Germline stop-gain variants in SAMD9L encode truncated gain-of-function proteins that result in a multisystem human disorder with autoinflammatory features. The interferon-inducible dominant proteins interfere with global protein synthesis via inhibition of translational elongation.
Abstract
SAMD9L is an interferon-induced tumor suppressor implicated in a spectrum of multisystem disorders, including risk for myeloid malignancies and immune deficiency. We identified a heterozygous de novo frameshift variant in SAMD9L in an infant with B cell aplasia and clinical autoinflammatory features who died from respiratory failure with chronic rhinovirus infection. Autopsy demonstrated absent bone marrow and peripheral B cells as well as selective loss of Langerhans and Purkinje cells. The frameshift variant led to expression of a truncated protein with interferon treatment. This protein exhibited a gain-of-function phenotype, resulting in interference in global protein synthesis via inhibition of translational elongation. Using a mutational scan, we identified a region within SAMD9L where stop-gain variants trigger a similar translational arrest. SAMD9L variants that globally suppress translation had no effect or increased mRNA transcription. The complex-reported phenotype likely reflects lineage-dominant sensitivities to this translation block. Taken together, our findings indicate that interferon-triggered SAMD9L gain-of-function variants globally suppress translation.
Introduction
IFN-inducible tumor suppressor genes, including SAMD9 and SAMD9L (Davidsson et al., 2018), represent frequent targets of virus-encoded virulence factors (Muñoz-Fontela et al., 2007). Little is known, however, regarding the normal function of SAMD9 family proteins. Haploinsufficiency at the locus encoding both SAMD9 and SAMD9L (-7/-7q) is strongly associated with myeloid malignancies. Consistent with this finding, heterozygous and homozygous Samd9l-null mice develop myeloid leukemia (Nagamachi et al., 2013). Separately, germline SAMD9L stop-gain variants were reported in association with autoinflammatory features, such as panniculitis and basal ganglia calcifications, yet the IFN gene signature was minimal and the mechanism unclear (de Jesus et al., 2020). In contrast, germline heterozygous SAMD9L missense mutations result in a disparate spectrum of disorders, including ataxia and bone marrow (BM) failure, that predispose to myeloid leukemia (Chen et al., 2016) or, alternatively, immunodeficiency syndromes with cytopenias, variable neurological features, and predisposition to myelodysplasia (Tesi et al., 2017). These observations have led to the hypothesis that SAMD9L gain-of-function mutations predispose to leukemia by restricting hematopoiesis, leading ultimately to the outgrowth of pathological clones following loss of the mutant allele (Collin, 2017).
SAMD9 proteins are implicated in response to a broad range of viruses (Wang et al., 2014; Tanaka et al., 2010; Li et al., 2013; OhAinle et al., 2018). Both SAMD9 and SAMD9L are host antiviral restriction factors targeted by virus-encoded virulence factors (Liu and McFadden, 2015). Viruses rely on host ribosomes for protein synthesis, and viral-specific translation represents a key target for host IFN-stimulated gene (ISG) intervention (Schneider et al., 2014; Walsh et al., 2013). SAMD9 induces the formation of nonconventional cytoplasmic antiviral granules (Nounamo et al., 2017) shown to inhibit postreplicative viral mRNA translation (Meng and Xiang, 2019; Sivan et al., 2018). In contrast, blocking conventional stress responses failed to alleviate the block in viral protein translation, suggesting a novel mechanism (Sivan et al., 2018).
Whether SAMD9L might affect host protein synthesis also remains unknown. Dysregulation of host cell protein synthesis is frequently pathogenic, including malignancies, BM failure, and neurological defects (Scheper et al., 2007; Ruggero and Shimamura, 2014; Kapur and Ackerman, 2018; Lehmkuhl and Zarnescu, 2018). Therefore, based on these unanswered questions, we tested the potential role(s) for SAMD9L in translation and the impact of gain-of-function SAMD9L variants on these events.
Results and discussion
Clinical characterization
Here, we comprehensively studied a female infant with clinical features of an interferonopathy, namely basal ganglia calcifications and dermal skin inflammation. She also had unique cytopenias, including B cell aplasia and severe thrombocytopenia (Fig. 1; see clinical description in Materials and methods). At birth, she had a suspected congenital infection with a diffuse blanching, papular, erythematous rash, yet she was also vigorous and afebrile. Bilateral basal ganglia calcifications and hepatosplenomegaly were found on computerized tomography scan, but extensive infectious testing was negative. Cerebrospinal fluid (CSF) revealed elevated protein but no pleocytosis or organismal growth. Genetic testing for Aicardi-Goutières syndrome revealed no mutations in TREX-1, RNASEH2 A/B/C, and SAMHD1, genes associated with interferonopathies. No malignant clones were detected. Lymphocyte subset analysis revealed the absence of circulating B cells, despite normal absolute naive CD45RA+ T cells and natural killer cells. Peripheral blood showed poor proliferative responses to both PHA and anti-CD3 stimulation.
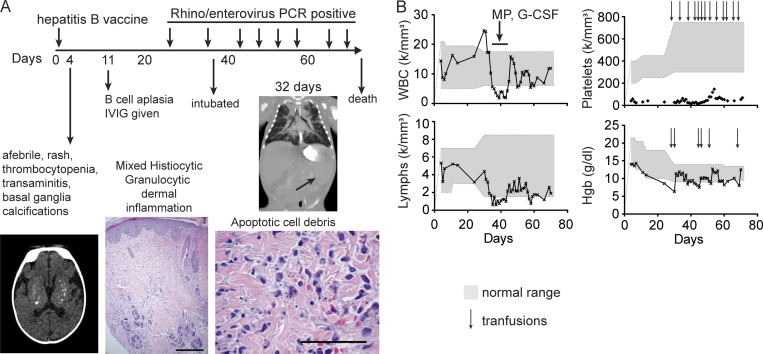
Figure 1.
Clinical course demonstrating persistent organ involvement from birth. (A) Time course of clinical features. Representative images show brain basal ganglia calcifications (bottom left) and skin biopsy at 4 d old, showing deep dermal inflammation (bottom middle). Diffuse dermal infiltrate comprising histiocytes and granulocytes permeates between collagen fibers and around adnexae and vessels (H&E, 40×; scale bar, 0.5 mm). Higher magnification (bottom right) demonstrates abundant apoptosis of inflammatory infiltrate resulting in admixed nuclear debris (H&E, 400×; scale bar, 100 µm). Computerized tomography scan revealed interstitial lung infiltrate and continued splenomegaly at 32 d old. (B) Complete blood counts plotted versus clinical course (shaded = normal range), with treatments or transfusions indicated with arrows. IVIG, intravenous Ig; MP, methylprednisolone.
At 3 wk, the patient developed rhinorrhea and intermittent cough and tested positive for human rhinovirus (HRV)/enterovirus. At that point, she developed progressive signs of BM failure (WBC, 1.9 K/mm3; absolute neutrophil count, 936/mm3; absolute lymphocyte count, 756/mm3; platelets, 28 K/mm3; hematocrit, 28.2%) despite G-CSF and was subsequently transfusion dependent. At 4 wk, the patient required intubation and showed irregular lung opacities prominent in the upper lobes (Fig. 1 A). She remained persistently rhinovirus positive by nasopharyngeal swab PCR testing (n = 8). We initiated HLA typing in anticipation of BM transplantation, but she expired at 9 wk from respiratory failure and cerebral atrophy. No significant pulmonary fibrosis or histological evidence of lower-tract rhinoviral disease was observed on autopsy; however, the lungs contained significant pulmonary hemorrhage. Extensive postmortem bacterial culture/molecular testing (DNA and RNA encephalitis/bacterial DNA sequencing panel) revealed only the known HRV infection. Her persistent clinical features included thrombocytopenia, basal ganglia calcifications, dermal infiltrate with cellular apoptosis, and B cell aplasia. The patient expired from progressive BM failure together with respiratory failure that accompanied the HRV infection.
Histological and immunological analysis revealed a unique constellation of cytopenias
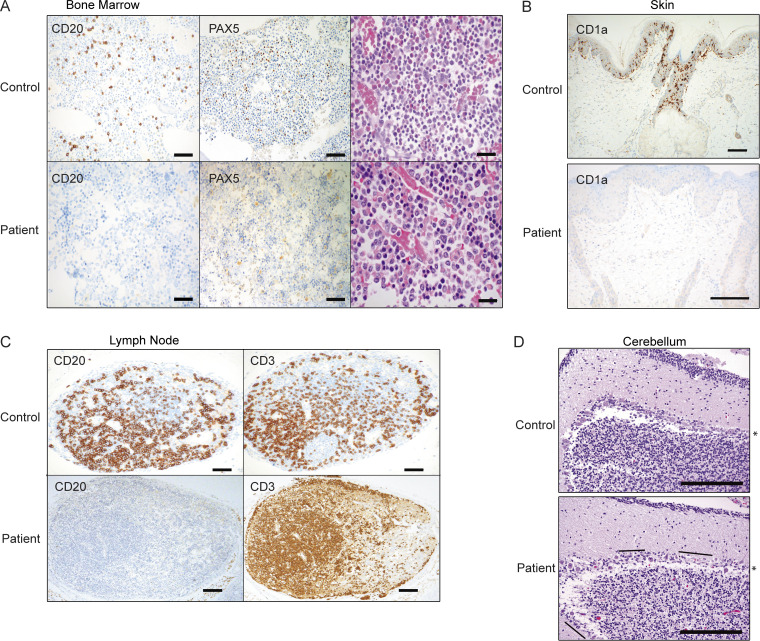
The patient had an unusual combination of B cell aplasia, absence of Langerhans cells, and nonimmune thrombocytopenia. Severe thrombocytopenia preceded the loss of other lineages (Fig. 1 B). Decreased megakaryocytes were observed with a prominent myeloid shift (myeloid:erythroid ratio 20:1; normal 3:1) but normal BM cellularity (Fig. 2 A). Skin biopsies (n = 3) revealed an absence of CD1a+ (or CD207+) Langerhans cells (Fig. 2 B) in the setting of mixed dermal inflammation and abundant apoptotic debris that were both interstitial and perivascular in distribution (Fig. 1 A). No viral cytopathic effects, fibrinoid necrosis, or vascular thrombosis were identified. Lymph nodes and BM showed a complete lack of B cells (CD20+), despite normal CD3+ T cell staining compared with age-matched control tissue (Fig. 2 C). The BM also lacked PAX5+ cells, markers for B cell progenitors (Fig. 2 A). Peripheral blood showed a complete absence of CD19+ B cells throughout her clinical course. Lastly, targeted cerebellum involvement was observed with mild and patchy loss of Purkinje cells (Fig. 2 D). Basal ganglia and insular cortex showed gliosis and activated microglia with multiple foci of perivascular calcifications. At the same time, cortical sections demonstrated normal lamellation and lacked necrosis or demyelination. The disparate cell lineages targeted in this subject were unusual for known autoinflammatory disorders.
