Abstract
ORF8 is a highly variable genomic region of SARS-CoV-2. Although non-essential and the precise functions are unknown, it has been suggested that this protein assists in SARS-CoV-2 replication in the early secretory pathway and in immune evasion. We utilized the binding partners of SARS-CoV-2 proteins in human HEK293T cells and performed genome-wide phylogenetic profiling and clustering analyses in 446 eukaryotic species to predict and discover ORF8 binding partners that share associated functional mechanisms based on co-evolution. Results classified 47 ORF8 binding partner proteins into 3 clusters (groups 1-3), which were conserved in vertebrates (group 1), metazoan (group 2), and eukaryotes (group 3). Gene ontology analysis indicated that group 1 had no significant associated biological processes, while groups 2 and 3 were associated with glycoprotein biosynthesis process and ubiquitin-dependent endoplasmic reticulum-associated degradation pathways, respectively. Collectively, our results classified potential genes that might be associated with SARS-CoV-2 viral pathogenesis, specifically related to acute respiratory distress syndrome, and the secretory pathway. Here, we discuss the possible role of ORF8 in viral pathogenesis and in assisting viral replication and immune evasion via secretory pathway, as well as the possible factors associated with the rapid evolution of ORF8.
Keywords: ORF8, phylogenetic profiling, functional mechanism
Introduction
The coronavirus disease 19 (COVID-19) pandemic was still rampant around the world after a year from its first detection in late December 2019.1 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, is the seventh coronavirus that has infected the human population. This virus is rapidly spreading across the globe and has caused a considerable burden on the global economy and human health. The SARS-CoV-2 genome is characterized as a positive-sense single-stranded RNA with a 5′ cap structure and a poly-A 3′ tail and encodes accessory and nonstructural proteins including spike (S), nucleocapsid (N), membrane (M), envelope (E), ORF1a, ORF1b, ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9a, ORF9b, and ORF10.2
The ORF8 protein has interesting properties as it is encoded by one of the most variable regions of SARS-CoV-2.2-4 This region was also identified as one of the hotspot areas for mutations and deletions that were detected during the early onset of infection via person-to-person transmission in late January.5 A 382-nucleotide (nt) deletion in SARS-CoV-2, which truncates both ORF7b and ORF8 and deletes the ORF8 transcriptional regulatory sequence, eliminating ORF8 transcription, has been reported.6,7 Infection of the ORF8 deleted variant in 29 patients showed a milder clinical illness as no patients develop hypoxia requiring supplemental oxygen.6 ORF8 mutations and deletions were also previously observed in SARS-CoV during the 2003 to 2004 SARS epidemic, and were experimentally determined to be associated with reduced fitness for both replication and viral attenuation.8 Conversely, the 382-nt deleted SARS-CoV-2 virus showed significantly higher replication ability than the wild type in vitro; while no difference in viral load was observed in patients, showing that the 382-nt deleted variant retained its replication suitability.7 The nonsense mutations and 3 major deletions significantly altered or deleted the SARS-CoV-2 ORF8 protein, respectively, suggesting that SARS-CoV-2 can survive without the functional ORF8 protein.9
Of note, no reliable sequence identity was detected with any sequence of known proteins for SARS-CoV-2 ORF8.10 As such, the role of SARS-CoV-2 ORF8 is difficult to determine through homology studies. Previously, the interaction landscape between SARS-CoV-2 proteins and human proteins has been mapped using affinity purification mass spectrometry assays.11 Interestingly, ORF8 showed the highest number of interactors, including proteins related with endoplasmic reticulum quality control, glycosylation, glycosaminoglycan synthesis, and extracellular matrix organization.11 However, since mass spectrometry is incapable of determining the direct and indirect interactions between SARS-CoV-2 and host proteins, the mechanism of action of ORF8 with respect to those interactors is obscure. In this study, we focused on ORF8-interacting proteins and aimed to elucidate their evolution and functional mechanisms based on phylogenetic profiling. Phylogenetic profiling may reveal novel functional links or predict functions of unknown genes. This method is based on the assumption that a group of functionally coupled genes, such as those that develop protein complexes, regulatory modules, or metabolic cascades, may undergo coevolution.12-16 Here, we discuss the role of ORF8 in the pathogenesis of SARS-CoV-2, with respect to the regulation of immune evasion via the secretory pathway as well as to the rapid evolutionary factors.
