ABSTRACT
Our understanding of how the obligate intracellular bacterial pathogen Chlamydia trachomatis reprograms the function of infected cells in the upper genital tract is largely based on observations made in cell culture with transformed epithelial cell lines. Here, we describe a primary organoid system derived from endometrial tissue to recapitulate epithelial cell diversity, polarity and ensuing responses to Chlamydia infection. Using high-resolution and time-lapse microscopy, we catalog the infection process in organoids from invasion to egress, including the reorganization of the cytoskeleton and positioning of intracellular organelles. We show this model is amenable to screening C. trachomatis mutants for defects in the fusion of pathogenic vacuoles, the recruitment of intracellular organelles and inhibition of cell death. Moreover, we reconstructed a primary immune cell response by co-culturing infected organoids with neutrophils, and determined that effectors like CPAF (also known as CT858) and TepP (also known as CT875) limit the recruitment of neutrophils to infected organoids. Collectively, our model can be applied to study the cell biology of Chlamydia infections in three-dimensional structures that better reflect the diversity of cell types and polarity encountered by Chlamydia in their animal hosts.
KEY WORDS: Chlamydia, Pathogen, Epithelial, Cytoskeleton, Neutrophils
Summary: 3D endometrial organoids are used to model Chlamydia infection and the role of secreted virulence factors in reprogramming host epithelial cells and immune cell recruitment.
INTRODUCTION
Chlamydia trachomatis is a clinically important pathogen, responsible for the majority of sexually transmitted bacterial infections (WHO, 2018). In the female upper genital tract (UGT), chronic and untreated asymptomatic infections can lead to pelvic inflammatory disease, tubal scarring and infertility (Haggerty et al., 2010), and are associated with cancers of the cervix and endometrium (Koskela et al., 2000). Yet, despite intense interest, the molecular mechanisms by which C. trachomatis disrupts epithelial tissue structure and induces pathology in the UGT are not well understood.
The genus Chlamydia is comprised of nine Chlamydia species that infect at least eleven different vertebrate hosts, including the human pathogen C. trachomatis and the rodent-adapted C. muridarum (Horn, 2008; Schachter, 1978). Based on sequence similarity and disease manifestations, C. trachomatis is further classified into serovars and biovars that target the genital tract (serovars D–K) and ocular epithelia (serovars A–C), the etiological cause of the inflammatory disease trachoma (Stephens et al., 2009). The lymphogranuloma venereum (LGV) biovar (serovars L1–L3) causes an invasive infection in the urogenital or anorectal tracts that disseminates to the lymph nodes (Mabey and Peeling, 2002). Of all the serovars, C. trachomatis LGV serovar L2 is the most tractable to molecular genetic manipulation (Mueller et al., 2016; Wang et al., 2011).
All Chlamydiae transition between two developmental forms: the environmentally stable and infectious elementary body (EB), and the non-infectious but replication-competent reticulate body (RB) (Abdelrahman and Belland, 2005). To invade cells, EBs use a type III secretion (T3S) system to deliver effector proteins directly into host epithelial cells. These invasion-associated early T3S effectors stimulate rearrangements of filamentous actin (F-actin) at attachment sites to promote bacterial entry and facilitate the formation of an intracellular membrane vacuole termed the ‘inclusion’ (reviewed in Elwell et al., 2016). Additional effectors are then secreted and inserted into the nascent inclusion membrane. These effectors, termed inclusion membrane (Inc) proteins (Rockey et al., 1995), co-opt microtubule-based trafficking to aid in the migration of the inclusion to the microtubule organizing center, subvert intracellular organelles – including the Golgi apparatus, endoplasmic reticulum, peroxisomes and lipid droplets – and establish membrane contact sites to presumably facilitate the acquisition of nutrients from the host (Agaisse and Derré, 2014; Boncompain et al., 2014; Derré et al., 2011; Dumoux et al., 2012; Grieshaber et al., 2003; Kumar et al., 2006; Mital et al., 2015; Moore et al., 2008). Within the inclusion, EBs differentiate into RB forms, replicate and expand along the limiting membrane of the inclusion. The inclusion itself is wrapped in a network of actin and intermediate filaments, microtubules, and septins (Dumoux et al., 2015; Kumar and Valdivia, 2008; Volceanov et al., 2014; Wesolowski et al., 2017). RBs asynchronously differentiate back into EBs and eventually escape from the cell through an F-actin-mediated extrusion of the inclusion or host cell lysis to initiate the next round of infections (Hybiske and Stephens, 2007).
Chlamydia maintains an intracellular replicative niche by suppressing cell-autonomous innate immunity (reviewed in Finethy and Coers, 2016). For example, Chlamydia-driven rearrangements of the cytoskeleton promote inclusion stability, limiting detection and the activation of innate immune responses (Kumar and Valdivia, 2008). Chlamydia Inc proteins counteract host defenses by deubiquitylating proteins at the inclusion, inhibiting the lytic activity of interferon-induced factors (e.g. guanylate-binding proteins) and suppressing cell death by subverting membrane trafficking to inhibit host surveillance programs and apoptosis (Coers et al., 2008; Faris et al., 2019; Fischer et al., 2017; Sixt et al., 2017). Despite these activities, Chlamydia is still partially recognized by the host cell leading to a pro-inflammatory response (Rasmussen et al., 1997), including the expression and secretion of chemokines, such as the neutrophil attractants interleukin-6 (IL-6) and interleukin-8 (IL-8) (Buchholz and Stephens, 2008; Rasmussen et al., 1997).
Our current understanding of how Chlamydia reprograms host cellular functions is based largely on studies in two-dimensional (2D) cell culture systems with transformed cervical epithelial cell lines like HeLa cells. Although these models are experimentally tractable, they do not accurately represent the natural sites of infection; polarized UGT epithelial cells have a distinct spatial organization of the cytoskeleton, organelle positioning and signaling modules that differ from those present in transformed non-polarized epithelial cancer cells (Dolat and Valdivia, 2019; Rodriguez-Boulan and Macara, 2014). Studies in polarized epithelia have uncovered unique aspects of the interaction between Chlamydia and its target cell. For instance, C. trachomatis replication is enhanced in polarized epithelial cells, and the inclusion preferentially intercepts vesicles destined for the basolateral side of infected cells (Guseva et al., 2007; Moore et al., 2008). Infections in primary polarized human ecto- and endo-cervical epithelial explants reveal that Chlamydia alters epithelial structure by inducing epithelial-to-mesenchymal transition (Zadora et al., 2019). Furthermore, long-term Chlamydia infections in fallopian tube organoids, self-renewing primary three-dimensional (3D) epithelial tissue-like cultures, promote epithelial stemness, cellular proliferation, and modifications to genome organization often associated with ageing tissues (Kessler et al., 2019). These observations may explain some of the associations observed between Chlamydia infections and cervical and ovarian cancers (Shanmughapriya et al., 2012; Zhu et al., 2016).
3D organoids have emerged as a compelling ex vivo system to study epithelial cell biology, because they better recapitulate aspects of the cellular diversity, structure and function of epithelial tissues (Rossi et al., 2018). Isolated human and mouse glandular endometrial epithelia cultured in a chemically defined medium were recently reported to form endometrial organoids (EMOs) comprised of hormonally responsive polarized epithelial subtypes (e.g. ciliated, secretory) that mimic aspects of the estrous cycle and implantation (Boretto et al., 2017; Turco et al., 2017). EMOs exhibit long-term genetic stability and are amenable to high-throughput screens (Boretto et al., 2019). Here, we describe an infection model that uses EMOs and microinjection to deliver Chlamydia to the apical surface of a polarized epithelium, thus mimicking the natural route of infection. Using both the rodent-adapted C. muridarum and the genetically-tractable human pathogen C. trachomatis serovar L2, we demonstrate key aspects of the infection cycle and cellular activities driven by C. trachomatis effectors. We further show that we can recapitulate aspects of cellular immunity in infected EMOs by monitoring the recruitment and behavior of neutrophils. Using C. trachomatis mutants that alter immunity-related signaling pathways, including the expression of neutrophil chemoattractants, we further demonstrate that this system can be used to monitor the manipulation of host immune responses by Chlamydia virulence factors.
RESULTS
Cells harvested from the upper genital tract develop into endometrial organoids of cellular composition similar to that of primary tissues
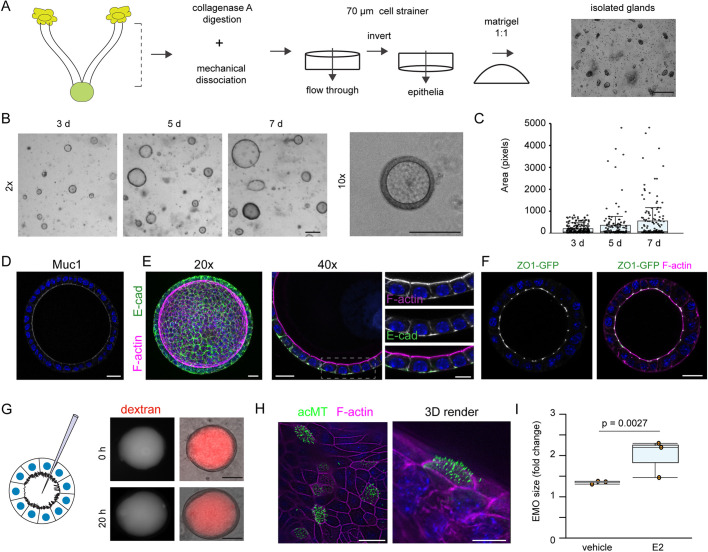
We isolated mouse endometrial epithelial glands, which contain stem cells that regenerate the luminal epithelia during the estrous cycle (Gargett et al., 2016; Jin, 2019), and cultured them in a 3D Matrigel matrix in the presence of conditioned medium derived from L-WRN cells (Fig. 1A). L-WRN cells stably express and secrete Wnt-3A, R-spondin-3, and noggin, which are sufficient to generate organoids from various mouse tissues with little batch-to-batch variation (Miyoshi and Stappenbeck, 2013; VanDussen et al., 2019). Indeed, EMOs generated from endometrial tissues readily formed over the course of one week and grew into large, spherical structures (Fig. 1B,C).
Fig. 1.
Generation of primary murine-derived polarized endometrial epithelial organoids. (A) Workflow for the isolation of mouse endometrial epithelial glands and culture in a 3D extracellular Matrigel plug. Scale bar: 100 µm. (B,C) Cells in endometrial organoids (EMOs) proliferate in the presence of Wnt-3a, R-spondin-3, noggin and EGF. (B) Brightfield images of organoids forming over a 7 day period are shown, including a single EMO at higher magnification, and (C) quantification of the relative surface area at each time point (n>100 EMOs per time point). Boxplots show the median and interquartile range (IQR), with whiskers indicating Q3+1.5×IQR. Scale bars: 100 µm. (D–F) EMOs express MUC1 and establish tight and adherens junctions. (D) Immunofluorescence microscopy of MUC1 (white) and DNA (blue) in an EMO. Scale bar: 20 µm. (E) Polarized EMOs establish adherens junctions. Immunolocalization of the epithelial marker E-cadherin (E-cad). Confocal images of EMOs stained for DNA (blue), F-actin and E-cadherin, and imaged using a 20× objective (left, maximum projection) and a 40× objective (right, medial section). Dashed box indicates region shown in enlarged images on the right. Scale bars: 20 µm. (F) EMOs establish tight junctions. Medial section of confocal microscopy images of EMOs derived from a ZO1–GFP knock-in mouse and stained for F-actin and DNA (blue). Scale bar: 20 µm. (G) EMO epithelia maintain barrier function. Left, cartoon of endometrial organoid microinjection system. Right, EMO microinjected with Texas Red–dextran and imaged immediately post injection (0 h) and 20 h later. Scale bars: 50 µm. (H) Immunofluorescence microscopy of an EMO treated with estrogen. EMOs were treated with 20 nM estrogen for 4 days prior to imaging. Confocal image and 3D render of an EMO stained for acetylated-tubulin (acMT), F-actin and DNA (blue) are shown. Scale bars: 10 µm. (I) Estrogen promotes EMO growth. Quantification of fold change in EMO size with and without estrogen (E2) for 5 days (n=3). Boxplots show the median and IQR, with whiskers indicating the minimum and maximum values. Statistical significance was measured using a Welch's t-test.