Figure 2.
Histological evidence of selective cytopenias. Biopsy or autopsy samples were analyzed and compared with cases controlled for equal stress or treatments. Data are representative. (A) Proband BM with left-shifted myelopoiesis and similar cellularity as control BM. All cell lines are present, although megakaryocytes are decreased. Immunohistochemical stains (CD20 and PAX5) confirm the presence of scattered B cells in the control sample but none in the patient (diaminobenzidine, 100×; scale bar, 200 µm; H&E, 200×; scale bar, 50 µm). (B) Langerhans cells are highlighted with CD1a in the epidermis of the control skin sample and absent in the patient (diaminobenzidine, 100× [top] and 200× [bottom]; scale bars, 100 µm). (C) Control lymph node has abundant CD20+ and CD3+ lymphocytes, with few primary follicles. The CD3+ T cell population is intact in the patient, but no B cells are identified with CD20 (diaminobenzidine, 40×; scale bar, 300 µm). (D) Cerebellum sections were stained with H&E, highlighting the layer of confluent Purkinje cells (*) in a control subject and regions of Purkinje cell loss (lines) in the patient (200×; scale bars, 100 µm).
Genetic evaluation and variant validation
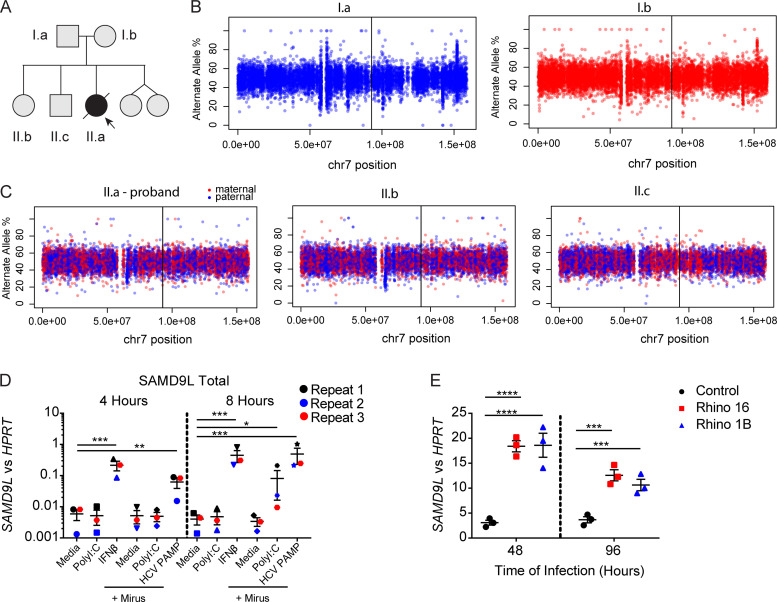
Whole-genome sequencing (WGS) was performed on the proband and healthy family members, including both parents and two siblings (Fig. 3 A). Analysis of WGS data identified a heterozygous de novo SAMD9L truncating variant (GenBank accession no. NM_152703.2:c.2658_2659del; p.F886Lfs*11) in the proband (Fig. 3 B). The same heterozygous mutation was confirmed in skin and liver genomic DNA samples from autopsy, and no other rare variants were found in SAMD9 or SAMD9L. The healthy family members demonstrated no evidence of uniparental disomy or gene conversion (Fig. S1). The proband’s mutation, p.F886Lfs*11, is located near one of the first described ataxia pancytopenia syndrome (ATXPC)–dominant variants (p.H880Q; Fig. 3 C; Chen et al., 2016) and subsequently reported together with a cluster of de novo truncating variants in this small region of SAMD9L (de Jesus et al., 2020).
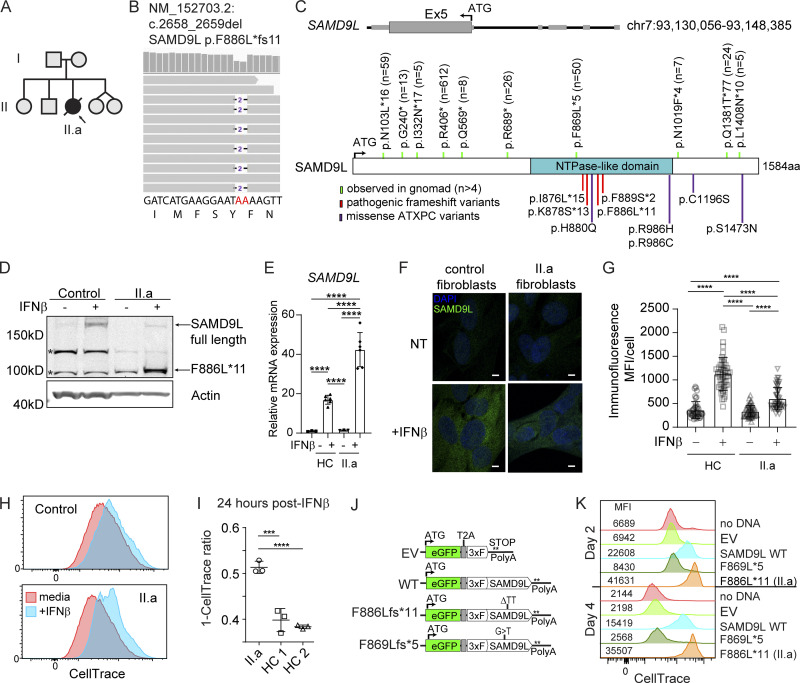
Figure 3.
SAMD9L truncation variants are expressed and restrict growth. (A) Pedigree with affected deceased proband (II.a). (B) WGS reads for proband with heterozygous germline deletion. (C) Diagram of the SAMD9L genomic locus with stop-gain or frameshift variants in the gnomAD version 2.1.1 database (green, n = allele count), published pathogenic truncation frameshift variants (red), and missense ATXPC variants (purple). ATG indicates start codon. (D–H) Comparing healthy control (HC) or proband primary fibroblasts. (D) Western blot analysis using anti-SAMD9L N-terminal monoclonal antibody shows IFN-inducible truncation variant (*, nonspecific bands). Representative data from n = 3 experiments. (E) Relative mRNA expression of SAMD9L or HPRT measured by qPCR following IFN-β treatment. Data are pooled means ± SD from three experiments (****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test). (F and G) Immunofluorescence staining for SAMD9L and costained with DAPI (scale bar, 5 µm). Representative images and MFI quantified for n = 50 cells per group from n = 2 experiments (****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test). NT, not treated. (H and I) CellTrace growth assay to internally compare media alone to IFN-β treatment growth restriction in proband (II.a) fibroblasts versus two separate HCs. Representative data from n = 2 experiments. Representative flow cytometry plots and graphical display of the difference measured by change in MFI when treated with IFN-β (***, P < 0.001; ****, P < 0.0001 by one-way ANOVA). (J) Diagram of expression vectors. (K) 293T cells labeled with CellTrace proliferation dye were transiently transfected with either EV or SAMD9L variant-expressing constructs. Representative flow cytometry with MFI indicated at days 2 and 4 after labeling (n = 3 experiments).
Figure S1.
Chromosomal analysis of the proband and unaffected relatives and regulation of SAMD9L expression. (A) Pedigree from the family annotated. (B and C) Allele frequency of phased heterozygous variants on chromosome (chr) 7 as calculated from the allelic read depth from WGS data. (C) The variants are color coded based on the parental origin for siblings from the respective parents shown in B. The vertical lines indicate the position of the SAMD9L proband variant (GenBank accession no. NM_152703.2:c.2658_2659del). (D) A549 lung epithelial cells were treated with media alone (1.25 µg/ml), high-molecular-weight poly(I:C), or human IFN-β (100 U/ml) or transfected with 1.25 µg high-molecular-weight poly(I:C) or 1 μg hepatitis C virus pathogen-associated molecular pattern using Mirus Bio lipofection reagents following the manufacturer protocol for 4 or 8 h before isolation of mRNA. RT-PCR for SAMD9L mRNA transcripts compared with HPRT mRNA transcript means ± SD from n = 3 pooled experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA with Tukey’s multiple comparison test). (E) Ex vivo–cultured lower airway epithelial cells from healthy children (n = 3) were cultured at baseline and during infection with HRV-A16 or -1B (Rhino 16 and 1B, respectively) for 48 and 96 h before mRNA isolation. Representative from n = 2 independent experiments (***, P < 0.001; ****, P < 0.0001 by one-way ANOVA and Tukey’s multiple comparison test).