Results
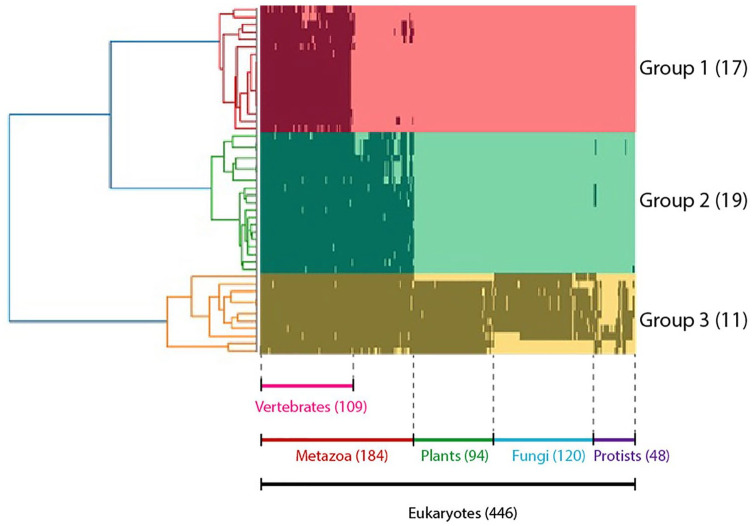
We developed a binary matrix based on the presence (1) and absence (0) of ORF8 binding partner homologues in 446 eukaryotic species for phylogenetic profiling and cluster analyses to illuminate the degree of co-evolution among the ORF8-interacting proteins. As a result, 47 proteins were classified into 3 groups: 1 to 3. Groups 1, 2, and 3 were conserved across vertebrates (17 proteins), animalia (19 proteins), and eukaryotes (11 proteins), respectively (Figure 1; Supplemental Table S1). The position of each protein with respect to the tree of phylogenetic profiling can be inspected in Supplemental Table S1.
Figure 1.
Phylogenetic profile analysis of proteins interacting with ORF8: The horizontal axis represents 446 eukaryotes whose entire genome has been deciphered, and the vertical axis represents 46 proteins that interact with ORF8. Proteins are shown in black if orthologs are present in each species. Proteins were classified into 3 groups, 1-3, based on the results of the cluster analysis. These groups were conserved in vertebrates, animals, and eukaryotes.
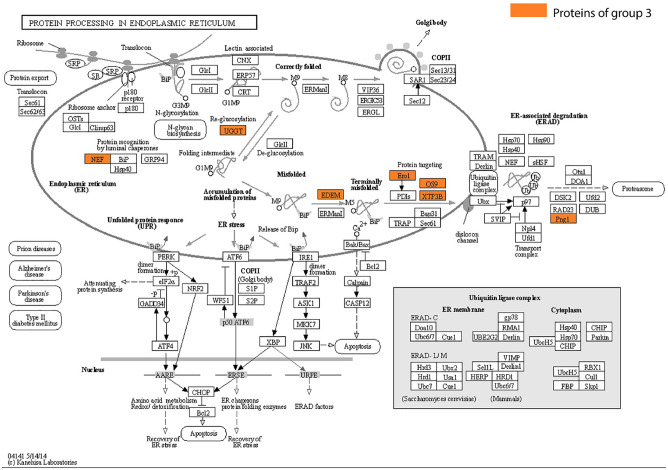
Further, the associated biological functions among each group of phylogenetic profile results were examined through gene ontology (GO) enrichment analysis. Group 1 comprises proteins with varied functionalities, including those related to immunity, such as interleukin-17 receptor A (IL17RA) and poliovirus receptor (PVR). However, the ORF8-interacting proteins in this group have no significant associated biological functions based on GO enrichment analysis. Numerous proteins within group 1, including IL17RA, PVR, milk fat globule-EGF factor 8 (MFGE8) or lactadherin, A disintegrin and metalloproteinase with thrombospondin motifs 1 (ADAMTS1), collagen alpha-1(VI) chain (COL6A1), protein-lysine 6-oxidase (LOX), neuronal pentraxin-1 (NPTX1), and plasminogen activator tissue type (PLAT), had nearly identical bit pattern profiles indicating co-evolution, and therefore, were suspected to have coupled functionality. Conversely, both groups 2 and 3 had significant GO enrichment results, with the former being related to the glycoprotein biosynthetic process (FDR 1.95E-03) and the latter to the ubiquitin-dependent ERAD pathway (FDR 1.24E-02), endoplasmic reticulum mannose trimming (FDR 4.39E-02), and downregulation of protein dislocation from endoplasmic reticulum (ER) (FDR 1.70E-02). Additionally, we mapped the functionalities of proteins from each group through Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY database for metabolic pathway analysis. The results indicated that 7 proteins in group 3, which are conserved across eukaryotes, were associated with protein processing in the ER (Figure 2). In addition, 5 of the 7 proteins were involved in the proteasome transport pathway. None of the proteins expressed in the ER in the other groups were mapped here.