Using high-resolution confocal microscopy, we determined that EMOs are comprised of a single layer of polarized epithelial cells surrounding a large, hollow lumen. The epithelia express and secrete mucin (MUC1) (Fig. 1D) and establish adherens and tight junctions (Fig. 1E,F), respectively marked by E-cadherin (also known as CDH1) and zonula occludens-1 (ZO-1, also known as TJP1). To assess the integrity of the spheroids and the functionality of the cell–cell junctions, we microinjected fluorescent dextran (3 kDa) into the organoid lumen. The fluorescent tracers remained in the lumen, indicating that barrier function is maintained over prolonged periods of time (>20 h) (Fig. 1G). Endometrial tissues are responsive to sex hormones, increasing cellular proliferation and altering gene expression and cell type abundance in the presence of estrogen and progesterone. EMOs are similarly responsive to hormones as assessed by the increase in size upon exposure to estrogen for 5 days, as compared to untreated EMOs (1.98× versus 1.34×), and the formation of multi-ciliated epithelial cells as marked by acetylated microtubules (Fig. 1H,I). Overall, the method produces hormonally responsive polarized EMOs with intact barrier function and a distinct lumen.
Chlamydia infection of endometrial epithelial cells leads to cytoskeletal reorganization and disruption of cell–cell junctions
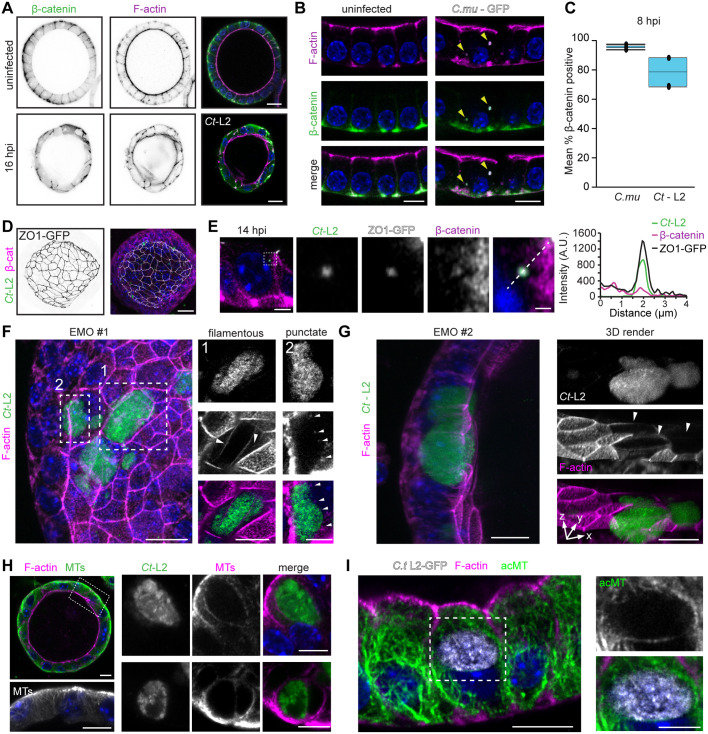
To mimic the natural route of infection in EMOs we microinjected Chlamydia into the lumen of the organoids where the EBs interact with the apical surface of the epithelia. EMOs reached a size amenable for infections by 7–10 days, and based on the ejected volume from the microinjection needle, we calculated the number of infectious units delivered into each EMO. We microinjected GFP-expressing C. trachomatis serovar L2 or C. muridarum CM006, a high pathology-inducing isolate (Poston et al., 2018), into the EMO lumen and monitored the subcellular localization of F-actin and β-catenin (CTNNB1), which are reported to localize to the inclusion (Kessler et al., 2012) at early timepoints (8–16 h) post injection. EBs readily invaded the polarized epithelial cells, promoted the assembly of actin filaments as the nascent inclusion formed and recruited β-catenin (Fig. 2A–C). Consistent with a previous observation in infected ex vivo fallopian tubes (Kessler et al., 2012), infected epithelia showed a loss of apical F-actin and cell–cell junction integrity, demarcated by more diffusive β-catenin localization, indicating that epithelial polarity and barrier function are compromised during invasion. We observed β-catenin recruitment to inclusions only at very early time points, with no significant recruitment to mature inclusions by 48 h post injection (Fig. S1A,B). More recently, Chlamydia has been shown to target tight junctions in a mouse oviduct epithelial cell line, altering the expression of tight junction proteins and reducing transepithelial electrical resistance (Kumar et al., 2019). Using EMOs derived from ZO1–GFP-expressing transgenic mice (Foote et al., 2013), we observed that ZO-1 was recruited to EBs during invasion, along with β-catenin, which may further underlie the disruption to epithelial barrier functions (Fig. 2D,E). Taken together, these data show that Chlamydia invasion remodels the F-actin network and cell-cell junction organization in a manner that disrupts cell polarity.
Fig. 2.
Chlamydia infection leads to the remodeling of the endometrial epithelial cytoskeleton and cell–cell junctions. (A–C) C. trachomatis (Ct-L2) and C. muridarum (C.mu) infection disrupt the organization of endometrial epithelial cell–cell junctions. Confocal images of EMOs uninfected or infected with for (A) 16 h (Ct-L2) or (B) 8 h (C.mu) and stained for F-actin, β-catenin and DNA (blue). Arrowheads in B indicate nascent inclusions. Scale bars: A, 20 µm; B, 10 µm. (C) Quantification of β-catenin recruitment to nascent inclusions at 8 h post infection (hpi; n=3–4 EMOs, 31 inclusions). Boxplots show the median and the interquartile range, with whiskers indicating the minimum and maximum values. (D,E) C. trachomatis early inclusions recruit the tight junction protein ZO-1. (D) Maximum projection confocal images of an EMO derived from ZO1–GFP-expressing mice and stained for β-catenin, and (E) a single confocal slice after infection with mCherry-expressing Ct-L2 for 14 h and stained for β-catenin and DNA (blue). The dashed box indicates the region presented in the enlarged panels, which show a single inclusion and the recruitment of ZO-1 and β-catenin. The dashed line indicates the section quantified in the fluorescence line profile of Ct-L2–mCherry, ZO-1 and β-catenin (right). Scale bars: D, 20 µm; E, 10 µm and 1 μm (enlarged panels). (F,G) C. trachomatis reorganizes the actin cytoskeleton of EMO cells. Confocal images of EMOs infected with GFP-expressing Ct-L2 for 24 h and stained for F-actin (left panels, maximum projections). (F) Dashed boxes indicate regions presented on the right as single confocal slices (top, Ct-L2; middle, F-actin; bottom, merge), showing different organization of actin (filamentous versus punctate). Arrowheads indicate filamentous actin around the inclusion (left panels) and actin puncta around the inclusion (right panels). (G) Images on the right show a reduction in F-actin signal on the apical membrane, as assessed by 3D rendering of Ct-L2 (GFP)-infected cells in EMO #2. Arrowheads indicate infected cells lacking apical F-actin. Scale bars: left panels, 20 µm; right panels, 10 µm. (H,I) Organization of microtubules (MTs) in EMOs. (H) Confocal images of an uninfected EMO stained for microtubules (MT) and F-actin (left). Dashed box indicates the region shown in the enlarged image beneath. Right, enlarged images of microtubules rearranged at the periphery of Ct-L2 inclusions at 24 h post infection. Scale bars: 10 µm. (I) C. trachomatis promotes the assembly of stable microtubules around the inclusion in EMOs, as assessed by immunostaining for acetylated microtubules (acMT). Confocal images of acetylated microtubules in an EMO infected with GFP-expressing Ct-L2 for 24 h. Dashed box indicates the region shown in enlarged panels on the right. Scale bars: 10 µm. For all images, DNA was stained with 2 µg/ml Hoechst (blue).
During the mid and late stages of infection, Chlamydia remodels the cytoskeleton of HeLa cells at the periphery of inclusions (Kumar and Valdivia, 2008). We monitored the localization of various cytoskeletal elements in infected EMOs at mid-cycle (24 h post infection) by probing for F-actin and immunostaining microtubules, cytokeratins and septins – all of which localize to the inclusion periphery (Chin et al., 2012; Kumar and Valdivia, 2008; Volceanov et al., 2014). In polarized epithelial cells the cytoskeletal architecture is markedly different from non-polarized and transformed cells. For example, the F-actin network is enriched at the apical membrane, microvilli and cell–cell junctions rather than a dense network of bundled actin stress fibers (Fig. 2A). Moreover, in contrast to 2D culture settings, microtubule organizing centers localize proximal to the apical membrane and generate microtubules that extend towards the basolateral domain, often along the lateral membrane (Pickett et al., 2019). We observed F-actin structures around the inclusion, either as filamentous-like rings or discrete puncta, which may depend on the stage of the infection cycle (Fig. 2F). Moreover, F-actin signal on the apical membrane was markedly reduced or absent in infected epithelia compared to that in uninfected neighboring cells (Fig. 2G), suggesting that infection and/or inclusion growth promotes the loss of microvilli. In uninfected EMOs, microtubules ran along the apico–basolateral axis and terminated at the basolateral membrane (Fig. 2H). During infection, we observed microtubules and acetylated microtubules prominently assembled around the inclusion (Fig. 2H,I). Because Chlamydia effectors, such as CT288 and IPAM (also known as CT223), target centrosomal proteins (Almeida et al., 2018; Dumoux et al., 2015), the EMO model can be used to explore effector functions in microtubule assembly during infection.
The organization and dynamics of F-actin and microtubules are regulated directly by septins, a conserved family of GTPases that assemble into heteropolymers and filaments (Mostowy and Cossart, 2012; Spiliotis, 2018). In EMOs, septin 2 (Sept2) localized to the apical and lateral membranes and partially colocalized with F-actin at the basolateral surface (Fig. S1C). In infected EMOs, Sept2 underwent a dramatic reorganization to form filamentous rings around the inclusion (Fig. S1D), as has been previously observed in non-polarized cells (Volceanov et al., 2014).
Finally, we probed the endometrial epithelia for reorganization of intermediate filaments. Both keratins and vimentin localize around the inclusion in transformed epithelia, and their absence disrupts inclusion stability (Kumar and Valdivia, 2008; Tarbet et al., 2018). As expected, EMO epithelia expressed keratins (Fig. S1E) which relocalized to the C. trachomatis inclusion (Fig. S1F). However, unlike transformed epithelia, EMO epithelial cells did not express vimentin (Fig. S1G), and its expression was not induced during infection with C. muridarum (Fig. S1H). In contrast, primary endometrial stromal fibroblasts infected with C. muridarum and C. trachomatis displayed prominent vimentin filament cages surrounding inclusions (Fig. S1I).
Overall, these findings indicate that Chlamydia likely employs conserved mechanisms to reorganize cytoskeletal structures in transformed, non-polarized epithelial cells and endometrial organoid epithelia during cell entry and the formation of inclusions.
Live imaging of inclusion expansion, dynamics and exit
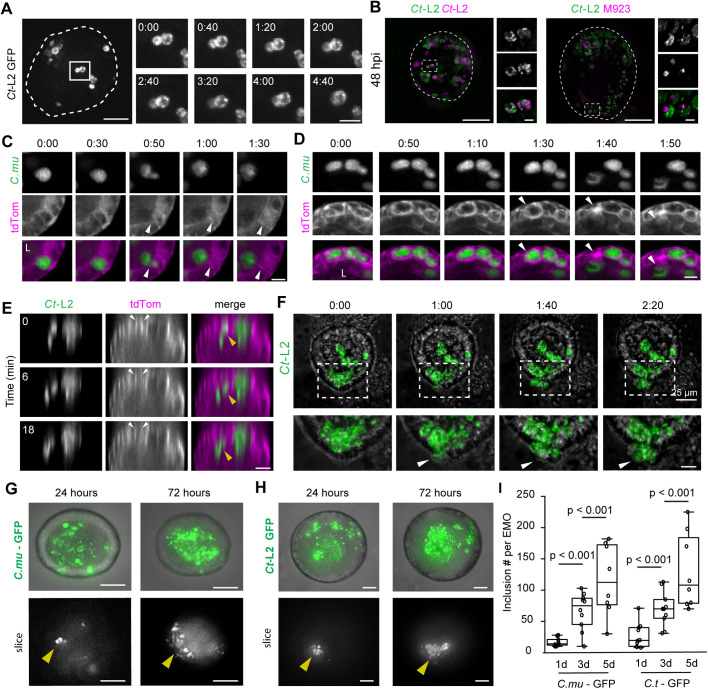
We developed an imaging platform to monitor inclusion growth, dynamics and exit using time-lapse 3D spinning-disk confocal microscopy. EMOs were infected for 24 h with C. trachomatis serovar L2 expressing GFP and were subsequently imaged live for an additional 16–18 h. We noted inclusion expansion and dynamics, including the apparent fusion of adjacent inclusions (Fig. 3A; Movie 1). We next tested whether inclusion fusion is a phenomenon that occurs in EMOs by co-infecting them with C. trachomatis L2 expressing GFP or mCherry, or with a C. trachomatis mutant (M923) lacking the fusogenic factor IncA (Kokes et al., 2015; Sixt et al., 2017). Inclusions containing both GFP and mCherry bacteria were readily apparent in EMOs co-infected with wild-type strains (Fig. 3B) but not in EMOs co-infected with the IncA mutants (Fig. 3B), indicating that the mechanisms guiding inclusion fusogenicity are conserved in EMOs.
