Abstract
Grafting can improve the resistance of watermelon to soil-borne diseases. However, the molecular mechanism of defense response is not completely understood. Herein, we used a proteomic approach to investigate the molecular basis involved in grafted watermelon leaf defense against Fusarium oxysporum f.sp. niveum (FON) infection. The bottle gourd rootstock-grafted (RG) watermelon seedlings were highly resistant to FON compared with self-grafted (SG) watermelon plants, with a disease incidence of 3.4 and 89%, respectively. Meanwhile, grafting significantly induced the activity of pathogenesis-related proteases under FON challenge. Proteins extracted from leaves of RG and SG under FON inoculation were analyzed using two-dimensional gel electrophoresis. Thirty-nine differentially accumulated proteins (DAPs) were identified and classified into 10 functional groups. Accordingly, protein biosynthetic and stress- and defense-related proteins play crucial roles in the enhancement of disease resistance of RG watermelon seedlings, compared with that of SG watermelon seedlings. Proteins involved in signal transduction positively regulated the defense process. Carbohydrate and energy metabolism and photosystem contributed to energy production in RG watermelon seedlings under FON infection. The disease resistance of RG watermelon seedlings may also be related to the improved scavenging capacity of reactive oxygen species (ROS). The expression profile of 10 randomly selected proteins was measured using quantitative real-time PCR, among which, 7 was consistent with the results of the proteomic analysis. The functional implications of these proteins in regulating grafted watermelon response against F. oxysporum are discussed.
Keywords: bottle gourd, Citrullus lanatus, Fusarium oxysporum f.sp. niveum, proteomics, rootstock grafting
Introduction
Watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai] is an important fruit crop and contributes 11.98% of the world fruit production (FAO, 2018)1. Fusarium wilt, caused by the soil-borne fungus Fusarium oxysporum f. sp. niveum (FON), is the most serious production-limiting disease in watermelon-growing areas all over the world (Zhang et al., 2015a). The pathogen’s survival in the infected field can be extended for over 10 years (Martyn, 1987) and causes up to 100% yield losses. Resistance to FON has been widely described, and resistance locus Fo-1.3 (Lambel et al., 2014; Meru and McGregor, 2016) to FON race 1 and locus qFon2-2 (Branham et al., 2017) to FON race 2 have been identified. Currently, the watermelon reference genome (Guo et al., 2013) and the whole-genome resequencing (Guo et al., 2019) developed a series of potential genes resistant to Fusarium wilt. These results should be useful for further elucidating the mechanism of resistance to Fusarium wilt and in the development of molecular markers for breeding programs of watermelon.
Grafting is an environmentally friendly and economic technique that is currently being adopted globally in watermelon to cope with soil-borne disease (Lee, 1994). Grafted watermelon was documented with enhanced Fusarium wilt resistance (Huh et al., 2002) as well as higher abiotic stress tolerance (Shi et al., 2019) and increased fruit weight and total yield (Davis et al., 2008). Grafting could also facilitate the uptake and utilization of nutrition (Zhang et al., 2012a; Huang et al., 2013). During the interaction between grafted watermelon and FON, previous reports have shown changes in physiological (Zhang et al., 2015b) and histological (Zhang et al., 2012b) aspects, indicating that grafting could induce higher levels of defense enzymes and immediately form tyloses in the infected xylem vessels soon after FON infection. Ling et al. (2013) found that chlorogenic and caffeic acids in root exudates from rootstock-grafted (RG) inhibited FON conidial germination and growth. Further experiment evidenced that grafting can shift the root-secreted protein profile and thus increased FON resistance (Song et al., 2016). Recently, a series of comparative proteomic analyses was conducted to explore the cold/chilling (Xu et al., 2016; Shi et al., 2019) or salt (Yang et al., 2012) stress-induced mechanisms in grafted watermelon seedlings. It is noteworthy that most of the previous studies on grafted watermelon resistance assessment were focused on root tissues, whereas few reports have discussed the FON-resistant mechanisms mediated by RG plants especially the above-ground leaf tissues. Herein, we hypothesized that the leaf protein profile has been altered after watermelon grafting and plays an essential role in resistance to Fusarium wilt.
To gain insight into the molecular mechanisms involved in RG watermelon against FON infection, a two-dimensional gel electrophoresis (2-DE) technique in combination with matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS) was used to investigate the above-ground leaf proteome profiles of RG and self-grafted (SG) under FON challenge. Proteomic analysis identified 39 differentially accumulated proteins (DAPs), including 11 specific to RG and 4 unique to SG. Thirty-nine DAPs are mainly involved in plant metabolism and energy, defense and stress, protein biosynthesis, and degradation. The expression patterns at the transcriptional level of 10 randomly selected DAPs were validated using real-time PCR. Meanwhile, the functional implications of 39 DAPs especially those specifically accumulated in RG plants were discussed.
Materials and Methods
Plant Materials
A FON-susceptible watermelon cv. “Sumi 1” (Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences, China) and a FON-resistant bottle gourd rootstock cv. “Chaofengkangshengwang” (a commercial rootstock from Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, China) were used as scion and rootstock, respectively. An “insertion grafting” method (Lee, 1994) was employed to create the grafting combinations. Scions were grafted onto itself (SG) and bottle gourd rootstock (RG), respectively, and the grafting plants were cultivated in a growth chamber at 28/18°C under an 18/6 h light/dark cycle with a relative humidity of 70%. Grafting seedlings at the three-true-leaf stage were used for FON inoculation. SG and RG seedlings with FON inoculation were named as SG-FON and RG-FON, and their corresponding control seedlings inoculated with distilled water were designated as SG-C and RG-C, respectively. Three replicates were conducted with 30 seedlings for each replication. Ten leaves from various seedlings were pooled for each sample at 240 hours post-inoculation (hpi), when wilting symptoms were observed above the ground and were immediately frozen in liquid nitrogen and stored at −80°C until further use. Three biological replicates were sampled.
Preparation of Inoculum and Inoculation of Plants
The FON race 1 (Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences, China) was used for plant inoculation. FON was maintained on potato dextrose agar (PDA) at 25°C for 7 days and then inoculated in liquid potato dextrose medium in a 250 ml triangular flask at 25°C on a rotary shaker at 150 rpm for 7 days. Fungal suspension with a concentration of 106 conidia per ml was used to inoculate plants (Chang et al., 2008). For inoculation, grafting seedlings at the three-true-leaf stage were carefully removed from the seedling-raising pot, followed by a rinse with tap water to exclude the soil particles. Roots of the seedlings were immersed in freshly prepared fungal suspension for 30 min, and then seedlings were re-planted in the seedling-raising pot with peat moss + vermiculite + perlite (6/1/3, v/v/v). Seedlings inoculated with sterile distilled water were treated as a control.