The coding sequence of SAMD9L is contained in a single large exon and, thus, theoretically permits protein expression from early truncation mutations. Notably, SAMD9L truncating variants are relatively common in public databases (Fig. 3 C). The observed-to-expected ratio in gnomAD version 2.1.1 (o/e = 0.58 [90% confidence interval: 0.44 – 0.78]) shows that stop-gain mutations are well tolerated (Fig. 3 C, green: frequent variants [n > 5] in gnomAD). Additionally, if found earlier in the coding region, SAMD9L cis-truncation mutations have reversed a strong proliferative block induced by pathogenic gain-of-function variants (Tesi et al., 2017; Wong et al., 2018). In contrast, truncating variants are rare in the region with in silico–predicted nucleoside triphosphatases homology (Mekhedov et al., 2017). Notably, this domain likely lacks enzymatic hydrolysis activity as the Walker motif sequences are not conserved; thus, the function remains untested.
To validate the pathogenic effect of the proband’s de novo truncating variant, we verified protein expression and cellular localization in primary patient cells. First, we treated proband or healthy control fibroblasts with IFN-β, revealing induced expression of full-length SAMD9L (184 kD) from all samples tested; in addition, the proband fibroblasts expressed high levels of a truncated anti-SAMD9L–reactive protein (predicted 104 kD) after IFN-β treatment (Fig. 3 D). Minimal transcript or protein expression of SAMD9L was observed at baseline in both of the samples (Fig. 3, D–G). Following IFN-β treatment, augmented SAMD9L transcripts were observed for both fibroblast lines (Fig. 3 E). Relative protein expression level was reduced in proband cells as assessed using a monoclonal antibody to the N-terminus (Fig. 3, F and G). SAMD9L staining was localized to the cytosol in both fibroblast lines. Lung epithelial cells (A549) exposed to either IFN-β or cytosolic polyinosinic:polycytidylic acid (polyI:C) strongly and rapidly upregulated SAMD9L expression (Fig. S1). Primary bronchial airway epithelial cells from healthy donors (n = 3) showed minimal baseline SAM9DL transcription but exhibited marked upregulation in response to several HRV strains.
Next, as a functional screen, we examined cellular growth using a cell-permeant dye where dilution indicates cell division. Fibroblasts from the proband or healthy controls were labeled with dye and cultured in media alone or treated with IFN-β for 24 h. IFN treatment is well known to restrict cellular growth (Platanias, 2005) and is also necessary to induce SAMD9L expression. When cell division in proband fibroblast cells treated with IFN-β was compared with untreated cells, there was substantial reduction in growth compared with a similar analysis in healthy donor fibroblasts (Fig. 3, H and I), indicating that the truncation variant may be pathogenic.
To directly test the effect of SAMD9L variants on cell proliferation, 293T cells were dye labeled and transfected with WT or variant SAMD9L proteins. We tested the proband’s variant (p.F886Lfs*11) as well as a nearby truncating variant, p.F869Lfs*5 (rs773878792), found in the healthy Latino population (n = 50 per 280,888 exomes or genomes). We selected this variant as one of our controls for the remainder of the article. We used a bicistronic vector expressing enhanced GFP and N-terminal triple-FLAG epitope-tagged (3xF) SAMD9L cDNAs separated by the viral 2A (T2A) peptide (Fig. 3 J). To ensure similar vector size and mRNA stability, the SAMD9L variants were each created in the full-length SAMD9L cDNA-containing vector with site-directed changes and compared with an empty vector (EV) control. Following transfection, cells were monitored by flow cytometry for GFP expression and dye dilution, indicating cell division. We found a pronounced proliferative block in cells transiently expressing the proband’s p.F886Lfs*11 variant at both 2 and 4 d (Fig. 3 K). WT SAMD9L demonstrated a mild cell growth restriction when overexpressed. In contrast, the p.F869Lfs*5 variant showed no restriction in cell growth based on similar dye dilution compared with cells transfected with EV. Transfection efficiency and cell death rates were comparable across the constructs, as shown by viability dye and a caspase activation probe, respectively (Fig. S2). Together, these findings support a model wherein IFN-induced expression of the patient truncation variant confers a gain-of-function phenotype leading to restricted cellular growth.
Figure S2.
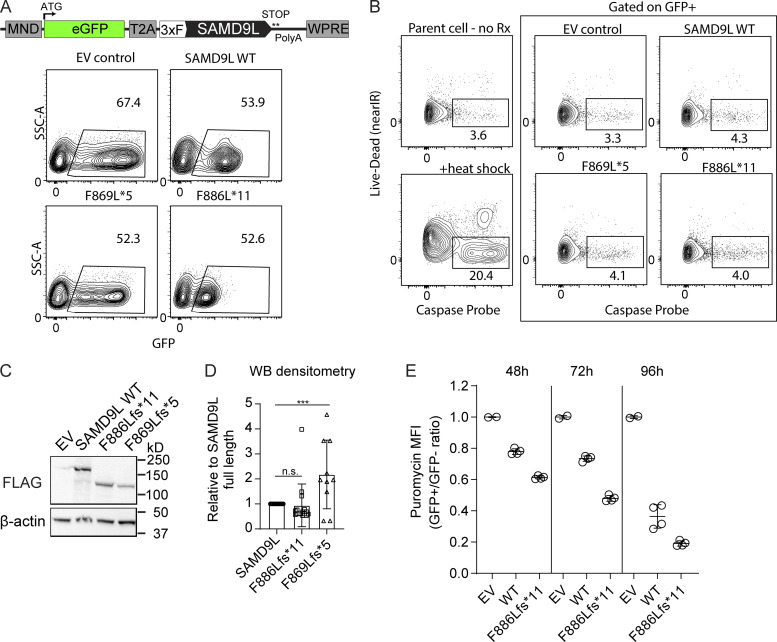
Characterization of the transient expression of SAMD9L variants in 293T cells. (A–F) All experiments presented were performed in 293T cells transiently transfected with Mirus Bio reagent with either no DNA, EV, or vectors containing SAMD9L full length (WT), F886Lfs*11, or F869Lfs*5 and grown for 18–24 h before experiments. ATG indicates start codon. (A) Schematic of the lentiviral vector. Representative flow cytometry for the GFP versus side scatter area (SSC-A) 18 h following transient transfection, showing equivalent transfection efficiency from five independent experiments. (B) Flow cytometry plots staining cells with live/dead marker (LIVE/DEAD Fixable Near-IR Stain) following incubation with a broad caspase probe (BD PharMingen Violet Live Cell Caspase Probe). Parental cells were heat shocked at 55°C for 30 min for a positive control. No difference in cell death or caspase activation was observed. Representative from two independent experiments. (C and D) Representative Western blot (WB) for FLAG-tagged SAMD9L variants in 293T lysates and densitometry of replicates normalized to SAMD9L WT and pooled with WT (n = 16), F886Lfs*11 (n = 16), and F869Lfs*5 (n = 10) represented. ***, P < 0.001 by one-way ANOVA. (E) Cells were transiently transfected with EV, WT, or F886Lfs*11 variant and grown for 48, 72, and 96 h, respectively, before 10-min puromycin pulse. The MFI of puromycin was compared with the GFP+ population divided by the puromycin MFI in the GFP− population in the same well then normalized to the EV control to combine the different days. Data were pooled from n = 4 independent experiments. Rx, treatment.
SAMD9L truncation mutations block translation without altering transcription
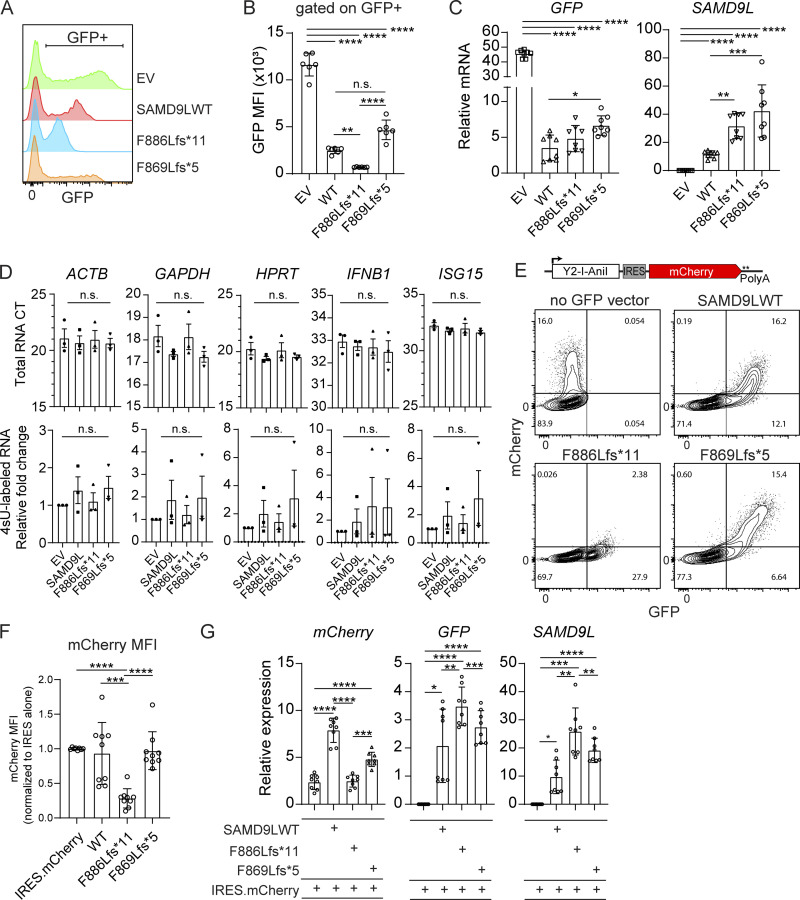
In performing our transient expression studies, we observed that fibroblasts with various SAMD9L constructs had striking differences in the fluorescence intensity of GFP (Fig. 4, A and B). GFP median fluorescence intensity (MFI) was measured by flow cytometry at 18–24 h following liposomal-based transient transfection of each construct. Cells transfected with EV exhibited equal transfection efficiency compared with other constructs but displayed increased GFP transcription and expression due to the large difference in vector size (EV 7.3 kb; SAMD9L plasmids 12.1 kb) as previously described (Fig. S2 and Fig. 4 C; Kreiss et al., 1999; Bochkov and Palmenberg, 2006; Hornstein et al., 2016). In contrast, while the SAMD9L variant constructs differed by only two nucleotides, there were marked differences in GFP protein levels that did not correlate with GFP mRNA transcript levels (Fig. 4, A–C). Transient transfection of the proband mutation (p.F886Lfs*11) resulted in blunted GFP MFI compared with transfection of the WT SAMD9L, while cells expressing SAMD9L p.F869Lfs*5 expressed augmented GFP relative to the other variants. The SAMD9L WT and F886Lfs*11 variant proteins were similarly expressed via Western blot, while expression of the SAMD9L p.F869Lfs*5 was enhanced (Fig. S2). These findings demonstrated that the proband variant was uniquely associated with reduced GFP expression.
