to confer bacterial resistance to β-lactam antibiotics. It is not surprising that rapidly emerging, highly antibiotic-resistant bacterial strains possess mechanisms to overcome the effect of CP on the functionality of these enzymes (vide infra).
Lipocalins
The mammalian lipocalin-2 (Lcn-2; siderocalin, NGAL) functions in nutritional immunity and, like CP, not only restricts pathogen access to transition metal ions but also activates the inflammatory response (Figures 1B and 4A). Lcn-2 restricts FeIII by sequestering the archetypical circular tris-catecholate siderophore Enterobactin (Ent), produced by members of the family Enterobacteria-ceae for acquiring iron, characterized by the highest FeIII affinity among all known siderophores [39]. Known siderophores are collectively characterized by an impressive diversity of chemical structures far beyond Ent that provide bacteria multiple ways to scavenge (and solubilize) this essential nutrient without directly interacting with human iron-transport proteins [40]. Additional host-driven evolutionary pressures may in fact have selected against Ent [41]. Consistently, hypervirulent bacterial strains encode multiple siderophores, and successful pathogens have leveraged an impressive diversity of biosynthesis pathways [42,43] and FeIII-uptake mechanisms to enhance Fe acquisition and thus pathogenesis. A well known evolutionary adaptation against FeIII–Ent sequestration by Lcn-2 is the modification of Ent through the addition of glucose groups to the C5 carbon to produce salmochelin (Sal) [44], which binds weakly to Lcn-2 (Figure 4A). Lcn-2 is also implicated in the regulation of the inflammatory response as it modulates the expression of cytokines in neutrophils and macrophages [45,46].
Figure 4. (A) Left: Neutrophil-Produced Siderocalin (Lcn-2, in Complex with Enterobactin, Ent; PDB: 3K3L) and Bacterial Response Exemplified with the Production of Ent and Salmochelin (Sal), Which Is Not Recognized by Lcn-2.
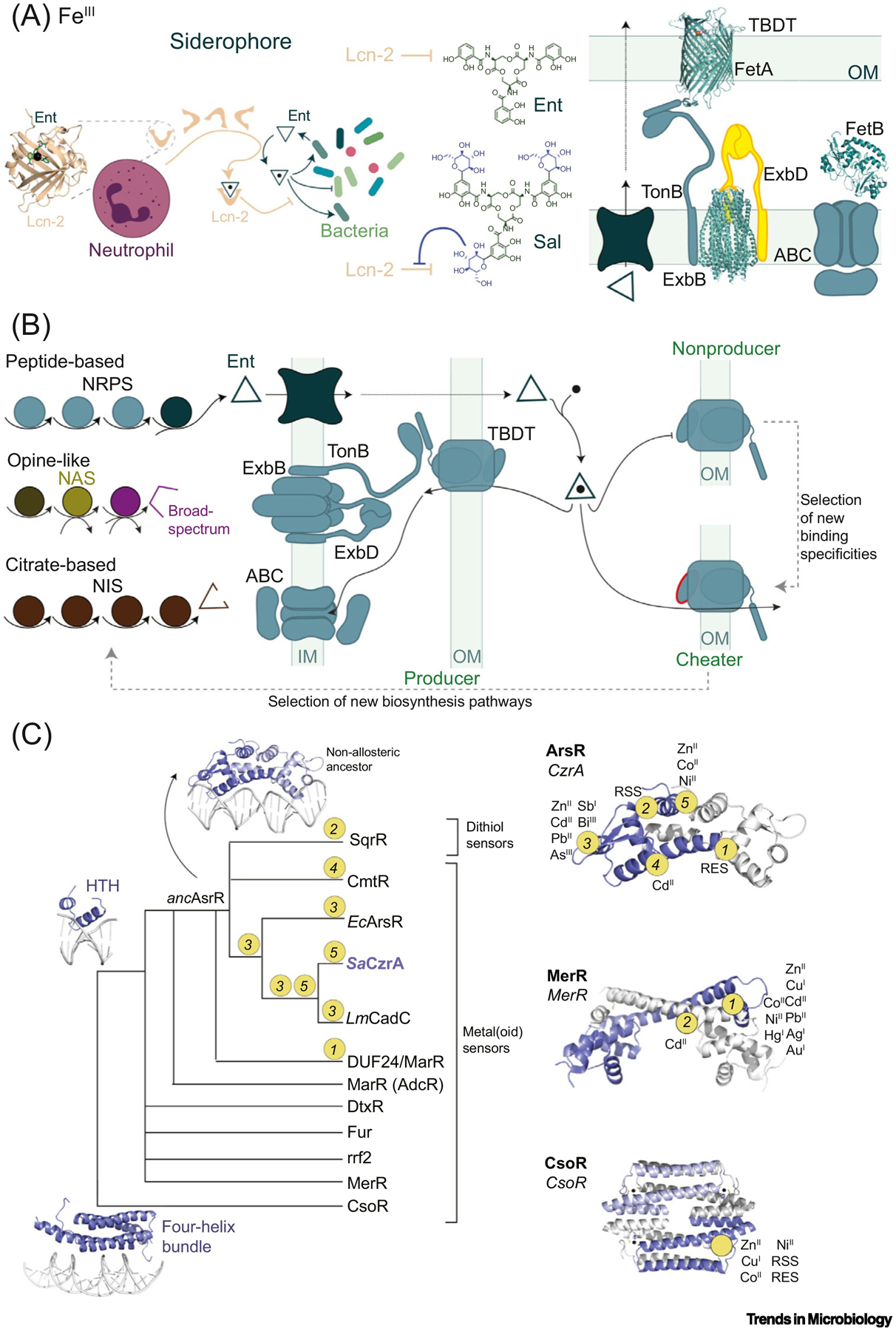
Right: Gram-negative bacterial import system exemplified by the TonB-dependent transporter (TBDT) ZnuD (PDB: 4RVW), ExbB/ExbD complex (PDB: 5SV1), and an ATP-binding cassette (ABC) transporter (showing the structure of solute-binding protein FepB (PDB: 2M6L). (B) Siderophore or metallophore production (left) and cellular uptake (right). In Gram-negative bacteria, siderophores are imported by TBDTs into the periplasm and then into the cytoplasm by ABC transporters. In non-siderophore-producing bacteria (cheaters), new binding affinities and specificities in TBDTs are selected for, allowing for adaptation to new niches [87]. This, in turn, exerts a selective pressure on siderophore-producing bacteria, resulting in new biosynthesis pathways that expand siderophore diversity. (C) Schematic representation of the evolution of transcriptional regulator families, as suggested by some studies [99,100,106,129]. Apart from the CsoR superfamily (which may have arisen from a four-helix bundle ancestor), most metallosensors share the helix-turn-helix (HTH) topology in a common ancestor. In the arsenic repressor (ArsR) superfamily, the five sensory sites evolved from a nonallosteric ancestor with distinct metal or reactive species affinities and reactivities, respectively. The approximate positions of inducer recognition sites and sensing specificities are shown in representatives of the ArsR (2M30), MerR (5CRL), and CsoR (4M1P) superfamilies [103,130]. Abbreviations: NIS, NRPS-independent siderophore; NRPS, non-ribosomal peptide synthesis; OM, outer membrane; RSS, reactive sulfur species; RES, reactive electrophile species.