Measurements of Physiological Traits
A total of 0.5 g leaf samples from RG or SG inoculated with FON were extracted with 2 ml citrate buffer (0.1 M, pH 5.0) to obtain the crude enzyme extract and used for enzyme activity assay. Chitinase extraction and measurements were performed according to the method described by Boller et al. (1983). In brief, assay mixture (1 ml) containing 0.1 ml of sodium acetate buffer (0.05 M, pH 5.0), 0.3 μM of sodium azide, 1 mg of colloidal chitin, and 0.4 ml of crude enzyme extract was incubated at 37°C for 3 h. Then, 0.4 ml of the assay mixture was mixed with 0.2 ml of sodium borate buffer (0.8 M, pH 9.1) and heated at 100°C for 3 min, followed by added 3 ml of 1% 4-dimethylaminobenzaldehyde (DMAB) solution and incubated for 15 min at 37°C. The absorbance was recorded at 585 nm. Three replicates were conducted.
The β-1,3-glucanase activity was measured as presented by Shu et al. (2006). In brief, assay mixture (1 ml) containing 0.48 ml of sodium acetate buffer (0.1 M, pH 5.2), 1 mg of laminarin, and 0.3 ml of crude enzyme extract was incubated at 50°C for 3 h. Then, 1 ml of 3,5-dinitrosalicylic acid (DNS) was added and heated at 100°C for 5 min. The absorbance was recorded at 540 nm. The amount of glucose was calculated from the standard curve of glucose. Three replicates were conducted.
Protein Extraction
Total proteins were extracted from leaves of SG and RG seedlings as described by Zhang et al. (2018a). In brief, protein pellets were dissolved in an extraction buffer containing 9.5 M urea, 4% w/v 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 65 mM dithiothreitol (DTT), and 2% v/v immobilized pH gradient (IPG) buffer pH 4–7. The protein concentration was determined using the Bradford method (Bradford, 1976) with bovine serum albumin (BSA) as a standard. Three biological replicates were prepared.
2-DE Analysis
Two-dimensional gel electrophoresis was performed using the GE Healthcare 2-DE system as described by Zhang et al. (2018a). Then, 800 μg protein for each sample was loaded to rehydration strip (pH gradient of 4–7, 13 cm) and separated on a Multiphor electrophoresis unit (GE Healthcare, Tokyo, Japan) with the following parameters: 30 V for 12 h, 1 h step from 200 to 500 V, 1 h gradient from 500 to 1,000 V, 30 min gradient from 1,000 to 4,000 V, 30 min gradient from 4,000 to 8,000 V, and 5 h at 8,000 V. After isoelectric focusing, the strips were equilibrated in equilibration buffer I [containing 6 M urea, 50 mM Tris–HCl (1.5 M stocking buffer, pH 8.8), 30% glycerol, 2% sodium dodecyl sulfate (SDS), 1% DTT] with gentle agitation for 15 min, followed by equilibration buffer II [containing 6 M urea, 50 mM Tris–HCl (1.5 M stocking buffer, pH 8.8), 30% glycerol, 2% SDS, 4% iodoacetamide] for 15 min. The strips were transferred to a 12.5% SDS-polyacrylamide gel for protein separation using a Hoefer SE 600 Ruby system (GE Healthcare, Tokyo, Japan). The electrophoresis was carried out at a constant current of 5 mA/gel for 15 min and 10 mA/gel for 6 h.
Gel Staining and Image Analysis
Gels were stained with Coomassie Brilliant Blue R-250 (Solarbio, Beijing, China). Gel images were acquired using a TyphoonTM 9400 imager (GE Healthcare, Tokyo, Japan) and analyzed by ImageMaster 2D Platinum 6.0 software (GE Healthcare, Tokyo, Japan). For image analysis, manual editing was carried out after automated detection and matching to correct any mismatched and unmatched spots. Proteins were considered to be differentially accumulated if their percent volume ratio was ≥1.5 and ANOVA test value was ≤0.05.
In-Gel Digestion and MALDI-TOF/TOF MS Analysis
The DAP spots were excised from the 2-DE gel, washed with 50% (v/v) acetonitrile in 0.1 M NH4HCO3, and digested with modified trypsin (Promega, United States). The digested peptides were further analyzed by MALDI-TOF/TOF MS. MS/MS spectra were searched against the NCBInr database with a Viridiplantae (green plants) restriction and Cucurbitaceae database using the Mascot search tool (Matrix Science, London, United Kingdom).
Quantitative Real-Time RT-PCR
Total RNA was extracted from leaves of SG and RG seedlings inoculated with FON by using an RNApure Plant kit (with DNase I) (CWBiotech, China). Reverse transcription with 1 μg RNA and the oligo dT primer was performed through a BU-Superscript RT kit (Biouniquer, China) according to the manufacturer’s instructions. Real-time PCR was carried out using the 1× SYBR Green PCR Master Mix (PE Applied Biosystems, United States) on the GeneAmp® 7300 Sequence Detection System (PE Applied Biosystems, United States) according to the manufacturer’s instructions. 18SrRNA (GenBank accession no. AB490410) was used as the internal control for normalization. The relative quantization of gene expressions was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Three replicates were performed. Primers used in this work were listed in Supplementary Table 1.
Bioinformatics Analysis
Protein subcellular localization was identified using WoLF PSORT2. The possible interaction between the DAPs was carried out through STRING3.
Results
Comparison of Disease Phenotypes and Physiological Changes of Grafting Seedlings Under FON Infection
Plant growth was significantly influenced by bottle gourd rootstock grafting under FON infection. SG seedlings exhibited visual wilting symptoms in cotyledons at 240 hpi, whereas RG seedlings grew well, and no cotyledon wilting was observed. RG plants showed high resistance to FON with 3.4% of infested plants, whereas SG plants were relatively highly susceptible with 89% of disease incidence at 21 days post-inoculation. FON infection induced the accumulation of both β-1,3-glucanase and chitinase, whereas their activities showed differences in RG and SG seedlings. β-1,3-Glucanase was significantly higher in RG than in SG (Figure 1A), whereas chitinase remarkably accumulated more in SG than in RG (Figure 1B). These results indicate that rootstock grafting could improve plants’ resistance by accumulating activity of PR proteases to prevent watermelon seedlings from FON infection.
FIGURE 1.
Physiological responses of the grafted watermelon seedlings infected with FON. Changes of β-1,3-glucanase (A) and chitinase (B) activities in the leaves of RG and SG plants. Vertical bars labeled with different letters are significantly different at p < 0.05. Error bars are based on three biological replicates.
Proteome Analysis of Grafting Plants With FON Infection
Leaf samples of RG and SG seedlings inoculated with FON at 240 hpi were analyzed to investigate the FON responsive proteins using 2-DE. More than 1,340 protein spots were generated on each gel, and 917 were reproducibly detected. Proteins with fold change >1.5 at p-value < 0.05 were considered to be differentially accumulated (DAP). Accordingly, 39 DAPs were identified (Table 1 and Figures 2, 3), of which 14 up-accumulated proteins were shared in both RG and SG; 10 and 2 proteins were up-accumulated only in RG and SG, respectively. Six down-accumulated proteins were overlapped in both RG and SG; 1 and 2 proteins were down-accumulated only in RG and SG, respectively (Figure 3). Three proteins were up-accumulated in RG but down-accumulated in SG; 1 protein was up-accumulated in SG but down-accumulated in RG (Figure 3).
