Abstract
A Belgian ring trial for pan-TRK immunohistochemistry (IHC) staining was organised to harmonise pan-TRK IHC staining protocols and interpretation. As a reference method, the VENTANA pan-TRK Assay (clone EPR17341) on the Benchmark Ultra platform was selected. Six samples were selected: 2 negative, 2 fusion positive and 2 samples with wild-type endogenous TRK expression. Each participating laboratory stained the slides using their routine pan-TRK IHC and reported their results. In addition, they were asked to return one TRK-stained slide from each case. The coordinating lab evaluated these slides, compared them with the reference method and scored them. Two clones were used during the ring trial: A7H6R (Cell Signaling) and EPR17341 (Abcam/Ventana). Seven protocols achieved a sufficient performance mark, and three labs were advised to further optimise the protocol. Interpretation of pan-TRK IHC proved to be challenging in cases with physiological TRK expression. In addition, depending on the NTRK fusion partner, the staining can vary strongly in both intensity and staining pattern. Labs using the Ventana ready-to-use system based on the EPR17341 clone and using the recommended protocol settings scored best. However, given some small optimisation, all labs scored well on the technical staining and the succeeding evaluation.
Keywords: Immunohistochemistry, NTRK fusions, Ring trial, Cancer screening, TRK inhibitor
Introduction
The neurotrophic tyrosine receptor kinases (NTRK, or commonly used TRK) are a family of transmembrane tyrosine kinases. TRKA, TRKB and TRKC proteins are encoded by the proto-oncogenes NTRK1, NTRK2, and NTRK3 respectively and are physiologically expressed in the testes, smooth muscle and central and peripheral nervous system [1, 2]. Oncogenic fusions involving the kinase domain of the NTRK genes have been identified with high prevalence in certain rare cancers like infantile fibrosarcoma or secretory carcinoma of the breast [3]. The most common form of NTRK fusion gene, ETV6-NTRK3, is present in about 70% of infantile fibrosarcoma, making it a defining diagnostic feature [4]. More recently, NTRK fusions have also been identified in a small percentage of common cancers [5, 6], like soft tissue sarcomas [7], gliomas [8] and carcinomas of the lung [9], colon [10] and thyroid [11]. Farago et al. estimated the frequency of NTRK fusions in non-small-cell lung cancer (NSCLC) to be 0.23% [9]. A similar result was found in a cohort of 11,500 patients with various solid tumours, where only 0.27% harboured NTRK fusions [12]. Less than 0.31% of colorectal carcinomas are found to be NTRK fusion positive, but rates are substantially higher in the high microsatellite instability (MSI-H) phenotype [13]. Typically, the 5′ region of a partner gene fuses with the 3′ region of an NTRK gene, resulting in ligand-independent receptor activation [14].
TRK inhibitors have shown great promise as therapy for patients with tumours harbouring NTRK gene fusions. Clinical trials revealed high response rates, durable responses and favourable safety profiles. Furthermore, clinical responses were seen regardless of patient age, fusion partner, NTRK gene and tumour type [15, 16]. This has led to fast approval by the Food and Drug Administration (FDA) of TRK inhibitors for NTRK fusion positive solid tumours instead of for specific tumour types. First approved in 2019 in Japan, the multikinase inhibitor entrectinib exhibits activity against NTRK, ROS1 and ALK oncogenic fusions. Larotrectinib, a highly selective TRK inhibitor received approval by the European Medicines Agency (EMA) in 2019.
As a consequence, identification of NTRK fusions has become vital for therapeutic management, and in some tumour types for diagnostic purposes. However, the presence of three different NTRK genes, combined with a high number of potential fusion partners and several possible breakpoints, makes the detection of NTRK fusions rather complex. A variety of techniques, like next-generation-sequencing (NGS), DNA and RNA-based assays or fluorescence in situ hybridisation (FISH) can be used to detect these fusions at the DNA, RNA or protein level [17]. In contrast to these assays, the use of immunohistochemistry (IHC) provides several benefits like a quick turnaround time, lower cost, wide availability and use of very limited tissue. Antibodies for IHC can be directed against specific TRK proteins [18] or can target an amino acid sequence common to TRKA, TRKB and TRKC (pan-TRK antibodies) [19]. Especially in low probability cancers (frequency < 5%), the use of a two-step approach is often suggested for NTRK fusion detection: The first step involves IHC as a screening or enrichment tool; the second step is to confirm the presence of a fusion by an RNA-based NGS analysis [20, 21].