Figure 4.
The proband SAMD9L truncation variant dominantly blocks protein expression without impacting transcription. (A–D) 293T cells were transfected with EV control or SAMD9L variants and analyzed at 24 h after transfection. (A and B) Representative flow cytometry histograms of GFP intensity and GFP MFI graphically displayed (n = 6 transfections/genotype; **, P < 0.01; ****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test). (C) qPCR for relative expression of GFP (left) and SAMD9L (right) for each genotype. Data pooled from n = 2 experiments (n = 8 per genotype; *, P < 0.05; **P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test). (D) Average CT values of input RNA (top) or the relative fold change of 4sU-labeled RNA (bottom). Data analyzed using a one-way ANOVA with Dunnet’s multiple comparison test (versus EV; n = 3 experiments). (E–G) 293T cells were transfected with an IRES.mCherry expression vector alone (schematic) or cotransfected with the indicated bicistronic GFP.T2A.SAMD9L vectors. (E) Representative flow cytometry plots (bottom) from n = 3 experiments. (F) mCherry MFI quantified graphically with data pooled from n = 3 experiments and normalized to the average MFI for the IRES.mCherry vector alone for each experiment (***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test). (G) Relative mRNA expression for mCherry, GFP, and SAMD9L transcripts as assessed by qPCR (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test).
We next tested whether the observed GFP reduction in cells expressing the proband variant was independent of transcription. First, we tested whether GFP and SAMD9L transcripts were present at equimolar ratios as would be expected for T2A-based vectors (Sharma et al., 2012). Compared with full-length SAMD9L-transfected cells, GFP and SAMD9L transcripts increased in parallel in cells transfected with the truncation variants (Fig. 4 B). Second, we performed 4-thiuridine (4sU) metabolic labeling of newly synthesized host mRNA in transfected cells. We compared levels of total to newly synthesized RNA to assess global transcription (Garibaldi et al., 2017; Forero et al., 2019). As shown in Fig. 4 D, expression of WT SAMD9L or the variants did not impact the total RNA levels (top panels) or RNA synthesis rates (bottom panels) across a range of cellular RNAs, including ACTB, GAPDH, and others.
Finally, we cotransfected cells with an mCherry reporter plasmid together with the SAMD9L bicistronic vectors to test whether the proband variant could manifest a dominant effect on protein and/or RNA expression. We transfected equal amounts of bicistronic GFP-expressing vectors together with a construct containing an internal ribosome entry site (IRES)–driven mCherry (Fig. 4 E). Despite equivalent transfection efficiencies, we found that both GFP and mCherry MFI were reduced in cells expressing the proband truncation variant (p.F886Lfs*11), indicating a dominant effect (Fig. 4, E and F). In contrast, full-length SAMD9L and p.F869Lfs*5 truncation variants had a similar MFI compared with the IRES-mCherry vector alone. In parallel, we performed transcript analysis (RT-PCR) and found no difference in mCherry mRNA levels in cells cotransfected with the SAMD9L F886Lfs*11 variant versus cells transfected with mCherry vector alone (Fig. 4 F). There was a modest increase in GFP and SAMD9L transcription in the presence of the proband truncation variant compared with cells coexpressing WT SAMD9L. mCherry mRNA levels were increased in the presence of full-length SAMD9L or F869Lfs*5 variant, but these transcript levels did not correlate with a significant change in mCherry expression (Fig. 4 E). Thus, reduced protein expression from both plasmids in the presence of the SAMD9L F886Lfs*11 truncation variant was not a result of reduced transcripts.
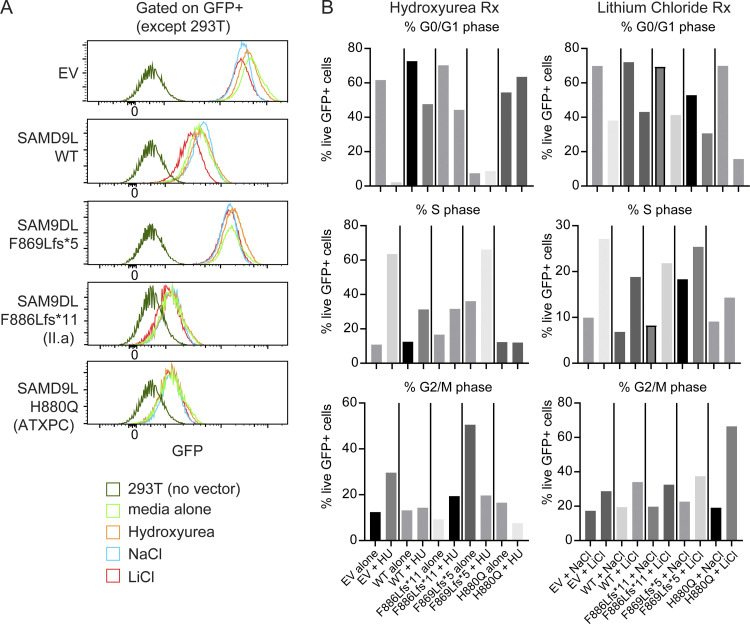
SAMD9L variants have been shown to block cells in the G0/G1 phase of the cell cycle (Wong et al., 2018). To ensure that the translational effect was not altered by cell cycle events, we plated 293T cells transfected with full-length or mutant SAMD9L constructs in reduced serum media with hydroxyurea to enrich in the S phase or with lithium chloride to enrich for G2/M phase to promote different stages of the cell cycle (Fig. S3). Irrespective of skewing to a cell cycle stage, we found similar GFP expression level results. Taken together, these results demonstrate that the proband truncation variant broadly blocks protein expression without impacting mRNA transcription in transfected cell lines using a plasmid expression model.
Figure S3.
Translational block is not altered by cell cycle. (A and B) 293T cells transiently transfected with Mirus Bio reagent with either no DNA, EV, or vectors containing SAMD9L full-length (WT), F886Lfs*11, F869Lfs*5, or H880Q missense variant and grown in either reduced serum media alone or together with 10 μM hydroxyurea (HU) to arrest at the G1/S boundary and accumulate. Alternatively, cells were cultured with 10 mM LiCl to arrest in G2/M phase or 10 mM NaCl as high-salt control conditions. Cells were stained with propidium iodide for cell cycle analysis. (A) Representative flow cytometry histograms of GFP intensity are displayed for each construct (n = 3 experiments). (B) The percentage of live GFP+ cells in the G0/G1 phase (top), S phase (middle) or G2/M phase (bottom) following treatment (Rx) for 18 h with hydroxyurea (left) or LiCl (right). Representative from n = 3 independent experiments.
SAMD9L truncation mutants uniquely block protein elongation
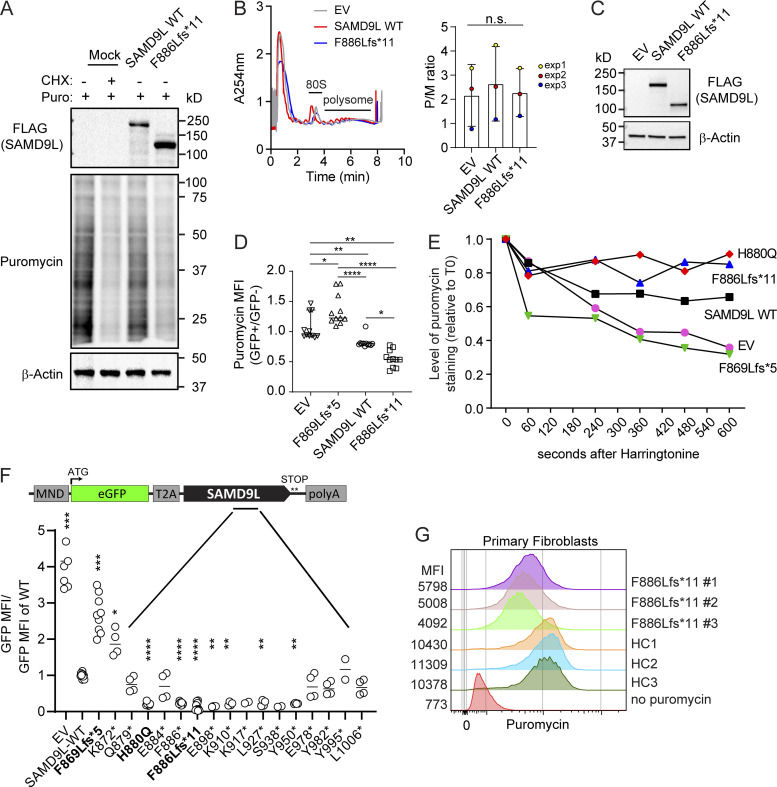
The profound differences in GFP expression led us to hypothesize that SAMD9L was playing a role in mRNA translation. Protein synthesis occurs first with initiation followed by elongation and termination (Robichaud et al., 2019; Sriram et al., 2018). Regulation of translation can have targeted, global, or selective impacts on translation (Cheng et al., 2019; Khajuria et al., 2018). To directly test global protein synthesis rates, we labeled nascently synthesized peptides with puromycin, a transfer RNA analogue incorporated into elongating polypeptides causing early chain termination. Cycloheximide pretreatment served as a negative control (Schneider-Poetsch et al., 2010). After 18–24 h posttransfection with SAMD9L variant–expressing vectors, we found that expression of SAMD9L p.F886Lfs*11 resulted in a dramatic reduction in puromycin incorporation by Western blot, similar to cycloheximide (Fig. 5 A). In contrast, transient expression of SAMD9L full length did not dramatically change the puromycin intensity compared with mock-transfected cells.