Fig. 3.
Time-lapse microscopy of inclusion dynamics and exit from infected endometrial epithelia. (A,B) C. trachomatis inclusions are fusogenic in EMOs. (A) Maximum projections of live-cell fluorescence spinning-disk microscopy of an EMO infected with GFP-expressing C. trachomatis (Ct-L2). Box marks the region presented in enlarged frames, showing two inclusions fusing over time. Dashed line outlines the EMO. Time display (h:min). Scale bars: 25 µm (left) 10 µm (right). (B) Fusion is blocked when co-infections are performed with an incA mutant (M923) expressing mCherry. Maximum projections of EMOs infected for 48 h and imaged by 3D widefield deconvolution microscopy. Dashed boxes indicate regions shown in enlarged panels on the right (top, GFP; middle, mCherry; bottom, merge). Dashed lines mark the EMO outline. Scale bars: 100 µm. (C,D) Apical extrusion of C. muridarum inclusions and infected host cells in EMOs. EMOs expressing a membrane anchored TdTomato were infected with GFP-expressing C. muridarum (C.mu) for 24 h and imaged by time-lapse 3D spinning-disk confocal microscopy. (C) Single confocal slices at the indicated timepoints show an inclusion released into the lumen (L). White arrowheads denote an infected epithelial cell that remains intact. (D) Single confocal slices at the indicated timepoints show lysis of the infected epithelial cell along with release of the inclusion into the EMO lumen (arrowheads). Time display (h:min). Scale bars: C, 5 µm; D, 10 µm. (E,F) C. trachomatis L2 inclusions extrude apically and basolaterally. (E) Orthogonal view of a tdTomato-expressing EMO infected with GFP-expressing Ct-L2 and imaged by time-lapse spinning-disk microscopy. Yellow arrowhead denotes an inclusion extruding into the inclusion lumen and white arrowheads indicate intact cell boundaries. Scale bar: 20 µm. (F) Maximum projections from time-lapse spinning-disk microscopy of an EMO infected with GFP-expressing C. trachomatis (Ct-L2). Dashed box denotes the region shown enlarged in the lower panels. White arrowheads denote cells and inclusions that bulge basolaterally into the matrix. Time display (h:mim). Scale bars: top, 25 µm; bottom, 10 µm. (G–I) C. muridarum and C. trachomatis re-infect cells within the EMO. (G) Maximum projections (top panels) and single sections (bottom panels) of the same EMO and region infected with GFP-expressing C. muridarum and imaged at 24 h and 72 h. (H) Maximum projections (top panels) and single sections (bottom panels) of the same EMO and region infected with GFP-expressing C. trachomatis and imaged at 24 h and 72 h. Arrowheads indicate the same area of each EMO and show an expansion of inclusion number. Scale bars: 50 µm. (I) Quantification of inclusion number per EMO (n=8–10 EMOs). Boxplots show the median and interquartile range, with whiskers indicating the minimum and maximum values. Statistical significance was measured using a Welch's t-test.
We next monitored inclusion exit using EMOs derived from ROSAmTmG mice that express a membrane-targeted tdTomato, which enables tracking of the plasma membrane of individual cells. First, EMOs were infected with either GFP-expressing C. muridarum for 24 h or C. trachomatis L2 for 48 h and imaged live for an additional 16 h. We observed that C. muridarum inclusions were released exclusively into the organoid lumen via lysis or extrusion, which left the infected cell intact, suggesting that apical exit may be the natural mechanism by which C. muridarum escapes the host cell (Fig. 3C,D; Movies 2,3). In contrast, C. trachomatis L2 inclusions were observed to extrude both apically into the lumen (Fig. 3E) and basolaterally into the extracellular matrix (Fig. 3E,F; Movie 4).
To determine whether new rounds of infection occurred after the initial challenge, we microinjected ten organoids with either GFP-expressing C. muridarum or C. trachomatis L2, imaged at 1, 3 or 5 days post infection, and quantified the number of inclusions per organoid. We observed a significant increase in the number of inclusions for both strains, indicating that Chlamydia completes its lifecycle within the endometrial epithelia and that released bacteria infect other cells (Fig. 3G–I).
The reorganization of the Golgi apparatus in infected endometrial epithelia requires the inclusion membrane protein InaC
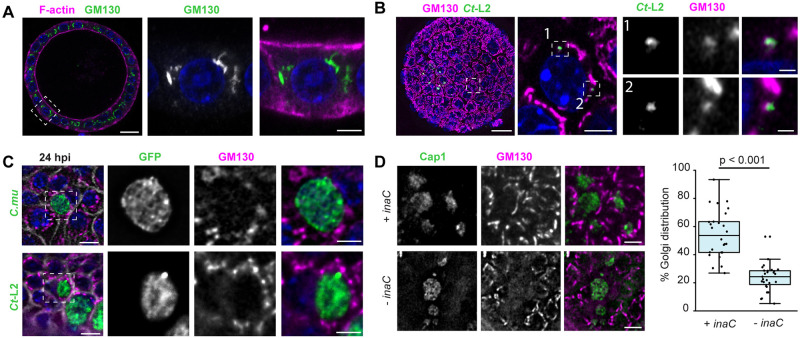
Host cell organelles are recruited to Chlamydia inclusions and are tethered in complexes to intercept lipid-rich vesicles and acquire nutrients (Elwell and Engel, 2012). The inclusion membrane proteins IncD, InaC (also known as CT813) and Cdu1 target Golgi-resident proteins and potentially regulate and/or intercept polarized trafficking (Agaisse and Derré, 2014; Derré et al., 2011; Kokes et al., 2015; Pruneda et al., 2018). During epithelial polarization, the Golgi apparatus migrates and expands at sites proximal to the apical membrane, where it regulates the polarized sorting of vesicles and proteins to distinct membrane domains (Bacallao et al., 1989). Using high-resolution confocal microscopy, we observed that the Golgi apparatus was more widely dispersed in EMO epithelial cells than in HeLa cells, often surrounding the nucleus and extending toward the apical surface (Fig. 4A). Consistent with previous studies (Heuer et al., 2009; Kokes et al., 2015; Pokrovskaya et al., 2012; Pruneda et al., 2018; Rejman Lipinski et al., 2009), we found that the Golgi apparatus was recruited and reorganized at the periphery of early inclusions (Fig. 4B).
Fig. 4.
Chlamydia infections causes reorganization of the Golgi apparatus in endometrial epithelia. (A,B) The organization of the Golgi apparatus in EMOs. (A) EMO stained for F-actin and GM130, as a Golgi marker, and imaged by confocal microscopy. Box denotes region shown in enlarged panels. Scale bars: 20 µm (left), 5 µm (right). (B) EMO infected with GFP-expressing C. trachomatis L2 (Ct-L2) for 20 h and stained for GM130, as a Golgi marker, imaged by confocal microscopy. Left, maximum intensity projection. Dashed box denotes region of interest showing relative localization of GM130 with respect to small, early inclusions in a single confocal section (middle). Numbered boxes indicate regions shown in enlarged panels on the right. Scale bars: left, 20 µm; middle, 5 µm; right, 1 µm. (C) The Golgi apparatus relocalizes to the periphery of C. muridarum (C.mu) and C. trachomatis inclusions at mid infection cycle. 3D deconvolution microscopy of tdTomato-expressing EMOs infected with GFP-expressing C. muridarum (top) or C. trachomatis L2 (bottom) for 24 h and stained for GM130. Scale bars: 10 µm. Enlarged region of interest (dashed box) shows relative localization of GM130 with respect to the inclusion. Scale bars: 5 µm. (D) InaC is required for Golgi repositioning in infected EMO cells. EMOs were infected with an inaC mutant M407 complemented with a plasmid expressing InaC (+ inaC) or transformed with an empty vector (− inaC) for 24 h. Samples were stained for GM130 as a Golgi marker and Cap1 for the inclusion. Scale bars: 10 µm. The percentage of Golgi distribution around the inclusion perimeter was quantified (n=26–27 inclusions). Boxplots show the median and interquartile range (IQR), with the bottom whiskers indicating the minimum value and the top whiskers indicating Q3+1.5×IQR. Statistical significance was measured using an unpaired, two-tailed Student's t-test. For all images, DNA was stained using 2 µg/ml Hoechst (blue).
Both C. muridarum and C. trachomatis inclusions recruited Golgi fragments to the inclusion periphery (Fig. 4C). Because the recruitment of Golgi stacks to the inclusion in C. trachomatis and is regulated by effector proteins such as InaC, we next determined whether this phenotype is recapitulated in inaC mutants. EMOs were infected with the C. trachomatis mutant M407 (bearing an InaC truncation, InaCQ103*), M407 complemented with inaC on a plasmid, or an empty vector control (Kokes et al., 2015). The InaC-deficient C. trachomatis failed to promote the redistribution of Golgi fragments around the inclusion of infected EMO cells, whereas infection with the complemented mutant resulted in a distribution similar to that of the wild-type-infected EMO cells (Fig. 4D). Collectively, these data show that despite the difference of Golgi complex morphology in polarized EMO epithelia, this organelle is still repositioned at the inclusion periphery and that this process is mediated by the same effectors.
CpoS protects endometrial epithelia from Chlamydia-induced cell death
Pathogen-mediated interference with the function of organelles can disrupt the activation of cell-autonomous immunity. For example, host recognition of microbial DNA induces the translocation of STING (stimulator of interferon genes) from the ER to the Golgi, to initiate signaling events important for type I interferon responses (Ishikawa et al., 2009). Chlamydia-infected cells activate STING (Barker et al., 2013; Prantner et al., 2010), but its translocation from the ER to post-Golgi compartments is inhibited by the Inc CpoS (also known as CT229; Sixt et al., 2017). In addition, CpoS is among a subset of Incs recently identified to promote inclusion integrity and the maintain the viability of infected cells (Sixt et al., 2017; Weber et al., 2017).
To monitor infection-dependent cell death in EMOs, we microinjected EMOs with C. trachomatis L2 in the presence of propidium iodide (PI) and imaged live at 24 h post infection. In both uninfected and infected EMOs we observed little to no PI staining, indicating that Chlamydia efficiently blocks the induction of cell autonomous cell death-mediated defense (Fig. 5A). We next tested whether C. trachomatis cpoS mutants would lead to cell death in EMOs by microinjection of the C. trachomatis mutant M007, encoding a truncated CpoSQ31*. Endometrial epithelial cells containing CpoS-deficient inclusions frequently stained positive for PI, indicating the activation of cell death (Fig. 5B). We measured the extent of PI-positive cells in each infected EMO and the percentage of PI-positive inclusions, both of which were consistently higher in EMOs infected with cpoS mutants (Fig. 5C,D). We next performed time-lapse microscopy of an EMO infected with GFP-expressing cpoS mutants in the presence of PI. Consistent with previous findings (Sixt et al., 2017), we observed sequential lysis of the inclusion followed by swelling of the infected cell, which was then extruded into the lumen before losing membrane integrity (Fig. 5E; Movie 5), and inclusions gradually becoming labeled with PI (Fig. 5E; Movie 6). These observations highlight that the molecular mechanisms used by Chlamydia to protect infected cells from the activation of cell death-based defense responses is conserved in endometrial epithelial cells.
Fig. 5.