The lipocalin superfamily encompasses secreted proteins that bind a wide array of small hydro-phobic ligands with high affinity, despite low sequence identity, and perform a transport function in the vascular system of pluricellular organisms [47,48]. To the best of our knowledge, no phylogenetic studies have reconstructed an ancestral Lcn-2, so ancestral or derived biological functions cannot be inferred, as has been done with CP. However, there is emerging evidence that Lcn-2 has a host-protective function in kidney epithelial cells, beyond its known antimicrobial activity, by binding simple catechols secreted by gut commensal bacteria for endogenous iron transport in aseptic injuries [49]. This suggests that the antimicrobial activity of Lcn-2 may have evolved from an ancestral commensal interaction with a nonpathogenic enteric bacterium.
Rapid Evolution of Antimicrobial Resistance in the Context of Nutritional Immunity
Multidrug-resistant bacteria represent one of the major threats to modern human civilization, as antibiotic-resistance genes are spreading around the world at an alarming rate. Hydrolysis (inactivation) of β-lactam antibiotics by bacterial β-lactamases (BLs) is the most prevalent mechanism of resistance [50,51]. The extensive use and misuse of these life-saving drugs exert a strong evolutionary pressure on bacteria that results in the selection of BLs capable of providing enhanced resistance. The evolution of BLs uncovered by molecular epidemiology therefore reports on the different bacterial microenvironments that define evolutionary restraints (Box 1) [52]. BLs can be grouped into two large families: serine β-lactamases (SBLs) and zinc-dependent or metallo-β-lactamases (MBLs) [51,53]. While SBLs have been used as a model to study protein evolution, in particular TEM-1 [54–57], molecular evolution of MBLs is not fully understood and no MBL inhibitor has been identified or developed [58,59] for these rapidly evolving enzymes [60]. Our current understanding of SBLs cannot be translated to MBLs since these enzymes are evolutionarily unrelated, displaying different folds, active sites, and catalytic mechanisms [51]. MBLs are specifically affected by host nutritional immunity since ZnII ions are essential for substrate binding [61] and hydrolysis [62] (Figure 3A,C,D). MBLs belong to an ancient superfamily of ZnII and FeII metalloenzymes with diverse activities, largely hydrolases and oxygenases [63]. There are three subclasses of MBLs (B1, B2, and B3) which differ in the ZnII coordinating residues of the active site and ZnII stoichiometries. Here we discuss the molecular evolution of B1 MBLs, the most extensively studied and clinically relevant among these enzymes. B1 MBLs bind two ZnII ions in a conserved coordination motif (Figure 3C), where a Cys residue and second sphere ligands play key roles in tuning the ZnII affinity [64,65].
Directed evolution experiments (Box 1) have provided significant insights into how mutations affect the catalytic efficiency and broad substrate specificity of MBLs [66,67]. Evolved variants of the Bacillus cereus MBL (BcII) that gain flexibility in the mobile loops flanking the active site display a broader substrate range than the native enzyme, without sacrificing the activity towards original substrates [68] (Figure 3C). This suggests that conformational dynamics is an evolutionary trait that can be optimized in MBL evolution (Box 2). These evolutionary lineages also allowed for the identification of an additional evolutionary trait, as evolved variants conferred enhanced resistance under conditions of metal starvation. Furthermore, strong epistatic interactions between coevolving residues dictate evolutionary outcomes under selective pressure, as mutations have pleiotropic effects, simultaneously impacting catalytic efficiency, protein stability, and ZnII-binding affinity [69]. What is relevant here is that the ZnII-binding affinity in the periplasm is an essential characteristic that defines the fitness landscape of MBLs.
Box 2. Evolution in Motion: The Role of Dynamics in Protein Function.
The sequences, structures, and functions of modern proteins are the result of an evolutionary process exerted upon sequence–structure–function space. Mutations constantly erode protein sequences, generating variants that may affect the biological activity of a protein, subjecting it to natural selection. An understanding of how changes in protein sequence are ultimately capable of generating new functions remains an elusive goal. The ‘one structure, one function’ paradigm suggests that function-shifting mutations should induce a measurable change in protein structure. However, this vision is rendered obsolete by the striking divergence in functions observed in many families of structurally homologous proteins. The native state of a protein is more accurately represented by an ensemble of conformations accessible at thermal equilibrium. This distribution of accessible states is encoded in protein dynamics. Changes in protein sequences impact on this equilibrium by restricting conformations necessary for their functions. Thus, altering the intrinsic dynamics of a protein is a mechanism by which new functions evolve.
Recent convergent developments in NMR spectroscopy, X-ray crystallography, and computational methods open up the possibility of merging structure and dynamics to obtain true atomic-resolution integrative descriptions of biomolecules [126]. Protein dynamics play a fundamental role in the most basic requirement for protein function: the capacity to bind to a particular ligand to the exclusion of all others. This tuning is generally difficult to accomplish from evolutionary perturbations of the few amino acids that directly interact with the ligand. Thus, specificity of a protein is controlled through conformational changes in groups of residues far beyond the ligand-binding site, allowing proteins to evolve allosteric connectivities and adapt to new selective pressures [127]. This is evidenced by multiple studies performed on β-lactamases and transcriptional regulators, where mutations occurring at sites far from the active site are responsible for altering protein function with minimal impact on the conformational ensemble [69,98,114]. Furthermore, this has been observed in directed evolution assays, where distant substitutions rescue native-like protein dynamics and restore enzymatic activity on functionally impaired proteins [67]. A recurring mechanism that seems to be employed by evolution of protein function is one that involves the ‘tuning’ of protein dynamics, or entropy redistribution. By enhancing the flexibility of certain sites while stiffening others, proteins retain the same overall fold but these substitutions impact their function, allowing either the acquisition of new functions or allosteric modulation of their activities [98,114,128].