TABLE 1.
Identification of differentially accumulated proteins in leaves of grafted watermelon under FON infection.
| aSpot ID | Watermelon accession no. | Protein description | bMr (KD)/pI | cSC (%) | Score | Cell compartment | dAbundance vol% |
| C metabolism | |||||||
| L14 | Cla97C05G082860 | Transketolase, putative | 80.62/6.37 | 15 | 272 | Cytoplasm |  |
| L26 | Cla97C09G167630 | Beta-xylosidase 2 | 83.30/8.63 | 8 | 227 | Vacuole |  |
| L33 | Cla97C02G032410 | Malate dehydrogenase | 43.60/7.75 | 33 | 380 | Chloroplast |  |
| N metabolism | |||||||
| L8 | Cla97C09G166940 | Glutamine synthetase | 39.19/5.99 | 14 | 365 | Cytoplasm |  |
| L9 | Cla97C05G093490 | Cytosolic glutamine synthetase | 39.22/5.90 | 11 | 313 | Cytoplasm |  |
| ROS metabolism | |||||||
| L7 | Cla97C02G045130 | Peroxidase | 34.58/8.32 | 36 | 283 | Chloroplast |  |
| L17 | Cla97C09G162960 | Germin-like protein | 21.51/6.50 | 29 | 291 | Chloroplast |  |
| L23 | Cla97C08G148570 | L-Ascorbate peroxidase T | 49.69/7.96 | 9 | 274 | Chloroplast |  |
| L37 | Cla97C02G046770 | L-Ascorbate peroxidase | 27.78/5.82 | 9 | 245 | Chloroplast |  |
| Energy metabolism | |||||||
| L13 | Cla97C08G146490 | ATP synthase subunit delta, chloroplastic | 25.77/9.78 | 11 | 218 | Mitochondria |  |
| L24 | Cla97C04G076580 | Enolase | 47.77/5.55 | 13 | 343 | Cytoplasm |  |
| L27 | Cla97C02G030740 | ATP synthase subunit alpha | 54.26/4.69 | 36 | 569 | Cytoplasm: cytoskeleton |  |
| L30 | Cla97C03G055870 | ATP synthase gamma-subunit | 41.25/5.99 | 21 | 492 | Chloroplast |  |
| Protein biosynthetic | |||||||
| L1 | Cla97C05G084830 | Cyanate hydratase | 18.89/6.53 | 25 | 100 | Cytoplasm |  |
| L10 | Cla97C11G219440 | Cysteine synthase | 40.97/7.46 | 22 | 165 | Chloroplast |  |
| L11 | Cla97C02G049790 | Enoyl reductase | 40.91/9.57 | 13 | 414 | Chloroplast |  |
| L12 | Cla97C07G132520 | 60S acidic ribosomal protein P0 | 34.21/4.77 | 17 | 165 | Cytoplasm |  |
| L21 | Cla97C10G187570 | Hsp70–Hsp90 organizing protein 3-like | 65.14/5.91 | 8 | 249 | Cytoplasm |  |
| L25 | Cla97C08G153070 | 6,7-Dimethyl-8-ribityllumazine synthase | 24.20/8.09 | 19 | 157 | Chloroplast |  |
| L28 | Cla97C06G109750 | Arginase | 47.99/7.36 | 20 | 380 | Chloroplast |  |
| L29 | Cla97C06G127160 | Elongation factor Tu, chloroplastic | 51.24/6.37 | 25 | 779 | Chloroplast |  |
| L34 | Cla97C05G092190 | Sulfurtransferase | 49.09/9.22 | 15 | 151 | Chloroplast |  |
| L39 | Cla97C02G042600 | RNA-binding KH domain-containing protein | 67.74/6.00 | 18 | 341 | Cytoplasm |  |
| Defends and stress | |||||||
| L2 | Cla97C05G088120 | Universal stress protein | 18.08/5.96 | 34 | 135 | Cytoplasm |  |
| L4 | Cla97C11G207000 | Jasmonate-induced protein | 23.69/6.39 | 37 | 314 | Cytoplasm |  |
| L6 | Cla97C08G158200 | Thioredoxin h | 12.99/5.62 | 57 | 235 | Cytoplasm |  |
| L15 | Cla97C01G003090 | Thaumatin-like protein | 24.49/6.97 | 26 | 194 | Chloroplast |  |
| L16 | Cla97C01G003090 | Thaumatin-like protein | 24.49/6.97 | 36 | 237 | Chloroplast |  |
| L22 | Cla97C11G207000 | Jasmonate-induced protein | 23.69/6.39 | 26 | 315 | Cytoplasm |  |
| L35 | Cla97C02G034000 | Isoflavone reductase-like protein/ | 33.75/7.77 | 32 | 321 | Cytoplasm |  |
| Translation | |||||||
| L3 | Cla97C05G097300 | Eukaryotic translation initiation factor 5A (eIF5A) | 17.56/5.87 | 35 | 220 | Cytoplasm |  |
| L20 | Cla97C05G084840 | Multiple organellar RNA editing factor 2 | 27.26/8.97 | 17 | 102 | Mitochondria |  |
| Photosystem | |||||||
| L31 | Cla97C10G185280 | Ferredoxin–NADP reductase | 40.30/8.17 | 23 | 371 | Chloroplast |  |
| L32 | Cla97C10G185280 | Ferredoxin–NADP reductase | 40.30/8.17 | 20 | 336 | Chloroplast |  |
| L36 | Cla97C11G213660 | Thylakoid lumenal 15 kDa protein 1, chloroplastic | 22.61/6.52 | 27 | 373 | Chloroplast |  |
| L38 | Cla97C07G137540 | Oxygen-evolving enhancer protein 2, chloroplastic | 28.35/8.04 | 24 | 256 | Chloroplast |  |
| Signal transduction | |||||||
| L18 | Cla97C05G080010 | Nucleoside diphosphate kinase | 16.43/6.81 | 42 | 418 | Cytoplasm |  |
| Transport | |||||||
| L5 | Cla97C07G142420 | Nuclear transport factor 2 | 13.51/6.50 | 34 | 168 | Nucleus |  |
| L19 | Cla97C02G027850 | Cytochrome b6-f complex iron–sulfur subunit | 24.22/8.28 | 23 | 421 | Chloroplast |  |
a Spot ID corresponding to spots in Figure 2. bMr, molecular weight; pI, isoelectric point. cSC, sequence coverage. dThe relative volumes of three replicates (±SD) are presented. White column means RG seedlings; black column means SG seedlings. From left to right, samples were RG-C, SG-C, RG-FON, and SG-FON.
FIGURE 2.
Representative 2-DE gels of proteins extracted from leaves of grafted watermelon seedlings. Proteins extracted from leaves of (A) SG-C, (B) RG-C, (C) SG-FON at 240 hpi, and (D) RG-FON at 240 hpi were focused on IPG strips (11 cm, pH 4–7 NL) and separated by SDS-PAGE (12.5%). Arrows mean the differentially accumulated proteins among RG and SG responding to FON infection.