The C. trachomatis effector CpoS blocks cell death in infected endometrial epithelia. (A) EMO cells remain viable after C. trachomatis infection. Live spinning-disk confocal images of an uninfected EMO and an EMO infected with GFP-expressing C. trachomatis (Ct-L2) for 24 h in medium containing propidium iodide (PI). Scale bars: 50 µm. (B–D) EMOs infected with a C. trachomatis cpoS mutant induce cell death. (B) EMOs were infected with a GFP-expressing cpoS mutant (M007), incubated with PI for 22 h and imaged by live 3D deconvolution microscopy. Dashed box indicates the region shown in the zoom image. Dashed line outlines the EMO. Scale bars: 50 µm; zoom, 5 µm. (C) Maximum projection images of PI staining from 3D spinning-disk confocal images of EMOs infected as in B with either CT-L2 or M007. Dashed lines outline the EMOs. Scale bars: 50 µm. (D) Boxplots showing the number of PI puncta per EMO and the percentage of PI-positive inclusions (n=10 EMOs per replicate). Boxplots show the median and interquartile range, with whiskers indicating the minimum and maximum values. For the L2 data in the right-hand plot, only the median line is shown; because most of the values are zero, this line overlaps with the x axis. Statistical significance was measured for each replicate using a Welch's t-test. (E) Live imaging of cell death in EMOs. Time-lapse spinning-disk confocal microscopy of an EMO infected with GFP-expressing cpoS mutants (M007) cultured in the presence of PI. Left panels: maximum projections at the beginning (top) and end (bottom) of image acquisition. Dashed boxes mark regions of interest. Scale bars: 50 µm. Right panels: enlarged regions of interest at the indicated time points showing (1) an inclusion lysing before the infected cell becomes PI-positive (white arrowheads), and (2) an inclusion gradually becoming positive for PI (white arrowheads). Time display (h:min). Scale bars: 20 µm.
Reconstitution of immune cell recruitment to Chlamydia-infected EMOs
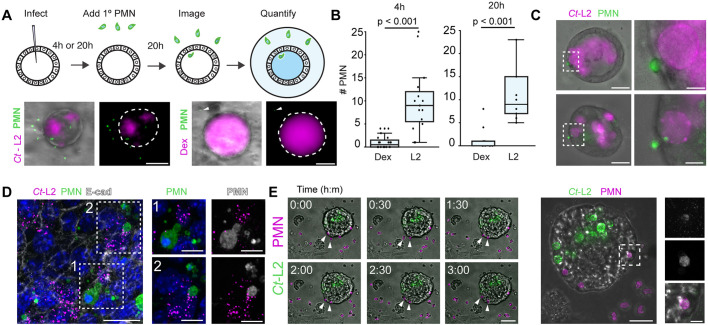
Immune cells, including neutrophils, are recruited to Chlamydia-infected sites where they can interact with and even invade epithelia to make direct contact with inclusions (Rank et al., 2011). Neutrophil recruitment and activation at infection foci can damage the UGT (Lacy et al., 2011; Lee et al., 2010a; Lijek et al., 2018), but experimentally tractable models to investigate the dynamics of their behavior are lacking. We extended the utility of the Chlamydia–EMO infection model by reconstituting the engagement of immune cells. We co-cultured infected EMOs with primary bone marrow-derived neutrophils and used high-resolution and time-lapse microscopy to quantify and visualize their recruitment to EMOs and subsequent interactions with infected epithelial cells. EMOs were microinjected with mCherry-expressing C. trachomatis L2 or fluorescently labeled dextran. At 4 or 20 h post infection, fluorescently labeled neutrophils (using CellTracker) were added to the medium for an additional 20 h prior to imaging (Fig. 6A). Neutrophils infiltrated the Matrigel and migrated specifically to infected EMOs (Fig. 6A,B). EMOs microinjected with fluorescent dextran failed to recruit a significant number of neutrophils, indicating that the transient damage from the microinjection does not induce a chemoattracting response (Fig. 6B). We observed that a subset of neutrophils made contact directly with infected EMO epithelial cells, and even inclusions, but did not appear to undergo death or NETosis (Fig. 6C,D). Using time-lapse spinning-disk microscopy, we visualized neutrophil dynamics at infected EMOs and again observed a subset of neutrophils interacting with the infected EMO and making contact with inclusions (Fig. 6E; Movie 7).
Fig. 6.
Neutrophils are recruited to infected organoids. (A) Schematic representation of EMO infections and addition of neutrophils/polymorphonuclear (PMN) cells. Representative widefield microscopy images (bottom) of EMOs infected with C. trachomatis (Ct-L2, left) or microinjected with Texas Red–dextran (Dex, right), cultured for 20 h and subsequently incubated with 5-chloromethylfluorescein diacetate (CFMDA)-labeled primary neutrophils (PMN) for an additional 20 h. Dashed lines outline the EMOs. Arrowheads indicate a single PMN proximal to the EMO. Scale bars: 50 µm. (B–D) PMNs are specifically recruited to infected EMOs and make contact with infected epithelial cells and intracellular inclusions. EMOs were either injected with Dex or infected with Ct-L2 for either 4 h or 20 h, co-cultured with PMNs for an additional 20 h, then imaged by widefield microscopy. (B) Quantification of PMN recruitment to the periphery of infected EMOs at the indicated times (4 h post infection, n=15–20 EMOs; 20 h post infection, n=10 EMOs). Boxplots show the median and interquartile range (IQR), with the bottom whiskers indicating the minimum value and the top whiskers indicating Q3+1.5×IQR. Statistical significance was measured using an unpaired, two-tailed Student's t-test. (C) Widefield microcopy images of tdTomato-expressing PMNs (green) making contact with infected epithelial cells. Dashed boxes indicate regions shown in enlarged panels (right). Scale bars: left, 25 µm; right, 5 µm. (D) EMOs infected with Ct-L2 (magenta) and co-cultured with PMNs (green) were stained for E-cadherin (E-cad) and imaged by confocal microscopy. Boxes indicate regions of interest shown on the right. Scale bars: left, 20 µm; right, 10 µm. (E) Left panels: time-lapse microscopy of PMN recruitment to infected EMOs. EMOs were infected with GFP-expressing Ct-L2, co-cultured with tdTomato-expressing PMNs for 20 h, and imaged by spinning-disk confocal microscopy. Arrowheads denote PMNs moving towards and interacting with an infected EMO. Scale bar: 50 µm. Middle: maximum projection of higher magnification images of an infected EMO. Dashed box denotes region of interest shown enlarged on the right. Scale bar: 20 µm. Right: region of interest showing PMN colocalizing with Ct-L2 within the EMO. Scale bar: 5 µm.
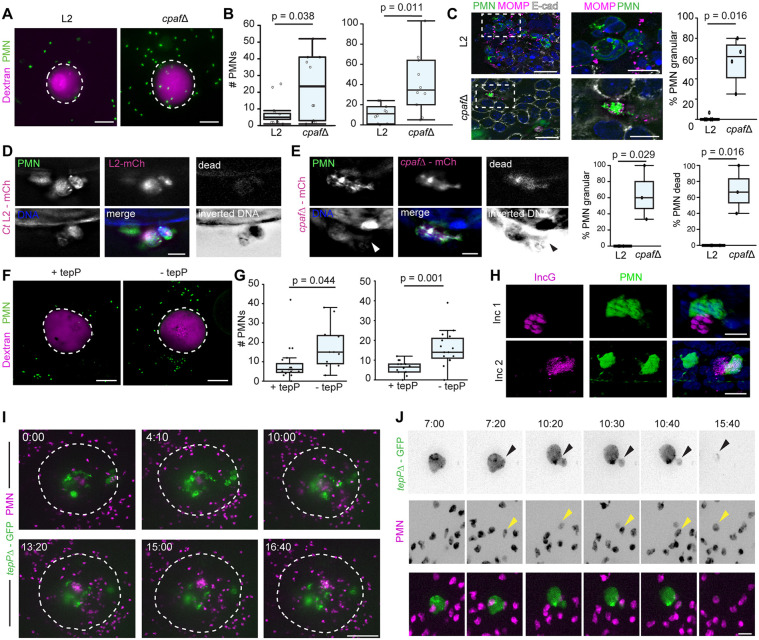
Chlamydia has evolved strategies to limit the function of neutrophils. For example, C. trachomatis can inhibit human and mouse neutrophil activation and cell death using the secreted effector chlamydial protease-like activating factor (CPAF) (Zhong et al., 2001), which targets and cleaves the neutrophil surface receptor formyl peptide receptor 2 (FPR2) to inhibit neutrophil chemotaxis and NETosis (Rajeeve et al., 2018). Thus, we first tested whether CPAF-deficient C. trachomatis (Schott et al., 2020) affects neutrophil recruitment, localization and activation in our model. Indeed, EMOs infected with the cpaf mutants showed enhanced neutrophil recruitment compared to wild-type Chlamydia (Fig. 7A,B; Movie 8). Using high-resolution microscopy, we found that neutrophils in contact with wild-type Chlamydia maintained their polymorphic nuclei (Fig. 7C), whereas neutrophils in contact with CPAF-deficient inclusions displayed a granular morphology including large blebs (Fig. 7D). To test whether neutrophils undergo NETosis and cell death, we labeled EMO co-cultures in situ with a DNA stain and a fixable live/dead marker. We determined that neutrophils in contact with inclusions from cpaf mutants exhibited diffusive DNA signal and had lost membrane integrity (Fig. 7E).
Fig. 7.
Chlamydia secreted effectors dampen the recruitment and activation of neutrophils. (A–C) CPAF limits the recruitment of neutrophils/polymorphonuclear cells (PMNs) to infected EMOs. EMOs were infected with either wild-type C. trachomatis (L2) or a cpaf mutant (cpafΔ) and Texas Red–dextran as a marker of microinjection. EMOs were then co-cultured with CFMDA-labeled PMNs at 4 h post infection and imaged by 3D deconvolution widefield microscopy 20 h later. (A) Representative maximum projection images show PMNs recruited to infected EMOs. Dashed lines outline the EMOs. Scale bars: 100 µm. (B) Quantification of two independent experiments as described in A (n=10 EMOs per infection). (C) EMOs infected with wild-type C. trachomatis L2 or cpaf mutant and co-cultured with CFMDA-labeled PMNs at 48 h post infection. Widefield deconvolution images (left) of EMOs stained for E-cadherin and Chlamydia stained for MOMP. Dashed boxes indicate regions shown in enlarged images (middle). Scale bars: left, 20 µm; middle 10 µm. Right, quantification of granular phenotype (n=4–6 EMOs, 4–25 PMNs per EMO). (D,E) PMNs undergo NETosis when in contact with CPAF-deficient Chlamydia. EMOs infected with mCherry (mCh)-expressing C. trachomatis L2 (Ct L2) (D) or cpaf mutants (E) were labeled in situ with Hoechst (to stain DNA) and a live/dead marker (dead, grayscale) at 68 h post infection and fixed. Arrowhead indicates diffuse DNA signal. Scale bars: 10 µm. Right: the granular phenotype and percentage of dead PMNs were quantified and plotted (n=3 EMOs, 4–14 PMNs per EMO) (F–H) TepP limits the recruitment of PMNs to infected EMOs. EMOs were infected with a TepP mutant complemented with an empty vector (−tepP) or a vector expressing TepP (+tepP), and Texas Red–dextran was used as marker of microinjection. EMOs were then co-cultured with CFMDA-labeled PMNs at 4 h post infection and imaged by 3D spinning-disk confocal microscopy 20 h later. (F) Representative maximum projection images show PMNs recruited to infected EMOs. Dashed lines outline the EMOs. Scale bars: 100 µm. (G) Quantification of PMN recruitment for two independent experiments. Left, n=at least 15 EMOs per infection; right, n=10 EMOs per infection. (H) Two 3D reconstructions of confocal images showing PMNs (green) in contact with inclusions of a Chlamydia tepP mutant complemented with empty vector (labeled by staining for IncG, magenta) at 48 h post infection in EMOs labeled for DNA (Hoechst). Scale bars: 10 µm (I,J) Live imaging of neutrophil recruitment to EMOs infected with Chlamydia tepP mutants. EMOs were infected with GFP-expressing tepP mutants (tepPΔ–GFP), co-cultured with tdTomato-expressing PMNs (magenta) and imaged by time-lapse spinning-disk microscopy at 48 h post infection. (I) Maximum projection images of PMNs swarming and entering the lumen of an organoid infected with a GFP-expressing tepP mutant. Dashed line outlines the EMO. Time display (h:min). Scale bar: 100 µm. (J) Maximum projection images of a GFP-expressing tepP mutant inclusion budding, and interacting with and being engulfed by PMNs (magenta). Arrowheads point to PMNs embracing the inclusion and remaining in contact with the inclusion as it buds. Time display (h:min). Scale bar: 20 µm. Boxplots show the median and interquartile range, with whiskers indicating the minimum and maximum values. Statistical significance was measured using a Welch's t-test.