New Delhi metallo-β-lactamases (NDMs) have recently arisen as potent, rapidly disseminating, broad-spectrum MBLs that show remarkable resistance to host-derived ZnII starvation [70]. While most MBLs are readily degraded in the absence of ZnII, NDM-1 accumulates in the periplasm in the apo form [71]. This is due to the fact that NDM-1 is lipidated at a Cys residue at the N terminus in a post-translational event after transport to the periplasm, which results in lipid anchoring to the outer membrane. Membrane anchoring renders NDM-1 resistant to degradation. This is a general phenomenon since engineering of the lipidation site into soluble MBLs also results in stabilization of these proteins in the absence of ZnII. The soluble domain of NDM-1 has also been shaped by evolution to favor the interaction with the membrane. Thus, membrane anchoring is a novel mechanism by which MBLs can achieve resistance to metal chelation induced by CP [71,72] (Figure 3D). Remarkably, 29 allelic variants of NDM have now been reported in the clinic [73], and all accumulate mutations far from the active site; most confer enhanced resistance under conditions of zinc deprivation, relative to NDM-1 [74]. Although the specific effects and potential epistatic interactions of each mutation remain to be dissected, these mutations enhance resistance by increasing the ZnII binding affinity, and/or augmenting the capacity of the apo forms to resist degradation in the periplasm, not by enhancing catalytic activity [74–76] (Figure 3D). The evolution of NDM β-lactamases represents a case study on how host-mediated nutritional immunity is driving the evolution of MBLs to enhance resistance to another pathogen challenge, antibiotics.
Evasion Mechanisms in Bacteria for Essential Nutrient Acquisition
A common feature of most successful pathogens is that they employ multiple, nonredundant and complementary mechanisms to acquire metal nutrients, notably Fe, to overcome host-mediated metal restriction. For example, while many streptococci possess only a single high-affinity Mn transporter [77], complementary mechanisms for Mn uptake in S. aureus have been shown to allow survival against the synergistic actions of nutritional immunity and other host defenses [78]. In Vibrio spp., different environmental signals allow for the expression of the more efficient Fe uptake mechanism in a certain niche [79]. Fe uptake mediated by siderophores constitutes a unique example of molecular evolution that shapes bacterial communities, due to the need to encode the biosynthetic pathways and/or the uptake systems required to utilize this form of Fe as a nutritional Fe source. Some organisms encode the capacity to biosynthesize more than one siderophore, which may be suitable for one host tissue over another, as recently observed in S. aureus in murine models of infection [80]. Other bacteria simply do not pay the biosynthetic costs to synthesize their own siderophores but instead express only the acquisition machinery, likely acquired via horizontal gene transfer, often for more than one siderophore class, in a process termed siderophore cheating. Other evasion mechanisms include the secretion or utilization of bacterially produced or host-endogenous bis- or monomeric Fe-chelating siderophores with lower FeIII stabilities, for example, simple catecholamines or Ent hydrolysis products, that cannot necessarily outcompete Tf or Lf, but employ sacrificial redox chemistry [32] or enhanced cell permeability to acquire Fe from the surrounding tissues [81]. Mechanisms for the direct uptake of FeII are emerging in importance as a bacterial strategy that avoids competition with the host for FeIII altogether; the host, in turn, is then able to deploy CP in an effort to sequester FeII from the pathogen (Figure 1B and Figure 4A).
Much of the chemical diversity of FeIII-chelating siderophores derives from the evolution of different combinations of chain initiation, elongation/tailoring, and termination steps in non-ribosomal peptide synthetases (NRPS) [43]; in addition, four structural classes of NRPS-independent siderophore (NIS) synthetases that synthetize derivatives of citric acid are known [42]. The chemical diversity of these molecules is remarkable (with catecholate, hydroxamate, carboxylate, and mixed-type FeIII coordinating donors); however, this diversity only allows for the uptake of FeIII. Broad-spectrum metal-binding molecules, termed metallophores, employ a distinct biosynthetic pathway characterized by low levels of chemical diversity, at least as far as is currently known [82–84] (Figure 4B). The uptake of both siderophores and metallophores requires high-affinity membrane transporters to actively import these scarce nutrients at considerable energetic cost. In Gram-negative bacteria, nutrient import into the periplasm is mediated by porin-like transporters in the outer membrane (OM) that are coupled to the proton-motive force through interaction with the TonB complex, tethered to the inner membrane (IM). The ligand specificity of a TonB-dependent transporter (TBDT) is strictly dictated by the globular ‘plug’ domain and the extracellular loops of these 22-stranded transmembrane β-barrels [85] (Figure 4A). Once inside the periplasm, nutrients are imported into the cytoplasm by ATP-binding cassette (ABC) transporters, in which the periplasm-facing solute binding proteins (SBPs) bind the nutrient cargo; these tend to be more promiscuous than the TBDTs [86]. In Gram-positive bacteria that lack an OM, the exquisite specificity of nutrient acquisition is dictated by the solute-binding lipoproteins themselves.
A suppressor screen in Bradyrhizobium japonicum mapped a handful of gain-of-function nutrient-adaptive SNPs and deletion mutations in a TBDT under conditions of iron deprivation and supplementation with a synthetic siderophore [87]. In other cases, the TBDT has some natural level of promiscuity, allowing transport of distinct endogenous and artificial siderophores [88]. TBDT plasticity derived from productive mutations or from functional group recognition also buttresses siderophore cheating, thus endowing the siderophore producer with a competitive advantage, while also serving a cooperative good [89]. TBDT plasticity may also constitute significant selective pressure to diversify siderophore biosynthesis pathways [90], as substantial fitness benefits of cooperation might not be evolutionarily stable [91]. In the context of an infection, bacterial cheater communities can be selected over siderophore producers [4], as has been demonstrated in Pseudomonas communities in lung tissues [92] (Figure 4B). The most extreme specialization is probably the synthesis, uptake, and production of membrane-anchored siderophores in Mycobacterium tuberculosis [91]. The evolutionary origin of siderophore secretion is unknown, but their soluble nature argues that, despite the self-directed benefits, the evolution of chemical diversity in siderophores may be determined in part by their indirect benefits to the community [91]. Future research in polymicrobial communities in the infected host will ultimately elucidate how the evolution of siderophore synthesis, secretion, and uptake has been shaped by both commensal and pathogenic bacteria [93].