Figure 5.
SAMD9L truncation variants suppress translation via blocking protein elongation. Untransfected or 293T cells transfected with EV or SAMD9L variants for 24 h were analyzed for effects on translation. (A) Puromycin-labeled peptides were assessed by Western blot in lysates from mock (positive control) or cycloheximide (CHX; negative control). Representative blot is shown from n = 4 experiments. (B and C) Polysome analysis from cells transfected with EV, SAMD9L WT, or SAMD9L proband F886Lfs*11 variant displaying the 80S peak and polysome peaks. (B) Representative plots are shown and the P/M ratio for each of three independent experiments plotted (one-way ANOVA). (C) Representative Western blot performed to ensure equal transfection from n = 3 experiments. (D) Puromycin-labeled peptides were quantified at the single-cell level by intracellular flow cytometry following transfection with EV, SAMD9L WT, or variants (n = 11 per genotype; n = 3 experiments), with ratio of puromycin MFI in GFP+/GFP– cells from each transfection well (*, P < 0.05; **, P < 0.01; ****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test). (E) Pretreatment of cells for various times with HHT blocks translation initiation followed by puromycin pulse to plot the elongation runoff rate by quantifying puromycin-labeled peptides over time. Representative experiment shown from n = 3 experiments. (F) Site-directed mutagenesis of the full-length SAMD9L bicistronic vector introduced stop-gain mutations along the region flanking known variants. GFP MFI was normalized across experiments to the GFP MFI of WT condition (n = 3–6 experiments per construct). p.H880Q (ATXPC), F869Lfs*5, and p.F886Lfs*11 (proband) variants were included for comparison (bold; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test); ATG indicates start codon. (G) Primary proband fibroblast subcultures (n = 3) or healthy control (HC) donor fibroblast lines were treated for 18 h with IFN-β (100 ng/ml) for puromycin incorporation analyzed by flow cytometry. The MFI is displayed to the left of representative flow histograms. Representative from n = 2 experiments. T0, time 0.
Next, we used polysome profiling to separate mRNAs actively engaged in translation from untranslated ones. If translation initiation is altered, we hypothesized that the polysome-to-monosome (P/M) ratio would be reduced. We performed polysome analysis from cytoplasmic lysates separated by sucrose gradients from fibroblasts transfected with full-length or proband-truncated SAMD9L (Fig. 5, B and C). Monosome (80S) and polysome peaks measured by absorbance in sucrose fractions were comparable across conditions tested. The overlay and compiled P/M ratios (Fig. 5 B) show that transient expression of the SAMD9L variants did not broadly alter the ribosomal profiles.
To quantify this effect at a single-cell level, we next used flow cytometric staining for intracellular puromycin (Fig. 5 D). We gated on transfected GFP+ cells to directly compare puromycin MFI by flow cytometry to the nontransfected GFP– cells within the same culture well. Cells transfected with EV showed puromycin incorporation similar to mock-transfected cells (GFP+:GFP− ratio ∼1.0; Fig. 5 D). In contrast, the cells expressing the SAMD9L full length had a mild reduction in puromycin that was not appreciated in the bulk Western blot assay. The proband truncation mutant (p.F886Lfs*11) showed ∼50% reduction in puromycin incorporation compared with the GFP– cells in the same well. Intriguingly, the common p.F869Lfs*5 variant resulted in elevated puromycin MFI, indicating a higher translation rate. SAMD9L variant expression over 2–4 d resulted in more pronounced translational block in cells expressing p.F886Lfs*11, while the WT SAMD9L displayed an intermediate phenotype (Fig. S2). These results suggested the presence of a possible competitive binding domain, such as a guanosine triphosphate–binding motif, that may be lost with further truncation.
We hypothesized that if initiation was not significantly altered, elongation may be blocked in the presence of the proband truncation mutant (p.F886Lfs*11). We measured elongation rates using the “SunRiSE” (SUnSET-based ribosome speed of elongation) method, which combines puromycin detection by flow cytometry with the addition of harringtonine (HHT), an initiation-specific translation inhibitor (Argüello et al., 2018). Treatment with HHT promotes ribosome runoff that, when coupled with a puromycin pulse, can detect the speed of elongation directly (Ingolia et al., 2011). We treated transiently transfected fibroblasts with HHT (2 μg/ml) for 0–10 min followed by a puromycin pulse (Fig. 5 E). In cells expressing the p.F869Lfs*5 variant or the control (EV), there was the expected steady decline in the ratio of puromycin incorporation with increasing HHT exposure compared with untreated cells. In contrast, cells expressing the proband truncation variant (p.F886Lfs*11) or the known ATXPC variant p.H880Q displayed a flat profile over time, with little change compared with time 0, consistent with an elongation block. We found an intermediate phenotype in cells transfected with SAMD9L full-length vectors, suggesting that this may be an important physiological role for SAMD9L during a viral infection.
To test whether this was a regional variant effect, we introduced stop-gain mutations in the region flanking the proband variant and measured GFP expression. We started at the known truncation variant (p.F869Lfs*5) found in healthy individuals in gnomAD and introduced single-nucleotide variant (SNV) stop-gain mutations downstream, as these were predicted to be more probable genetic events to encounter clinically. To keep the same vector size and distance from the 3′ untranslated region elements, we again used site-directed mutagenesis of the bicistronic SAMD9L full-length vector. Each potential stop-gain variant was compared for GFP MFI in transfected cells at 18–24 h (Fig. 5 F). Testing multiple SNVs, we identified seven additional truncation variants (p.F886*, p.E898*, p.K910*, p.K917*, p.L927*, p.S938*, p.Y950*) with stop-gain mutations in the region from aa.886–950, resulting in dominant blockade of GFP expression. In contrast, the flanking variants (p.Q879*, p.E884* and p.E978*, p.Y982*, p.Y995* or p.L1006*) led to GFP expression levels similar to WT. Of note, we included the known p.H880Q ATXPC variant in our screen. This disease-associated variant also severely depressed GFP expression, indicating that missense as well as truncation variants could suppress translation. The proband frameshift (p.F886Lfs*11) introduces an 11-residue-long neopeptide before a stop codon. We therefore introduced a stop-gain mutation at p.F886* to focus on the truncation. This variant also profoundly blocked GFP expression. In contrast, a few variants did not restrict GFP expression. For example, the p.872* and the p.F869Lfs*5 truncation variant displayed GFP expression similar to the EV, suggesting a range of translational effects likely representing loss-of-function alleles. Taken together, these findings identify a region within SAMD9L where truncating variants exhibit gain-of-function activity in blocking elongation, thereby leading to a dramatic reduction in GFP reporter expression. These findings also suggest that this phenotype is not unique to the nucleotide changes present in our proband subject.
Lastly, we used primary fibroblasts from our patient harboring the p.F886Lfs*11 variant and performed a puromycin pulse assay. We isolated three separate sublines grown from our patient, each genotyped at the time of the assay to ensure no reversion variants or loss of the pathogenic allele. These lines were compared with healthy control fibroblasts for puromycin incorporation as measured by intracellular flow cytometry after treating with IFN-β for 24 h. In agreement with our results in 293T cells, the puromycin MFI was reduced by ∼50% in cells harboring the p.F886Lfs*11 variant compared with healthy control fibroblasts (Fig. 5 G). Therefore, these results indicate that the p.F886LFs*11 variant of SAMD9L is a gain-of-function variant capable of triggering a global translational reduction in primary cells under endogenous expression conditions. Overall, we describe the clinical features of an infant with a de novo truncating variant in SAMD9L who succumbed to respiratory failure. The clinical features represent SAMD9L-associated autoinflammatory disease but overlap considerably with the ATXPC and with patients with other gain-of-function SAMD9L variants that present with immune deficiency. Our findings suggest a common mechanism for these clinically distinct disorders. Notably, transient overexpression of SAMD9L full-length protein, and/or, more profoundly, gain-of-function truncation variants, decrease translation in the absence of IFN treatment. We did not find that SAMD9L variants drive an IFN signature, but more likely, we find that IFN is an inducer of SAM9DL. SAMD9L is an ISG; thus, an IFN trigger (such as a viral infection) may be required to initiate disease mediated by the p.F886Lfs*11 variant. Future studies should investigate the specific mechanisms controlling the induction of SAMD9L and if these mechanisms differ by cell or tissue type, as these might yield insights into the tissue and cell type–specific manifestations of disease in patients with these gain-of-function truncation variants. Our findings also suggest that the degree of translational block triggered by SAMD9L variants is likely to correlate with clinical severity. Although our screen was not exhaustive, our data highlight a critical region within SAMD9L wherein eight novel truncation variants, including our proband variant, induce a similar translational block. This finding represents an important feature of the single coding exon genomic structure. Our data revealed a similar, not previously reported, translational block for an ATXPC variant. Further testing of reported SAMD9L disease variants is warranted and will help clarify this possible common mechanism of disease.