Infected epithelial cells secrete IL-6, IL-8, and members of the CXC chemokine family that can drive the recruitment of neutrophils towards Chlamydia-infected cells (Dessus-Babus et al., 2000; Frazer et al., 2011; Rasmussen et al., 1997). In mice, the absence of CXCR2, the CXC chemokine receptor, reduces acute inflammation and pathology in the UGT without affecting bacterial burden (Lee et al., 2010b). A transcriptional analysis of infected endocervical epithelial cells indicated that the Chlamydia effector TepP enhances early type I interferon responses and dampens the expression of IL-6 and CXCL3 (Chen et al., 2014), chemokines that promote neutrophil chemotaxis (Fielding et al., 2008; Wright et al., 2014). We tested whether TepP played a role in in regulating neutrophil migration by infecting EMOs with a TepP-deficient C. trachomatis mutant generated by targeted insertion of an aadA cassette into the tepP locus (Fig. S2). This mutant was then transformed with a plasmid expressing TepP under its native promote (+tepP) or an empty vector (−tepP), and the resulting strains were microinjected into EMOs followed by co-culturing with primary neutrophils at 4 h post microinjection. Infected EMOS were imaged after 20 h. C. trachomatis strains lacking TepP showed significantly more neutrophil recruitment to the EMO, suggesting that TepP dampens the influx of neutrophils to infections sites (Fig. 7F,G). In contrast to the cpaf mutant, high-resolution microscopy showed that neutrophils in contact with tepP mutant inclusions were morphologically indistinguishable from those that made contact with wild-type inclusions (Fig. 7H). We next performed live imaging of neutrophils with EMOs infected with a GFP-expressing tepP mutant at 48 h post infection. Long-term time-lapse microscopy showed two interesting behaviors not observed with wild-type or CPAF-deficient C. trachomatis: (1) neutrophils swarming towards the infected EMO, transmigrating into the lumen and aggregating as they contact inclusions and/or EBs extruded into the lumen of organoids (Fig. 7I; Movie 9); and (2) neutrophils engaging intracellular inclusions and releasing inclusion fragments that are then engulfed by a neutrophil (Fig. 7J; Movie 10).
Collectively, these data show that we can reconstitute the infiltration of innate immune cells to infected epithelia and address the role of Chlamydia virulence factors regulating this process.
DISCUSSION
We developed a primary organoid model system to characterize the interactions between Chlamydia and epithelial cells in a cellular context that better mimics the architecture and diversity of the UGT epithelia. The UGT is a complex tissue with a unique immunoregulatory system because of its role in tolerance during pregnancy and because of the proliferative capacity and altered physiology of cells in response to hormonal stimulation. Although some aspects of this complexity can be reconstituted in EMOs, how hormonal changes impact the cell biology of Chlamydia–host cell interactions in the UGT is largely unexplored. Despite differences in the cytoskeletal and endomembrane organization in EMO cells compared to those in traditional 2D tissue culture systems, we found remarkable conservation in the type of intracellular processes targeted by Chlamydia and in the effector proteins used. Our observations complement other recently reported ex vivo models, including a fallopian tube organoid infection model (Kessler et al., 2019) and an endometrial organoid infection model that shows Chlamydia replicates within EMO epithelia and is amenable to inducible expression of Chlamydia Incs (Bishop et al., 2020). These models, however, do not recapitulate infections via the apical side of polarized epithelial cells within organoids, which precludes the ability to monitor the natural route of Chlamydia invasion.
We provide evidence that Chlamydia reprograms the epithelial cytoskeleton and disrupts epithelial structure, at least transiently, during apical invasion and inclusion formation. These results are in agreement with infections in fallopian tube explants where Chlamydia infection disrupts epithelial architecture, altering the organization of polarity proteins in infected and uninfected neighboring cells (Kessler et al., 2012). Although we observed marked recruitment of β-catenin to early inclusions and Chlamydia entry sites, this localization did not occur in mid-to-late stage inclusions. Nevertheless, β-catenin promotes Chlamydia infection, possibly through Wnt-based signaling, as pharmacological inhibition of Wnt signaling in transformed endometrial epithelial cells reduces Chlamydia growth (Kintner et al., 2017).
An advantage of EMOs lies in the ability to generate organoids from a variety of transgenic mouse lines deficient for host proteins of interest or expressing fluorescently tagged proteins. For instance, we used EMOs from ROSAmTmG mice to better delineate single cell boundaries when following the dynamics of inclusions exiting infected cells and ZO-1–GFP to track tight junctions. Whereas all of the C. muridarum inclusions exited apically, C. trachomatis L2 inclusions were able to exit through the basolateral membrane, reminiscent of observations made in polarized tamoxifen-treated endometrial cancer cells infected with C. trachomatis serovar E (Hall et al., 2011). It is unclear whether the exit strategy depends on the organization of polarized epithelial cells, cell-type-specific responses or serovar-specific functions. Because EMOs are comprised of multiple epithelial subtypes (Fitzgerald et al., 2019), this model can be used to identify cell-type-specific responses to infection.
We extended the application of EMOs to monitor the interaction between primary immune cells and the infected epithelium. The rapid and specific recruitment of neutrophils to infected EMOs indicates that this model can be applied to study the dynamics of immune cell interactions with Chlamydia-infected cells and the role played by host factors that regulate the innate immune responses. Neutrophils appear to have a limited role in clearing experimental infections, but rather influence the adaptive immune response by promoting the recruitment of T cells (Lacy et al., 2011). Studies using a combination of host and Chlamydia genetics can further dissect the interaction between Chlamydia effectors and the innate and adaptive immune responses. For instance, CPAF-dependent inhibition of neutrophils (Rajeeve et al., 2018) and the suppression of the T cell chemoattractant CXCL10 (Schott et al., 2020) can be modeled using organoids and immune cells from mice harboring mutations in CPAF targets.
How TepP influences neutrophil behaviour, and whether neutrophils can more effectively clear TepP mutants in vivo, remains to be determined, and these studies will need to account for the likely possibility that Chlamydia employs redundant factors to limit neutrophil responses. Because neutrophil infiltration is regulated by type I and type III interferons (Blazek et al., 2015; Seo et al., 2011), TepP-dependent regulation of the interferon response and chemokine expression may function in part to reduce immune cell infiltration and promote survival in the UGT. Our studies lay the foundation to investigate how TepP regulates neutrophil swarming and aggregation, behaviors that are dependent upon leukotriene signaling (Lämmermann et al., 2013). Of note, C. trachomatis is a human-adapted pathogen that differs from the rodent-adapted C. muridarum in its ability to counteract cell-autonomous immunity (Finethy and Coers, 2016). Future studies using C. muridarum mutants will be needed to better define the role of virulence factors in regulating immune responses.
In conclusion, we provide a high-resolution blueprint to study the cell biology of Chlamydia infections in primary endometrial organoids. This foundation will provide a framework for future studies that target host and pathogens to better identify how Chlamydia subverts epithelial biology and the innate immune response.
MATERIALS AND METHODS
Ethics statement
All animal experiments were approved and performed in accordance to the Duke University Institutional Animal Care and Use Committee.
Cell lines and conditioned medium
Vero cells were purchased from ATCC (CCL-81) and cultured in Dulbecco's Modified Eagle's Medium (DMEM; Life Technologies) containing 10% fetal bovine serum (FBS; Sigma-Aldrich). L-WRN cells were purchased from ATCC (CRL-3276) and cultured in DMEM containing 0.5 mg/ml geneticin (Gibco) and 0.5 mg/ml hygromycin B (Thermo Fisher Scientific) at 37°C with 5% CO2. Prior to generating conditioned medium, the cells were passaged twice in medium without antibiotics. Conditioned medium was generated as previously described (Miyoshi and Stappenbeck, 2013). In brief, cells were plated in T175 tissue culture treated flasks and grown to greater than 90% confluence. 25 ml of 1:1 DMEM/F12 (Gibco) medium was collected every 24 h, centrifuged at 500 g for 5 min, and stored at 4°C. At the end of 5 days, the media were combined, sterile filtered and stored at −80°C. All cells were cultured in the presence of ciprofloxacin for two passages (∼1 week) prior to cryopreservation and PCR-verified to be free of Mycoplasma contamination.
Chlamydia strains, propagation and transformation
C. trachomatis strains L2/434/Bu (CTL2; ATCC VR-902B), the chemical mutants M007, M407 and M923 (Kokes et al., 2015; Sixt et al., 2017), and a Δcpaf::cat mutant (Schott et al., 2020) were propagated in Vero cells, harvested at 44–48 h post infection by water lysis and sonication, then were diluted in SPG (sucrose-phosphate-glutamate) buffer to 1× concentration (75 g/l sucrose, 0.5 g/l KH4PO4, 1.2 g/l Na2HPO4 and 0.72 g/l glutamic acid, pH 7.5) and stored as single use aliquots at −80°C. The C. muridarum strain CM006 was a gift from Catherine O'Connell (University of North Carolina, Chapel Hill, NC), and was propagated in Vero cells and harvested at 36 h post infection as above. CM006 was transformed with pSW2NiggCDS2 (Wang et al., 2014; gift from Dr Ian Clarke, University of Southampton, Southampton, UK) as follows: Approximately 107 inclusion-forming units (IFU) were incubated with 10 µg DNA in buffer containing 0.9 mM calcium chloride for 30 min, added to confluent Vero cells in a six-well plate and centrifuged at 1500 g for 30 min at 10°C. At 12 h post infection, 1 U/ml penicillin was added. The infections were passaged every 36 h until inclusions were present and fluorescent. Transformants were subsequently plaque-purified to obtain a clonal strain.
To generate the spectinomycin-resistant TepP-deficient C. trachomatis, CTL2 was transformed using the TargeTron gene disruption system (Sigma-Aldrich) as previously described (Carpenter et al., 2017) with the exception that an aadA cassette was inserted using the same tepP targeting sequences. Transformants were expanded in Vero cells in the presence of 150 µg/ml spectinomycin, as described above, plaque purified, and gene disruptions were verified by PCR analysis using primers flanking the insertion site and the aadA cassette. The ΔtepP::aadA strain was complemented with the E.coli–Chlamydia GFP-expressing, shuttle vector p2TK2-SW2 (Agaisse and Derré, 2013) containing a bla cassette and the TepP open reading frame and its upstream promoter sequence (Carpenter et al., 2017) or p2TK2-SW2 alone. In brief, 108 IFU were incubated with 10 µg DNA in buffer containing 0.9 mM calcium chloride for 30 min, added to confluent Vero cells in a six-well plate, and centrifuged at 1500 g for 30 min at 10°C. At 12 h post infection, 1 U/ml penicillin was added. The infections were passaged every 48 h until inclusions were present, at which point the penicillin concentration was increased to 10 U/ml. Transformants were subsequently plaque-purified in the presence of both spectinomycin and penicillin to obtain a clonal strain and verified by PCR using primers targeting both the aadA and bla cassette and the full-length tepP sequence. The Δcpaf::cat mutant (Schott et al., 2020) was transformed as above with the mCherry-expressing p2TK2-SW2 (Agaisse and Derré, 2013).
Western blotting
For western blots, Vero cells were infected with the indicated strains for 48 h, water lysed, sonicated and boiled in Laemmli buffer for 10 min. The lysates were centrifuged at 9600 g for 5 min, and equal volumes were resolved using a 10% SDS–PAGE gel. The resolved proteins were transferred to a nitrocellulose membrane (Amersham), blocked with a phosphate-buffered saline solution containing 5% milk for 1 h at 25°C, and sequentially probed with primary anti-TepP (1:500; Chen et al., 2014) and anti-Slc1 (1:1000; Chen et al., 2014) antibodies and Li-COR infrared dye-conjugated secondary antibodies (1:10,000). Membranes were imaged using a Li-COR Odyssey system (Li-COR Biosciences).
EMO generation and hormone stimulation
C57BL/6J (strain no. 000664) and B6.129(Cg)-Gt(ROSA)26SORtm4(ACTB-tdTomato,-EGFP)Luo/J (strain no. 007676) mice were purchased from The Jackson Laboratory. ZO-1 GFP knock-in mice (Foote et al., 2013) were a generous gift from Dr Terry Lechler (Duke University, Durham, USA). The endometria from ∼6–12-week-old females were dissected, washed in cold Dulbecco's phosphate-buffered saline (PBS; Gibco) on ice, cleaned of vascular and fat tissue, minced into ∼2 mm pieces, transferred to DMEM (Gibco) containing 0.2% collagenase A (Thermo Fisher Scientific), 10% FBS and 1 U/ml penicillin/streptomycin (Gibco), then incubated on a shaker at 37°C for 2.5–3 h. The tissue was washed three times with cold PBS and mechanically disrupted by shaking in cold 10 ml PBS containing 0.1% bovine serum albumin (BSA; Fraction V, Equitech-Bio) for 1 min. After allowing the tissue to settle for one minute, the supernatant was collected and passed through a 70 µm strainer (Falcon). The mechanical disruption was repeated once more, and the strainer was subsequently inverted over a new 50 ml conical tube and the epithelia were washed off the strainer three times with 10 ml PBS containing 0.1% BSA. The epithelial fraction was centrifuged at 600 g for 5 min at 10°C, resuspended in cold DMEM/F12 (Gibco), mixed with low-hormone Matrigel (Corning) at a 1:1 ratio and pipetted in 35 µl drops in a 24 well plate or 125 µl drops in a 35 mm glass-bottom dish (for microinjections). The plates were incubated at 37°C for 40 min before gently overlaying 2 ml 1:1 DMEM/F12 medium containing 50% L-WRN conditioned medium, 50 µg/ml gentamicin (Gibco) and 50 ng/ml EGF (StemCell Technologies). The medium was changed every 2–3 days; the gentamicin concentration was reduced to 10 µg/ml after the second medium change. For hormonal stimulation studies, EMOs were cultured in the presence of 20 nM 17-β-estradiol (E2, diluted in ethanol; Sigma-Aldrich) for 5 days.