Transcriptional Regulators Coordinate a Systems-Level Metallostasis Response
Metallostasis controls the expression of all nutrient acquisition processes described above in response to transition metal bioavailability, generally in the cytoplasm [94]. Both protein- and RNA riboswitch-based transcriptional regulation and post-transcriptional small regulatory RNA (sRNA)-mediated processes are operative [95,96] so as to avoid large fluctuations in metal levels, which could result in undermetalation or mismetalation of the proteome [31,97]. Protein-based transcriptional activators and repressors are bioinorganic allosteric switches and primary arbiters of bacterial metallostasis, as changes in gene expression result from changes in the metalation status of these metallosensors. Among the more than 20 structural classes of so-called ‘one-component’ sensors in bacteria, where direct ligand (metal) binding allosterically regulates DNA operator binding, no fewer than 12 have metallosensors [98]. Most metallosensors are helix-turn-helix and winged helix superfamily members proposed to be monophyletic [99], with the prominent exception of the CsoR (copper-sensitive operon repressor) protein superfamily, proposed to have evolved from a four-helix-bundle RNA-binding protein [100] (Figure 4C).
The capacity of bacterial pathogens to infect their hosts depends on the precise spatiotemporal regulation of gene expression, and it is this tight regulation that enables the evolution and diversification of the metallostasis response. Thus, the metal specificity and functional diversity of these metallosensors could serve as excellent model systems to explore the molecular evolution of metallostasis. However, in most cases the evolutionary origin of transition metal specificity remains elusive, and almost certainly derives from contributions of the first (primary) and second metal coordination shells [100–102]. In addition, a high degree of sequence divergence over a relatively small domain structure (≈80–130 residues) hinders robust reconstruction methods that could be used to identify a primordial sensor for each metallosensor family. In many cases, even when sequence similarity networks suggest a common evolutionary origin [103], the structure of extant proteins with similar functional specificities, yet distinct metalloregulatory sites, suggests that many metal-binding sites in metallosensors may have convergently evolved [104] (Figure 4C).
As the bacterial genome size and developmental lifestyles increase in complexity, the number of individual members of a metallosensor family tends to increase [103,105,106]. Diversification of a metallosensor family may occur through gene duplication events that expand the evolutionary space of a protein without compromising an essential primordial function. In the case of MarR (multiple antibiotic resistance repressor), an ancestral family of transcriptional regulators that pre-dates the divergence of Bacteria and Archaea, gene duplication events are thought to have been the primordial means by which this family diversified [107]. On the other hand, horizontal gene transfer in polymicrobial communities has likely played an important role in the evolution of the metal-sensing motifs in metallosensors [103]. The metal specificity profile of a metallosensor is almost certainly evolutionarily linked to adaptation to a particular microenvironment, whether an obligate human pathogen, for example, M. tuberculosis, or an environmental organism, for example, Methanocella arvoryzae. For example, the actinomycetes Streptomyces coelicolor, a soil bacterium, and M. tuberculosis encode 13 and 10 arsenic repressor (ArsR) members, respectively, and 5 CsoR family members, far more than virtually any other organism. Moreover, the multiple metal-sensing sites of global uptake regulators for iron and zinc, Fur and Zur (ferric and zinc uptake repressors, respectively) [28], may define an evolutionarily acquired trait that enables a more nu-anced ‘graded’ response to distinct degrees of metal starvation, particularly important as Fur and Zur regulons expand [108,109].
Given the complications of studying the natural evolution of diverse metallosensor superfamilies, comparative studies of functionally distinct sensors from the same structural class have been undertaken to obtain ‘snapshots’ into the evolution of different biological outputs (functional diversity). In some metallosensor families new biological outputs are proposed to have arisen by missense substitutions that ‘remodel’ a single sensing site while maintaining an evolutionarily conserved allosteric connection(s) [101,105,110,111]. However, there are clear exceptions to this simple view. CdII sensing by Pseudomonas putida CadR (cadmium regulator), a MerR (mercury resistance regulator) family transcriptional activator [112], requires the unanticipated cooperation between a canonical thiolate-rich site I, remodeling of which generally impacts MerR family metal specificity, and a unique histidine-rich site II [113], thus repurposing a MerR into an exquisitely selective cadmium-sensing activator dependent on the evolution of a new allosteric connection. Similarly, in the ZnII uptake repressor AdcR, a MarR family member from S. pneumoniae, the canonical ‘cleft’ that defines ligand recognition [105] and allosteric inhibition of DNA binding in other MarRs has been repurposed by the insertion of a flexible loop in the DNA-binding domain; this evolutionary insertion entropically impairs DNA binding by apo-AdcR, and activates repression upon ZnII coordination to primary and secondary metal sites, thus allowing for a more finely tuned ‘graded’ response to Zn starvation [114,115]. Finally, CuI-dependent inhibition and ZnII-dependent activation of DNA binding in the copper sensor CopY from S. pneumoniae has evolved on the same molecular scaffold by repurposing the remarkable structural plasticity of BlaI, the β-lactamase repressor from S. aureus [116].