Materials and methods
Human subjects
The study was approved by the local ethical institutional review board with written informed consent for phenotypic, functional, and genetic analysis according to the requirements of the review board (Seattle Children’s Hospital Institutional Review Board #11738). Autopsies were performed according to standard practices.
WGS
DNA from peripheral blood of family members or fibroblast and liver DNA from the proband were used for WGS. Libraries for next-generation sequencing were generated using the TruSeq PCR-Free library kit (Illumina). WGS was performed using an Illumina HiSeq 4000 instrument with 100-bp paired-end reads. Analysis of WGS data was performed in collaboration with the University of Washington, Center for Mendelian Genomics. Reads were aligned to human genome build GRCh37 using BWA-MEM (Burrows-Wheeler Alignment–maximal exact matches) algorithm. SNVs and small insertions and deletions were identified using the Genome Analysis Toolkit Unified Genotyper version 3.2-2. The resulting multisample variant call format was decomposed (VT version 0.57), annotated (VEP version 87), and filtered for rare variants (allele frequency <0.005) according to the expected de novo inheritance model using GEMINI. Large structural variation from WGS data was detected using LUMPY (Layer et al., 2014). The quality of detected variants was visually inspected using the Integrated Genome Viewer (Robinson et al., 2017).
Cells, treatments, and reagents
Primary fibroblasts were expanded from skin biopsy and stimulated with recombinant human IFN-β (300-02BC; PeproTech), poly(I:C) (InvivoGen), or hepatitis C virus pathogen-associated molecular pattern (Saito et al., 2008). 293T cells (CRL-3216; American Type Culture Collection [ATCC]) and A549 (CCL-185; ATCC) were grown in DMEM with 10% FCS plus GlutaMAX or in reduced serum in Opti-MEM (Thermo Fisher Scientific) alone in 24-well culture for 24 h or together with 10 mM LiCl or 10 mM NaCl as control to arrest in G2/M phase or with 10 μM hydroxyurea to arrest at the G1/S boundary. Cell cycle was determined using propidium iodide staining. Fixable Near-IR viability dye (L34975; Thermo Fisher Scientific) and Violet Live Cell Caspase Probe (565521; BD Biosciences) were used to assess cell death, and CellTrace Violet (C34557; Molecular Probes) membrane dye was used to assess cell proliferation. For transient expression, 293T cells were seeded in 6-well plates and allowed to adhere overnight. Plasmids were then transfected using TransIT-LT1 or TransIT-X2 (Mirus Bio) for the flow cytometric and puromycin pulse Western blot experiments, respectively.
Vectors
Bicistronic vector with MND promoter (Astrakhan et al., 2012) driving transcription of enhanced GFP followed by T2A sequence and 3xF. The vector had either no gene or the human SAMD9L (GenBank accession no. NM_152703) sequence (Origene) inserted downstream of the T2A and N-terminal 3xF using In-Fusion cloning (Takara Bio). The coding sequence ends with a stop codon and the woodchuck hepatitis virus posttranscriptional regulatory element proximal to the polyadenylation site. Site-directed mutagenesis was performed using QuikChange II (Agilent). The pCVL.SFFV.Y2 I-AniI.IRES.mCherry vector contains the spleen focus-forming virus promoter (Aubert et al., 2011). pUC19 (Takara Bio) DNA was used as the carrier DNA for the mCherry-alone condition.
Western blot
Primary fibroblasts were trypsinized, and radioimmunoprecipitation assay lysates (89900; Thermo Fisher Scientific) were run on NuPAGE 3–8% Tris-Acetate Protein gels (EA0375; Thermo Fisher Scientific), transferred to nitrocellulose blocked using Odyssey Blocking Buffer (927-40000; LI-COR), and stained using mouse anti–FLAG-M2 (F1804; Sigma-Aldrich), rabbit anti-SAMD9L (#HPA019465; Sigma-Aldrich), rabbit anti–β-actin (5125S; Cell Signaling Technology), mouse anti–β-actin (3700; Cell Signaling Technology), and anti-puromycin (clone 12D10; Millipore). Secondary detection was with anti-mouse, anti-rat, or anti-rabbit IgG (H+L) IRDye 680CW or 800CW secondary reagents (Odyssey).
Translation assays
For puromycin pulse experiments, 100 μg/ml cycloheximide was added at 37°C for 15 min or media alone. Following this, 10 μg/ml puromycin was added at 37°C for 10 min. Cells were lysed and processed for Western blotting as described below. “SUnSET” (surface sensing of translation; Schmidt et al., 2009) was performed by culturing cells in puromycin dihydrochloride (10 μg/ml; P8833; Sigma-Aldrich) for 10 min before fixation and permeabilization (BD Cytofix/Cytoperm Kit; 554714; BD Biosciences) and detection with Alexa Fluor 647–labeled anti-puromycin antibody, clone 12D10 (MABE343-AF647; Millipore). The SUnRiSE approach used HHT (2 μg/ml) preincubation for variable times (0–10 min) before adding puromycin (Argüello et al., 2018).
Polysome analysis
Cytoplasmic lysates were prepared from cells in hypotonic lysis buffer, as follows: 25 mM Tris-HCl (pH 7.5; AM9850G; Ambion), 150 mM KCl (9640G; Ambion), 15 mM MgCl2 (AM9530G; Ambion), 1 mM dithiothreitol (10197777001; Ambion), 1% NP-40 (28324; Thermo Fisher Scientific), 100 μg/ml cycloheximide (11836170001; Sigma-Aldrich), 100 U/ml SUPERase•In RNase Inhibitor (AM2694; Ambion), 25 U/ml TURBO DNase (AM2238; Ambion), and cOmplete EDTA-free Protease Inhibitor (11836170001; Sigma-Aldrich) in nuclease-free water. Nuclei were removed by centrifugation at 13,000 g for 10 min at 4°C. Cytoplasmic lysates were layered onto a linear sucrose gradient (15–60% sucrose; #S5-12; Thermo Fisher Scientific) and centrifuged in an SW 41 Ti rotor (Beckman) for 2 h at 35,000 rpm at 4°C. Fractions were collected by an Auto-Densi Flow pump attached to a Foxy R1 fraction collector. Continuous absorbance at 254 nm was read using a UA-6 Absorbance Detector (Teledyne ISCO) to obtain polysome traces.
Immunofluorescence
Primary human fibroblasts were seeded on poly-L-ornithine hydrobromide–coated (P3655; Sigma-Aldrich) coverslips and allowed to adhere overnight. Cells were treated with 100 U/ml IFN-β (11415-1; PBL Assay Science) for 24 h. At 24 h after treatment/expression, cells were washed with PBS and then fixed in PBS containing 4% PFA (15710; Thermo Fisher Scientific) for 10 min at room temperature, washed with PBS, and then permeabilized with PBS containing 0.1% Triton X-100 for 10 min at room temperature. Cells were washed with PBS and then resuspended in PBS containing 3% BSA (BP1600-100; Thermo Fisher Scientific) blocking solution for 1 h. After blocking, cells were stained with rabbit anti-SAMD9L (HPA019465; Sigma-Aldrich) in PBS containing 1% BSA and 0.3% Triton X-100 for 1 h in the dark at room temperature. Cells were washed three times with PBS and then stained with goat anti-rabbit IgG Alexa Fluor 488 (A11008; Thermo Fisher Scientific) in PBS containing 1% BSA and 0.3% Triton X-100 for 1 h in the dark at room temperature. Samples were washed three times with PBS and then mounted with ProLong Diamond Antifade mounting media (P36962; Thermo Fisher Scientific). Samples were cured in the dark at room temperature for 24–48 h before imaging. Samples were imaged with a Nikon Eclipse Ti laser scanning confocal microscope using a 60× oil immersion lens and 4× digital zoom. Images were processed and analyzed using NIS-Elements software.
4sU labeling of RNA
At 24 h after transfection, HEK 293T cells were incubated in media containing 500 mM 4sU (16373; Cayman Chemical) for 10 min. Total RNA was harvested using the NucleoSpin RNA extraction kit (740948; Clontech). RNAs metabolically labeled with 4sU were biotinylated and separated as described previously (Garibaldi et al., 2017). RNA was biotinylated by mixing 100 μg RNA with HPDP-biotin (A35390; Thermo Fisher Scientific) resuspended at a concentration of 1 mg/ml in dimethylformamide. The biotinylation reaction was incubated at room temperature for 2 h with gentle rotation. After, RNA was extracted with a solution of chloroform:phenol:isoamyl alcohol (15593031; Thermo Fisher Scientific) twice followed by ethanol precipitation of the RNA. Magnetic separation of the biotinylated RNA was performed using Dynabeads M280 Streptavidin (11205D; Thermo Fisher Scientific) followed by washing four times. The RNA was eluted using 100 mM dithiothreitol (646563; Sigma-Aldrich) and purified through ethanol precipitation. PrimeScript cDNA synthesis kit (RR037B; Takara Bio) was used to generate cDNA from RNA. Real-time quantitative PCR (qPCR) was performed using the TaqMan Universal Master Mix II, no UNG (4440048; Thermo Fisher Scientific), and a ViiA7 qPCR system. Probe-based assays in this study were from Integrated DNA Technologies (GAPDH, assay ID: Hs.PT.39a.22214836I; HPRT, assay ID: Hs.PT.58v.45621572; ACTB, assay ID: Hs.PT.39a.22214847; IFN-B1, assay ID: Hs.PT.58.39481063; ISG15, assay ID: HS.PT.58.39185901). For the fold enrichment calculation, the input cycle threshold (CT) is normalized to the input to that of the pulldown (1 μg RNA for input and 100 μg for pulldown). Input adjustment calculation is [CT input − log2 (dilution factor)]. The ΔCT is calculated (adjusted input CT − immunoprecipitation CT), and the percent input is calculated by (100 × 2^ΔCT) and normalized to the EV value by dividing experimental values by EV values.