Figure 2.
ORF8-interacting proteins expressed in the endoplasmic reticulum. The ORF8-interacting proteins expressed in the KEGG PATHWAY “PROTEIN PROCESSING IN ENDOPLASMIC RETICULUM” are highlighted in yellow. All of these proteins belong to group 3.
The 3 groups obtained by phylogenetic profile analysis (Figure 1) were classified based on intracellular localization and their ratios calculated. We focused on intra- and extra-cellular localization, determining the ratios of extracellular and secretory proteins compared to ER proteins (Figure 3). The results showed that group 1 had a higher ratio of extracellular and secreted proteins than ER-proteins (52.9%). This was in contrast to groups 2 and 3, which had higher ratios of ER-proteins than extracellular and secreted proteins, at 52.6% and 63.3%, respectively. Most secreted molecules are glycosylated and extracellular matrices of multicellular organisms are rich in glycans and glycoconjugates.
Figure 3.
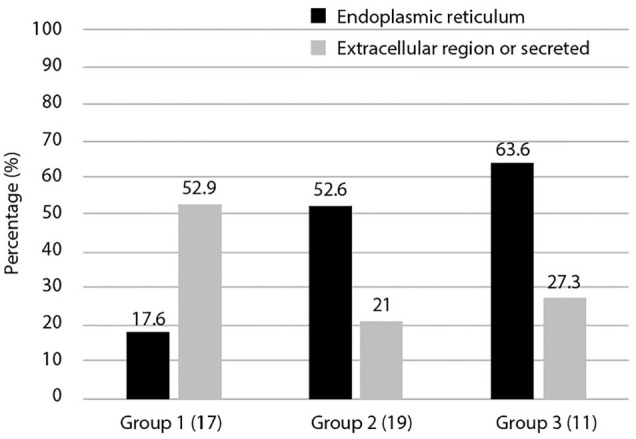
Cellular localization of 3 groups of ORF8-interacting proteins. The localization of proteins in the dataset that interact with ORF8 is shown. Duplication of intracellular localization was included. The vertical axis represents the ratio for each group. Here, we focused on the more characteristic extracellular and secretory systems and the endoplasmic reticulum. More detailed cellular localization is shown in Supplemental Table S1.
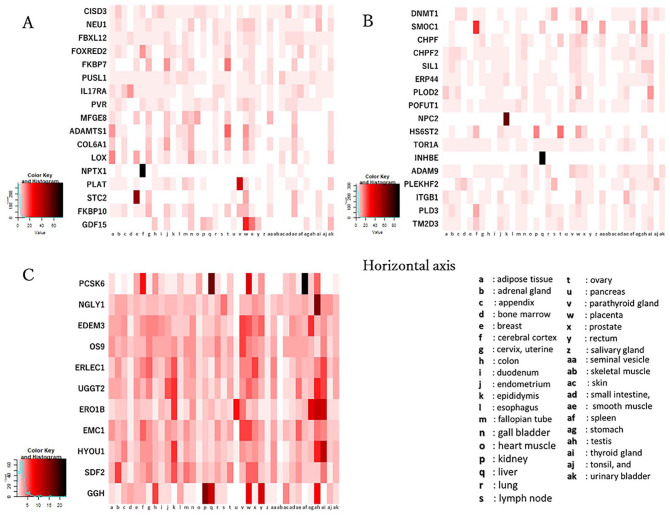
For 47 ORF8 interaction proteins, RNA expression levels in 37 human tissues were obtained in transcripts per million (TPM) from The Human Protein Atlas.17 The TPM values of 2 proteins, Protein O-glucosyltransferase 2 (POGLUT2) and Protein O-glucosyltransferase 3 (POGLUT3), were not included in The Human Protein Atlas. Figure 4 shows the tissue expression pattern of each protein in groups 1 to 3, defined based on the above-mentioned phylogenetic profile analysis (Supplemental Table S2). Eight proteins; NPTX1, PLAT, stanniocalcin 2 (STC2), growth/differentiation factor 15 (GDF15), SPARC related modular calcium binding 1 (SMOC1), NPC intracellular cholesterol transporter 2 (NPC2), inhibin subunit beta E (INHBE), and proprotein convertase subtilisin/kexin Type 6 (PCSK6), were identified to be specifically expressed in human tissues, including the cerebral cortex, parathyroid gland, breast, placenta, cerebral cortex, epididymis, liver, and spleen, respectively. However, here we did not find any significance about the tissue expression pattern among the 3 groups.