FIGURE 3.
Venn diagram of differentially accumulated proteins (DAPs) in RG and SG infected with FON. DAPs were analyzed based on SG-FON vs. SG-C and RG-FON vs. RG-C libraries, respectively.
Classification of FON Responsive DAPs
Thirty-nine DAP spots were excised from the gel and subjected for MALDI-TOF/TOF MS analysis. All the 39 DAPs were successfully identified (Table 1), of which 90% of proteins were identified with >10% sequence coverage showing high confidence. The DAPs were functionally categorized mainly in 10 different pathways: C metabolism-related (3), N metabolism (2), energy metabolism (4), protein biosynthetic (10), photosystem (4), defense and stress (7), reactive oxygen species (ROS) metabolism (4), translation (2), signal transduction (1), and transport (2) (Table 1 and Figure 4A). In addition, it was reported that subcellular localization of the proteins was closely related to their physiological functions in plants (Sun et al., 2014). Thirty-nine DAPs were further localized to the chloroplast (19), cytoplasm (15), mitochondria (2), nucleus (1), cytoskeleton (1), and vacuole (1) (Figure 4B), indicating that chloroplast located proteins (49%) play crucial roles in plant defense against FON in grafting seedlings.
FIGURE 4.
Functional category (A) and subcellular location (B) of DAPs in leaves of grafted watermelon in response to FON infection.
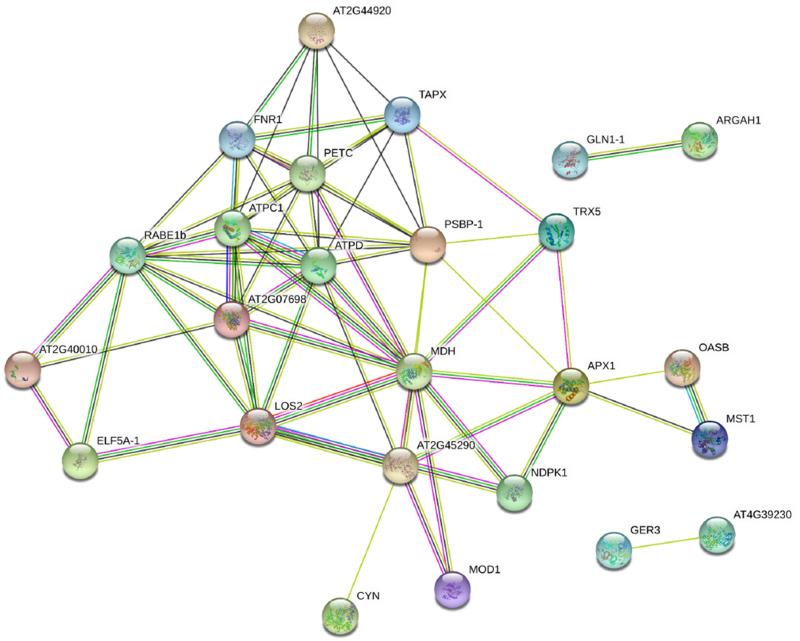
Protein–Protein Interaction Network
A predicted protein–protein interaction network was generated using STRING 9.0 to reveal functional links between proteins differentially accumulated in watermelon leaves in response to F. oxysporum infection. As expected, one protein interacted with another one to constitute a complex interaction network (Figure 5 and Supplementary Table 2). The main cluster revealed strong interaction among TAPX-FNR1-RABE1b-LOS2-MDH. These proteins belong to ROS metabolism, photosystem, translation, energy metabolism, and C metabolism. This observation indicated that these proteins function cooperatively to prevent plants from FON infection in grafted watermelon seedlings.
FIGURE 5.
Protein–protein interaction (PPI) network as elucidated through the STRING 11.0 online software with a confidence score of 0.4 using Arabidopsis thaliana. The network nodes represent proteins, and the edges indicate the predicted functional associations. The clusters mean the highly interacting proteins involved in photosystem, energy metabolism, translation, and C metabolism.
Analysis of the Expression Profiles of the mRNAs of Some Identified Proteins by Real-Time PCR
Ten DAPs were selected for transcript level to validate the proteomic data. In our proteome work, protein spots L2 (USP), L22 (JIP), and L35 (IRL) were up-accumulated in both SG and RG. Protein spot L31 (FNR) was down-accumulated in both SG and RG. Protein spot L21 (HOP) was up-accumulated in RG but down-accumulated in SG. Protein spots L6 (TrxH), L23 (APXT), and L28 (ARG) were only up-accumulated in RG but no response in SG. Protein spots L9 (GS) and L15 (thaumatin-like protein, TLP) were only up-accumulated in SG but no response in RG. Quantitative reverse transcription-PCR (qRT-PCR) (Figure 6) showed that the gene transcription level of seven proteins (USP, TrxH, GS, HOP, JIP, APXT, and ARG) was correlated with MS-based level supporting the reliability of the proteomic approach, whereas the gene expression profile of the other three proteins (TLP, FNR, and IRL) was not correlated with their protein level.
FIGURE 6.
Relative expression level of genes corresponding to 10 randomly selected DAPs in leaves of grafted watermelon in response to FON infection. The value of the relative expression level was normalized to 18SrRNA. Error bars were based on three technical replicates. USP (spot L2), universal stress protein; TrxH (spot L6), thioredoxin h; GS (spot L9), glutamine synthetase; TLP (spot L15), thaumatin-like protein; HOP (spot L21), Hsp70–Hsp90 organizing protein; JIP (spot L22), jasmonate-induced protein; APXT (spot L23), L-ascorbate peroxidase T; ARG (spot L28), arginase; FNR (spot L31), ferredoxin–NADP reductase; IRL (spot L35), isoflavone reductase-like protein.
Discussion
Energy and Metabolism Proteins
Defense mechanisms are employed to minimize pathogenic damage, among which plant metabolism and energy supply are the key factors for the defense of plants against pathogens (Zaynab et al., 2019). Pathogen infection leads to dramatic changes in the carbohydrate metabolism of the infected plant tissue that was supported in our work by the alteration of C metabolism-related proteins (spots L14, L26, L33) in grafted watermelon seedlings infected with FON. Specifically, the abundance of malate dehydrogenase (MDH, spot L33) was up-accumulated extremely in RG under FON challenge. MDH is the key tricarboxylic acid (TCA) cycle enzyme that reversibly catalyzes the interconversion of malate and oxaloacetate (Musrati et al., 1998). Many studies have evaluated the function of MDH in plant defense against various abiotic stresses either by maintaining energy homeostasis (Selinski et al., 2014) or by enhancing organic acid synthesis (Tesfaye et al., 2001). We therefore speculated that the improved FON tolerance of RG may be related to the up-representation of MDH (spot L33) that enhanced the TCA cycle.