Currently, there is limited experience with NTRK gene fusion testing in Belgium. The lack of an external quality assessment (EQA) for the pan-TRK IHC staining prompted us to organise a pan-TRK IHC ring trial. The key aim was to evaluate the reproducibility of TRK staining across different institutions using the same well-characterised samples and to provide feedback aimed at standardising the implementation and interpretation of TRK staining protocols. In addition, we also assessed the inter-observer variability in the evaluation of TRK IHC among pathologists.
Materials and methods
Sample selection
A total number of six formalin-fixed paraffin-embedded (FFPE) tissues were selected from the archives of the Antwerp University Hospital (UZA). The samples represented a mix of tumours, NTRK fusion positive and NTRK fusion negative samples. Besides tumour tissue harbouring a NTRK fusion, also tissue with endogenous TRK expression was included. The NTRK fusion status and possible fusion partner of the samples were confirmed by NGS testing. Targeted RNA-based NGS was conducted with the Oncomine Focus Assay (OFA) panel (Thermo Fisher Scientific, San Francisco, CA) on an S5 instrument, according to the manufacturer’s recommendations.
Pan-TRK IHC
As a reference method we used the VENTANA pan-TRK assay (clone EPR17341) performed according to the instructions of the vendor on a Benchmark Ultra (Ventana Medical Systems, Tucson, AZ). This widely used EPR17341 clone is reactive with a conserved proprietary peptide sequence from the C-terminus of TRKA, TRKB and TRKC and is therefore reactive with any of the oncogenic TRK proteins. Tumours are considered positive if ≥ 1% of tumour cells exhibit staining at any intensity above background [12]. In addition, the different subcellular staining patterns (cytoplasmic, membranous, nuclear and peri-nuclear) are all considered to be positive. Staining intensity was quoted as negative (0), weak (1+), moderate (2+) or strong (3+).
Design ring trial
The ring trial was coordinated by UZA and was conducted according to the Declaration of Helsinki, and ethical committee approval 18/49/577 was obtained on 07 January 2019 from the ethical committee of UZA. A total of 9 Belgian hospital labs participated in the ring trial. One laboratory participated with 2 protocols; therefore, the results will be discussed as if 10 labs participated.
The ring trial was setup according to the following steps:
The lead institute prepared blank slides from 6 resection cases. The first, middle and last slides were stained for pan-TRK to ensure similar staining patterns throughout the tissue.
Each participating laboratory received two blank coated slides to stain using their routine testing procedure. Labs were free to select their antibody and detection system of preference but were asked to provide information on the antibody, protocol and scoring method. Upon TRK staining, one or two pathologists of each centre reviewed and interpreted the staining. The laboratories were requested to return one TRK-stained slide per sample to the coordinating lab together with results of scoring and interpretation.
Once returned to the lead institute, the slide stained by the laboratories was compared with the reference staining and evaluated by a team of two experienced pathologists. Feedback was provided to the laboratories, including a technical evaluation and a comparison of the evaluation by the participating laboratories and by the team of trial-designated pathologists. Hereby, staining intensity, percentage of positive tumour cells and background staining were taken into account.
Statistical analysis
To quantify the degree of agreement between the different observers, Cohen’s Kappa statistics (K) was used. It measures the inter-rater agreement for qualitative items and takes the possibility of the agreement occurring by chance into account. Kappa’s coefficient can range from − 1 to 1, where 0 represents the amount of agreement that can be expected from random chance. All statistical analyses were produced using SPSS statistics version 24 (IBM, Brussels, Belgium). The interpretation of Kappa’s coefficient is based on the Koch and Landis scale [22].