Relative mRNA quantification
RNA was isolated using Qiagen RNEasy Mini Kit, with RNA input normalized across samples before cDNA reaction. Samples were pretreated with dsDNase and reverse transcribed using Maxima First Strand cDNA synthesis (Thermo Fisher Scientific). All qPCRs were analyzed using a Bio-Rad CFX96 96-well optical cycler. To assess SAMD9L expression, primers targeted either human SAMD9L mRNA (forward: 5′-CTGGGATTCTGAGCATCCTTTC-3′, reverse: 5′-CACATGCTCTTTGGTCCAGT-3′) or SAMD9L plasmid (forward: 5′-GCTGGGCAGAGTCTCCTAAA-3′, reverse: 5′-CACAAGGAGAAGCACAGGAA-3′), and expression was compared with HPRT mRNA (forward: 5′-GTTGGATATAAGCCAGACTTTGTTG-3′, reverse: 5′-AGGGAACTGATAGTCTATAGGCT-3′). qPCR was performed using Power SYBR Green (Thermo Fisher Scientific) and 400 nM primers and cycled according to manufacturer instructions. To assess mCherry and GFP mRNA expression, Taqman gene expression assays (Thermo Fisher Scientific) with Taqman Fast Advanced Master Mix (Thermo Fisher Scientific) were used according to manufacturer instructions. GFP FAM-MGB (assay ID: Mr04097229_mr; Thermo Fisher Scientific) or mCherry FAM-MGB (assay ID: Mr07319438_mr; Thermo Fisher Scientific) were multiplexed with ACTB VIC-MGB (assay ID: Hs01060665_g1; Thermo Fisher Scientific) as a loading control.
Clinical description
The patient was born at 39 wk gestation by spontaneous vaginal delivery to a G4P4 mother. The patient’s mother was rubella immune. Prenatal laboratories were negative for rapid plasma reagin, and there was no known history of HSV. She had tested positive for group B streptococcus with three prior pregnancies but negative at week 38 of this pregnancy. She did not receive intrapartum antibiotics. There was concern for low amniotic fluid at the time of medical rupture of membranes. There was mild fetal distress, with heart rate decelerations during labor. The child had Apgar scores of 5 and 9. There were no other delivery complications. The birth weight was 6 lb 0.5 oz. Hepatitis B vaccination #1 was given. Mother was found to be cytomegalovirus (CMV) IgG+, and thus, breastfeeding was stopped after 4 d with transition to formula. Infant was immediately discharged home with her mother and father and three siblings (2, 3, and 10 yr old).
Upon admission to Seattle Children’s Hospital at day 4, the infectious testing was negative for adenovirus, CMV, HSV, EBV, enterovirus, parechovirus, parvovirus B19, and HIV1 RNA. Toxoplasmosis and rapid plasma reagin for syphilis testing was negative. The electrocardiogram of this child was normal. X-rays showed no osseous manifestations of congenital infection, specifically looking for syphilis or rubella. Dilated ophthalmological exam revealed no abnormality. CSF revealed elevated protein at 266 mg/dl, glucose 40 mg/dl, 22,550 RBCs, and 7 nucleated cells (25% polys). She was empirically started on i.v. ampicillin, cefotaxime, and meningitis dosing of acyclovir during the workup.
Blood, CSF, and urine cultures remained negative as well as extensive infectious molecular and serologic testing. She regained her birthweight and remained afebrile off antimicrobial therapy and was discharged for outpatient follow-up care. Subsequently, viral fluorescent antibody testing was negative, and nasal wash PCR testing for CMV, EBV, human herpesvirus 6 (HHV6), parainfluenza, influenza, respiratory syncytial virus, coronavirus, adenovirus, and bocavirus were all negative. The infant developed oral ulcers, but oral, rectal, throat, and skin swabs were negative for HSV. She remained on empirical acyclovir throughout her clinical course. She spiked a fever at 7 wk old and started meropenem and vancomycin for 2 d, but the infectious workup was negative again and the medications discontinued.
At 21 d, the child was admitted with respiratory symptoms and tested positive for HRV infection by semiquantitative viral respiratory real-time PCR for respiratory viruses by nasal wash. The HRV assay detects rhinovirus 5′ noncoding region gene. She was negative for CMV, EBV, HHV6, parainfluenza, influenza, respiratory syncytial virus, coronavirus, adenovirus, and bocavirus in the same assay. She was noted to have oral thrush and was started on nystatin and then switched to fluconazole. T cell proliferative responses to mitogen activation were poor, so she was started on trimethoprim/sulfamethoxazole prophylaxis against Pneumocystis jirovecii out of concern for a cellular immunodeficiency. Fluorescent antibody testing for Pneumocystis continued to be negative. Repeat nasal wash revealed that she was persistently HRV+ (tested eight times over a 33-d period) by qualitative PCR testing. She did not receive testing during the last 15 d, but lung tissue from the autopsy was sent for viral culture, and no virus was isolated. IFN-α testing on CSF was <5 IU/ml. Postmortem lung culture grew Staphylococcus aureus, but she had no overt histopathology to indicate a significant bacterial pneumonia. Total nucleic acid was extracted from CSF and brain (cerebrum and basal ganglia) tissue and buccal and rectal swab samples from interim and autopsy specimens using the NucliSENS easyMAG system, and MassTag Multiplex PCR was performed; all samples were negative for the agents detected by the DNA and RNA encephalitis panels (Center for Infection and Immunity at Columbia University; see below for testing included).
RNA encephalitis panel
Testing included enterovirus/rhinovirus/poliovirus, influenza A virus, Japanese encephalitis virus, West Nile virus, St. Louis encephalitis virus, LaCrosse virus, lymphocytic choriomeningitis virus, mumps virus, Nipah virus, parvovirus, rabies virus, Eastern equine encephalitis virus, and Venezuelan equine encephalitis virus.
DNA encephalitis panel
Testing included adenovirus, Candida albicans, Cryptococcus neoformans, CMV, HHV6, EBV, HSV1, HSV2, Haemophilus influenzae, Leptospira interrogans, Mycobacterium tuberculosis, Neisseria meningitidis, Streptococcus pneumoniae, Toxoplasma gondii, and vesicular stomatitis virus. All samples were also negative for enterovirus, S. pneumoniae, rabies virus, Western equine encephalitis, and HSV1 via consensus PCR technique.
Clinical history of control autopsy samples
Two control cases were selected for the histological examination of this case to control for age, lung disease, exposure to corticosteroids, and need for intensive care with intubation. A 2-wk term male infant expired due to a large intracranial immature teratoma, chronic neonatal lung disease, and generalized inanition. The other was a 5-wk term male infant with left diaphragmatic hernia expired from severe respiratory compromise from hypoplastic lung, pulmonary hypertension, and right heart failure following extracorporeal membrane oxygenation for 2.5 wk. There was severe thymic stress involution and acute tubular necrosis noted as well. Full autopsies were performed previously for standard of care, and tissue blocks were prepared similar to our case.
Online supplemental material
Fig. S1 demonstrates lack of uniparental disomy or gene conversion in the proband or healthy relatives. The figure also demonstrates the rapid SAMD9L transcription following interferon or virus treatment in human airway cells. Fig. S2 depicts comparable transfection efficiency and cell death rates across the constructs, as well as protein expression levels and the puromycin incorporation with the different SAMD9L variants. Fig. S3 demonstrates the GFP expression intensity associated with various SAMD9L variants was not altered by skewing the cell cycle.
Acknowledgments
We thank Richard James, Marshall Horwitz, and Wendy Raskind for insightful comments.
Sequencing was provided by the University of Washington Center for Mendelian Genomics and was funded by National Human Genome Research Institute and National Heart, Lung, and Blood Institute grants UM1 HG006493 and U24 HG008956 and by the Office of the Director, National Institutes of Health, under award number S10 OD021553. This work was supported by the National Institutes of Health under award numbers NIH T32 GM007270 and NIH F31 AI140530 (to F. Soveg), NIH 5R21 AI143227 and NIH 1F32 AI145283-01A1 (to L. So), and NIH 1K08 DK114568-01 (to E.J. Allenspach). Additional support through a pilot grant from Brotman Baty Institute for Precision Medicine (to E.J. Allenspach), the Seattle Children’s Research Institute Center for Immunity and Immunotherapies Program for Cell and Gene Therapy, the Children’s Guild Association Endowed Chair in Pediatric Immunology (to D.J. Rawlings), and the Hansen Investigator in Pediatric Innovation Endowment (to D.J. Rawlings). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions: E.J. Allenspach designed and performed most of the experiments, analyzed the data, performed genetic analysis, and wrote the manuscript. L.S. Finn performed the autopsy analysis and made critical histological observations. F. Soveg and L. So performed polysome profiling, immunoprecipitation, and immunofluorescence experiments. J.A. Gorman and A.B.I. Rosen helped with the cloning, biochemistry, flow cytometry, and experiment execution. S. Skoda-Smith clinically treated the patient. M.M. Wheeler helped to perform genetic analysis. J.S. Debley designed the primary airway epithelial cell experiments performed by K.A. Barrow and L.M. Rich. M.J. Bamshad and D.A. Nickerson supervised the genomic analysis. R. Savan helped to design the translation analysis experiments, analyzed data, supervised the work, and wrote the manuscript. T.R. Torgerson provided samples, performed clinical diagnoses, helped to analyze data, supervised the work, and wrote the manuscript. D.J. Rawlings helped to analyze data, supervised the work, and wrote the manuscript. All authors reviewed and approved the manuscript.