Photosynthesis is one of the plant metabolisms that could be involved in defense against pathogens. Most studies have shown that pathogen invasion locally reduced the rates of photosynthesis that could be interpreted as for freeing up resources utilized for the defense response (Somssich and Hahlbrock, 1998) or protecting the photosynthetic apparatus against light-induced damage (Niyogi, 2000). In contrast, stimulated rates of photosynthesis have been described in several compatible plant–pathogen interactions. As an example, in the tomato plants infected with Pseudomonas syringae and Botrytis cinerea, Berger et al. (2004) found the expected inhibition of photosynthesis in the infection sites as well as distinct stimulation of photosynthesis in the surrounding circular areas. The authors presumed that the enhanced photosynthesis could be part of the defense strategy for plants to produce assimilates for defense reactions that ultimately helps to confine pathogen growth. In this study, three ATP synthase proteins (spots L13, L27, L30) and two ferredoxin–NADP reductase (FNR) proteins (spots L31, L32) were downregulated in abundance in both RG and SG plants in response to FON infection. One of the possible explanations might be the plant’s attempt to maintain the intactness of the membranes or repair them for carrying on the electron transport reactions (Dinakar et al., 2012).
Proteins Involved in Defense and Stress
Activation of plant defense systems was evidenced by an altered abundance of defense-related proteins, such as universal stress protein 1 (spot L2), jasmonate-induced protein (spots L4, L22), isoflavone reductase-like protein (spot L35), and pathogenesis-related protein PR-5 (TLP, spots L15, L16). The density of TLP (spot L15) was up-accumulated in SG, whereas there remained no changes in RG. Conversely, the abundance of TLP (spot L16) was significantly activated in RG, whereas it was repressed in SG. A wealth of evidence suggests that the accumulation of TLPs induces systemic acquired resistance that boosts the plant’s resistance against fungal pathogens (Dong, 2001). Sun et al. (2020) proved that TLP performs different functions in other physiological processes. They found that Pe-TLP played a role as an elicitor of other anti-fungal proteins that cause the activation of other defense pathways. Meanwhile, fungal pathogens employed different mechanisms simultaneously or in succession to facilitate successful infection during the plant–fungus interaction. It was proven that TLP could either be bound to Trichoderma virens Alt a 1 protein (Kumar and Mukherjee, 2020) or be absorbed by fungal mycelia (Marcato et al., 2017) and hence suppressing plant defense. Based on these observations, we presume that different mechanisms may be deployed during the interaction of grafted watermelon and FON.
Among ROS-scavenging enzymes, thioredoxin h (spot L6) and peroxidase (spot L7) were identified in grafted watermelon leaves infected with FON. The abundance of these proteins was up-accumulated while exhibiting different profiles in RG and SG seedlings. Thioredoxin h (spot L6) was observed to be up-represented in RG seedlings, whereas there remained no changes in SG. A series of reports documented that increased production of thioredoxin h was triggered by accumulation of ROS and misfolded proteins in ROS homeostasis in defense against fungal (Zhang et al., 2017) and viral infections (Das et al., 2019) and abiotic stress (Shi et al., 2019). Thioredoxin h functions in the protection of infected plants against oxidative stress by reducing disulfide bonds on selected target proteins (Jacquot et al., 1997) or by acting as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome (Du et al., 2015). Based on these observations, elevated thioredoxin h protein in RG might be the result of maintaining ROS homeostasis in defense of RG watermelon plants against FON and thus allows the RG seedlings to survive.
The abundance of peroxidase (spot L7) was higher in SG than in RG (Table 1 and Figure 1), indicating that SG seedlings need more peroxidase to reduce the ROS accumulation in their intracellular system (Das et al., 2019). Peroxidase activity was also proven as key players particularly in cell wall modifications by catalyzing lignification (Passardi et al., 2004), implicating that structures were produced around the sites of potential FON ingress to establish physical barriers to prevent the progress of pathogen in SG plants (Zaynab et al., 2019). Additionally, the differential representation of antioxidant proteins was observed in grafted watermelon infected with F. oxysporum. Two L-ascorbate peroxidase (APX, spots L23, L37) were identified. APX removes potential harmful H2O2 from plant cells by detoxificating H2O2 into water utilizing ascorbate as electron donor (Shigeoka et al., 2002). Overproduction of APX in this work might be the result of the enhancement of active oxygen scavenging system in grafted watermelon (Sarowar et al., 2005) and thus result in F. oxysporum resistance. However, other studies have suggested that chloroplastic APX is highly sensitive to inactive to excessive ROS, namely, high contents of ROS under extreme stress conditions may repress APX, which was interpreted as that APX deficiency could activate a compensatory mechanism to protect plants against oxidative stress (Caverzan et al., 2014) and therefore explained the low density of APX (spot L23) in SG plants in this work.
Germin-like protein (GLP) is a ubiquitous water-soluble glycoprotein characterized by various enzymatic activities and is known to play crucial roles in plants’ resistance to fungal pathogens due to its antioxidant potential (Wang et al., 2013). GLP-overexpressing plants exhibited enhanced resistance to bacterial blight and fungal pathogens, which was explained by promoting ROS accumulation (Beracochea et al., 2015) or regulating the expression of plant defense-related genes (Liu et al., 2016) in transgenic plants. The up-representation of GLP (spot L17) in the current work suggests that GLP functions as a positive regulator of grafted watermelon resistance to FON.
Protein Biosynthesis and Degradation
Changes in protein synthesis and degradation were evidenced by the altered abundance of 10 proteins (Table 1). Earlier reports showed the increased amino acid triggered plants’ resistance to salt stress resulting from the strong nitrogen uptake ability by the root system of the bottle gourd rootstock (Martinez-Ballesta et al., 2010). In our study, two proteins, cyanate hydratase (spot L1) and sulfurtransferase (spot L34), were up-accumulated in both RG and SG; four proteins (spots L11, L21, L25, L28) were up-represented specifically in RG, whereas they were down-accumulated in SG, especially for Hsp70–Hsp90 organizing protein (HOP/Sti 1 spot L21). HOP/Sti 1 is a co-chaperone that could bind both Hsp70 and Hsp90 chaperones and enables complex formation at the same time. HOP/Sti 1 functions in host physiological processes linked to disease states and roles in aiding protein folding and avoiding or rescuing misfolded proteins (Tiroli-Cepeda and Ramos, 2011). The differential alteration of HOP/Sti 1 protein abundance (spot L21) in this work implicates its crucial role in maintaining proteostasis in RG and SG under F. oxysporum challenge.
Four proteins (spots L10, L39, L12, L29) involved in protein synthesis were under-represented in both RG and SG. EF-Tu (spot 29) was suppressed strikingly in RG, whereas it remained unchanged in SG. EF-Tu plays a vital role in mRNA decoding by proofreading each aminoacyl-tRNA (Morse et al., 2020). Although speculation exists that EF-Tu is involved in the other cell functions (Choi et al., 2000), the contribution of the lower abundance of EF-Tu to the RG plant’s resistance to F. oxysporum remains to be explored.