Results
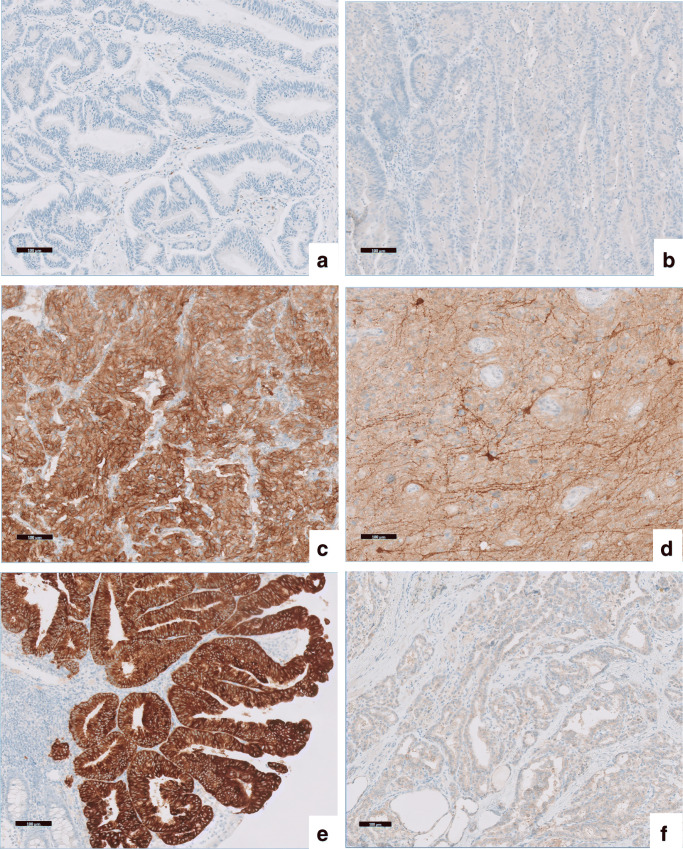
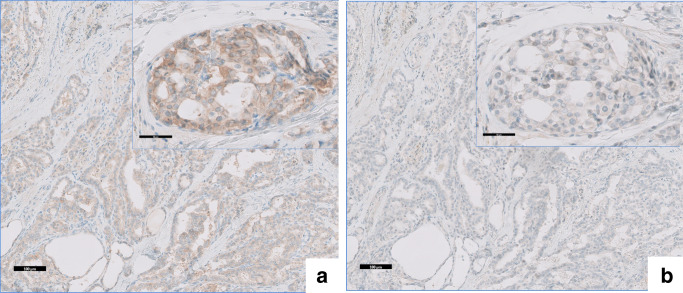
The ring trial consisted of 6 samples: 2 negative samples, 2 samples with endogenous TRK expression and 2 samples with TRK fusion expressing tumour cells (Table 1). Stained with the VENTANA pan-TRK assay (clone EPR17341), the two negative samples did not show any expression (score 0; Fig. 1 a and b). The staining of the endogenous TRK expression of the pheochromocytoma case was predominantly granular cytoplasmic with membranous accentuation at variable intensities (score 1–3; Fig. 1c). Also, the glioma sample showed weak, diffuse staining of pre-existing brain tissue. The tumour cells however showed no staining (score 0; Fig. 1d). Both fusion-positive tumours demonstrated a cytoplasmic staining pattern: homogenous and strong staining in the microsatellite instability (MSI) positive colon carcinoma (score 3+; Fig. 1e) and diffuse and weak granular staining was present in the thyroid carcinoma (score 1+; Figs. 1f and 2).
Table 1.
Overview samples
| Sample | Cancer type | Result reference TRK staining |
|---|---|---|
| 1 | Colorectal adenocarcinoma | No TRK staining |
| 2 | Colorectal adenocarcinoma | No TRK staining |
| 3 | Pheochromocytoma | Endogenous TRK expression |
| 4 | Glioma | Endogenous TRK expression |
| 5 | Colorectal MSI positive adenocarcinoma | Strong cytoplasmic staining of 100% of tumour cells (TPM3–NTRK1 fusion) |
| 6 | Papillary thyroid carcinoma | Weak to moderate cytoplasmic staining of 90% of tumour cell (TPR–NTRK1 fusion) |
Fig. 1.
pan-TRK staining of ring trial samples. The scale bar indicates 100 μm. a Colorectal carcinoma. b Colorectal carcinoma. c Pheochromocytoma. d Glioma. e Colorectal MSI positive carcinoma. f Papillary thyroid carcinoma
Fig. 2.