These examples seem to argue that bacterial pathogens have relied not only on the ability of allosteric systems to evolve new biology by permutation of a single ligand recognition site, but also the rapid evolution of new allosteric connectivities. A hallmark example of a metallosensor superfamily with diverse stressor recognition sites is the ArsR family of proteins (Figure 4C). ArsRs are characterized by the greatest metal-site diversity among all metallosensors in terms of the number of distinct sites and variety of metals, metalloids, and reactive small molecules that can be sensed by individual ArsRs. This functional diversity is anchored by at least five structurally distinct sensory sites that have evolved throughout a conserved molecular scaffold and that are proposed to have arisen from a single primordial CdII sensor (Figure 4C) [103]. Recently reported conformational dynamics experiments on a single ArsR protein reveal that allosteric connectivity is facilitated by an ‘entropy reservoir’ that stabilizes the DNA-bound form of the repressor, and metal binding simply prevents access to this reservoir [98]. Although the degree to which these features characterize other ArsR family regulators is unknown, this model of evolution of new allosteric connections suggests that internal dynamics may create conserved latent allosteric connections, enabling new functionalities in the evolution of other metallosensor families [117] (Box 2). The existence of such latent allosteric connections within the physical scaffold of a functionally highly diverse metallosensor family suggests that it would be possible to rewire allostery of non-native inducer recognition sites in a directed evolution experiment [118,119], where the complications of natural evolution of an ancestral system are avoided.
Concluding Remarks
Microbial pathogens and vertebrate hosts engage in a perpetual struggle for supremacy. Nutritional immunity is among the first responses a pathogen encounters in a new host. Therefore, nutritional immunity has shaped, through natural selection, those bacterially derived molecules responsible for their virulence and adaptation to nutrient-deficient conditions. This adaptation has, in turn, resulted in selective pressures in host-derived proteins that function in nutritional immunity, which evolved new traits to counteract bacterial infections. Insights into this complex interplay, beautifully illustrative of the Red Queen hypothesis [4], have been gained only recently, with the application of directed evolution, ASR, and high-throughput sequencing methods. Here, we have highlighted this interplay in our discussion of the sequence variability of human iron-transport proteins, which originated with the evolutionary pressure from bacterial receptors that directly interact with them. Similarly, nutritional immunity exerts a selective pressure in bacterial metal-import systems resulting in diversification of siderophores by bacterial producers and in the emergence of new affinities of importers such as TBDTs from bacterial ‘cheaters’. By contrast, the evolution of S100 proteins seems to have been driven by gene duplication events that gave rise to a clade that evolved high-affinity transition metal-binding sites. In this area, ASR provided key insights into ancestral calgranulin function and helped to directly assess the emergence of new immune functions during evolution. We believe that this approach could yield breakthroughs in our understanding of not only the evolution of our immune response but also in our understanding of bacterial resistance and its emergence, particularly in the evolution of bacterial metalloregulators. Because of this, we hope to see ASR applied more consistently across the field. We believe that the evolution of regulation and antibiotic resistance in the context of nutritional immunity defines an exciting new field of research ripe for discovery, as protein dynamics and allostery have likely facilitated the rapid rise of new functionalities in bacterially encoded regulatory proteins [98,114] (see Outstanding Questions). As bacterial resistance becomes a major threat to humankind, understanding the molecular evolution of the systems reviewed here will play a critical role in preventing deadly pandemics in the postantibiotic era.
Outstanding Questions.
The emergence of a transferrin paralog allowed for accumulation of mutations that led to the antimicrobial activity of lactoferrin. Did gene duplication events also drive the emergence of new variants of bacterial hemoglobin receptors such as IsdB, which confers enhanced pathogenicity in Staphylococcus spp.?
Heterodimeric calprotectin acts as a potent antimicrobial transition metal chelator that forms tetramers of high stability, while S100A8 and S100A9 homodimers function as proinflammatory cytokines that are rapidly degraded by proteases. This prevents substantial damage to the host during inflammation. How is the production of these distinct complexes modulated at infection sites to maximize a systemic antimicrobial response?
The functional importance of an ‘entropy reservoir’ has been documented for the staphylococcal zinc efflux repressor, CzrA. Is the entropy reservoir a conserved evolutionary feature of the arsenic repressor (ArsR) protein family? If so, to what extent is this feature responsible for the emergence of new biological outcomes in this ubiquitous protein family?
What is the mechanism by which mutations far removed from the catalytic dinuclear ZnII site in metallo-β-lactamases enhance the ZnII binding affinity?
Why does membrane association provide enhanced periplasmic stability to apo-metallo-β-lactamases?
Highlights.
The interaction between human iron-transport proteins and bacterial receptors acts as a driving force for the diversification of both families of proteins.
Calprotectin, a pivotal nutritional immunity protein, evolved from a family of proinflammatory proteins that developed high-affinity binding sites for transition metals.
Plasticity of siderophore importers allows for nutritional iron acquisition by bacterial cheaters, which, in turn, selects for diversification of siderophore biosynthetic pathways in producers.
Comparative studies of inorganic sensors from the same structural class provide insights into the evolution of functional diversity in regulators of metallostasis.
ZnII-binding affinity and apo-protein stability of periplasmic metallo-β-lactamases are key evolutionary traits of these enzymes in the context of nutritional immunity.
Acknowledgments
This work was supported by grants from the US National Institutes of Health (R35 GM118157 to D.P.G., R01 AI100560 to A.J.V.), and the Pew Foundation, USA, Bunge & Born, Argentina, and Williams Foundations, Argentina (to D.A.C.). D.A.C and A.J.V. are Staff Members from CONICET. G.T.A. is supported by a graduate fellowship provided by CONICET, Argentina.
