Figure 4.
Time series data of a GRN's evaluation for associative memory
This trace describes the run time state changes in evaluating associative memory of a mammalian cell cycle network.
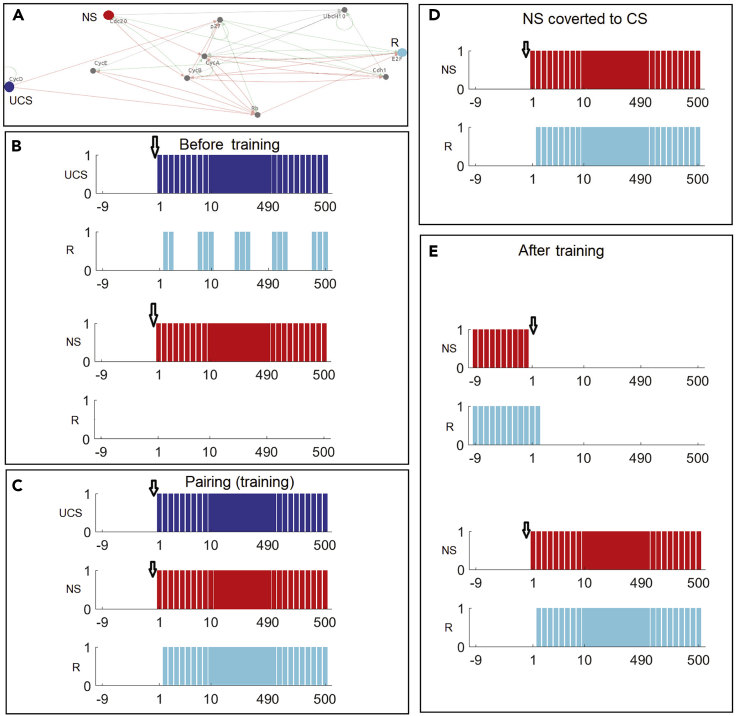
(A) In the mammalian cell cycle network 2006, the genes used as UCS, NS/CS, and R are highlighted with blue, red, and cyan respectively. With these respective colors the states of UCS, NS/CS, and R in different plots are defined. A downward arrow in each plot shows the start of the activation of the corresponding stimuli. In each panel, we show the 10 past states of a stimulus to depict its state change upon the activation at time 0.
(B) The resultant states of R, observed from activation of UCS and NS, respectively, before training: R gets activated with onset of UCS but NS cannot trigger R.
(C) Pairing (training) experiment shows the successful activation of R.
(D) After training, activation of the previously neutral stimulus causes R to be activated, confirming that the experience of paired stimuli has converted the NS node to a CS.
(E) As further confirmation of stable causality established between CS and R by training, we first deactivate CS, to see if R gets deactivated, and then reactivate the CS to ensure that it can activate R again.