Papillary thyroid carcinoma. The scale bar represents 100 μm; the scale bar of the insert represents 50 μm. a Pan-TRK staining, b Negative control staining
Each of the participating laboratories stained the six slides with their routine testing procedure. After staining and analysing samples, they reported back to the coordinating lab. Each case was expected to be classified as either negative, positive or unclear. In addition, information was asked on staining percentage, intensity and clinical conclusion (fusion positive, fusion negative or further testing was required). The stained slides were afterwards sent back to the coordinating lab for evaluation by the designated trial pathologists.
Technical evaluation
Pathologists from the coordinating lab evaluated the slides of the participating laboratories based on staining intensity, percentage of positive tumour cells and background staining (Table 2). These were then compared with the reference method and scored according to the following categories:
Table 2.
Evaluation different protocols: I: intensity, %: percentage of positive tumour cells, B: background staining, 0: no staining, 1: weak staining, 2: moderate staining, 3: strong staining
| Protocol | Samples | Result | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||||||||||||||
| I | % | B | I | % | B | I | % | B | I | % | B | I | % | B | I | % | B | ||
| Reference | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 1–2 | 90 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 2–3 | 100 | 1 | Good |
| 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 2 | 100 | 1 | Good |
| 3 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 1–2 | 100 | 0 | Optimal |
| 4 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 1–2 | 90 | 1 | Good |
| 5 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 0–1 | 70 | 0 | Borderline |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 1–2 | 100 | 0 | 0 | 0 | 0 | 2–3 | 100 | 0 | 0 | 0 | 0 | Poor |
| 7 | 0 | 0 | 0 | 0 | 0 | 2 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 3 | 100 | 1 | Good |
| 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 90 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 0–1 | 80 | 1 | Borderline |
| 9 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 1–2 | 100 | 0 | Optimal |
| 10 | 0 | 0 | 0 | 0 | 0 | 1 | 1–3 | 100 | 0 | 0 | 0 | 0 | 3 | 100 | 0 | 1–2 | 90 | 0 | Optimal |
Optimal: The staining method scores identical to the reference method.
Good: The staining method shows an increased background staining, but diagnosis was correct.
Borderline: The staining method shows a decreased intensity of staining in tumour cells, but diagnosis was correct.
Poor: The staining method shows a decreased intensity of staining in tumour cells, generating a false negative result.
Compared with the pan-TRK assay reference method, seven protocols achieved a sufficient mark (optimal or good). Three labs were advised to further optimise their protocol (Table 3). Only two different antibody clones were used during the ring trial: A7H6R (Cell Signaling Technology) and EPR17341 (Abcam/Ventana). EPR17341 is available in a concentrated form (Abcam) or as a ready-to-use (RTU) assay by Ventana. Four labs (40%) used the RTU antibody, of which three followed the recommended in vitro diagnostic (IVD) settings and achieved the optimal mark (this protocol was also used as the reference method). The fourth lab added an amplification step, causing a slightly increased background staining (Table 3). The Ventana BenchMark was the most popular platform, being used by 80% of the labs. The remaining labs (20%) used the Dako Omnis platform. Both lab developed tests on Dako Omnis did not receive a sufficient mark, due to weak staining (Table 3). Seven participating laboratories (70%) used lab-developed tests. Because of increased background staining or decreased staining intensity, these lab-developed tests scored lower than the RTU assays (Tables 2 and 3).
Table 3.