Figure 4.
Expression pattern of ORF8-interacting proteins grouped based on phylogenetic profile in human tissues. The vertical axes represent the target proteins, and the horizontal axes represent 37 human tissues. The expression level of each protein in human tissues is shown with the white, red and black colors indicating low, intermediate and high tissue specificity, respectively. A; Group 1 has 17 proteins, B; group 2 has 17 proteins (POGLUT2 and POGLUT3 have no data), C; group 3 has 11 proteins.
Discussion
Phylogenetic profiling is a completely independent technique to infer functional associations between proteins via the correlation of the occurrence across a set of genomes or so-called profiles that are clustered with various available approaches. This method relies on the idea that genes that function together are gained and lost together during evolution.12 Since the phylogenetic profile shows the degree of co-evolution, it is considered that proteins with close relationship are related based on common functionality, share common pathways, and contribute to corresponding diseases.14,15,18,19 The use of this method has led to the discovery of cilia genes, the detection of potential genomic determinants of hyperthermophily, and the identification of small RNA pathway-related genes.20-22 Of note, the development of phylogenetic profiling also allows us to depict the evolutionary roots of individual genes.23
Our phylogenetic profiling results on the ORF8-interacting proteins showed that the evolutionary of these proteins can be classified into 3 groups, groups 1, 2, and 3, which were conserved across vertebrates (17 proteins), metazoan (19 proteins), and eukaryotes (11 proteins), respectively. GO enrichment results on those groups showed that groups 2 and 3 had significant biological function toward glycoprotein biosynthetic process and ubiquitin-dependent ERAD pathway, respectively. Accordingly, some members of groups 2 and 3, which previously have no record toward glycoprotein biosynthetic processes and the ubiquitin-dependent ERAD pathway, respectively, here are predicted to have the functional associations. However, the comprehensive pathways among proteins in groups 2 and 3 were not defined; additionally, group 1 was not associated with any annotated functionality. In this case, the possible incompleteness of the used dataset, the inherent incompleteness of biological databases (such as those in GO) and KEGG pathways and the limitations of phylogenetic profiling inference, may be associated with these results. In fact, ORF8 is not the sole SARS-CoV-2 viral protein able to interact with host proteins associated with either glycoprotein biosynthetic processes or the ubiquitin-dependent ERAD pathway; other SARS-CoV-2 viral proteins with such ability include ORF9c, NSP7, NSP13, M, ORF3a, NSP2, S, and NSP4.11 Of note, even considering such limitations, our results can still be used and considered from the viewpoint of phylogenetic profiling; however, the insights provided here should be interpreted cautiously. Therefore, follow-up studies of a similar methodology using a more comprehensive dataset including all SARS-CoV-2 proteins interactors are needed to shed light on the coordinated process among SARS-CoV-2 proteins in host cell. Additionally, group 1 had a higher ratio of extracellular and secreted proteins than did ER-proteins compared to groups 2 and 3. Most secreted molecules are glycosylated, and extracellular matrices of multicellular organisms are rich in glycans and glycoconjugates. Since our data were retrieved from a study that employs mass spectrometry, the direct and indirect interaction between SARS-CoV-2 ORF8 and host proteins is obscure.11 It is possible that the secretion of some proteins in group 1 are modulated by proteins in groups 2 and 3, further investigation is needed to elucidate this possibility.