DAPs Involved in Signal Transduction
The abundance of a nucleoside diphosphate kinase (NDPK, spot L18) was identified to be significantly up-represented in RG infected with F. oxysporum. A similar result was observed in tolerant Arabidopsis thaliana line upon challenge with Alternaria brassicae (Sharma et al., 2007). The authors speculated the NDPK role in mediating plant defense to A. brassicae via the ROS-mediated signaling pathway. NDPK, a ubiquitous and highly conserved enzyme, plays a role in the primary metabolism for maintaining the nucleotide balance in the cell (Sweetlove et al., 2001), as well as functions in signal transduction pathway for mediating plant defense against abiotic stress (Moon et al., 2003), hormone responses (Galvis et al., 2001), and pathogen (Sharma et al., 2007). Overexpression of AtNDPK2 reduced the accumulation of ROS and thereby conferred enhanced tolerance to multiple environmental stresses (Moon et al., 2003). Further evidence showed that the enhancement of plant defense resulted from the association of AtNDPK2 with H2O2-mediated MAPK signaling in plants. Our current observation that NDPK (spot L18) abundance is strikingly increased in RG suggests that NDPK appears to play a vital regulatory role via ROS-mediated signaling in mediating RG plants’ response to FON challenge.
Translation and Transport-Related Proteins
Two proteins, eukaryotic translation initiation factor 5A (eIF5A, spot L3) and multiple organellar RNA editing factor 2 (MORF2, spot 20), associated with translation were identified in grafted watermelon infected with FON. The abundance of eIF5A (spot L3) was up-accumulated in both RG and SG plants under pathogen challenge. eIF5A is involved in the initial process of protein translation and can be induced by the pathogen (Campo et al., 2004). In the present work, the up-representation of eIF5A (spot L3) following F. oxysporum challenge may be responsible for regulating proteins important for pathogen attack. Rather, MORF2 up-represented extremely in RG while with low intensity in SG. MORF2 is an essential component of the plant RNA editosome and is a major player as an editing factor regulating the RNA editing efficiency at multiple sites (Bentolila et al., 2013). The function of MORF2 during plant and pathogen interaction remains unknown, although observation exists that plastid-signaling defective mutant gun1, directly interacting with MORF2, showed a deficiency of rapid RNA multiplication that results in more severe oxidative stress (Zhang et al., 2011).
A nuclear transport factor 2 (NTF2, spot L5) protein was identified in this work as shown to be markedly accumulated in RG, whereas it remained unchanged in SG under FON challenge. The up-representation of NTF2 was observed previously in the compatible interaction of Verticillium dahliae with tomato (Hu et al., 2019). Furthermore, the silencing of TaNTF2 reduced the resistance of wheat to different avirulent isolates of the Puccinia striiformis, which speculated that NTF2 acted as a critical regulator in the Ran-mediated signal transduction in the plant immune system (Zhang et al., 2018b).
Conclusion
In conclusion, we reported here that bottle gourd rootstock grafting can significantly improve watermelon resistance against FON. Proteomic analysis revealed 39 DAPs in leaves of grafted watermelon plants under FON challenge. These reprogrammed proteins were classified into 10 different biological functional categories, among which, protein biosynthesis (26%) and defense and stress (18%) are the two major affected biological functional processes, indicating that they act as a positive regulator in defense responses triggered by FON in grafted watermelon. Notably, we identified six proteins, i.e., MDH (spot L33), HOP/Sti 1 (spot L21), MORF2 (spot L20), NTF2 (spot L5), NDPK (spot L18), and thioredoxin h (spot L6) were accumulated in abundance only in RG in response to FON infection. These proteins presented in different functional categories while interacted with each other closely and play crucial roles in rootstock grafting-mediated resistance. These specifically accumulated proteins will be further characterized to elucidate their roles in rootstock grafting-induced resistance during watermelon and FON interaction. Overall, the proteomic data provide us with new insight into a better understanding of the molecular defense mechanisms in rootstock-grafted watermelon.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XYN and MZ designed the experiments. MZ performed 2-DE and SDS-PAGE experiment. RR, LL, and JiaX contributed to the data analysis. GL and XYO contributed to the enzyme activity analysis. JinX and XYN contributed to revise the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Ministry of Agriculture of China and Jiangsu Provincial Department of Science and Technology for financial support.
Funding. This work was funded by the National Industrial Technology System for Watermelon & Melon (CARS-25), the Earmarked Fund for Jiangsu Agricultural Industry Technology System (JATS [2018]254), and the Natural Science Foundation of Jiangsu Province (BK20171323).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.632758/full#supplementary-material
Primer sequences for encoding mRNA corresponding to 12 selected proteins quantified using real-time RT-PCR.
Abbreviations of the specific protein names in the protein–protein interaction network.
References
- Beracochea V. C., Almasia N. I., Peluffo L., Nahirnak V., Hopp E. H., Paniego N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell. Rep. 34 1717–1733. 10.1007/s00299-015-1819-4 [DOI] [PubMed] [Google Scholar]
- Bentolila S., Oh J., Hanson M. R., Bukowski R. (2013). Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9:e1003584. 10.1371/journal.pgen.1003584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Papadopoulos M., Schreiber U., Kaiser W., Roitsch T. (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plantarum. 122, 419–428. 10.1111/j.1399-3054.2004.00433.x [DOI] [Google Scholar]
- Boller T., Gehri A., Mouch F., Vogli U. (1983). Chitinase in bean leaves: induction by ethylene, purification, properties and possible function. Planta 157 22–31. 10.1007/bf00394536 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Branham S. E., Levi A., Farnham M. W., Wechter W. P. (2017). A GBS-SNP-based linkage map and quantitative trait loci (QTL) associated with resistance to Fusarium oxysporun f.sp. niveum race 2 identified in Citrullus lanatus var. citroides. Theor. Appl. Genet. 130 319–330. 10.1007/s00122-016-2813-0 [DOI] [PubMed] [Google Scholar]
- Campo S., Carrascal M., Coca M., Abián J., Segundo B. S. (2004). The defense response of germinating maize embryos against fungal infection: a proteomics approach. Proteomics 4 383–396. 10.1002/pmic.200300657 [DOI] [PubMed] [Google Scholar]