REFERENCES
- Argüello, R.J., Reverendo M., Mendes A., Camosseto V., Torres A.G., Ribas de Pouplana L., van de Pavert S.A., Gatti E., and Pierre P.. 2018. SunRiSE - measuring translation elongation at single-cell resolution by means of flow cytometry. J. Cell Sci. 131:jcs214346. 10.1242/jcs.214346 [DOI] [PubMed] [Google Scholar]
- Astrakhan, A., Sather B.D., Ryu B.Y., Khim S., Singh S., Humblet-Baron S., Ochs H.D., Miao C.H., and Rawlings D.J.. 2012. Ubiquitous high-level gene expression in hematopoietic lineages provides effective lentiviral gene therapy of murine Wiskott-Aldrich syndrome. Blood. Blood. 119:4395–4407. 10.1182/blood-2011-03-340711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert, M., Ryu B.Y., Banks L., Rawlings D.J., Scharenberg A.M., and Jerome K.R.. 2011. Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI. PLoS One. 9:e16825. 10.1371/journal.pone.0016825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov, Y.A., and Palmenberg A.C.. 2006. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques. 41:283–292. 10.2144/000112243 [DOI] [PubMed] [Google Scholar]
- Chen, D.-H., Below J.E., Shimamura A., Keel S.B., Matsushita M., Wolff J., Sul Y., Bonkowski E., Castella M., Taniguchi T., et al. 2016. Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am. J. Hum. Genet. 98:1146–1158. 10.1016/j.ajhg.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Mugler C.F., Keskin A., Hodapp S., Chan L.Y.-L., Weis K., Mertins P., Regev A., Jovanovic M., and Brar G.A.. 2019. Small and large ribosomal subunit deficiencies lead to distinct gene expression signatures that reflect cellular growth rate. Mol. Cell. 73:36–47.e10. 10.1016/j.molcel.2018.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin, M. 2017. I am SAMD9L: 7q regulator I am. Blood. 129:2210–2212. 10.1182/blood-2017-03-770198 [DOI] [PubMed] [Google Scholar]
- Davidsson, J., Puschmann A., Tedgård U., Bryder D., Nilsson L., and Cammenga J.. 2018. SAMD9 and SAMD9L in inherited predisposition to ataxia, pancytopenia, and myeloid malignancies. Leukemia. 32:1106–1115. 10.1038/s41375-018-0074-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus, A.A., Hou Y., Brooks S., Malle L., Biancotto A., Huang Y., Calvo K.R., Marrero B., Moir S., Oler A.J., et al. 2020. Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J. Clin. Invest. 130:1669–1682. 10.1172/JCI129301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero, A., Ozarkar S., Li H., Lee C.H., Hemann E.A., Nadjsombati M.S., Hendricks M.R., So L., Green R., Roy C.N., et al. 2019. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity. 51:451–464.e6. 10.1016/j.immuni.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibaldi, A., Carranza F., and Hertel K.J.. 2017. Isolation of newly transcribed RNA using the metabolic label 4-thiouridine. Methods Mol. Biol. 1648:169–176. 10.1007/978-1-4939-7204-3_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein, B.D., Roman D., Arévalo-Soliz L.M., Engevik M.A., and Zechiedrich L.. 2016. Effects of circular DNA length on transfection efficiency by electroporation into HeLa cells. PLoS One. 11:e0167537. 10.1371/journal.pone.0167537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia, N.T., Lareau L.F., and Weissman J.S.. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 147:789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur, M., and Ackerman S.L.. 2018. mRNA translation gone awry: translation fidelity and neurological disease. Trends Genet. 34:218–231. 10.1016/j.tig.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria, R.K., Munschauer M., Ulirsch J.C., Fiorini C., Ludwig L.S., McFarland S.K., Abdulhay N.J., Specht H., Keshishian H., Mani D.R., et al. 2018. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 173:90–103.e19. 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiss, P., Cameron B., Rangara R., Mailhe P., Aguerre-Charriol O., Airiau M., Scherman D., Crouzet J., and Pitard B.. 1999. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 27:3792–3798. 10.1093/nar/27.19.3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer, R.M., Chiang C., Quinlan A.R., and Hall I.M.. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15:R84. 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmkuhl, E.M., and Zarnescu D.C.. 2018. Lost in translation: evidence for protein synthesis deficits in ALS/FTD and related neurodegenerative diseases. Adv. Neurobiol. 20:283–301. 10.1007/978-3-319-89689-2_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Ding S.C., Cho H., Chung B.C., Gale M. Jr., Chanda S.K., and Diamond M.S.. 2013. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio. 4:e00385–e13. 10.1128/mBio.00385-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and McFadden G.. 2015. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J. Virol. 89:1925–1931. 10.1128/JVI.02262-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhedov, S.L., Makarova K.S., and Koonin E.V.. 2017. The complex domain architecture of SAMD9 family proteins, predicted STAND-like NTPases, suggests new links to inflammation and apoptosis. Biol. Direct. 12:13. 10.1186/s13062-017-0185-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X., and Xiang Y.. 2019. RNA granules associated with SAMD9-mediated poxvirus restriction are similar to antiviral granules in composition but do not require TIA1 for poxvirus restriction. Virology. 529:16–22. 10.1016/j.virol.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela, C., García M.A., Collado M., Marcos-Villar L., Gallego P., Esteban M., and Rivas C.. 2007. Control of virus infection by tumour suppressors. Carcinogenesis. 28:1140–1144. 10.1093/carcin/bgm048 [DOI] [PubMed] [Google Scholar]
- Nagamachi, A., Matsui H., Asou H., Ozaki Y., Aki D., Kanai A., Takubo K., Suda T., Nakamura T., Wolff L., et al. 2013. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell. 24:305–317. 10.1016/j.ccr.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Nounamo, B., Li Y., O’Byrne P., Kearney A.M., Khan A., and Liu J.. 2017. An interaction domain in human SAMD9 is essential for myxoma virus host-range determinant M062 antagonism of host anti-viral function. Virology. 503:94–102. 10.1016/j.virol.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle, M., Helms L., Vermeire J., Roesch F., Humes D., Basom R., Delrow J.J., Overbaugh J., and Emerman M.. 2018. A virus-packageable CRISPR screen identifies host factors mediating interferon inhibition of HIV. eLife. 7:e39823. 10.7554/eLife.39823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias, L.C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375–386. 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- Robichaud, N., Sonenberg N., Ruggero D., and Schneider R.J.. 2019. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 11:a032896. 10.1101/cshperspect.a032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., and Mesirov J.P.. 2017. Variant Review with the Integrative Genomics Viewer. Cancer Res. 77:e31–e34. 10.1158/0008-5472.CAN-17-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D., and Shimamura A.. 2014. Marrow failure: a window into ribosome biology. Blood. 124:2784–2792. 10.1182/blood-2014-04-526301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., Owen D.M., Jiang F., Marcotrigiano J., and M. Gale, Jr.. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 454:523–527. 10.1038/nature07106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper, G.C., van der Knaap M.S., and Proud C.G.. 2007. Translation matters: protein synthesis defects in inherited disease. Nat. Rev. Genet. 8:711–723. 10.1038/nrg2142 [DOI] [PubMed] [Google Scholar]
- Schmidt, E.K., Clavarino G., Ceppi M., and Pierre P.. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 6:275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Schneider, W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch, T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., and Liu J.O.. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6:209–217. 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P., Yan F., Doronina V.A., Escuin-Ordinas H., Ryan M.D., and Brown J.D.. 2012. 2A peptides provide distinct solutions to driving stop-carry on translational recoding. Nucleic Acids Res. 40:3143–3151. 10.1093/nar/gkr1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan, G., Glushakow-Smith S.G., Katsafanas G.C., Americo J.L., and Moss B.. 2018. Human host range restriction of the vaccinia virus C7/K1 double deletion mutant is mediated by an atypical mode of translation inhibition. J. Virol. 92:e01329-18. 10.1128/JVI.01329-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram, A., Bohlen J., and Teleman A.A.. 2018. Translation acrobatics: how cancer cells exploit alternate modes of translational initiation. EMBO Rep. 19:e45947. 10.15252/embr.201845947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., Shimbo T., Kikuchi Y., Matsuda M., and Kaneda Y.. 2010. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int. J. Cancer. 126:1982–1991. 10.1002/ijc.24965 [DOI] [PubMed] [Google Scholar]
- Tesi, B., Davidsson J., Voss M., Rahikkala E., Holmes T.D., Chiang S.C.C., Komulainen-Ebrahim J., Gorcenco S., Rundberg Nilsson A., Ripperger T., et al. 2017. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 129:2266–2279. 10.1182/blood-2016-10-743302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, D., Mathews M.B., and Mohr I.. 2013. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 5:a012351. 10.1101/cshperspect.a012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Zhai Y.-Y., Dai J.-H., Li K.-Y., Deng Q., and Han Z.-G.. 2014. SAMD9L inactivation promotes cell proliferation via facilitating G1-S transition in hepatitis B virus-associated hepatocellular carcinoma. Int. J. Biol. Sci. 10:807–816. 10.7150/ijbs.9143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J.C., Bryant V., Lamprecht T., Ma J., Walsh M., Schwartz J., Del Pilar Alzamora M., Mullighan C.G., Loh M.L., Ribeiro R., et al. 2018. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight. 3:e121086. 10.1172/jci.insight.121086 [DOI] [PMC free article] [PubMed] [Google Scholar]