The acquisition of genes during evolution occurs through various processes, such as duplication, de novo acquisition from a stretch of non-coding DNA, and horizontal gene transfer (HGT) involving the movement of transposable elements between different species.24,25 HGT is easily distinguished in phylogenetic profiling as the method shows an intermittent presence of a gene, usually between distantly related species, during the evolution. Meanwhile, duplication and de novo acquisition are showed similarly in phylogenetic profiling commonly as a normal acquisition of gene from the void, hence the determination between those events requires further investigation. In this study, our phylogenetic profiling result showed that the evolutionary relatedness of proteins in a group is determined by the evolutionary acquisition of most members; that is, groups 1, 2, and 3 were mostly conserved across vertebrates (17 proteins), metazoan (19 proteins), and eukaryotes (11 proteins), respectively. Some proteins in all groups however exhibit an intermittent presence beyond the determined taxon of their group, displaying an HGT phenomenon (eg, group 1 has some proteins intermittently present between vertebrates and metazoan), complicating the functional relevance of a protein in that group. This case also restricts the reliability of our phylogenetic profiling result to predict the functional association of the dataset.
Viruses that exploit host glycosylation to glycosylate their own proteins during replication are well known to mediate immune evasion by camouflaging immunogenic protein epitopes, propagation, and viral tropism.26,27 For SARS-CoV-2, ORF8 has been suggested to play a crucial role as it can bind to the pivotal enzymes involved in this mechanism, such as POGLUT2 and POGLUT3, 2 O-glucosyltransferases, and POFUT1, an O-fucosyltransferase.28 Additionally, the ubiquitin-dependent ERAD pathway, a key pathway for limiting protein condensation and aggregation in the early secretory compartment, together with the glycosylation biosynthetic process, are associated with the secretory pathway, which is essential for virus transport in the host cell.28 In fact, a study to identify the binding partners of SARS-CoV-2 proteins in human HEK293T cells using affinity purification mass spectrometry assays showed that ORF8 interacts with the most host proteins related to the secretory pathway.11 It was suggested that ORF8 helps SARS-CoV-2 replication by reducing ER stress and enhancing protein folding, promoting chaperone production, and increasing the production of suitably glycosylated S proteins.28 Since ORF8 is not an essential protein for the SARS-CoV-2 life cycle, ORF8 is likely to functionally associate with other SARS-CoV-2 components to assist with SARS-CoV-2 replication and assembly in host cells, but, the coordinated process of this mechanism is still unknown. Additionally, a comprehensive evolutionary analysis of angiotensin-converting enzyme 2 (ACE2), the key receptor for SARS-CoV-2 entry mechanism to cell, identified that the protein is conserved across the majority of metazoan, suggesting that most metazoan is a potential reservoir for SARS-CoV-2.29 Combined with the insight of the conservation of ACE2, our results on the evolutionary roots of genes in groups 2 and 3 suggest that the SARS-CoV-2 ability to hijack the glycosylation biosynthetic processes and ubiquitin-dependent ERAD pathway to mediate immune evasion may be conserved among metazoans. This phylogenetic profiling approach can also be used to predict the potential intermediate hosts of a viral pathogen by comparing the evolutionary roots of host cell receptor and host interactors of the essential protein of a viral pathogen.