Primary stromal fibroblast culture and infections
To culture primary stromal fibroblasts, the cell suspension that was collected through the 70 µm strainer was centrifuged at 600 g for five minutes, resuspended in 50% conditioned medium containing 50 µg/ml gentamicin, and plated on 12 mm #1 coverslips (Bellco Glass, Product No. 1943-10012A). Prior to plating, the coverslips were incubated with 30 µg/ml Type I rat collagen (Thermo Fisher Scientific; A1048301) diluted in 0.1% acetic acid for five minutes and washed twice with medium. When the cells reached confluence, GFP-expressing C. muridarum (CM006) or CTL2 were diluted in medium at an MOI<1, vortexed for 15 s, centrifuged onto the cells at 1500 g for 20 min at 10°C, and re-fed with warm conditioned medium.
EMO microinjection
Borosilicated capillary glass needles with filament (Sutter Instrument; BF100-58-10) were pulled with a WPI PUL-1000 system (World Precision Instruments). Chlamydia strains were diluted in PBS to a final concentration of 5×105–5×106 IFU/ml, vortexed for 30 s, and pipetted into the glass needle. Microinjections were performed using an Eppendorf FemtoJet 4× coupled with a Stereo Microscope (Nikon). Prior to microinjections, the EMOs were re-fed with media without gentamicin. EMOs were punctured once using a steep vertical angle and manually injected with equal volumes. When organoids were injected alone or co-injected with 3 kDa Texas-Red–dextran (Invitrogen), dextran was used at a final concentration of 0.01 mg/ml. The EMOs were re-fed with media containing 10 µg/ml gentamicin 1 h post injection.
Primary neutrophil isolation and co-culture with infected EMOs
To isolate primary neutrophils, mouse femurs were dissected, cleaned with an ethanol-soaked Kimwipe to remove excess tissue, dipped in 70% ethanol then twice in RPMI medium (Gibco). Both ends of the femur were cut and 10 ml RPMI medium was passed through the bone with a 25 G needle (BD Biosciences). The supernatant was centrifuged at 600 g for 5 min, resuspended in 5 ml red cell lysis buffer (Millipore), and incubated for 5 min. An additional 5 ml of RPMI medium was added to the cells before centrifugation at 600 g for 5 min. The cells were resuspended gently in 0.5 ml RPMI medium before following the negative-selection EasySep Mouse Neutrophil Enrichment kit (StemCell Technologies, Cat No. 19762). Neutrophils were labeled with 2.5–5 µM CellTracker (5-chloromethylfluorescein diacetate, CFMDA; Thermo Fisher Scientific) for 10 min at 37°C, centrifuged at 300 g for 10 min at 4°C and resuspended in 0.5 ml cold RPMI. Neutrophils (3×105 cells) were added directly to the medium and cultured for an additional 20 h. For time-lapse microscopy, 6×105 neutrophils were added.
Antibodies
For immunofluorescence microscopy, the following antibodies were used: mouse anti-GM130 (BD Biosciences, cat no. 610822; 1:200), anti-α-tubulin (Sigma-Aldrich, cat no. T5618, clone B-5-1-2; 1:200), anti-MUC1 (Cell Signaling Technologies, cat no. VU4H5; 1:100), anti-β-catenin (BD Biosciences, cat no. 610153, clone 14; 1:400), anti-MOMP (Santa Cruz, cat no. 57678; 1:500), anti-pan-keratin (Sigma-Aldrich, cat no. C2562; 1:100), anti-vimentin (Sigma-Aldrich, cat no. V5255, clone VIM-13.2; 1:100) and rabbit anti-Slc1 (generated and validated in Chen et al., 2014), anti-E-cadherin (Cell Signaling Technologies, cat no. 3195S, clone 24E10; 1:200), anti-acetylated-α-tubulin (Cell Signaling Technologies, cat no. 5335S; 1:200), anti-SEPT2 (Protein Tech, cat no. 11397-1-AP; 1:100), anti-Cap1 (gift from Agathe Subtil, Institut Pasteur, Paris, France; 1:250) and anti-IncG (gift from Ted Hackstadt, Rocky Mountain Laboratories, NIAID, Hamilton, USA; 1:250). The following secondary antibodies from Thermo Fisher Scientific were used: cross-adsorbed goat anti-mouse (H+L) and goat anti-rabbit (H+L) conjugated to Alexa Fluor 488 (A-11029; A-11008), Alexa Fluor 555 (A-21428; A21422) or Alexa Fluor 647 (A-21244; A-21236.) All secondary antibodies were diluted 1:1000, vortexed gently and centrifuged at 8000 g for 1 min.
Brightfield and immunofluorescence microscopy
EMO growth was monitored by brightfield microscopy using the EVOS FL Cell Imaging System (Thermo Fisher Scientific) equipped with 2×/0.06 and 10×/0.25 NA objectives and a CCD camera. For fixed samples, EMOs were rinsed twice with warm PBS and incubated with warm PBS containing 3% formaldehyde (Sigma-Aldrich) for 20 min. The fixative was removed gently, and the organoids were resuspended in 0.25% ammonium chloride (Sigma-Aldrich) and transferred to a 1.5 ml tube, centrifuged at 600 g for 5 min, resuspended gently in 2% BSA (Sigma-Aldrich) containing 0.1% Triton X-100 and incubated with gentle rocking for 30 min. EMOs were centrifuged again at 500 g for 5 min, incubated with the indicated primary antibodies diluted in 0.5 ml 2% BSA containing 0.1% Triton X-100, and incubated at 25°C for 2–3 h or overnight at 4°C with gentle rocking. EMOs were washed once with 2% BSA, centrifuged at 500 g for 5 min, and incubated with secondary antibodies diluted in 1.0 ml 2% BSA containing 0.1% Triton X-100 for 1.5 h at 25°C with gentle rocking. Phalloidin conjugated to Alexa Fluor 555 (1:500; Acti-stain, Cytoskeleton, Inc) and Hoechst (2 µg/ml; Thermo Fisher Scientific) were added for the final 20 min of incubation with the secondary antibodies. The EMOs were centrifuged at 600 g for 5 min and resuspended in∼35 µl Vectashield (Vector Labs; H-1000) using a cut pipette tip and pipetted onto a glass slide. A coverslip was overlaid gently and sealed with nail polish.
Organoids were imaged using an inverted confocal laser scanning microscope (LSM 880; Zeiss) equipped with a motorized stage; Airyscan detector (Hamamatsu); and diode (405 nm), argon ion (488 nm), double solid-state (561 nm) and helium-neon (633 nm) lasers. Images were acquired using a 20× Plan Apochromatic NA 0.8 air (Zeiss) or 40× C-Apochromatic NA 1.2 water objective (Zeiss) and were deconvolved using automatic Airyscan Processing in the Zen Software (Zeiss). Images were opened in ImageJ (NIH) or Imaris (Oxford Instruments) software, and exported TIFFs were rendered in the Adobe suites (Photoshop and Illustrator). Only linear adjustments were made to fluorescence intensity. For 3D deconvolution imaging, organoids were imaged on an inverted microscope (Ti2-E; Nikon Instruments) equipped with a motorized stage, an ORCA Flash 4.0 V3 sCMOS camera (Hamamatsu) and a SOLA solid-state white light illumination (Lumencor). Images were acquired using a 20× Plan Apochromatic NA 0.75 air objective or a 60× Plan Apochromatic NA 1.40 oil objective and deconvolved in the NIS-Elements Software (Nikon).
Stromal fibroblasts were rinsed twice with warm PBS, fixed with warm PBS containing 3% formaldehyde for 20 min, washed three times with 0.25% ammonium chloride, permeabilized with PBS containing 0.1% Triton X-100 for 10 min and blocked with 2% BSA for 20 min. All images were rendered in ImageJ (NIH) or Imaris. Exported TIFFs were reconstructed in the Adobe suites (Photoshop and Illustrator).
Time-lapse microscopy of inclusion dynamics, cell death and neutrophil recruitment
EMOs cultured in 35 mm glass-bottom dishes (Mat-Tek) were imaged live using an inverted microscope (Zeiss AxioObserverZ.1) equipped with a motorized stage containing a heated Insert P environmental chamber (Zeiss) and XLIGHT V2 spinning-disk unit (Crest Optics). Images were acquired using a 60× NA 1.4 oil objective (Olympus), a 20× NA 0.8 air objective (Olympus) or a 10× NA 0.4 air objective (Olympus); an LDI multiline laser (89 North); and an sCMOS ORCA Flash 4.0 V3 camera (Hamamatsu). Using the same microscope, neutrophil recruitment to infected EMOs was imaged using 5 μm confocal sections and spanning ∼100 µm above and below each organoid. Time-lapse microscopy of neutrophil motility was performed using a 20×/0.8 NA objective (Olympus). For all time-lapse microscopy, confocal sections (5 µm) were acquired every 10 min for the duration of ∼16 h, with the exception of Fig. 6E where confocal sections (2 µm) were acquired every 3 min for the duration of 3 h. All images were rendered in ImageJ. Exported TIFFs were reconstructed in the Adobe suites (Photoshop and Illustrator).
Analysis of cell death
To assess cell death with the cpoS mutant, organoids were microinjected with wild-type L2 or cpoS mutant (M007) C. trachomatis, incubated with medium containing 0.75 µM propidium iodide (PI; Immunochemistry Technologies, product no. 638) at 2 h post injection, and imaged using the spinning-disk microscope described above. Time-lapse imaging of an EMO infected with a GFP-expressing M007 mutant C. trachomatis was performed at 18 h post infection in the presence of 0.75 µM PI, and also imaged using the spinning-disk microscope described above; 5 µm confocal stacks were acquired every 10 min for ∼15 h.
To assess cell death in organoids co-cultured with neutrophils, as shown in Fig. 7, co-cultures were incubated with Fixable Viability Dye eFluor 660 (Thermo Fisher Scientific), a membrane impermeant dye that reacts with free amines, at the manufacturer's recommended dilution (1:1000) for 30 min at 37°C. Organoid co-cultures were washed twice with warm PBS and fixed with warm 3% formaldehyde diluted in PBS for 20 min. The formaldehyde solution was removed, and the organoids were gently washed twice with 0.5 ml ammonium chloride and twice with PBS. The co-cultures were imaged directly on the 35 mm glass-bottom dish using the 3D deconvolution microscope described above.
Image analysis
To quantify EMO size, microscopy images were imported into ImageJ and converted to 8-bit TIFFs. EMO borders were identified using the Max Entropy threshold algorithm. EMOs were individually selected, and the perimeters were measured. Assessment of β-catenin recruitment to early inclusions was performed manually in ImageJ. Inclusions with associated β-catenin signal were marked positive. Golgi reorganization around the inclusion was quantified in ImageJ as previously described (Pruneda et al., 2018). Microscopy images were imported and converted to 8-bit TIFFs. Four to five sections were projected to capture the entire inclusion. The inclusion edges were manually traced to measure the perimeter length. Using the segmented line tool, the GM130 signal directly adjacent to the inclusion was selected and traced to measure its length. To quantify cell death, microscopy images of EMOs incubated with PI were quantified in ImageJ. Microscopy images were imported and converted to 8-bit TIFFs. Maximum projection images were background subtracted using the rolling ball radius (value=50), and puncta were identified using the find maxima algorithm. Neutrophil recruitment to infected EMOs was quantified in ImageJ. Maximum projection images were background subtracted as above. To identify proximal neutrophils, a circle was placed around the inclusion 200 µm from the EMO basolateral edge. The number of neutrophils was identified using the find maxima algorithm. In Fig. 7, neutrophils were scored as granular based on morphology and as dead based upon fluorescence with the fixable viability dye. The number of granular and/or dead polymorphonuclear cells (PMNs) in contact with inclusions were counted and divided by the total number of PMNs in contact with inclusions. All measurements and values were exported to Microsoft Excel. Boxplots and scatterplots were generated in R software.