- Caverzan A., Bonifacio A., Carvalho F. E. L., Andrade C. M. B., Passaia G., Schünemann M., et al. (2014). The knockdown of chloroplastic ascorbate peroxidases reveals its regulatory role in the photosynthesis and protection under photo-oxidative stress in rice. Plant Sci. 214 74–87. 10.1016/j.plantsci.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Chang P. F. L., Hsu C. C., Lin Y. H., Chen K. S., Huang J. W., Liou T. D. (2008). Histopathology comparison and phenylalanine ammonia lyase (PAL) gene expressions in Fusarium wilt infected watermelon. Aust. J. Agr. Res. 59 1146–1155. 10.1071/ar08066 [DOI] [Google Scholar]
- Choi K. R., Roh K. S., Kim J. K., Sim W. S. (2000). Genomic cloning and characterization of mitochondrial elongation factor Tu (EF-Tu) gene (tufM) from maize (Zea mays L.). Gene 257 233–242. 10.1016/s0378-1119(00)00404-2 [DOI] [PubMed] [Google Scholar]
- Das P. P., Lin Q. S., Wong S. M. (2019). Comparative proteomics of tobacco mosaic virus-infected Nicotiana tabacum plants identified major host proteins involved in photosystems and plant defence. J. Proteomics 194 191–199. 10.1016/j.jprot.2018.11.018 [DOI] [PubMed] [Google Scholar]
- Davis A. R., Perkins-Veazie P., Sakata Y., Lopez-Galarza S., Maroto J. V., Lee S. G., et al. (2008). Cucurbit grafting. Crit. Rev. Plant. Sci. 27 50–74. [Google Scholar]
- Dinakar C., Djilianov D., Bartels D. (2012). Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci. 182 29–41. 10.1016/j.plantsci.2011.01.018 [DOI] [PubMed] [Google Scholar]
- Dong X. (2001). Genetic dissection of systemic acquired resistance. Curr. Opin. Plant Biol. 4 309–314. 10.1016/s1369-5266(00)00178-3 [DOI] [PubMed] [Google Scholar]
- Du H., Kim S., Hur Y. S., Lee M. S., Lee S. H., Cheon C. I. (2015). A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome. Mol. Cell 38 187–194. 10.14348/molcells.2015.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2018). Available online at: http://www.fao.org/faostat/en (accessed December 22, 2020). [Google Scholar]
- Galvis M. L. E., Marttila S., Hakansson G., Forsberg J., Knorpp C. (2001). Heat stress response in pea involves interaction of mitochondrial nucleoside diphosphate kinase with a novel 86-kilodalton protein. Plant Physiol. 126 69–77. 10.1104/pp.126.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Zhang J., Sun H., Salse J., Lucas W. J., Zhang H., et al. (2013). The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45 51–58. [DOI] [PubMed] [Google Scholar]
- Guo S. G., Zhao S. J., Sun H. H., Wang X., Wu S., Lin T., et al. (2019). Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 51 1616–1623. 10.1038/s41588-019-0518-4 [DOI] [PubMed] [Google Scholar]
- Hu X. P., Puri K. D., Gurung S., Klosterman S. J., Wallis C. M., Britton M., et al. (2019). Proteome and metabolome analyses reveal differential responses in tomato-Verticillium dahlia-interactions. J. Proteomics 207:103449. 10.1016/j.jprot.2019.103449 [DOI] [PubMed] [Google Scholar]
- Huang Y., Li J., Hua B., Liu Z. X., Fan M. L., Bie Z. L. (2013). Grafting onto different rootstocks as a means to improve watermelon tolerance to low potassium stress. Sci. Hort. 149 80–85. 10.1016/j.scienta.2012.02.009 [DOI] [Google Scholar]
- Huh Y. C., Om Y. H., Lee J. M. (2002). Utilization of Citrullus germplasm with resistance to Fusarium wilt (Fusarium oxysporum f. sp. niveum) for watermelon rootstocks. Acta Hort. 588 127–132. 10.17660/actahortic.2002.588.18 [DOI] [Google Scholar]
- Jacquot J. P., Lancelin J. M., Meyer Y. (1997). Thioredoxins: structure and function in plant cells. New Phytol. 136 543–570. 10.1046/j.1469-8137.1997.00784.x [DOI] [PubMed] [Google Scholar]
- Kumar R., Mukherjee P. K. (2020). Trichoderma virens Alt a 1 protein may target maize PR5/thaumatin-like protein to suppress plant defence: an in silico analysis. Physiol. Mol. Plant Pathol. 112:101551. 10.1016/j.pmpp.2020.101551 [DOI] [Google Scholar]
- Lambel S., Lanini B., Vivoda E., Fauve J., Patrick Wechter W., Harris-Shultz K. R., et al. (2014). A major QTL associated with Fusarium oxysporum race 1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor. Appl. Genet. 127 2105–2115. 10.1007/s00122-014-2363-2 [DOI] [PubMed] [Google Scholar]
- Lee J. M. (1994). Cultivation of grafted vegetables. I. Current status, grafting methods, and benefits. HortScience 29 235–239. 10.21273/hortsci.29.4.235 [DOI] [Google Scholar]
- Ling N., Zhang W., Wang D., Mao J., Huang Q., Guo S., et al. (2013). Root exudates from grafted-root watermelon showed a certain contribution in inhibiting Fusarium oxysporum f. sp. niveum. PLoS One 8:e63383. 10.1371/journal.pone.0063383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yang J., Yan S., Zhang S., Zhao J., Wang W., et al. (2016). The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol. Biol. 92 411–423. 10.1007/s11103-016-0521-4 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marcato R., Sella L., Lucchetta M., Vincenzi S., Odorizzi S., Curioni A., et al. (2017). Necrotrophic fungal plant pathogens display different mechanisms to counteract grape chitinase and thaumatin-like protein. Physiol. Mol. Plant. Pathol. 99 7–15. 10.1016/j.pmpp.2016.09.005 [DOI] [Google Scholar]
- Martinez-Ballesta M. C., Alcaraz-López C., Muries B., Mota-Cadenas C., Carvajal M. (2010). Physiological aspects of rootstock-scion interactions. Sci. Hortic. 127 112–118. 10.1016/j.scienta.2010.08.002 [DOI] [Google Scholar]
- Martyn R. D. (1987). Fusarium oxysporum f. sp. niveum race 2: a highly aggressive race new to the United States. Plant Dis. 71 233–236. 10.1094/pd-71-0233 [DOI] [Google Scholar]
- Meru G., McGregor C. (2016). Genotyping by sequencing for SNP discovery and genetic mapping of resistance to race 1 of Fusarium oxysporum in watermelon. Sci. Hort. 209 31–40. 10.1016/j.scienta.2016.06.005 [DOI] [Google Scholar]
- Moon H., Lee B., Choi G., Shin D., Prasad D. T., Lee O., et al. (2003). NDP kinase 2 interacts with two oxidative stress activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. U.S.A. 100 358–363. 10.1073/pnas.252641899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse J. C., Girodat D., Burnett B. J., Holm M., Altman R. B., Sanbonmatsu K. Y., et al. (2020). Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection. Proc. Natl. Acad. Sci. U.S.A. 117 3610–3620. 10.1073/pnas.1904469117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musrati R. A., Kollárova M., Mernik N., Mikulášová D. (1998). Malate dehydrogenase: distribution, function and properties. Gen. Physiol. Biophys. 17 193–210. [PubMed] [Google Scholar]
- Niyogi K. K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant. Biol. 3 455–460. 10.1016/s1369-5266(00)00113-8 [DOI] [PubMed] [Google Scholar]