As ORF8-interacting proteins have been shown to be expressed in lung tissue,11 functional inhibition or modulation of ORF8-interacting proteins by ORF8 may contribute to the onset of the pathological features in the respiratory systems of COVID-19 patients. Acute respiratory distress syndrome (ARDS) is a rapidly progressive respiratory failure symptom. It causes fluid to leak into the lungs, making it difficult to transfer oxygen into the blood. Although our GO enrichment analysis identified no significant associated biological functions for group 1, almost half of the group 1 proteins were characterized as extracellular or secreted proteins. Therefore, it would be interesting to identify the functional association of SARS-CoV-2 ORF8 with human membrane-associated and secreted extracellular matrix molecules related to coevolution in vertebrates. Of note, among group 1 proteins, members of the collagen superfamily and of the ADAMTS family, collagen 6A1, and ADAMTS1, respectively, are those that have specific roles in vertebrates.30-32 When we focused on the respiratory system-related functions of these molecules, we observed inhibition of IL-17RA signaling and PAI-1, a plasminogen activator inhibitor, in the ARDS-related virus-infected cells and patients.33,34 Thus, ARDS-like symptoms of SARS-CoV-2 infection may cause the functional inhibition of IL-17RA signaling and prevention of plasminogen-dependent blood clot. We identified that PVR, MFGE8, ADAMTS1, COL6A1, and LOX have identical evolutionary profile with IL-17RA and are expressed in lung. Owing to the fact that amyloid formation is one of the ARDS symptoms and MFGE8 has been correlated with amyloid formation in the tissues outside of the brain,35 we speculate that the interaction between ORF8 and PVR, MFGE8, ADAMTS1, COL6A1, LOX, and IL-17RA might be related to the ARDS-like symptoms and/or amyloid formation in COVID-19. Despite this hypothesis, another potential mechanism that may explain the contribution of ORF8 to the SARS-CoV-2 virulence is the ability to disrupt antigen presentation via the downregulation of the expression of MHC-1 molecules, hindering the viral clearance by cytotoxic cells.36,37
Despite the fact that ORF8 is one of the most divergent genes in SARS-CoV-2, a recent study indicated that the positive selection of ORF8 as well as of ORF3a characterized the early evolution of SARS-CoV-2 during the COVID-19 pandemic.38 Numerous factors have been suggested to offer variants in a virus population the opportunity to compete favorably with the predominating one. These include the interference by defective interfering particles, different host or cell types, transfer from vectors to hosts or vice versa, and immune selection.39 Although the conservation of ACE2 suggests that most metazoans serve as a potential reservoir for SARS-CoV-2,29 our analysis indicated that only vertebrates that hold an identical profile to human ORF8-interacting proteins. It would be interesting to investigate whether the varied profile of ORF8 binding partners across metazoans contributes to ORF8 heterogeneity. Immune selection is another potential explanation of the rapid evolution of ORF8. Previously, a study exploring the landscape of antibody responses to SARS-CoV-2 found that N, ORF8 and ORF3b elicit the strongest specific antibody responses.40 As mentioned previously, the dynamic evolution of SARS-CoV-2 during the COVID-19 pandemic is characterized by the positive selection of ORF8 and ORF3a.38 Therefore, it is likely that immunological pressure (e.g., antibody-mediated) drives the rapid evolution of ORF8. Additionally, it is likely that ORF8 rapid evolution is also driven by the non-essential role of ORF8 for replication as SARS-CoV-2 can survive without the functional ORF8 protein.9
Although this is an in silico study based on the theory of functionally coupled gene co-evolution, here we provide insight into the importance of SARS-CoV-2 ORF8 from a novel perspective of target-gene co-evolution. We discuss the possible role of ORF8 in the secretory pathway and ARDS, as well as the possible factors associated with the rapid evolution of ORF8. Indeed, the utilization of linear prediction is inferior if not accompanied by experimental validation; hence, we suggest that the hypotheses proposed here should be proved in a future study.
Materials and Methods
Datasets
The information of ORF8-interacting protein in human was retrieved from a study11 which expressed SARS-CoV-2 individual protein in HEK293T cells and identified targeted host proteins by affinity purification and mass spectrometry; 47 proteins were identified to bind with SARS-CoV-2 ORF8.11 Information such as amino acid sequence, protein function, cellular localization, and KEGG-ID were added to the dataset from UniProt KB/Swiss-Prot, which was manually annotated in UniProt (release 2020_04).41
Phylogenetic profile analysis
Phylogenetic profile analyses of the 47 human ORF8-interacting proteins were performed on 446 genome-decoded species registered in KEGG.42 The KEGG Ortholog Cluster was used to determine the ortholog of the target protein in each species.43 The KEGG Ortholog Cluster is a tool that aligns each amino acid sequence using the Similarity-Waterman algorithm and classifies it is an ortholog when the score fulfills specific criteria (score ⩾ 150 and symmetric similarity measures).43 The generated phylogenetic profile was calculated using Euclidean distance with or without an ortholog of protein distance, and then hierarchically clustered using Ward’s method. The results obtained via clustering were characterized for gene expression patterns and intracellular localization in tissues and cells. Phylogenetic profile analysis is a bit pattern that indicates the presence or absence of target protein orthologs in other species, and is a method for predicting protein functions, interactions, and co-evolution from the perspective of phylogenetic evolution.
Metabolic pathway analysis
Metabolic pathway data from the KEGG PATHWAY database were extracted using the KEGG Mapper.42,44 The results of the hierarchical clustering classification were then mapped to metabolic pathways and evolutionary analysis of the metabolic pathways of proteins that interact with ORF8 was performed.