Statistical analyses
All statistical analyses were performed in either Microsoft Excel or R Studio. In R, datasets were measured for equal variance using the Levene test. Datasets with normal variance were further analyzed for significance using a two-tailed Student's t-test. Datasets showing unequal variance were further analyzed for significance using a Welch's t-test.
Supplementary Material
Acknowledgements
We thank Dr Lisa Cameron at the Duke Light Microscopy Core Facility and Dr David Tobin for assistance with spinning-disk microscopy, Dr Alison Criss and members of the Valdivia lab for critical feedback on this project, Dr Jorn Coers and Dr Ryan Finethy for assistance with mice and immune cell isolations, Dr Terry Lechler for the ZO-1 GFP knock-in mice, Dr Agathe Subtil for the Cap1 antibody, Dr Ian Clarke for the pSW2NiggCDS2 plasmid and Alyson Barnes for assistance with Chlamydia transformations.
Footnotes
Competing interests
R.H.V. is co-founder at Bloom Sciences (San Diego, CA). The company did not sponsor any of the shown work nor has financial interest in the outcome of these studies.
Author contributions
Conceptualization: L.D., R.H.V.; Validation: L.D.; Formal analysis: L.D.; Investigation: L.D., R.H.V.; Writing - original draft: L.D., R.H.V.
Funding
This work was supported by National Institutes of Health (F32AI138372 to L.D. and AI134891 to R.H.V.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.252403.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.252403.reviewer-comments.pdf
References
- Abdelrahman, Y. M. and Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949-959. 10.1016/j.femsre.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Agaisse, H. and Derré, I. (2013). A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS ONE 8, e57090. 10.1371/journal.pone.0057090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse, H. and Derré, I. (2014). Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect. Immun. 82, 2037-2047. 10.1128/IAI.01530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, F., Luís, M. P., Pereira, I. S., Pais, S. V. and Mota, L. J. (2018). The human centrosomal protein CCDC146 binds chlamydia trachomatis inclusion membrane protein CT288 and is recruited to the periphery of the chlamydia-containing vacuole. Front. Cell Infect. Microbiol. 8, 254. 10.3389/fcimb.2018.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacallao, R., Antony, C., Dotti, C., Karsenti, E., Stelzer, E. H. and Simons, K. (1989). The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J. Cell Biol. 109, 2817-2832. 10.1083/jcb.109.6.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, J. R., Koestler, B. J., Carpenter, V. K., Burdette, D. L., Waters, C. M., Vance, R. E. and Valdivia, R. H. (2013). STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. MBio 4, e00018-13. 10.1128/mBio.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, R. C., Boretto, M., Rutkowski, M. R., Vankelecom, H. and Derré, I. (2020). Murine endometrial organoids to model chlamydia infection. Front. Cell Infect. Microbiol. 10, 416. 10.3389/fcimb.2020.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek, K., Eames, H. L., Weiss, M., Byrne, A. J., Perocheau, D., Pease, J. E., Doyle, S., McCann, F., Williams, R. O. and Udalova, I. A. (2015). IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med. 212, 845-853. 10.1084/jem.20140995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain, G., Müller, C., Meas-Yedid, V., Schmitt-Kopplin, P., Lazarow, P. B. and Subtil, A. (2014). The intracellular bacteria Chlamydia hijack peroxisomes and utilize their enzymatic capacity to produce bacteria-specific phospholipids. PLoS ONE 9, e86196. 10.1371/journal.pone.0086196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boretto, M., Cox, B., Noben, M., Hendriks, N., Fassbender, A., Roose, H., Amant, F., Timmerman, D., Tomassetti, C., Vanhie, A.et al. (2017). Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775-1786. 10.1242/dev.148478 [DOI] [PubMed] [Google Scholar]
- Boretto, M., Maenhoudt, N., Luo, X., Hennes, A., Boeckx, B., Bui, B., Heremans, R., Perneel, L., Kobayashi, H., Van Zundert, I.et al. (2019). Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 21, 1041-1051. 10.1038/s41556-019-0360-z [DOI] [PubMed] [Google Scholar]
- Buchholz, K. R. and Stephens, R. S. (2008). The cytosolic pattern recognition receptor NOD1 induces inflammatory interleukin-8 during Chlamydia trachomatis infection. Infect. Immun. 76, 3150-3155. 10.1128/IAI.00104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, V., Chen, Y.-S., Dolat, L. and Valdivia, R. H. (2017). The Effector TepP Mediates Recruitment and Activation of Phosphoinositide 3-Kinase on Early Chlamydia trachomatis Vacuoles. mSphere 2, e00207-17. 10.1128/mSphere.00207-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-S., Bastidas, R. J., Saka, H. A., Carpenter, V. K., Richards, K. L., Plano, G. V. and Valdivia, R. H. (2014). The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog. 10, e1003954. 10.1371/journal.ppat.1003954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, E., Kirker, K., Zuck, M., James, G. and Hybiske, K. (2012). Actin recruitment to the Chlamydia inclusion is spatiotemporally regulated by a mechanism that requires host and bacterial factors. PLoS ONE 7, e46949. 10.1371/journal.pone.0046949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers, J., Bernstein-Hanley, I., Grotsky, D., Parvanova, I., Howard, J. C., Taylor, G. A., Dietrich, W. F. and Starnbach, M. N. (2008). Chlamydia muridarum evades growth restriction by the IFN-γ-inducible host resistance factor Irgb10. J. Immunol. 180, 6237-6245. 10.4049/jimmunol.180.9.6237 [DOI] [PubMed] [Google Scholar]
- Derré, I., Swiss, R. and Agaisse, H. (2011). The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 7, e1002092. 10.1371/journal.ppat.1002092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessus-Babus, S., Knight, S. T. and Wyrick, P. B. (2000). Chlamydial infection of polarized HeLa cells induces PMN chemotaxis but the cytokine profile varies between disseminating and non-disseminating strains. Cell. Microbiol. 2, 317-327. 10.1046/j.1462-5822.2000.00058.x [DOI] [PubMed] [Google Scholar]
- Dolat, L. and Valdivia, R. H. (2019). A renewed tool kit to explore Chlamydia pathogenesis: from molecular genetics to new infection models. [version 1; peer review: 3 approved]. F1000Res. 8, F1000 Faculty Rev-935. 10.12688/f1000research.18832.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoux, M., Clare, D. K., Saibil, H. R. and Hayward, R. D. (2012). Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic 13, 1612-1627. 10.1111/tra.12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoux, M., Menny, A., Delacour, D. and Hayward, R. D. (2015). A Chlamydia effector recruits CEP170 to reprogram host microtubule organization. J. Cell Sci. 128, 3420-3434. 10.1242/jcs.169318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell, C. A. and Engel, J. N. (2012). Lipid acquisition by intracellular Chlamydiae. Cell. Microbiol. 14, 1010-1018. 10.1111/j.1462-5822.2012.01794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell, C., Mirrashidi, K. and Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385-400. 10.1038/nrmicro.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris, R., Merling, M., Andersen, S. E., Dooley, C. A., Hackstadt, T. and Weber, M. M. (2019). Chlamydia trachomatis CT229 Subverts Rab GTPase-Dependent CCV Trafficking Pathways to Promote Chlamydial Infection. Cell Rep. 26, 3380-3390.e5. 10.1016/j.celrep.2019.02.079 [DOI] [PubMed] [Google Scholar]
- Fielding, C. A., McLoughlin, R. M., McLeod, L., Colmont, C. S., Najdovska, M., Grail, D., Ernst, M., Jones, S. A., Topley, N. and Jenkins, B. J. (2008). IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J. Immunol. 181, 2189-2195. 10.4049/jimmunol.181.3.2189 [DOI] [PubMed] [Google Scholar]
- Finethy, R. and Coers, J. (2016). Sensing the enemy, containing the threat: cell-autonomous immunity to Chlamydia trachomatis. FEMS Microbiol. Rev. 40, 875-893. 10.1093/femsre/fuw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, A., Harrison, K. S., Ramirez, Y., Auer, D., Chowdhury, S. R., Prusty, B. K., Sauer, F., Dimond, Z., Kisker, C., Hefty, P. S.et al. (2017). Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. eLife 6, e21465. 10.7554/eLife.21465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, H. C., Dhakal, P., Behura, S. K., Schust, D. J. and Spencer, T. E. (2019). Self-renewing endometrial epithelial organoids of the human uterus. Proc. Natl. Acad. Sci. USA 116, 23132-23142. 10.1073/pnas.1915389116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote, H. P., Sumigray, K. D. and Lechler, T. (2013). FRAP analysis reveals stabilization of adhesion structures in the epidermis compared to cultured keratinocytes. PLoS ONE 8, e71491. 10.1371/journal.pone.0071491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, L. C., O'Connell, C. M., Andrews, C. W., Zurenski, M. A. and Darville, T. (2011). Enhanced neutrophil longevity and recruitment contribute to the severity of oviduct pathology during Chlamydia muridarum infection. Infect. Immun. 79, 4029-4041. 10.1128/IAI.05535-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargett, C. E., Schwab, K. E. and Deane, J. A. (2016). Endometrial stem/progenitor cells: the first 10 years. Hum. Reprod. Update 22, 137-163. 10.1093/humupd/dmv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshaber, S. S., Grieshaber, N. A. and Hackstadt, T. (2003). Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 116, 3793-3802. 10.1242/jcs.00695 [DOI] [PubMed] [Google Scholar]
- Guseva, N. V., Dessus-Babus, S., Moore, C. G., Whittimore, J. D. and Wyrick, P. B. (2007). Differences in Chlamydia trachomatis serovar E growth rate in polarized endometrial and endocervical epithelial cells grown in three-dimensional culture. Infect. Immun. 75, 553-564. 10.1128/IAI.01517-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty, C. L., Gottlieb, S. L., Taylor, B. D., Low, N., Xu, F. and Ness, R. B. (2010). Risk of sequelae after Chlamydia trachomatis genital infection in women. J. Infect. Dis. 201 Suppl. 2, S134-S155. 10.1086/652395 [DOI] [PubMed] [Google Scholar]
- Hall, J. V., Schell, M., Dessus-Babus, S., Moore, C. G., Whittimore, J. D., Sal, M., Dill, B. D. and Wyrick, P. B. (2011). The multifaceted role of oestrogen in enhancing Chlamydia trachomatis infection in polarized human endometrial epithelial cells. Cell. Microbiol. 13, 1183-1199. 10.1111/j.1462-5822.2011.01608.x [DOI] [PubMed] [Google Scholar]
- Heuer, D., Rejman Lipinski, A., Machuy, N., Karlas, A., Wehrens, A., Siedler, F., Brinkmann, V. and Meyer, T. F. (2009). Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 457, 731-735. 10.1038/nature07578 [DOI] [PubMed] [Google Scholar]
- Horn, M. (2008). Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62, 113-131. 10.1146/annurev.micro.62.081307.162818 [DOI] [PubMed] [Google Scholar]
- Hybiske, K. and Stephens, R. S. (2007). Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 104, 11430-11435. 10.1073/pnas.0703218104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, H., Ma, Z. and Barber, G. N. (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788-792. 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S. (2019). Bipotent stem cells support the cyclical regeneration of endometrial epithelium of the murine uterus. Proc. Natl. Acad. Sci. USA 116, 6848-6857. 10.1073/pnas.1814597116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, M., Zielecki, J., Thieck, O., Mollenkopf, H.-J., Fotopoulou, C. and Meyer, T. F. (2012). Chlamydia trachomatis disturbs epithelial tissue homeostasis in fallopian tubes via paracrine Wnt signaling. Am. J. Pathol. 180, 186-198. 10.1016/j.ajpath.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Kessler, M., Hoffmann, K., Fritsche, K., Brinkmann, V., Mollenkopf, H.-J., Thieck, O., Teixeira da Costa, A. R., Braicu, E. I., Sehouli, J., Mangler, M.et al. (2019). Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat. Commun. 10, 1194. 10.1038/s41467-019-09144-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner, J., Moore, C. G., Whittimore, J. D., Butler, M. and Hall, J. V. (2017). Inhibition of Wnt signaling pathways impairs chlamydia trachomatis infection in endometrial epithelial cells. Front. Cell Infect. Microbiol. 7, 501. 10.3389/fcimb.2017.00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokes, M., Dunn, J. D., Granek, J. A., Nguyen, B. D., Barker, J. R., Valdivia, R. H. and Bastidas, R. J. (2015). Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe 17, 716-725. 10.1016/j.chom.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela, P., Anttila, T., Bjørge, T., Brunsvig, A., Dillner, J., Hakama, M., Hakulinen, T., Jellum, E., Lehtinen, M., Lenner, P.et al. (2000). Chlamydia trachomatis infection as a risk factor for invasive cervical cancer. Int. J. Cancer 85, 35-39. [DOI] [PubMed] [Google Scholar]