Assessment marks depending on antibody, platform and protocol: conc. AB: concentrated antibody, RTU AB: ready-to-use antibody, LDT: lab developed test, IVD: in vitro diagnostic test
| Percentage (number) | No. of optimal | No. of good | No. of borderline | No. of poor | ||
|---|---|---|---|---|---|---|
| Antibody | A7H6R | 30 (3) | 0 | 2 | 1 | 0 |
| EPR17341 conc. AB (Abcam) | 30 (3) | 0 | 1 | 1 | 1 | |
| EPR17341 RTU AB (Ventana) | 40 (4) | 3 | 1 | 0 | 0 | |
| Platform | Ventana | 80 (8) | 3 | 4 | 1 | 0 |
| Dako Omnis | 20 (2) | 0 | 0 | 1 | 1 | |
| Protocol | LDT with conc. AB | 60 (6) | 0 | 3 | 2 | 1 |
| LDT with RTU AB | 10 (1) | 0 | 1 | 0 | 0 | |
| IVD assay | 30 (3) | 3 | 0 | 0 | 0 |
Evaluation of the analysis
To assess the inter-observer variability in the evaluation of pan-TRK IHC, the testing laboratory reported their results back to UZA and the stained slides of each lab were evaluated by two experienced pathologists. When comparing these results (being positive or negative), only one case gave a false positive discordant result, probably due to mild background staining (Fig. 1b). When interpreting the slides, nobody missed a positive case. Therefore, the false negative rate was 0%. To quantify the degree of agreement between the different observers, Cohen’s Kappa statistics (K) was used. In this case Kappa’s coefficient = 0.925 (p < 0.001), indicating an almost perfect degree of agreement between the different observers. However, besides positive or negative scores, the pathologist was also asked whether further testing was needed. When indicated this was necessary, the answer was always considered to be correct.
Discussion
Oncogenic NTRK fusions are seen in many cancer types, but with the exception of some very rare tumour types, their incidence remains very low. These fusions have important therapeutic implications for patients with advanced cancers, making their routine detection a priority. Following the remarkable and often durable responses to TRK tyrosine kinase inhibitors, a wide range of techniques became available to detect the presence of NTRK fusions. Immunohistochemistry is a fast, cost-effective and widely available technique and provides an effective approach to screen for tumours harbouring NTRK fusions [23]. Especially in cases with a low probability of NTRK gene fusions, pan-TRK IHC can be performed as an enrichment strategy to select tumours for subsequent (RNA-based) NGS analysis. In contrast, for the rare subtypes that commonly harbour NTRK fusions (like infantile fibrosarcoma and secretory carcinomas of the breast and salivary glands) a histology-based triage followed by RNA-level fusion testing is suggested [23, 24]. The detection of RNA-level fusions provides direct evidence of functional transcription. In addition, splicing out of introns simplifies the technical requirements for detection of NTRK fusions, making RNA-based sequencing the preferred approach [20].
EPR17341 is a widely investigated pan-TRK clone and has demonstrated to be an efficient and reliable screening method for NTRK fusions [23]. Studies have shown sensitivities ranging from 75 to 92.5% and specificities between 81.1 and 100% [12, 19, 23, 25]. The A7H6R clone is also reactive with any of the TRK proteins but is less investigated than EPR17341. One study compared these two pan-TRK IHC clones in advanced melanoma tumour samples. Different staining results suggest that the clones do not target the same epitopes in the TRK proteins. In addition, the authors hypothesised that EPR17341 might be more specific—but perhaps also less sensitive—than A7H6R. Due to a lack of NTRK-rearranged tumours, no real comparison could be made [26]. In a recent study by Guibourg et al., a total of 71 salivary gland tumours were stained with the two pan-TRK IHC antibody clones. Only one case was fusion-positive, and this NTRK3-rearranged salivary secretory carcinoma was found to be positive with both the EPR17341 and the A7H6R clone [27]. Finally, in a large cohort of over 4000 colorectal cancer samples, both antibodies demonstrated similar staining characteristics and showed diffuse strong cytoplasmic staining in all nine fusion-positive cases. In addition, there was also complete concordance between the two observers in interpreting both antibodies [13].
Based on the results of this ring trial, the EPR17341 and A7H6R clones are both highly recommendable antibodies for pan-TRK IHC. Labs using the Ventana ready-to-use system based on the EPR17341 clone and following the recommended protocol settings scored best. However, given some small optimisation, labs using a lab developed test or labs using the A7H6R clone can also achieve a sufficient or optimal mark.