- Passardi F., Penel C., Dunand C. (2004). Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9 534–540. 10.1016/j.tplants.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Sarowar S., Kim E. N., Kim Y. J., Ok S. H., Kim K. D., Hwang B. K., et al. (2005). Overexpression of a pepper ascorbate peroxidase-like 1 gene in tobacco plants enhances tolerance to oxidative stress and pathogens. Plant Sci. 169 55–63. 10.1016/j.plantsci.2005.02.025 [DOI] [Google Scholar]
- Selinski J., König N., Wellmeyer B., Hanke G. T., Linke V., Neuhaus H. E., et al. (2014). The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol. Plant 7 170–186. 10.1093/mp/sst151 [DOI] [PubMed] [Google Scholar]
- Sharma N., Rahman M. H., Strekov S., Thiagarajah M., Bansal V. K., Kav N. N. V. (2007). Proteome-level changes in two Brassica napus lines exhibiting differential responses to the fungal pathogen Alternaria brassicae. Plant Sci. 172 95–110. 10.1016/j.plantsci.2006.07.016 [DOI] [Google Scholar]
- Shi X. F., Wang X. B., Cheng F., Cal H. S., Liang H., Lu J. Y., et al. (2019). iTRAQ-based quantitative proteomics analysis of cold stress-induced mechanisms in grafted watermelon seedlings. J. Proteomics 192 311–320. 10.1016/j.jprot.2018.09.012 [DOI] [PubMed] [Google Scholar]
- Shigeoka S., Ishikawa T., Tamoi M., Miyagawa Y., Takeda T., Yabuta Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53 1305–1319. 10.1093/jexbot/53.372.1305 [DOI] [PubMed] [Google Scholar]
- Shu C. H., Xu C. J., Lin E. S. (2006). Production, purification and partial characterization of a novel endo-β-1,3-glucanase from Agaricus brasiliensis. Process Biochem. 5 1229–1233. 10.1016/j.procbio.2005.12.011 [DOI] [Google Scholar]
- Somssich I. E., Hahlbrock K. (1998). Pathogen defence in plants-a paradigm of biological complexity. Trends Plant Sci. 3 86–90. 10.1016/s1360-1385(98)01199-6 [DOI] [Google Scholar]
- Song Y., Ling N., Ma J. H., Wang J. C., Zhu C., Raza W., et al. (2016). Grafting Resulted in a distinct proteomic profile of watermelon root exudates relative to the un-grafted watermelon and the rootstock plant. J. Plant Growth Regul. 35 778–791. 10.1007/s00344-016-9582-5 [DOI] [Google Scholar]
- Sun W. B., Zhou Y., Movahedi A., Wei H., Zhuge Q. (2020). Thaumatin-like protein (Pe-TLP) acts as a positive factor in transgenic poplars enhanced resistance to spots disease. Physiol. Mol. Plant. Pathol. 112 101512. 10.1016/j.pmpp.2020.101512 [DOI] [Google Scholar]
- Sun Y., Wang C., Yang B., Wu F., Hao X., Liang W., et al. (2014). Identifcation and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J. Exp. Bot. 65 2171–2188. 10.1093/jxb/eru092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L. J., Mowday B., Hebestreit H. F., Leaver C. J., Millar A. H. (2001). Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett. 508 272–276. 10.1016/s0014-5793(01)03069-1 [DOI] [PubMed] [Google Scholar]
- Tesfaye M., Temple S. J., Allan D. L., Vance C. P., Samac D. A. (2001). Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol. 127 1836–1844. 10.1104/pp.127.4.1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiroli-Cepeda A. O., Ramos C. H. I. (2011). An overview of the role of molecular chaperones in protein homeostasis. Protein Pept. Lett. 18 101–109. 10.2174/092986611794475093 [DOI] [PubMed] [Google Scholar]
- Wang T., Chen X., Zhu F., Li H., Li L., Yang Q., et al. (2013). Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS One 8:e61722. 10.1371/journal.pone.0061722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. H., Zhang M., Liu G., Yang X. P., Hou X. L. (2016). Comparative transcriptome profiling of chilling stress responsiveness in grafted watermelon seedlings. Plant Physiol. Biochem. 109 561–570. 10.1016/j.plaphy.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Wang L. P., Tian J., Li J., Sun J., He L. Z., et al. (2012). Proteomic study participating the enhancement of growth and salt tolerance of bottle gourd rootstock-grafted watermelon seedlings. Plant Physiol. Biochem. 58 54–65. 10.1016/j.plaphy.2012.05.026 [DOI] [PubMed] [Google Scholar]
- Zaynab M., Fatima M., Sharif Y., Zafar M. H., Ali H., Khan K. A. (2019). Role of primary metabolites in plant defense against pathogens. Microb. Pathogen. 137 103728. 10.1016/j.micpath.2019.103728 [DOI] [PubMed] [Google Scholar]
- Zhang L., Meng X. X., Liu N., Yang J. H., Zhang M. F. (2012a). Effects of grafting on phosphorus uptake and utilization of watermelon at early stage under low phosphorus stress. J. Fruit Sci. 29 120–124. [Google Scholar]
- Zhang M., Cheng S. T., Wang H. Y., Wu J. H., Luo Y. M., Wang Q., et al. (2017). iTRAQ-based proteomic analysis of defence responses triggered by the necrotrophic pathogen Rhizoctonia solani in cotton. J. Proteomics 152 226–235. 10.1016/j.jprot.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Zhang M., Xu J. H., Liu G., Yao X. F., Li P. F., Yang X. P. (2015a). Characterization of the watermelon seedling infection process by Fusarium oxysporum f.sp. niveum. Plant Pathol. 64 1076– 1084. [Google Scholar]
- Zhang M., Xu J. H., Liu G., Yao X. F., Ren R. S., Yang X. P. (2018a). Proteomic analysis of responsive root proteins of Fusarium oxysporum-infected watermelon seedlings. Plant Soil 422 169–181. 10.1007/s11104-017-3294-x [DOI] [Google Scholar]
- Zhang M., Yang X. P., Xu J. H., Liu G., Yao X. F., Li P. F. (2015b). Physiological responses of watermelon grafted onto bottle gourd to Fusarium oxysporum f.sp. niveum infection. Acta Hort. 1086 107–111. 10.17660/actahortic.2015.1086.12 [DOI] [Google Scholar]
- Zhang M., Yang X. P., Liu G., Xu J. H., Zhu L. L., Gao C. Z., et al. (2012b). “Histological differences between watermelon grafted onto bottle gourd rootstock and self-rooted seedlings inoculated with Fusarium oxysporum f.sp. niveum,” in Proceedings of the Xth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, October 15-18th, eds Sari N., Solmaz I., Aras V. Antalya, 289–295. [Google Scholar]
- Zhang Q., Wang B., Wei J. P., Wang X. J., Han Q. M., Kang Z. S. (2018b). TaNTF2, a contributor for wheat resistance to the stripe rust pathogen. Plant Physiol. Biochem. 123 260–267. 10.1016/j.plaphy.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Zhang Z. W., Yuan S., Xu F., Yang H., Chen Y. E., Yuan M., et al. (2011). Mg-protoporphyrin, haem and sugar signals double cellular total RNA against herbicide and high-light-derived oxidative stress. Plant Cell Environ. 34 1031–1042. 10.1111/j.1365-3040.2011.02302.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences for encoding mRNA corresponding to 12 selected proteins quantified using real-time RT-PCR.
Abbreviations of the specific protein names in the protein–protein interaction network.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.