Glossary
- Bacterial cheaters
non-siderophore-producing bacteria that benefit from the siderophores synthesized by other bacteria (called xenosiderophores)
- Entropy reservoirs
a set of residues in a protein that gain degrees of freedom (increased conformational entropy) upon a binding event, for example, in transition from an apo to a ligand-bound state
- Epistasis
a generic term used to define a context-dependent effect of a particular mutation. Hence, the fitness effect of such mutation depends on the genetic background in which is introduced, that is, the presence or absence of another mutation
- Metallosensors
allosteric transcriptional regulators that bind a specific transition metal(s) in a way that modulates DNA binding affinity and/or the expression of regulated metallostasis genes
- Metallostasis
a systemic process by which cells control the intracellular bioavailability of functionally required transition metal ion cofactors, avoiding both metal ion starvation and toxicity. It is an abbreviated expression for bacterial ‘transition metal homeostasis’
- Nutritional immunity
host innate immune response that involves the withholding of metal nutrients to prevent bacterial growth. One of the most relevant nutritional immunity proteins is calprotectin, which sequesters virtually every transition metal available at an infection site
- Pleiotropy
a phenomenon in which a single mutation is responsible for affecting more than one molecular trait in a protein or enzyme, for example, protein stability and transition metal affinity
References
- 1.Pittman KJ et al. (2016) The legacy of past pandemics: common human mutations that protect against infectious disease. PLoS Pathog. 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olive AJ and Sassetti CM (2016) Metabolic crosstalk between host and pathogen: Sensing, adapting and competing. Nat. Rev. Microbiol 14, 221–234 [DOI] [PubMed] [Google Scholar]
- 3.Rohmer L et al. (2011) Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber MF and Elde NC (2015) Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet. 31, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg ED (1975) Nutritional immunity. JAMA 231, 39. [DOI] [PubMed] [Google Scholar]
- 6.Sheldon JR and Skaar EP (2019) Metals as phagocyte antimicrobial effectors. Curr. Opin. Immunol 60, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cellier MF (2012) Nramp: From sequence to structure and mechanism of divalent metal import. Curr. Top. Membr 69, 249–293 [DOI] [PubMed] [Google Scholar]
- 8.Ehrnstorfer IA et al. (2014) Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol 21, 990–996 [DOI] [PubMed] [Google Scholar]
- 9.Iatsenko I et al. (2020) Iron sequestration by transferrin 1 mediates nutritional immunity in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A 117, 7317–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montemiglio LC et al. (2019) Cryo-EM structure of the human ferritin–transferrin receptor 1 complex. Nat. Commun 10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdizadeh H et al. (2017) Computational approaches for deciphering the equilibrium and kinetic properties of iron transport proteins. Metallomics 9, 1513–1533 [DOI] [PubMed] [Google Scholar]
- 12.Lambert LA (2012) Molecular evolution of the transferrin family and associated receptors. Biochim. Biophys. Acta 1820, 244–255 [DOI] [PubMed] [Google Scholar]
- 13.Pogoutse AK and Moraes TF (2017) Iron acquisition through the bacterial transferrin receptor. Crit. Rev. Biochem. Mol. Biol 52, 314–326 [DOI] [PubMed] [Google Scholar]
- 14.Noinaj N et al. (2012) Structural basis for iron piracy by pathogenic Neisseria. Nature 483, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barber MF and Elde NC (2014) Escape from bacterial iron piracy through rapid evolution of transferrin. Science 346, 1362–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barber MF et al. (2016) Antimicrobial functions of lactoferrin promote genetic conflicts in ancient primates and modern humans. PLoS Genet. 12, e1006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choby JE et al. (2018) Molecular basis for the evolution of species-specific hemoglobin capture by Staphylococcus aureus. mBio 9, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikkelsen JH et al. (2020) The human protein haptoglobin inhibits IsdH-mediated heme-sequestering by Staphylococcus aureus. J. Biol. Chem 295, 1781–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis-Guardiola K et al. (2020) The Staphylococcus aureus IsdH receptor forms a dynamic complex with human hemoglobin that triggers heme release via two distinct hot spots. J. Mol. Biol 432, 1064–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler LC et al. (2016) Multiple evolutionary origins of ubiquitous Cu2+ and Zn2+ binding in the S100 protein family. PLoS One 11, e016474027764152 [Google Scholar]
- 21.Gonzalez LL et al. (2020) Role of S100 proteins in health and disease. Biochim. Biophys. Acta Mol. Cell Res 1867, 118677. [DOI] [PubMed] [Google Scholar]
- 22.Ehrchen JM et al. (2009) The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol 86, 557–566 [DOI] [PubMed] [Google Scholar]
- 23.Loes AN et al. (2018) Coevolution of the Toll-like receptor 4 complex with calgranulins and lipopolysaccharide. Front. Immunol 9, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zygiel EM and Nolan EM (2018) Transition metal sequestration by the host-defense protein calprotectin. Annu. Rev. Biochem 87, 621–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozzi AT and Nolan EM (2020) Avian MRP126 restricts microbial growth through Ca(II)-dependent Zn(II) sequestration. Biochemistry 59, 802–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunden LS et al. (2016) Calcium ions tune the zinc-sequestering properties and antimicrobial activity of human S100A12. Chem. Sci 7, 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunden LS and Nolan EM (2018) Bioinorganic explorations of Zn(II) sequestration by human S100 host-defense proteins. Biochemistry 57, 1673–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunden LS et al. (2017) Biochemical and functional evaluation of the intramolecular disulfide bonds in the zinc-chelating antimicrobial protein human S100A7 (Psoriasin). Biochemistry 56, 5726–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zackular JP et al. (2015) Nutritional immunity: S100 proteins at the host–pathogen interface. J. Biol. Chem 290, 18991–18998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikari J et al. (2020) Calcium binding to the innate immune protein human calprotectin revealed by integrated mass spectrometry. J. Am. Chem. Soc 142, 13372–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan MR et al. (2020) Multi-metal nutrient restriction and crosstalk in metallostasis systems in microbial pathogens. Curr. Opin. Microbiol 55, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zygiel EM et al. (2019) The human innate immune protein calprotectin induces iron starvation responses in Pseudomonas aeruginosa. J. Biol. Chem 294, 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harman JL et al. (2020) Evolution of multifunctionality through a pleiotropic substitution in the innate immune protein S100A9. eLife 9, 865493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephan JR and Nolan EM (2016) Calcium-induced tetramerization and zinc chelation shield human calprotectin from degradation by host and bacterial extracellular proteases. Chem. Sci 7, 1962–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogl T et al. (2018) Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J. Clin. Invest 128, 1852–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Besold AN et al. (2018) Antimicrobial action of calprotectin that does not involve metal withholding. Metallomics 10, 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kammerman MT et al. (2020) Molecular insight into TdfH-mediated zinc piracy from human calprotectin by Neisseria gonorrhoeae. mBio 11, e00949–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stork M et al. (2013) Zinc piracy as a mechanism of Neisseria meningitidis for evasion of nutritional immunity. PLoS Pathog. 9, e1003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnstone TC and Nolan EM (2017) Determination of the molecular structures of ferric enterobactin and ferric enantioenterobactin using racemic crystallography. J. Am. Chem. Soc 139, 15245–15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y et al. (2020) Iron acquisition by bacterial pathogens: beyond tris-catecholate complexes. ChemBioChem 21, 1955–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holden VI and Bachman MA (2015) Diverging roles of bacterial siderophores during infection. Metallomics 7, 986. [DOI] [PubMed] [Google Scholar]