Comparative analysis of RNA expression in human tissues
The RNA expression levels of 47 ORF8-interacting proteins in human tissues were obtained from The Human Protein Atlas (Retrieved 2019_4, Version 18.1).17 This database contains a collection of expression data of all human proteins in cells, tissues, and organs generated using various omics techniques such as antibody-based imaging, mass spectrometry-based proteomics, and transcriptomics. RNA expression level data were obtained as a TPM value acquired by correcting the gene length and the RNA expression level in each tissue by a ratio based on the RNA-seq data for 37 types of human tissues (Supplemental Table S2).
Supplemental Material
Supplemental material, sj-xlsx-1-evb-10.1177_11769343211003079 for The Functional Classification of ORF8 in SARS-CoV-2 Replication, Immune Evasion, and Viral Pathogenesis Inferred through Phylogenetic Profiling by Muhamad Fahmi, Hiromu Kitagawa, Gen Yasui, Yukihiko Kubota and Masahiro Ito in Evolutionary Bioinformatics
Supplemental material, sj-xlsx-2-evb-10.1177_11769343211003079 for The Functional Classification of ORF8 in SARS-CoV-2 Replication, Immune Evasion, and Viral Pathogenesis Inferred through Phylogenetic Profiling by Muhamad Fahmi, Hiromu Kitagawa, Gen Yasui, Yukihiko Kubota and Masahiro Ito in Evolutionary Bioinformatics
Acknowledgments
We wish to thank Ms. Natsumi Ota, Mr. Tasuku Hamasaki, and Mr. Arashi Esaki for their support and helpful comments.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by the MEXT-Supported Program for the Strategic Research Foundation at Private Universities, grant number S1511028, and the Takeda Science Foundation.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Conceptualization, M.I.; methodology, H.K., G.Y., and M.I.; software, H.K., G.Y., and M.I.; validation, M.F. and M.I.; formal analysis, M.F., H.K., G.Y., and M.I.; investigation, M.F., H.K., G.Y., and M.I.; resources, M.I.; data curation, H.K. and G.Y.; writing—original draft preparation, M.F., H.K, G.Y., Y.K., and M.I; writing—review and editing, M.F., Y.K., and M.I.; visualization, H.K. and G.Y.; supervision, M.I.; project administration, M.I.; funding acquisition, M.I.
ORCID iD: Masahiro Ito  https://orcid.org/0000-0001-8364-8440
https://orcid.org/0000-0001-8364-8440
Supplemental material: Supplemental material for this article is available online.
References
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fahmi M, Kubota Y, Ito M. Nonstructural proteins NS7b and NS8 are likely to be phylogenetically associated with evolution of 2019-nCoV. Infect Genet Evol. 2020;81:104272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boni MF, Lemey P, Jiang X, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5:1408-1417. [DOI] [PubMed] [Google Scholar]
- 5. Vankadari N. Overwhelming mutations or SNPs of SARS-CoV-2: a point of caution. Gene. 2020;752:144792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Young BE, Fong S-W, Chan Y-H, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su YCF, Anderson DE, Young BE, et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11:e01610-e01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muth D, Corman VM, Roth H, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep. 2018;8:15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect Genet Evol. 2020;85:104525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. 2021;118:e2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pellegrini M. Using phylogenetic profiles to predict functional relationships. In: van Helden J, Toussaint A, Thieffry D, eds. Bacterial Molecular Networks. Springer; 2012:167-177. [DOI] [PubMed] [Google Scholar]
- 13. Nakamura T, Fahmi M, Tanaka J, Seki K, Kubota Y, Ito M. Genome-wide analysis of whole human glycoside hydrolases by data-driven analysis in silico. Int J Mol Sci. 2019;20:6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tabach Y, Golan T, Hernández-Hernández A, et al. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol Syst Biol. 2013;9:692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sherill-Rofe D, Rahat D, Findlay S, et al. Mapping global and local coevolution across 600 species to identify novel homologous recombination repair genes. Genome Res. 2019;29:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fahmi M, Yasui G, Seki K, et al. In silico study of rett syndrome treatment-related genes, MECP2, CDKL5, and FOXG1, by evolutionary classification and disordered region assessment. Int J Mol Sci. 2019;20:5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 18. von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Date SV, Marcotte EM. Discovery of uncharacterized cellular systems by genome-wide analysis of functional linkages. Nat Biotechnol. 2003;21(9):1055-1062. [DOI] [PubMed] [Google Scholar]