Interpretation of IHC TRK staining may seem straightforward, but can be more challenging than anticipated. The staining can vary strongly in both intensity and staining pattern, which often correlates with the subcellular localisation of the fusion partners. In contrast to the membrane-associated expression of native TRK, the fusion partner can direct the fusion protein to localise to other cell compartments. As a consequence, the staining pattern can be cytoplasmic, nuclear, perinuclear or membranous [19]. Currently, approximately 80 different 5′ NTRK gene fusion partners have been identified in a wide array of tumour types [14]. Also, the percentage of tumour cells with positive staining can vary among the tumours. This variation in staining pattern, intensity and percentage was illustrated by the two TRK positive samples included in the ring trial (Fig. 1 e and f). Both samples harbour an NTRK1 fusion, resulting in a cytoplasmic staining pattern, but with very different intensity. In contrast to the strong, uniform staining of the MSI positive colon carcinoma, the papillary thyroid carcinoma showed only very weak diffuse staining. As a consequence, one of the sub-optimal staining protocols from the ring trial led to a false negative result. In these unclear borderline cases, including a negative control (IHC staining without addition of the primary antibody) can facilitate correct interpretation (Fig. 2 a and b).
Another potential pitfall in the interpretation of pan-TRK IHC is the occurrence of physiological TRK expression. Under normal circumstances wild-type TRK is expressed in smooth muscles, testes and neural components. Particularly challenging is the interpretation of glioma samples because of the positivity of normal brain tissue (Fig. 1d). In addition, also, a subset of tumours, like pheochromocytoma (a neural crest-derived tumour), express TRK, but without the presence of an NTRK gene rearrangement (Fig. 1c). The usefulness of TRK IHC in these two samples from the ring trial is limited, illustrating that selection of the appropriate assay for NTRK fusion detection also depends on tumour type. Especially in tumours with neural differentiation, specificity of pan-TRK IHC may be insufficient and RNA-based NGS analysis should be considered [23].
As reported in literature, an additional limitation for IHC analysis is the reduced sensitivity for NTRK3 fusions [12, 23]. While sensitivities close to 100% (96% for NTRK1 and 100% for NTRK2) were demonstrated, only 79% sensitivity was found for NTRK3. In some of these cases, staining was found to be weak and focal, increasing the chance of false negative results [23]. Therefore, it would be of great interest for a future EQA to include an NTRK3 fusion positive sample.
There was an almost perfect degree of agreement (Kappa's coefficient 0.925, p < 0.001) in TRK IHC scoring between the pathologists of the participating laboratories and the trial-designated pathologists. Only one slide was interpreted differently by the observers. This is partly due to the—always considered correct—option to indicate extra testing was needed. Most labs scored the pheochromocytoma and glioma samples (Fig. 1 c and d) as positive, but also indicated the need for further testing. The available drugs (larotrectinib and entrectinib) are only useful in NTRK-rearranged tumours; no responses are seen in NTRK mutated or amplified tumours [5, 28]. Therefore, further testing to identify the cause of IHC staining is often needed. However, some of the labs even indicated the need for further testing, even in the absence of (tumoural) staining. In these labs, the efficiency of screening by IHC to detect TRK positive cases was limited.
The main purpose of this ring trial was to harmonise pan-TRK IHC staining protocols and subsequent interpretation. Despite the use of different antibodies and detection systems, TRK IHC staining results were comparable between the various laboratories, proving the robustness of these procedures. Care should be taken when endogenous expression occurs in the sample, in which case RNA-based NGS analysis is needed. To conclude, the use of IHC as a screening tool, followed by molecular testing to confirm the fusion partners, seems to be an effective approach, especially in case of tumour types with a low incidence of NTRK fusions. Finally, by including more diverse staining patterns and different NTRK fusion partners (especially NTRK3 fusion positive samples), the educational value of future ring trials could increase.
Acknowledgments
Contributions
KDW, LS, SL, VS, PP and KZ have contributed to the design. KDW, VS, GB, FD, ND, LH, LL, JVH, BW, KW and PP have contributed to the execution of the experiment. KDW, LS and KZ have contributed to the data analysis. KDW and KZ have prepared the manuscript.
Code availability
Not applicable. No patient data were requested or shared with the participating centres.
Funding
This study was funded by Bayer.
Data availability
All data has been presented in the manuscript.
Compliance with ethical standards
Conflicts of interest
Author PP received research grants from Bayer. Author BW received research grants from Bayer. The other authors, KDW, LS, SL, VS, GB, FD, ND, LH, LL, JVH, KW and KZ, declare that they have no conflict of interest.
Ethical approval
The study has been approved by the ethical committee of UZA (reference number 18/49/577).
Consent to participate
Not applicable. This study was performed on rest material from the archive of the Antwerp University Hospital (UZA).