- 42.Challis GL (2005) A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 6, 601–611 [DOI] [PubMed] [Google Scholar]
- 43.Reitz ZL et al. (2019) Genomic analysis of siderophore β-hydroxylases reveals divergent stereocontrol and expands the condensation domain family. Proc. Natl. Acad. Sci. U. S. A 116, 19805–19814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abergel RJ et al. (2006) Microbial evasion of the immune system: structural modifications of enterobactin impair siderocalin recognition. J. Am. Chem. Soc 128, 10998–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu F et al. (2019) Functions and regulation of lipocalin-2 in gut-origin sepsis: a narrative review. Crit. Care 23, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flo TH et al. (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 47.Deriu E et al. (2013) Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flower DR (1996) The lipocalin protein family: structure and function. Biochem. J 318, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao G et al. (2010) Iron traffics in circulation bound to a siderocalin (Ngal)-catechol complex. Nat. Chem. Biol 6, 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonomo RA (2017) β-Lactamases: a focus on current challenges. Cold Spring Harb. Perspect. Med 7, a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tooke CL et al. (2019) β-Lactamases and β-lactamase inhibitors in the 21st century. J. Mol. Biol 431, 3472–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bush K and Bradford PA (2020) Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev 33, e00047–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crowder MW et al. (2006) Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res 39, 721–728 [DOI] [PubMed] [Google Scholar]
- 54.Weinreich DM (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 [DOI] [PubMed] [Google Scholar]
- 55.Sideraki V (2001) A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc. Natl. Acad. Sci. U. S. A 98, 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bershtein S et al. (2006) Robustness–epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444, 929–932 [DOI] [PubMed] [Google Scholar]
- 57.Wang X et al. (2002) Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol 320, 85–95 [DOI] [PubMed] [Google Scholar]
- 58.Ju L-C et al. (2018) The Continuing challenge of metallo-β-lactamase inhibition: mechanism matters. Trends Pharmacol. Sci 39, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palacios AR et al. (2020) Metallo-β-lactamase inhibitors inspired on snapshots from the catalytic mechanism. Biomolecules 10, 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mojica MF et al. (2019) Population structure, molecular epidemiology, and β-lactamase diversity among Stenotrophomonas maltophilia isolates in the United States. mBio 10, e00405–e00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasia RM and Vila AJ (2004) Structural determinants of substrate binding to Bacillus cereus metallo-β-lactamase. J. Biol. Chem 279, 26046–26051 [DOI] [PubMed] [Google Scholar]
- 62.Lisa M-N et al. (2017) A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-β-lactamases. Nat. Commun 8, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettinati I et al. (2016) The chemical biology of human metallo-β-lactamase fold proteins. Trends Biochem. Sci 41, 338–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.González JM et al. (2012) Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat. Chem. Biol 8, 698–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.González LJ et al. (2014) Host-specific enzyme-substrate interactions in SPM-1 metallo-β-lactamase are modulated by second sphere residues. PLoS Pathog. 10, e1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomatis PE et al. (2005) Mimicking natural evolution in metallo-β-lactamases through second-shell ligand mutations. Proc. Natl. Acad. Sci. U. S. A 102, 13761–13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomatis PE et al. (2008) Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc. Natl. Acad. Sci. U. S. A 105, 20605–20610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.González MM et al. (2016) Optimization of conformational dynamics in an epistatic evolutionary trajectory. Mol. Biol. Evol 33, 1768–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meini M-R et al. (2015) Quantitative description of a protein fitness landscape based on molecular features. Mol. Biol. Evol 32, 1774–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh TR et al. (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis 11, 355–362 [DOI] [PubMed] [Google Scholar]
- 71.González LJ et al. (2016) Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat. Chem. Biol 12, 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prunotto A et al. (2020) Molecular bases of the membrane association mechanism potentiating antibiotic resistance by New Delhi metallo-β-lactamase 1. bioRxiv Posted online June 1, 2020. 10.1101/2020.06.01.126664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naas T et al. (2017) Beta-lactamase database (BLDB)-structure and function. J. Enzyme Inhib. Med. Chem 32, 917–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bahr G et al. (2017) Clinical evolution of new delhi metallo-β-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob. Agents Chemother 62, e01849–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng Z et al. (2018) Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J. Biol. Chem 293, 12606–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stewart AC et al. (2017) Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect. Dis 3, 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lisher JP et al. (2013) Physical characterization of the manganese-sensing pneumococcal surface antigen repressor from Streptococcus pneumoniae. Biochemistry 52, 7689–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Radin JN et al. (2018) Synergy between nutritional immunity and independent host defenses contributes to the importance of the MntABC manganese transporter during Staphylococcus aureus infection. Infect. Immun 87, e00642–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Payne SM et al. (2016) Vibrio iron transport: evolutionary adaptation to life in multiple environments. Microbiol. Mol. Biol. Rev 80, 69–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perry WJ et al. (2019) Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc. Natl. Acad. Sci. U. S. A 116, 21980–21982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bailey DC et al. (2018) Structural and functional delineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae. J. Biol. Chem 293, 7841–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ghssein G et al. (2016) Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 352, 1105–1109 [DOI] [PubMed] [Google Scholar]
- 83.Lhospice S et al. (2017) Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep 7, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mastropasqua MC et al. (2017) Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Mol. Microbiol 106, 543–561 [DOI] [PubMed] [Google Scholar]