- 20. Avidor-Reiss T, Maer AM, Koundakjian E, et al. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527-539. [DOI] [PubMed] [Google Scholar]
- 21. Makarova KS, Wolf YI, Koonin EV. Potential genomic determinants of hyperthermophily. Trends Genet. 2003;19:172-176. [DOI] [PubMed] [Google Scholar]
- 22. Tabach Y, Billi AC, Hayes GD, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Škunca N, Dessimoz C. Phylogenetic profiling: how much input data is enough? PLoS One. 2015;10:e0114701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472-482. [DOI] [PubMed] [Google Scholar]
- 25. Baalsrud HT, Tørresen OK, Solbakken MH, et al. De novo gene evolution of antifreeze glycoproteins in codfishes revealed by whole genome sequence data. Mol Biol Evol. 2017;35:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11:527-534. [DOI] [PubMed] [Google Scholar]
- 27. Watanabe Y, Bowden TA, Wilson IA, Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863:1480-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sicari D, Chatziioannou A, Koutsandreas T, Sitia R, Chevet E. Role of the early secretory pathway in SARS-CoV-2 infection. J Cell Biol. 2020;219:e202006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun M, Sharon E, Unterman I, et al. ACE2 co-evolutionary pattern suggests targets for pharmaceutical intervention in the COVID-19 pandemic. iScience. 2020;23:101384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ricard-Blum S, Dublet B, Van der Rest M. Unconventional Collagens: Types VI, VII, VIII, IX, X, XII, XIV, XVI, and XIX. Oxford University Press on Demand; 2000. [Google Scholar]
- 31. Dubail J, Apte SS. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 2015;44-46:24-37. [DOI] [PubMed] [Google Scholar]
- 32. Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biology. 2015;16:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orlov M, Wander PL, Morrell ED, Mikacenic C, Wurfel MM. A case for targeting Th17 cells and IL-17A in SARS-CoV-2 infections. J Immunol. 2020;205:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Whyte CS, Morrow GB, Mitchell JL, Chowdary P, Mutch NJ. Fibrinolytic abnormalities in acute respiratory distress syndrome (ARDS) and versatility of thrombolytic drugs to treat COVID-19. J Thromb Haemost. 2020;18:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westermark GT, Westermark P. Localized amyloids important in diseases outside the brain – lessons from the islets of Langerhans and the thoracic aorta. FEBS J. 2011;278:3918-3929. [DOI] [PubMed] [Google Scholar]
- 36. Zhang Y, Zhang J, Chen Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv. 2020:2020.05.24.111823. [Google Scholar]
- 37. Park MD. Immune evasion via SARS-CoV-2 ORF8 protein? Nat Rev Immunol. 2020;20:408-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Velazquez-Salinas L, Zarate S, Eberl S, Gladue DP, Novella I, Borca MV. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front Microbiol. 2020;11:550674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steinhauer DA, Holland J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409-431. [DOI] [PubMed] [Google Scholar]
- 40. Hachim A, Kavian N, Cohen CA, et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat Immunol. 2020;21:1293-1301. [DOI] [PubMed] [Google Scholar]
- 41. Consortium TU. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2009;38(suppl 1):D142-D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016;45:D353-D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakaya A, Katayama T, Itoh M, et al. KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2013;41:D353-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kanehisa M, Sato Y. KEGG mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-evb-10.1177_11769343211003079 for The Functional Classification of ORF8 in SARS-CoV-2 Replication, Immune Evasion, and Viral Pathogenesis Inferred through Phylogenetic Profiling by Muhamad Fahmi, Hiromu Kitagawa, Gen Yasui, Yukihiko Kubota and Masahiro Ito in Evolutionary Bioinformatics
Supplemental material, sj-xlsx-2-evb-10.1177_11769343211003079 for The Functional Classification of ORF8 in SARS-CoV-2 Replication, Immune Evasion, and Viral Pathogenesis Inferred through Phylogenetic Profiling by Muhamad Fahmi, Hiromu Kitagawa, Gen Yasui, Yukihiko Kubota and Masahiro Ito in Evolutionary Bioinformatics