- Kumar, Y. and Valdivia, R. H. (2008). Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4, 159-169. 10.1016/j.chom.2008.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Y., Cocchiaro, J. and Valdivia, R. H. (2006). The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16, 1646-1651. 10.1016/j.cub.2006.06.060 [DOI] [PubMed] [Google Scholar]
- Kumar, R., Gong, H., Liu, L., Ramos-Solis, N., Seye, C. I. and Derbigny, W. A. (2019). TLR3 deficiency exacerbates the loss of epithelial barrier function during genital tract Chlamydia muridarum infection. PLoS ONE 14, e0207422. 10.1371/journal.pone.0207422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy, H. M., Bowlin, A. K., Hennings, L., Scurlock, A. M., Nagarajan, U. M. and Rank, R. G. (2011). Essential role for neutrophils in pathogenesis and adaptive immunity in Chlamydia caviae ocular infections. Infect. Immun. 79, 1889-1897. 10.1128/IAI.01257-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann, T., Afonso, P. V., Angermann, B. R., Wang, J. M., Kastenmüller, W., Parent, C. A. and Germain, R. N. (2013). Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371-375. 10.1038/nature12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. Y., Schripsema, J. H., Sigar, I. M., Murray, C. M., Lacy, S. R. and Ramsey, K. H. (2010a). A link between neutrophils and chronic disease manifestations of Chlamydia muridarum urogenital infection of mice. FEMS Immunol. Med. Microbiol. 59, 108-116. 10.1111/j.1574-695X.2010.00668.x [DOI] [PubMed] [Google Scholar]
- Lee, H. Y., Schripsema, J. H., Sigar, I. M., Lacy, S. R., Kasimos, J. N., Murray, C. M. and Ramsey, K. H. (2010b). A role for CXC chemokine receptor-2 in the pathogenesis of urogenital Chlamydia muridarum infection in mice. FEMS Immunol. Med. Microbiol. 60, 49-56. 10.1111/j.1574-695X.2010.00715.x [DOI] [PubMed] [Google Scholar]
- Lijek, R. S., Helble, J. D., Olive, A. J., Seiger, K. W. and Starnbach, M. N. (2018). Pathology after Chlamydia trachomatis infection is driven by nonprotective immune cells that are distinct from protective populations. Proc. Natl. Acad. Sci. USA 115, 2216-2221. 10.1073/pnas.1711356115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabey, D. and Peeling, R. W. (2002). Lymphogranuloma venereum. Sex. Transm. Infect. 78, 90-92. 10.1136/sti.78.2.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mital, J., Lutter, E. I., Barger, A. C., Dooley, C. A. and Hackstadt, T. (2015). Chlamydia trachomatis inclusion membrane protein CT850 interacts with the dynein light chain DYNLT1 (Tctex1). Biochem. Biophys. Res. Commun. 462, 165-170. 10.1016/j.bbrc.2015.04.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, H. and Stappenbeck, T. S. (2013). In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471-2482. 10.1038/nprot.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, E. R., Fischer, E. R., Mead, D. J. and Hackstadt, T. (2008). The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9, 2130-2140. 10.1111/j.1600-0854.2008.00828.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy, S. and Cossart, P. (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183-194. 10.1038/nrm3284 [DOI] [PubMed] [Google Scholar]
- Mueller, K. E., Wolf, K. and Fields, K. A. (2016). Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. MBio 7, e01817-15. 10.1128/mBio.01817-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, M. A., Naturale, V. F. and Feldman, J. L. (2019). A polarizing issue: diversity in the mechanisms underlying apico-basolateral polarization in vivo. Annu. Rev. Cell Dev. Biol. 35, 285-308. 10.1146/annurev-cellbio-100818-125134 [DOI] [PubMed] [Google Scholar]
- Pokrovskaya, I. D., Szwedo, J. W., Goodwin, A., Lupashina, T. V., Nagarajan, U. M. and Lupashin, V. V. (2012). Chlamydia trachomatis hijacks intra-Golgi COG complex-dependent vesicle trafficking pathway. Cell. Microbiol. 14, 656-668. 10.1111/j.1462-5822.2012.01747.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston, T. B., O'Connell, C. M., Girardi, J., Sullivan, J. E., Nagarajan, U. M., Marinov, A., Scurlock, A. M. and Darville, T. (2018). T cell-independent gamma interferon and B cells cooperate to prevent mortality associated with disseminated Chlamydia muridarum genital tract infection. Infect. Immun. 86, e00143-18. 10.1128/IAI.00143-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner, D., Darville, T. and Nagarajan, U. M. (2010). Stimulator of IFN gene is critical for induction of IFN-β during Chlamydia muridarum infection. J. Immunol. 184, 2551-2560. 10.4049/jimmunol.0903704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda, J. N., Bastidas, R. J., Bertsoulaki, E., Swatek, K. N., Santhanam, B., Clague, M. J., Valdivia, R. H., Urbé, S. and Komander, D. (2018). A Chlamydia effector combining deubiquitination and acetylation activities induces Golgi fragmentation. Nat. Microbiol. 3, 1377-1384. 10.1038/s41564-018-0271-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeeve, K., Das, S., Prusty, B. K. and Rudel, T. (2018). Chlamydia trachomatis paralyses neutrophils to evade the host innate immune response. Nat. Microbiol. 3, 824-835. 10.1038/s41564-018-0182-y [DOI] [PubMed] [Google Scholar]
- Rank, R. G., Whittimore, J., Bowlin, A. K. and Wyrick, P. B. (2011). In vivo ultrastructural analysis of the intimate relationship between polymorphonuclear leukocytes and the chlamydial developmental cycle. Infect. Immun. 79, 3291-3301. 10.1128/IAI.00200-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, S. J., Eckmann, L., Quayle, A. J., Shen, L., Zhang, Y. X., Anderson, D. J., Fierer, J., Stephens, R. S. and Kagnoff, M. F. (1997). Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Invest. 99, 77-87. 10.1172/JCI119136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman Lipinski, A., Heymann, J., Meissner, C., Karlas, A., Brinkmann, V., Meyer, T. F. and Heuer, D. (2009). Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 5, e1000615. 10.1371/journal.ppat.1000615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey, D., Heinzen, R. and Hackstadt, T. (1995). Cloning and characterization of a Chlamydia psittaci gene coding for a protein locaiized in the inclusion membrane of infected cells. Mol. Microbiol. 15, 617.- . 10.1111/j.1365-2958.1995.tb02371.x [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan, E. and Macara, I. G. (2014). Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225-242. 10.1038/nrm3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, G., Manfrin, A. and Lutolf, M. P. (2018). Progress and potential in organoid research. Nat. Rev. Genet. 19, 671-687. 10.1038/s41576-018-0051-9 [DOI] [PubMed] [Google Scholar]
- Schachter, J. (1978). Chlamydial infections (first of three parts). N. Engl. J. Med. 298, 428-435. 10.1056/NEJM197802232980805 [DOI] [PubMed] [Google Scholar]
- Schott, B. H., Antonia, A. L., Wang, L., Pittman, K. J., Sixt, B. S., Barnes, A. B., Valdivia, R. H. and Ko, D. C. (2020). Modeling of variables in cellular infection reveals CXCL10 levels are regulated by human genetic variation and the Chlamydia-encoded CPAF protease. Sci. Rep. 10, 18269. 10.1038/s41598-020-75129-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, S.-U., Kwon, H.-J., Ko, H.-J., Byun, Y.-H., Seong, B. L., Uematsu, S., Akira, S. and Kweon, M.-N. (2011). Type I interferon signaling regulates Ly6C(hi) monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog. 7, e1001304. 10.1371/journal.ppat.1001304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmughapriya, S., Senthilkumar, G., Vinodhini, K., Das, B. C., Vasanthi, N. and Natarajaseenivasan, K. (2012). Viral and bacterial aetiologies of epithelial ovarian cancer. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2311-2317. 10.1007/s10096-012-1570-5 [DOI] [PubMed] [Google Scholar]
- Sixt, B. S., Bastidas, R. J., Finethy, R., Baxter, R. M., Carpenter, V. K., Kroemer, G., Coers, J. and Valdivia, R. H. (2017). The chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe 21, 113-121. 10.1016/j.chom.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis, E. T. (2018). Spatial effects-site-specific regulation of actin and microtubule organization by septin GTPases. J. Cell Sci. 131, jcs207555. 10.1242/jcs.207555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, R. S., Myers, G., Eppinger, M. and Bavoil, P. M. (2009). Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol. Med. Microbiol. 55, 115-119. 10.1111/j.1574-695X.2008.00516.x [DOI] [PubMed] [Google Scholar]
- Tarbet, H. J., Dolat, L., Smith, T. J., Condon, B. M., O'Brien, E. T., III, Valdivia, R. H. and Boyce, M. (2018). Site-specific glycosylation regulates the form and function of the intermediate filament cytoskeleton. eLife 7, e31807. 10.7554/eLife.31807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco, M. Y., Gardner, L., Hughes, J., Cindrova-Davies, T., Gomez, M. J., Farrell, L., Hollinshead, M., Marsh, S. G. E., Brosens, J. J., Critchley, H. O.et al. (2017). Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568-577. 10.1038/ncb3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDussen, K. L., Sonnek, N. M. and Stappenbeck, T. S. (2019). L-WRN conditioned medium for gastrointestinal epithelial stem cell culture shows replicable batch-to-batch activity levels across multiple research teams. Stem Cell Res. 37, 101430. 10.1016/j.scr.2019.101430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volceanov, L., Herbst, K., Biniossek, M., Schilling, O., Haller, D., Nölke, T., Subbarayal, P., Rudel, T., Zieger, B. and Häcker, G. (2014). Septins arrange F-actin-containing fibers on the Chlamydia trachomatis inclusion and are required for normal release of the inclusion by extrusion. MBio 5, e01802-14. 10.1128/mBio.01802-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Cutcliffe, L. T., Skilton, R. J., Ramsey, K. H., Thomson, N. R. and Clarke, I. N. (2014). The genetic basis of plasmid tropism between Chlamydia trachomatis and Chlamydia muridarum. Pathog. Dis. 72, 19-23. 10.1111/2049-632X.12175 [DOI] [PMC free article] [PubMed]
- Wang, Y., Kahane, S., Cutcliffe, L. T., Skilton, R. J., Lambden, P. R. and Clarke, I. N. (2011). Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7, e1002258. 10.1371/journal.ppat.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M. M., Lam, J. L., Dooley, C. A., Noriea, N. F., Hansen, B. T., Hoyt, F. H., Carmody, A. B., Sturdevant, G. L. and Hackstadt, T. (2017). Absence of specific chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Rep. 19, 1406-1417. 10.1016/j.celrep.2017.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowski, J., Weber, M. M., Nawrotek, A., Dooley, C. A., Calderon, M., St Croix, C. M., Hackstadt, T., Cherfils, J. and Paumet, F. (2017). Chlamydia hijacks ARF gtpases to coordinate microtubule posttranslational modifications and golgi complex positioning. MBio 8, e02280-16. 10.1128/mBio.02280-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2018). Report on Global Sexually Transmitted Infection Surveillanc. World Health Organization. [Google Scholar]
- Wright, H. L., Cross, A. L., Edwards, S. W. and Moots, R. J. (2014). Effects of IL-6 and IL-6 blockade on neutrophil function in vitro and in vivo. Rheumatology 53, 1321-1331. 10.1093/rheumatology/keu035 [DOI] [PubMed] [Google Scholar]
- Zadora, P. K., Chumduri, C., Imami, K., Berger, H., Mi, Y., Selbach, M., Meyer, T. F. and Gurumurthy, R. K. (2019). Integrated phosphoproteome and transcriptome analysis reveals chlamydia-induced epithelial-to-mesenchymal transition in host cells. Cell Rep. 26, 1286-1302.e8. 10.1016/j.celrep.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Zhong, G., Fan, P., Ji, H., Dong, F. and Huang, Y. (2001). Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193, 935-942. 10.1084/jem.193.8.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Shen, Z., Luo, H., Zhang, W. and Zhu, X. (2016). Chlamydia trachomatis infection-associated risk of cervical cancer: a meta-analysis. Medicine 95, e3077. 10.1097/MD.0000000000003077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.