Consent for publication
Not applicable. This study was performed on rest material from the archive of the Antwerp University Hospital (UZA).
Footnotes
This article is part of the Topical Collection on Quality in Pathology
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holtzman DM, Kilbridge J, Li Y, Cunningham ET, Jr, Lenn NJ, Clary DO, Reichardt LF, Mobley WC. TrkA expression in the CNS: evidence for the existence of several novel NGF-responsive CNS neurons. J Neurosci. 1995;15(2):1567–1576. doi: 10.1523/JNEUROSCI.15-02-01567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond Ser B Biol Sci. 2006;361(1473):1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasudev P, Onuma K. Secretory breast carcinoma: unique, triple-negative carcinoma with a favorable prognosis and characteristic molecular expression. Arch Pathol Lab Med. 2011;135(12):1606–1610. doi: 10.5858/arpa.2010-0351-RS. [DOI] [PubMed] [Google Scholar]
- 4.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18(2):184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 5.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R (2018) Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol 2018. 10.1200/PO.18.00183 [DOI] [PMC free article] [PubMed]
- 7.Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada-Bernal A, Keysar S, Jimeno A, Varella-Garcia M, Aisner DL, Li Y, Stephens PJ, Morosini D, Tuch BB, Fernandes M, Nanda N, Low JA. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5(10):1049–1057. doi: 10.1158/2159-8290.CD-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambella A, Senetta R, Collemi G, Vallero SG, Monticelli M, Cofano F, Zeppa P, Garbossa D, Pellerino A, Ruda R, Soffietti R, Fagioli F, Papotti M, Cassoni P, Bertero L (2020) NTRK fusions in central nervous system tumors: a rare, but worthy target. Int J Mol Sci 21(3). 10.3390/ijms21030753 [DOI] [PMC free article] [PubMed]
- 9.Farago AF, Taylor MS, Doebele RC, Zhu VW, Kummar S, Spira AI, Boyle TA, Haura EB, Arcila ME, Benayed R, Aisner DL, Horick NK, Lennerz JK, Le LP, Iafrate AJ, Ou SI, Shaw AT, Mino-Kenudson M (2018) Drilon A (2018) Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol:1–12. 10.1200/PO.18.00037 [DOI] [PMC free article] [PubMed]
- 10.Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, Hechtman JF, Christiansen J, Novara L, Tebbutt N, Fuca G, Antoniotti C, Kim ST, Murphy D, Berenato R, Morano F, Sun J, Min B, Stephens PJ, Chen M, Lazzari L, Miller VA, Shoemaker R, Amatu A, Milione M, Ross JS, Siena S, Bardelli A, Ali SM, Falcone A, de Braud F, Cremolini C (2017) ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst 109(12). 10.1093/jnci/djx089 [DOI] [PubMed]
- 11.Ricarte-Filho JC, Li S, Garcia-Rendueles ME, Montero-Conde C, Voza F, Knauf JA, Heguy A, Viale A, Bogdanova T, Thomas GA, Mason CE, Fagin JA. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest. 2013;123(11):4935–4944. doi: 10.1172/JCI69766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019;32(1):147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 13.Chou A, Fraser T, Ahadi M, Fuchs T, Sioson L, Clarkson A, Sheen A, Singh N, Corless CL, Gill AJ. NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas-a study of 4569 cases. Mod Pathol. 2019;33:924–932. doi: 10.1038/s41379-019-0417-3. [DOI] [PubMed] [Google Scholar]