- 85.Noinaj N et al. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol 64, 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delepelaire P (2019) Bacterial ABC transporters of iron containing compounds. Res. Microbiol 170, 345–357 [DOI] [PubMed] [Google Scholar]
- 87.Chatterjee A and O’Brian MR (2018) Rapid evolution of a bacterial iron acquisition system. Mol. Microbiol 108, 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rey-Varela D et al. (2019) The outer membrane protein fstc of Aeromonas salmonicida subsp. salmonicida acts as receptor for amonabactin siderophores and displays a wide ligand plasticity. Structure–activity relationships of synthetic amonabactin analogues. ACS Infect. Dis 5, 1936–1951 [DOI] [PubMed] [Google Scholar]
- 89.Griffin AS et al. (2004) Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 90.Stilwell P et al. (2018) The effect of cheats on siderophore diversity in Pseudomonas aeruginosa. J. Evol. Biol 31, 1330–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arnold FM et al. (2020) The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 580, 413–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andersen SB et al. (2018) Privatisation rescues function following loss of cooperation. eLife 7, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Celis AI and Relman DA (2020) Competitors versus collaborators: micronutrient processing by pathogenic and commensal human-associated gut bacteria. Mol. Cell 78, 570–576 [DOI] [PubMed] [Google Scholar]
- 94.Wang J et al. (2019) Metal ion homeostasis. Ref. Modul. Chem. Mol. Sci. Chem. Eng Published online 30 August 2019. 10.1016/B978-0-12-409547-2.14675-X [DOI] [Google Scholar]
- 95.Capdevila DA et al. (2017) Metallochaperones and metalloregulation in bacteria. Essays Biochem. 61, 177–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin JE et al. (2019) A Mn-sensing riboswitch activates expression of a Mn2+/Ca2+ ATPase transporter in Streptococcus. Nucleic Acids Res. 47, 6885–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Osman D et al. (2019) Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol 15, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Capdevila DA et al. (2017) Entropy redistribution controls allostery in a metalloregulatory protein. Proc. Natl. Acad. Sci. U. S. A 114, 4424–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosinski JA and Atchley WR (1999) Molecular evolution of helix-turn-helix proteins. J. Mol. Evol 49, 301–309 [DOI] [PubMed] [Google Scholar]
- 100.Liu T et al. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol 3, 60–68 [DOI] [PubMed] [Google Scholar]
- 101.Higgins KA and Giedroc D (2014) Insights into protein allostery in the CsoR/RcnR family of transcriptional repressors. Chem. Lett 43, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Campanello GC et al. (2013) Allosteric inhibition of a zinc-sensing transcriptional repressor: insights into the arsenic repressor (ArsR) family. J. Mol. Biol 425, 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roy R et al. (2018) In silico identification and characterization of sensory motifs in the transcriptional regulators of the ArsR-SmtB family. Metallomics 10, 1476–1500 [DOI] [PubMed] [Google Scholar]
- 104.Prabaharan C et al. (2019) Structures of two ArsR As(III)-responsive transcriptional repressors: implications for the mechanism of derepression. J. Struct. Biol 207, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deochand DK and Grove A (2017) MarR family transcription factors: dynamic variations on a common scaffold. Crit. Rev. Biochem. Mol. Biol 52, 595–613 [DOI] [PubMed] [Google Scholar]
- 106.Santos CL et al. (2009) A phylogenomic analysis of bacterial helix–turn–helix transcription factors. FEMS Microbiol. Rev 33, 411–429 [DOI] [PubMed] [Google Scholar]
- 107.Will WR and Fang FC (2020) The evolution of MarR family transcription factors as counter-silencers in regulatory networks. Curr. Opin. Microbiol 55, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma Z et al. (2011) Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39, 9130–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pi H and Helmann JD (2017) Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A 114, 12785–12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ibáñez MM et al. (2015) A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J. Bacteriol 197, 1606–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Osman D et al. (2016) The effectors and sensory sites of formaldehyde-responsive regulator FrmR and metal-sensing variant. J. Biol. Chem 291, 19502–19516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hobman JL (2007) MerR family transcription activators: similar designs, different specificities. Mol. Microbiol 63, 1275–1278 [DOI] [PubMed] [Google Scholar]
- 113.Liu X et al. (2019) Selective cadmium regulation mediated by a cooperative binding mechanism in CadR. Proc. Natl. Acad. Sci. U. S. A 116, 20398–20403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Capdevila DA et al. (2018) Tuning site-specific dynamics to drive allosteric activation in a pneumococcal zinc uptake regulator. eLife 7, e37268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu R et al. (2017) Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation. Proc. Natl. Acad. Sci. U. S. A 114, 13661–13666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glauninger H et al. (2018) Metal-dependent allosteric activation and inhibition on the same molecular scaffold: the copper sensor CopY from Streptococcus pneumoniae. Chem. Sci 9, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pillai AS et al. (2020) Origin of complexity in haemoglobin evolution. Nature 581, 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ellefson JW et al. (2018) Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nat. Chem. Biol 14, 361–367 [DOI] [PubMed] [Google Scholar]
- 119.Machado LFM et al. (2019) Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. J. Biol. Eng 13, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hochberg GKA and Thornton JW (2017) Reconstructing ancient proteins to understand the causes of structure and function. Annu. Rev. Biophys 46, 247–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Merkl R and Sterner R (2016) Ancestral protein reconstruction: techniques and applications. Biol. Chem 397, 1–21 [DOI] [PubMed] [Google Scholar]
- 122.Kimura M (1977) Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267, 275–276 [DOI] [PubMed] [Google Scholar]
- 123.Packer MS and Liu DR (2015) Methods for the directed evolution of proteins. Nat. Rev. Genet 16, 379–394 [DOI] [PubMed] [Google Scholar]
- 124.Fowler DM and Fields S (2014) Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morrison MS et al. (2020) The developing toolkit of continuous directed evolution. Nat. Chem. Biol 16, 610–619 [DOI] [PubMed] [Google Scholar]
- 126.van den Bedem H and Fraser JS (2015) Integrative, dynamic structural biology at atomic resolution – it’s about time. Nat. Methods 12, 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rivoire O (2019) Parsimonious evolutionary scenario for the origin of allostery and coevolution patterns in proteins. Phys. Rev. E 100, 032411. [DOI] [PubMed] [Google Scholar]
- 128.Campitelli P et al. (2020) The role of conformational dynamics and allostery in modulating protein evolution. Annu. Rev. Biophys 49 annurev-biophys-052118–115517 [DOI] [PubMed] [Google Scholar]
- 129.Mukherjee D et al. (2014) Crystal structure of HlyU, the hemolysin gene transcription activator, from Vibrio cholerae N16961 and functional implications. Biochim. Biophys. Acta 1844, 2346–2354 [DOI] [PubMed] [Google Scholar]
- 130.Saha RP et al. (2017) Metal homeostasis in bacteria: the role of ArsR–SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. BioMetals 30, 459–503 [DOI] [PubMed] [Google Scholar]