- 14.Amatu A, Sartore-Bianchi A, Bencardino K, Pizzutilo EG, Tosi F, Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol. 2019;30(Suppl_8):viii5. doi: 10.1093/annonc/mdz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, Bauer TM, Farago AF, Wheler JJ, Liu SV, Doebele R, Giannetta L, Cerea G, Marrapese G, Schirru M, Amatu A, Bencardino K, Palmeri L, Sartore-Bianchi A, Vanzulli A, Cresta S, Damian S, Duca M, Ardini E, Li G, Christiansen J, Kowalski K, Johnson AD, Patel R, Luo D, Chow-Maneval E, Hornby Z, Multani PS, Shaw AT, De Braud FG. Safety and Antitumor Activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7(4):400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, Turpin B, Dowlati A, Brose MS, Mascarenhas L, Federman N, Berlin J, El-Deiry WS, Baik C, Deeken J, Boni V, Nagasubramanian R, Taylor M, Rudzinski ER, Meric-Bernstam F, Sohal DPS, Ma PC, Raez LE, Hechtman JF, Benayed R, Ladanyi M, Tuch BB, Ebata K, Cruickshank S, Ku NC, Cox MC, Hawkins DS, Hong DS, Hyman DM. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon JP, Hechtman JF. Detection of NTRK Fusions: Merits and Limitations of Current Diagnostic Platforms. Cancer Res. 2019;79(13):3163–3168. doi: 10.1158/0008-5472.CAN-19-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatematsu T, Sasaki H, Shimizu S, Okuda K, Shitara M, Hikosaka Y, Moriyama S, Yano M, Brown J, Fujii Y. Investigation of neurotrophic tyrosine kinase receptor 1 fusions and neurotrophic tyrosine kinase receptor family expression in non-small-cell lung cancer and sensitivity to AZD7451 in vitro. Mol Clin Oncol. 2014;2(5):725–730. doi: 10.3892/mco.2014.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, Arcila ME, Dogan S, Klimstra DS, Ladanyi M, Jungbluth AA. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41(11):1547–1551. doi: 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchio C, Scaltriti M, Ladanyi M, Iafrate AJ, Bibeau F, Dietel M, Hechtman JF, Troiani T, Lopez-Rios F, Douillard JY, Andre F, Reis-Filho JS. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol. 2019;30(9):1417–1427. doi: 10.1093/annonc/mdz204. [DOI] [PubMed] [Google Scholar]
- 21.Murphy DA, Ely HA, Shoemaker R, Boomer A, Culver BP, Hoskins I, Haimes JD, Walters RD, Fernandez D, Stahl JA, Lee J, Kim KM, Lamoureux J, Christiansen J. Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol. 2017;25(7):513–523. doi: 10.1097/PAI.0000000000000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landis JR, Koch GG. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics. 1977;33(2):363–374. doi: 10.2307/2529786. [DOI] [PubMed] [Google Scholar]
- 23.Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, Jungbluth AA, Zehir A, Benayed R, Drilon A, Hyman DM, Ladanyi M, Sireci AN, Hechtman JF. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020;33(1):38–46. doi: 10.1038/s41379-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solomon JP, Benayed R, Hechtman JF, Ladanyi M. Identifying patients with NTRK fusion cancer. Ann Oncol. 2019;30(Suppl_8):viii16–viii22. doi: 10.1093/annonc/mdz384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell D, Ferrarotto R, Liang L, Goepfert RP, Li J, Ning J, Broaddus R, Weber RS, El-Naggar AK. Pan-Trk immunohistochemistry reliably identifies ETV6-NTRK3 fusion in secretory carcinoma of the salivary gland. Virchows Arch. 2020;476(2):295–305. doi: 10.1007/s00428-019-02640-7. [DOI] [PubMed] [Google Scholar]
- 26.Bourhis A, Redoulez G, Quintin-Roue I, Marcorelles P, Uguen A. Screening for NTRK-rearranged tumors using immunohistochemistry: comparison of 2 different pan-TRK clones in melanoma samples. Appl Immunohistochem Mol Morphol. 2020;28(3):194–196. doi: 10.1097/PAI.0000000000000708. [DOI] [PubMed] [Google Scholar]
- 27.Guibourg B, Cloarec E, Conan-Charlet V, Quintin-Roue I, Grippari JL, Le Flahec G, Marcorelles P, Uguen A (2020) EPR17341 and A7H6R pan-TRK immunohistochemistry result in highly different staining patterns in a series of salivary gland tumors. Appl Immunohistochem Mol Morphol. 10.1097/PAI.0000000000000825 Publish Ahead of Print [DOI] [PubMed]
- 28.Ricciuti B, Genova C, Crino L, Libra M, Leonardi GC. Antitumor activity of larotrectinib in tumors harboring NTRK gene fusions: a short review on the current evidence. Onco Targets Ther. 2019;12:3171–3179. doi: 10.2147/OTT.S177051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data has been presented in the manuscript.